Abstract
Context: Fatty liver is an important complication of obesity; however, regulatory mechanisms mediating altered gene expression patterns have not been identified.
Objective: The aim of the study was to identify novel transcriptional changes in human liver that could contribute to hepatic lipid accumulation and associated insulin resistance, type 2 diabetes, and nonalcoholic steatohepatitis.
Design: We evaluated gene expression in surgical liver biopsies from 13 obese (nine with type 2 diabetes) and five control subjects using Affymetrix U133A microarrays. PCR validation was performed in liver biopsies using an additional 16 subjects. We also tested thyroid hormone responses in mice fed chow or high-fat diet.
Setting: Recruitment was performed in an academic medical center.
Participants: Individuals undergoing elective surgery for obesity or gallstones participated in the study.
Results: The top-ranking gene set, down-regulated in obese subjects, was comprised of genes previously demonstrated to be positively regulated by T3 in human skeletal muscle (n = 399; P < 0.001; false discovery rate = 0.07). This gene set included genes related to RNA metabolism (SNRPE, HNRPH3, TIA1, and SFRS2), protein catabolism (PSMA1, PSMD12, USP9X, IBE2B, USP16, and PCMT1), and energy metabolism (ATP5C1, COX7C, UQCRB). We verified thyroid hormone regulation of these genes in the liver after injection of C57BL/6J mice with T3 (100 μg/100 g body weight); furthermore, T3-induced increases in expression of these genes were abolished by high-fat diet. In agreement, expression of these genes inversely correlated with liver fat content in humans.
Conclusions: These data suggest that impaired thyroid hormone action may contribute to altered patterns of gene expression in fatty liver.
Impaired thyroid hormone action may contribute to altered patterns of gene expression in humans with obesity and fatty liver.
Hepatic lipid accumulation contributes to insulin resistance, hyperglycemia, and hyperlipidemia (1,2,3). However, the cellular mechanisms mediating hepatic lipid accumulation in obese humans remain unclear. Liver may serve as an alternative depot for lipid storage when adipose capacity is exceeded, as in obesity or adipocyte dysfunction (e.g. lipodystrophy) (4). Furthermore, decreased mitochondrial oxidative capacity may contribute to lipid accumulation in both liver and skeletal muscle (5,6). These defects, together with adipose-derived inflammatory signals (e.g. TNFα), can contribute to increased production of reactive oxygen species and development of nonalcoholic steatohepatitis (NASH) (7).
Previous reports of gene expression in liver of patients with nonalcoholic fatty liver disease and/or NASH suggest both increased de novo lipogenesis and lipid oxidation (8,9). Genes regulating lipid synthesis (e.g. ACC1, FASN, DGAT1, and SREBP1c) and mitochondrial β-oxidation (e.g. LCAD, HADHA, UCP2, and ACOX) are up-regulated in nonalcoholic fatty liver disease (8). Furthermore, altered expression and activation of proinflammatory genes (e.g. IL1R family) and genes regulating fibrotic responses (TGFB1, FGFR2) may contribute to the development of NASH and cirrhosis (10,11).
We aimed to identify novel alterations in gene expression related to hepatic lipid accumulation in obese patients with and without type 2 diabetes mellitus (T2DM). Thus, we analyzed 18 liver samples from 13 obese subjects and five lean controls, using Affymetrix U133A expression arrays. Gene Set Enrichment Analysis (GSEA) (12) revealed that the top-ranking gene set related to obesity was a set of genes positively regulated by T3 (13), the biologically active thyroid hormone.
Subjects and Methods
Study subjects
Human studies were performed in accord with the Helsinki Declaration and were approved by Institutional Review Boards of Joslin Diabetes Center and Beth Israel Deaconess Medical Center. Informed consent was obtained from all subjects. Participants had normal hepatic and thyroid function, no history of excessive alcohol intake, and no other chronic illness. Although subjects had no history of impaired glucose tolerance or T2DM at the time of preoperative medical clearance, all obese participants underwent a 2-h, 75-g oral glucose tolerance test (14); based on these results, nine obese subjects were diagnosed preoperatively with T2DM. Fasting samples were obtained for glucose, insulin, hemoglobin A1c (HbA1c), and lipid profiles in all subjects. Insulin resistance was determined by homeostasis model assessment (HOMA-IR); in obese subjects, iv glucose tolerance tests were performed preoperatively, with minimal model analysis (15).
Liver biopsies
Open liver biopsies were performed in the fasting state during elective cholecystectomy from lean control individuals without diabetes (n = 5) and during gastric bypass from obese subjects (n = 13; Table 1). Biopsies were immediately frozen in liquid nitrogen and stored at −80 C. A portion was paraffin-embedded, sectioned, and stained with hematoxylin and eosin and Trichrome for analysis of histology, fibrosis, and NASH score (16). Intrahepatic fat (percentage of area) was quantified by an observer blinded to group (ImagePro). PCR validation was performed in liver from 24 subjects (eight from original cohort plus 16 additional); these included six lean, seven obese, and 11 obese individuals with diabetes (Supplementary Table 1).
Table 1.
Characteristics of study subjects (microarray analysis)
| Lean controls | Obese | Obese T2DM | |
|---|---|---|---|
| No. of subjects in group | 5 | 4 | 9 |
| Gender (males/females) | 0/5 | 1/3 | 3/6 |
| Age (yr) | 36.2 ± 11.5 | 39.0 ± 10.9 | 46.1 ± 12.4 |
| Body mass index (kg/m2) | 26.7 ± 7.5 | 51.5 ± 4.4b | 52.4 ± 6.7c |
| HbA1c | 5.2 ± 0.3 | 5.2 ± 0.1 | 7.9 ± 2.1b,d |
| Fasting glucose (mmol/liter) | 4.7 ± 0.8 | 4.8 ± 0.5 | 9.3 ± 3.5b,d |
| Fasting insulin (pmol/liter) | 43.8 ± 38.8 | 152.1 ± 144.4a | 307.6 ± 272.9a |
| HOMA-IR | 1.5 ± 1.4 | 5.0 ± 5.4a | 20.3 ± 23.6a |
| Si (insulin sensitivity) | NA | 0.9 ± 1.5 | 0.2 ± 0.2 |
| Sg (glucose effectiveness) | NA | 0.016 ± 0.003 | 0.040 ± 0.006 |
| Cholesterol (mmol/liter) | NA | 4.6 ± 1.0 | 4.6 ± 0.6 |
| HDL-cholesterol (mmol/liter) | NA | 1.1 ± 0.1 | 1.1 ± 0.2 |
| Triglycerides (mmol/liter) | NA | 1.0 ± 0.5 | 2.0 ± 1.0 |
| ALT (U/liter) | NA | 19 ± 7 | 32 ± 20 |
| AST (U/liter) | NA | 20 ± 4 | 29 ± 17 |
| Liver fat content (%) | 2.4 ± 3.1 | 9.8 ± 17.6 | 14.3 ± 9.3a |
| NASH score (no. of subjects) | |||
| 0 | 4 | 2 | 5 |
| 1 | 1 | 0 | 1 |
| 2 | 0 | 2 | 2 |
| 4 | 0 | 0 | 1 |
Mean ± sd. To convert to mg/dl, divide glucose by 0.0555, cholesterol by 0.0259, and triglycerides by 0.0113. To convert insulin to mU/liter, divide by 6.945. ALT, Alanine aminotransferase; AST, aspartate aminotransferase; HDL, high-density lipoprotein; NA, not assessed.
P < 0.05.
P < 0.01.
P < 0.001 vs. lean controls.
P < 0.01 vs. obese subjects with diabetes.
Analytical methods
Glucose was measured by glucose oxidation; fasting cholesterol, and high-density lipoprotein by cholesterol esterase; triglycerides via hydrolysis to glycerol and FFA (Synchron CX3delta/CX9; Beckman, Brea, CA); HbA1c by HPLC (Tosoh, San Francisco, CA); and insulin by duplicate immunoassays (Diagnostic Systems Laboratories, Webster, TX).
Expression analysis
Total RNA was isolated with Trizol (Invitrogen, Carlsbad, CA), and cRNA was generated and hybridized to Affymetrix HG-U133A arrays (Affymetrix, Inc. Santa Clara, CA) (17). Differential expression was analyzed using Bioconductor multtest (www.bioconductor.org). GSEA 2.0 (www.broad.mit.edu/gsea) and MAPPFinder 2.0 (www.genmapp.org) were used to identify differentially expressed gene sets/pathways (12), with threshold false discovery rate (FDR) less than 0.25 (GSEA) and adjusted P < 0.05 (MAPPFinder). Correlations with clinical variables were assessed for each probe set and for gene sets (12) (Supplementary Table 2). Because of nonnormal distribution of liver fat content, Spearman correlation was used. All array data are posted at Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; gene accession no. GSE15653).
cDNA was synthesized using random hexamers (Advantage; Clontech, Mountain View, CA); PCR products were detected using SYBR Green (ABI Prism 7900; Applied Biosystems, Inc., Foster City, CA). Expression was normalized to the mean of three controls (TBP, cyclophilin, and 18S ribosomal RNA). Primer sequences are provided in Supplementary Table 3. Mouse PCR data were normalized to TBP.
Deiodinase assays
Human liver samples were homogenized in cold 0.25 m sucrose/10 mm dithiothreitol (DTT) and sonicated briefly. For D1 assay, 10 μg of protein were incubated for 3 h in 0.1 m phosphate/1 mm EDTA (pH 6.9) with 100–200,000 cpm 125I-labeled rT3 with 10 mm DTT, 1 nm cold rT3, and 1 mm propylthiouracil for background calculation. For D2 assay, 100–300 μg of protein were incubated for 2 h with 100–200,000 cpm 125I-labeled T4, 20 mm DTT, 1 mm propylthiouracil, and 0.1 nm cold T4, or 100 nm cold T4 for background calculation. Deiodination was calculated as release of free 125iodide after precipitation with serum and trichloroacetic acid (18).
Animal care
All protocols were approved by Joslin Institutional Animal Care and Use Committee. Mice were housed four per cage in an Office of Laboratory Animal Welfare-certified facility with a 12-h light cycle. Six-week-old male C57BL/6J mice were fed chow (17% calories from fat) or high-fat diet (HFD) (42% calories from milk fat) (Harlan Teklad, Madison, WI) for 6 wk. Mice were injected with 100 μg/100 g body weight T3 or saline; tissues were harvested 20 h later after pentobarbital anesthesia.
Statistical analysis
Basic data analysis was performed with SPSS/Win (version 10.0, SPSS Inc., Chicago, IL). P < 0.05 was considered significant. Data are presented as mean ± sd.
Results
Characteristics of five lean control and 13 obese subjects (array cohort) are shown in Table 1. Obese subjects were divided into subgroups with (n = 9) and without T2DM (n = 4). Groups were similar for age, lipids, and liver enzymes. Obese subjects were more insulin resistant, with higher fasting insulin and HOMA-IR. Liver fat was markedly higher in obese subjects with T2DM compared with lean (P = 0.005) and intermediate in obese nondiabetic subjects.
Thyroid-responsive gene set is down-regulated in liver of obese subjects
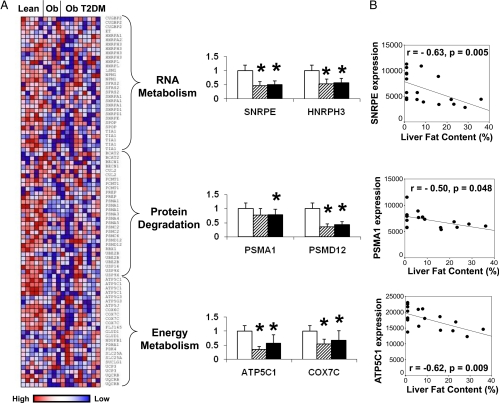
The top-ranking gene set in obese subjects without diabetes, enriched with down-regulated genes, was a set of genes previously identified as responsive to thyroid hormone treatment (T3, 75 μg/d for 14 d) in human skeletal muscle (q = 0.07; Table 2 and Supplementary Table 2) (13). Sixty-one of 399 probes in this set were down-regulated in obese subjects (nominal P < 0.05), particularly within the following ontology subsets (designated in Ref. 13 and shown in Supplementary Fig. 1): 1) RNA metabolism (five of 34 probe sets, genes SNRPE, HNRPH3, TIA1, and SFRS2); 2) protein catabolism, particularly proteasomal degradation (nine of 32, genes PSMA1, PSMD12, USP9X, IBE2B, USP16, and PCMT1); and 3) energy metabolism, specifically mitochondrial oxidative phosphorylation (seven of 25, genes ATP5C1, COX7C, and UQCRB; Fig. 1A). PCR confirmed that expression of representative genes was reduced in both obesity and obesity accompanied by diabetes (Fig. 1A). Furthermore, expression of these same genes correlated negatively with liver fat (Fig. 1B). Similarly, MAPPFinder analysis identified electron transport and proteasomal degradation pathways as overrepresented in genes correlating negatively with liver fat (adjusted P = 0.001 and 0.003, not shown).
Table 2.
Gene sets dysregulated in liver of obese or obese diabetic subjects compared to lean subjects or correlated with clinical phenotypes using GSEA
| Size | ES | NES | Nominal P value | FDR Q value | |
|---|---|---|---|---|---|
| Gene sets enriched with altered genes in study groups | |||||
| Downregulated in obese subjects | |||||
| Regulated_Thyroidhormone_Muscle | 365 | 0.43 | 1.86 | <0.001 | 0.071 |
| mRNA processing | 107 | 0.43 | 1.87 | <0.001 | 0.142 |
| Melton_Stemness | 254 | 0.41 | 1.79 | <0.001 | 0.185 |
| Leptin pathway | 23 | 0.60 | 1.73 | <0.001 | 0.217 |
| Histidine metabolism | 34 | 0.51 | 1.74 | 0.030 | 0.239 |
| Upregulated in obese diabetic subjects | |||||
| Glycolysis pathway | 30 | 0.72 | 1.97 | 0.042 | 0.170 |
| Enriched with genes correlation with clinical phenotypes | |||||
| Positive correlation with HOMA-IR | |||||
| T_ALL_Downing_ALL | 116 | 0.58 | 2.23 | <0.001 | <0.001 |
| Eosinophils pathway | 19 | 0.72 | 1.95 | <0.001 | 0.065 |
Gene sets with FDR <0.25 are shown. No gene sets were enriched (FDR Q <0.25) for upregulation in obese subjects or downregulation in obese diabetic subjects, or for the correlation with liver fat content, fasting glucose, fasting insulin, or HbA1c. ES, Enrichment score; NES, normalized enrichment score.
Figure 1.
Thyroid-responsive gene set is enriched with down-regulated genes in liver of obese humans. A, Heatmap showing liver expression of genes in different subcategories of the T3-regulated gene set (genes regulated by T3 in human skeletal muscle). Each column presents data for an individual subject, whereas each row represents an individual probe set. Expression values are colored by row-normalized values. Graphs demonstrate PCR verification of representative genes in liver of lean (open bars), obese (striped), and obese diabetic (black) subjects (mean ± sd). *, P < 0.05. Data for all genes are shown in Supplementary Table 2. B, Spearman correlation of liver fat content with expression of representative genes.
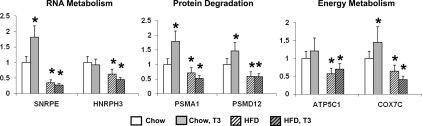
To verify that genes within the thyroid-responsive gene set are indeed T3-regulated in liver, euthyroid C57BL/6J mice were injected with T3 (100 μg/100 g body weight). T3 treatment stimulated expression of genes that were down-regulated in liver from obese humans (Fig. 2). Furthermore, basal expression of these genes was down-regulated by high-fat feeding, a pattern similar to that in obese humans. Importantly, T3 injection did not increase expression of these genes in mice fed a HFD (Fig. 2), indicating that HFD inhibited both basal and T3-stimulated gene expression (T3 resistance).
Figure 2.
Genes in RNA metabolism, protein degradation, and energy metabolism categories of the thyroid-responsive gene set are up-regulated by T3 in chow-fed mice. Basal levels of expression, as well as T3-stimulated responses, are abolished by HFD in mice. Mean ± sd shown in bar graphs. *, P < 0.05.
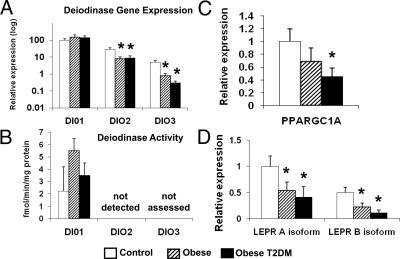
To evaluate potential mechanisms mediating impaired thyroid hormone action in liver of obese individuals, we analyzed expression of genes regulating intracellular T3 levels or T3-mediated effects on gene expression. Whereas expression of DIO1, the predominant liver deiodinase, was not changed, expression of the less abundant DIO2 and DIO3 was decreased in obesity (all P < 0.05; Fig. 3A). However, DIO1 activity did not vary among the three groups, and DIO2 activity was below detection (Fig. 3B). We observed no change in expression of thyroid hormone receptors THRA (both α1 and α2 isoforms) or THRB (data not shown).
Figure 3.
Potential regulators of thyroid hormone action. A, Expression of DIO2 and DIO3 is decreased in liver of obese subjects. B, DIO1 activity is not significantly altered in obese vs. lean humans. Expression of PPARGC1A (C) and LEPR (D) isoforms, as determined by PCR, was decreased in subjects with obesity (striped bars) and T2DM (black bars). Mean ± sd is shown in bar graphs. *, P < 0.05.
Another potential mechanism responsible for down-regulation of thyroid-responsive genes could be altered expression of coactivators/corepressors known to interact with TRβ (http://www.hprd.org). Array expression levels for JMJD1C, PPARGC1A (PGC1α gene), COPS2, MED21, and ZNHIT3 were significantly decreased in liver of obese subjects (Supplementary Table 4). We analyzed PPARGC1A further because it is a known coactivator of thyroid hormone receptor (THR) (19), and reductions in PPARGC1A expression have been linked to reduced skeletal muscle expression of mitochondrial oxidative metabolism genes in humans with diabetes and obesity (20,21) and after cellular lipid accumulation (17). Although expression of PPARGC1A was reduced by 55% in obese diabetic subjects (PCR; P < 0.05; Fig. 3C), expression of PPARGC1A was inversely correlated only with the energy metabolism subset of the thyroid-responsive gene set (e.g. with ATP5C1; Supplementary Fig. 2).
We next evaluated differential expression within the leptin pathway, given its down-regulation in obese liver in this study (Table 2) and earlier observations demonstrating decreased expression of leptin receptor (LEPR) in liver of obese humans (22) and altered thyroid hormone deiodination in leptin- and leptin receptor-deficient mice (23). Seven of 23 probes within this pathway were down-regulated, including five for LEPR (Supplementary Table 2). Expression of both A and B isoforms of LEPR was decreased (by 40–70%) in obese subjects with and without T2DM (P < 0.05 for both). The functionally active B isoform was decreased more than the A isoform in obese subjects (P = 0.034), and even further in obese diabetic subjects (P = 0.012), as previously observed (22). Total LEPR expression was inversely correlated with liver fat (r = 0.67; P = 0.002), but not significantly with other clinical phenotypes (Supplementary Table 2). Expression of LEPR correlated negatively with the energy metabolism subset of the thyroid-responsive gene set (Supplementary Fig. 2) and positively with PPARGC1 (r = 0.65; P = 0.004).
Correlation between gene expression and clinical phenotypes
We next assessed correlation between gene expression and liver fat content, fasting glucose and insulin, HOMA-IR, and HbA1c. We observed robust positive correlation between HOMA-IR, an index incorporating both fasting glucose and insulin, and expression of inflammatory genes, including HLA class II molecules (Table 2; T_ALL_Downing_ALL; full list in Supplementary Table 2). This appeared to be largely accounted for by glucose because 13 of 28 probe sets for HLA class II molecules were positively correlated with fasting glucose, whereas only five of 28 were correlated with fasting insulin (Supplementary Table 2).
Gene sets related to T2DM
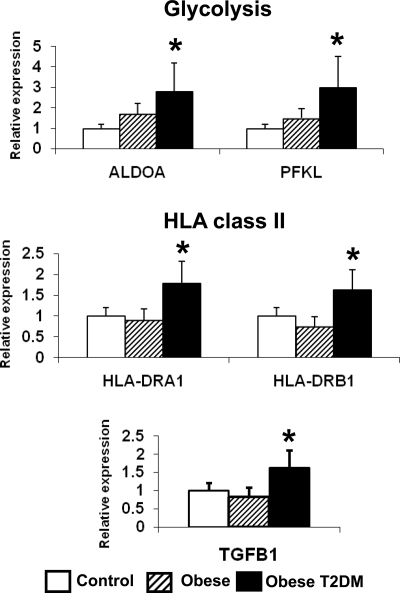
Pathway analysis revealed that the single gene set significantly enriched (up-regulated) in patients with T2DM was the glycolysis pathway (Table 2), including pyruvate kinase (PKLR), phosphofructokinase (PFKL), aldolase (ALDOA/ALDOB), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, PCR in Fig. 4, and Supplementary Table 2). HLA class II genes were also specifically up-regulated in patients with T2DM (Fig. 4), in accord with the HOMA-IR correlation analyses. In addition, expression of TGFB1, a potential regulator of liver fibrosis (24), was increased in patients with T2DM (Fig. 4). Expression of these genes correlated positively with fasting glucose (Supplementary Table 2). However, expression of these T2DM-regulated genes (ALDOA, HLA-DRA1, and TGFB1) was not altered by in vivo T3 treatment in mice, indicating that changes observed specifically in patients with T2DM are unlikely to be related to altered thyroid hormone action in liver (data not shown).
Figure 4.
Expression of genes regulating glycolysis, HLA class II molecules, and TGBF1 is increased in liver of subjects with T2DM. Mean ± sd is shown in bar graphs. *, P < 0.05.
Other differentially expressed candidate genes
In addition to our pathway-based analyses, we also analyzed expression differences at a per-gene level, concentrating on genes regulating lipid synthesis and/or oxidation, given that lipid accumulation is a hallmark of hepatic insulin resistance (Supplementary Table 5). Expression of FASN, DGAT1, and DGAT2 was significantly increased in obese subjects (Supplementary Fig. 3) and correlated positively with liver fat (r > 0.5; P < 0.05). Expression of classic lipogenic T3 target genes ME and SPOT14 was not altered between groups (data not shown). In contrast to some previous reports, we did not observe up-regulation of genes regulating fatty acid degradation (8) or oxidation (9) in either obese or diabetic subjects (data not shown). Interestingly, expression of TCF7L2, a high-ranking T2DM candidate gene (25), was down-regulated by 35% (P = 0.01–0.001) in obese individuals (Supplementary Fig. 3).
Discussion
We sought to identify novel transcriptional changes in human liver that could contribute to or result from liver fat accumulation in obese humans. The most striking finding was that the top-ranking gene set was a group of genes previously shown to be positively regulated by the biologically active thyroid hormone T3 in human skeletal muscle (13) (Table 2). Although this set includes genes regulating a wide array of metabolic functions, the most consistent changes were observed in subsets regulating RNA metabolism, protein degradation, and energy metabolism (Fig. 1A). Expression of these thyroid hormone-responsive genes also correlated inversely with liver fat in humans (Fig. 1B) and was similarly decreased in mice with obesity induced by high-fat feeding (Fig. 2). Interestingly, acute T3 injection increased expression of these same genes in liver of chow-fed mice, but not in high fat-fed mice, suggesting that expression of these genes was resistant to T3 regulation in obesity (Fig. 2).
What is the evidence that thyroid hormones may regulate hepatic gene expression? To our knowledge, there are no prior studies investigating either liver gene expression in hypo- or hyperthyroid humans or the effects of thyroid hormone treatment on gene expression in human liver. Prior studies in hypothyroid mice reveal distinct acute and long-term effects of thyroid hormone on subsets of target gene(s) (26). For example, among the classical gene targets directly regulated by thyroid hormone in hypothyroid mice are lipogenic genes; 2 h after T3 treatment, expression of ME, SPOT14, and FASN increases by 3- to 6-fold (26). However, more chronic treatment with T3 (5 d) results in only modest up-regulation of ME and SPOT14 (<3-fold increase for both) and 30% down-regulation of FASN; by contrast, mitochondrial oxidative genes remain up-regulated (e.g. CYCS, COX7A, and UQCRH). It is important to note that the thyroid-responsive gene set used in this study and differentially regulated in obesity was initially identified during a more chronic (2-wk) T3 treatment protocol in humans (13). Most notably, this gene set did not include ME and SPOT14 because their expression was not altered significantly in human skeletal muscle in response to T3 (13). Thus, these data support the hypothesis that patterns of gene expression in obese liver may result from chronic impairments in thyroid hormone action, with only partial overlap with acute effects of T3. In further support of this hypothesis, we demonstrate that thyroid hormone effects on subsets of thyroid-responsive genes are abolished in mice with diet-induced obesity (Fig. 2).
Potential mechanisms mediating changes in thyroid hormone-responsive gene expression include alterations in thyroid function, deiodinase activity, or the THR transcriptional complex. Firstly, although our subjects were euthyroid, TSH levels, even within the euthyroid range, have been positively correlated with body mass index in both men and women (27), suggesting that modest subclinical impairment in thyroid hormone levels and/or action could contribute to altered gene expression patterns in obesity. More interestingly, in a large population study (n = 10,292), decreased thyroid hormone levels were inversely related to liver enzymes, even in euthyroid subjects (28). In fact, in preliminary analysis including 15 subjects with available data, TSH values correlated positively with genes in inflammation-related gene sets (T_ALL_ DOWNING_ALL, CTLPATHWAY, TCRAPATHWAY, and CTLA4PATHWAY) (GSEA, FDR <0.25 for all gene sets; data not shown), suggesting a potential link between thyroid function and proinflammatory responses.
We next considered the possibility that activity of deiodinases regulating intracellular concentrations of active T3 could be altered in obesity (Fig. 3, A and B). Although decreased expression of DIO2 in obese liver suggests that intracellular conversion of T4 to active T3 could be reduced, expression of DIO1 was not altered, and the simultaneous down-regulation of inactivating DIO3 makes prediction of net intracellular T3 concentrations difficult. Furthermore, we found no significant difference in DIO1 activity and no detectable DIO2 activity. Thus, despite the limitations of our relatively small sample size and frozen tissue analysis, it is unlikely that altered deiodinase activity is a major contributor to reduced expression of thyroid hormone-responsive genes in human liver.
Impaired thyroid hormone-responsive gene expression in human liver (Fig. 1) and resistance to T3 in vivo in mice (Fig. 2) may be due to alterations in the complex of THRs, interacting nuclear receptors, and/or coactivator/corepressors. Indeed, alterations in thyroid signaling can produce major hepatic metabolic phenotypes; introduction of the human THRB mutation (P499H) into the THRA gene in mice results in visceral obesity and hepatic steatosis, via altered PPARα signaling (29). Moreover, TRβ agonists are effective in reducing hepatic triglyceride accumulation (30,31). Interestingly, we observed changes in expression of other genes encoding proteins known to interact with THRB (the major isoform in the liver) (Supplementary Table 4). PPARGC1A (PGC1α gene), a known coactivator of THRs, was reduced by 55% in obese diabetic human liver (Fig. 3C). Thus, decreased PGC1α expression in obese liver could contribute to impaired thyroid hormone signaling, via impaired coactivation of THR (32). Conversely, impaired expression of PGC1α could also result from decreased thyroid hormone action because T3 can induce PGC1α expression in rat liver (33). PPARGC1A expression correlated with expression of genes in the energy metabolism subset of the thyroid-responsive gene set, but not with other subsets of thyroid-responsive genes (e.g. proteasome) (Supplementary Fig. 2). Moreover, PGC1α or β overexpression in isolated hepatocytes does not fully recapitulate these patterns (34). Thus, altered activity of other nuclear receptors and coactivators/repressors is likely to contribute to these patterns (35). Given the limitations of human liver biopsy samples, we could not study THR complex transcriptional complex activity.
LEPR expression was clearly decreased in liver from obese individuals (40–50% decrease in both isoforms; P < 0.05), as previously described (22). This finding may be secondary to obesity and liver fat accumulation, as observed during HFD in rats (29). It is unlikely to be a direct consequence of liver insulin resistance because LEPR expression is increased in liver of insulin receptor-deficient mice (36). Animal models of leptin deficiency (ob/ob mouse) have increased expression of genes regulating fatty acid and triglyceride synthesis and oxidative stress (37), findings concordant with our data and others (8).
Given reductions in both PGC1α and LEPR expression, we queried public expression databases to determine whether altered PGC1 expression or impaired leptin action might produce similar thyroid hormone-responsive expression patterns as observed in humans. As seen in Supplementary Fig. 4, neither PGC1α nor PGC1β overexpression in primary hepatocytes produces a pattern reciprocal to that observed in human obesity, nor does leptin deficiency (ob/ob mice) recapitulate the human pattern. Although isolated hepatic insulin resistance can cause dyslipidemia (38), thyroid-regulated gene expression does not differ in experimental liver-specific insulin resistance (Supplementary Fig. 4). Therefore, down-regulation of the thyroid hormone-responsive gene set in human obesity cannot be fully accounted for by defects within these metabolic pathways.
Our data also suggest that hyperglycemia per se may contribute specifically to gene expression, including inflammatory and profibrotic responses, as demonstrated by the positive correlation between fasting glucose and expression of HLA class II genes and TGFB1 (Supplementary Table 2). Interestingly, glucose has been shown to stimulate hepatic stellate cells to proliferate and produce collagen through free radical mediated-activation of mitogen-activated protein kinase (ERK1/2) (39). Our findings are concordant with earlier observations linking hepatic inflammation and fibrosis with hyperglycemia and diabetes (10,11). We recognize that interpretation of inflammatory expression profiles in our whole-tissue biopsy samples requires caution. Although weak activation of class II molecule expression has been demonstrated in hepatocytes (40), the expression pattern in our samples could also arise from infiltrating inflammatory cells. Expression of 1) other T or B cell markers, including CD3 subunits (δ, ε, γ, and ζ), CD4, CD8, or IL-2; 2) the dendritic cell marker cd11c; and 3) the macrophage marker cd11b was not elevated in patients with T2DM (data not shown). Similarly, markers of stellate cells were not altered (data not shown). However, definitive assessment of cell type-specific expression will require detailed immunohistochemical analysis in the future.
We recognize several limitations of our study that primarily relate to the difficulties in obtaining liver biopsies from metabolically characterized humans. First, our findings in a morbidly obese population undergoing gastric bypass may differ from a less obese cohort. For example, we did not observe increased expression of β-oxidation genes in obesity, as previously observed in studies including less obese subjects (8,9). Second, verification of T3 responsiveness of identified genes is not possible in humans because liver biopsy would be unethical in the absence of a clear clinical indication. Thus, we used a mouse model to demonstrate that obesity is associated with resistance to T3-mediated expression. We acknowledge that our sample size is relatively small for correlation analysis, and thus may not have been adequately powered to identify less robust correlations between clinical phenotypes, histopathology, and gene expression. Finally, our study groups were not optimally balanced for gender and age. The majority of study subjects were females, likely a consequence of the female preponderance of the patient population undergoing either cholecystectomy or gastric bypass surgery. However, all differences in gene expression reported for PCR verification in Fig. 1 remained significant when adjusted for sex and age (data not shown).
In conclusion, we demonstrate decreased expression of genes potentially regulated by thyroid hormone in liver samples from obese humans. Furthermore, in a mouse model of diet-induced obesity, we observed similar patterns of gene expression and resistance to acute T3 treatment. Thus, we hypothesize that impaired thyroid hormone action in the liver may contribute to multiple metabolic alterations, including derangements of both leptin and PGC1α signaling, liver fat accumulation, and potentially to NASH risk. Furthermore, our data support the concept that selective enhancement of thyroid hormone signaling in liver may have beneficial effects on not only lipid metabolism, but also liver fat accumulation (30,31).
Supplementary Material
Acknowledgments
We gratefully acknowledge research support from the National Institutes of Health, the Diabetes Genome Anatomy Project, the Graetz Family Foundation, and the General Clinical Research Center and Diabetes and Endocrinology Research Center, Joslin Diabetes Center. We also thank Joyclyn Yee and Martha Vokes for assistance with microarray analysis.
Footnotes
This work was supported by National Institutes of Health Grants DK062948 (to M.E.P.) and DK060837 (Diabetes Genome Anatomy Project, to M.E.P. and A.B.G.), DK70648 (to A.B.G.), M01 RR001032 (General Clinical Research Center), and D36836 (to Diabetes and Endocrinology Research Center, Joslin Diabetes Center). J.P. also received support from the Sigrid Juselius Foundation, Maud Kuistila Foundation, Northern Savo Cultural Foundation, and Viipuri Tuberculosis Foundation.
Disclosure Summary: The authors declare no competing financial interests.
First Published Online June 23, 2009
Abbreviations: DTT, Dithiothreitol; FDR, false discovery rate; GSEA, Gene Set Enrichment Analysis; HbA1c, hemoglobin A1c; HFD, high-fat diet; HOMA-IR, homeostasis model of assessment for insulin resistance; LEPR, leptin receptor; NASH, nonalcoholic steatohepatitis; T2DM, type 2 diabetes mellitus; THR, thyroid hormone receptor.
References
- Browning JD, Horton JD 2004 Molecular mediators of hepatic steatosis and liver injury. J Clin Invest 114:147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adiels M, Taskinen MR, Packard C, Caslake MJ, Soro-Paavonen A, Westerbacka J, Vehkavaara S, Häkkinen A, Olofsson SO, Yki-Järvinen H, Borén J 2006 Overproduction of large VLDL particles is driven by increased liver fat content in man. Diabetologia 49:755–765 [DOI] [PubMed] [Google Scholar]
- Kotronen A, Seppälä-Lindroos A, Bergholm R, Yki-Järvinen H 2008 Tissue specificity of insulin resistance in humans: fat in the liver rather than muscle is associated with features of the metabolic syndrome. Diabetologia 51:130–138 [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Garg A 2006 Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet 7:175–199 [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI 2004 Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Carreras M, Del Hoyo P, Martín MA, Rubio JC, Martín A, Castellano G, Colina F, Arenas J, Solis-Herruzo JA 2003 Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 38:999–1007 [DOI] [PubMed] [Google Scholar]
- Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML, Clore JN 2001 Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120:1183–1192 [DOI] [PubMed] [Google Scholar]
- Kohjima M, Enjoji M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Takayanagi R, Nakamuta M 2007 Re-evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int J Mol Med 20:351–358 [PubMed] [Google Scholar]
- Misu H, Takamura T, Matsuzawa N, Shimizu A, Ota T, Sakurai M, Ando H, Arai K, Yamashita T, Honda M, Yamashita T, Kaneko S 2007 Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia 50:268–277 [DOI] [PubMed] [Google Scholar]
- Chiappini F, Barrier A, Saffroy R, Domart MC, Dagues N, Azoulay D, Sebagh M, Franc B, Chevalier S, Debuire B, Dudoit S, Lemoine A 2006 Exploration of global gene expression in human liver steatosis by high-density oligonucleotide microarray. Lab Invest 86:154–165 [DOI] [PubMed] [Google Scholar]
- Younossi ZM, Gorreta F, Ong JP, Schlauch K, Del Giacco L, Elariny H, Van Meter A, Younoszai A, Goodman Z, Baranova A, Christensen A, Grant G, Chandhoke V 2005 Hepatic gene expression in patients with obesity-related non-alcoholic steatohepatitis. Liver Int 25:760–771 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP 2005 Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément K, Viguerie N, Diehn M, Alizadeh A, Barbe P, Thalamas C, Storey JD, Brown PO, Barsh GS, Langin D 2002 In vivo regulation of human skeletal muscle gene expression by thyroid hormone. Genome Res 12:281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1979 Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. National Diabetes Data Group. Diabetes 28:1039–1057 [DOI] [PubMed] [Google Scholar]
- Bergman RN 1989 Lilly Lecture 1989. Toward physiological understanding of glucose tolerance. Minimal-Model Approach. Diabetes 38:1512–1527 [DOI] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ 2005 Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41:1313–1321 [DOI] [PubMed] [Google Scholar]
- Crunkhorn S, Dearie F, Mantzoros C, Gami H, da Silva WS, Espinoza D, Faucette R, Barry K, Bianco AC, Patti ME 2007 Peroxisome proliferator activator receptor γ coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem 282:15439–15450 [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli C, Gereben B, Zavacki AM, Kim BW, Huang S, Harney JW, Larsen PR, Bianco AC 2003 In vivo dimerization of types 1, 2, and 3 iodothyronine selenodeiodinases. Endocrinology 144:937–946 [DOI] [PubMed] [Google Scholar]
- Wu Y, Delerive P, Chin WW, Burris TP 2002 Requirement of helix 1 and the AF-2 domain of the thyroid hormone receptor for coactivation by PGC-1. J Biol Chem 277:8898–8905 [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ 2003 Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100:8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC 2003 PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273 [DOI] [PubMed] [Google Scholar]
- Le D, Marks D, Lyle E, Corless CL, Diggs BS, Jobe BA, Kay T, Deveney CW, Wolfe BM, Roberts Jr CT, O'Rourke RW 2007 Serum leptin levels, hepatic leptin receptor transcription, and clinical predictors of non-alcoholic steatohepatitis in obese bariatric surgery patients. Surg Endosc 21:1593–1599 [DOI] [PubMed] [Google Scholar]
- Kaplan MM, Young JB 1987 Abnormal thyroid hormone deiodination in tissues of ob/ob and db/db obese mice. Endocrinology 120:886–893 [DOI] [PubMed] [Google Scholar]
- Gressner AM, Weiskirchen R, Breitkopf K, Dooley S 2002 Roles of TGF-β in hepatic fibrosis. Front Biosci 7:d793–d807 [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K 2006 Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–323 [DOI] [PubMed] [Google Scholar]
- Flores-Morales A, Gullberg H, Fernandez L, Ståhlberg N, Lee NH, Vennström B, Norstedt G 2002 Patterns of liver gene expression governed by TRβ. Mol Endocrinol 16:1257–1268 [DOI] [PubMed] [Google Scholar]
- Fox CS, Pencina MJ, D'Agostino RB, Murabito JM, Seely EW, Pearce EN, Vasan RS 2008 Relations of thyroid function to body weight. Arch Intern Med 168:587–592 [DOI] [PubMed] [Google Scholar]
- Targher G, Montagnana M, Salvagno G, Moghetti P, Zoppini G, Muggeo M, Lippi G 2008 Association between serum TSH, free T4, and serum liver enzyme activities in a large cohort of unselected outpatients. Clin Endocrinol (Oxf) 68:481–484 [DOI] [PubMed] [Google Scholar]
- Liu YY, Heymann RS, Moatamed F, Schultz JJ, Sobel D, Brent GA 2007 A mutant thyroid hormone receptor α antagonizes peroxisome proliferator-activated receptor α signaling in vivo and impairs fatty acid oxidation. Endocrinology 148:1206–1217 [DOI] [PubMed] [Google Scholar]
- Cable EE, Finn PD, Stebbins JW, Hou J, Ito BR, van Poelje PD, Linemeyer DL, Erion MD 2009 Reduction of hepatic steatosis in rats and mice after treatment with a liver-targeted thyroid hormone receptor agonist. Hepatology 49:407–417 [DOI] [PubMed] [Google Scholar]
- Bryzgalova G, Effendic S, Khan A, Rehnmark S, Barbounis P, Boulet J, Dong G, Singh R, Shapses S, Malm J, Webb P, Baxter JD, Grover GJ 2008 Anti-obesity, anti-diabetic, and lipid lowering effects of the thyroid receptor β subtype selective agonist KB-141. J Steroid Biochem Mol Biol 111:262–267 [DOI] [PubMed] [Google Scholar]
- Crunkhorn S, Patti ME 2008 Links between thyroid hormone action, oxidative metabolism, and diabetes risk? Thyroid 18:227–237 [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Hamann S, Jauk M, Lacey M, Filbry A, Radtke C, Iwen KA, Kutz S, Harneit A, Lizardi PM, Seitz HJ 2003 Hepatic gene expression patterns in thyroid hormone-treated hypothyroid rats. J Mol Endocrinol 31:291–303 [DOI] [PubMed] [Google Scholar]
- Spiegelman B 2004 Effect of PGC-1α and PGC-1β on gene expression in myocytes and hepatocytes. http://www.diabetesgenome.org/chipperdb/expt.cgi?id=73 [Google Scholar]
- Astapova I, Lee LJ, Morales C, Tauber S, Bilban M, Hollenberg AN 2008 The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci USA 105:19544–19549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SE, Kokkotou E, Biddinger SB, Kondo T, Gebhardt R, Kratzsch J, Mantzoros CS, Kahn CR 2007 High circulating leptin receptors with normal leptin sensitivity in liver-specific insulin receptor knock-out (LIRKO) mice. J Biol Chem 282:23672–23678 [DOI] [PubMed] [Google Scholar]
- Ferrante Jr AW, Thearle M, Liao T, Leibel RL 2001 Effects of leptin deficiency and short-term repletion on hepatic gene expression in genetically obese mice. Diabetes 50:2268–2278 [DOI] [PubMed] [Google Scholar]
- Biddinger SB, Hernandez-Ono A, Rask-Madsen C, Haas JT, Alemán JO, Suzuki R, Scapa EF, Agarwal C, Carey MC, Stephanopoulos G, Cohen DE, King GL, Ginsberg HN, Kahn CR 2008 Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab 7:125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto R, Enjoji M, Kohjima M, Tsuruta S, Fukushima M, Iwao M, Sonta T, Kotoh K, Inoguchi T, Nakamuta M 2005 High glucose stimulates hepatic stellate cells to proliferate and to produce collagen through free radical production and activation of mitogen-activated protein kinase. Liver Int 25:1018–1026 [DOI] [PubMed] [Google Scholar]
- Ballardini G, Bianchi FB, Mirakian R, Fallani M, Pisi E, Bottazzo GF 1987 HLA-A,B,C, HLA-D/DR and HLA-D/DQ expression on unfixed liver biopsy sections from patients with chronic liver disease. Clin Exp Immunol 70:35–46 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.