Abstract
In the recent past, several thousand noncoding RNA (ncRNA) genes have been predicted within eukaryal genomes. However, for their functional analysis only a few high-throughput methods are currently available to knock down selected ncRNA species, such as microRNAs, which are targeted by antisense probes, termed antagomirs. We thus compared the efficiencies of four knockdown strategies, previously mainly employed for the analysis of protein-coding genes, to study the function of ncRNAs, in particular, small nucleolar RNAs (snoRNAs). Thereby, the class of snoRNAs represents one of the most abundant ncRNA species. The majority of snoRNAs has been shown to mediate nucleotide modifications by targeting ribosomal RNAs (rRNAs) through complementary antisense elements. However, some snoRNAs, termed “orphan snoRNAs,” lack telltale complementarities to rRNAs and thus their function remains elusive. We therefore applied RNA interference (RNAi), locked nucleic acid (LNA), or peptide nucleic acid antisense approaches, as well as a ribozyme-based strategy to knock down a snoRNA. As a proof of principle, we targeted the canonical U81 snoRNA, which has been shown to mediate modification of nucleotide A391 within eukaryal 28S rRNA. Our results demonstrate that while RNAi is an unsuitable tool for snoRNA knockdown, a ribozyme-based strategy, as well as an LNA-antisense oligonucleotide approach, resulted in a decrease of U81 snoRNA expression levels up to 60%. However, no concomitant decrease in enzymatic activity of U81 snoRNA was observed, indicating that improvement of more efficient knockdown techniques for ncRNAs will be required in the future.
Keywords: noncoding RNA, knockdown, snorbozyme, antisense, functional analysis
INTRODUCTION
The number of newly discovered small noncoding RNAs (ncRNAs) has increased exponentially in recent years (Hüttenhofer et al. 2005). By experimental, as well as computational approaches, an unexpected large number of small ncRNAs, in particular, small nucleolar RNAs (snoRNAs), has been identified in different model organisms (Washietl et al. 2005; Lung et al. 2006; Madej et al. 2007; Jochl et al. 2008).
For functional characterization of protein-coding genes, knockdown as well as knockout approaches have been employed. As a conventional gene knockout is highly time consuming; knockdown approaches—such as RNA interference (RNAi)—have become the method of choice to investigate the function of protein-coding genes. In contrast, knockdown approaches have not routinely been employed for the functional characterization of ncRNAs. As an exception, for knock down of microRNAs (miRNAs), chemically engineered oligonucleotides, termed “antagomirs,” as well as locked nucleic acids (LNAs), have been employed in vivo to inactivate their function (Krutzfeldt et al. 2005; Orom et al. 2006). In addition, RNAi was shown to serve as a useful tool to knock down 7SK RNA in the nucleus of human cells (Robb et al. 2005).
Furthermore, Liang et al. (2003) showed knockdown of four mature snoRNAs, but not of snoRNA precursors, in Leptomonas collosoma, Leishmania major, and Trypanosoma brucei species. However, knockdown efficiencies by small interfering RNAs (siRNAs) varied significantly, depending on the snoRNA species, suggesting different accessibilities within target sites. More importantly, for two additional snoRNAs no knockdown could be observed.
SnoRNAs are located in a subnuclear compartment, the nucleolus, and therefore, might represent one of the most difficult ncRNA species to target. They exhibit sizes between 60 and 300 nucleotides (nt) and act as so-called “guide RNAs,” which mediate modifications within ribosomal RNAs (rRNA) or small nuclear RNAs (snRNA), respectively.
Based on the presence of conserved sequence and structure motifs, snoRNAs are divided into two major classes, box C/D or H/ACA snoRNAs, respectively (Hüttenhofer et al. 2002). Both snoRNA classes associate with a common set of four core proteins each, and assemble into small ribonucleo-protein particles (RNPs), designated as snoRNPs. By specific base pairing of short antisense elements, located within snoRNAs, to a complementary RNA-target snoRNAs direct either site-specific 2′-O-ribose methylation (box C/D snoRNAs) or pseudouridylation (box H/ACA snoRNAs) (Bachellerie et al. 2002; Decatur and Fournier 2002).
For numerous snoRNA species, designated as “orphan snoRNAs,” no known target sites within ribosomal or small nuclear RNAs could be identified, as of now (Hüttenhofer et al. 2001; Bachellerie et al. 2002). Hence, they might target other RNA species, such as messenger RNAs, and thus function as regulators of gene expression. As an example, the orphan snoRNA MBII-52 (HBII-52 in humans) has been postulated to target the serotonin receptor 5-HT2C mRNA, thereby influencing alternative splicing or RNA editing (Cavaille et al. 2000; Vitali et al. 2005; Kishore and Stamm 2006). Hence, in this study, we compared (1) the efficiency of RNA interference, (2) a ribozyme-based approach, and (3) LNA or peptide nucleic acid (PNA) antisense oligonucleotides (ASOs) as molecular tools to knock down snoRNAs.
The efficiency of RNA interference
RNA interference (RNAi) has recently emerged as an efficient method to silence expression of protein-coding genes in mammalian cells (Elbashir et al. 2002; Tijsterman and Plasterk 2004). Thereby, small 19–21-nt-long RNA oligonucleotide duplexes, i.e., siRNAs, are incorporated into the RNA-induced silencing complex (RISC) and mediate the sequence-specific degradation of the complementary RNA target (Elbashir et al. 2002; Serganov and Patel 2007). Besides delivering siRNA as oligonucelotide duplexes, RNAi-mediated knockdown can also be initiated by expression of short hairpin RNAs (shRNAs) (Paddison et al. 2002). ShRNAs are typically transcribed, as stem–loop structures, by RNA polymerase III, and are subsequently cleaved by the enzyme Dicer. Even though RNA interference has initially been reported within the cytoplasm, recent reports suggest that the RNAi machinery might also exert its function within the nucleus of mammalian cells (Robb et al. 2005).
A ribozyme-based approach
“Ribozymes” are RNA-based enzymes that catalyze chemical reactions in the absence of proteins. Naturally occurring ribozymes serve as catalysts for numerous biological reactions, such as RNA cleavage, RNA splicing RNA ligation, and protein synthesis (Birikh et al. 1997; Cech 2000). The hammerhead ribozyme (HHrz) catalyzes the site-specific cleavage of a phosphodiester bond yielding a 2′,3′-cyclic phosphate and a 5′-hydroxyl product (Murray et al. 1998; Warashina et al. 2001; Blount and Uhlenbeck 2002). Besides their pivotal role in different biological processes, ribozymes are also employed as molecular tools for sequence-specific inhibition of gene expression through intermolecular cleavage of target mRNA molecules (Bertrand and Rossi 1994; Lebedeva and Stein 2001).
LNA or PNA antisense oligonucleotides as molecular tools to knock down snoRNAs
Antisense oligonucleotides are 15–25-nt-long, single-stranded DNA or RNA analogs that are designed to specifically bind to essential sites within target RNAs (Lauritsen and Wengel 2002). So-called “gapmer ASOs” are composed of two LNA segments (Koshkin et al. 1998; Kurreck et al. 2002) flanking a central DNA core (Scherer and Rossi 2003). When binding to the RNA target, the DNA/RNA heteroduplex is recognized by RNase H, which mediates degradation of the RNA moiety of the heteroduplex (Egholm et al. 1993). In contrast to LNA gapmers, PNA ASOs exhibit a N-(2-aminoethyl)-glycine backbone (Nielsen et al. 1991), upon binding to a target RNA essential sites within the RNA are blocked, thus resulting in the inhibition of the function.
In this study, we compared four different knockdown strategies (i.e., RNAi, a ribozyme-based strategy, as well as two different antisense approaches by PNA or LNA, respectively) for their efficiency to inhibit the function of the canonical U81 snoRNA. Due to their small size, nucleolar localization, and complex structure, snoRNAs might represent the most challenging knockdown targets within an eukaryal cell.
RESULTS AND DISCUSSION
RNA interference is not a general suitable tool for snoRNA knockdown
To investigate whether siRNAs are able to knock down snoRNAs, we generated two vectors, expressing shRNAs encoding siRNAs designed to trigger cleavage of the U81 snoRNA at positions 32 and 58, respectively (see Fig. 1A). These two sites within U81 snoRNA optimally fulfill the requirements for efficient cleavage according to siRNA target-design rules (Elbashir et al. 2002). In addition, the cleavage site at position 32 is located within the D′ antisense element of U81 snoRNA, which has been reported to guide methylation of A391 within 28S rRNA (Maden 2001), and thus, is likely to represent one of the most accessible sites within U81 snoRNA.
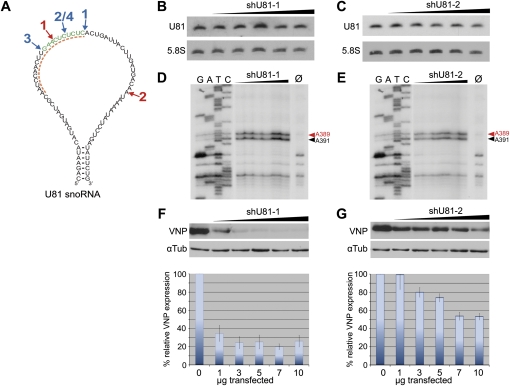
FIGURE 1.
(A) Secondary structure of the U81 snoRNA. Snorbozyme targeting sites (indicated in blue, RZ1-4), shRNA targeting sites (indicated in red, shU81-1 and shU81-2), and the ASO target site (indicated by a dashed orange line) are shown. (B,C) Analysis of U81 snoRNA expression and methylation activity in HEK-293T cells, transfected with increasing amounts (1–10 μg) of shRNA-expressing plasmids. (B,C) Northern blot analysis: two shRNAs, both targeting U81 snoRNA (shU81-1 and shU81-2, respectively), do not reduce U81 snoRNA expression levels. (D,E) Primer extension analysis of nucleotide A391 within 28S rRNA investigating the 2′-O-methylation activity of U81 snoRNA within HEK-293T cells, transfected with increasing amounts (1–10 μg) of shRNA-expressing plasmids (see above). Methylation of A391 is indicated by a black arrow; a second 2′-O-methylation at position A389, which is guided by U26 snoRNA, is indicated by a red arrow and serves as an internal loading control. Ø: reverse transcription control reaction at 0.5 mM dNTP lacking a stop signal at A391 and A389, respectively (see text). (F,G) Top: Western blot analysis showing reduction in the expression levels of VNP fluorescent protein; target sites for shU81-1 or shU81-2 were inserted in the VPN gene as described in Materials and Methods. Bottom: FACS analysis: upon transfection of increasing amounts of shRNA expressing plasmids (see above), VNP expression in HEK-293T cells is reduced by about 80% (shU81-1) or 50% (shU81-2), respectively.
To determine the efficiencies of siRNAs to knock down U81 snoRNA, increasing amounts of plasmid DNA (i.e., from 1 to 10 μg) of each vector construct (shU81-1, shU81-2) were transfected into HEK-293T cells. Subsequently, expression levels of U81 snoRNA were analyzed by Northern blotting, while its enzymatic activity was assayed investigating the methylation efficiency of the U81 snoRNP at position A391 within 28S rRNA by primer extension analysis. In this assay, reverse transcription of 28S rRNA, employing an oligonucleotide primer in close vicinity to the modified nucleotide at position A391 results in a stop of the reverse transcriptase at the methylation site (Rebane et al. 2002). This is only observed at low dNTP concentrations (i.e., at 0.04 mM) whereas at high dNTP concentrations (i.e., at 0.5 mM dNTP), the reverse transcription elongates without pausing (see Materials and Methods).
Northern blotting, as well as primer extension analysis, showed that neither expression levels of U81 snoRNA nor its enzymatic activity (assayed by the methylation status of A391) changed upon transfection of shRNA-expressing plasmids (Fig. 1B–E). A plausible explanation might be that snoRNAs (and snoRNA precursors) are located within the nucleolus and the nucleus, respectively, whereas shRNAs are known to be exported to the cytoplasm by Exportin-5 (Zeng and Cullen 2004). Thus, a significant co-localization between siRNA and its U81 snoRNA target seems unlikely.
To assess shRNA function, we demonstrated in vitro cleavage of U81 snoRNA by incubation of radiolabeled U81 snoRNA with an S100 extract, derived from HEK-293T, cells together with the shRNA, as shown previously for other targets (Elbashir et al. 2001; Meister et al. 2004; data not shown). Additionally, we introduced the region of U81 snoRNAs, which harbors the target sites for shU81-1 and shU81-2 RNAs, respectively, downstream of the start codon of the VNP fluorescent protein. Subsequently, shRNA expression vectors, as well as the respective VNP construct, were co-transfected into HEK-293T cells. Expression levels of recombinant VNP protein were assessed by Western blot analysis (Fig. 1F,G, top) and flow cytometry (Fig. 1F,G, bottom). As expected, both shRNA constructs showed a significant reduction of VNP protein expression levels. The most efficient decrease of VNP protein (to about 20%) was observed at low yields (1.5 μg) of shU81-1 plasmid; higher plasmid amounts did not increase knockdown efficiencies (Fig. 1F, bottom). In contrast to the observations of Liang et al. (2003), our data suggest that RNA interference might not be a useful general tool for knock down of ncRNA species, located in the nucleolus of a cell.
Snorbozymes reduce U81 snoRNA expression levels, but do not affect its enzymatic activity
For a ribozyme-based snoRNA knockdown approach we generated “snorbozymes” (Samarsky et al. 1999; Michienzi et al. 2000), which represent a hybrid between a HHrz that targets a snoRNA, and a carrier snoRNA (U16 or U20) that transports the ribozyme into the nucleolus. Upon co-localization with the snorbozyme, the target snoRNA will be cleaved and subsequently digested by exonucleases (Samarsky et al. 1999).
We generated four different hammerhead riboyzmes targeting U81 snoRNA at three sites within its antisense element (Fig. 1A) and inserted them either in the apical loop of U16 snoRNA (U16 snorbozymes) or within the antisense box of U20 (U20 snorbozymes) (Supplemental Fig. S1B). Subsequently, efficiencies of snorbozyme constructs were assessed by in vitro cleavage of U81 snoRNA (Supplemental Fig. S1A). With the exception of RZ3, all snorbozymes constructs showed efficient cleavage of the U81 snoRNA target. Thus, we refrained from further analyzing the RZ3 constructs. For expression in mammalian cells, controlled by the H1 RNA polymerase III promoter, snorbozymes were ligated into the pENTR-THT (U191) vector. Since colocalization between the ribozyme and its snoRNA target is essential for cleavage, we investigated the subcellular localization of snorbozymes within HEK-293T cells by fluorescent in situ hybridization. Indeed, U16, as well as U20, snorbozymes localized to the nucleolus of cells together with the nucleolar marker U3. (Fig. 2A; and data not shown).
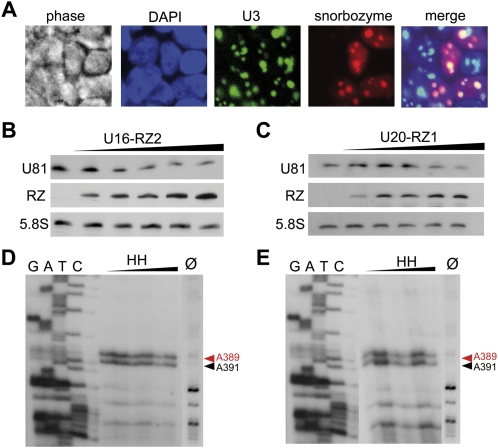
FIGURE 2.
“Snorbozyme” mediated knock down of U81 snoRNA. (A) Fluorescent in situ hybridization of U16-RZ2 or U20-RZ1 snorbozymes transfected into HEK-293T cells: snorbozyme expression was detected by employing a Cy3-labeled (red) antisense oligonucleotide. U3 snoRNA was used as a nucleolar marker and was visualized by employing an Oregon Green 488-labeled antisense oligonucleotide; nuclei of HEK-293T cells were stained with DAPI (blue). (B,C) Northern blot analysis of U81 snoRNA and snorbozyme expression levels: upon transfection of increasing amounts (1–10 μg) of U16-RZ2 and U20-RZ1 snorbozyme expressing plasmids, U81 expression levels were reduced by 60% and 30%, respectively. (D,E) Primer extension analysis of nucleotide A391 within 28S rRNA investigating the 2′-O-methylation activity of U81 snoRNA within HEK-293T cells, transfected with U16-RZ2 or U20-RZ1 snorbozyme constructs. Methylation of A391 is indicated by a black arrow; a second 2′-O-methylation at position A389, which is guided by U26 snoRNA, is indicated by a red arrow and serves as an internal loading control. Ø: reverse transcription control reaction at 0.5 mM dNTP lacking a stop signal at A391 and A389, respectively (see the text).
To investigate whether snorbozymes are able to mediate U81 cleavage in vivo, we transfected increasing amounts of selected snorbozyme-expressing plasmids (see above) into HEK-293T cells. After 72 h, U81 expression, as well as the methylation status of A391 of 28S rRNA, was determined. Northern blot analysis demonstrated that U16-RZ2, as well as U20-RZ1, snorbozyme constructs were able to mediate U81 snoRNA degradation (Fig. 2B,C). However, even though U81 expression levels were reduced up to 60%, as assessed by Northern blotting, methylation levels of A391 within 28S ribosomal RNA did not change (Fig. 2D,E).
Likely, remaining amounts of the uncleaved U81 snoRNA are sufficient to mediate wild-type levels of methylation at A391 within 28S rRNA. A detectable change in methylation levels at position A391, however, will only be expected to be observed after several cell divisions, since at the time point of transfection A391 within 28S rRNA is methylated to wild-type levels. Hence, upon inactivation of U81 snoRNA, only newly synthesized ribosomes will appear unmethylated at this position. Since HEK-293T cells divide about once within 24 h, the maximum decrease in A391 methylation, which can be expected 72 h post-transfection, is about 87%, based on a 100% knockdown efficiency.
LNAs, but not PNAs, mediate U81 snoRNA degradation
As a third strategy to inhibit ncRNA function, we compared inhibition efficiencies of two antisense oligonucleotide approaches, i.e., LNA or PNA ASOs complementary to the antisense box of U81 snoRNA. Apart from an unmodified control PNA ASO, which is unable to cross the cellular membrane, we employed modified PNA ASOs fused to either a peptide encoding nuclear localization signal (NLS), a tri-lysine or a nitro modification at its N-terminus. These modifications are expected to either enhance binding of the PNA to its RNA target (tri-lysine and nitro modification) or, in case of the addition of a NLS, are thought to enhance transport of the PNA ASO to the nucleus.
We first investigated the intracellular localization of Cy5-labeled LNAs or Lissamin-labeled PNA (lissamin) ASOs by confocal microscopy (Fig. 3A). Thereby, LNAs localized mainly to the cytoplasm, but also to some exten to nucleoli, whereas PNAs appeared as dot-like structures within the cytoplasm. Notably, we could not observe localization of PNA ASOs either to the nucleoplasm or to nucleoli. However, independent on their N-terminal modification, all PNAs that entered the cell showed a “dot-like,” cytoplasmic distribution (Fig. 3A).
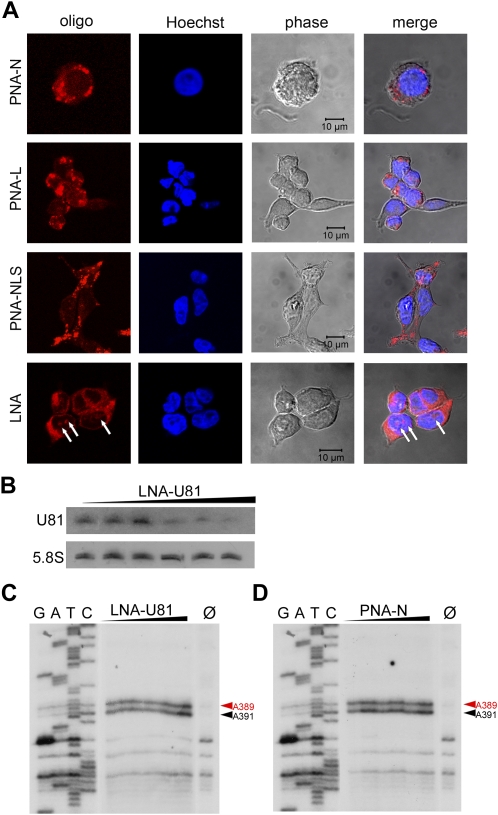
FIGURE 3.
Investigation of knockdown efficiencies of antisense oligonucleotides (ASOs) targeting U81 snoRNA. Intracellular localization of the LNA ASO and PNA ASO was determined by confocal microscopy. (A) The LNA ASO shows predominatly cytoplasmatic localization and, to a lower extent, a nucleolar distribution (indicated by white arrows); in contrast, all PNAs show a dot-like distribution within the cytoplasm, but do not locate to nucleoli; nuclei were stained with Hoechst 33342. (B) Northern blot analysis of U81 expression levels in HEK-293T cells, transfected with the LNA-U81 ASO: transfection was performed with increasing amounts of LNA-U81 ASO (12.5, 25, 50, 100, and 200 mM). At highest LNA concentrations, a reduction of about 60% was observed. (C,D) Primer extension analysis of nucleotide A391 within 28S rRNA investigating the 2′-O-methylation activity of U81 snoRNA within HEK-293T cells, transfected with LNA or PNA ASOs, respectively. Methylation of A391 is indicated by a black arrow; a second 2′-O-methylation at position A389, which is guided by U26 snoRNA, is indicated by a red arrow and serves as an internal loading control. Ø: reverse transcription control reaction at 0.5 mM dNTP lacking a stop signal at A391 and A389, respectively (see the text).
To examine the inhibition efficiencies of LNA or PNA ASOs, we transfected increasing amounts of LNAs into HEK-293T cells. For PNAs, which passively cross the cellular membrane, we added an eightfold amount to the cell culture media to reach equivalent concentration levels as for LNA-transfected cells. After 72 h of incubation with ASOs, cells were analyzed for expression levels of U81 snoRNA and the methylation status of A391 within 28S rRNA. U81 expression levels were only assessed for LNA ASOs since PNA ASOs were not expected to reduce RNA levels by RNase H cleavage.
Northern blot analysis demonstrated that, in general, LNAs were able to partially knock down snoRNAs in an in vivo cell culture system. A significant reduction in the expression levels of U81 snoRNA could be observed (Fig. 3B). We note, however, that reduction of U81 expression strongly depended on transfection efficiencies and varied from 10% to 60%. This reduction of expression was likely mediated by RNase H, since the LNAs employed in this study consisted of a central DNA portion, flanked by LNA nucleotides. Following binding of this mixmer oligonucleotide to its RNA target, the DNA–RNA heteroduplex was predicted to be recognized by RNase H and cleaved within its RNA portion. We confirmed this assumption by an in vitro assay, in which an LNA–U81 heteroduplex was digested by RNase H (data not shown). As observed for snorbozymes, reduction of expression levels of U81 snoRNAs by LNA ASOs did not result in a concomitant reduction of A391 methylation (Fig. 3C).
In cells exposed to PNA ASOs no decrease in methylation of A391, mediated by U81 snoRNA, could be observed (Fig. 3D). This is not unexpected due to the observation that PNAs accumulate within the cytoplasm and, thus, are not able to interact with snoRNAs or their precursors located within the nucleus or the nucleolus. Since PNAs are known to inhibit their RNA targets by blocking accessible sites and not by degradation of targeted RNAs, we refrained from analyzing expression levels of U81 snoRNA.
CONCLUSION
All of the four employed strategies for knock down of U81 snoRNA are based on the sequence-specific inhibition of the complementary target by either RNA cleavage or blocking a functional site within the targeted RNA. These knockdown strategies were aimed to target the RNA component of the snoRNP particle by Watson–Crick base pairing. Although significant knockdown efficiencies of up to 60% were achieved, no detectable changes in methylation activity of the U81 snoRNP could be observed. This is likely due to the fact that the remaining, uncleaved snoRNA levels suffice to retain full enzymatic snoRNP function. For further studies on the biological roles of orphan snoRNAs, it will thus be required that knockdown efficiencies are improved significantly. In contrast, for some of the numerous eukaryal ncRNA candidates of unknown functions, the obtained knockdown efficiencies might indeed suffice to elucidate their biological roles.
Our studies demonstrate that, currently, knock down of eukaryal ncRNA candidates, exhibiting unknown functions, might most efficiently be achieved by either a ribozyme-based approach or by employing LNA oligonucleotides. However, our study also implicates that novel and more efficient knockdown techniques will have to be established for future analysis of all ncRNAs exhibiting unkown functions. As the design and generation of effective ribozymes is rather time consuming, LNA ASOs might turn out as the method of choice to effectively inhibit the function of ncRNA candidates. Modifications of LNA oligonucleotides that specifically target these molecules to subcellular compartments will be a further step to enhance their knockdown efficiencies.
MATERIALS AND METHODS
Plasmid constructs
Plasmids expressing shRNAs targeting U81 snoRNA (Fig. 1A) were generated by annealing phosphorylated oligonucleotides, followed by ligation into a BglII/HindIII digested pENTR-THT-U191 vector (Ploner et al. 2008). Recombinant Venus-NLS-PEST (VNP) fluorescent protein (Nagoshi et al. 2004) was generated by PCR using primers containing the siRNA target sequence. Employing Invitrogen's Gateway recombination system, PCR products were recombined into pDONR-207 (Invitrogen) and subsequently into pDelta-T-Flag-Dest vector (kindly provided by Stephan Geley, Biocenter Innsbruck), thereby generating mammalian expression vectors.
“Snorbozymes” were cloned into a BglII/HindIII digested pENTR-THT-U191 vector. For the insertion of various hammerhead ribozymes, a multiple cloning site (NdeI/StuI/NcoI) was inserted either in the apical loop of U16 or instead of the antisense box of U20 snoRNA (Supplemental Fig. S1B). Subsequently, ribozymes were cloned into the NdeI and NcoI restriction sites.
Antisense oligonucleotides
LNA gapmer oligonucleotides (5′-GAGAGAGTTCAAGTTGGA-3′), comprised of 40% LNA monomers and 60% DNA monomers, targeting U81 snoRNA within its antisense element, have been designed and generated by Exiqon (Vedbaek; www.exiqon.com). To investigate their localization within HEK-293T cells, LNA oligonucleotides were Cy5-labeled at the 5′ end. LNAs were transfected using Metafectene (Biontex) transfection reagent.
Cell membrane crossing oligonucleotides (CMCOs) harboring the identical sequence as LNA oligomers have been synthesized by Ugichem (Innsbruck; www.ugichem.at). CMCOs are modified PNAs that passively enter cells without the addition of transfection reagents. To enhance solubility and binding kinetics of the ASOs, a tri-lysine (PNA-L) or a nitro (PNA-N) modification was added to the N-terminus of the PNA molecule. In addition, a PNA-variant harboring a nuclear localization signal (PNA NLS) was synthesized. As a control, a PNA oligonucleotide, which was not able to cross the cellular membrane, was also used. For in vivo detection, PNAs were labeled with the fluorescent dye lissamin, emitting red light at a wavelength of 586 nm.
Cell culture
HEK-293T cells were cultured in DMEM (PAA) containing 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (PAA) at 37°C and 5% CO2. For transient expression, cells were grown in six-well plates to 80%–90% confluency and further transfected with 1–10 μg of plasmid DNA or antisense oligonulceotides employing Metafectene as a transfection reagent (Biontex). Cells were harvested 72 h after transfection and subjected to further analysis.
Fluorescent in situ hybridization
In situ hybridization was performed according to the Singer Lab protocol (see http://singerlab.aecom.yu.edu/protocols/). For detection we used the following amino-modified thymidine nucleotides (C2-amino-linker) containing primers:
AP128-Insitu-U3: 5′-GT*TCTCTCCCTCT*CACTCCCCAAT*ACGGAGAGAAGAACGAT*CATCAATGGCT*G-3′; and
AP129-Insitu-HH-RZ2: 5′-CAT*GGAACTTGAACT*TTCGCCGCGAACGGCT*CATCAGTCTCACCAT*A-3′.
T* indicates the amino-modified thymidine nucleotides. The specific oligonculeotide for U3 was chemically conjugated with Oregon green 488 (Invitrogen), the HHrz probe was chemically conjugated to the Cy3 fluorophore (CyTM3 monofunctional reactive dye; Amersham Pharmacia). Nucleoli were stained by DAPI (0.1 μg/mL) (Invitrogen). Localization of the HHrz was analyzed by fluorescent microscopy using a Zeiss Axiovert 200M microscope.
RNA isolation and Northern blot analysis
Total RNA was isolated according to the TRIzol (Gibco BRL) manual; 10–20 μg of total RNA were denatured for 3 min at 95°C in 1X RNA loading dye (47.5% formamide, 0.0125% SDS, 0.0125% bromophenol blue) and separated on a denaturing 8% polyacrylamide gel (7 M urea, 1× TBE). Electrophoresis was performed at 10 [W]. Transfer onto Hybond-N+ membrane (Amersham Biosciences) was performed by electroblotting using a semidry blotting apparatus (TransblotSD, Bio-Rad) at 400 mA, 15 V (max) for 45 min in 0.5 TBE buffer. RNA was immobilized by UV-cross-linking with 120 mJ in a UV-Stratalinker (Stratagene). Oligonucleotides were 5′-labeled with [γ-32P] ATP using T4 polynucleotide kinase (Promega). Hybridization was carried out at 45°C in 1 M sodium phosphate buffer (pH 6.2), 7% (w/v) SDS for 12 h. Subsequently, blots were washed for 10 min at RT in washing solution I (2X SSC, 0.1% [w/v] SDS) followed by a second washing step at RT for 10 min with washing solution II (0.2X SSC, 0.1% [w/v] SDS). To increase specificity, blots were additionally washed at 58°C in washing solution II for 1–3 min. Membranes were either exposed to Kodak MS-1 film, using an intensifier screen or to a PhosphorImager screen (Bio-Imaging Analyzer) overnight (16 h). As a loading control, membranes were hybridized with a DNA oligonucleotide complementary to 5.8S ribosomal RNA.
In vitro snorbozyme reaction
For in vitro snorbozyme reactions, 15 pmol of 32P body labeled U81 snoRNA were incubated with 75 pmol snorbozyme in 40 mM Tris/HCl at pH 7.5 and 10 mM MgCl2 for 1 h at 37°C. Subsequently, the reactions were dissolved on a denaturing polyacrylamide gel, and cleavage efficiencies were determined using a PhosphorImager screen.
Primer extension analysis
To detect 2′-O-methylation of riboses on ribosomal RNA, a primer extension assay was performed. Two micrograms of total RNA were heat denatured for 5 min at 96°C in the presence of 0.1 pmol 5′ 32P end-labeled oligodeoxynucleotide primer. Following a 30 min hybridization step at 42°C, reverse transcription was carried out in buffer containing 122.5 mM Tris/HCl at pH 8.4, 11 mM MgCl2, 15 mM KCl, 11 mM DTT, 0.5/0.04 mM dNTPs, and 1 unit AMV reverse transcriptase (Promega). For sequencing reactions, dideoxynucleotides were added to a final concentration of 0.06 mM. cDNA products were ethanol precipitated and primer extension products were resolved on a 8% denaturing polyacrylamide gel with sequencing reactions and visualized by autoradiography.
Western blot analysis
For preparation of whole cell extracts, 5 × 106–1 × 107 cells were washed once in PBS and resuspended in 60 μL RIPA buffer containing 50 mM Tris at pH 7.5, 150 mM NaCl, 1 mM ETDA, 0.1% SDS, 0.5% sodium deoxycholate, and 1% NP-40. Prior to usage, a protease inhibitor mix was added. Samples were incubated for 1 h on ice and cleared by centrifugation (15,000g, 10 min, 4°C). Supernatants were collected and the amount of protein was determined by Bradford analysis. Twenty micrograms of total protein were mixed with the appropriate amount of 4XSSB-buffer containing 5% β-mercaptoethanol. Protein samples were denatured at 98°C for 5 min and size fractionated on 12.5% SDS/polyacrylamide gels in a CBS minigel chamber at 200 V/50 mA. Proteins were transferred by electroblotting onto nitrocellulose membranes (0.45 μm) by a Bio-Rad semidry transfer apparatus (I = 0.8 mA/cm2, constant). Transfer efficiency was assessed by Ponceau-S-red staining. Membranes were incubated in Western blot blocking buffer (PBS/1% NP-40, 5% MP) for 2 h and with a rabbit GFP antibody detecting VNP fluorescent protein (kindly provided by Stephan Geley, Biocenter Innsbruck) overnight at 4°C. Membranes were washed three times in PBS/1% NP-40 for 10 min each. The anti-rabbit horseradish peroxidase-conjugated secondary antibody was diluted 1:1000 in blocking buffer and added to the membranes for 45 min at room temperature. Thereafter, the membranes were washed three times with PBS/1% NP-40 and incubated with ECL chemiluminescence substrate according to the manufacturer's instructions (Amersham). The blots were exposed to AGFA Curix X-ray films.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Setphan Geley for providing expression vectors, and Matthias Erlacher, Christoph Jöchl, Norbert Polacek, and Mathieu Rederstorff for critically reading of the manuscript. We also thank Holger Bock and Thomas Lindthorst (Ugichem) for providing PNAs. This work was supported by an EU grant D-110420-011-011 of the 6th Framework Program (6FP), an Austrian genome research grant (Gen-Au grant D 110420-011-013) to A.H., and a grant from the Austrian Science Fund (SFB-F021).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1740009.
REFERENCES
- Bachellerie JP, Cavaille J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Bertrand EL, Rossi JJ. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birikh KR, Heaton PA, Eckstein F. The structure, function, and application of the hammerhead ribozyme. Eur J Biochem. 1997;245:1–16. doi: 10.1111/j.1432-1033.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC. The hammerhead ribozyme. Biochem Soc Trans. 2002;30:1119–1122. doi: 10.1042/bst0301119. [DOI] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. Structural biology. The ribosome is a ribozyme. Science. 2000;289:878–879. doi: 10.1126/science.289.5481.878. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: An experimental approach that identifies 201 candidates for novel, small, nonmessenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Brosius J, Bachellerie JP. RNomics: Identification and function of small, nonmessenger RNAs. Curr Opin Chem Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Schattner P, Polacek N. Noncoding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Hüttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with antagomirs. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen A, Wengel J. Oligodeoxynucleotides containing amide-linked LNA-type dinucleotides: Synthesis and high-affinity nucleic acid hybridization. Chem Commun (Camb) 2002:530–531. doi: 10.1039/b111000d. [DOI] [PubMed] [Google Scholar]
- Lebedeva I, Stein CA. Antisense oligonucleotides: Promise and reality. Annu Rev Pharmacol Toxicol. 2001;41:403–419. doi: 10.1146/annurev.pharmtox.41.1.403. [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc Natl Acad Sci. 2003;100:7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Hüttenhofer A. Identification of small noncoding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej MJ, Alfonzo JD, Hüttenhofer A. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. 2007;35:1544–1554. doi: 10.1093/nar/gkm004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BE. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Michienzi A, Cagnon L, Bahner I, Rossi JJ. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc Natl Acad Sci. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG. The structural basis of hammerhead ribozyme self-cleavage. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, Kofler R. The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia. 2008;22:370–377. doi: 10.1038/sj.leu.2405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebane A, Roomere H, Metspalu A. Locations of several novel 2′-O-methylated nucleotides in human 28S rRNA. BMC Mol Biol. 2002;3:1. doi: 10.1186/1471-2199-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Samarsky DA, Ferbeyre G, Bertrand E, Singer RH, Cedergren R, Fournier MJ. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc Natl Acad Sci. 1999;96:6609–6614. doi: 10.1073/pnas.96.12.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer LJ, Rossi JJ. Approaches for the sequence-specific knock down of mRNA. Nat Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Ribozymes, riboswitches, and beyond: Regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Plasterk RH. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117:1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Hüttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warashina M, Kuwabara T, Kato Y, Sano M, Taira K. RNA–protein hybrid ribozymes that efficiently cleave any mRNA independently of the structure of the target RNA. Proc Natl Acad Sci. 2001;98:5572–5577. doi: 10.1073/pnas.091411398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]