Abstract
Bacteria spend their lives buffeted by changing environmental conditions. To adapt to and survive these stresses, bacteria have global response systems that result in sweeping changes in gene expression and cellular metabolism. These responses are controlled by master regulators, which include: alternative sigma factors, such as RpoS and RpoH; small molecule effectors, such as ppGpp; gene repressors such as LexA; and, inorganic molecules, such as polyphosphate. The response pathways extensively overlap and are induced to various extents by the same environmental stresses. These stresses include nutritional deprivation, DNA damage, temperature shift, and exposure to antibiotics. All of these global stress responses include functions that can increase genetic variability. In particular, up-regulation and activation of error-prone DNA polymerases, down-regulation of error-correcting enzymes, and movement of mobile genetic elements are common features of several stress responses. The result is that under a variety of stressful conditions, bacteria are induced for genetic change. This transient mutator state may be important for adaptive evolution.
Keywords: adaptive mutation, stationary-phase mutation, SOS response, general stress response, error-prone polymerase, polyphosphate
INTRODUCTION
In a stimulating but controversial paper published in 1988, Cairns et al. (1988) described experiments suggesting that a population under selection had access to a process that could direct mutagenic change to the very genes that would relieve the selective pressure. Such a process, of course, would be a great boon to adaptive evolution (Fitch, 1982). After nearly 20 years of research, evidence now suggests that various types of stresses induce responses that have mutagenic consequences, and that sometimes this essentially random process can appear to be directed. Originally it was also thought that there was just one mechanism for all cases of adaptive mutation. This idea also proved to be wrong–it seems there are many mechanisms by which genetic change can be produced when organisms are under stress.
This review concentrates on stress-induced mutagenesis in Escherichia coli and related bacteria with a few examples from other organisms. The mutagenic phenomena are variously called adaptive, stationary-phase, stress-induced, or starvation-induced mutagenesis (see Table 1). In the first section, I describe some of the global responses to stress in bacteria, and how each can result in increased mutation rates. In the second section, I give some specific examples of stress-induced mutagenesis (see Table 2), and then discuss in greater detail what we have learned from studying a particular case, adaptive mutation to Lac+ in E. coli. I conclude with a discussion of the potential impact that stress-induced mutagenesis may have on survival and adaptation.
TABLE 1.
Definition of terms
| Term | Definition |
|---|---|
| Directed mutation* | The appearance among cells under selection of mutations that relieve the selective pressure in the absence of the appearance of nonselected mutations. |
| Adaptive mutation | The appearance among cells under selection of mutations that relieve the selective pressure whether or not other nonselected mutations are also produced. |
| Stress-induced mutation | The induction of a general mutagenic state in response to stress. If the stress is starvation, then the equivalent term is “starvation-induced mutation”. |
| Transient mutation | A temporary state during which the mutation rate is increased, resulting in the accumulation of mutations throughout the genome. Could be the result of various inducible processes. |
| Hypermutation | A state during which the mutation rate is increased to extremely high, potentially lethal, levels resulting in the accumulation of mutations throughout the genome. |
| Stationary-phase mutation | A general mutagenic state that occurs in cells in stationary phase. |
| Stationary phase | The period of laboratory batch culture when the cells have run out of one or more nutrients. Also used to describe nutrient-limitation in natural populations. |
| Long-term stationary phase | The period of laboratory batch culture after the cells have entered stationary phase and after the majority (e.g., 99% or more) have died. Characterized by balanced growth and death of the population. |
| Starvation | A general term describing the lack of one or more nutrients required for growth. Starvation can result from the normal depletion of nutrients during batch culture, or from a population being artificially suspended in nutrient-free medium. |
| Nutrient-limitation; nutritional-deprivation | Same as starvation, but often implies that only one nutrient is lacking. |
| Hunger | The state of cells when they are growing with suboptimal levels of nutrients, as for example in nutrient-limited chemostats. |
| Nutritional selection | A nutrient limitation that can be overcome by mutation; e.g., Lac− cells incubated with lactose as the only carbon and energy source. |
| Adaptive evolution | Evolution that proceeds by selection for improved fitness, in contrast to evolutionary changes caused by neutral mutations and genetic drift. |
Mutation and mutagenesis are used interchangeably.
TABLE 2.
Examples of Stress-induced Mutation
| Name† | Organisms | Type of Mutation | Selected Phenotype | Genetic Requirements | References |
|---|---|---|---|---|---|
| Starvation-induced Mu-mediated fusions | E. coli, Mu | Transposition | Growth on arabinose plus lactose | RpoS, ClpP, HNS* | (Cairns et al., 1988; Foster et al., 1994a; Gomez-Gomez et al., 1997; Lamrani et al., 1999; Maenhaut-Michel et al., 1994; Maenhaut-Michel et al., 1997; Mittler et al., 1990; Shapiro, 1984) |
| Resting organisms in a structured environment (ROSE) mutagenesis | E. coli | Base substitutions | Resistance to rifampicin | CyaA. RecA. LexA*, UvrB, Pol I, Not Pol V, Not RecBCD, Not RpoS | (Taddei et al., 1995; Taddei et al., 1997a) |
| Mutagenesis in aging colonies (MAC) | E. coli | Base substitutions | Resistance to rifampicin | RpoS, Crp, CyaA, RecA, MMR*, Pol II, Not RecBCD, Not Pol I | (Bjedov et al., 2003) |
| SOS-dependent spontaneous mutagenesis | E. coli | Base substitutions | Tryptophan prototrophy | RecA, Pol V | (Bhamre et al., 2001; Timms et al., 1999) |
| Stationary-phase mutagenesis | P. putida | Frameshifts, base substitutions, transposition | Growth on phenol | Pol IV, Pol V, RpoS, MutY, Not RecA, Not MMR* | (Ilves et al., 2001; Kasak et al., 1997; Saumaa et al., 2002; Saumaa et al., 2006; Tark et al., 2005; Tegova et al., 2004) |
| Stationary-phase mutagenesis | B. subtilis | Base substitutions | Amino acid prototrophy | ComA, ComK, Pol IV, MMR*, Mfd, Not RecA, Not RpoS analog | (Pedraza-Reyes et al., 2004; Ross et al., 2006; Sung et al., 2002; Sung et al., 2003) |
| Adaptive mutation | E. coli | Frameshifts | Growth on lactose | Pol IV, RecA, RecBCD, RpoS, GroE, Ppk, Not Pol V | See text |
Usually the name used by the authors of the work.
Loss or inactivation.
GLOBAL STRESS RESPONSES
The SOS Response
The stress that most obviously and directly results in genetic change is exposure to DNA damaging agents. E. coli and other bacteria have an extensive and highly effective response to DNA damage, the SOS response, that minimizes the lethal and mutagenic consequences of such exposure (for reviews, see Friedberg et al., 2006; Kelley, 2006). When the cell’s DNA is damaged, regions of single-stranded DNA arise, either directly as a result of the damage or indirectly when the DNA is repaired or when DNA replication is stalled. The ssDNA is recognized and bound by RecA, the bacterial recombinase, forming nucleoprotein complexes. These complexes stimulate the self-cleavage of the LexA repressor. The destruction of LexA results in the derepression of the SOS genes, many of which encode enzymes that promote DNA repair, recombination, and DNA synthesis past lesions that block replication. Approximately 30 genes are known to be repressed by LexA in E. coli (Fernandez de Henestrosa et al., 2000; Wade et al., 2005); Bacillus subtilis has a similar number, but only eight are homologs of the E. coli genes (Au et al., 2005).
Three of the genes induced as part of the SOS response in E. coli encode DNA polymerases that have the ability to replicate damaged DNA (for reviews, see Fuchs et al., 2004; Lehmann, 2006; Nohmi, 2006; Tippin et al., 2004). These polymerases temporarily take the place of the replicative polymerase (Pol III) and synthesize past blocking lesions in the DNA template. One of the SOS polymerases is Pol II, a conventional polymerase that under most circumstances is accurate and processive. Pol II can bypass some DNA lesions and can extend synthesis when lesions are bypassed by other polymerases; its major role may be to restart stalled replication forks. Two of the SOS polymerases belong to the widely distributed Y-family of specialized polymerases. Members of this family are characterized not only by their ability to replicate damaged DNA, but also by their lack of processivity and low fidelity when copying undamaged template. Pol V, encoded by the umuDC genes, can replicate past many types of DNA lesions, and is error-prone on both damaged and undamaged DNA. Pol IV, encoded by the dinB gene, is also error-prone, but the repertoire of lesions that it can bypass is limited. Pol IV readily extends DNA synthesis from a mispaired or misaligned primer-template terminus, and thus, like Pol II, may restart stalled replication forks and extend replication after lesion-bypass by another polymerase. In such situations, Pol IV and Pol II may be in competition.
In normally growing, undamaged cells neither Pol IV (Kuban et al., 2004; McKenzie et al., 2003; Strauss et al., 2000; Wolff et al., 2004) nor Pol V (Kuban et al., 2006) contributes much to spontaneous mutation rates. Without SOS induction the cellular levels of Pol V are undetectable (Woodgate et al., 1991), so it is not surprising that its loss has no phenotype. In contrast, uninduced cells have about 250 molecules of Pol IV (Kim et al., 2001), a fairly large number for a polymerase (for example, there are 10 to 20 copies of Pol III holoenzyme; Wu et al., 1984). With that many copies it is not clear why, under most conditions, Pol IV has so little mutational effect. However, Pol IV does contribute to spontaneous mutagenesis of genes on the large conjugal plasmid, F (Kuban et al., 2004) and on the bacteriophage lambda chromosome (Brotcorne-Lannoye et al., 1986). In addition, Pol IV is active in nongrowing cells (discussed below).
When induced via the SOS response or other pathways (see below), both Pol IV and Pol V increase the cell’s spontaneous mutation rate even in the absence of DNA damage. This phenotype can be observed in the laboratory using mutant strains with a constitutively activated SOS response. In such strains Pol IV accounts for about a 3-fold, and Pol V accounts for about a 10-fold, increase in mutation frequencies (Kuban et al., 2006). The mutations are presumably due to errors made by the polymerases replicating undamaged DNA (i.e., they are “untargeted” mutations), although spontaneous DNA damage may also contribute. In the absence of SOS induction, overproduction of Pol IV is also a powerful mutator–simply supplying the dinB gene on a plasmid increases mutation frequencies as much as 100-fold (Kim et al., 1997; Wagner et al., 2000). Pol V cannot be overproduced so simply because to be active, UmuC (the polymerase subunit) must associate with two UmuD subunits that have undergone a RecA-facilitated autocleavage reaction; RecA is additionally required to stimulate Pol V activity (Schlacher et al., 2006). However, Pol V homologues on many naturally-occurring plasmids are not so stringent. For example, the RumAB protein, originally found on a conjugal R (antibiotic resistance) plasmid, increases the spontaneous mutation frequency of an uninduced E. coli strain four-fold when on a low-copy plasmid; in a strain constitutively expressing SOS, it has 50-fold effect (Mead et al., 2007). Many of these Pol V homologues are not as active when in their native locations, suggesting that there are additional functions on plasmids in nature that regulate their mutagenic activity (Koch et al., 2000).
Exposure to exogenous DNA damaging agents is not the only way that the SOS response can be induced. DNA damage from metabolic intermediates or mistakes in replication, recombination, and chromosome segregation can be powerful SOS stimuli if not repaired (O’Reilly et al., 2004). The SOS response will also be induced, at least partially, whenever the levels of active LexA decline. For example, LexA is inactivated in vitro under slightly alkaline conditions (Little, 1991), and in vivo in aging colonies (Taddei et al., 1995) and when cells reach saturation in rich medium (Dri et al., 1994). Other, seemingly unrelated, stresses can activate the SOS response; these include exposure to antibiotics (see below) and high pressure (1000-fold higher than atmospheric) (Aertsen et al., 2006; Bjedov et al., 2003). Interestingly, high-pressure SOS-induction is triggered by double-strand breaks induced by one of E. coli’s restriction endonucleases acting on its own DNA (Aertsen et al., 2005).
The General Stress Response
When E. coli and its relatives enter stationary phase or experience nutrient limitation they induce the “general stress response” (for a review see Hengge-Aronis, 2002). The master controller of this response is the sigma factor RpoS, also called σ38, σS, or KatF. Sigma factors are the subunits of RNA polymerase (RNAP) that direct transcription to particular classes of promoters. During exponential growth under aerobic conditions, most genes are transcribed by RNAP complexed with the vegetative sigma factor, σ70, also called RpoD (the holoenzymes are designated as Eσ70 or Eσ38). As cells enter stationary phase the amount of active RpoS dramatically increases, resulting in the formation of Eσ38 that transcribes genes required for survival during starvation and other stresses. In E. coli, over 200 genes are, directly or indirectly, under RpoS control (Patten et al., 2004; Weber et al., 2005). In addition to nutrient limitation and starvation, the general stress response is induced by growth-rate reduction, high osmotic pressure, low pH, and extreme shifts in temperature. The induction and activation of RpoS is controlled at several levels by a variety of cis- and trans-acting regulatory elements; in addition, the RpoS regulon overlaps extensively with other global response networks. This complex regulation ensures that the stressed cells are prepared for a variety of contingencies.
There are two ways currently known by which the RpoS-dependent general stress response can increase the spontaneous mutation rate of stressed cells. The first is by up-regulating the error-prone DNA polymerase, Pol IV. About 10 hours after E. coli enters stationary phase, the cellular levels of Pol IV increase about three-fold. Surprisingly, this increase is independent of the SOS response but is, instead, RpoS-dependent. In the absence of RpoS Pol IV is rapidly lost, so that after 48 hours in stationary phase, RpoS-proficient cells have 30 to 50 times the amount of Pol IV as do RpoS-deficient cells (Layton et al., 2003). The Pol IV protein is unstable, and its instability is not affected by RpoS (as measured by quantitative immunoblots after protein synthesis inhibition; Storvick et al., 2007); therefore, continuous transcription of the dinB gene under RpoS control is required to maintain the high levels of Pol IV during stationary phase. This RpoS-dependent induction of Pol IV is required for Lac+ adaptive mutation (see below). In addition, maintenance of Pol IV transcription is required for survival and for adaptive changes in E. coli populations during long-term stationary phase (Yeiser et al., 2002).
The second way that RpoS enhances spontaneous mutation rates during stress is by down-regulating enzymes that are responsible for mismatch repair (MMR). In E. coli and other bacteria, the MMR complex recognizes base mismatches in the DNA and excises the newly synthesized DNA strand; the old strand then serves as template for new synthesis. E. coli normally discriminates between the newly synthesized (presumably error-containing) strand and the template (correct) strand by their methylation state–only newly-synthesized DNA is unmethylated (for a review, see Kunkel et al., 2005). Although MMR remains active in stationary-phase cells (Foster, 1999a; Reddy et al., 1997), the levels of two of the MMR proteins, MutS and MutH, are down-regulated in an RpoS-dependent manner (Feng et al., 1996; Tsui et al., 1997). Two types of evidence indicate that this down-regulation can, at least under some circumstances, increase mutation rates: (1) the frequency of mutation is decreased if one or more of the MMR proteins is overproduced (Bjedov et al., 2003; Foster et al., 1995a; Harris et al., 1999; Zhao et al., 2000); and (2) the frequency of mutation is not increased if MMR is genetically eliminated (Bjedov et al., 2003). Which MMR protein becomes limiting in stationary phase differs among experiments, experimenters, and (perhaps) genetic backgrounds (Bjedov et al., 2003; Foster, 1999a; Harris et al., 1999; Zhao et al., 2000). The contribution that MMR decline makes to stationary-phase mutation is also debated. As discussed below, in nutrient-limited populations mutations do arise in MMR proficient cells (Rosche et al., 1999). In addition, loss of the MMR system cannot produce mutations de novo but can only preserve errors made during previous DNA synthesis.
RpoS has also been implicated in stress-induced mutation in ways that are less well understood than the above examples. Epistasis tests show that in addition to up-regulating Pol IV, RpoS has a second role in Lac+ adaptive mutation (Layton et al., 2003). RpoS is also required for amplification of the lac region during lactose selection (Lombardo et al., 2004). RpoS is required for base substitution mutations in aging E. coli colonies (Bjedov et al., 2003), frameshift mutations in starved Pseudomonas putida cells (Saumaa et al., 2006), and transposition and transposon-induced rearrangements in starving E. coli (Gomez-Gomez et al., 1997; Lamrani et al., 1999) and P. putida (Ilves et al., 2001). Although the exact pathways by which RpoS exerts its effects may not be the same in all of these examples, these various mutagenic events are consequences of the RpoS-dependent general stress response.
Polyphosphate-Mediated Nutrient-Limitation Response
Polyphosphate (polyP) consists of chains of phosphates, tens to hundreds of residues long, linked together by high energy phosphoanhydride bonds. Although polyP is found in organisms from all domains life, in most cases its function remains a mystery (for reviews, see Kornberg et al., 1999; Kulaev et al., 2000; Shiba et al., 2000). PolyP is made by polyphosphate kinase (Ppk), which transfers the gamma phosphate of ATP onto the growing phosphate chain. PolyP accumulates when cells enter stationary phase or undergo nutritional deprivation. In E. coli, a large part of this accumulation is due to the inhibition of the major enzyme that degrades polyP, exopolyphosphatase (Ppx), by (p)ppGpp, which is induced by the stringent response (discussed below) (Kuroda et al., 1997). Since polyP has profound effects on cell physiology, loss of Ppk or overproduction of Ppx has pleiotropic effects. Of particular relevance here, polyP is required for the transcriptional induction of RpoS in stationary phase (Shiba et al., 1997), and of RecA after DNA damage (Tsutsumi et al., 2000).
Some of the effects of PolyP may be due to the fact that it acts as a DNA mimic and binds to DNA binding proteins. PolyP can inhibit the in vitro reactions of restriction enzymes, ligase, and Taq polymerase (Rodriguez, 1993). However, polyP-binding may also serve a regulatory function. For example, RNA polymerase isolated from E. coli cells in stationary phase, but not from cells in exponential phase, was found to be complexed with polyP (Kusano et al., 1997). Kusano et al. (1997) further showed that at low salt concentrations polyP inhibited the in vitro transcriptional activity of both Eσ70 and EσS; but, at high salt concentrations polyP stimulated transcription by EσS and not Eσ70. These results suggest that, at least at high osmolarity, polyP helps RNA polymerase to distinguish between vegetative and stationary-phase promoters (Kusano et al., 1997). Lon, which is a major ATP-dependent protease, is also a DNA-binding protein. PolyP binds to purified Lon and stimulates it to degrade ribosomal proteins, which also bind polyP (Kuroda et al., 2001).
Recently we discovered that polyP regulates the activity of E. coli’s Y-family polymerases (Stumpf et al., 2005). Pol IV-dependent Lac+ adaptive mutation is reduced by 65 to 80% in a ppk mutant strain. Overproduction of Pol IV in exponentially growing cells is profoundly mutagenic, and loss of Ppk inhibits this mutagenesis as well. Yet, in both of these cases the amount of Pol IV is not affected, but only its activity. The pathway by which Ppk activates Pol IV is independent of Pol IV’s regulation by RpoS or RecA. Pol V-dependent UV-induced mutations are also reduced in a ppk mutant strain (Stumpf et al., 2005); likewise, Pol IV’s lesion-bypass activity is lost in a ppk mutant strain (Stumpf et al., 2007a). Although we favor the hypothesis that polyP is the active agent, we have not ruled out the possibility that it is the Ppk enzyme itself that is exerting these effects. Taken together, however, these results suggest that the build-up of polyP in stationary phase and during nutritional deprivation stimulates the mutagenic and lesion bypass activities of the Y-family polymerases.
The Stringent Response
When E. coli and other bacteria are starved for amino acids or experience nutritional down-shift, the synthesis of stable RNA species (tRNA and rRNA) is rapidly down-regulated. This reaction, called the “stringent response,” is mediated by the unusual guanosine nucleotides pppGpp and ppGpp, together known as (p)ppGpp. Decades of research has revealed the complex genetic control of the stringent response and much of the physiological effects of (p)ppGpp accumulation. The stringent response is global, encompassing not only down-regulation of stable RNA, but also down-regulation of protein synthesis, up-regulation of protein turnover, and activation of amino acid biosynthesis (for recent reviews, see Braeken et al., 2006; Magnusson et al., 2005).
(p)ppGpp (together with the co-effector DskA) regulates transcription by binding to RNAP and changing the distribution of its transcriptional activity at various promoters (for a review see Gourse et al., 2006). One promoter that is positively, but probably indirectly, regulated by (p)ppGpp is RpoS; in addition, many RpoS-dependent genes require (p)ppGpp for their induction (Kvint et al., 2000). Thus, the RpoS-dependent general stress response, discussed above, and the ppGpp-dependent stringent response are co-induced and overlapping (Chang et al., 2002; Traxler et al., 2006). (p)ppGpp also positively regulates a large number of genes encoding enzymes for the biosynthesis and uptake of amino acids (Cashel et al., 1996; Paul et al., 2005). Because amino acid biosynthetic genes tend to be regulated by end-product repression, they will be transcriptionally activated both by the stringent response and by derepression during starvation for amino acids.
Transcription creates a bubble of single-stranded DNA and single-stranded DNA is more susceptible than double-stranded DNA to a variety of spontaneous damage. For example, cytosine deaminates to uracil 100 times faster in single stranded DNA than in double stranded DNA (although the half-life of a cytosine in single stranded DNA is still 200 years) (Frederico et al., 1990). Since actively transcribed genes have more single-stranded DNA, they would be expected to sustain more potentially mutagenic damage. In addition, DNA repair is recruited to transcribed genes (for a review, see Mellon, 2005); while this transcription-coupled repair would be expected to reduce mutations, the resulting more frequent DNA synthesis could also create mutations. If transcription is intrinsically mutagenic, during nutrient deprivation mutations would be “directed” to actively transcribed genes that could be the very ones that, when mutated, might relieve the stress (Davis, 1989; Fitch, 1982; Wright, 2004). For example, the transcriptional activation of amino acid biosynthetic genes would make them potentially more vulnerable to mutation during amino acid starvation (Wright et al., 1999).
Active transcription of a gene has been reported to increase its spontaneous mutation rate in E. coli (Beletskii et al., 1996; Klapacz et al., 2002; Klapacz et al., 2005; Wright et al., 1999), B. subtilis (Rudner et al., 1999), and in the yeast, Saccharomyces cerevisiae (Datta et al., 1995). In some experiments, the increase in mutation was dependent on the bacteria being competent for the stringent response (Reimers et al., 2004; Rudner et al., 1999). Interestingly, stationary-phase mutation in B. subtilis cells is dependent on Mfd, the factor that couples DNA repair to transcription (Ross et al., 2006) (although apparently this is not the case in E. coli; Bridges, 1995b). This last result suggests that transcription-coupled DNA repair may, at least in some circumstances, stimulate error-prone DNA synthesis.
While the results referenced above indicate that transcription can be mutagenic, in other cases it has been shown not to be. For example, gratuitous induction of the lac operon in the absence lactose did not increase the number of Lac+ revertants subsequently recovered (Foster et al., 1992). Revertants of a histidine auxotrophy did not accumulate when Salmonella typhimurium cells were incubated in the absence of histidine (Hughes et al., 1997). In one case, the number of Trp+ revertants increased during starvation of E. coli for tryptophan in the absence of transcription of the gene (Barionovi et al., 2003). Clearly more research is needed before the impact of transcription on mutation rates is understood.
Induction of the stringent response causes DNA replication to halt, presumably to await the restoration of nutrients (Gourse et al., 2007). However, the way in which DNA replication is arrested appears to differ among bacteria. In E. coli, (p)ppGpp inhibits replication initiation (Schreiber et al., 1995), whereas in B. subtilis (p)ppGpp inhibits replication elongation (Autret et al., 1999). Recently it was found that in B. subtilis, (p)ppGpp binds to RNA primase, inhibiting its activity and thus halting the replication fork (Wang et al., 2007). The forks are not disrupted, at least in the short term–they do not recruit RecA and are thus not perceived by the cells as needing repair. However, after a prolonged arrest replication might be restarted and, if the process involved error-prone polymerases, be a source of mutations (Wang et al., 2007).
What is Nutritional Stress?
The hallmark of adaptive mutation is that mutations occur not when the bacteria are merely starving, but only when they are incubated under a specific nutritional selection, e.g., Lac− cells incubated on lactose medium. Indeed, several studies have shown that mutations do not usually accumulate when cells are incubated without any nutrients at all (Cairns et al., 1988; Cairns et al., 1991; Foster, 1997; Hughes et al., 1997) (although this is not true of movement of IS elements; see below). In some cases of starvation-induced mutagenesis, the starving population may be dynamic, with some cells dying and others slowly proliferating (Saumaa et al., 2006). Even when cells are incubated for long periods in spent medium, they are not without nutrients–mutant bacteria able to survive on the waste products of their neighbors periodically appear and sweep the population (Finkel, 2006).
The failure to observe mutations in the absence of nutritional selection suggests that some small amount of energy is required for the mutational process. For example, in the case of adaptive mutation to Lac+ the mutant lac allele is slightly leaky, making about two units of β-galactosidase, an amount that is not sufficient for cell proliferation. As discussed below, our model for adaptive mutation postulates that a double-strand break causes a replication fork to collapse, initiating the recombination that leads to Lac+ mutation. We propose that the role of lactose is simply to supply a slow influx of energy that allows occasional DNA replication (Stumpf et al., 2007b). In support of this hypothesis, lactose is not required if double-strand breaks are artificially induced (Ponder et al., 2005).
When cells are not starving but growing in nutrient-limited chemostats they express what has been called “the hunger response” (for a review, see Ferenci, 2001). This response is characterized by up-regulation of scavenging functions, and is distinguished from response to starvation by its independence of RpoS. Indeed, the hunger response is antagonized by RpoS, probably because RpoS competes with RpoD or other sigma factors required for transcription of the scavenging pathway genes (Ferenci, 2001; Notley-McRobb et al., 2002). In contrast, starvation-induced or adaptive mutation usually requires RpoS function (see below), suggesting that “hungry” bacteria do not indulge in such mutagenesis.
Since natural populations of E. coli most likely experience frequent periods of nutrient scarcity, their periodic requirement for scavenging functions may explain why E. coli populations are heterogenic for rpoS alleles. Attenuation of RpoS function is frequently selected in long-term stationary-phase cultures, during which cells evolve heightened abilities to scavenge various micronutrients (Finkel, 2006). But the fact that null alleles of RpoS are not selected suggests that additional RpoS-regulated functions that are dispensable in nutrient-limited chemostats are useful to cells during long-term stationary phase.
The Heat-Shock Response
When subjected to a rapid increase in external temperature, all organisms respond by inducing proteins that increase their thermo-tolerance. In E. coli and other bacteria, the heat-shock response is mainly under control of the sigma factor RpoH (σ32); another sigma factor, RpoE (σ24), also participates in response to extreme temperature elevation and envelope stress (for reviews see Gruber et al., 2003; Rosen et al., 2002; Yura et al., 2000). The RpoH-regulon is induced, in whole or in part, by conditions that result in unfolded proteins and cytoplasmic stress. In addition to heat, these include DNA damage, oxidative stress, exposure to antibiotics or heavy metals, phage infection, and starvation for carbon source or amino acids. Thus, induction of the RpoH-regulon is a general stress response.
The heat-shock proteins include molecular chaperones that aid in protein folding and proteases that degrade unwanted or damaged proteins. The molecular chaperone GroE (E. coli’s HSP60) interacts with at least 250 E. coli proteins; 85 of these, including 13 essential proteins, require GroE for activity (Kerner et al., 2005). Thus, GroE is essential at all temperatures. UV mutagenesis is GroE-dependent because GroE interacts with UmuC, the polymerase subunit of Pol V, and protects it from degradation (Donnelly et al., 1989; Liu et al., 1990). GroE is likely also to interact with Pol IV. Cells that are deficient for GroE have only 10% of Pol IV compared with GroE proficient cells, and GroE is required for Pol IV-dependent adaptive mutation (Layton et al., 2005). Presumably GroE protects Pol IV from degradation, although there may be an additional Pol IV-independent role for GroE in adaptive mutation (Layton et al., 2005). Thus, during stress conditions that induce the RpoH-regulon, both Y-family DNA polymerases, Pol IV and Pol V, will be protected from degradation by GroE, and thus able to perform their mutagenic functions.
Exposure to Antibiotics
Antibiotics of the quinolone class, such as ciprofloxacin and nalidixic acid, target gyrase and topoisomerase IV, enzymes that coil and uncoil DNA. These antibiotics ‘trap’ the topoisomerase-DNA complex after the DNA strands have been cleaved but before they are rejoined. Thus a double-strand break is produced that, after RecBCD produces a region of single-stranded DNA, induces the SOS response (for a review, see Drlica et al., 1997). Mutations in the genes that encode gyrase and topo IV can convey resistance to the antibiotics. The quinolone antibiotics are clearly mutagenic (for example, see Mamber et al., 1993), so it would not be surprising if resistance to the antibiotics were induced by exposure to them. Indeed, recently it was shown using a mouse-infection model that pathogenic E. coli became resistant to ciprofloxacin after exposure to it (Cirz et al., 2005). In a laboratory reconstruction, all three SOS-induced DNA polymerases (Pol II, IV, and V) were involved in this mutational response (Cirz et al., 2005). Interestingly, when E. coli cells were plated on medium containing a bacteriostatic concentration of ciprofloxacin, resistant mutants appeared over the course of 2 weeks–conditions very similar to those used in adaptive mutation experiments (see below). In addition to simply inducing mutations, exposure to quinolone antibiotics can produce antibiotic resistances in other ways. For example, exposure of Vibrio cholerae to ciprofloxacin can induce the movement of integrating conjugative elements (ICEs), some of which carry genes that confer antibiotic resistances (Beaber et al., 2004).
Toxin-antitoxin systems are often encoded on large, low copy plasmids to ensure plasmid transmission. Typically the toxin is long-lived and the antitoxin is short-lived, so a cell that does not inherit the plasmid will die. Several of the toxins, such as microcin B17 and CcdB, are topoisomerase inhibitors with activities similar to the quinolones, and these toxins similarly induce the SOS response (Couturier et al., 1998; Drlica et al., 1997). Recently it was reported that transcription of CcdB increases as cells enter stationary phase, and that CcdB enhances adaptive mutation to Lac+. Thus, plasmids carrying such toxins may increase the genetic diversity of their host cells during times of nutritional deprivation (Aguirre-Ramirez et al., 2006).
β–lactam antibiotics, such as penicillin, also induce the SOS response but do so by a completely different pathway (Miller et al., 2004). These antibiotics act by binding to the penicillin binding proteins (PBPs) and disrupting cell wall synthesis. Thus, cells that are not actively dividing are resistant to killing by β-lactams. Inactivation of PBP3, either by exposure to β-lactam antibiotics or genetically, induces the dpiBA operon, which encodes a response-effector two-component system (for a review of two-component systems, see Mascher et al., 2006). DpiA, the effector, binds to the chromosomal replication origin, inhibiting replication and inducing the SOS response (Miller et al., 2003). One of the SOS genes induced, sulA (also known as sfiA), encodes an inhibitor of cell division, and inhibition of cell division protects the cell, at least temporarily, from killing by the β-lactam antibiotic (Miller et al., 2004). While this protection is likely the selective advantage that shaped the dpiBA response, a consequence of inducing the SOS response is an increase in genetic variability (Miller et al., 2004). Exposure to β-lactam antibiotics also induces the dinB gene via a SOS-independent pathway (Perez-Capilla et al., 2005). Thus, exposure to both β-lactam and quinolone antibiotics appears to produce a state of global genetic instability in bacteria.
Sublethal doses of streptomycin produce a mutator state that is independent of SOS mutagenic functions (Balashov et al., 2002). Streptomycin and other amino-glycoside antibiotics interfere with ribosome function and inhibit translation. Exposure to sublethal doses of streptomycin has long been known to result in inaccurate translation (Balashov et al., 2003; Rosset et al., 1969). As first proposed by Ninio (1991), mistranslation of DNA repair and replication proteins can create transient mutator states. For example, mutations that result in certain mutant tRNAs have mutator phenotypes because the proofreading subunit of the DNA polymerase III holoenzyme is mistranslated, making replication error-prone (Al Mamun et al., 2002; Slupska et al., 1998). The mutator phenotype induced by exposure to streptomycin is likely to have a similar cause.
STRESS-INDUCED MUTAGENESIS
In the sections below, I describe examples of what appear to be stress-induced mutagenesis (summarized in Table 2). This survey is not intended to be comprehensive, but instead to be an exploration of the ways that stress can increase genetic variability. Hence, I have chosen examples from E. coli and other bacteria that reveal the genetic factors coupling stress to mutagenic mechanisms. In the last part of this section I discuss the well studied Lac+adaptive mutation in E. coli.
Starvation-Induced Mutation on Solid Surfaces
Bacteria living on solid surfaces display some phenotypes that are not exhibited by planktonic bacteria (Ben-Jacob et al., 1998; DiLuzio et al., 2005; Kolter et al., 2006; Shapiro, 1995). Taddei et al. (1995; 1997a) discovered that the SOS response was induced in colonies of E. coli as they aged for a week on rich agar medium; concomitantly, the frequency of rifampicin resistant (RifR) mutants within the colonies increased10-fold. Calling the phenomenon ROSE, for “resting organisms in a structured environment”, Taddei et al. (1995) found that cyclic AMP was required for both SOS induction and ROSE mutagenesis, suggesting that declining energy reserves signal the response. However ROSE was independent of RpoS. ROSE mutagenesis was completely dependent on RecA, UvrB (encoding a component of the nucleotide excision repair complex), and DNA polymerase I (Pol I) but independent of Pol V and only slightly dependent on RecBCD. ROSE mutagenesis required the expression of at least one LexA-repressed gene in addition to RecA (which could be UvrB). These results suggested that ROSE mutagenesis results from error-prone DNA synthesis, perhaps by Pol I, stimulated by excision repair (Taddei et al., 1997a).
Because the frequency of RifR mutants did not increase when E. coli was incubated for a week in liquid rich medium (Taddei et al., 1997a), even though the SOS response is induced under such conditions (Taddei et al., 1995), ROSE mutagenesis suggests that the special conditions inside a colony result in DNA damage. A number of studies have found that inactivation of the enzymes for repair of oxidative and alkylation damage has severe mutagenic consequences in nondividing cells on agar surfaces (Benov et al., 1996; Bharatan et al., 2004; Bridges, 1995a; Bridges et al., 1998; Bridges et al., 2001; Foster et al., 1992; Mackay et al., 1994; Rebeck et al., 1991). Thus, DNA damage by endogenous oxidative and alkylating agents appears to be rampant during prolonged incubation on solid surfaces.
Most studies of stress-induced mutagenesis use laboratory strains of bacteria that may have diverged substantially from their free-living ancestors. Bjedov et al. (2003) collected 787 E. coli isolates from various environments around the world and assayed them for mutagenesis using the ROSE method. Because the mutagenic phenomenon appeared to have different genetic requirements than ROSE (see Table 2), they named it MAC for “mutagenesis in aging colonies.” Most isolates showed an increase in the frequency of RifR mutants within their colonies during 7 days of incubation; 40% had increases that were 10 times the median and 13% had increases that were 100 times the median. About 3% of the isolates were constitutive mutators (i.e. a high frequency of RifR mutants on day one). Interestingly, there was a negative correlation between an isolate’s frequency of RifR mutants on day one and its fold-increase in this frequency by day 7, suggesting that bacteria have either high constitutive mutation rates, or high inducible mutation rates, but not both. In a sample of ten isolates, MAC proved to be dependent on carbon source starvation, aerobic conditions, and incubation on a solid substrate. The genetic requirements for MAC of one isolate were explored. In this isolate, MAC was dependent on RpoS, cyclic-AMP, the cAMP receptor protein (required for catabolite repression and activation), and RecA; MAC was independent of LexA inactivation. Overproduction of MutS inhibited MAC, suggesting that MMR declines in the aging cells. In contrast to ROSE (see above), MAC was not dependent on nucleotide excision repair or DNA polymerase I, but was dependent on DNA polymerase II.
This study by Bjedov et al. (2003) is by far the most far-reaching analysis of stress-induced mutagenesis in bacteria to date. Since the isolates were from various environments and hosts, and included both commensal and pathogenic strains, it is clear that the phenomenon is widespread. There was a tendency for pathogenic isolates to have high constitutive mutation frequencies and for nonpathogenic isolates to have high MAC frequencies, suggesting that pathogens experience rapidly changing conditions, whereas commensals experience rather static environments punctuated by periods of stress.
Mutagenesis Due to Spontaneous DNA Damage
As mentioned above, exogenous and endogenous agents may cause extensive DNA damage in nongrowing cells. The mutations that result will be dependent not only on the nature of the damage, but also on how active the enzymes that repair the damage are in nongrowing cells. Although some types of damage, such as alkyl-lesions, may cause mutations directly, most damage-induced mutations depend on induction of the SOS response.
Many alleles of the E. coli trp genes revert only via specific mutations (Yanofsky et al., 1966; Yanofsky et al., 1987) and are useful for mutational studies (for examples, see Eisenstadt, 1987). Some of these mutant alleles revert to Trp+ continuously during several days of selection for tryptophan prototrophy. Bhamre et al. (2001) found that the revertants of trpA23 that accumulate during ten days of tryptophan selection were predominantly AT base pair transversions at the trpA23 site. This accumulation was dependent on SOS induction of Pol V, and was enhanced if the strain had a plasmid carrying the MucAB homologs of Pol V. In contrast, Timms et al. (1999) found that in a mutY mutant strain, Pol V-dependent GC to CG mutations that reverted trpA23 accumulated during selection (in addition there was a substantial level of SOS- and Pol V- independent reversion during these experiments). Because MutY encodes a glycosylase that removes A paired with 8-oxoG, Timms et al. (1999) concluded that 8-oxoG was a major spontaneous mutagenic lesion in nutritionally deprived cells. However, this may only be true in a mutY mutant strain, as the mutations at AT base pairs found by Bhamre et al. (2001) in MutYproficient cells are not easily explained by 8-oxoG lesions. Nonetheless, oxidative damage has been implicated in other cases of mutation in nutritionally deprived cells (Benov et al., 1996; Bharatan et al., 2004; Bridges, 1995a; Bridges et al., 1998; Bridges et al., 2001). Using a system designed to distinguish between mutations occurring in nongrowing and growing cells, Bhratan et al. (2004) found that MutY, MutT (which hydrolyzes 8-oxoGTP), and glycosylases that remove oxidized pyrimidines, were active in preventing mutations in nongrowing cells. Given the large contribution that oxidative damage makes to spontaneous mutation in growing cells (Bhamre et al., 2001; Miller, 1996; Sakai et al., 2006), and the oxygen-induced damage that dormant cells suffer (Eisenstark et al., 1992), it is not surprising that oxidative damage produces major mutagenic damage in nongrowing cells.
DNA alkylation by endogenous alkylating agents is an important source of spontaneous DNA damage, and cells have several repair activities to cope with this damage (for a review, see Sedgwick et al., 2007). Two enzymes, Ada and Ogt, remove alkyl groups from the O6 position of guanine and the O4 position of thymine. Loss of either or both of these enzymes dramatically increases spontaneous mutation in nongrowing cells (Bharatan et al., 2004; Foster et al., 1992; Mackay et al., 1994; Rebeck et al., 1991). The mutagenic lesions are likely to be produced by methylation agents created by the enzymatic nitrosation of amides (Taverna et al., 1996). Ogt is constitutively expressed, but Ada is induced by alkylation damage and, in stationary phase, under control of RpoS (Taverna et al., 1996). In nongrowing cells, the levels of Ada are 20-fold higher than in rapidly growing cells, suggesting that aging cells are particularly subject to alkylation damage (Taverna et al., 1996).
Stress-induced Mutation in Other Bacteria
This review focuses on E. coli and its close relatives, but studies of other bacteria and microorganisms have revealed that the phenomenon of stress-induced or stationary-phase mutagenesis is general. Below I describe experiments characterizing mutation in nongrowing cells of Pseudomonas putida and Bacillus subtilis.
P. putida cells carrying plasmid-encoded promoterless pheAB genes cannot grow on phenol, but Phe+ mutations accumulate during 10 days incubation on plates with phenol as the only carbon source. The Phe+ phenotype is due to mutations that activate transcription by creating a new promoter sequence (Kasak et al., 1997). During incubation on the phenol plates, the number of viable cells detected by plating was constant, but vital staining revealed that many cells had died; presumably surviving cells had proliferated on nutrients from the dead ones (Saumaa et al., 2006). Thus, the population on the phenol selective plates appears to be dynamic, resembling the conditions that prevail in long-term stationary phase experiments (e.g., Finkel et al., 1999). These population dynamics probably explain why the accumulation of Phe+ mutations over time is not linear.
Several types of mutations, including base substitutions, small deletions, and the insertion of transposable elements, can create a good promoter sequence for the pheAB genes. During phenol selection all of these different types of mutations occurred, but by different mechanisms (Saumaa et al., 2002). For example, the Phe+ mutations that appeared early during phenol selection (three to four days after plating) were dominated by a particular C to A base substitution; in growing cells MutY prevents these mutations from occurring, but the levels of MutY declined early during selection. Later during selection, deletions creating the Phe+ phenotype appeared, and these required RpoS (Saumaa et al., 2002). Transposition of a transposon into the promoter region also depended on RpoS (Ilves et al., 2001). The occurrence of single base-pair deletions, but not base substitutions, was dependent on Pol IV but not on RecA (Tegova et al., 2004). In P. putida, Pol IV is poorly induced by DNA damage, and although transcription of Pol IV increases in stationary phase, it is not under RpoS control (Tegova et al., 2004). The appearance of Phe+ mutations during phenol selection was not due to a decline in mismatch repair because the spectrum of stationary-phase mutations changed if MMR was defective (Saumaa et al., 2006).
When an auxotrophic strain of B. subtilis is incubated without its required amino acid, prototrophic mutants appear at a constant rate for about 9 days (Sung et al., 2002). The authors called these stationary-phase mutations. The appearance of these mutations did not depend on RecA or B. subtilis’ RpoS analogue. Interestingly, the stationary-phase mutagenesis depended, at least partially, on the Com factors that regulate the development of natural competence (Sung et al., 2002). In addition, some, but not all, of the stationary-phase mutations depended on a Pol IV homologue, YdjH (Sung et al., 2003). Loss of MMR increased and overexpression of MutS reduced stationary-phase mutagenesis, suggesting that MMR was deficient in some, but not all, of the cells (Pedraza-Reyes et al., 2004). As was found in E. coli (see below), about 1% of the revertants carried second, nonselected mutations. This result, plus the involvement of the Com factors, suggests that the population under selection included a hypermutating subpopulation (Sung et al., 2002).
Mfd, also known as transcription repair coupling factor (TRCF), is required for the repair of DNA that is being actively transcribed. Mfd recognizes a stalled RNAP, dislodges it, and recruits DNA repair enzymes to the site (for a review see Saxowsky et al., 2006). As mentioned above, in B. subtilis loss of Mfd reduced stationary-phase mutation (Ross et al., 2006). In addition, loss of Mfd increased the proportion of revertants that were due to suppressor tRNAs relative to true revertants. This result suggested that Mfd targeted the mutational process to genes that were being actively transcribed, which under most conditions would be those that were under selection. Thus, Mfd would be an agent for “directed mutagenesis.” Ross et al. (2006) hypothesized that Mfd could promote this mutagenesis by facilitating RNAP’s ability to synthesize pass a lesion in the DNA template, producing a mutant transcript. The mutant transcript might be translated into a protein active enough to allow some metabolism, and the cell would then replicate its DNA without repairing the damage, immortalizing the mutation. This scheme, called by some “retromutagenesis’ (Saxowsky et al., 2006), has been proposed before (Bridges, 1994; Davis, 1989; Fitch, 1982) and is a variation of that suggested by Cairns et al. (1988) for producing directed mutations. In support of this hypothesis, RNAP can bypass lesions and produce mutant transcripts; however, Mfd does not facilitate, but opposes this bypass (Bregeon et al., 2003; Saxowsky et al., 2006). Thus a more likely explanation for the role of Mfd in stationary-phase mutagenesis is that the DNA synthesis associated with transcription-coupled repair is error-prone.
Movement of Mobile Genetic Elements
The various mobile genetic elements found in bacteria–insertion elements, transposons, integrating bateriophage, integrating conjugative elements, self-splicing group II introns, retrons, integrons–constitute a flexible gene pool that can be exchanged among individuals, and even between species, by horizontal gene transfer (Hacker et al., 2001). Given the selfish nature of the elements, it is not surprising that their mobility would be linked to bacterial stress responses. In addition, given that the movement of mobile elements can induce DNA breaks and rearrangement, it is not surprising that hosts would attempt to regulate the movements of their resident elements. The dynamic nature of mobile element evolution and host responses predicts a plethora of stress-induced phenomena.
As mentioned above, induction of the SOS response in Vibrio cholerae causes integrating conjugative elements (ICEs) to move (Beaber et al., 2004; Quinones et al., 2005). ICEs are related to bateriophage, and their induction resembles the well-known UV-induced induction of prophages. The SOS response has been reported to both stimulate (Kuan et al., 1991) and inhibit (Weinreich et al., 1991) transposition of transposons. Recently, a random search for mutations affecting transposition revealed that dinD, a SOS gene in E. coli, enhanced transposition of insertion element IS903 and transposon Tn10; however, transposition of other elements was not stimulated (Twiss et al., 2005). Thus SOS induction appears to affect different classes of mobile genetic elements in different ways.
Transposition requires the formation of protein-DNA complexes, and can be regulated by host factors that interact with the chromosome and modify its structure. The amounts and activities of two of these factors, IHF and H-NS, are modulated by stress responses (Haniford, 2006; Twiss et al., 2005). That various types of nutritional stress cause small insertion elements (ISs) to move is well documented. For example, a strain of E. coli stored for 30 years in an agar stab was found to have experienced both constant and episodic IS movement (Naas et al., 1994). The cryptic bgl operon in E. coli can be activated by IS movement into the control region, allowing growth on the β-glucosides salicin or arbutin (Reynolds et al., 1981); such Bgl+ mutants appear at increasing frequencies over several days when the cells have arbutin as their only carbon source (Hall, 1998a). Incubation of a Phe− strain of Pseudomonas putida with phenol as the only carbon source stimulates activation of the phenol-utilization operon by insertion of a transposon into the promoter region (Ilves et al., 2001). In a particular strain of E. coli, excision of a Mu element produces an active lacZ gene by fusing the araB regulatory region to the lacZ and lacY genes; fusion is stimulated by incubating the Lac− parent cells with lactose as the only carbon source and arabinose as inducer (Cairns et al., 1988; Shapiro, 1984), or by prolonged aerobic incubation without a carbon source (Foster et al., 1994a; Maenhaut-Michel et al., 1994; Mittler et al., 1990; Sniegowski, 1995). Interestingly, the structures of the ara-lac fusions that are induced by incubation on lactose-arabinose or by aerobic starvation are different (Maenhaut-Michel et al., 1994; Maenhaut-Michel et al., 1997). Because fusion formation is a function of bateriophage Mu induction and transposition, the genetic requirements are complex (Gomez-Gomez et al., 1997; Lamrani et al., 1999). However, the link to environmental stress is obvious, as fusion formation requires RpoS and is inhibited by H-NS (Gomez-Gomez et al., 1997; Lamrani et al., 1999).
Adaptive Mutation in E. coli strain FC40
For over a decade adaptive mutation in FC40 has been the paradigm for mutagenesis in nutritionally stressed bacteria. Nonetheless, there is still controversy about the mechanism by which the adaptive mutations are produced. Below, I describe the phenomenology and genetics of adaptive mutation in this particular strain of E. coli, and then discuss the various competing models that attempt to explain the phenomenon.
The Characteristics of E. coli Strain FC40
The lineage of strain FC40 dates back to Jacob and Monod. Its immediate parent, P90C, also known as CSH142 (Miller, 1992), has a chromosomal deletion, originally called Δ(lac-proB)XIII but now known as Δ(gpt-lac)5, that removes approximately 2.5 minutes of the chromosome including the lac and proAB operons (but not the dinB gene). The episome is a large conjugal plasmid that carries about 3 minutes of the E. coli chromosome including the lac and proAB operons (and the dinB gene). can be mated into and stably maintained in P90C and its derivatives by selecting for proline prototrophy. P90C carrying the episome with an up-promoter mutation (lacIQ) in the lacI gene promoter, and a down-promoter mutation (L8) in the lac operon promoter, is known as GM1 (Coulondre et al., 1977), or CSH100 (Miller, 1992); this strain has figured prominently in the extensive mutational studies by Miller and colleagues.
The immediate parent of FC40 is C36, which is a spontaneous RifR mutant of P90C. The version of in FC40 has a fusion of the lacI gene to the lacZ gene, Φ(lacI-lacZ), originally isolated by Müller-Hill and colleagues (Brake et al., 1978). The fusion eliminates all of the lac regulatory region, the last four residues of lacI and the first 23 residues of lacZ. Transcription of the fusion initiates at the lacIQ promoter and is constitutive. The particular allele of the fusion that we use, Φ(lacI33-lacZ), has a +1 frameshift at the 320th codon of lacI, changing CCC to CCCC (Calos et al., 1981). This and similar alleles have been used by Miller and colleagues to study various mutational mechanisms (Miller, 1985; Tlsty et al., 1984). The episome carrying Φ(lacI33-lacZ) (obtained from J.H. Miller) was mated into C36 to give FC40 (Cairns et al., 1991). FC40 is Lac− but the allele is slightly leaky, probably due to ribosomal frameshifting, and makes about 2 Miller units of β-galactosidase. The lacI33 mutation is polar on lacY, so FC40 is also defective for lactose permease. When fully reverted, FC40 makes about 200 Miller units of β-galactosidase, which is enough to make it Lac+, but much less than the 1000 units made by fully-induced wild-type Lac+ bacteria (Miller, 1992).
Lac− FC40 cannot grow on lactose. When incubated in liquid minimum lactose medium, the Lac− population is completely static for at least 4 days (Foster, 1994) (Figure 1). However, when plated on minimal lactose plates, FC40 can double several times over the course of a few days. This growth is due to the impurities in the agar or, possibly, breakdown products of lactose, and can be prevented by adding an excess of Lac− “scavenger” cells to the plates. The scavengers, FC29 (Cairns et al., 1991), are P90C carrying an episome that is deleted for lacZ, and so cannot revert to Lac+ (obtained from J. Beckwith). In a typical adaptive mutation experiment, 108 FC40 cells are mixed with 109 FC29 cells, plated on minimal plates with lactose as the only carbon source, and incubated for five days. Because of the low amount of β-galactosidase made by Lac+ revertants of FC40, Lac+ colonies take 2 days to become visible. Therefore, most of the colonies that appear on day two are due to mutations that occurred during growth prior to plating. Thereafter, from day 3 to day 5, new Lac+ colonies appear at a constant rate. These late-arriving revertants are due to mutations that occurred after the cells were on lactose, and are called adaptive mutations.
FIGURE 1.
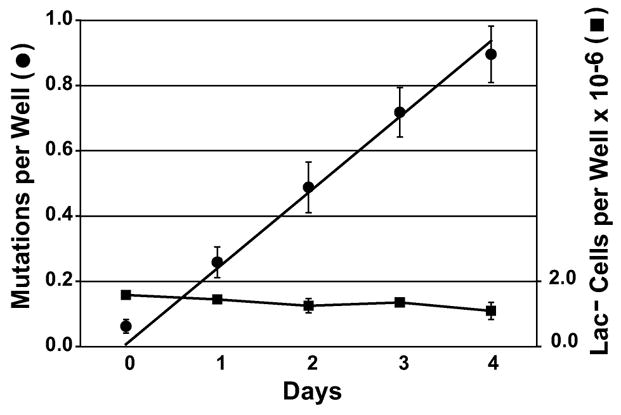
The accumulation of Lac+ mutations in liquid minimum lactose medium. Eight independent cultures of FC40 were diluted into minimum lactose medium and 100 μl aliquots containing approximately 106 cells were dispensed into 726 microtiter dish wells. Turbid wells were counted daily. The total number of Lac− cells was determined in cultures incubated in parallel. The minimum lactose medium had been scavenged of non-lactose carbon sources by incubating it with 109 Δ(lac) cells per mL for three hours; these cells were removed by centrifugation followed by filtration. Because it takes 2 days for a well to become turbid after a Lac+ mutation occurs, the mutations per well have been shifted two days to the left. (Reprinted with permission from Foster, 1994. Genetics. 138:256. The Genetics Society of America. www.genetics.org.)
The Phenomenology of Adaptive Mutation in E. Coli Strain FC40
The basic phenomenology of adaptive mutation in FC40 is as follows:
During growth under non-selective conditions, Lac+ mutations occur at the rate of about one in 109 cells per generation. During lactose selection, Lac+ mutations appear at the rate of 20 to 50 per 108 cells per day (Cairns et al., 1991; Foster, 1994).
Lac− cells can be incubated on lactose minimal medium without growth or death for several days (Cairns et al., 1991; Foster, 1994; Rosche et al., 2001). During this time, the rate at which Lac+ mutations appear remains constant.
Lac+ adaptive mutation is not restricted to agar plates, but, as shown in Figure 1, occurs at the same rate in liquid medium as on plates (Foster, 1994). Thus, adaptive mutation may occur equally well in structured and non-structured environments (with the caveat that the cells in the liquid medium were not agitated and settled to the bottom.)
Lac+ mutations do not accumulate when FC40 is incubated without a carbon source, but immediately begin to appear when lactose is added to the starving cells. Thus mere starvation in the absence of selection does not induce Lac+ mutations–lactose is required (Cairns et al., 1991; Foster, 1994).
While incubating on lactose, Lac− cells accumulate nonselected mutations. The rate at which these appear is high for genes on the episome (Foster, 1997) and low for genes on the chromosome (Bull et al., 2000a; Foster, 1994). Thus mutations are not “directed” to lac.
While incubating on lactose, Lac+ revertants accumulate nonselected mutations at frequencies 20-to 100-fold higher than the frequencies at which these mutations appear among the Lac− population (Rosche et al., 1999; Torkelson et al., 1997). Since after isolation the Lac+ revertants with second mutations do not have a higher than normal mutation rate, some proportion of the cells must experience a transient hypermutation during lactose selection.
About 2% of the Lac+ colonies that appear after 5 days on lactose plates are composed of cells that have amplified the Lac− allele (Foster, 1994; Foster et al., 1995a). After 10 days on lactose plates, this proportion increases to 60% (Hastings et al., 2000; Powell et al., 2001). These colonies appear late because they are slow to develop.
Genetic Requirements for Adaptive Mutation in FC40
Adaptive mutation in FC40 has genetic requirements that distinguish it from mutation during normal growth. These are:
-
The mutations that give rise to Lac+ revertants during selection are different than those that give rise to Lac+ revertants during nonselected growth. The mutations that occur during lactose selection are almost all one base-pair deletions in runs of the same base, whereas the mutations that occur during growth include deletions, duplications, and larger frameshifts (Foster et al., 1994b; Rosenberg et al., 1994).
The −1 frameshifts produced during lactose selection are exactly the mutations that would be expected to be made by DNA polymerases in general, and Pol IV in particular (Kim et al., 1997; Wagner et al., 2000). During nonselected growth, most of these potential −1 frameshifts mutations are corrected by MMR (Longerich et al., 1995). Thus, it has been argued that the difference in the spectrum of Lac+ mutations during growth and during lactose selection is due a failure of MMR in the latter case (Longerich et al., 1995). However, it is equally as likely that because Pol IV is active in the nondividing cells, there are simply more −1 frameshifts being made.
The Lac+ mutations that appear during lactose selection occur by a different mechanism than the Lac+ mutations that appear during growth. Enzymes for the recombinational repair of double-strand breaks, namely RecA, RecBCD, and RuvABC, are required for Lac+ mutations during lactose selection but not during growth (Cairns et al., 1991; Foster, 1993; Foster et al., 1996; Harris et al., 1994; Harris et al., 1996).
The high level of adaptive reversion to Lac+ seen in FC40 requires that the lac allele be on the episome. When the lac allele is in its normal place on the chromosome, the rate of reversion to Lac+ during lactose selection falls 100-fold and it is no longer dependent on recombination functions (Foster et al., 1995b; Radicella et al., 1995).
-
The high level of adaptive reversion to Lac+ also requires that the episome’s conjugal functions be expressed. If conjugal functions are defective, the rate of adaptive mutation falls 10-fold, but it is still largely dependent on recombination functions (Foster et al., 1995b).
The most likely role for the conjugal functions is to initiate recombination by producing a nick at the conjugal origin (Ponder et al., 2005; Rodriguez et al., 2002). However, defects in several of the conjugal tra genes, not just traI, the gene that encodes the nicking enzyme, have the same inhibitory effect on adaptive mutation (Foster et al., 1995b; Rodriguez et al., 2002). This is likely because the Tra proteins form a multi-protein complex (Lawley et al., 2003), and defects in one protein can disrupt the function of the whole complex.
-
Fifty to 80% of adaptive Lac+ mutations are due to DNA polymerase IV (Foster, 2000; McKenzie et al., 2001). Pol II is also active, but does not contribute to adaptive mutations (Foster et al., 1995a); in fact, Pol II appears to compete with and limit the mutational potential of Pol IV (Foster, 2000). Pol V is not involved (Cairns et al., 1991; McKenzie et al., 2000).
In the absence of Pol IV, the remaining Lac+ mutations are most likely produced by Pol III (Foster, 2000; McKenzie et al., 2001). Pol IV is also required for transient hypermutation (see below) (Tompkins et al., 2003).
Adaptive mutation to Lac+ is reduced about 10-fold in the absence of RpoS (Layton et al., 2003; Lombardo et al., 2004). Most of this reduction is due to failure to induce Pol IV (discussed above), but RpoS also has a small Pol IV-independent role in adaptive mutation (Layton et al., 2003; Lombardo et al., 2004).
Adaptive mutation to Lac+ is reduced 10- to 20-fold if GroE is deficient (Layton et al., 2005). The amount of Pol IV declines 10-fold in GroE deficient cells (discussed above), and this is sufficient to explain the reduction in adaptive mutation in wild-type cells. However, in recG mutant cells GroE has a small effect on adaptive mutation that is independent of Pol IV (Layton et al., 2005).
Adaptive mutation to Lac+ is reduced about five-fold in the absence of polyphosphate kinase, Ppk (Stumpf et al., 2005). As discussed above, PolyP (or Ppk itself) is required for the maximum mutagenic and lesion bypass activity of both Pol IV and Pol V (Stumpf et al., 2005, 2007a). In addition, Ppk has a small Pol IV-independent role in adaptive mutation (Stumpf et al., 2005).
-
The rate of adaptive mutation to Lac+ is increased up to 100-fold by loss of RecG (Foster et al., 1996; Harris et al., 1996) or DNA Pol II (Foster et al., 1995a). In strains defective for recG or polB (encoding Pol II), the levels of Pol IV are increased 5- to 30- fold (Layton et al., 2005), which may account for the increase in mutation rate.
recG mutant cells are partially induced for the SOS response (Lloyd et al., 1991; McCool et al., 2004), and a similar induction may explain the elevated amounts of Pol IV in polB mutant cells. In addition, the errors leading to Lac+ adaptive mutations in recG mutant cells appear to be poorly corrected by MMR, possibly because the MMR system is overwhelmed (Foster et al., 1996). Whether these two effects are sufficient to explain the large increases in adaptive mutation in recG and polB mutants remains to be seen.
-
The number of Lac+ revertants appearing during lactose selection is increased 10- to 30-fold in a recD mutant strain (Harris et al., 1994). However, most, if not all, of this increase is due to the accumulation of extra copies of the episome and, consequently, of the lac allele (Foster et al., 1999).
RecD is the exonuclease subunit of RecBCD and recD mutants have a hyper-recombination phenotype. But, loss of RecD also causes plasmids to over-replicate (for example, see Seelke et al., 1987). recD mutants of FC40 have extra copies of the episome and, because of their extra lac alleles, can proliferate on lactose. The combination of over-replication and cell-growth accounts for an ever increasing accumulation of Lac+ revertants during lactose selection (Foster et al., 1999). This nonlinear accumulation, which is expected when the number of lac alleles increases, is not characteristic of adaptive mutation to Lac+ in wild-type strains (Foster, 1994).
-
Lac+ adaptive mutation is increased about 100-fold if MMR is defective. MMR deficiency can be produced by inactivating one of the MMR proteins (Foster et al., 1992), or by overproducing the Dam methylase, which eliminates strand-discrimination (Foster et al., 1996). These results show that, during lactose selection, MMR is active and the DNA is methylated.
MMR could be inactive during lactose selection in a subpopulation of cells. This hypothesis is supported by the fact that overproduction of the MMR enzymes MutS, MutL, or both, reduces Lac+ adaptive mutation (Foster et al., 1995a; Foster, 1999a; Harris et al., 1997). However, overproduction of the MMR enzymes can also reduce mutations in growing cells, suggesting that some components of MMR are always limiting (Foster et al., 1995a; Foster, 1999a). Disagreement over the meaning of these various results is still unresolved (Foster, 1999a; Harris et al., 1999). As discuss below, loss of MMR does not increase the frequency at which other, nonselected mutations appear in Lac+ cells, indicating that MMR is inactive in those cells that are hypermutators (Rosche et al., 1999).
Models for Adaptive Mutation in FC40
Currently there are two alternative models for how Lac+ mutations arise during lactose selection–recombination-dependent mutation (RDM) and amplification-dependent mutation (ADM), each of which is described below. While these models are not mutually exclusive, what animates the disagreement about the models is that they incorporate different beliefs about the basic process by which mutations are produced. The RDM model proposes that a special mutagenic mechanism is at work in cells during selection, whereas the ADM model proposes that mutations occur by an ordinary mechanism among a growing subpopulation. More detailed discussions of the models, the evidence supporting each, and personal opinions pro and con, can be found in references Foster, 2004; Rosenberg et al., 2004; and, Roth et al., 2004.
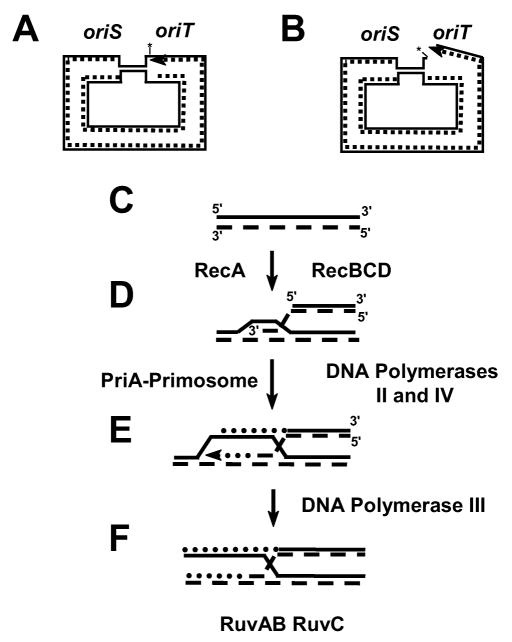
The initiating event for RDM is production of a double-strand break in the DNA, which occurs at a high frequency on the episome because the conjugal origin is subjected to continuous nicking even in the absence of active conjugation. While the Lac− cells are on lactose, the slow influx of energy produced by the leaky lac allele allows replication to be occasionally initiated from one of the episome’s vegetative origins (Figure 2A). When the moving replication fork encounters the persistent nick at oriT, a double strand end is produced (Figure 2B) and this initiates double-strand break repair. This RecA-and RecBCD- dependent repair proceeds via strand invasion of either the homologous duplicated portion of the same episome or of another copy of the episome (Figure 2D). DNA synthesis primed by the 3′ end of the invading strand is initiated by the PriA-dependent primosome (Figure 2E). The new DNA is synthesized by either Pol II, in which case it is mostly error-free, or by Pol IV, in which case it has a high frequency of errors. Mistakes can also be made by Pol III, which is recruited to continue replication (Figure 2F). The Holiday junction produced during recombination is then resolved by the RuvAB/RuvC enzymes. If the region of new DNA includes the lac region, then Lac+ mutations can arise; if other regions of the episome are included, then the genes in those regions can have mutations. While the recombination-dependent mechanism occurs at a high rate on the episome, it can also produce mutations on any replicon, including the chromosome, whenever the replication fork encounters a nick, or when a double-strand break is produced in some other way.
FIGURE 2.
A model for recombination-dependent mutation. A and B: the replication fork initiated at oriS collapses when it encounters the nick at oriT, creating a double-strand end. C and D: RecA and RecBCD catalyze invasion of the broken arm into the homologous region of the sister chromosome or a different episome copy. E: the PriA-primosome initiates DNA synthesis from the invading 3′ end, using either Pol IV or Pol II; the four-stranded Holliday junction is formed. F: the replication fork is re-established with Pol III. Not shown: the Holliday junction is resolved by RuvABC. Dashed lines are newly synthesized DNA; dotted lines are DNA synthesized after recombination. The arrow head indicates the 3′ end of the leading DNA strand; * at oriT indicates TraI. (Reprinted, with permission, from Foster 1999, Ann Rev Gene 33:62. Annual Reviews. www.annualreviews.org)
The initiating event for ADM is a duplication of a region of DNA that includes the lac region. These duplications arise during nonselective growth before lactose selection at a frequency of about one in every 1000 cells. On lactose medium, cells with lac duplications have enough energy to slowly proliferate and further amplify their lac regions via a RecA, RecBCD-dependent mechanism. Eventually a true Lac+ revertant arises among the amplified arrays, allowing the cell with the Lac+ copy to proliferate. Within this clone the amplified arrays deamplify, leaving the single Lac+ copy, and these Lac+ cells overgrow the remaining slower-growing precursors.
The disagreements about which of these models is correct reflect real disagreements among researchers about the actual phenomenology of the experiment. Is there a slow-growing subpopulation (Hendrickson et al., 2002), or not (Foster, 1994)? Does every Lac+ colony contain cells that are unstably Lac+ (Hendrickson et al., 2002), or not (Slack et al., 2006)? Are all sectored colonies the result of amplification (Andersson et al., 1998), or not (Slack et al., 2006)? Can the amplification model describe the dynamics of Lac+ mutant appearance (Pettersson et al., 2005), or not (Stumpf et al., 2007b)? Does the amplification model account for all of the genetic requirements (Roth et al., 2004), or not (Hastings et al., 2004)?
While all researchers agree that both true reversion and pseudo-reversion by amplification occur while FC40 is under lactose selection, it remains contentious whether amplification is a separate pathway or a necessary precursor for true reversion. The question could be settled by inhibiting amplification and determining if Lac+ reversion is reduced. This was attempted by Hendrickson et al. (2002), who placed a transposon encoding tetracycline resistance (TetR) on the episome at various distances from the lac region. Although the TetR genes normally make cells resistant to tetracycline, if they are in multiple copy cells become sensitive to tetracycline because overproduction of the efflux transporter encoded by tetA is toxic (Chopra et al., 1981; Eckert et al., 1989). Thus, if the lac region is amplified during lactose selection, TetR genes close to the lac region should be co-amplified and kill the amplifying cells. If amplification is a necessary precursor to Lac+ reversion, the closer the TetR genes are to lac, the more the rate of Lac+ reversion should be reduced. While it was true that the rate of adaptive mutation to Lac+ was low in the presence of tetracycline when the TetR genes were to close to lac (2002), further analysis showed that this was also true when tetracycline was not present, i.e., when the transposons were close to lac the rate of lac reversion was low for some reason other than toxicity (Stumpf et al., 2007b). In addition, a similar effect was seen when the transposon tetA genes were mutant. The rate at which the mutant tetA genes reverted to TetR during lactose selection was independent of the distance between tetA and lac; indeed, TetR reversion occurred even when tetA and lac were on different replicons. Thus reversion of a gene during lactose selection does not require that it is co-amplified with the lac region (Stumpf et al., 2007b).
In addition, the ADM model simply does not account for the fact that during lactose selection the appearance of Lac+ revertants is nearly linear with time (Figure 1). Like other “growth of an intermediate” models (Lenski et al., 1989), the ADM model predicts that the rate at which Lac+ revertants appear should increase with time. The ADM model further predicts a rapid increase in the appearance of Lac+ colonies when the number of Lac− alleles reaches a critical threshold. In confirmation, computer simulations and laboratory reconstructions of ADM show that nearly all the Lac+ revertants arise almost simultaneously after five days on lactose (Pettersson et al., 2005; Slechta et al., 2003). Yet, in actual adaptive mutation experiments, hundreds of Lac+ colonies appear at a constant rate from day 2 to 5.
Transient Hypermutation
As mentioned above, a significant fraction of the Lac+ revertants that appear during lactose selection also carry mutations that were not being selected (Rosche et al., 1999; Slechta et al., 2002; Torkelson et al., 1997). When isolated and tested, these Lac+ revertants do not have elevated mutation rates, meaning that they are not constitutive mutators. Therefore, they must have experienced a transient increase in their mutation rate during lactose selection. This phenomenon is called hypermutation, and was originally proposed by Hall (1990) to explain why mutations appear to be directed. While all researchers agree that hypermutation is a feature of adaptive mutation in FC40, they disagree about its causes and consequences.
The percentage of Lac+ revertants that have a second mutant phenotype increases linearly with the time that the population is under lactose selection. For example, over 5 days the proportion of the Lac+ revertants that is also defective for motility (Mot−) increases from 0% to 2%. This increase indicates either that the hypermutating cells do not die, and thus continue to accumulate mutations, or that the proportion of cells that are progressing into the hypermutator state increases with time (Rosche et al., 1999). Second mutations appear at a much lower frequency among Lac+ revertants if Pol IV is defective, suggesting that both the Lac+ and the nonselected mutations are due to the activity of Pol IV (Slechta et al., 2003; Tompkins et al., 2003). Loss of MMR increases the rate at which adaptive Lac+ revertants appear about 100-fold, but only increases the frequency of second mutations in the Lac+ revertants by about 70%, meaning that the hypermutating cells are wholly or partially MMR defective (Rosche et al., 1999).
The high mutation rate of hypermutators cannot be due solely to loss of MMR. Lac+ revertants with a second mutation must be derived from the hypermutating population, and the frequency of a third mutation among these Lac+ double mutants is about 10-fold higher than the frequency of a second mutation among MMR− Lac+ cells (Rosche et al., 1999). This means that loss of MMR is not sufficient to account for hypermutation. A likely additional factor is overexpression of Pol IV, which could occur, for example, in cells expressing both the RpoS-dependent and SOS responses. Loss of MMR and hyper-induction of Pol IV could be independent, or MMR could be saturated by errors due to very high levels of Pol IV activity in a subpopulation of cells (McKenzie et al., 2001; Tompkins et al., 2003).
An alternative hypothesis for hypermutation is that it occurs only in the subpopulation of cells that have amplified the dinB gene along with the lac region (Slechta et al., 2003). The original recombination event that created placed dinB much closer to lac than it is on the chromosome, and dinB could be included when the lac region is amplified (Kofoid et al., 2003). However, in a strain with Φ(lacI33-lacZ) in its normal place on the chromosome, 2% of the chromosomal Lac+ revertants had second, nonselected mutations (Rosche et al., 1999); since adaptive mutation in this case is not recombination-dependent (Foster et al., 1995b), hypermutation is unlikely to be due to co-amplification of dinB.
Although it has been proposed that all Lac+ revertants occur in hypermutating cells (Torkelson et al., 1997), simple logic dictates that there are at least two mutating populations. All available data indicate that the frequency of second mutations in Lac+ single mutants is only a tenth of the frequency of third mutations among Lac+ double mutants (Rosche et al., 1999; Torkelson et al., 1997). Since the Lac+ double mutants arose in hypermutators, some fraction of the Lac+ single mutants must have arisen in a population that has a lower mutation rate than the hypermutators (Cairns, 2000; Foster, 2004).
Based on ratios of the frequencies of single, double, and triple mutants, Rosche et al. (1999) estimated that about 0.1% of the Lac− population were hypermutators and their mutation rate was 200-fold higher than the rest of the population. If so, then they account for only 10% of the adaptive Lac+ mutants (but virtually all of the double and triple mutants), and 90% the Lac+ mutations are produced in cells that do not have an extraordinary increase in mutation rate (although there could, in fact, be a continuum of mutation rates within the population) (Foster, 2004). Note, this conclusion does not mean that the mechanism by which mutations are produced must differ among the populations. Rosenberg and coworkers also estimated the hypermutating subpopulation to be 0.01 to 0.1% of the total, but they hypothesize that all Lac+ revertants come from this population, and so the mutation rate must be much higher (Bull et al., 2000b; Torkelson et al., 1997). However, Roth et al. (2003) argue that to produce all the observed number of Lac+ mutants the mutation rate within this tiny population would have to be so high (more than 105-fold higher than normal) that the hypermutators would be unlikely to survive. This problem mostly disappears if the mutation rate on the episome, which carries the lac allele in FC40, is higher than on the chromosome, where the genes essential for life reside (Cairns et al., 2003).
The calculations of the size of the hypermutating subpopulation and its contribution to Lac+ adaptive mutations depend on the frequencies of single, double, and triple mutants, and the number of triple mutants is very small (Rosche et al., 1999). In addition, the calculations also depend on the frequency of the same nonselected mutations in Lac− cells, and this frequency is low and may not be due to mutations that occurred during selection. So the magnitude of the contribution of hypermutators to adaptive mutation to Lac+ is still open. The theoretical relationship among the population of the Lac− cells that are hypermutators (p), their increase in mutation rate (M), and the proportion of Lac+ revertants that occur in the hypermutators (R), is plotted in Figure 3.
FIGURE 3.
The relationship between the proportion of Lac+ revertants that occur in the hypermutating subpopulation (Y axis), the proportion of cells that are hypermutators (X axis), and the elevation in mutation rate in the hypermutating cells (M). The curves are base on the derivation by Cairns (1998).
Are There Any Directed Mutations?
As pointed out by Hall (1998b), there are two ways that mutations could be directed–selective generation or selective capture. The only examples of selective generation appear to be transcription-induced mutations in genes that are highly induced by the selective conditions (Wright, 2004) (see above). The fact that F′ has a higher mutation rate than the chromosome gives the appearance of selective generation, but nonselected mutations on the episome readily occur during selective conditions (Foster, 1997). Thus it seems that organisms only occasionally have access to a selective generation process.
Selective capture, on the other hand, may be a constant feature of mutation during selection. Indeed, it may be unavoidable. The idea is as follows: Cells under selective pressure experience potentially mutagenic alterations to their DNA from a variety of causes. If a sequence change does not have a useful result, it is repaired or discarded. However, if a sequence change gives rise to an immediately expressed useful phenotype, the cell will begin to proliferate, fail to repair the damage, and immortalize the mutation. Of course, other premutations existing at the moment the cell begins to grow may also be immortalized, but the population at large will not accumulate unwanted mutations. Variations of this “trial and error” idea have been discussed for years (Boe, 1990; Cairns et al., 1988; Stahl, 1988, 1992). Hall’s original “hypermutator state” model, which proposed that hypermutating cells die if a useful mutation does not occur (Hall, 1990), can be considered an extreme form of “trial and error.” However, evidence strongly suggesting a trial and error mechanism has only recently been generated. When LacI+ cells are given phenyl-galactoside (Pgal) as a carbon source, the mutations that allow them to grow are those that inactivate the lacI gene or destroy the ability of the lac operator to bind LacI. lacI− mutations can be recessive or dominant whereas the operator mutations, designated lacOc, are always dominant. Bharatan et al. (2004) found that when cells were placed under Pgal selection, early appearing mutations were almost all recessive, but the proportion of dominant mutations increased with time; after 10 days, almost all the mutations that appeared were dominant. They proposed that this bias reflected a need for “instantaneous gratification” in order to capture the mutagenic sequence change. The “instantaneous gratification” mechanism may prove to be universal, but previously overlooked because most mutations that have been studied in nongrowing cells are dominant.
CONCLUDING REMARKS
It has become almost de rigueur to begin reviews of mutagenesis in natural populations by declaring that bacteria have a “feast and famine” lifestyle (Koch, 1971). Since without genetic variation there is no evolution, the ability to mutate yourself out of famine and into feast would be adaptive. In recent years, it has become apparent that among isolates of bacteria from natural environments, several percent have constitutive mutation rates high enough to qualify them as mutators compared to laboratory strains (Matic et al., 1997). Laboratory experiments have demonstrated the ease at which constitutive mutators can be enriched by selecting for novel phenotypes (Mao et al., 1997; Miller et al., 1999) or competitive fitness (Cox et al., 1974; Sniegowski et al., 1997). Modeling and simulations have confirmed that mutator alleles can contribute to adaptive evolution in changing environments (Leigh, 1973; Taddei et al., 1997b). However, the mutator allele is always doomed–when a successful mutation is achieved, individuals that continue to mutate are at a disadvantage. Thus, continuous feast-full conditions should select for low mutation rates, and they apparently do (Trobner et al., 1984).
However, if mutation rates increased only when mutations would be advantageous, and then returned to normal when success was achieved, the gene that conferred a transient mutator phenotype would not be strongly selected against. The individual that achieved a successful mutation would be burdened only with the extra mutations that occurred during the mutator state. Simulations have shown that if a population is undergoing periodic stress, stress-inducible mutator genes can be fixed more readily than constitutive mutator genes; and, importantly, under these conditions stress-inducible genes can speed up adaptive evolution (Bjedov et al., 2003).
Stress-induced mutagenesis is neither a theoretical construct nor a laboratory artifact. Among natural isolates of E. coli, 40% had at least a 10-fold increase in mutant frequency, and 13% had at least a 100-fold increase in mutant frequency, when they were incubated on agar plates for 7 days (Bjedov et al., 2003). Among the same group of isolates, only 3% and 1% had equivalently increased constitutive mutant frequencies, respectively (Bjedov et al., 2003). These numbers suggest that inducible mutators are widespread, and that they make a greater contribution to natural variation within a population than do constitutive mutators.
No matter how adaptive the consequences, a question remains–have inducible mutator genes been retained to achieve an increase in variation during stressful times, or is the mutator state simply an unavoidable side effect of cellular activities that have been selected for other purposes? For example, all three SOS-inducible DNA polymerases contribute to competitive survival in long-term stationary phase cultures (Yeiser et al., 2002). The conservative interpretation of this result is that the three polymerases improve fitness by allowing damaged DNA to be replicated. However, they may also be advantageous because they create mutations, allowing the evolution of the “growth advantage in stationary phase” (GASP) phenotype (Finkel, 2006). Likewise, since the MMR system sacrifices a piece of newly synthesized DNA thousands of bases long to repair one mismatch, down-regulation of the MMS repair system in stationary phase may simply be a way to save energy. Nonetheless, the result of MMR decline is that any errors made by DNA polymerases in stationary phase may not be efficiently corrected. What may be more important, since MMR is normally a barrier to recombination between non identical DNA, down-regulation of MMR could enhance variation by allowing genetic exchange of diverged alleles (Rayssiguier et al., 1989). Up-regulation of error-producing DNA polymerases and down-regulation of error-correcting systems may have evolved for other purposes, but yet may be retained in the population because they increase diversity when it is advantageous to do so.
Acknowledgments
I thank John Cairns for his continuing collaboration and for critically reading this manuscript. I thank past and present members of my laboratory for their ideas, discussions, and research contributions. Work in my laboratory is supported by grant GM065175 from the U.S. National Institutes of Health.
References
- Aertsen A, Michiels CW. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Molec Microbiol. 2005;58:1381–1391. doi: 10.1111/j.1365-2958.2005.04903.x. [DOI] [PubMed] [Google Scholar]
- Aertsen A, Michiels CW. Upstream of the SOS response: figure out the trigger. Trends Microbiol. 2006;14:421–423. doi: 10.1016/j.tim.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Aguirre-Ramirez M, Ramirez-Santos J, Van ML, Gomez-Eichelmann MC. Expression of the F plasmid ccd toxin-antitoxin system in Escherichia coli cells under nutritional stress. Can J Microbiol. 2006;52:24–30. doi: 10.1139/w05-107. [DOI] [PubMed] [Google Scholar]
- Al Mamun AA, Marians KJ, Humayun MZ. DNA polymerase III from Escherichia coli cells expressing mutA mistranslator tRNA Is error-prone. J Biol Chem. 2002;277:46319–46327. doi: 10.1074/jbc.M206856200. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, et al. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autret S, Levine A, Vannier F, Fujita Y, Seror SJ. The replication checkpoint control in Bacillus subtilis: identification of a novel RTP-binding sequence essential for the replication fork arrest after induction of the stringent response. Molec Microbiol. 1999;31:1665–1679. doi: 10.1046/j.1365-2958.1999.01299.x. [DOI] [PubMed] [Google Scholar]
- Balashov S, Humayun MZ. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J Molec Biol. 2002;315:513–527. doi: 10.1006/jmbi.2001.5273. [DOI] [PubMed] [Google Scholar]
- Balashov S, Humayun MZ. Escherichia coli cells bearing a ribosomal ambiguity mutation in rpsD have a mutator phenotype that correlates with increased mistranslation. J Bacteriol. 2003;185:5015–5018. doi: 10.1128/JB.185.16.5015-5018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barionovi D, Ghelardini P, Di LG, Paolozzi L. Mutations arise independently of transcription in non-dividing bacteria. Mol Genet Genomics. 2003;269:517–525. doi: 10.1007/s00438-003-0857-8. [DOI] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature (London) 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- Beletskii A, Bhagwat AS. Transcription-induced mutations: increases in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Jacob E, Cohen I, Gutnick DL. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol. 1998;52:779–806. doi: 10.1146/annurev.micro.52.1.779. [DOI] [PubMed] [Google Scholar]
- Benov L, Fridovich I. The rate of adaptive mutagenesis in Escherichia coli is enhanced by oxygen (superoxide) Mutat Res. 1996;357:231–236. doi: 10.1016/0027-5107(96)00128-5. [DOI] [PubMed] [Google Scholar]
- Bhamre S, Gadea BB, Koyama CA, White SJ, Fowler RG. An aerobic recA-, umuC-dependent pathway of spontaneous base-pair substitution mutagenesis in Escherichia coli. Mutat Res. 2001;473:229–247. doi: 10.1016/s0027-5107(00)00155-x. [DOI] [PubMed] [Google Scholar]
- Bharatan SM, Reddy M, Gowrishankar J. Distinct signatures for mutator sensitivity of lacZ reversions and for the spectrum of lacI/lacO forward mutations on the chromosome of nondividing Escherichia coli. Genetics. 2004;166:681–692. doi: 10.1534/genetics.166.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- Boe L. Mechanism for induction of adaptive mutations in Escherichia coli. Molec Microbiol. 1990;4:597–601. doi: 10.1111/j.1365-2958.1990.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006;14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Brake AJ, Fowler AV, Zabin I, Kania J, Müller-Hill B. β-galactosidase chimeras: primary structure of a lac repressor-β-galactosidase protein. Proc Natl Acad Sci USA. 1978;75:4824–4827. doi: 10.1073/pnas.75.10.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- Bridges BA. mutY ‘directs’ mutation? Nature (London) 1995a;375:741. doi: 10.1038/375741a0. [DOI] [PubMed] [Google Scholar]
- Bridges BA. Starvation-associated mutation in Escherichia coli strains defective in transcription repair coupling factor. Mutat Res. 1995b;329:49–56. doi: 10.1016/0027-5107(95)00016-c. [DOI] [PubMed] [Google Scholar]
- Bridges BA, Foster PL, Timms AR. Effect of endogenous carotenoids on “adaptive” mutation in Escherichia coli FC40. Mutat Res. 2001;473:109–119. doi: 10.1016/s0027-5107(00)00144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA, Timms AR. Effect of endogenous carotenoids and defective RpoS sigma factor on spontaneous mutation under starvation conditions in Escherichia coli: evidence for the possible involvement of singlet oxygen. Mutat Res. 1998;403:21–28. doi: 10.1016/s0027-5107(98)00013-x. [DOI] [PubMed] [Google Scholar]
- Bridges BA. Starvation-associated mutation in Escherichia coli: a spontaneous lesion hypothesis for “directed” mutation. Mutat Res. 1994;307:149–156. doi: 10.1016/0027-5107(94)90287-9. [DOI] [PubMed] [Google Scholar]
- Brotcorne-Lannoye A, Maenhaut-Michel G. Role of RecA protein in untargeted UV mutagenesis of bacteriophage lambda: evidence for the requirement for the dinB gene. Proc Natl Acad Sci USA. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull HJ, McKenzie GJ, Hastings PJ, Rosenberg SM. Evidence that stationary-phase hypermutation in the Escherichia coli chromosome is promoted by recombination. Genetics. 2000a;154:1427–1437. doi: 10.1093/genetics/154.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull HJ, McKenzie GJ, Hastings PJ, Rosenberg SM. Response to John Cairns: The contribution of transiently hyper-mutable cells to mutation in stationary phase. Genetics. 2000b;156:925–926. [Google Scholar]
- Cairns J. Mutation and cancer: The antecedents to our studies of adaptive mutation. Genetics. 1998;148:1433–1440. doi: 10.1093/genetics/148.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. The contribution of bacterial hypermutators to mutation in stationary phase. Genetics. 2000;156:923. doi: 10.1093/genetics/156.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Foster PL. The risk of lethals for hypermutating bacteria in stationary phase. Genetics. 2003;165:2317–2318. doi: 10.1093/genetics/165.4.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature (London) 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Calos MP, Miller JH. Genetic and sequence analysis of frameshift mutations induced by ICR-191. J Molec Biol. 1981;153:39–66. doi: 10.1016/0022-2836(81)90525-8. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Chang DE, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Molec Microbiol. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- Chopra I, Shales SW, Ward JM, Wallace LJ. Reduced expression of Tn10-mediated tetracycline resistance in Escherichia coli containing more than one copy of the transposon. J Gen Microbiol. 1981;126:45–54. doi: 10.1099/00221287-126-1-45. [DOI] [PubMed] [Google Scholar]
- Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C, Miller JH. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Molec Biol. 1977;117:525–575. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Couturier M, Bahassi EM, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- Cox EC, Gibson TC. Selection for high mutation rates in chemostats. Genetics. 1974;77:169–184. doi: 10.1093/genetics/77.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Davis BD. Transcriptional bias: A non-Lamarckian mechanism for substrate-induced mutations. Proc Natl Acad Sci USA. 1989;86:5005–5009. doi: 10.1073/pnas.86.13.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLuzio WR, Turner L, Mayer M, Garstecki P, Weibel DB, Berg HC, Whitesides GM. Escherichia coli swim on the right-hand side. Nature (London) 2005;435:1271–1274. doi: 10.1038/nature03660. [DOI] [PubMed] [Google Scholar]
- Donnelly CE, Walker GC. groE mutants of Escherichia coli are defective in umuDC-dependent UV mutagenesis. J Bacteriol. 1989;171:6117–6125. doi: 10.1128/jb.171.11.6117-6125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dri AM, Moreau PL. Control of the LexA regulon by pH: evidence for a reversible inactivation of the LexA repressor during the growth cycle of Escherichia coli. Molec Microbiol. 1994;12:621–629. doi: 10.1111/j.1365-2958.1994.tb01049.x. [DOI] [PubMed] [Google Scholar]
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Molec Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B, Beck CF. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J Bacteriol. 1989;171:3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstadt E. Analysis of mutagenesis. In: Neidhardt FC, Ingra-ham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, DC: ASM Press; 1987. pp. 1016–1033. [Google Scholar]
- Eisenstark A, Miller C, Jones J, Leven S. Escherichia coli genes involved in cell survival during dormancy: role of oxidative stress. Biochem Biophys Res Commun. 1992;188:1054–1059. doi: 10.1016/0006-291x(92)91338-q. [DOI] [PubMed] [Google Scholar]
- Feng G, Tsui HCT, Winkler ME. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J Bacteriol. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. Hungry bacteria - definition and properties of a nutritional state. Environ Microbiol. 2001;3:605–611. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- Fernandez de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Molec Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Micro. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. The challenges to Darwinism since the last centennial and the impact of molecular studies. Evolution. 1982;36(6):1133–1143. doi: 10.1111/j.1558-5646.1982.tb05484.x. [DOI] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation: The uses of adversity. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Population dynamics of a Lac− strain of Escherichia coli during selection for lactose utilization. Genetics. 1994;138:253–261. doi: 10.1093/genetics/138.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Are adaptive mutations due to a decline in mismatch repair? The evidence is lacking. Mutat Res. 1999;436:179–184. doi: 10.1016/s1383-5742(98)00023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation in Escherichia coli. Cold Spring Harbor Symp Quant Biol. 2000;65:21–29. doi: 10.1101/sqb.2000.65.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Adaptive Mutation in Escherichia coli. J Bacteriol. 2004;186:4846–4852. doi: 10.1128/JB.186.15.4846-4852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Cairns J. Mechanisms of directed mutation. Genetics. 1992;131:783–789. doi: 10.1093/genetics/131.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Cairns J. The occurrence of heritable Mu excisions in starving cells of Escherichia coli. EMBO J. 1994a;13:5240–5244. doi: 10.1002/j.1460-2075.1994.tb06855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Gudmundsson G, Trimarchi JM, Cai H, Goodman MF. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA. 1995a;92:7951–7955. doi: 10.1073/pnas.92.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Rosche WA. Increased episomal replication accounts for the high rate of adaptive mutation in recD mutants of Escherichia coli. Genetics. 1999;152:15–30. doi: 10.1093/genetics/152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994b;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc Natl Acad Sci USA. 1995b;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM, Maurer RA. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochem. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellen-berger T. DNA Repair and Mutagenesis. 2. Washington, D.C: ASM Press; 2006. [Google Scholar]
- Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez JM, Blazquez J, Baquero F, Martinez JL. H-NS and RpoS regulated emergence of Lac Ara+ mutants of Es-cherichia coli MCS2. J Bacteriol. 1997;179:4620–4622. doi: 10.1128/jb.179.14.4620-4622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Keck JL. Magic spots cast a spell on DNA primase. Cell. 2007;128:823–824. doi: 10.1016/j.cell.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Rutherford ST. General pathway for turning on promoters transcribed by RNA polymerases containing alternative σ Factors. J Bacteriol. 2006;188:4589–4591. doi: 10.1128/JB.00499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Spontaneous point mutations that occur more often when they are advantageous than when they are neutral. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Activation of the bgl operon by adaptive mutation. Molec Biol Evol. 1998a;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- Hall BG. Adaptive mutagenesis: a process that generates almost exclusively beneficial mutations. Genetica. 1998b;102/103:109–125. [PubMed] [Google Scholar]
- Haniford DB. Transpososome dynamics and regulation in Tn10 transposition. Crit Rev Biochem Mol Biol. 2006;41:407–424. doi: 10.1080/10409230600987415. [DOI] [PubMed] [Google Scholar]
- Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Hastings PJ, Winkler ME, Rosenberg SM. Mismatch repair is diminished during stationary-phase mutation. Mutat Res. 1999;437:51–60. [PubMed] [Google Scholar]
- Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Winkler ME, Rosenberg SM. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Harris RS, Ross KJ, Rosenberg SM. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Bull HJ, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2004;2:e399. doi: 10.1371/journal.pbio.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc Natl Acad Sci USA. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, Andersson DI. Carbon starvation ofSalmonella typhimurium does not cause a general increase of mutation rates. J Bacteriol. 1997;179:6688–6691. doi: 10.1128/jb.179.21.6688-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves H, Horak R, Kivisaar M. Involvement of σS in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J Bacteriol. 2001;183:5445–5448. doi: 10.1128/JB.183.18.5445-5448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasak L, Horak R, Kivisaar M. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc Natl Acad Sci USA. 1997;94:3134–3139. doi: 10.1073/pnas.94.7.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Molec Microbiol. 2006;62:1228–1238. doi: 10.1111/j.1365-2958.2006.05444.x. [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA Pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- Klapacz J, Bhagwat AS. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J Bacteriol. 2002;184:6866–6872. doi: 10.1128/JB.184.24.6866-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapacz J, Bhagwat AS. Transcription promotes guanine to thymine mutations in the non-transcribed strand of an Escherichia coli gene. DNA Repair. 2005;4:806–813. doi: 10.1016/j.dnarep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Koch AL. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microb Physiol. 1971;6:147–217. doi: 10.1016/s0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- Koch WH, Fernandez de Henestrosa AR, Woodgate R. Identification of mucAB-like homologs on two IncT plasmids, R394 and Rts-1. Mutat Res. 2000;457:1–13. doi: 10.1016/s0027-5107(00)00134-2. [DOI] [PubMed] [Google Scholar]
- Kofoid E, Bergthorsson U, Slechta ES, Roth JR. Formation of an F′ plasmid by recombination between imperfectly repeated chromosomal Rep sequences: a closer look at an old friend (F′128pro lac) J Bacteriol. 2003;185:660–663. doi: 10.1128/JB.185.2.660-663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature (London) 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kuan CT, Liu SK, Tessman I. Excision and transposition of Tn5 as an SOS activity in Escherichia coli. Genetics. 1991;128:45–57. doi: 10.1093/genetics/128.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Jonczyk P, Gawel D, Malanowska K, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J Bacteriol. 2004;186:4802–4807. doi: 10.1128/JB.186.14.4802-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, Jonczyk P, Fijalkowska IJ. Mutator phenotype resulting from DNA Polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J Bacteriol. 2005;187:6862–6866. doi: 10.1128/JB.187.19.6862-6866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Banach-Orlowska M, Schaaper RM, Jonczyk P, Fi-jalkowska IJ. Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J Bacteriol. 2006;188:7977–7980. doi: 10.1128/JB.01088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaev I, Kulakovskaya T. Polyphosphate and phosphate pump. Annu Rev Microbiol. 2000;54:709–734. doi: 10.1146/annurev.micro.54.1.709. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Murphy H, Cashel M, Kornberg A. Guano-sine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon pro-tease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- Kusano S, Ishihama A. Functional interaction of Escherichia coli RNA polymerase with inorganic polyphosphate. Genes to Cells. 1997;2:433–441. doi: 10.1046/j.1365-2443.1997.13203301320330.x. [DOI] [PubMed] [Google Scholar]
- Kvint K, Farewell A, Nystrom T. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σs. J Biol Chem. 2000;275:14795–14798. doi: 10.1074/jbc.C000128200. [DOI] [PubMed] [Google Scholar]
- Lamrani S, Ranquet C, Gama MJ, Nakai H, Shapiro JA, Toussaint A, Maenhaut-Michel G. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Molec Microbiol. 1999;32:327–344. doi: 10.1046/j.1365-2958.1999.01352.x. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is regulated by the molecular chaperon GroE in Escherichia coli. J Bacteriol. 2005;187:449–457. doi: 10.1128/JB.187.2.449-457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR. New functions for Y family polymerases. Mol Cell. 2006;24:493–495. doi: 10.1016/j.molcel.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Leigh EG. The evolution of mutation rates. Genetics Suppl. 1973;73:1–18. [PubMed] [Google Scholar]
- Lenski RE, Slatkin M, Ayala FJ. Mutation and selection in bacterial populations: alternatives to the hypothesis of directed mutation. Proc Natl Acad Sci USA. 1989;86:2775–2778. doi: 10.1073/pnas.86.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- Liu SK, Tessman I. groE genes affect SOS repair in Escherichia coli. J Bacteriol. 1990;172:6135–6138. doi: 10.1128/jb.172.10.6135-6138.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longerich S, Galloway AM, Harris RS, Wong C, Rosenberg SM. Adaptive mutation sequences reproduced by mismatch repair-deficiency. Proc Natl Acad Sci USA. 1995;92:12017–12020. doi: 10.1073/pnas.92.26.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay WJ, Han S, Samson LD. DNA alkylation repair limits spontaneous base substitution mutations in Escherichia coli. J Bacteriol. 1994;176:3224–3230. doi: 10.1128/jb.176.11.3224-3230.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Blake CE, Leach DR, Shapiro JA. Different structures of selected and unselected araB-lacZ fusions. Molec Microbiol. 1997;23:1133–1145. doi: 10.1046/j.1365-2958.1997.3031666.x. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Shapiro JA. The roles of starvation and selection in the emergence of araB-lacZ fusion clones. EMBO J. 1994;13:5229–5244. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Mamber SW, Kolek B, Brookshire KW, Bonner DP, Fung-Tomc J. Activity of quinolones in the Ames Salmonella TA102 mutagenicity test and other bacterial genotoxicity assays. Antimicrob Agents Chemother. 1993;37:213–217. doi: 10.1128/aac.37.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Molec Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Molec Microbiol. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Harris RS, Lee PL, Rosenberg SM. The SOS response regulates adaptive mutation. Proc Natl Acad Sci USA. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Molecular Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Magner DB, Lee PL, Rosenberg SM. The dinB operon and spontaneous mutation in Escherichia coli. J Bacteriol. 2003;185:3972–3977. doi: 10.1128/JB.185.13.3972-3977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S, Vaisman A, Valjavec-Gratian M, Karata K, Vandewiele D, Woodgate R. Characterization of polVR391: a Y-family polymerase encoded by rumA’B from the IncJ conjugative transposon, R391. Molec Microbiol. 2007;63:797–810. doi: 10.1111/j.1365-2958.2006.05561.x. [DOI] [PubMed] [Google Scholar]
- Mellon I. Transcription-coupled repair: a complex affair. Mutat Res. 2005;577:155–161. doi: 10.1016/j.mrfmmm.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Miller C, Ingmer H, Thomsen LE, Skarstad K, Cohen SN. DpiA binding to the replication origin of Escherichia coli plasmids and chromosomes destabilizes plasmid inheritance and induces the bacterial SOS response. J Bacteriol. 2003;185:6025–6031. doi: 10.1128/JB.185.20.6025-6031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- Miller JH. Mutagenic specificity of ultraviolet light. J Molec Biol. 1985;182:45–65. doi: 10.1016/0022-2836(85)90026-9. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- Miller JH, Suthar A, Tai JYA, Truong C, Stewart JL. Direct selection for mutators in Escherichia coli. J Bacteriol. 1999;181:1576–1584. doi: 10.1128/jb.181.5.1576-1584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler JE, Lenski RE. New data on excisions of Mu from E. coli MCS2 cast doubt on directed mutation hypothesis. Nature (London) 1990;344:173–175. doi: 10.1038/344173a0. [DOI] [PubMed] [Google Scholar]
- Naas T, Blot M, Fitch WM, Arber W. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics. 1994;136:721–730. doi: 10.1093/genetics/136.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Transient mutators: A semiquantitative analysis of the influence of translation and transcription errors on mutation rates. Genetics. 1991;129:957–962. doi: 10.1093/genetics/129.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol. 2002;184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186:7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schell-horn HE. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics. 2004;272:580–591. doi: 10.1007/s00438-004-1089-2. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza-Reyes M, Yasbin RE. Contribution of the mismatch DNA repair system to the generation of stationary-phase-induced mutants of Bacillus subtilis. J Bacteriol. 2004;186:6485–6491. doi: 10.1128/JB.186.19.6485-6491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Capilla T, Baquero MR, Gomez-Gomez JM, Ionel A, Martin S, Blazquez J. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol. 2005;187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson ME, Andersson DI, Roth JR, Berg OG. The amplification model for adaptive mutation—simulations and analysis. Genetics. 2005;169:1105–1115. doi: 10.1534/genetics.104.030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Powell SC, Wartell RM. Different characteristics distinguish early versus late arising adaptive mutations in Escherichia coli FC40. Mutat Res. 2001;473:219–228. doi: 10.1016/s0027-5107(00)00149-4. [DOI] [PubMed] [Google Scholar]
- Quinones M, Kimsey HH, Waldor MK. LexA cleavage is required for CTX prophage induction. Mol Cell. 2005;17:291–300. doi: 10.1016/j.molcel.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Radicella JP, Park PU, Fox MS. Adaptive mutation in Escherichia coli: A role for conjugation. Science. 1995;268:418–420. doi: 10.1126/science.7716545. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature (London) 1989;342:396–400. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Rebeck GW, Samson LD. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M, Gowrishankar J. A genetic strategy to demonstrate the occurrence of spontaneous mutations in non-dividing cells within colonies of Escherichia coli. Genetics. 1997;147:991–1001. doi: 10.1093/genetics/147.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K-12. Nature (London) 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez C, Tompkin J, Hazel J, Foster PL. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J Bacteriol. 2002;184:5599–5608. doi: 10.1128/JB.184.20.5599-5608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RJ. Polyphosphate present in DNA preparations from filamentous fungal species of Colletotrichum inhibits restriction en-donucleases and other enzymes. Anal Biochem. 1993;209:291–297. doi: 10.1006/abio.1993.1122. [DOI] [PubMed] [Google Scholar]
- Rosche WA, Foster PL. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA. 1999;96:6862–6867. doi: 10.1073/pnas.96.12.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche WA, Foster PL. 2001 Unpublished results. [Google Scholar]
- Rosen R, Ron EZ. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom Rev. 2002;21:244–265. doi: 10.1002/mas.10031. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Hastings PJ. Adaptive point mutation and adaptive amplification pathways in the Escherichia coli Lac system: stress responses producing genetic change. J Bacteriol. 2004;186:4838–4843. doi: 10.1128/JB.186.15.4838-4843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J Bacteriol. 2006;188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset R, Gorini L. A ribosomal ambiguity mutation. J Molec Biol. 1969;39:95–112. doi: 10.1016/0022-2836(69)90336-2. [DOI] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Adaptive mutation: how growth under selection stimulates Lac+ reversion by increasing target copy number. J Bacteriol. 2004;186:4855–4860. doi: 10.1128/JB.186.15.4855-4860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Kofoid E, Roth FP, Berg OG, Seger J, Andersson DI. Regulating general mutation rates: examination of the hypermutable state model for Cairnsian adaptive mutation. Genetics. 2003;163:1483–1496. doi: 10.1093/genetics/163.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner R, Murray A, Huda N. Is there a link between mutation rates and the stringent response in Bacillus subtilis? Ann NY Acad Sci. 1999;870:418–422. doi: 10.1111/j.1749-6632.1999.tb08917.x. [DOI] [PubMed] [Google Scholar]
- Sakai A, Nakanishi M, Yoshiyama K, Maki H. Impact of reactive oxygen species on spontaneous mutagenesis in Escherichia coli. Genes Cells. 2006;11:767–778. doi: 10.1111/j.1365-2443.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- Saumaa S, Tarassova K, Tark M, Tover A, Tegova R, Kivisaar M. Involvement of DNA mismatch repair in stationary-phase mutagenesis during prolonged starvation of Pseudomonas putida. DNA Repair. 2006;5:505–514. doi: 10.1016/j.dnarep.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Saumaa S, Tover A, Kasak L, Kivisaar M. Different spectra of stationary-phase mutations in early-arising versus late-arising mutants of Pseudomonas putida: involvement of the DNA repair enzyme MutY and the stationary-phase sigma factor RpoS. J Bacteriol. 2002;184:6957–6965. doi: 10.1128/JB.184.24.6957-6965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional muta-genesis? Chem Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Pham P, Cox MM, Goodman MF. Roles of DNA polymerase V and RecA Protein in SOS damage-induced mutation. Chem Rev. 2006;106:406–419. doi: 10.1021/cr0404951. [DOI] [PubMed] [Google Scholar]
- Schreiber G, Ron EZ, Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Seelke R, Kline B, Aleff R, Porter RD, Shields MS. Mutations in the recD gene of Escherichia coli that raise the copy number of certain plasmids. J Bacteriol. 1987;169:4841–4844. doi: 10.1128/jb.169.10.4841-4844.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Observations on the formation of clones containing araB-lacZ cistron fusions. Molec Gen Genet. 1984;194:79–90. doi: 10.1007/BF00383501. [DOI] [PubMed] [Google Scholar]
- Shapiro JA. The significances of bacterial colony patterns. Bioes-says. 1995;17:597–607. doi: 10.1002/bies.950170706. [DOI] [PubMed] [Google Scholar]
- Shiba T, Tsutsumi K, Ishige K, Noguchi T. Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochem (Moscow) 2000;65:315–323. [PubMed] [Google Scholar]
- Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao NN, Kornberg A. Inorganic polyphosphate and the induction of rpoS expression. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta ES, Bunny KL, Kugelberg E, Kofoid E, Andersson DI, Roth JR. Adaptive mutation: General mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci USA. 2003;100:12847–12852. doi: 10.1073/pnas.1735464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta ES, Liu J, Andersson DI, Roth JR. Evidence that selected amplification of a bacterial lac frameshift allele stimulates Lac+ reversion (adaptive mutation) with or without general hypermutability. Genetics. 2002;161:945–956. doi: 10.1093/genetics/161.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska MM, King AG, Lu LI, Lin RH, Mao EF, Lackey J-HC, Baikalov C, Miller JH. Examination of the role of DNA polymerase proofreading in the mutator effect of miscoding tRNAs. J Bacteriol. 1998;180:5712–5717. doi: 10.1128/jb.180.21.5712-5717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD. A test of the directed mutation hypothesis in Escherichia coli MCS2 using replica plating. J Bacteriol. 1995;177:1119–1120. doi: 10.1128/jb.177.4.1119-1120.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature (London) 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Stahl FW. A unicorn in the garden. Nature (London) 1988;335:112–113. doi: 10.1038/335112a0. [DOI] [PubMed] [Google Scholar]
- Stahl FW. Unicorns revisited. Genetics. 1992;132:865–867. doi: 10.1093/genetics/132.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storvick KM, Foster PL. 2007 Unpublished results. [Google Scholar]
- Strauss BS, Roberts R, Francis L, Pouryazdanparast P. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J Bacteriol. 2000;182:6742–6750. doi: 10.1128/jb.182.23.6742-6750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf JD, Foster PL. 2007a Unpublished results. [Google Scholar]
- Stumpf JD, Foster PL. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Molec Microbiol. 2005;57:751–761. doi: 10.1111/j.1365-2958.2005.04724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf JD, Poteete AR, Foster PL. Amplification of lac cannot account for adaptive mutation to Lac+ in Escherichia coli. J Bacteriol. 2007b;189:2291–2299. doi: 10.1128/JB.01706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HM, Yasbin RE. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J Bacteriol. 2002;184:5641–5653. doi: 10.1128/JB.184.20.5641-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HM, Yeamans G, Ross CA, Yasbin RE. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J Bacteriol. 2003;185:2153–2160. doi: 10.1128/JB.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Halliday JA, Matic I, Radman M. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Molec Gen Genet. 1997a;256:277–281. doi: 10.1007/s004380050570. [DOI] [PubMed] [Google Scholar]
- Taddei F, Matic I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature (London) 1997b;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- Tark M, Tover A, Tarassova K, Tegova R, Kivi G, Horak R, Kivisaar M. A DNA polymerase V homologue encoded by TOL plasmid pWW0 confers evolutionary fitness on Pseudomonas putida under conditions of environmental stress. J Bacteriol. 2005;187:5203–5213. doi: 10.1128/JB.187.15.5203-5213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegova R, Tover A, Tarassova K, Tark M, Kivisaar M. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J Bacteriol. 2004;186:2735–2744. doi: 10.1128/JB.186.9.2735-2744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms AR, Muriel W, Bridges BA. A UmuD,C-dependent pathway for spontaneous G:C to C:G transversions in stationary phase Escherichia coli. Mutat Res. 1999;435:77–80. doi: 10.1016/s0921-8777(99)00035-x. [DOI] [PubMed] [Google Scholar]
- Tippin B, Pham P, Goodman MF. Error-prone replication for better or worse. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Tlsty DT, Albertini AM, Miller JH. Gene amplification in the lac region of E. coli. Cell. 1984;37:217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- Tompkins JD, Nelson JE, Hazel JC, Leugers SL, Stumpf JD, Foster PL. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J Bacteriol. 2003;185:3469–3472. doi: 10.1128/JB.185.11.3469-3472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Chang DE, Conway T. Guanosine 3′, 5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobner W, Piechocki R. Selection against hypermutability in Escherichia coli during long term evolution. Mol Gen Genet. 1984;198:177–178. doi: 10.1007/BF00328720. [DOI] [PubMed] [Google Scholar]
- Tsui HCT, Feng G, Winkler ME. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi K, Munekata M, Shiba T. Involvement of inorganic polyphosphate in expression of SOS genes. Biochim Biophys Acta. 2000;1493:73–81. doi: 10.1016/s0167-4781(00)00165-2. [DOI] [PubMed] [Google Scholar]
- Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Molec Microbiol. 2005;57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich MD, Makris JC, Reznikoff WS. Induction of the SOS response in Escherichia coli inhibits Tn5 and IS50 transposition. J Bacteriol. 1991;173:6910–6918. doi: 10.1128/jb.173.21.6910-6918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Molec Gen Genet. 1991;266:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- Wright BE, Longacre A, Reimers JM. Hypermutation in derepressed operons of E. coli K12. Proc Natl Acad Sci USA. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. Stress-directed adaptive mutations and evolution. Molec Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Wu YH, Franden MA, Hawker JR, Jr, McHenry CS. Monoclonal antibodies specific for the alpha subunit of the Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 1984;259:12117–12122. [PubMed] [Google Scholar]
- Yanofsky C, Crawford IP. The tryptophan operon. In: Neidhardt FC, Ingraham JL, Magasanik B, Low KB, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1987. pp. 1453–1472. [Google Scholar]
- Yanofsky C, Ito J, Horn V. Amino acid replacements and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:151–162. doi: 10.1101/sqb.1966.031.01.023. [DOI] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci USA. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Kanemori M, Morita MT. The heat shock response: regulation and function. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington DC: ASM Press; 2000. pp. 3–18. [Google Scholar]
- Zhao J, Winkler ME. Reduction of GC to TA transversion mutation by overexpression of MutS in Escherichia coli K-12. J Bacteriol. 2000;182:5025–5028. doi: 10.1128/jb.182.17.5025-5028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]