SUMMARY
Histone acetyltransferases (HATs) play important roles in gene regulation and DNA repair by influencing the accessibility of chromatin to transcription factors and repair proteins. Here we show that deletion of Gcn5 leads to telomere dysfunction in mouse and human cells. Biochemical studies reveal that depletion of Gcn5 or ubiquitin specific protease 22 (Usp22), which is another bona fide component of the Gcn5-containing SAGA complex, increases ubiquitination and turnover of TRF1, a primary component of the telomeric shelterin complex. Inhibition of the proteasome or over expression of USP22 opposes this effect. The USP22 deubiquitinating module requires association with SAGA complexes for activity, and we find that depletion of Gcn5 compromises this association in mammalian cells. Thus, our results indicate that Gcn5 regulates TRF1 levels through effects on Usp22 activity and SAGA integrity.
INTRODUCTION
Gcn5 (also known as KAT2 (Allis et al., 2007) ) was the first transcription-related HAT to be identified (Brownell et al., 1996), and it is highly conserved across evolution in structure and enzymatic specificity (Candau and Berger, 1996; Candau et al., 1996; Roth et al., 2001). Mammals contain two highly related proteins homologous to yeast Gcn5, Gcn5 (KAT2A) (encoded by the GCN5l2 gene in mice; hereafter referred to as Gcn5) and PCAF (KAT2B) (p300 CBP Associated Factor) (Xu et al., 1998; Yang et al., 1996). The functions of Gcn5 in yeast are well-studied (Grant et al., 1997; Trievel et al., 1999; Zhang et al., 1998), but the functions of this HAT in mammalian cells are less well defined. Genetic studies in mice indicate that Gcn5 is essential during early embryogenesis (Bu et al., 2007; Lin et al., 2007; Lin et al., 2008; Xu et al., 2000). Deletion of PCAF has little effect on mouse embryos or adults (Xu et al., 2000; Yamauchi et al., 2000), but deletion of Gcn5 results in embryonic lethality. At E8.5 Gcn5 null embryos are small and exhibit defects in presomitic mesoderm. These defects are associated with increased apoptosis but are not due to generalized defects in gene transcription (Xu et al., 2000). Interestingly, embryos expressing catalytically inactive Gcn5 survive significantly longer than Gcn5 null embryos and do not show increased apoptosis (Bu et al., 2007), indicating that Gcn5 has important functions during embryogenesis that are independent of its HAT activity.
Both Gcn5 and PCAF function within the context of large, multisubunit complexes that are remarkably similar to the yeast SAGA complex, harboring homologues of the Ada2, Ada3, Spt3, Tra1 (PAF400 or TRRAP) and TAF proteins (Lee and Workman, 2007). These complexes show a high degree of similarity not only in subunit content but also in overall structure and subdomain organization (Nagy and Tora, 2007; Wu et al., 2004). Additional biochemical studies subsequently identified a ubiquitin-specific protease, Ubp8, as a component of SAGA that removes ubiquitin from histone H2B K123 to facilitate transcription (Daniel et al., 2004; Henry et al., 2003; Ingvarsdottir et al., 2005; Powell et al., 2004; Sanders et al., 2002). Human STAGA/TFTC (hereafter hSAGA) and Drosophila SAGA complexes harbor orthologs of Ubp8 (USP22 and Nonstop, respectively) that confer deubiquitination (deUB) activity to these complexes as well (Weake et al., 2008; Zhao et al., 2008). Notably, the deubiquitination activity of Ubp8 in SAGA depends on the presence of other subunits within the complex and the proper organization of the whole deUB module. In yeast, Ubp8 association with SAGA requires Sgf1 1 (Ingvarsdottir et al., 2005; Lee et al., 2005), and the deUB activity of SAGA is modulated by Sus1 (Kohler et al., 2006; Rodriguez-Navarro et al., 2004). The human orthologs of Sgf1 1 and Sus1, ATX7L3 and ENY2, are likewise required for integration of USP22 into hSAGA and facilitation of deUB activity (Zhao et al., 2008). Accordingly, the ability of purified USP22 to remove the ubiquitin moiety from histone H2B in vitro is significantly decreased relative to that of the intact hSAGA complex (Zhang et al., 2008).These data suggest a conserved mode of regulation of the Ubp8/USP22 module within yeast and human SAGA complexes, even though the location of the deUB module within SAGA is not known.
Although the activity of Ubp8 and USP22 towards histones is well-documented, it is not known whether these enzymes and SAGA also affect ubiquitination of other proteins. We report here that deletion of Gcn5 in mice leads to telomere dysfunction and that these defects are linked to decreased levels of two telomere-associated proteins, TRF1 and POT1a. Gcn5 loss does not affect TRF1 mRNA expression and the steady state levels of this protein are not altered in cells expressing catalytically inactive Gcn5, indicating a HAT independent function of Gcn5 or SAGA is required for post-transcriptional maintenance of TRF1. Previous work by others demonstrated that TRF1 levels are controlled through ubiquitin-mediated proteolysis (Chang et al., 2003; Lee et al., 2006b). Notably, we find that depletion of USP22 has the same impact on TRF1 protein levels as does loss of Gcn5 and that this effect is prevented by inhibition of the proteasome. Furthermore depletion of GCN5 and USP22 as well as the mammalian ortholog of Sgf1l, ATXN7L3, leads to increased levels of ubiquitinated TRF1. Over expression of wild type USP22, but not a catalytically inactive mutant, restores TRF1 levels. Our data suggest that GCN5, USP22, and SAGA contribute to telomere maintenance through control of TRF1 turnover by deubiquitination of this substrate. More globally, these results indicate that Gcn5 and SAGA affect gene expression at multiple levels in the cell, from gene transcription to protein stability.
RESULTS
Deletion of Gcn5 induces chromosomal fusions and formation of dysfunctional telomere induced foci (TIFs)
We reported previously that deletion of Gcn5 leads to early embryonic lethality in mice (Xu et al., 2000). Gcn5 null embryos die at day 8.5 in part due to extensive apoptosis in mesodermal lineages. This apoptosis is p53 dependent and double Gcn5 and p53 knockout embryos survive significantly longer (Bu et al., 2007) than do Gcn5 null embryos. To further investigate this phenotype, we examined cells isolated from E8.5 Gcn5 null embryos for overt chromosomal abnormalities that might induce p53 and trigger apoptosis in these cells. Examination of fifty giemsa-stained metaphase nuclei from E8.5 wild type embryos (n=11) and homozygous null littermates (n=5) revealed a striking increase in chromosomal end associations and apparent end-to-end fusions in nuclei from Gcn5 null embryos (Figure 1A, Figure S1). A large portion (28–48%; data not shown) of the nuclei from Gcn5 null embryos contained chromosomal end associations or fusions, whereas a much smaller fraction (0–14%; data not shown) of nuclei from wild type or heterozygous littermates exhibited these chromosomal aberrations.
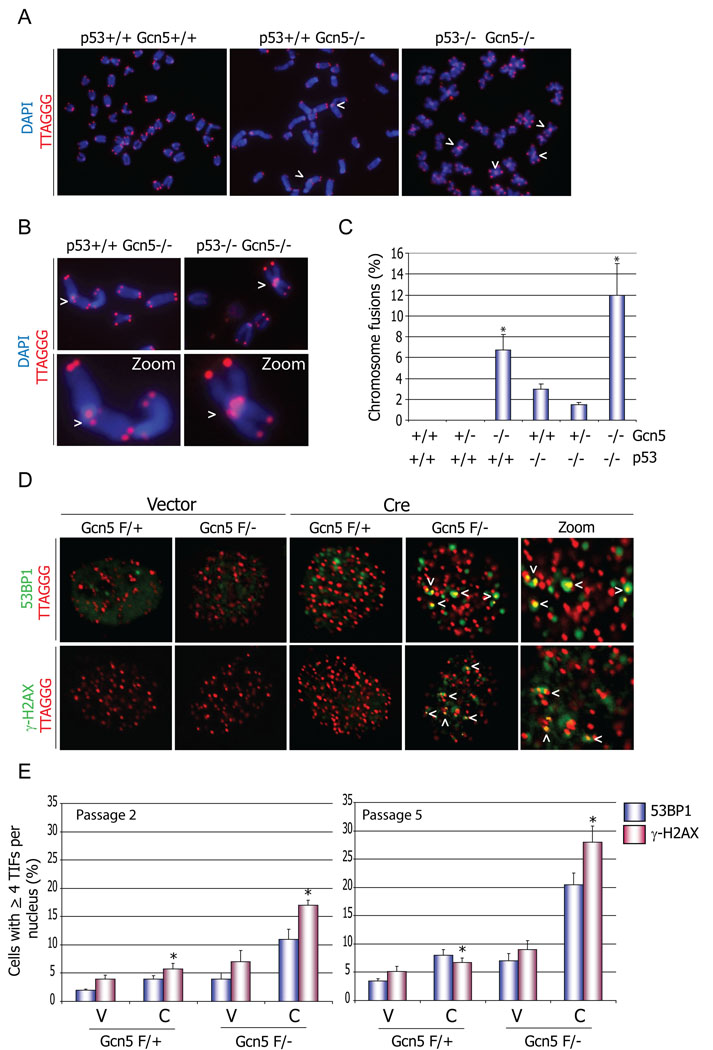
Figure 1. Loss of Gcn5 impairs proper telomere maintenance.
(A) Loss of Gcn5 leads to telomere end-to-end fusions in mouse embryonic cells. Cells were isolated from E8.5 embryos with indicated genotypes, and metaphase spreads were subjected to FISH analysis. Chromosomal DNA was stained with DAPI (blue) and telomeres were labeled with a TRITC-conjugated telomere specific probe (TTAGGG). Arrow heads indicate chromosomal end-to-end fusions in cells isolated from Gcn5 null embryos. (B) Close up view of chromosome fusions represented in (A). (C) Quantification of the data represented in A and B. 30 non-apoptotic nuclei from each embryo genotype (n=2) were examined, and the number of chromosome fusions in each metaphase was scored. * p values <0.001 relative to the wild type sample, as determined by students two-tailed t-test (D) Depletion of Gcn5 induces a DNA damage response on the telomeres in MEF cells. Telomere localization of the DNA damage signal, indicated with the arrows, was verified after in situ hybridization with TRITC-conjugated telomere specific probe as above. (E) Quantification of data represented in (D). Approximately one hundred cells from each genotype were counted two and five passages after the initial Vector or Cre transfection. Cells with more than 4 TIFs per nucleus were scored. (error bars in (C) and (E) indicate S.E.M., asterisk, p<0.05 based on a two tailed Student’s t-test). Cells from three independent embryo isolates were used. V = vector and C = Cre.
We then asked whether the abnormal chromosomal structures observed in Gcn5 null nuclei involve telomeres by performing fluorescent in situ hybridization (FISH) and DAPI staining of metaphase chromosomes in cells from E8.5 Gcn5 null, p53 −/− Gcn5 −/− double null, and control embryos (Figure 1). FISH using a telomere-specific probe confirmed that telomere sequences were present at the sites of chromosomal end-to-end fusions in Gcn5 null cells and in the double mutants (Figure 1 A , B). Quantitation of at least 30 (non-apoptotic) nuclei per embryo confirmed an increased incidence of end-to-end fusions in cells from Gcn5 null embryos (Figure 1C). No fusions were found in nuclei from Gcn5 heterozygous or wild type E8.5 embryos (n=4), but 7–12% of nuclei from Gcn5 null embryos (n=2) contained chromosomal end fusions (Figure 1C). An even greater incidence of chromosome end-to-end associations and telomere fusion was observed in nuclei from p53 Gcn5 double null embryos (Figure 1 A and C). The above data collectively suggest that induction of p53 in cells from Gcn5 null embryos with telomere fusions triggers apoptosis of the damaged cells.
Chromosome end associations and fusions are characteristic of telomere dysfunction (de Lange, 2002). Telomeres are protected by a six-protein containing complex called shelterin (de Lange, 2005), which is thought to stabilize t-loop structures at telomeres (de Lange, 2005; Griffith et al., 1998; Stansel et al., 2001). When shelterin functions are diminished, the unprotected telomeres become targets of the DNA repair machinery, leading to formation of telomere damage induced foci (TIFs). These foci can be visualized by immunostaining of DNA repair proteins such as γH2AX and 53BP1, followed by hybridization with fluorophore-coupled peptide nucleic acid probes specific for telomere sequences [PNA-FISH;(d'Adda di Fagagna et al., 2003)]. To test this possibility, we derived embryonic fibroblasts (MEFs) from mice in which exons 3 to 18 in one allele of Gcn5 are flanked by loxP sites (Lin et al., 2007). Expression of Cre recombinase in these cells leads to almost complete deletion of the Gcn5 coding sequence (Figure S2A). Gcn5 protein levels in Gcn5flox/Δ cells were undetectable seventy-two hours after initial Cre expression (hereafter referred to as Gcn5 null cells) compared to Gcn5flox/wt MEFs (hereafter referred to as Gcn5 heterozygous cells) (Figure S2B).
To determine whether Gcn5 loss compromises the protection of the telomeres, we monitored TIF formation in early passage cells Gcn5 null MEFs and found that approximately 11% of these cells showed ≥ 4 TIFs per nucleus detected by 53BP1 staining and approximately 17% exhibited TIFs detected by γH2AX staining (Figure 1D, E). In contrast, Gcn5 heterozygous cells were only minimally positive for either of these markers. The number of TIF positive Gcn5 null cells increased with time after Gcn5 depletion, such that by five passages after the initial Cre treatment, ~21% of the cells were positive for 53BP1 TIFs and more than 25% were positive for γH2AX TIFs (Figure 1E). These results confirm that telomere structures, and likely Shelterin functions, are altered upon loss of Gcn5.
Gcn5 deletion does not have significant impact on DNA Double Strand Break Repair
HATs play a role during DNA repair processes by altering chromatin states to make the damaged sites more accessible to the repair machinery (Ikura et al., 2000; Morrison and Shen, 2006). TRRAP, another member of the SAGA complex as well as the Tip60 HAT complex, is also indispensable during mouse embryogenesis (Herceg et al., 2001) and plays an important role in DNA double strand break repair (Murr et al., 2006). We reasoned, then, that the increased apoptosis levels, chromosomal aberrations, and the increased telomere DNA damage signal observed in Gcn5 null cells and embryos might be due to altered efficiency of DNA double strand break repair. To test this possibility, we monitored the appearance and disappearance of γH2AX and 53BP1 foci induced by gamma irradiation in Gcn5 null or Gcn5 heterozygous MEFs. γH2AX and 53BP1 foci are generated at DNA double strand breaks immediately after their formation and disappear upon their repair. Both Gcn5 null and heterozygous cells were exposed to 1Gy of gamma irradiation and then subjected to γH2AX and 53BP1 immunostaining. One hundred cells of two independent isolates of each MEF genotype were counted at different time points after irradiation, and nuclei containing 10 or more repair foci were scored (Figure S3A). The results revealed that the kinetics of both γH2AX and 53BP1 appearance and disappearance were similar in Gcn5 containing and Gcn5 deficient cells (Figure S3B). Based on these results we concluded that the repair efficiency of DNA strand breaks is not significantly altered upon Gcn5 deletion (Figure S3). These data also fit our previous findings that Gcn5 null ES cells do not exhibit increased sensitivity to a variety of DNA damaging agents (Lin et al., 2007). Although these results do not rule out a role for Gcn5 in double strand break repair, they strongly indicate it is not essential for this process. Therefore, the chromosomal aberrations and the damage induced signals at telomeres we observe in cells isolated from Gcn5 null embryos or Gcn5 null MEFs likely reflect altered telomere structures rather than general defects in the efficiency of the DNA double strand break repair in these cells.
GCN5 depletion alters steady state levels of TRF1 and POT1a proteins
We next asked whether the expression levels of the shelterin components were altered upon Gcn5 deletion. Immunoblots revealed that TRF1 (Figure 2A, compare lanes 2 and 4 to lanes 1 and 3) and Pot1a (Figure 2B, lanes 5 and 6) protein levels were substantially decreased in Gcn5 null cells compared to heterozygous MEFs, whereas the levels of the other shelterin components such as RAP1 and TRF2 (de Lange, 2005; Wu et al., 2006) were not altered. These effects are due to deletion of Gcn5 upon Cre treatment, as they were not observed in cells transfected with empty vector (Figure 2B, lanes 3, 4). Interestingly, cells expressing catalytically inactive Gcn5 (Bu et al., 2007) do not show any alterations in shelterin component protein levels (Figure 2B lanes 1, 2,). These findings are consistent with a lack of increased apoptosis (Bu et al., 2007) or obvious telomere defects (data not shown) in Gcn5 hat/hat mice. Together these studies indicate that the functions of Gcn5 in telomere maintenance are likely due to effects on the integrity of the shelterin complex but are independent of Gcn5 acetyltransferase functions.
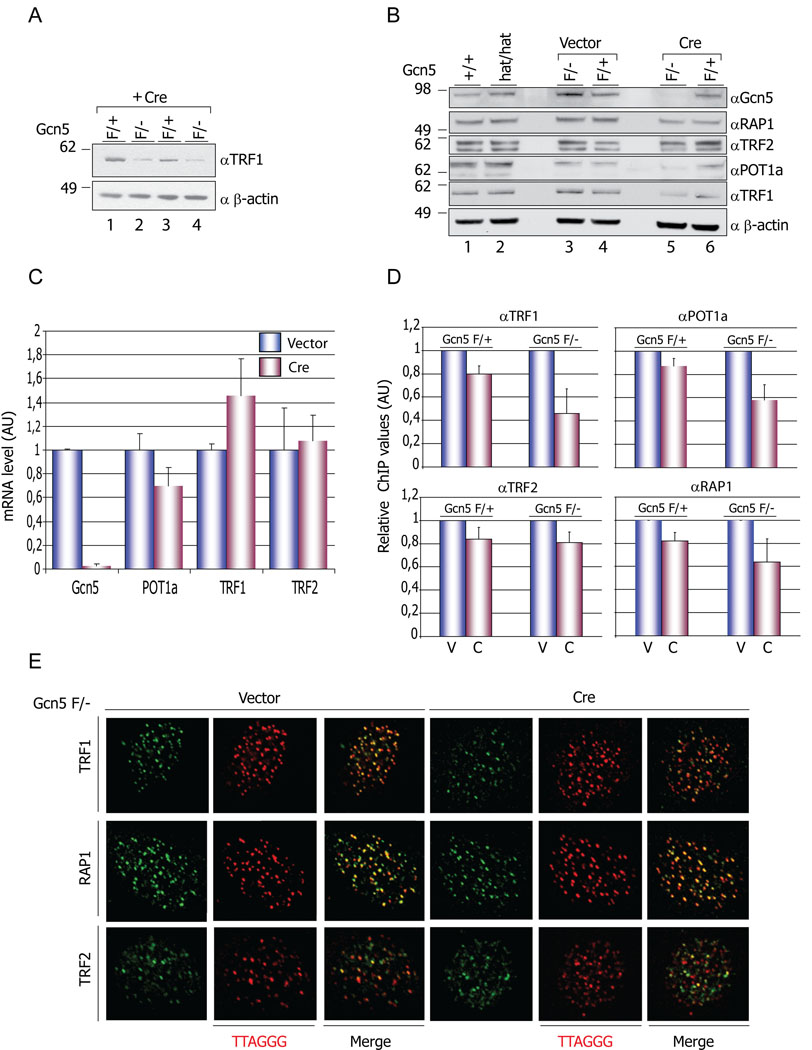
Figure 2. Decreased levels of Shelterin components in Gcn5 null cells.
(A) Decreased levels of TRF1 protein in Gcn5 null MEFs. Protein levels of TRF1 are decreased in Gcn5F/− (lanes 2 and 4) compared to Gcn5F/+ (lanes 1 and 3) MEFs following Cre treatment. (B) Immunoblot data showed decreased TRF1 and POT la protein levels in Gcn5F/− (after Cre treatment) but not in Gcn5F/+ or Gcn5hat/hat cells (lane 5 compared to lanes 1, 2, 3, 4, and 6). Levels of the other shelterin components monitored do not show significant variations in any of the genotypes examined. (C) mRNA level of shelterin components tested by real-time RT-PCR. Experiments were performed with 2 individual MEF cell lines. mRNA level data were normalized by that of GAPDH and vector-treated group values were set as 1. Error bars represent standard deviation from the mean, (D) Decreased amounts on TRF1 and POT la on telomeres. Chromatin immunoprecipitations (ChIPs) were performed using the indicated antibodies and slot blots hybridized with a γ32P-ATP end-labeled TTAGGG probe. The membranes were exposed to Phospholmager screens and the signals quantified with ImageQuant software. Error bars - S.E.M, n=3 independent ChIP experiments. V=vector, C=Cre. (E) Proper localization of TRF1, TRF2 and RAP1 on telomeres. Vector or Cre transfected Gcn5F/− MEFs were immunostained with the indicated antibodies and telomere localization of TRF1, TRF2 and RAP1 was monitored after hybridization with TRITC-conjugated TTAGGG probe.
Gcn5 is known to serve as a transcriptional coactivator, so the decreased levels of TRF1 and POT 1a proteins might reflect decreased transcription of these genes upon Gcn5 loss. However, we were unable to find any significant differences in the mRNA levels of TRF1 as measured by quantitative reverse transcriptase PCR (Figure 2C). POT 1a mRNA levels were decreased slightly, but this decrease was not statistically significant (p>0.28). These data indicate that Gcn5 loss affects TRF1 expression, and likely POT 1a expression, at a post-transcriptional step.
Diminished levels of TRF1 and Pot1a at telomeres upon loss of Gcn5
We next asked whether decreased levels of TRF1 and POT1a in Gcn5 null cells reflect loss of these proteins from telomeres, or whether the diminished levels of these proteins affect the association of other shelterin components with telomeres. Telomeric chromatin immunoprecipitation (ChIP) assays using shelterin specific antibodies showed substantial decreases in levels of TRF1 and POT1a at the telomeres in Gcn5 null cells compared to Gcn5 heterozygous cells (Figure 2D, pink bars and Figure S4). RAP1 levels were also diminished at the telomeres despite the fact that we did not find significant changes in the protein levels of this shelterin component upon Gcn5 depletion (Figure 2B lanes 5 and 6). Perhaps TRF1 and POT1a further stabilize RAP1 on telomeres.
Immunofluorescence staining of TRF1, TRF2 and RAP1 followed by telomere PNA-FISH confirmed that these proteins exhibit proper telomere localization in both Gcn5 null and Gcn5 heterozygous cells (Figure 2E). We could not detect Gcn5 at telomeres by ChIP assays in wild type cells (Figure S4), suggesting either that Gcn5 associates with these regions very transiently or that the effects of Gcn5 on TRF1 and POT 1a are mediated at a point when these proteins are not telomere-associated.
Inhibition of the proteasome restores TRF1 levels in Gcn5 null cells
TRF1 protein levels are regulated via ubiquitination and degradation by the proteasome following eviction from the telomere (Chang et al., 2003). Therefore we next considered the possibility that the decreased levels of TRF 1 and POT1 a observed in Gcn5 null cells reflect increased turnover of these proteins. Indeed, treatment of Gcn5 null cells with the proteasome inhibitor MG132 resulted in stabilization of TRF1 (Figure 3A, compare lanes 2 and 4). Similarly, levels of exogenous FLAG-mTRF1 were decreased significantly in Gcn5 null cells but were stabilized upon MG132 treatment (Figure 3 B, lanes 1 and 4). We examined effects of Gcn5 loss on a POT1a-Myc fusion construct (Wu et al., 2006). Exogenous POT1a-Myc was barely detectable in cell lysates (Figure S5), but was easily detected after immunoprecipitation (Figure 3C). Levels of POT1a-Myc were decreased upon deletion of Gcn5 (Figure 3C, lanes 1–3), consistent with the effects on the endogenous protein observed above, but were stabilized upon inhibition of the proteasome by MG132 (Figure 3C, lanes 4–6). These data indicate that Gcn5 is required to protect TRF1 and POT 1a from proteasomal degradation.
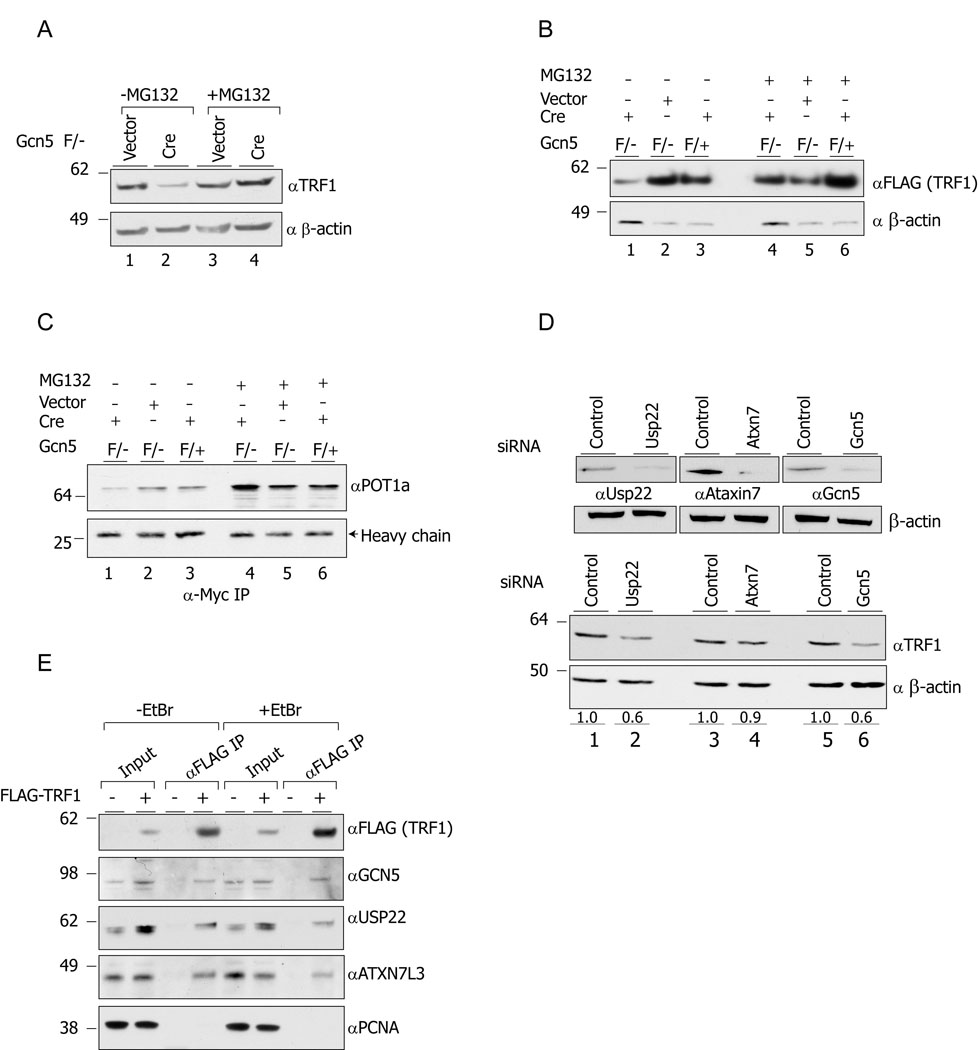
Figure 3. Gcn5 loss alters TRF1 and POT1a protein turnover.
(A) Treatment of the cells with the proteasome inhibitor MG132 stabilizes TRF1 protein levels in Gcn5 null cells (lanes 2 and 4). Lysates from Gcn5 null and Gcn5 heterozygous MEFs, treated with MG132 or DMSO only, were resolved by SDS-PAGE, and TRF1 protein levels were monitored by immunoblot. (B and C) Decreased levels of exogenous FLAG-mTRFl and POT1a-Myc in Gcn5 depleted cells. MEFs with the indicated genotypes were transfected with mTRFl-FLAG expressing vector or infected with POT1a-Myc expressing retroviral vectors. After MG132 or DMSO treatment, lysates were prepared and the exogenous TRF1-FLAG levels were monitored by using anti-FLAG antibody. For monitoring POT1a-Myc protein levels, anti-Myc IP was performed. (D) Depletion of Usp22 has a similar impact on TRF1 protein levels as does Gcn5 depletion (compare lanes 1 and 2 to lanes5 and 6). (E) TRF1 co-immunoprecipitates with GCN5, USP22 and ATXN7L3.
USP22 interacts with TRF1 and is required for TRF1 stability
Gcn5 is part of multisubunit complexes including hSAGA and ATAC (Cavusoglu et al., 2003; Lee and Workman, 2007; Martinez et al., 2001; Wang et al., 2008). Interestingly, the human and fly SAGA complexes possess a module that houses ubiquitin hydrolase activity mediated by the USP22 (human) and Nonstop (fly) proteins (Weake et al., 2008; Zhang et al., 2008; Zhao et al., 2008). This deubiquitinating (deUB) module is similar to one identified in yeast SAGA that consists of four proteins, Ubp8, Sgf1 1, Sgf73, and Sus1 (Ingvarsdottir et al., 2005; Kohler et al., 2008; Rodriguez-Navarro et al., 2004). In yeast, flies, and human cells, this module deubiquitinates histones H2A and H2B, but only when associated with SAGA (Kohler et al., 2008; Weake et al., 2008; Zhao et al., 2008). However, mutations in nonstop lead to increased ubiquitination of multiple unidentified proteins (Poeck et al., 2001), and both human USP22 and mouse Usp22 proteins remove ubiquitin moieties from non-histone substrates in vitro (Lee et al., 2006a), indicating that this deUB module may have non-histone substrates in vivo as well.
Given the known regulation of TRF1 by ubiquitination (Chang et al., 2003), we hypothesized that the SAGA complex might facilitate Usp22-dependent deubiquitination of TRF1 and that loss of Gcn5 might alter this activity by compromising complex integrity. To test this idea, we first used siRNAs to deplete endogenous Usp22, Ataxin7 (Atxn7), which is homologous to yeast Sgf73 (Kohler et al., 2008) or Gcn5 in mouse 3T3 cells. The endogenous levels of these proteins were efficiently depleted after the corresponding specific siRNA treatments (Figure 3D). Immunoblots indicated that TRF1 protein levels were decreased in Usp22 depleted cells (Figure 3E, lanes 1 and 2) to almost the same extent as in Gcn5 depleted cells (Figure 3E, lanes 5 and 6), 36% and 40%, respectively. TRF1 levels were also slightly decreased after Atxn7 depletion but not to the same extent as observed upon depletion of GCN5 or USP22 (Figure 3D). These results demonstrate that the observed TRF1 decrease is not limited to Gcn5 null MEFs and that TRF1 stability is affected by both Gcn5 and Usp22 loss.
We next asked whether the deubiquitination module of SAGA interacts physically with TRF1. We over expressed FLAG tagged-hTRF1 in human 293T cells and monitored the presence of USP22, GCN5 and ATXN7L3 in the anti-FLAG precipitated fractions. ATXN7L3 is related to both hATXN7 and to Sgf1 1 family proteins, and it was shown to be indispensable for the USP22 activity toward ubiquitinated histones H2A and H2B (Zhao et al., 2008). All three of these SAGA proteins co-precipitated with TRF1, whereas another relatively abundant nuclear protein, PCNA, did not (Figure 3E). These interactions are unlikely to be indirect due to DNA associations since addition of ethidium bromide to the immunoprecipitation reactions did not alter the association of TRF1 with these hSAGA components.
The above data suggest that USP22 might affect TRF1 levels through deubiquitination. If so, then ubiquitin-conjugated TRF1 (ub-TRF1) levels should be increased upon depletion of GCN5, USP22 or ATXN7L3. To test this idea, we generated 293T cells stably expressing shRNA against GCN5, USP22 or ATXN7L3 (Figure 4A). Co-expression of hTRF1-FLAG and 6xHis-tagged ubiquitin in these cells revealed significant increases in ub-TRF 1 levels upon loss of the SAGA components relative to the control cells (Figure 4 B, compare lanes 2, 3 and 4 to lane 1). The ub-TRF1 levels were most abundant in USP22 depleted cells compared to GCN5 and ATXN7L3 depleted cells (Figure 4 B, compare lane 3 to lanes 2 and 4), consistent with the function of these proteins as cofactors for USP22 mediated deubiquitination (Zhao et al., 2008). The higher molecular weight species corresponding to ub-TRF1 were only observed in cells expressing His-tagged ubiquitin (Figure 4B, lane 5).
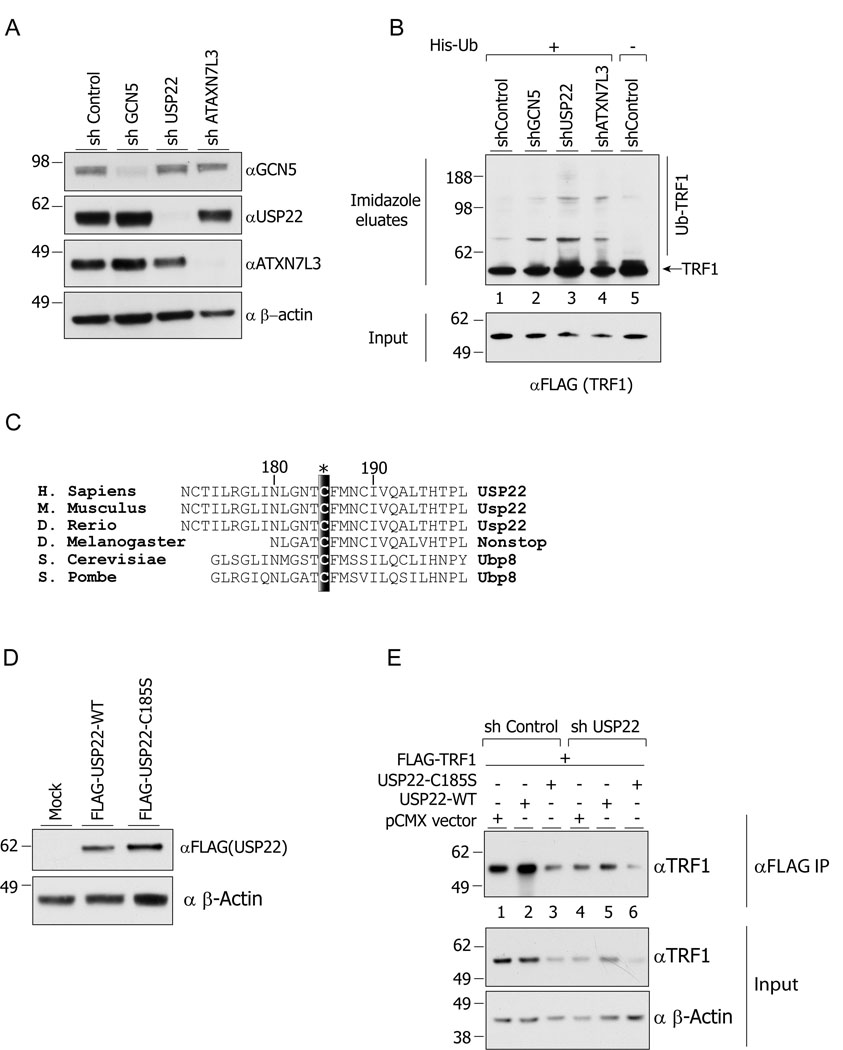
Figure 4. USP22 deUB activity regulates the ubiquitination and steady state levels of TRF1.
(A) Efficient depletion of the indicated proteins in 293T cells. (B) Increased ubiquitination of TRF1 upon depletion of GCN5, USP22 or ATXN7L3. Cells were transfected with 6xHis-ubiquitin and FLAG-TRF1 expression vectors and the ubiquitinated species were precipitated by Ni-NTA agarose from nuclear extracts. (C) Schematic representation of Cysteine 185 residue conservation among the USP22 orthologs in different species. (D) Expression of wt -USP22 and C185S–USP22 in HeLa cells. (E) Expression of wt-USP22 restores TRF1 protein levels in USP22 depleted cells, while over expression of C185S–USP22 decreases TRF1 protein levels.
To further verify that TRF1 protein levels are affected by USP22 dependent deubiquitination, we over expressed wild type USP22 (wt-USP22) or a mutant version of USP22 harboring a point mutation that converted cysteine 185 to serine (C185S–USP22). This cysteine residue is highly conserved across evolution (Figure 4 C), and point mutations at this position abolish the USP22 deUB activity toward in vitro substrates (Lee et al., 2006a). We verified that both the wild type and C185S mutant USP22 proteins were expressed to approximately equal levels in HeLa cells (Figure 4 D). Overexpression of wt-USP22 led to increased FLAG-TRF1 protein levels (Figure 4 E compare lanes 1 and 2). In contrast, over expression of the C185S–USP22 mutant decreased FLAG-TRF1 levels (Figure 4E compare lanes 1 and 3), further confirming that USP22 activity is required for TRF1 stabilization. The impact of USP22 expression was even more obvious after immunprecipitation of the FLAG-TRF 1 from cell lysates (Figure 4 E, upper panel). Furthermore, expression of wt-USP22, but not the C185S mutant, was able to partially restore TRF1 levels in cells depleted for endogenous USP22 (Figure 4 E, compare panels 4, 5 and 6). Collectively, these results confirm that TRF1 steady state levels are affected by USP22 expression and deUb activity.
TIF formation and telomere elongation upon depletion of GCN5, USP22, or ATXN7L3
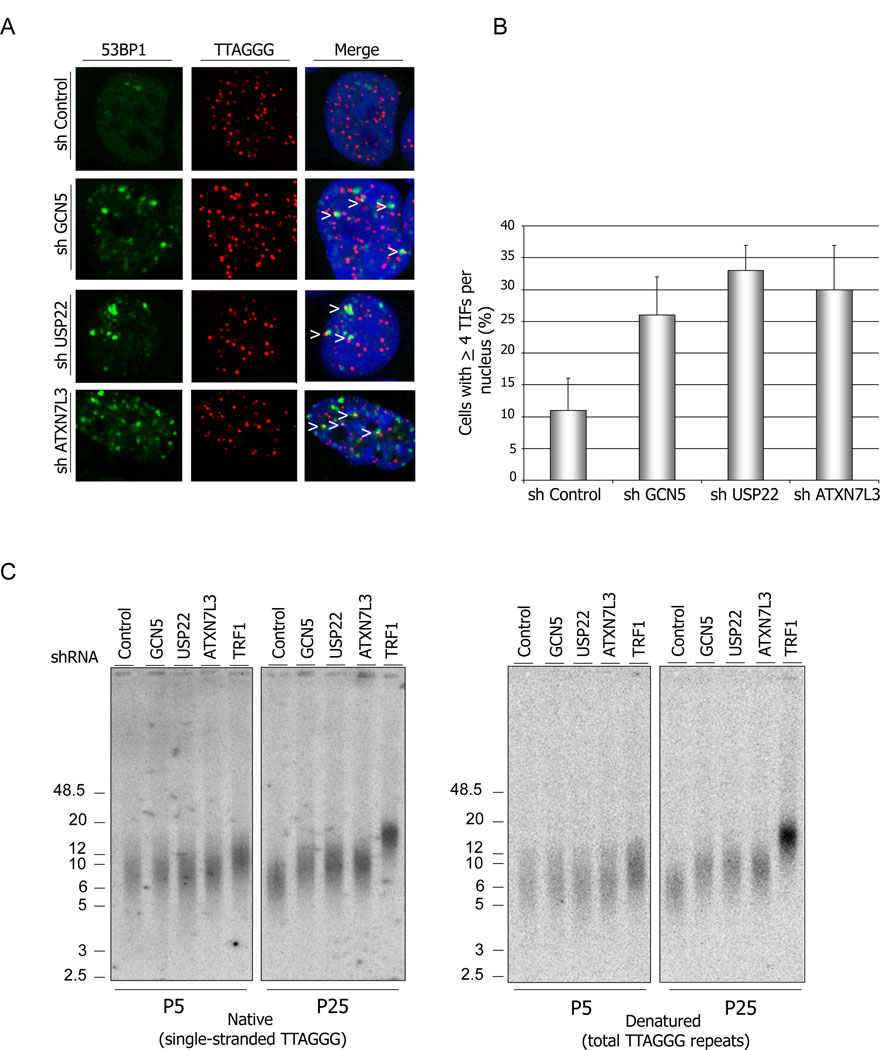
Since depletion of GCN5, USP22 or ATXN7L3 leads to increased ub-TRF1 levels, and Gcn5 or USP22 depletions impact TRF1 protein levels, we next asked whether USP22 and ATXNL3 depletions are sufficient to induce TIFs. We found that TIF formation was increased in 293T cells depleted for any one of these proteins (Figure 4A), such that approximately 33% of the USP22 depleted and ~ 26 and 30% of the GCN5 and ATXN7L3 depleted cells respectively showed ≥4 TIFs per nucleus. Only about 10% of the control cells exhibited TIFs (Figure 5 A and B). These results further confirm a role for the hSAGA deUb module in telomere maintenance.
Figure 5. Depletion of hSAGA deUB module induces TIFs and telomere elongation.
(A) GCN5, USP22 or ATXN7L3 depleted cells, as well as control shRNA treated cells, were subjected to immunostaining with anti-53BP1 antibody (green) followed by PNA-FISH (red). 100 cells of each sample were counted, and cells with ≥4 TIFs per nucleus (yellow signals indicated with arrowheads) were scored.
(B) Quantification of the data represented in (A). Error bars represent standard deviation of the mean (n=2) (C) Telomere elongation after depletion of GCN5, USP22 and ATXN7L3 in HeLA cells. Cells stably expressing denoted shRNAs were harvested at the indicated passages and used for telomere restriction fragment Southern analysis. In-gel hybridizations were done using 32P labeled (GGGTTA)4 or (CCCTAA)4 probes. The hybridized gels were exposed to phosphoimager screens.
Given the known function of TRF1 as a negative regulator of the telomere length (van Steensel and de Lange, 1997), we next asked whether depletion of the SAGA deUB module impacts telomere length. We examined telomere length in HeLa cells stably expressing shRNAs targeted against GCN5, USP22, ATX7L3, or TRF1, as well as cells expressing non-targeting, control shRNAs, by telomere restriction fragment (TRF) Southern analysis. The efficient depletion of the corresponding proteins was confirmed by immunoblot (data not shown). No changes in telomere length were observed at early passages (Passage 5) after depletion of the hSAGA proteins as measured by either the amount of the single-stranded TTAGGG repeats (G-strand overhang) (native conditions) or total TTAGGG repeats (denaturing conditions) (Figure 5C). However, 25 passages after depletion of GCN5, USP22 or ATXN7L3, telomere lengths were notably increased. As expected, TRF1 depletion resulted in a strong telomere lengthening phenotype. These results are in agreement with our observations above that GCN5, USP22 and ATXN7L3 depletions have substantial impact on steady state levels of TRF1.
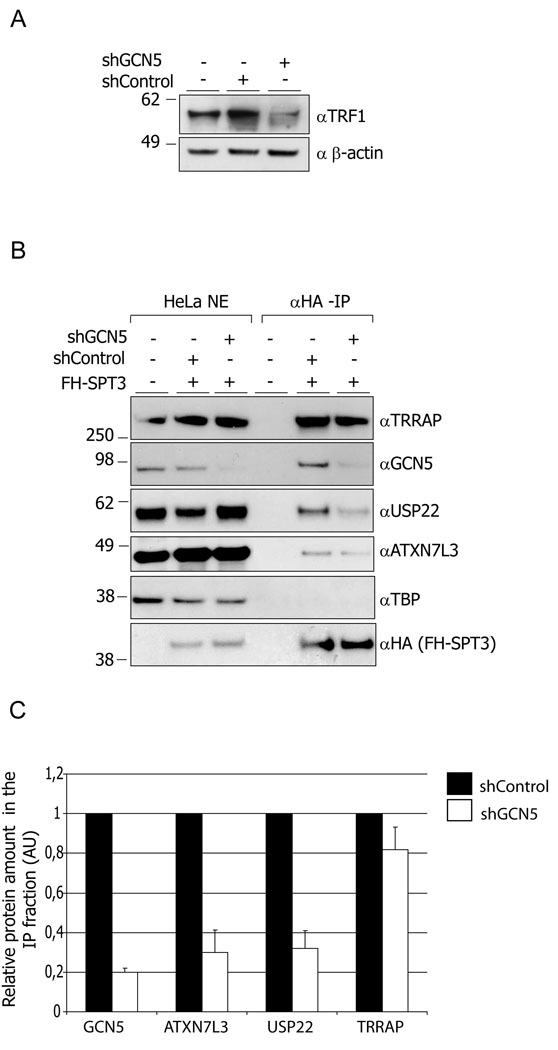
Depletion of GCN5 compromises association of USP22 with hSTAGA
Given that the human USP22 and the yeast Ubp8 deubiquitinating modules are most active when associated with SAGA complexes, we asked whether USP22 association with SAGA was affected by GCN5 loss. We purified hSAGA from HeLa S3 cells containing a FLAG-HA epitope-tagged SPT3 SAGA subunit (Martinez et al., 2001). As expected, TRF1 levels were decreased in these HeLa cells following knock down of GCN5 by shRNA treatment (Figure 6A). Overall levels of hSAGA components were not affected by Gcn5 loss (Figure 6B). SPT3 was isolated together with several hSAGA components, including USP22, ATXN7L3 and GCN5 from control treated cells (Figure 6B) as previously reported (Martinez et al., 2001). TBP did not co-purify with the hSAGA components, confirming the specificity of our HA-SPT3 isolations. However, prior depletion of GCN5 by shRNA in these cells led to a more than 2-fold decrease of USP22 and ATXN7L3 in the immunoprecipitated fractions (Figure 6B and C). TRRAP association with hSAGA was not affected, consistent with previous reports that loss of Gcn5 does not affect the overall integrity of the complex (Sterner et al., 1999). Thus, these results indicate that GCN5 plays an important role in supporting the association of the USP22 deubiquitination module with the SAGA complex.
Figure 6. Depletion of GCN5 impacts hSAGA integrity.
(A) Depletion of GCN5 in HeLa S3 cells by shRNA leads to lowered TRF1 protein levels.
(B) Depletion of GCN5 compromises association of USP22 and ATXN7L3 with SAGA. SAGA was purified from nuclear extracts prepared from FLAG-HA-SPT3 expressing HeLa S3 cells. Immunoprecipitated fractions as well as nuclear extracts were resolved by SDS-PAGE and blotted with the indicated antibodies. (C) Quantification of the data from multiple experiments like that shown in (B). X-ray films were scanned and the images were quantified using ImageQuant software. Error bars represent standard deviations of the mean (n=4)
DISCUSSION
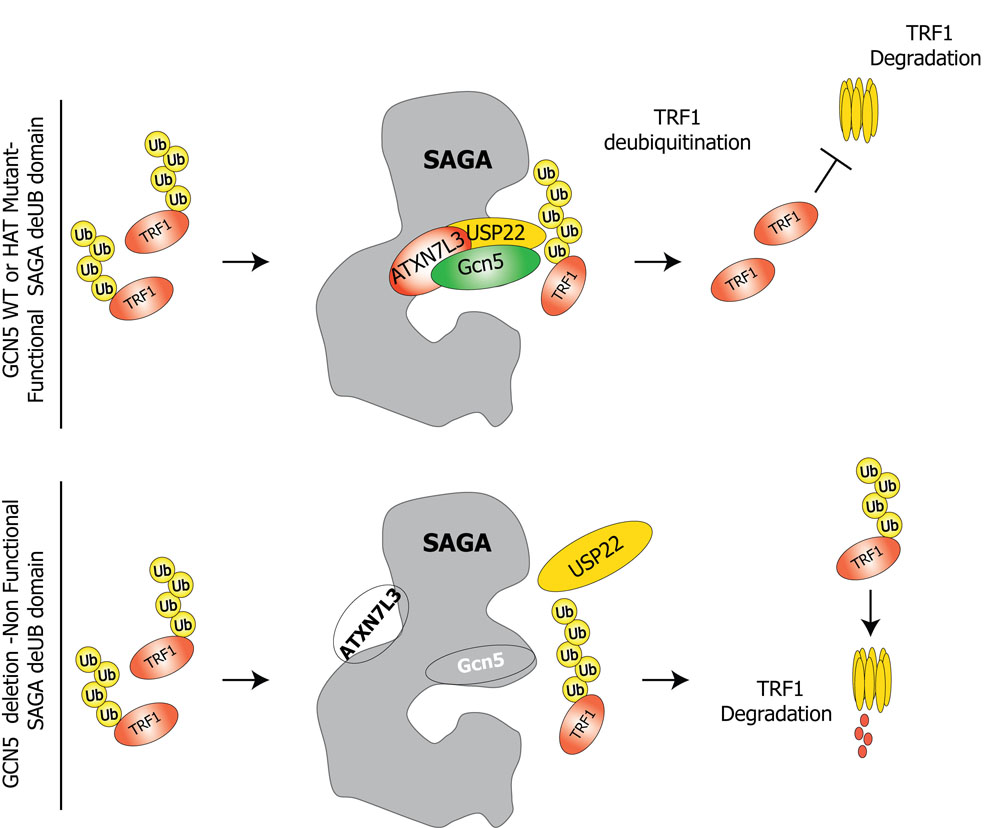
Our data reveal an unexpected link between Gcn5 and the SAGA histone modifying complex and telomere maintenance, most likely through affects on shelterin protein levels. This work also indicates for the first time that SAGA and USP22 regulate protein stability and leads to a model wherein loss of Gcn5 leads to loss of the USP22 module from SAGA in mammalian cells. This dissociation leads to lowered USP22 activity, which in turn leads to increased ubiquitination and turnover of TRF1 (Figure 7).
Figure 7.
A proposed model for the role of GCN5 and SAGA in shelterin protein turnover. SAGA deUb module stabilizes TRF1 protein levels by deubiquitination, thereby inhibiting degradation. Depletion of GCN5 or ATXN7L3 leads to destabilization of the deubiquitination module, which in turn leads to altered steady state levels of TRF1.
In human cells, TRF1 undergoes ubiquitin mediated proteolysis after its eviction from the telomeres (Chang et al., 2003), which is triggered by prior poly (ADP)-ribosylation mediated by tankyrase 1 (Smith et al., 1998). We do not yet know if Gcn5 and USP22 affect TRF1 before or after its eviction from telomeres. However, our observation of similar effects of Gcn5 and Usp22 on TRF1 in mouse and human cells indicates these effects are independent of the tankyrase pathway since mouse TRF 1 lacks a tankyrase 1 binding site and over expression of tankyrase 1 in mouse cells does not induce eviction of TRF1 form telomeres (Donigian and de Lange, 2007). Others have shown that depletion or over expression of the F-box protein FBX4, which promotes TRF1 ubiquitination, significantly impacts telomere structure by regulating the cellular levels of this shelterin component (Lee et al., 2006b). Also, the ubiquitin ligase RLIM regulates the telomere homeostasis by affecting TRF1 steady state levels promoting its ubiquitination (Her and Chung, 2009). Taken together these data suggest that TRF1 ubiquitination and degradation are tightly controlled and appear to provide an additional level of telomere homeostasis control. Future experiments will determine how, when, and where SAGA is directed to TRF 1, and why levels of this protein might be regulated by both ubiquitination and deubiquitination.
Our studies also revealed that Gcn5 depletion affects the steady state levels of another shelterin protein, POT 1a. Although it is not known whether POT 1a is ubiquitinated, it might also be affected in a similar way to TRF1 by Gcn5 loss. Alternatively, Pot1a may be destabilized as a secondary consequence of TRF1 depletion.
We do not formally know whether the telomere fusions we observe in Gcn5 null cells are linked to the decreased levels of TRF1 and Pot1a that we observe. However, our findings are in agreement with previous studies in mouse cells that show depletion of TRF1 leads to dysfunctional telomere induced DNA repair foci and a mild chromosome fusion phenotype (Iwano et al., 2004; Okamoto et al., 2008). Depletion of mTRF1 in mouse ES cells significantly diminishes the ability of another shelterin component, TIN2, to localize to telomeres. Over expression of chicken TRF1 in these cells can rescue some but not all of the TRF1 induced phenotypes and cannot restore telomeric localization of TIN2 (Okamoto et al., 2008). To our knowledge, no one has investigated the combined effects of diminished levels of both TRF1 and POT la, but it seems reasonable that loss of a second shelterin component would enhance TRF1-related telomere dysfunction phenotypes.
Previous studies by others indicate that SAGA complex integrity and proper subdomain organization are important for its functional activity (Ingvarsdottir et al., 2005; Lee et al., 2005; Rodriguez-Navarro et al., 2004; Weake et al., 2008; Zhao et al., 2008). In particular, the deubiquitination activity of the Ubp8 and USP22 modules towards H2B in yeast and towards H2B and H2A in mammalian cells requires other factors such as Sus1 and Sgf11 (Ingvarsdottir et al., 2005; Kohler et al., 2006; Lee et al., 2005) and their mammalian orthologues, ANY2 and ATXN7L3 (Zhao et al., 2008). Moreover, the protein structures of Ubp8/USP22 indicate that these enzymes cannot bind free ubiquitin, so the surrounding subunits may be required not only to facilitate association of the deUB enzymes with the complex but also to provide substrate binding and specificity (Bonnet et al., 2008). Interestingly, the mammalian complexes contain multiple proteins related to yeast Sgf73, which is required for Ubp8 incorporation into SAGA (Kohler et al., 2008). The functions of ATXN7, ATXNL2, and ATXNL3 are not known, but our data suggest that ATXNL3 is particularly important for USP22 functions, at least towards nonhistone substrates, since knock down of ATXN7 had much less effect on TRF1 levels than did knock down of ATXN7L3.
Gcn5 is closely associated with USP22 or its fly orthologue, Nonstop (Lee et al., 2005; Weake et al., 2008; Zhang et al., 2008; Zhao et al., 2008). However, the effects of Gcn5 loss on the deubiquitination activity of USP22 were not previously defined. In vitro experiments indicate that SAGA purified from gcn5Δ yeast is still capable of removing ubiquitin from H2B, but this activity may not be as efficient as in SAGA purified from GCN5 wild type cells (Lee et al., 2005), consistent with our findings. Importantly, yeast H2B was recently demonstrated to be polyubiquitinated and this modification was affected by deletion of Ubp8 or Ubp1O (Geng and Tansey, 2008). These results highlight the ability of Ubp8, and likely USP22, to deubiquitinate polyubiquitinated substrates, as indicated by our findings with TRF1.
Overall, our results indicate that Gcn5 and SAGA affect gene expression at multiple levels, from gene transcription to protein stability. These findings will likely foreshadow discovery of additional proteins that are destabilized upon depletion of the Gcn5 or the SAGA deUB module. USP22 is part of an 11 gene signature for highly metastatic cancers with poor prognosis (Glinsky, 2006), raising the possibility that alterations in its role in controlling protein stability may contribute to cancer progression.
EXPERIMENTAL PROCEDURES
Details regarding imunoblots, siRNA transfections, shRNA transductions, telomere Chromatin Immunoprecipitation (ChIP), real time reverse transcriptase -PCR analyzes, immunoprecipitations, TRF southern analysis, creation of USP22 point mutation, cellular transfections and expression vectors are presented in Supplemental Experimental Procedures due to space constraints.
Antibodies and TIF Analysis
Previously described antibodies used in this study include: Anti-USP22 (2391), anti-ATXN7L3 (2325), anti TRRAP (2TRR-2D5) (Zhao et al., 2008), anti-mPOT1a (1220), anti-mRAP1 (1252), anti-mTRF2 (1254) (Denchi and de Lange, 2007) and anti-mTRF1 (Iwano et al., 2004). Commercially available antibodies used here include : anti-Gcn5 (Cell Signaling), anti-hTRF1 (Alpha Diagnostics International Inc.), anti-FLAG (M2, Sigma), anti-HA (Roche), and anti-γ-H2AX (Trevigen). Anti-Gcn5 (N-18), anti-53BP1, anti-β-actin, anti-TBP and anti-PCNA (PC10) were all from Santa Cruz Biotechnoloy. PNA-FISH probes and TIF analyses were described before (Guo et al., 2007)
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Bill Morgan (Univ. Maryland) and Dr. David Roth (NYU) for help with the initial analyses of metaphase chromosomes from Gcn5 null embryos, presented in Suppl Figure 1, Dr. Titia de Lange (Rockefeller University) for providing TRF2, RAP1 and POT 1a specific antibodies, Dr. Yoichi Shinkai (Kyoto University) for the Flag-mTRFl construct and anti mTRFl antibody, and Dr. Albert R. La Spada for the FH-SPT3 HeLa S3 cell line. Y.A.E. was supported by a Sowell-Huggins Fellowship. This work was supported by a grants from the NIH (AG02888 and CA129037) to S.C. and (GM067718) to S.Y.R.D and by grants from the the Agence Nationale de la Recherche (ANR-05-MRAR-O29-01) from the Fondation de la Recherche Médicale (FRM, DLC20060206408) and from ANR (05-BLAN-0396-01; Regulome), European Community (HPRN-CT 00504228) to D.D and L.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Romier C, Tora L, Devys D. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem Sci. 2008;33:369–375. doi: 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Berger SL. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, Berger SL. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavusoglu N, Brand M, Tora L, Van Dorsselaer A. Novel subunits of the TATA binding protein free TAFII-containing transcription complex identified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry following one-dimensional gel electrophoresis. Proteomics. 2003;3:217–223. doi: 10.1002/pmic.200390030. [DOI] [PubMed] [Google Scholar]
- Chang W, Dynek JN, Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Donigian JR, de Lange T. The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J Biol Chem. 2007;282:22662–22667. doi: 10.1074/jbc.M702620200. [DOI] [PubMed] [Google Scholar]
- Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Griffith J, Bianchi A, de Lange T. TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol. 1998;278:79–88. doi: 10.1006/jmbi.1998.1686. [DOI] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her YR, Chung IK. Ubiquitin Ligase RLIM Modulates Telomere Length Homeostasis through a Proteolysis of TRF1. J Biol Chem. 2009;284:8557–8566. doi: 10.1074/jbc.M806702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet. 2001;29:206–211. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf1 1 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf1 1. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr Patterns. 2006a;6:277–284. doi: 10.1016/j.modgep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf1 1 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006b;281:759–768. doi: 10.1074/jbc.M509855200. [DOI] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY. Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev Dyn. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- Lin W, Zhang Z, Srajer G, Chen YC, Huang M, Phan HM, Dent SY. Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev Dyn. 2008;237:928–940. doi: 10.1002/dvdy.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. Chromatin modifications in DNA repair. Results Probl Cell Differ. 2006;41:109–125. doi: 10.1007/400_008. [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JL, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Iwano T, Tachibana M, Shinkai Y. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J Biol Chem. 2008;283:23981–23988. doi: 10.1074/jbc.M802395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck B, Fischer S, Gunning D, Zipursky SL, Salecker I. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29:99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. Potl deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Roth SY. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18:5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1 A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.