Abstract
Background
Jasmonates are ubiquitously occurring lipid-derived compounds with signal functions in plant responses to abiotic and biotic stresses, as well as in plant growth and development. Jasmonic acid and its various metabolites are members of the oxylipin family. Many of them alter gene expression positively or negatively in a regulatory network with synergistic and antagonistic effects in relation to other plant hormones such as salicylate, auxin, ethylene and abscisic acid.
Scope
This review summarizes biosynthesis and signal transduction of jasmonates with emphasis on new findings in relation to enzymes, their crystal structure, new compounds detected in the oxylipin and jasmonate families, and newly found functions.
Conclusions
Crystal structure of enzymes in jasmonate biosynthesis, increasing number of jasmonate metabolites and newly identified components of the jasmonate signal-transduction pathway, including specifically acting transcription factors, have led to new insights into jasmonate action, but its receptor(s) is/are still missing, in contrast to all other plant hormones.
Key words: Oxylipins, jasmonic acid, jasmonate metabolites, enzymes in biosynthesis and metabolism, signal function
INTRODUCTION
In recent years, large scale analyses of gene sequences (genomics), expression (transcriptomics), protein patterns (proteomics), metabolite profiles (metabolomics) and lipid compounds (lipidomics) have been performed and remain the focus of many studies. The tremendous amount of accumulated data has led to new insights into how plants respond to biotic or abiotic stress and how a growth and developmental program takes place in the plant life cycle. An essential link within such programming is provided by small molecules, such as plant hormones. Among these, jasmonates have attracted much interest over the last two decades.
In 1962, jasmonic acid methyl ester (JAME; see Appendix for a list of abbreviations) was isolated for the first time from the essential oil of Jasminum grandiflorum (Demole et al., 1962). The first physiological effects of JAME, however, and its free acid (JA) were only observed two decades later. In 1980, Ueda's group in Osaka, Japan, described a senescence-promoting effect of JA, and Sembdner's group in Halle, Germany, isolated and characterized these compounds as growth inhibitors (Ueda and Kato, 1980; Dathe et al., 1981). Inhibition of root growth became the most prominent assay for screening of mutants affected in respect of JA signalling. The biosynthesis of JA was elucidated by Vick and Zimmermann (1984). The functional analysis of the mode of action was initiated by the observation in Parthier's group in Halle that application of JA or its methyl ester to barley leaves leads to an altered protein pattern. The synthesis of abundantly accumulating proteins, so-called jasmonate- induced proteins (JIPs) and degradation of housekeeping proteins such as Rubisco were described for the first time in 1987 (Weidhase et al., 1987a, b). In 1990, another dramatic alteration of gene expression by JAME was observed in tomato plants (Farmer and Ryan, 1990). Here, a proteinase inhibitor (PIN2) was found to accumulate upon wounding (such as that caused by herbivore attack) or by treatment with airborne JAME. Subsequently, JA-induced alteration of gene expression and metabolite patterns became the focus of studies examining accumulation of vegetative storage proteins in Staswick's group (Staswick et al., 1992) and alkaloids in Zenk's group (Gundlach et al., 1992). In the latter case, an endogenous rise of JA was demonstrated upon elicitation of cell suspension cultures, thus indicating JA as a putative signal between the external stimulus and the response by cells of alkaloid formation. Other breakthroughs were the first cloning of a JA biosynthetic enzyme, allene oxide synthase (AOS) in Brash's lab (Song et al., 1993) and isolation of the first JA-insensitive mutant in Turner's lab (Feys et al., 1994). This mutant was screened by use of coronatine, a bacterial toxin with structural and functional similarity to JA (coronatine-insensitive1, coi1). Four years later, Turner's lab were able to clone the corresponding gene. COI1 codes for an F-box protein, which is part of the ubiquitin-mediated proteasome pathway (Xie et al., 1998). In the same year, the corresponding F-box protein in auxin signaling, TIR1 was cloned. Consequently, much effort was spent in trying to understand hormone signalling in molecular terms and this led to the identification of TIR1 as one of the auxin receptors (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). The JA receptor, however, is still unknown. Other important steps in understanding the action of jasmonates were the identification of mutants affected in JA-biosynthesis (cf. Turner et al., 2002), the identification of the first transcription factors regulating JA-responsive gene expression by Memelink's group (van der Fits and Memelink, 2001), the proof that fatty acid β-oxidation acts in JA biosynthesis (Li et al., 2005), and the cloning of JA amino acid conjugate synthase (JAR1) by Staswick's lab (Staswick et al., 2002; Staswick and Tiryaki, 2004). Subsequent to the latter, interest was shifted to JA metabolism and the action of JA metabolites.
A number of reviews emphasizing different aspects of JA physiology have appeared in recent years (e.g. Walling, 2000; Blée, 2002; Feussner and Wasternack, 2002; Wasternack and Hause, 2002; Farmer et al., 2003; Halitschke and Baldwin, 2004; Howe, 2004; Mithöfer et al., 2004; Pauw and Memelink, 2004; Pozo et al., 2004; Browse, 2005; Rosahl and Feussner, 2005; Schilmiller and Howe, 2005; Wasternack, 2006). The following review will cover some recent results in biosynthesis, metabolism, signal transduction and action of jasmonates. I apologize for references not cited due to space limitations.
THE LIPOXYGENASE PATHWAY
Lipid-derived signals
Membrane lipids consist of polyunsaturated fatty acids (PUFAs) esterified to glycerol in the sn1 and sn2 positions. The third position of glycerol is linked via an inorganic ester linkage to a phosphate group. The resulting lipid molecule phosphatidic acid (PA), and the cleavage product of phosphoinositol bisphosphate, inositol triphosphate (IP3), are known as signals in Ca2 + signalling and stress responses, respectively (Bargmann and Munnik, 2006). However, it is not only the main group of this class of lipids that may serve as secondary messengers, but their fatty acid residues, for example PUFAs, are also the source of further signals, such as oxylipins, which include jasmonates. Oxylipins originate from α-linolenic acid (18:3) (α-LeA) released from chloroplast membranes. Lipid hydrolyzing enzymes belong to at least five different families (Dörmann, 2005): (1) phospholipase A1, which cleaves in the sn1 position; (2) phospholipase A2, which cleaves in the sn2 position; (3) patatin-like acyl hydrolases with phospholipase and glycolipase activity; (4) DAD-like lipases, which degrade phospholipids and galactolipids; and (5) SAG (senescence-associated gene) 101-like acyl hydrolases.
So far a link to JA biosynthesis has only been shown for the wound-induced phospholipase A2 (Narváez-Vásquez et al., 1999) and a DAD-like phospholipase A1. The Arabidopsis mutant dad1 exhibits delayed anther dehiscence, shorter filament length and JA deficiency in flowers, indicating role of DAD1 in JA biosynthesis and of JA in filament elongation (Ishiguro et al., 2001; see below). The corresponding DAD-like lipases of leaves are unknown.
Oxylipins generated in the lipoxygenase pathway
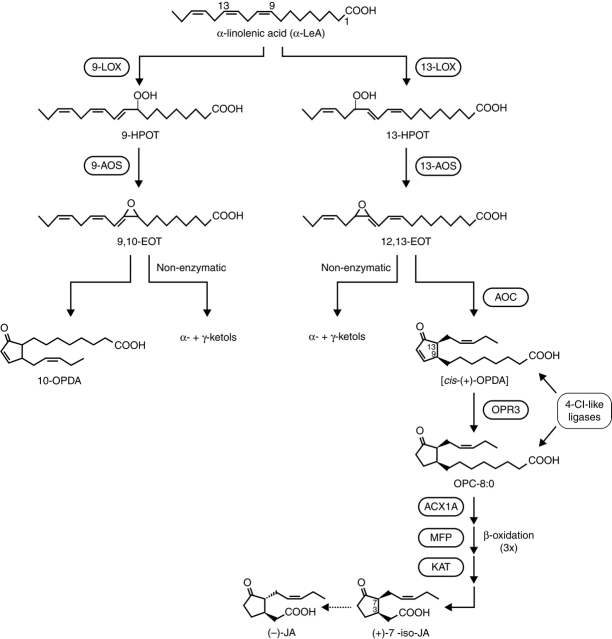
α-LeA released by lipase activity on chloroplast membranes is the substrate for numerous oxygenated compounds collectively called oxylipins including jasmonates, which comprise JA, JAME, JA amino acid conjugates and further JA metabolites (see below). The generation of oxylipins is initiated by lipoxygenases (LOXs), which form hydroperoxides from α-LeA (18:3) or linoleic acid (18:2) (Feussner and Wasternack, 2002). With α-LeA as the substrate, (13S)-hydroperoxyoctadecatrienoic acid (13-HPOT) or (9S)-hydroperoxyoctadecatrienoic acid (9-HPOT) are formed (Fig. 1). The corresponding products with linoleic acid as the substrate are (13S)-hydroperoxyoctadecadienoic acid (13-HPOD) and (9S)-hydroperoxyoctadecadienoic acid (9-HPOD). Although non-enzymatic oxygenation can take place by formation of phytoprostanes (Mueller, 2004), the LOX-catalysed reaction is indicated by the exclusive formation of the S-isomer. The increasing amount of LOX sequence data has allowed the construction of phylogenetic trees, which show gene families of different size for many plant species (Feussner and Wasternack, 2002). Analysis of the substrate-binding pockets by site-directed mutagenesis has revealed their role in the positional specificity of 9-LOXs and 13-LOXs (Liavonchanka and Feussner, 2006). A change in the histidine residues in the binding pocket of a cucumber lipid-body 13-LOX led to a 9-LOX activity (Hornung et al., 1999). In addition to these structural requirements for positional specificity, clear differences were found for the expression of genes coding for 9-LOXs and 13-LOXs. Consequently, a concept for discrete 9-LOX and 13-LOX pathways was proposed (Howe and Schilmiller, 2002) and may explain the occurrence of numerous oxylipins (Fig. 1). The LOX products 13-HPOT and 13-HPOD represent branch points within the LOX pathway. At least seven different groups of compounds are formed by individual enzyme families (Feussner and Wasternack, 2002).
Octadecanoids and jasmonates originating from 13-allene oxide synthase (13-AOSs) activity.
Epoxyhydroxy-PUFAs formed by peroxygenases.
Aldehydes, ω-oxo fatty acids and alcohols formed by hydroperoxy lyases (13-HPLs).
Divinyl ether-containing PUFAs synthesized by divinyl ether synthases.
Epoxyhydroxy PUFAs formed by epoxy alcohol synthases.
Keto-PUFAs formed by LOXs.
Hydroxy PUFAs formed by reductases.
Fig. 1.
The 9-LOX and 13-LOX pathways, and JA biosynthesis (modified after Schilmiller and Howe, 2005; Wasternack, 2006).
Increasing examples are known where there are corresponding reactions with the 9-LOX products 9-HPOT and 9-HPOD. Subsequently acting enzymes should be specific for 9-LOX and 13-LOX products. Indeed, 9-HPLs and 13-HPLs as well as 9-AOSs and 13-AOS have been characterized, thus supporting the concept of discrete 9-LOX and 13-LOX pathways (Stumpe and Feussner, 2006). In potato, there are two 13-AOSs and one 9-AOS (AOS3), which is highly specific for 9-hydroperoxides and leads in vitro to α- and γ-ketol formation (Stumpe et al., 2006). In below-ground organs, AOS3 is expressed in sprouting eyes, stolons, tubers and roots leading to α-ketol formation in vivo.
JA BIOSYNTHESIS
The AOS branch in the LOX pathway: JA biosynthesis
Conversion of 13-HPOT, the first step in JA biosynthesis, is carried out by 13-AOS, leading to an unstable allene oxide (Vick and Zimmermann, 1987). AOSs belong to the family of CYP74A enzymes, which are independent from molecular oxygen and NADPH, exhibit low affinity to CO and use the hydroperoxide group as a source for reducing equivalents and oxygen (Song et al., 1993; Feussner and Wasternack, 2002; Howe and Schilmiller, 2002). Based on structural information on AOS, an effective and selective inhibitor, JM-8686, has been designed (Oh et al., 2006). First cloned from flax seeds, more than 20 AOS sequences have been included in a phylogenetic tree analysis in which 9-AOSs and 13-AOSs were grouped separately (Stumpe and Feussner, 2006). With the exceptions of the AOS of guayule (Pan et al., 1995) and barley (Maucher et al., 2000), all of the 13-AOSs carry a plastid transit peptide. Plastid localization has been shown by immunolocalization and import studies, even for the barley AOSs that lack the transit peptides (Maucher et al., 2000; Froehlich et al., 2001). This compartmentation corresponds to the plastid location of 13-LOX, whereas 9-LOX activity was found in the cytosol (Feussner and Wasternack, 2002) (Fig. 2). The picture is, however, more complicated. The recently cloned potato 9-AOS is associated with amyloplasts and leucoplasts (Stumpe et al., 2006), and the recombinant AOS of A. thaliana exhibits dual 9-/13-specificity regulated by monomer-micelle association (Hughes et al., 2006). This raises the possibility that in vivo an AOS can be a versatile enzyme in respect to ‘substrate-specific’ membrane association and oligomeric state (Hughes et al., 2006).
Fig. 2.
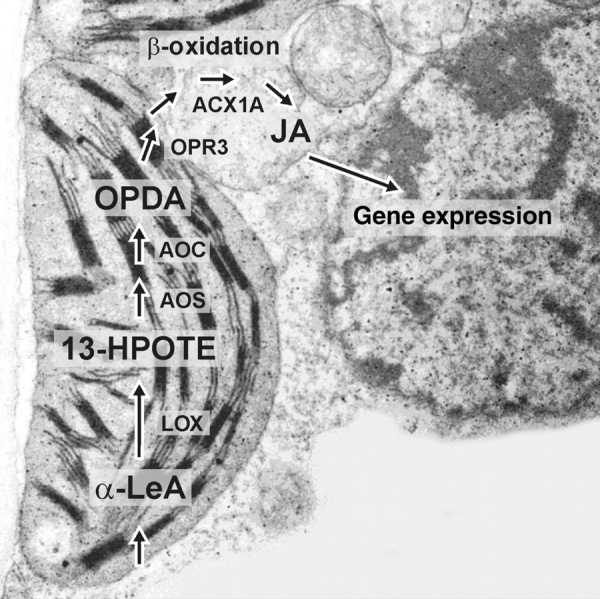
Intracellular location of enzymes and intermediates in JA biosynthesis, illustrated on a SEM of a barley mesophyll cell showing the associated cellular compartments (photograph: B. Hause).
The subsequent enzyme of the AOS branch, the allene oxide cyclase (AOC) is also plastid-located (Ziegler et al., 2000). Using the highly unstable allene oxide, the AOC establishes the enantiomeric structure at the cyclopentenone ring that occurs in JA. Non-enzymatic side-reactions in the absence of AOC are the formation of racemic OPDA and the cleavage to α- and γ-ketol (Fig. 1). Due to the instability of the allene oxide and the common location of AOS and AOC, both enzymes were thought to be in close contact. But so far there is no proof on an AOS–AOC protein interaction (Zerbe et al., 2007). The AOC product cis(+)-12-oxophytodienoic acid (OPDA) is the final product of the plastid-located part of JA biosynthesis. The subsequent step, reduction of the cyclopentenone ring, is catalysed by a peroxisomal OPDA reductase (OPR; Strassner et al., 2002). In Arabidopsis and tomato there are more than three enzymes, but only OPR3 exhibits specificity for cis(+)-OPDA (Schaller et al., 2000; Strassner et al., 2002).
OPRs reduce conjugated enone structures including that occurring in OPDA. For OPR1 there are hints that the enzyme might be responsible for the formation of 9-ketodienes, such as 9-KODE (Tani et al., 2006). The role of OPR3 in JA biosynthesis was shown by genetic evidence provided by the JA-deficient mutants opr3 and dde1, both affected in OPR3 (Sanders et al., 2000; Stintzi and Browse, 2000). All OPR3s cloned so far carry a peroxisomal target sequence, and a peroxisomal location of the OPR protein was shown by immunocytochemical techniques and by analysis of expression of OPR3 fused to a reporter gene (Strassner et al., 2002). Conversion of OPDA by the peroxisomal OPR3 raises the question as to how OPDA is released from the chloroplast and imported into peroxisomes. To date, an OPDA transporter of chloroplast membranes is unknown. For the import of OPDA or its CoA ester into peroxisomes, there is some evidence for transport by the ABC transporter COMATOSE (CTS), covering the PXA1 activity. The CTS mutant is partially JA-deficient, but is not male-sterile, a characteristic phenotype of complete JA-deficiency. Consequently, a parallel pathway of OPDA import by ion trapping has been suggested (Theodoulou et al., 2005).
Initial experiments with labelled OPDA revealed β-oxidative side-chain shortening as essential steps in JA biosynthesis (Miersch and Wasternack, 2000). More recently, for Arabidopsis and tomato genetic evidence has been described showing that enzymes of fatty acid β-oxidation, such as acyl-CoA oxidase of tomato (ACX1), a multifunctional protein (MFP) and a L-3-ketoacyl CoA thiolase all have a function in JA biosynthesis (Fig. 1) (Cruz Castillo et al., 2005; Li et al., 2005; Wasternack et al., 2006). Participation of ACX1 in JA biosynthesis was clearly shown by isolation of an acx1 mutant of tomato exhibiting JA deficiency but accumulating OPDA upon wounding (Li et al., 2005). In Arabidopsis only the double mutant acx1/5 shows less JA formation upon wounding, indicating redundancy among the five ACX genes (Schilmiller et al., 2007). Further proof for role of fatty acid β-oxidation enzymes in JA biosynthesis has come from analysis of the aim1 mutant, affecting one of the MFPs, and the pex6 mutant, affecting peroxisome biogenesis (C. Delker et al., unpubl. res.; Leibniz Institute of Plant Biochemistry, Halle/Saale, Germany). β-oxidative steps take place only with the corresponding CoA ester. The formation of the OPDA–CoA ester in the chloroplast has been suggested, but not proven experimentally. There are, however, several indications for synthesis of OPDA–CoA ester in peroxisomes by 4-coumarate:CoA ligase-like (4-Cl-like) enzymes encoded by a small gene family in A. thaliana (Schneider et al., 2005). Interestingly, this type of enzyme is also able to activate downstream intermediates in JA biosynthesis such as OPC-8 (Schneider et al., 2005; Koo et al., 2006) or OPC-6 (E. Kombrink, pers. comm.; Max Planck Institute for Plant Breeding Research, Cologne, Germany). These data suggest different import reactions (free OPDA versus OPDA–CoA) and activation of OPDA (or its products) where it carrys a different carboxylic acid side-chain to the corresponding CoA ester. It is, however, still unclear whether OPR3 can convert the OPDA–CoA ester. The existence of gene families for 4-CL-like enzymes and for genes encoding enzymes in β-oxidation, such as acyl CoA oxidase, suggest distinct and/or overlapping roles in β-oxidation of fatty acids and in the final steps of biosynthesis of JA. Similar β-oxidation steps were found for the final steps in auxin biosynthesis. Consequently, mutant analysis requires double or triple mutants in order to dissect the role in JA-dependent, auxin-dependent or β-oxidation (development)-dependent processes. These aspects have been recently reviewed (Baker et al., 2006).
Regulation of JA biosynthesis
All genes encoding enzymes of JA biosynthesis are JA-inducible (Wasternack, 2006), and promoters analysed so far increase their activity upon JA treatment (Kubigsteltig et al., 1999; I. Stenzel and C. Wasternack, unpubl. res.). This has led to the suggestion that JA biosynthesis is regulated by a positive feedback. Further experimental evidence for this has come from the following observations. (1) Mutants with constitutively elevated JA levels, such as cev, exhibit a phenotype characteristic for JA treatment (Ellis et al., 2002) and show elevated AOC expression (I. Stenzel and C. Wasternack, unpubl. res.). (2) JA-deficient mutants, such as opr3 or coi1, show decreased AOC expression (Stenzel et al., 2003b; I. Stenzel and C. Wasternack, unpubl. res.). Interestingly, as shown for tomato leaves by feeding experiments with deuterated OPDA, JA biosynthesis is not induced by endogenous jasmonates, which suggests extracellular perception of JA (Miersch and Wasternack, 2000). This has been substantiated by an autoradiographic study where applied labelled-JA was detected exclusively in the apoplast of tomato leaves (Bücking et al., 2004).
Gene expression data are only part of the information necessary to understand regulation of JA biosynthesis. For example, the fully developed leaves of A. thaliana carry LOX, AOS and AOC proteins abundantly (Stenzel et al., 2003b); however, JA-formation takes place only upon external stimuli, such as wounding. Furthermore, the wound-induced rise in JA is transient and appears before expression of LOX, AOS and AOC (Howe et al., 2000; Stenzel et al., 2003a, b), and plants over-expressing AOS or AOC constitutively do not show elevated JA levels before wounding or other stimuli (Laudert et al., 2000; Stenzel et al., 2003a). These data clearly show that JA biosynthesis is regulated by substrate availability. Finally, the tissue-specific location of JA biosynthetic enzymes contribute to the outcome in JA biosynthesis. In contrast to A. thaliana (Stenzel et al., 2003b), in tomato AOC is confined to vascular bundles (Hause et al., 2000) and even the sieve elements carry AOC protein (Hause et al., 2003); consequently, cell- and tissue-specific formation of JA has been considered to have a role in wound signalling (Schilmiller and Howe, 2005; Wasternack et al., 2006).
Arabidopsides: esterified oxylipins and protein-oxylipin adducts
Most of the substrates of many enzymes within the LOX pathway occur in free and esterified form. As well as the 18:2 and 18:3 PUFAs, their 9- and 13-hydroperoxides and 9- and 13-hydroxides have been found esterified in A. thaliana leaves and tomato flower organs (Stenzel et al., 2003b; Miersch et al., 2004).
Recently, esterified OPDA has received particular attention. First, esterified OPDA was found in galactolipids (MGDG) in the sn-1 position in untreated A. thaliana leaves (Stelmach et al., 2001). Subsequently, so-called arabidopsides A and B, derived from MGDG, and arabidopsides C and D, derived from DGDG, were found containing OPDA and dinor-OPDA, respectively (Hisamatsu et al., 2003, 2005). Finally, 13-OPDA-, 18- and 16-carbon ketol-containing MGDG, DGDG and PC species were found (Buseman et al., 2006). The diversity of esterified OPDA and the large amount found in lipid membranes (Stelmach et al., 2001) raise the question as to its biological functions. OPDA exhibits individual signalling in plant defence reactions (Stintzi et al., 2001), and there is a specific subset of genes that is specifically expressed in response to OPDA (Taki et al., 2005). Hence release of esterified OPDA would greatly influence defence reactions. Indeed, wounding leads to a dramatic increase of free OPDA (Stelmach et al., 2001; Buseman et al., 2006). A specific function in the hypersensitive response of A. thaliana was found for a new esterified OPDA, arabidopside E. Here, OPDA is bound to the sn1 and the sn2 position, and one dinor-OPDA to the galactose of MDGD (Andersson et al., 2006). This compound accumulates up to 7–8 % of the total lipid content in a race-specific defence reaction against bacterial pathogens and inhibits bacterial growth in vitro (Andersson et al., 2006). This suggests an important role of the 13-LOX pathway in the hypersensitive response- (HR) associated cell death. Furthermore, there was an increase in non-enzymatically formed hydroxy fatty acids and F1-phytoprostanes (reviewed by Mueller, 2004) being esterified in membranes upon infection with a virulent and with an avirulent strain of Pseudomonas syringae that was up to 30-fold greater in level than that of the corresponding free compounds (Grun et al., 2007).
Whilst induction of plant senescence has been known for a long time as a characteristic property of JA (Ueda and Kato, 1980; Parthier, 1990; Wasternack, 2004), recently a much stronger senescence-promotion effect was shown for arabidopside A (Hisamatsu et al., 2006). It will be interesting to see whether arabidopsides occur in species other than Arabidopsis thaliana and which other JA-related processes are affected by these compounds.
Many oxylipins generated by the LOX pathway display cell toxicity and antimicrobial activity (Rusterucci et al., 1999; Prost et al., 2005). Therefore, the observation that oxylipins can be trapped by binding to lipid transfer proteins (LTPs) is an exciting result in helping to understand cell injury and the role of LTPs (Bakan et al., 2006). LTPs are ubiquitously occurring plant lipid-binding proteins known for a long time to be associated with developmental and stress responses. But their function is still unknown. Direct inhibitory activity of LTPs on microbes was initially hypothesized, but the role in stress signalling is now the preferred suggestion. This is strongly supported by the identification of an abundantly occurring LTP1 in germinating barley seeds, which is covalently modified by the α-ketol 9-hydroxy-10-oxo-12(Z)-octadecanoic acid. This reactive oxylipin is formed by 9-LOX and AOS activity and is specifically trapped by covalent linkage to LTP1 (Bakan et al., 2006). It will be interesting to know whether other LTPs form adducts with other oxylipins or hydrophobic molecules. Another putative candidate is a JA-binding LTP occurring in JA-loaded cells (Buhot et al., 2004)
First crystal structures of enzymes in JA biosynthesis
AOS
So far, there is no crystal structure determined for a higher plant AOS. Interestingly, in corals a fusion protein with LOX and AOS domains has been found (Koljak et al., 1997). These organisms form eicosanoid compounds similar to mammals. The AOS domain has weak activity and a sequence homology to a catalase. Recent crystal-structure analysis has revealed that in the coral AOS the switch from activity of a catalase to that of an AOS is facilitated (Oldham et al., 2005). It is possible that the LOX–AOS fusion protein has been evolved by successive mutations in discrete but interacting LOX and AOS proteins.
AOC
Recently the first crystal structure has been described for the AOC2 of A. thaliana (Hofmann et al., 2006). This is of special interest for understanding JA biosynthesis, since it is in the AOC reaction that the correct enantiomeric form of naturally occurring jasmonates is established. Knowledge of the crystal structure has led to insights into the oxylipin cyclization reaction. The stereoselectivity occurring in AOC catalysis was found to be a steric restriction for the essential substrate isomerization. Surprisingly, AOC2 of A. thaliana was detected as a trimeric protein in crystals and in planta, and forms a hydrophobic binding cavity with two distinct polar patches (Hofmann et al., 2006). The folding pattern and sequence homology suggest that AOC2 is a member of the lipocalin family. This type of protein represents a widely distributed protein family characterized by a six- or eight-stranded β-barrel and a highly conserved motif with functions in the transport of small, hydrophobic molecules (Grzyb et al., 2006). Consequently, implications for functions in cell growth, membrane biogenesis, induction of apoptosis, environmental stress responses and regulation of immune stress responses have been suggested (Charron et al., 2005). The identification of AOC2 as a member of plant lipocalins is exciting and sheds new light on the diverse functions of lipid-based signalling and jasmonates.
OPR1 and OPR3
OPRs belong to a family of flavoproteins identified first with Warburg's old yellow enzyme (OYE). OYEs reduce the C = C double bond of α-,β-unsaturated carbonyl compounds, but only for OPR3 has a physiological function been identified. This enzyme reduces all four 1-stereoisomers of OPDA with preference for the only intermediate in JA biosynthesis, (9S,13S)-OPDA, whereas OPR1 reduces (9R,13R)-OPDA (Schaller et al., 1998, 2000). This specificity for enantiomers has now been explained by the crystal structures of OPR1 (Breithaupt et al., 2001) and OPR3 (Breithaupt et al., 2006). The overall structures of OPR1 and OPR3 are very similarly characterized by a (β/α) barrel fold. The substrate-binding cavity, however, reveals a narrow entrance for OPR1 and a more ‘released’ cavity for OPR3 (Breithaupt et al., 2006). Surprisingly, OPR3 was found as an extraordinary self-inhibited dimer in the crystal structure. A weak association in vitro but a strong dimerization in vivo by a putative phosphorylation at Tyr-364 has been suggested as a regulatory mechanism for OPR3 (Breithaupt et al., 2006). This scenario accords with an activation of pre-existing JA biosynthetic enzymes leading to the early rise in JA before expression of JA biosynthetic genes (Stenzel et al., 2003a, b).
ACX1
In the recently isolated tomato mutant acx1, a single point mutation of the conserved residue Thr138 is substituted by isoleucine (Li et al., 2005). The crystal structure of tomato ACX1 showed an unusual packing arrangement compared to other ACX enzymes (Powers et al., 2006); in tomato ACX1, three monomers form an asymmetric unit.
METABOLITES OF JA
Diversity among naturally occurring jasmonates and structure–activity relationships
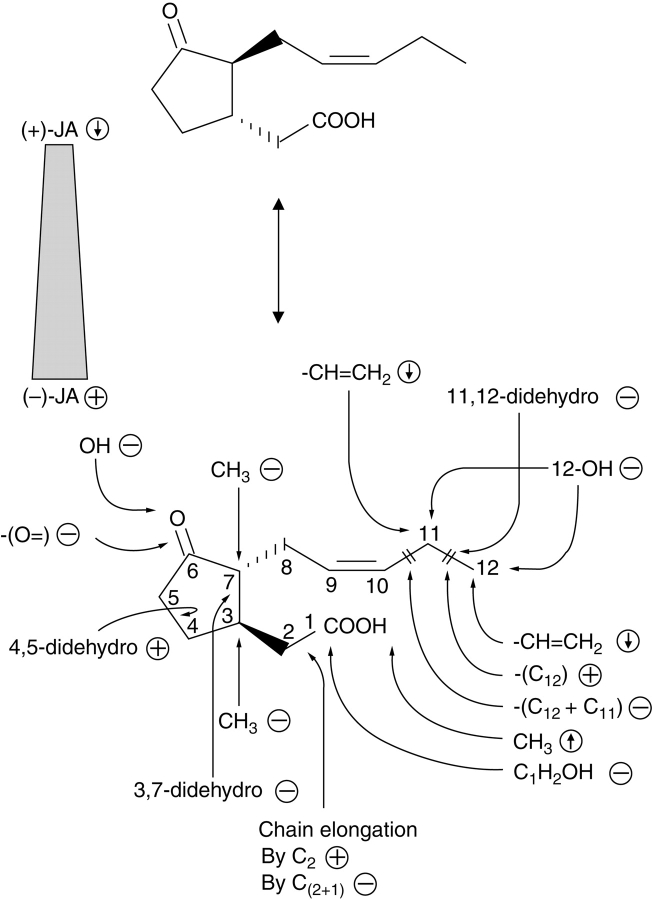
Since the first isolation of JAME as a constituent of the oil of Jasminum grandiflorum (Demole et al., 1962), numerous jasmonate compounds have been detected in diverse plant phyla (Meyer et al., 1984). Jasmonates occur in algae, mosses, fungi, gymnosperms and angiosperms. The capacity to form or to convert various jasmonates is remarkably high in fungi; for example, more than 20 jasmonates were detected in culture filtrate of Fusarium oxysporum (Miersch et al., 1999a), whilst Aspergillus niger grown on liquid medium could form more than 25 jasmonate compounds upon application of JA, 9,10-dihydro-JA and their methyl esters (Miersch et al., 1999c). Some fungi, such as Botryodiplodia theobromae, are able to accumulate up to 500 µg mL−1 of culture medium of (+)-7-iso-JA, the initial product in JA biosynthesis (Miersch et al., 1989). Whereas in fungi the biological function of various jasmonates is unknown, application of jasmonates and their structural analogs to higher plants led to the first insights into their structural requirements for biological activity. Based on different assays, such as alkaloid formation (Blechert et al., 1995), tendril coiling (Blechert et al., 1999), tuber formation (Koda, 1992), or gene expression (Miersch et al., 1999b), the following structural requirements have been found (Fig. 3).
(−)-JA and its derivatives are more active than (+)-derivatives.
An intact pentenyl side-chain is most active, whereas hydroxylation at C-11 or C-12 and reduction between C-11 and C-12 reduce biological activity.
Chain elongation of the carboxylic acid side-chain retains activity only with an even number of C-atoms.
Activity is retained or increased by methylation or conjugation to amino acids.
A cyclopentanone ring carrying a keto group at C-6 is essential.
Fig. 3.
Structure–activity relationships among jasmonates. The various structural analogs and enantiomers were tested with respect to JA-induced gene expression in barley (data from Miersch et al., 1999b). ↓, decreased expression; ↑, increased expression; –, inactive; +, active in gene expression.
More recently, highly active JA compounds such as coronalon (Schüler et al., 2004) and other 6-substituted 4-oxo-indanoyl-isoleucine conjugates have been synthesized (Mithöfer et al., 2004). The construction of these compounds was deduced by modelling based on the structure of the bacterial toxin coronatine. The extremely high biological activity of coronatine has been used as a tool and has led to isolation of the first JA-insensitive mutant of A. thaliana, coi1 (Feys et al., 1994). Recent data have revealed distinct responses for JA and coronatine (Uppalapati et al., 2005).
Metabolic fate of JA
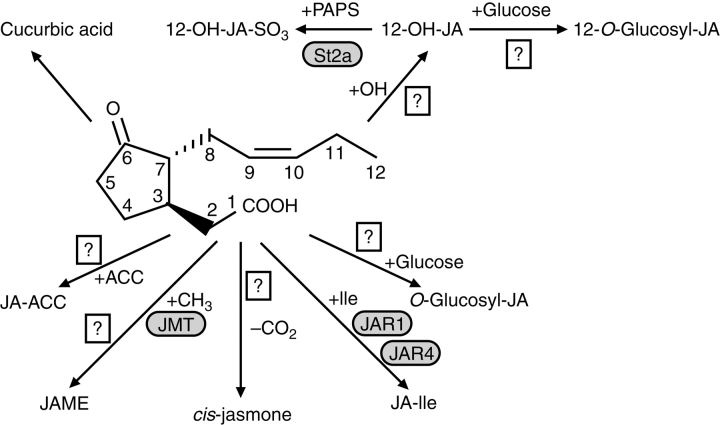
Homoestasis between various metabolites is a common mechanism in plants to sustain the level of active hormones such as auxins, cytokinines, gibberellins and jasmonates. In the case of JA, seven different metabolic routes have been suggested by identification of the corresponding products, but only a few enzymes have been cloned so far (Fig. 4).
Formation of amino acid conjugates by a JA conjugate synthase (JAR1) (Staswick and Tiryaki, 2004) upon adenylation at the carboxylic acid side-chain of JA by the AMP-transferase activity of JAR1.
Methylation of JA by a JA-specific methyl transferase (Seo et al., 2001).
Hydroxylation at C-11 or C-12 of the pentenyl side-chain and subsequent O-glucosylation (Sembdner and Parthier, 1993; Swiatek et al., 2004) or sulfation (Gidda et al., 2003).
Decarboxylation of JA to cis-jasmone (Koch et al., 1997).
Formation of cucurbic acids by reduction of the keto group of the cyclopentanone ring (Sembdner and Parthier, 1993).
Formation of jasmonoyl-1-β-glucose, jasmonoyl-1-β-gentiobiose and hydroxyjasmonoyl-1-β-glucose (Swiatek et al., 2004).
Conjugation of the ethylene precursor ACC to JA (Staswick and Tiryaki, 2004).
Fig. 4.
Metabolic fate of jasmonic acid. The carboxylic acid side-chain can be conjugated to the ethylene precursor 1-amino cyclopropane-1-carboxylic acid (ACC), methylated by JA methyl transferase (JMT), decarboxylated to cis-jasmone, conjugated to amino acids such as Ile by JA amino acid synthase (Arabidopsis, JAR1; tobacco, JAR4) or glucosylated. The pentenyl side-chain can be hydroxylated in positions C-11 or C-12. In the case of 12-OH-JA, glucosylation or sulfation are subsequent reactions. Reduction of the keto group of the pentenone ring can lead to cucurbic acid.
JA amino acid conjugates are permanent constituents of plant tissues (Sembdner and Parthier, 1993) and their levels increase upon osmotic stress (Kramell et al., 1995). In barley leaves applied JA amino acid conjugates are active per se without cleavage (Kramell et al., 1997). Many of them induce a subset of genes compared with JA or JAME (Kramell et al., 2000). Among the various conjugates, JA-Ile mainly accumulates in leaves (Kramell et al., 1997), flowers (Hause et al., 2000) and mycorrhizal roots (Hause et al., 2002, 2007). This preference is reflected by the enzymatic properties of the JA amino acid conjugate synthase (JAR1), and the jar1 mutant can be complemented by JA-Ile (Staswick and Tiryaki, 2004). JAR1 belongs to a large gene family encoding enzymes that adenylate a carboxylic acid-containing substrate followed by exchange with a second substrate. In the case of JAR1, AMP is exchanged with an amino acid (Staswick et al., 2002; Staswick and Tyriaki, 2004). Similar reactions are catalysed by auxin conjugate synthase (Staswick et al., 2005) and 4-CL-like CoA ligases activating OPDA or OPC-8 to the corresponding CoA esters (Schneider et al., 2005; Koo et al., 2006). The tobacco homologue of JAR1 has recently been cloned as JAR4 from N. attenuata (Kang et al., 2006). Interestingly, JAR4-silenced plants were shown to be more susceptible to Manduca sexta attack, indicating the role of JA-Ile in plant defence. Another JA metabolite highly active in plant–insect interactions via emission of volatiles is cis-jasmone (Birkett et al., 2000; Bruce et al., 2003).
ACTION OF JASMONATES
Jasmonates as novel anti-cancer agents
Whilst growth-inhibitory effects of salicylate on various types of cancer have been known for a long time, only recently has it been shown that cell lines of a wide spectrum of malignancies can exhibit selective cytotoxic effects of JAME (Flescher, 2005). These effects were independent of transcription, translation and p53 expression. Mitochondria seem to be one target. In hep3B hepatoma cells, JAME-induced swelling of mitochondria and release of cytochrome c was observed, this being characteristic for an apoptotic pathway (Kim et al., 2004; Rotem et al., 2005). The selectivity of jasmonates on cancer cells was surprisingly high. Another target of jasmonates was detected in human myeloid leukemia cells. Here, jasmonates exert a growth-inhibitory and differentiation-induced effect by activating a NO-activated protein kinase (MAPK) pathway in a cAMP-independent manner (Ishi et al., 2004). Among the various jasmonates tested, a methyl-4,5-didehydrojasmonate was 30-times more active than JAME. Jasmonates have not been detected so far in mammalian cells, but they are ubiquitous in plants. Some plant extracts have for a long time been used as therapeutic agents in cancer therapy. It will be interesting to see whether such plants contain high levels of jasmonates.
Jasmonates as signals in biotic and abiotic stresses
Solanaceae
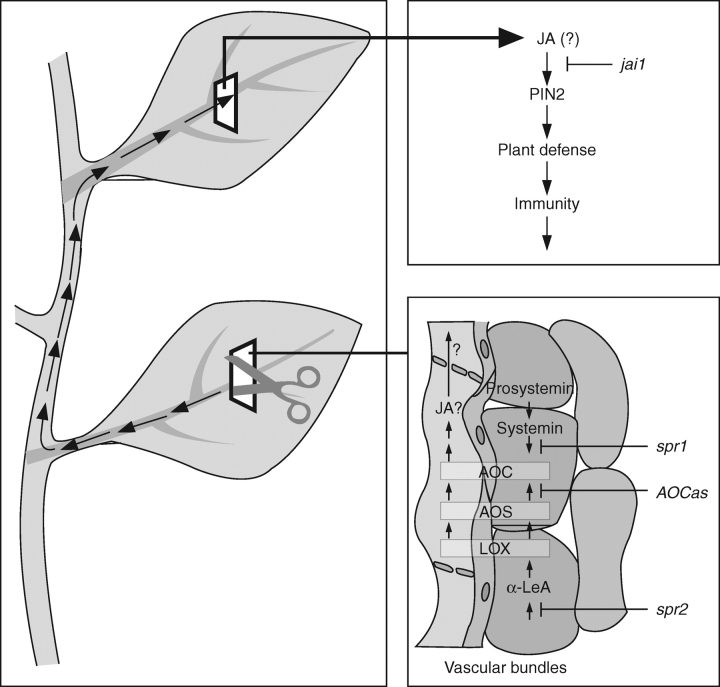
One of the best-studied signal-transduction pathways of jasmonates is that of wound-response, which was initially studied mainly in tomato. Briefly, a sequential action of the 18-aa peptide systemin, cleaved from prosystemin upon local wounding, activates JA biosynthetic enzymes such as AOC and leads to local rise in JA (Fig. 5). An amplification in JA formation has been suggested due to JA-dependent PROSYSTEMIN expression, a systemin-dependent AOC expression, and their common location in vascular bundles (Stenzel et al., 2003a; Narváez-Vásquez and Ryan, 2004). It is possible that JA acts as a systemic signal, leading also to systemic expression of genes encoding proteinase inhibitors (PINs) and other foliar compounds with negative effects on herbivore performance. Consequently the plant becomes immunized against a subsequent herbivore attack. Grafting experiments with JA-deficient and JA-signalling mutants in G. Howe's laboratory have shown that JA signalling – but not JA biosynthesis – is necessary in the systemic leaf. The numerous data on the wound response pathway have been reviewed repeatedly, including discussions on cross-talk to other signals, further elements of the wound-response pathway such as MAP kinases, and additional herbivore-induced compounds such as volatiles and their consequences for diverse plant–insect interactions, together with overlap to plant pathogen–interactions (Farmer et al., 2003, Howe, 2004; Schilmiller and Howe, 2005; Wasternack, 2006). Here, I want to mention only few important new developments.
Fig. 5.
Local and systemic wound response in tomato and cell-type-specific occurrence of AOC, AOS and LOX (after Hause et al., 2003; modified from Wasternack et al., 2006).
For a long time, an active systemin was found only in tomato (Solanum lycopersicon), but not in tobacco. Even in the closely related Solanum nigrum the systemin-homolog of Solanum lycopersicon is not active in defence responses (Schmidt and Baldwin, 2006). In tobacco, hydroxyproline-rich homologs of tomato systemin have been found that do not induce PIN expression in tomato (Pearce et al., 2001; Pearce and Ryan, 2003). Recently, peptides active in wound-signalling have been identified outside the Solanaceaen species. In A. thaliana a 23-aa peptide (AtPEP1) processed from a 92-aa precursor is part of the innate immune response (Huffaker et al., 2006). PROPEP1 expression is induced by wounding, JA and ethylene leading to H2O2 formation and PDF1·2 expression. Arabidopsis thaliana plants over-expressing PROPEP1 show altered root development and enhanced resistance to the root pathogen Pythium irregulare. PROPEP1 belongs to a small gene family, and AtPEP1 exhibits striking similarities in structure and function to tomato systemin (Huffaker et al., 2006). The receptor of AtPEP1 has been isolated and characterized as a functional LRR receptor, which is active even if transformed in tobacco (Yamaguchi et al., 2006). The link between defence against pathogens and wound signalling provided by AtPEP1 suggests that receptor-mediated defence-signalling peptides generated upon wounding may amplify defence against pathogens in A. thaliana (De Vos et al., 2006). Resistance against pathogens can also be increased by modified emission of green-leaf volatiles initially released upon herbivory (Shiojiri et al., 2006).
The JA-mediated wound response following herbivore attack leads to indirect (volatile emissions) and direct (formation of defence proteins and small compounds) defence (cf. reviews by Halitschke and Baldwin, 2004; Howe, 2004; Pieterse et al., 2006), and even mechanical wounding is sufficient to give a volatile pattern similar to that caused by herbivore attack (Mithöfer et al., 2005). Among the defence proteins formed in tomato there are PINs that are thought to inhibit proteolytic degradation in the midgut of herbivores. Herbivore-induced proteins in tomato also include leucine aminopeptidases, threonine deaminase (TD) and arginase (Walling, 2000; Chen et al., 2004). Their direct role in the digestive tract of herbivores has recently been shown (Chen et al., 2005). All these wound-induced plant proteins were detected in the midgut, and TD showed extremely high activity (Chen et al., 2005). TD catalyzes the initial step in isoleucine biosynthesis of plants and bacteria and is negatively regulated by Ile via a C-terminal regulatory domain. Interestingly, the TD occurring in the midgut of a herbivore following infestation on tomato leaves lacked its regulatory domain. Consequently, threonine degradation could take place in the presence of high Ile concentrations. Correspondingly, larvae infested on transgenic lines over-expressing arginase exhibited high arginase activity in their midgut and produced much less weight (Chen et al., 2005). Obviously, catabolism of higher amino acids in the midgut of herbivores following their infestation on leaves acts synergistically to the deterrent role of PINs. The appearance within the midgut of a distinct set of proteins with altered properties, as in case of TD, and their anti-nutritional effects by perturbing amino acid homoestasis may be evolved in plants such as Solanacean species attacked by herbivores. Dual strategies seem to occur. The TD can exert an anti-nutritive defence in the midgut of herbivores, and within the plant contributes to the formation of Ile as an essential substrate for synthesis of JA-Ile, a highly active wound-response signal. In TD-silenced tobacco plants, JA-Ile-dependent defences are impaired (Kang et al., 2006). It is possible that chemically based and developmentally based defence strategies co-evolved: the central element of JA-regulated gene expression, COI1, an F-box protein functioning in the SCF proteasome pathway, is essential for proper trichome development of tomato (Li et al., 2004), and PIN2 of Solanum americanum is constitutively expressed in glandular trichomes and positively regulates trichome density (Liu et al., 2006).
Arabidopsis
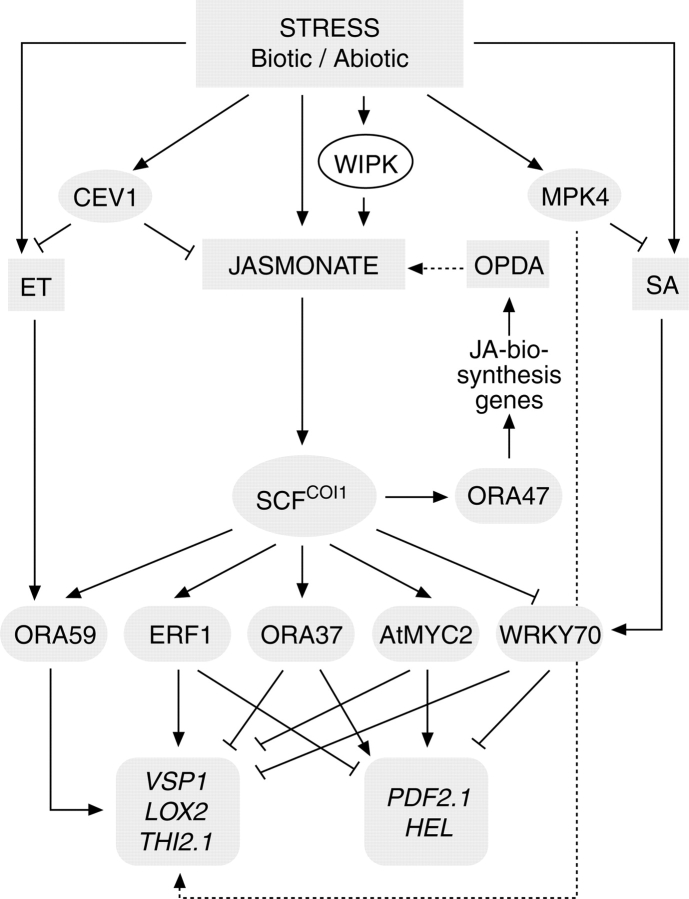
Analyses of transcript accumulation upon wounding or pathogenic attack have greatly improved our understanding of plant responses to biotic and abiotic stress (Pieterse et al., 2006). Mutants in JA biosynthesis and JA signalling have become an important tool in dissecting signalling pathways and in determining signalling properties of JA and related compounds (Turner et al., 2002; Lorenzo and Solano, 2005; Delker et al., 2006; Wasternack, 2006). Initial expression analyses with the JA-deficient but OPDA-accumulating opr3 mutant revealed OPDA-specific gene expression (Stintzi et al., 2001). Following the first large-scale transcription analysis, genes could be grouped into COI1-dependent and COI1-independent types (Reymond et al., 2000, 2004). Recent transcription analyses have shown distinct and overlapping groups of genes being expressed dependently and independently with respect to COI1, OPDA and JA (Devoto et al., 2005; Taki et al., 2005). Similar differences and overlaps in action have been found at the level of transcription factors acting in JA-/OPDA-dependent processes (Fig. 6). Among transcription factors acting downstream of JA in stress responses are the ethylene response factor 1 (ERF1), the bHLHzip-type transcription factor ATMYC2 (Lorenzo et al., 2004), WRKY70 and the newly found family of ORAs (identified by J. Memelink's group). ORAs are the Arabidospis homologs of ORCAs initially identified in Catharanthus roseus cell suspension cultures (Memelink et al., 2001). Among them, ORA47 is a COI1-dependent positive regulator of JA biosynthesis, whereas ORA59, ERF1, ORA37, MYC2 and WRKY70 act positively or negatively on different groups of defence genes (Fig. 6). The antagonistic action of MYC2 and ERF1 may cause the independence between wound signalling and pathogen-defence signaling (Lorenzo et al., 2004; Lorenzo and Solano, 2005).
Fig. 6.
Transcription factors involved in signalling pathways of JA and cross-talk to ethylene and SA (adapted from Pre, 2006, and Delker, 2007).
Jasmonates in development
Some aspects on role of jasmonates in developmental processes are summarized in Table 1.
Table 1.
Jasmonates in development
| Developmental process | Putative signal | Alteration/species | Reference for initial observation |
|---|---|---|---|
| Root growth | JA, JA-Ile | Inhibition | Dathe et al. (1981); Staswick et al. (1992) |
| Seed germination | JA | Inhibition | Corbineau et al. (1988) |
| Tuber formation | 12-OH-JA | Induction/potato | Yoshihara et al. (1989) |
| Tendril coiling | OPDA | Stimulation/Bryonia | Falkenstein et al. (1991) |
| Nyctinasty | 12-OH-JA Glu | Stimulation/Albizzia | Nakamura et al. (2006) |
| Trichome formation | JA | Induction/tomato | Li et al. (2004) |
| Senescence | JA | Stimulation | Ueda and Kato (1980) |
| Flower development | |||
| Anther development + dehiscence | JA | Induction/Arabidopsis | McConn and Browse (1996) |
| Female organ development | JA | Induction/tomato | Li et al. (2004) |
| Filament elongation | JA | Induction/Arabidopsis | Stintzi and Browse (2000) |
Root growth inhibition
This was one of the first physiological effects detected for JA (Dathe et al., 1981; Staswick et al, 1992). JA or its methyl ester supplemented to the growth medium are active in a picomolar range. Consequently, JA-insensitive mutants such as coi1, jin1, or jar1 are similar in root length upon JAME treatment compared with the untreated wild type (Fig. 7). In contrast, mutants such as cev1, cet1, joe2 or cex1 that are characterized by constitutive over-expression of JA-responsive genes due to constitutively elevated JA levels have reduced root length and a stunted growth phenotype similar to JA-treated plants (Ellis et al., 2002). Most mutants that are affected in components of JA- and COI-dependent signal transduction (including transcription factors such as MYC2, the SCF proteasom and its upstream-acting proteins such as AXR1, JAI4/SGT1b or MPK4) exhibit a reduction in root-growth inhibition (Fig. 6; cf. Browse, 2005; Wasternack, 2006).
Fig. 7.
Root growth inhibition by 100 µm methyl jasmonate in wild-type (Col) and the JA-insensitive mutant coi1-16 (photograph: C. Delker).
Tuber formation
This has been known for a long time to be induced by 12-OH-JA (tuberonic acid; Yoshihara et al., 1989), the glucoside of which has been suggested to be the transport form. Initially, 12-OH-JA was found mainly in Solanaceaen species. More recently, it has been detected in A. thaliana and identified as the substrate of a sulfotransferase (AtST2a), one of the 18 sulfotransferases occurring in the Arabidopsis genome (Gidda et al., 2003). A recent screening revealed abundant occurrence of 12-OH-JA in different tissues and plant species, for example seeds of Glycine max (O. Miersch and C. Wasternack, unpubl. res.).
Tendril coiling and touch
Touch-mediated processes in plants are initiated by specific organs, such as tactiles. In the case of tendril coiling in Bryonia, a highly sensitive JA-dependent mechanism has been found (Falkenstein et al., 1991; Weiler, 1997). Inspection of the response kinetics revealed that OPDA was more active than JA, and this was substantiated by the endogenous rise of OPDA upon touch (Stelmach et al., 1998; Blechert et al., 1999). More recently, an excellent approach combining cell biology, chemistry and biochemistry has given indications as to how nyctinasty takes place in Albizzia plants. The leaf movement of Albizzia was shown to be dependent on a specific enantiomer of 12-OH-JA-O-glucoside, which binds with cell-type-specificity to the motor cells of leaves of Albizzia julibrissica (Nakamura et al., 2006).
Flower development
This is essentially linked to intact JA biosynthesis and JA signalling. Clear evidence for a JA-dependent phenotype in flower development has come from Arabidospis mutants affected in JA biosynthesis, such as dad1, fad3-2fad7-2fad8, dde2-2, dde1, opr3 and aim1 as well as mutants in JA-signalling, such as coi1. All these mutants are male-sterile due to delayed anther development and incomplete anther dehiscence, or are impaired in filament elongation (cf. Turner et al., 2002; Delker et al., 2006). Surprisingly, in Arabidopsis an impaired F-box protein COI1 leads to male sterility, whereas its tomato homolog is essential for proper female fertility (Xie et al., 1998; Li et al., 2004).
The reduced filament elongation in JA-deficient mutant such as dad1 and opr3 corresponds to reduced JA levels in filaments of the double mutant arf6/arf8. These mutants are impaired in two auxin response factors responsible for proper filament elongation, and they indicate auxin signalling upstream of JA (Nagpal et al., 2005). Recently, a large-scale analysis of gene expression in opr3 stamen development following JA treatment has led to an excellent overview on JA-dependent processes in stamen development (Mandaokar et al, 2006). Among 13 transcription factors identified to function in stamen maturation, two of them, MYB21 and MYB24, are JA-inducible and mediate the jasmonate response during stamen development (Mandaokar et al, 2006).
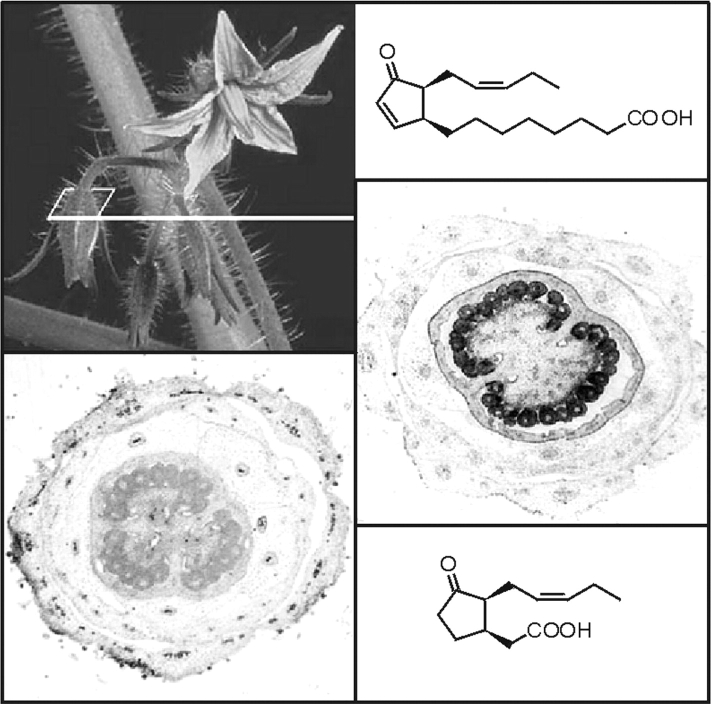
Specific roles for JA and other oxylipins are also suggested by the abundant occurrence of AOC protein in ovules of tomato flowers and by the distinct oxylipin signature in different tomato-flower organs (Hause et al., 2000) (Fig. 8). The total amount and the ratio between OPDA and JA can be increased by constitutive over-expression of AOC (Miersch et al., 2004). It will be interesting to see whether the oxylipin signature is responsible for distinct steps in flower development, flower abscission, embryo development and seed formation.
Fig. 8.
The allene oxide cyclase (AOC) that establishes the correct enantiomeric structure of the cyclopentanone ring of jasmonate is encoded by a single copy gene in tomato, which is specifically expressed in ovules of young flower buds as revealed by AOC promoter activity tests (staining, left bottom) and immunocytological detection of AOC protein (black staining, right). The structure of jasmonic acid (bottom) and its precursor 12-oxophytodienoic acid (top) is shown.
Senescence
One of the first biological activities observed for jasmonates was the senescence-promoting effect (Ueda and Kato, 1980; Parthier, 1990). Although it occurs ubiquitously following JA treatment, monocotyledonous plants are more sensitive. Senescence is the last stage in plant development. Consequently, genes expressed during leaf senescence (senescence-associated genes, SAGs) code for proteins with functions in sink/source relationships, photosynthesis, intermediary metabolism, proteolysis and plant defence (cf. Wasternack and Hause, 2002; Wasternack, 2004). The role of JA in senescence is linked to (1) downregulation of housekeeping proteins encoded by photosynthetic genes and (2) upregulation of genes active in defence reactions against biotic and abiotic stresses (Wasternack, 2004, 2006). Several microarray analyses have led to more detailed information, including cross-talk between plant hormones (Lin and Wu, 2004; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006). A direct role of JA in leaf senescence of A. thaliana was shown by expression-analysis of JA biosynthetic genes (He et al., 2002). Another molecular explanation for the role of JA in leaf senescence was given by identification of a novel nuclear-localized CCCH-type zinc finger protein in rice (Kong et al., 2006). This protein acts as a negative regulator of the JA pathway and leads to a delay in the onset of leaf senescence. Another negative regulator of senescence is ORE9, an F-box protein (Woo et al., 2001). However, there are also JA-independent processes in leaf senescence, as shown by identification of NAC family transcription factors that have an important role in leaf senescence (Guo and Gan, 2006).
CONCLUSIONS
In the last few years interesting new data on JA biosynthesis and action have appeared. Crystal structures for several JA biosynthetic enzymes now allow molecular explanations for substrate specificity. Fatty acid β-oxidation enzymes and 4-Cl-like CoA ligases have been identified to function in JA biosynthesis. New esterified oxylipin derivatives such as the arabidopsides suggest new functions in defence reactions. Occurrence and specific functions of JA metabolites have been detected. Transcription factors acting in JA signalling have shed new light on cross-talk in signalling pathways that are active following biotic and abiotic stresses, as well as in developmental processes. Finally, it is possible that a putative role of jasmonates in cancer formation may be substantiated.
Future work will address questions on specific actions of JA metabolites, on JA and oxylipin perception, on MAP kinases active in JA signalling, on the diversity of action of JA in plant–plant and plant–microbe interactions; and, finally, cell- and organ-specific functions of jasmonates in plant development will be analysed.
ACKNOWLEDGEMENTS
Financial support for the research of my group related to this review was given by the Deutsche Forschungsgemeinschaft (SPP 1067, WA 875/3–1/2/3; SFB 363, project C5; SFB 648, project C2). I thank C. Dietel for typing the manuscript and PD. Dr. B. Hause for critical reading.
APPENDIX
Abbreviations used in the text
| ACX | Acyl-CoA oxidase |
| α-LeA | α-linolenic acid |
| AOC | Allene oxide cyclase |
| AOS | Allene oxide synthase |
| ERF1 | Ethylene response factor 1 |
| HPL | Hydroperoxide lyase |
| HPOD | Hydroperoxyoctadecadienoic acid |
| HPOT | Hydroperoxyoctadecatrienoic acid |
| IP3 | Inositol triphosphate |
| JA | Jasmonic acid |
| JAME | Jasmonic acid methyl ester |
| JAR1 | JA conjugate synthase |
| JIP | Jasmonate-induced protein |
| LOX | Lipoxygenase |
| LTP | Lipid transfer protein |
| MFP | Multifunctional protein |
| OPDA | cis(+)-12-oxophytodienoic acid |
| OPR | OPDA reductase |
| OYE | Warburg's old yellow enzyme |
| PA | Phosphatidic acid |
| PIN | Proteinase inhibitor |
| PUFA | Polyunsaturated fatty acid |
| SAG | Senescence-associated gene |
| TD | Threonine deaminase |
LITERATURE CITED
- Andersson MX, Hamberg M, Kourtchenko O, Brunnström Å, McPhail KL, Gerwick WH, et al. Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: formation of a novel oxo-phytodienoic acid containing galactolipid, arabidopside E. Journal of Biological Chemistry. 2006;281:31528–31537. doi: 10.1074/jbc.M604820200. [DOI] [PubMed] [Google Scholar]
- Bakan B, Hamberg M, Perrocheau L, Maume D, Rogniaux H, Tranquet O, et al. Specific adduction of plant lipid transfer protein by an allene oxide generated by 9-lipoxygenase and allene oxide synthase. Journal of Biological Chemistry. 2006;281:38981–38988. doi: 10.1074/jbc.M608580200. [DOI] [PubMed] [Google Scholar]
- Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. Chewing the fat: β-oxidation in signalling and development. Trends in Plant Science. 2006;11:124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Munnik T. The role of phospholipase D in plant stress responses. Current Opinion in Plant Biology. 2006;9:515–522. doi: 10.1016/j.pbi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Birkett MA, Campbell CAM, Chamberlain K, Guerrieri E, Hick AJ, Martin JL, et al. New roles for cis-jasmone as an insect semiochemical and in plant defense. Proceedings of the National Academy of Sciences of the USA. 2000;97:9329–9334. doi: 10.1073/pnas.160241697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan TM, Mueller MJ, Xia Z-Q, Zenk MH. The octadecanoic pathway: signal molecules for the regulation of secondary pathways. Proceedings of the National Academy of Sciences of the USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert S, Bockelmann C, Füβlein M, von Schrader T, Stelmach BA, Niesel U, Weiler EW. Structure activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta. 1999;207:470–479. [Google Scholar]
- Blée E. Impact of phyto-oxylipins in plant defense. Trends in Plant Sciences. 2002;7:315–321. doi: 10.1016/s1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- Breithaupt C, Strasser J, Breitinger U, Huber R, Macheroux P, Schaller A. X-Ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE. Structure. 2001;9:419–429. doi: 10.1016/s0969-2126(01)00602-5. [DOI] [PubMed] [Google Scholar]
- Breithaupt C, Kurbauer R, Lilie H, Schaller A, Strassner J, Huber R, Macheroux P, Clausen T. Crystal structure of 12-oxophytodienoate reductase 3 from tomato: self-inhibition by dimerization. Proceedings of the National Academy of Sciences of the USA. 2006;103:14337–14342. doi: 10.1073/pnas.0606603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. Jasmonate: an oxylipin signal with many roles in plants.Vitamines and Hormones. 2005;72:431–456. doi: 10.1016/S0083-6729(05)72012-4. [DOI] [PubMed] [Google Scholar]
- Bruce TJ, Martin JL, Pickett JA, Pye BJ, Smart LE, Wadhams LJ. Cis-Jasmone treatment induces resistance in wheat plants against the grain aphi, Sitobion avenae (Fabricius) (Homoptera: Aphididae) Pest Managment Sciences. 2003;59:1031–1036. doi: 10.1002/ps.730. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Bücking H, Förster H, Stenzel I, Miersch O, Hause B. Applied jasmonates accumulate extracellularly in tomato, but intracellularly in barley. FEBS Letters. 2004;562:45–50. doi: 10.1016/S0014-5793(04)00178-4. [DOI] [PubMed] [Google Scholar]
- Buhot N, Gomès E, Milat M-L, Ponchet M, Marion D, Lequeu J, et al. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Molecular Biology of the Cell. 2004.;15:5047–5052. doi: 10.1091/mbc.E04-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buseman CM, Tamura P, Sparks AA, Baughman EJ, Maatta S, Zhao J, et al. Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiology. 2006;142:28–39. doi: 10.1104/pp.106.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J-BF, Quellet F, Pelletier M, Danyluk J, Chauve C, Sarhan F. Identification, expression, and evolutionary analyses of plant lipocalins. Plant Physiology. 2005;139:2017–2028. doi: 10.1104/pp.105.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaig BC, Melotto M, He SY, Howe GA. Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. Journal of Biological Chemistry. 2004;279:45998–46007. doi: 10.1074/jbc.M407151200. [DOI] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proceedings of the National Academy of Sciences of the USA. 2005;102:19237–19242. doi: 10.1073/pnas.0509026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbineau F, Rudnicki RM, Come D. The effects of methyl jasmonate on sunflower (Helianthus annuus L.) seed germination and seedling development. Plant Growth Regulation. 1988;7:157–169. [Google Scholar]
- Cruz Castillo M, Martínez C, Buchala A, Métraux JP, León J. Gene-specific involvement of β-oxidation in wound-activated responses in Arabidopsis. Plant Physiology. 2005;135:85–94. doi: 10.1104/pp.104.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe W, Rönsch H, Preiss A, Schade W, Sembdner G, Schreiber K. Endogenous plant hormones of the broad bean, Vicia faba L. (−)-Jasmonic acid, a plant growth inhibitor in pericarp. Planta. 1981;155:530–535. doi: 10.1007/BF00385537. [DOI] [PubMed] [Google Scholar]
- Delker C. Germany: Martin-Luther-Universität Halle-Wittenberg; 2007. Jasmonatbiosynthese in Arabidopsis thaliana – Charakterisierung der Allenoxidcyclase-Genfamilie und von Mutanten der Fettsäure-β-Oxidation. [Google Scholar]
- Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C. Jasmonate biosynthesis in Arabidopsis thaliana – enzymes, products, regulation. Plant Biology. 2006;8:297–306. doi: 10.1055/s-2006-923935. [DOI] [PubMed] [Google Scholar]
- Demole E, Lederer E, Mercier D. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant charactéristique de lèssence de jasmin. Helvetica Chimica Acta. 1962;45:675–685. [Google Scholar]
- De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ. Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiology. 2006;142:352–363. doi: 10.1104/pp.106.083907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang H-S, Chilcott C, Zhu T, Turner JG. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Molecular Biology. 2005;58:497–513. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dörmann P. Membrane lipids. In: Murphy DJ, editor. Plant lipids: biology, utilization and manipulation. Oxford: Blackwell Publishing Ltd; 2005. pp. 123–161. [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. The Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenstein E, Groth B, Mithöfer A, Weiler EW. Methyljasmonate and α-linolenic acid are potent inducer of tendril coiling. Planta. 1991;185:316–322. doi: 10.1007/BF00201050. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proceedings of the National Academy of Sciences of the USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Current Opinion in Plant Biology. 2003;6:372–378. doi: 10.1016/s1369-5266(03)00045-1. [DOI] [PubMed] [Google Scholar]
- Feussner I, Wasternack C. The lipoxygenase pathway. Annual Review of Plant Biology. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate and resistant to a bacterial pathogen. The Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher E. Jasmonates – a new family of anti-cancer agents. Anticancer Drugs. 2005;16:911–916. doi: 10.1097/01.cad.0000176501.63680.80. [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Itoh A, Howe GA. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450 involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiology. 2001;125:306–317. doi: 10.1104/pp.125.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda KS, Miersch O, Schmidt J, Wasternack C, Varin L. Biochemical and molecular characterization of a hydroxy-jasmonate sulfotransferase from Arabidopsis thaliana. Journal of Biological Chemistry. 2003;278:17895–17900. doi: 10.1074/jbc.M211943200. [DOI] [PubMed] [Google Scholar]
- Grun C, Berger S, Matthes D, Mueller M. Early accumulation of non-enzymatically synthesized oxylipins in Arabidopsis thaliana after infection with Pseudomonas syringae. Functional Plant Biology. 2007;34:65–71. doi: 10.1071/FP06205. [DOI] [PubMed] [Google Scholar]
- Grzyb J, Latowski D, Strzałka K. Lipocalins – a family portrait. Journal of Plant Physiology. 2006;163:895–915. doi: 10.1016/j.jplph.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Gundlach H, Müller MJ, Kutchan TM, Zenk MH. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proceedings of the National Academy of Sciences of the USA. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT. Jasmonates and related compounds in plant–insect interactions. Journal of Plant Growth Regulation. 2004;23:238–245. [Google Scholar]
- Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C. Tissue-specific oxylipin signature of tomato flowers – allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. The Plant Journal. 2000;24:113–126. doi: 10.1046/j.1365-313x.2000.00861.x. [DOI] [PubMed] [Google Scholar]
- Hause B, Maier W, Miersch O, Kramell R, Strack D. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiology. 2002;130:1213–1220. doi: 10.1104/pp.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Hause G, Kutter C, Miersch O, Wasternack C. Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant & Cell Physiology. 2003;44:643–648. doi: 10.1093/pcp/pcg072. [DOI] [PubMed] [Google Scholar]
- Hause B, Mrosk C, Isayenkov S, Strack D. Jasmonates in arbuscular mycorrhizal interactions. Phytochemistry. 2007;68:101–110. doi: 10.1016/j.phytochem.2006.09.025. [DOI] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiology. 2002;128:876–884. doi: 10.1104/pp.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu Y, Goto N, Hasegawa K, Shigemori H. Arabidopsides A and B, two new oxylipins from Arabidopsis thaliana. Tetrahedron Letters. 2003;44:5553–5556. [Google Scholar]
- Hisamatsu Y, Goto N, Sekiguchi M, Hasegawa K, Shigemori H. Oxylipins arabidopsides C and D from Arabidospis thaliana. Journal of Natural Products. 2005;68:600–603. doi: 10.1021/np0495938. [DOI] [PubMed] [Google Scholar]
- Hisamatsu Y, Goto N, Hasegawa K, Shigemori H. Senescence-promoting effect of arabidopside A. Zeitschrift für Naturforschung. 2006;61:363–366. doi: 10.1515/znc-2006-5-611. [DOI] [PubMed] [Google Scholar]
- Hofmann E, Zerbe P, Schaller F. The crystal structure of Arabidospis thaliana allene oxide cyclase: insights into the oxylipin cyclization reaction. The Plant Cell. 2006;18:3201–3217. doi: 10.1105/tpc.106.043984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung E, Walter M, Kühn H, Feussner I. Conversion of cucumber linoleate 13 − lipoxygenase to a 9-lipoxygenating species by site-directed mutagenesis. Proceedings of the National Academy of Sciences of the USA. 1999;96:4192–4197. doi: 10.1073/pnas.96.7.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA. Jasmonates as signals in the wound response. Journal of Plant Growth Regulation. 2004;23:223–237. [Google Scholar]
- Howe GA, Schilmiller AL. Oxylipin metabolism in response to stress. Current Opinion in Plant Biology. 2002;5:230–236. doi: 10.1016/s1369-5266(02)00250-9. [DOI] [PubMed] [Google Scholar]
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE. Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiology. 2000;123:711–724. doi: 10.1104/pp.123.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences of the USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KH, Belfield EJ, Ashton R, Fairhurst SA, Göbel C, Stumpe M, Feussner I, Casey R. Allene oxide synthase from Arabidopsis thaliana (CYP74A1) exhibits dual specificity that is regulated by monomer-micelle association. FEBS Letters. 2006;580:4188–4194. doi: 10.1016/j.febslet.2006.06.075. [DOI] [PubMed] [Google Scholar]
- Ishi Y, Kiyota H, Sakai S, Honma Y. Induction of differentiation of human myeloid leukemia cells by jasmonates, plant hormones. Leukemia. 2004;18:1413–1419. doi: 10.1038/sj.leu.2403421. [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kwai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation. The Plant Cell. 2001;13:2191–2209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT. Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. The Plant Cell. 2006;18:3303–3320. doi: 10.1105/tpc.106.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;26:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SY, Oh SY, Han SI, Park HG, Yoo MA, Kang HS. Methyl jasmonate induces apoptosis through induction of Bax/Bcl-XS and activation of caspase-3 via ROS production in A549 cells. Oncology Reports. 2004;12:1233–1238. [PubMed] [Google Scholar]
- Koch T, Bandemer K, Boland W. Biosynthesis of cis-jasmone: a pathway for the inactivation and the disposal of the plant stress hormone jasmonic acid to the gas phase? Helvetica Chimica Acta. 1997;80:838–850. [Google Scholar]
- Koda Y. The role of jasmonic acid and related compounds in the regulation of plant development. International Reviews in Cytology. 1992;135:155–199. doi: 10.1016/s0074-7696(08)62040-9. [DOI] [PubMed] [Google Scholar]
- Koljak R, Boutaud O, Shieh B-H, Samel N, Brash AR. Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiology. 2006;141:1376–1388. doi: 10.1104/pp.106.082941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Chung HS, Kobayashi Y, Howe GA. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. Journal of Biological Chemistry. 2006;281:33511–33520. doi: 10.1074/jbc.M607854200. [DOI] [PubMed] [Google Scholar]
- Kramell R, Atzorn R, Schneider G, Miersch O, Brückner C, Schmidt J, Sembdner G, Parthier B. Occurrence and identification of jasmonic acid and its amino acid conjugates induced by osmotic stress in barley leaf tissue. Journal of Plant Growth Regulation. 1995;14:29–36. [Google Scholar]
- Kramell R, Miersch O, Hause B, Ortel B, Parthier B, Wasternack C. Amino acid conjugates of jasmonic acid induce jasmonate-responsive gene expression in barley (Hordeum vulgare L.) FEBS Letters. 1997;414:197–202. doi: 10.1016/s0014-5793(97)01005-3. [DOI] [PubMed] [Google Scholar]
- Kramell R, Miersch O, Atzorn R, Parthier B, Wasternack C. Octadecanoid-derived alteration of gene expression and the ‘oxylipin signature’ in stressed barley leaves – implications for different signalling pathways. Plant Physiology. 2000;123:177–186. doi: 10.1104/pp.123.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubigsteltig I, Laudert D, Weiler EW. Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta. 1999;208:463–471. doi: 10.1007/s004250050583. [DOI] [PubMed] [Google Scholar]
- Laudert D, Schaller F, Weiler EW. Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta. 2000;211:163–165. doi: 10.1007/s004250000316. [DOI] [PubMed] [Google Scholar]
- Li C, Schilmiller AL, Liu GL, Lee GI, Jayanty S, Sageman C, et al. Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. The Plant Cell. 2005;17:987–999. doi: 10.1105/tpc.104.029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. The Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liavonchanka A, Feussner I. Lipoxygenases: occurrence, functions and catalysis. Journal of Plant Physiology. 2006;163:348–357. doi: 10.1016/j.jplph.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Lin J-F, Wu S-H. Molecular events in senescing Arabidopsis leaves. The Plant Journal. 2004;39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Xia K-F, Zhu J-C, Deng Y-G, Huang X-L, Hu B-L, Xu X, Xu Z-F. The nightshade proteinase inhibitor IIb gene is constitutively expressed in glandular trichomes. Plant & Cell Physiology. 2006;47:1274–1284. doi: 10.1093/pcp/pcj097. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Solano R. Molecular players regulating the jasmonate signalling network. Current Opinion in Plant Biology. 2005;8:532–540. doi: 10.1016/j.pbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, et al. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. The Plant Journal. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C. Allene oxide synthases of barley (Hordeum vulgare cv. Salome) – tissue-specific regulation in seedling development. The Plant Journal. 2000;21:199–213. doi: 10.1046/j.1365-313x.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- McConn M, Browse J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. The Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J, Verpoorte R, Kijne JW. ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends in Plant Sciences. 2001;6:212–9. doi: 10.1016/s1360-1385(01)01924-0. [DOI] [PubMed] [Google Scholar]
- Meyer A, Miersch O, Büttner C, Dathe W, Sembdner G. Occurrence of the plant growth regulator jasmonic acid in plants. Journal of Plant Growth Regulation. 1984;3:1–8. [Google Scholar]
- Miersch O, Wasternack C. Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: endogenous jasmonates do not induce jasmonate biosynthesis. Biological Chemistry. 2000;381:715–722. doi: 10.1515/BC.2000.092. [DOI] [PubMed] [Google Scholar]
- Miersch O, Sembdner G, Schreiber K. Occurrence of jasmonic acid analogues in Vicia faba. Phytochemistry. 1989;28:339–340. [Google Scholar]
- Miersch O, Bohlmann H, Wasternack C. Jasmonates and related compounds form Fusarium oxysporum. Phytochemistry. (a) 1999;50:517–523. [Google Scholar]
- Miersch O, Kramell R, Parthier B, Wasternack C. Structure-activity relations of substituted, deleted or stereospecifically altered jasmonic acid in gene expression of barley leaves. Phytochemistry. (b) 1999;50:353–361. [Google Scholar]
- Miersch O, Porzel A, Wasternack C. Microbial conversion of jasmonates – hydroxylations by Aspergillus niger. Phytochemistry. (c) 1999;50:1147–1152. doi: 10.1016/s0031-9422(98)00698-0. [DOI] [PubMed] [Google Scholar]
- Miersch O, Weichert H, Stenzel I, Hause B, Maucher H, Feussner I, Wasternack C. Constitutive overexpression of allene oxide cyclase in tomato (Lycopersicon esculentum cv. Lukullus) elevates levels of jasmonates and octadecanoids in flower organs but not in leaves. Phytochemistry. 2004;65:847–856. doi: 10.1016/j.phytochem.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Maitrejean M, Boland W. Structural and biological diversity of cyclic octadecanoids, jasmonates, and mimetics. Journal of Plant Growth Regulation. 2004;23:170–178. [Google Scholar]
- Mithöfer A, Wanner G, Boland W. Effects of feeding Spodoptera littoralis on lima bean leaves. II. Continuous mechanical wounding resembling insect feeding is sufficient to elicit herbivory-related volatile emission. Plant Physiology. 2005;137:1160–1168. doi: 10.1104/pp.104.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MJ. Archetype signals in plants: the phytoprostanes. Current Opinion in Plant Biology. 2004;7:441–448. doi: 10.1016/j.pbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploenase SE, Barkawi LS, Guilfoyle TJ, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Matsubara A, Miyatake R, Okada M, Ueda M. Bioactive substances to control nyctinasty of Albizzia plants and its biochemistry. Regulation of Plant Growth & Development. 2006;41(Supplement):44. [Google Scholar]
- Narváez-Vásquez J, Ryan CA. The cellular localization of prosystemin: a functional role for phloem parenchyma in systemic wound signaling. Planta. 2004;218:360–369. doi: 10.1007/s00425-003-1115-3. [DOI] [PubMed] [Google Scholar]
- Narváez-Vásquez J, Florin-Christensen J, Ryan CA. Positional specificity of a phospholipase A2 activity induced by wounding, systemin, and oligosaccharide elicitors in tomato leaves. The Plant Cell. 1999;11:2249–2260. doi: 10.1105/tpc.11.11.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K, Asami T, Matsui K, Howe GA, Murofushi N. Characterization of novel imidazole derivative, JM-8686, a potent inhibitor of allene oxide synthase. FEBS Letters. 2006;580:5791–5796. doi: 10.1016/j.febslet.2006.09.044. [DOI] [PubMed] [Google Scholar]
- Oldham ML, Brash AR, Newcomer ME. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proceedings of the National Academy of Sciences of the USA. 2005;102:297–302. doi: 10.1073/pnas.0406352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Durst F, Werck-Reichhart D, Gardner HW, Camara B, Cornish K, Backhaus RA. The major protein of guayule rubber particles is a cytochrome P450. Journal of Biological Chemistry. 1995;70:8487–8494. doi: 10.1074/jbc.270.15.8487. [DOI] [PubMed] [Google Scholar]
- Parthier B. Hormonal regulators or stress factors in leaf senescence? Journal of Plant Growth Regulation. 1990;9:1–7. [Google Scholar]
- Pauw B, Memelink J. Jasmonate-responsive gene expression. Journal of Plant Growth Regulation. 2004;23:200–210. [Google Scholar]
- Pearce G, Ryan CA. Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense signaling glycopeptide hormones coded in a single precursor gene. Journal of Biological Chemistry. 2003;278:30044–30050. doi: 10.1074/jbc.M304159200. [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA. Production of multiple plant hormones from a single polyprotein precursor. Nature. 2001;411:817–820. doi: 10.1038/35081107. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Schaller A, Mauch-Mani B, Conrath U. Signaling in plant resistance responses: divergence and cross-talk of defense pathways. In: Tuzun S, Bent E, editors. Multigenic and induced systemic resistance in plants. New York: Springer; 2006. pp. 166–196. [Google Scholar]
- Powers RA, Rife CL, Schilmiller AL, Howe GA, Garavito RM. Structure determination and analysis of acyl-CoA oxidase (ACX1) from tomato. Acta Crystallographica, SectionD: Biological Crystallography. 2006;62:683–6. doi: 10.1107/S0907444906014107. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Van Loon LC, Pieterse CMJ. Jasmonates – signals in plant microbe interactions. Journal of Plant Growth Regulation. 2004;23:211–222. [Google Scholar]
- Pre MR. Germany: University of Leiden; 2006. Functional analysis of jasmonate-responsive AP2/ERF-domain transcription factors in Arabidopsis thaliana. [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiology. 2005;139:1902–1913. doi: 10.1104/pp.105.066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Diamond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. The Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE. A conserved transcript pattern in response to a specialist and a generalist herbivore. The Plant Cell. 2004;16:3132–3147. doi: 10.1105/tpc.104.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl S, Feussner I. Oxylipins. In: Murphy DJ, editor. Plant lipids: biology, utilization and manipulation. Oxford, UK and Boca Raton, FL: Blackwell Publishing Ltd/CRC Press; 2005. pp. 329–354. [Google Scholar]
- Rotem R, Heyfets A, Fingrut O, Blickstein D, Shaklai M, Flescher E. Jasmonates: novel anticancer agents acting directly and selectively on human cancer cell mitochondria. Cancer Research. 2005;65:1984–1993. doi: 10.1158/0008-5472.CAN-04-3091. [DOI] [PubMed] [Google Scholar]
- Rusterucci C, Montillet JL, Agnel JP, Battesti C, Alonso B, Knoll A, et al. Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. Journal of Biological Chemistry. 1999;274:36446–36455. doi: 10.1074/jbc.274.51.36446. [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. The Plant Cell. 2000;12:1041–1061. doi: 10.1105/tpc.12.7.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Hennig P, Weiler EW. 12-Oxophytodienoate-19,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiology. 1998;188:1345–1351. doi: 10.1104/pp.118.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Biesgen C, Müssig C, Altmann T, Weiler EW. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 2000;210:979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. Systemic signaling in the wound response. Current Opinion in Plant Biology. 2005;8:369–377. doi: 10.1016/j.pbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Koo AJK, Howe GA. Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiology. 2007;143:812–824. doi: 10.1104/pp.106.092916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Baldwin IT. Systemin in Solanum nigrum. The tomato-homologous polypeptide does not mediate direct defense responses. Plant Physiology. 2006;142:1751–1758. doi: 10.1104/pp.106.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Kienow L, Schmelzer E, Colby T, Bartsch M, Miersch O, et al. A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity of activate biosynthetic precursors of jasmonic acid. Journal of Biological Chemistry. 2005;280:13962–13972. doi: 10.1074/jbc.M413578200. [DOI] [PubMed] [Google Scholar]
- Schüler G, Mithöfer A, Baldwin IT, Berger S, Ebel S, Santos JG, et al. Coronalon: a powerful tool in plant stress physiology. FEBS Letters. 2004;563:17–22. doi: 10.1016/S0014-5793(04)00239-X. [DOI] [PubMed] [Google Scholar]
- Sembdner G, Parthier B. The biochemistry and the physiological and molecular actions of jasmonates. Annual Review of Plant Physiology and Plant Molecular Biology. 1993;44:569–589. [Google Scholar]
- Seo HS, Song JT, Cheong J-J, Lee Y-H, Lee Y-W, Hwang I, Lee JS, Choi YD. Jasmonic acid carboxyl methyl transferase: a key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Sciences of the USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojiri K, Kishimoto K, Ozawa R, Kugimiya S, Urashimo S, Arimura G, et al. Changing green leaf volatile biosynthesis in plants: an approach for improving plant resistance against both herbivores and pathogens. Proceedings of the National Academy of Sciences of the USA. 2006;103:16672–16676. doi: 10.1073/pnas.0607780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WC, Funk CD, Brash KAR. Molecular cloning of an allene oxide synthase. A cytochrome P-450 specialized for metabolism of fatty acid hydroperoxides. Proceedings of the National Academy of Sciences of the USA. 1993;90:8519–8523. doi: 10.1073/pnas.90.18.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences of the USA. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe M. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. The Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW. Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry. 1998;47:539–546. doi: 10.1016/s0031-9422(97)00547-5. [DOI] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Gebhardt S, Schubert-Zsilavecz M, Weiler EW. A novel class of oxylipins, sn1-O-(12-Oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl diglycer ide, from Arabidopsis thaliana. Journal of Biological Chemistry. 2001;276:12832–12838. doi: 10.1074/jbc.M010743200. [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan C, Wasternack C. Allene oxide cyclase dependence of the wound response and vascular bundle specific generation of jasmonates in tomato – amplification in wound-signalling. The Plant Journal. (a) 2003;33:577–589. doi: 10.1046/j.1365-313x.2003.01647.x. [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Molecular Biology. (b) 2003;51:895–911. doi: 10.1023/a:1023049319723. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proceedings of the National Academy of Sciences of the USA. 2000;97:10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proceedings of the National Academy of Sciences of the USA. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. The Plant Journal. 2002;32:585–601. doi: 10.1046/j.1365-313x.2002.01449.x. [DOI] [PubMed] [Google Scholar]
- Stumpe M, Feussner I. Formation of oxylipins by CYP74 enzymes. Phytochemical Reviews. 2006;5:347–357. [Google Scholar]
- Stumpe M, Göbel C, Demchenko K, Pawlowski K, Feussner I. Identification of an allene oxide synthase (CYP74C) that leads to formation of α-ketols from 9-hydroperoxides of linoleic and linolenic acid in below ground organs of potato. The Plant Journal. 2006;47:883–896. doi: 10.1111/j.1365-313X.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- Swiatek A, Van Dongen W, Esmans EL, Van Onckelen H. Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiology. 2004;135:161–172. doi: 10.1104/pp.104.040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, et al. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology. 2005;139:1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T, Chujo T, Jikumaru Y, Okada K, Nojiri H, Yamane H. Biological functions of 12-oxophytodienoic acid reductase homologs in Arabodopsis thaliana. Regulation of Plant Growth & Development. 2006;41(Supplement):47. [Google Scholar]
- Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiology. 2005;137:835–840. doi: 10.1104/pp.105.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. The jasmonate signal pathway. The Plant Cell. 2002;Supplement:S153–S164. doi: 10.1105/tpc.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiology. 1980;66:246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. The Plant Journal. 2005;42:201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- Van der Fits L, Memelink J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. The Plant Journal. 2001;25:43–53. doi: 10.1046/j.1365-313x.2001.00932.x. [DOI] [PubMed] [Google Scholar]
- Van der Graaff E, Schwacke R, Schneider A, Deesimone M, Flügge U-I, Kunze R. Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology. 2006;141:776–92. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. Biosynthesis of jasmonic acid by several plant species. Plant Physiology. 1984;75:458–461. doi: 10.1104/pp.75.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. Pathways of fatty acid hydroperoxide metabolism in spinach leaf chloroplasts. Plant Physiology. 1987;85:1073–1078. doi: 10.1104/pp.85.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. The myriad plant responses to herbivores. Journal of Plant Growth Regulation. 2000;19:195–216. doi: 10.1007/s003440000026. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates – biosynthesis and role in stress responses and developmental processes. In: Nooden LD, editor. Plant cell death processes. New York: Elsevier/Academic Press; 2004. pp. 143–155. [Google Scholar]
- Wasternack C. Oxilipins: biosynthesis, signal transduction and action. In: Hedden P, Thomas S., editors. Plant hormone signaling. Annual Plant Reviews. Oxford: Blackwell Publishing Ltd; 2006. pp. 185–228. [Google Scholar]
- Wasternack C, Hause B. Jasmonates and octadecanoids: signals in plant stress responses and plant development. Progress in Nucleic Acid Research and Molecular Biology. 2002;72:165–221. doi: 10.1016/s0079-6603(02)72070-9. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, et al. The wound response in tomato – role of jasmonic acid. Journal of Plant Physiology. 2006;163:297–306. doi: 10.1016/j.jplph.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Weidhase RA, Lehmann J, Kramell H, Sembdner G, Parthier B. Degradation of ribulose-1,5-bisphosphate carboxylase and chlorophyll in senescing barley leaf segments triggered by jasmonic acid methyl ester, and counteraction by cytokinin. Physiologia Plantarum. (a) 1987;69:161–166. [Google Scholar]
- Weidhase RA, Kramell R, Lehmann J, Liebisch HW, Lerbs W, Parthier B. Methyl jasmonate-induced changes in the polypeptide pattern of senescing barley leaf segments. Plant Sciences. (b) 1987;51:177–186. [Google Scholar]
- Weiler E. Octadecanoid-mediated signal transduction in higher plants. Naturwissenschaften. 1997;84:340–349. [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. The Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidospis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences of the USA. 2006;103:10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T, Omer E-LA, Koshino H, Sakamura S, Kikuta Y, Koda Y. Structure of a tuber-inducing stimulus from potato leaves (Solanum tuberosum L.) Agriculture & Biological Chemistry. 1989;53:2835–2837. [Google Scholar]
- Zerbe P, Weiler EW, Schaller F. Preparative enzymatic solid phase synthesis of cis(+)-12-oxo-phytodienoic acid – physical interaction of AOS and AOC is not necessary. Phytochemistry. 2007;68:229–236. doi: 10.1016/j.phytochem.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Ziegler J, Stenzel I, Hause B, Maucher H, Miersch O, Hamberg M, et al. Molecular cloning of allene oxide cyclase: the enzyme establishing the stereochemistry of octadecanoids and jasmonates. Journal of Biological Chemistry. 2000;275:19132–19138. doi: 10.1074/jbc.M002133200. [DOI] [PubMed] [Google Scholar]