Abstract
In this communication, we describe the integration of microarray sensor technology with logic capability for screening combinations of proteins and DNA in a biological sample. In this system, we have demonstrated the use of a single platform amenable to both protein detection and protein-DNA detection using molecular logic gates. The pattern of protein and DNA inputs results in fluorescence outputs according to a truth table for AND and INHIBIT gates, thereby demonstrating the feasibility of performing medical diagnostics using a logic gate design. One possible application of this technique would be for the direct screening of various medical conditions that are dependent on combinations of diagnostic markers.
Molecular logic gates and molecular computational systems have used a variety of recognition mechanisms including proteins, 1-5 biochemical pathways in living cells, 6-15 and DNA16-31. Molecular logic and computation may also be applied to medical diagnostics. For example, if abnormal results were detected during medical examination, they could be interpreted using Boolean logic resulting in intelligent diagnostics. In this communication, we report the integration of microarray sensor technology with logic capability for screening combinations of proteins and DNA in a biological sample. In this system, the reporter and receptor molecules perform simple logic operations by coupling multiple molecular recognition inputs to a fluorescence signal output.
Previously developed molecular AND gates by the de Silva group allowed the creation of a “lab on a molecule” prototype, which responded to inputs of the electrolytes Na+, H+, and Zn2+ in water leading to an enhanced fluorescence signal as output.28 In addition, the NOTIF logic function was previously demonstrated in designed synthetic peptide networks that mimic some basic logic functions of more complex biological networks.32 In this communication, we demonstrate both AND and INHIBIT (NOTIF) logic gates that respond to the presence of both protein and DNA in a sample. Such a system potentially could be used for performing smart diagnostics. For instance, increased airway obstruction in patients with chronic obstructive pulmonary disease (COPD) or bronchial asthma is frequently associated with bacterial respiratory infections, especially Haemophilus influenzae (H. influenzae), and is accompanied by the secretion of proinflammatory cytokines, such as IL-8 protein.32-37 It is possible to describe the resulting logic network of bacterial DNA and IL-8 protein using a Boolean operator truth table (Figure 1A).
Figure 1.
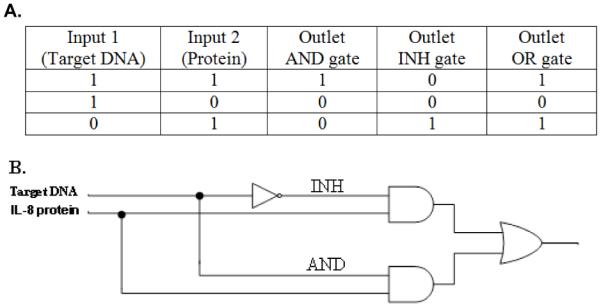
(A) Truth table for logic gates. (B) AND and INHIBIT logic gates.
Our system is designed to determine if both a protein and a nucleic acid sequence are present in a sample, or if only protein is present. A schematic illustration of the protein and protein-DNA sensors is shown in Figures 2A and B, respectively. The fiberoptic microarray was prepared by loading monoclonal antibody (MAb)-functionalized microspheres into micro-wells created by selectively etching the distal end of optical fiber bundles. 38 Every microsphere on the array is encoded with a unique optical barcode comprised of fluorescent dye incorporated into each microsphere. The identity of each microsphere in the random array is then determined using image-processing techniques to allow positional registration of the entire array (Figure 3 A, B (1)). MAb-functionalized microspheres were used as receptors for capturing protein on the surface followed by binding of a secondary antibody labeled with DNA (capture probe) via a biotin-avidin bridge (described in detail in the Supporting Information).
Figure 2.
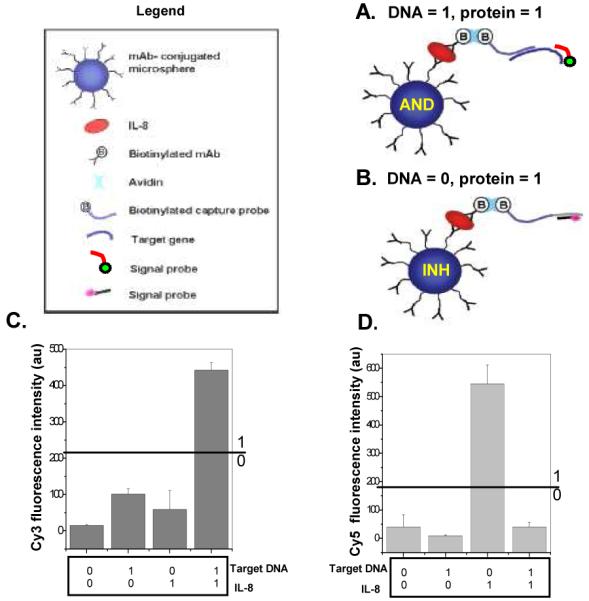
Schematic illustration for (A) protein-DNA detection and (B) protein detection. Output averaged fluorescence intensities in the absence (0) or in the presence (1) (protein (1) = 10 nM, Target DNA (1) = 50 nM) of different inputs; (C) Cy3 signal (D) Cy5 signal.
Figure 3.
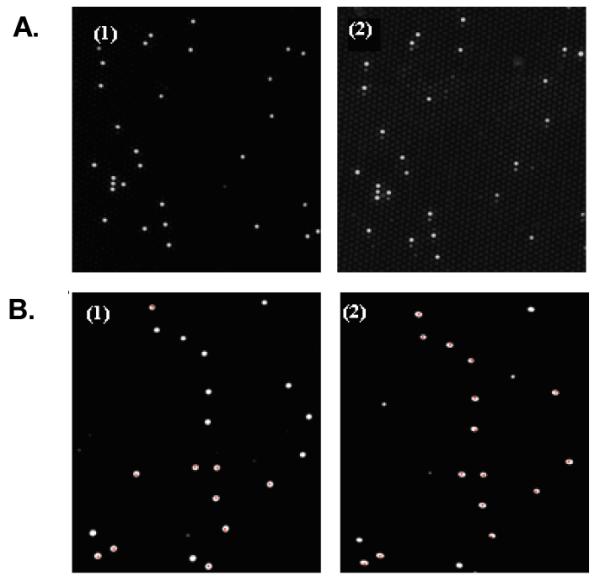
Fluorescence micrographs of a small section of a fiber-optic microarray (A, B) (1)) Positional registration of the entire Europium (Eu)-dye encoded microsphere array. (A (2)) Cy5 microarray signal image of IL8-protein (10 nM) and Target DNA (0 nM). (B (2)) Cy3 microarray signal image of IL8-protein (10 nM) and Target DNA (50 nM). Note the correspondence between the encoded microspheres with the Eu-dye and the microspheres showing fluorescence in the signal images.
To use this platform for both protein detection and protein-DNA detection, two fluorescent probes were designed for the detection step. If both DNA and protein are present in the sample, the Cy3-labeled signal probes hybridize to the complementary target DNA sequence (Figure 2A) while the remaining secondary antibodies free of target DNA hybridize to Cy5-signal probes (Figure 2B). No Cy5 fluorescence is obtained when the capture probe is saturated with a high concentration of target DNA (50 nM) (Figure 2D (1,1)). The detection of the specific DNA sequence in the designed system is possible only if the protein is also present. Therefore, Cy3 signal indicates the detection of both target DNA and protein. Conversely, if target DNA is not present in the sample solution, only the Cy5-labeled signal probes hybridize to the secondary antibody labeled with complementary sequence (Figure 2B).
This biochemical system can be described as a simple functional model of AND and INHIBIT (INH) logic gates (Figure 1B). The INHIBIT (INH) function can be interpreted as a particular integration of AND and NOT logic functions, where the output signal is inhibited by one of the active inputs. Therefore, the INH logic gate presented here queries protein presence or absence in a sample. If only protein is present in the sample (input (0,1)), the gate switches on and a Cy5 fluorescence emission is produced (output 1) (Figure 3A (2)). If protein is not present in the sample (inputs (0,0);(1,0)), then the fluorescent output is “inhibited”; consequently, no Cy5 fluorescence or Cy3 is produced (output 0). AND logic is represented by the situation where the output of an AND gate occurs only if both inputs are present. The AND logic gate presented here queries two biological species (protein and DNA) in a sample to determine in a single test if they are both present. Experiments were performed by testing the AND gate sensor with all four possible input combinations of protein and DNA (Figure 2C). In the presence of both protein and DNA inputs in a sample (1,1), the AND gate switches on and a Cy3 fluorescence emission is produced (output 1) (Figure 3B (2)). No Cy5 fluorescence is obtained for (1,1) since the capture probe is saturated with a high concentration of target DNA (50 nM) (Figure 2D (1,1)). If one (0,1; 1,0) or neither (0,0) of the analytes is present in the sample, no Cy3 fluorescence is produced (output 0). In our system, the absence of input protein has the power to disable the entire system regardless of the presence of the other input—DNA.
In summary, we have demonstrated the use of a single platform amenable to both protein detection and protein-DNA detection using molecular logic gates. The pattern of protein and DNA inputs to fluorescence outputs executed according to the truth table for AND and INHIBIT gates demonstrates the feasibility of performing medical diagnostics using a logic gate design. One possible application of this technique would be for the direct screening of various medical conditions that are dependent on combinations of diagnostic markers.
Supplementary Material
ACKNOWLEDGMENT
We thank the National Institute of Dental and Craniofacial Research (NIDCR) for their support of this work (Grant U01 DE017788). We further thank Dr. Timothy M. Blicharz, Dr. Christopher LaFratta, and Ryan B. Hayman from the Department of Chemistry at Tufts for their technical assistance.
REFERENCES
- (1).Hjelmfelt A, Weinberger ED, Ross J. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10983–10987. doi: 10.1073/pnas.88.24.10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hjelmfelt A, Weinberger ED, Ross J. Proc. Natl. Acad. Sci. U.S.A. 1992;89:383–387. doi: 10.1073/pnas.89.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dueber JE, Yeh BJ, Chak K, Lim WA. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- (4).Sivan S, Tuchman S, Lotan N. BioSystems. 2003;70:21–33. doi: 10.1016/s0303-2647(03)00039-x. [DOI] [PubMed] [Google Scholar]
- (5).Muramatsu S, Kinbara K, Taguchi H, Ishii N, Aida T. J. Am. Chem. Soc. 2006;128(11):3764–3769. doi: 10.1021/ja057604t. [DOI] [PubMed] [Google Scholar]
- (6).Margulies David, Melman Galina, Shanzer Abraham. J. Am. Chem. Soc. 2006;128(14):4865–4871. doi: 10.1021/ja058564w. [DOI] [PubMed] [Google Scholar]
- (7).Gardner T, Cantor C, Collins J. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- (8).Guet C, Elowitz M, Hsing W, Leibler S. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- (9).Hasty J, McMillen D, Collins JJ. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- (10).Atkinson Mariette R, Savageau Michael A, Myers Jesse T, Ninfa A. J. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- (11).Raymo FM. Adv. Mater. 2002;14:401–414. [Google Scholar]
- (12).De Silva AP, McClenaghan ND. Chem.Eur. J. 2004;10:574–586. doi: 10.1002/chem.200305054. [DOI] [PubMed] [Google Scholar]
- (13).Conrad M, Zauner KP, Sienko T, Adamatzky A, Rambidi N. Molecular Computing. MIT Press; 2003. pp. 1–31. chapter 1. [Google Scholar]
- (14).Strack G, Ornatska M, Pita M, Katz E. J. Am. Chem. Soc. 2008;130(13):4234–4235. doi: 10.1021/ja7114713. [DOI] [PubMed] [Google Scholar]
- (15).Nurse P. Nature. 2008;454:424–426. doi: 10.1038/454424a. [DOI] [PubMed] [Google Scholar]
- (16).Adleman LM. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- (17).Lipton RJ. Science. 1995;268:542–545. doi: 10.1126/science.7725098. [DOI] [PubMed] [Google Scholar]
- (18).Guarnieri F, Fliss M, Bancroft C. Science. 1996;273:220–223. doi: 10.1126/science.273.5272.220. [DOI] [PubMed] [Google Scholar]
- (19).Ouyang Q, Kaplan PD, Liu S, Libchaber A. Science. 1997;278:446–449. doi: 10.1126/science.278.5337.446. [DOI] [PubMed] [Google Scholar]
- (20).Faulhammer D, Cukras AR, Lipton RJ, Landweber LF. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1385–1389. doi: 10.1073/pnas.97.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liu Q, Wang L, Frutos AG, Condon AE, Corn RM, Smith LM. Nature. 2000;403:175–179. doi: 10.1038/35003155. [DOI] [PubMed] [Google Scholar]
- (22).Mao C, LaBean TH, Reif JH, Seeman NC. Nature. 2000;407:493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- (23).Benenson Y, Paz-Elizur T, Adar R, Keinan E, Livneh Z, Shapiro E. Nature. 2001;414:430–434. doi: 10.1038/35106533. [DOI] [PubMed] [Google Scholar]
- (24).Carbone A, Seeman NC. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12577–12582. doi: 10.1073/pnas.202418299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Stojanovic MN, Stefanovic D. J. Am. Chem. Soc. 2003;125:6673–6676. doi: 10.1021/ja0296632. [DOI] [PubMed] [Google Scholar]
- (26).Saghatelian A, Voelcker NH, Guckian KM, Lin VSY, Ghadiri MR. J. Am. Chem. Soc. 2003;125:346–347. doi: 10.1021/ja029009m. [DOI] [PubMed] [Google Scholar]
- (27).Stojanovic MN, Stefanovic D. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- (28).Yan H, Feng L, LaBean TH, Reif JH. J. Am. Chem. Soc. 2003;125:14246–14247. doi: 10.1021/ja036676m. [DOI] [PubMed] [Google Scholar]
- (29).Voelcker NH, Guckian KM, Saghatelian A, Ghadiri RM. Small. 2008;28(4):427–431. doi: 10.1002/smll.200700113. [DOI] [PubMed] [Google Scholar]
- (30).Cockroft SL, Chu J, Amorin M, Ghadiri MRJ. Am. Chem. Soc. 2008;130(3):818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Winfree Erik. ACM. 2008;42(2):1–1. [Google Scholar]
- (32).Ashkenasy G, Ghadiri RM. J. Am. Chem. Soc. 2004;126:11140–11141. doi: 10.1021/ja046745c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rutmam A, Dowling R, Wills P, Feldman C, Cole PJ, Wilson R. Antimicrobial agents and chemotherapy. 1998:772–778. doi: 10.1128/aac.42.4.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Khair OA, Devalia JL, Abdelaziz MM, Sapsford RJ, Davies RJ. Eur Respir J. 1995;8:1451–1457. [PubMed] [Google Scholar]
- (35).Tong HH, Chen Y, James M, Van Deusen J, Welling DB, Demaria TF. Infection and Immunity. 2001:3678–3684. doi: 10.1128/IAI.69.6.3678-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chung KF. Eur. Respir. J. 2001;18(34):50–59. [Google Scholar]
- (37).Pauwels R, Verschraegen G, Van Der Straeten M. Allergy. 1980;35(8):665–669. doi: 10.1111/j.1398-9995.1980.tb02019.x. [DOI] [PubMed] [Google Scholar]
- (38).LaFratta CN, Walt DR. Chem. Rev. 2008;108(2):614–637. doi: 10.1021/cr0681142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


