Abstract
N-linked protein glycosylation was originally thought to be specific to eukaryotes, but evidence of this post-translational modification has now been discovered across all domains of life: Eucarya, Bacteria, and Archaea. In all cases, the glycans are first assembled in a step-wise manner on a polyisoprenoid carrier lipid. At some stage of lipid-linked oligosaccharide synthesis, the glycan is flipped across a membrane. Subsequently, the completed glycan is transferred to specific asparagine residues on the protein of interest. Interestingly, though the N-glycosylation pathway seems to be conserved, the biosynthetic pathways of the polyisoprenoid carriers, the specific structures of the carriers, and the glycan residues added to the carriers vary widely. In this review we will elucidate how organisms in each basic domain of life synthesize the polyisoprenoids that they utilize for N-linked glycosylation and briefly discuss the subsequent modifications of the lipid to generate a lipid-linked oligosaccharide.
Keywords: isoprenoid, N-glycosylation, DOXP, MVA, dolichol, lipid-linked oligosaccharide
1. Introduction
Historically, it was thought that N-glycosylation was unique to eukaryotic organisms and that a single, defined pathway led to the biosynthesis of the polyisoprenoids that act as the lipid-linked oligosaccharide (LLO) donors in N-glycosylation. In the last few decades, however, both of these ideas have been disproven, evoking questions about how N-glycosylation occurs in different organisms, including what type of carrier lipids are used and how they are synthesized.
In 1976, Mescher and Strominger reported a protein from the cell envelope in Halobacterium salinarium that contained glycans covalently linked to asparagine residues [1]. This marked the first observation of N-glycosylation in what would later become the domain Archaea. With this discovery, the idea that N-linked glycosylation occurred only in eukaryotes was dispelled. More recently, evidence of N-glycosylation in bacteria was discovered by Szymanski et al. in the Gram-negative bacteria Campylobacter jejuni [2]. N-glycosylation is now thought to be a protein modification that is conserved across all three majors domains of life: Eucarya, Bacteria, and Archaea [2, 3].
The N-glycosylation pathways in each domain are similar in that they include the step-wise assembly of sugars, donated by nucleotide activated sugars or activated lipids, onto a polyisoprenoid carrier by specific glycosyltransferases to form a LLO [4, 5]. At some point during synthesis, the LLO is flipped across a membrane, and the oligosaccharide is transferred from the carrier lipid onto the protein by an oligosaccharide transferase (OST) enzyme or enzyme complex [4, 5]. The glycan is ultimately bound to an asparagine residue in the protein via a β-glycosylamide linkage [4].
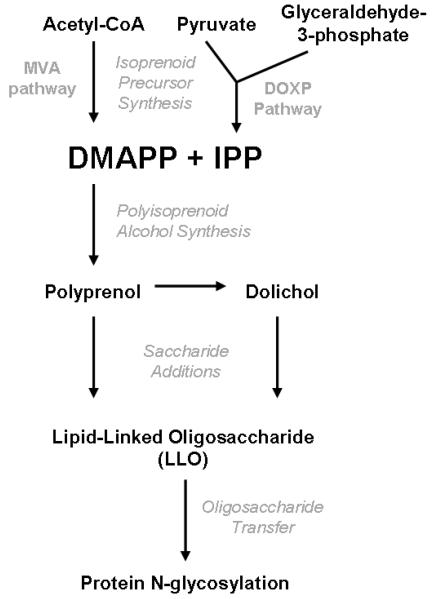
Although eukaryotes, bacteria, and archaea all seem to have certain characteristics of the N-glycosylation pathway in common, the identity of the lipid oligosaccharide carrier, its method of biosynthesis, and the structure of the attached glycans are sometimes strikingly different. This review will focus primarily on the synthesis of the polyisoprenoid alcohols that provide the carrier lipid portion of the LLO. Polyisoprenoid alcohols are hydrophobic polymers generated by the condensation of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), the universal 5-carbon precursors of all isoprenoids [6-8]. A schematic diagram of how polyisoprenoid alcohols are formed and utilized in N-glycosylation is shown in Fig. 1. The review will begin with an overview of the two possible pathways for generating the IPP and DMAPP building blocks. It will then discuss the formation of the polyisoprenoid alcohols, dolichol and polyprenol, from IPP and DMAPP. Next, the review will identify the polyisoprenoid alcohols known to be used in N-glycosylation for organisms in each of the three domains of life and will determine which biosynthetic pathways are responsible for providing the IPP and DMAPP building blocks in each case. Finally, the sugar modifications that result in generation of a completed LLO will be described briefly for each domain.
Fig 1.
Schematic overview polyisoprenoid alcohol formation and utilization in N-glycosylation. The MVA and DOXP pathways provide the isoprenoid precursors DMAPP and IPP, which are used to synthesize dolichol and polyprenol. Dolichol and polyprenol and modified by sugar additions into a completed LLO, then the oligosaccharide is transferred off of the carrier lipid and onto asparagine residues on the protein.
2. Isoprenoid biosynthesis
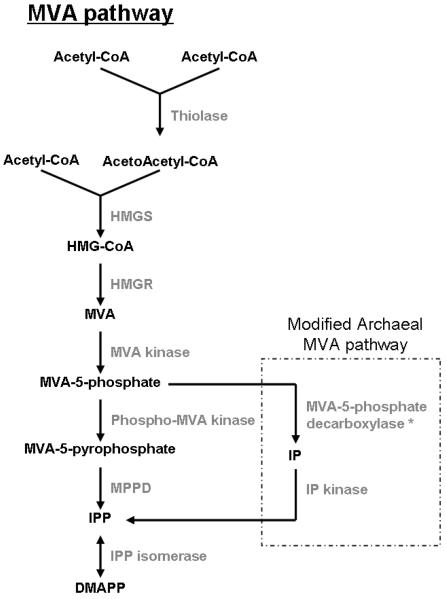
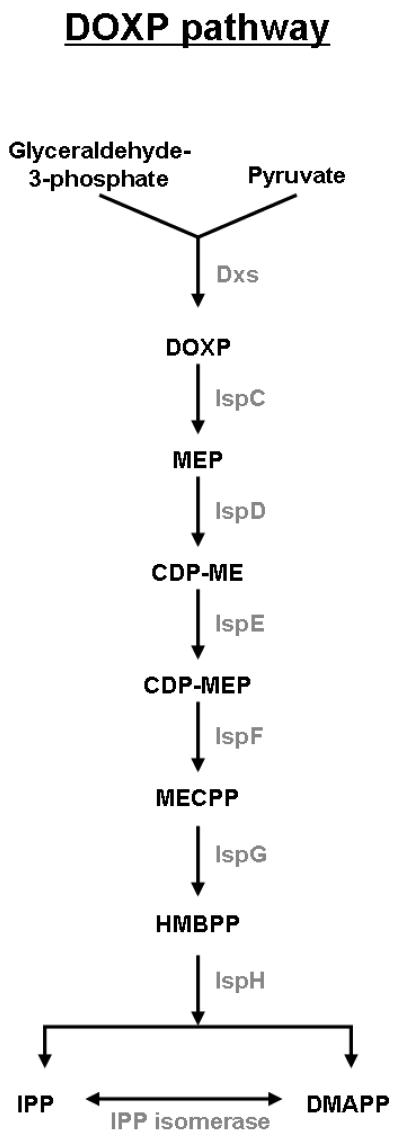
Originally, there was thought to be a single pathway responsible for the biosynthesis of IPP and DMAPP, the universal 5-carbon precursors of all isoprenoids [6-8]. This was called the mevalonate (MVA) pathway and it involves the formation of IPP from acetyl-CoA with mevalonate as an intermediate (Fig. 2). It was widely accepted that all organisms form DMAPP and IPP only through this pathway [7]. However, in 1993, Rohmer et al. provided evidence for a new pathway for DMAPP and IPP synthesis that did not involve mevalonate (Fig. 3) [9, 10]. This work began with the observation of unexpected labeling patterns in bacterial hapanoids using 13C-labeled acetate [11]. Independently, Arigoni et al. discovered the existence of a mevalonate-independent pathway in the plant, Ginkgo biloba, as well as in E. coli [12-14]. Since then, this pathway, called the 1-deoxy-D-xylulose-5 phosphate (DOXP) pathway, has been further elucidated and has proven to be an alterative to the MVA pathway for IPP and DMAPP synthesis [8, 15, 16].
Fig 2.
The MVA biosynthetic pathway for IPP and DMAPP. Acetyl-CoA is used to synthesize IPP and DMAPP with MVA as an intermediate. In the modified archaeal pathway, MVA-5-phosphate is first decarboxylated then phosphorylated to form IPP, the opposite of what occurs in the traditional MVA pathway. *Enzyme is theoretical and has yet to be discovered. For a review of the MVA pathway with chemical structures see [108].
Fig 3.
The DOXP biosynthetic pathway for IPP and DMAPP. Glyceraldehyde and pyruvate are used to synthesize IPP and DMAPP with DOXP as an intermediate. For a review of the DOXP pathway with chemical structures see [108].
2.1 The MVA pathway
The MVA pathway (Fig. 2) begins with the conversion of two molecules of acetyl-CoA to acetoacetyl-CoA by the enzyme acetyl-CoA acetyltransferase (also called thiolase) [6, 17-19]. A third molecule of acetyl-CoA and the acetoacetyl-CoA are then condensed to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) via HMG-CoA synthase (HMGS) [6, 17, 20]. HMG-CoA is reduced to mevalonate by HMG-CoA reductase (HMGR), in a NADPH-dependent reaction. HMGR is the key regulatory enzyme in the MVA pathway [21-23]. In the next step, MVA is phosphorylated by MVA kinase to form MVA-5-phosphate, which is further phosphorylated by phospho-MVA kinase to form MVA-5-pyrophosphate [24, 25]. Both of these phosphorylation reactions are ATP dependent [26, 27]. MVA-5-pyrophosphate undergoes a decarboxylation reaction via MVA-5-pyrophosphate decarboxylase (MPPD) to produce IPP [6, 28]. In the final step of this pathway, some of the IPP is converted to DMAPP by IPP isomerase [27, 29]. IPP isomerase is essential for the production of DMAPP via the MVA pathway [8].
2.2 The DOXP pathway
The DOXP pathway (Fig. 3, for review see [6]) does not require acetyl-CoA, but rather begins with the condensation of pyruvate and glyceraldehyle 3-phosphate into DOXP by DOXP synthase (Dxs) [30, 31]. This reaction requires thiamine pyrophosphate as a cofactor [6]. DOXP reductase (IspC) converts DOXP to 2-C-methyl-D-erythritol 4-phosphate (MEP) [32] which then reacts with cytidine triphosphate (CTP) to form 4-pyrophosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) with the help of CDP-ME synthase (IspD) [8, 33]. Next, 4-pyrophosphocytidyl-2C-methyl-D-erythritol kinase (IspE) catalyzes the phosphorylation of CDP-ME to CDP-ME-2-phosphate (CDP-MEP) [34]. CDP-MEP is then converted to 2C-methyl-D-erythritol-2,4-cyclopyrophosphate (MECPP) by MECPP synthase (IspF) in a reaction that also involves the elimination of cytidine monophosphate (CMP) [8, 35, 36]. The final stages in the DOXP synthesis pathway are the least understood. In the last two steps, MECPP undergoes reduction and elimination reactions catalyzed by two enzymes, designated IspG and IspH, with 1-hydroxy-2-methyl-2-(E)-butenyl-4-pyrophosphate (HMBPP) as an intermediate [15, 37, 38]. Both IPP and DMAPP are formed by these reactions, so IPP isomerase is not essential for production of DMAPP via the DOXP pathway. Nonetheless, as in the MVA pathway, IPP isomerase can interconvert DMAPP and IPP, allowing for adjustments in the ratio of IPP to DMAPP [8]. Not much is known about regulation of the DOXP pathway, however, IspF has been shown to interact with IPP, DMAPP, and other small isoprenoids that are downstream products of the DOXP pathway [39]. This data raises the possibility that these small isoprenoids could be involved in regulation of the DOXP pathway by a feedback mechanism with IspF as the point of control [6, 39]. There is also evidence that in plants, some of the enzymes in the DOXP pathway are regulated by light [40, 41]. This will be discussed further in section 3.3.
2.3 Polyisoprenoid Alcohols – Dolichol and Polyprenol
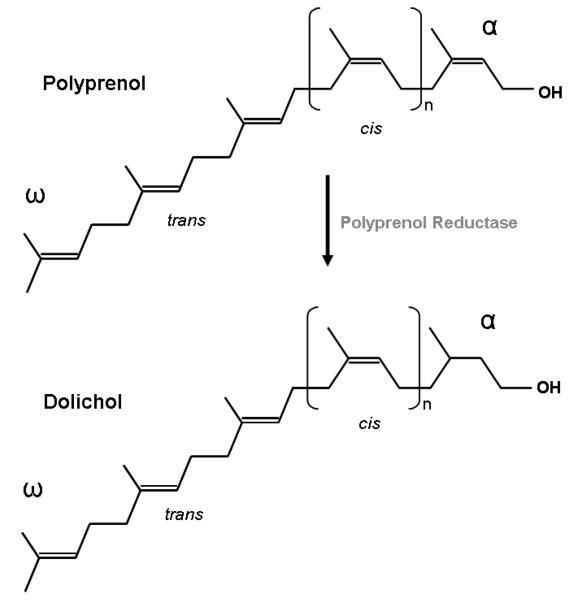
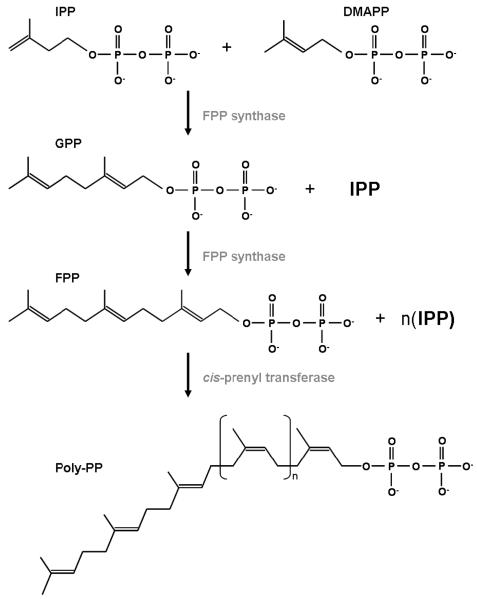
Polyisoprenoid alcohols are hydrophobic polymers that consist of a number of 5-carbon isoprene units linked head to tail. The polyisoprenoid alcohols involved in N-glycosylation can be from several up to about 25 isoprene units in length and have a hydroxyl group at one end (α) and a hydrogen atom at the other (ω) (Fig. 4) [21, 42]. Polyisoprenoid alcohols are formed via a condensation reaction between DMAPP and several molecules of IPP [21, 29, 43]. This reinforces the importance of the IPP isomerase in the MVA pathway, since in this pathway, DMAPP cannot be formed without IPP isomerase, and both IPP and DMAPP are required for polyisoprenoid synthesis. DMAPP acts as an acceptor for molecules of IPP, which are added sequentially via specific prenyl transferases to form chains of various lengths, such as geranyl pyrophosphate (GPP, C10), farnesyl pyrophosphate (FPP,C15), geranylgeranyl pyrophosphate (GGPP, C20), and farnesylgeranyl pyrophosphate (FGPP, C25) [21, 44-46]. Longer polyprenyl pyrophosphates (Poly-PP) are typically synthesized from FPP and IPP (Fig. 5) [46], and the overall chain length is determined by the specific prenyl transferases, most likely through a restriction in the size of the enzyme's active site [21, 47, 48]. The prenyl transferases also control lipid stereochemistry, as both cis- and trans-prenyl transferases exist in nature [46]. GPP and FPP are synthesized by a trans-prenyl transferase called FPP synthase [45, 49], however, cis-prenyl transferase is the more important enzyme in the context of N-glycosylation lipid donors (Fig. 5) [21, 48, 50].
Fig 4.
Polyprenol is reduced via polyprenol reductase to form dolichol. The structure of polyprenol and dolichol differ based on the saturation of the α-isoprene unit. The three isoprene units originating from farnesyl pyrophosphate (FPP) at the ω end of the molecule are in the trans configuration, while the remaining isoprene units are in the cis configuration.
Fig 5.
The biosynthetis of Poly-PP from IPP and DMAPP through the intermediates GPP and FPP. n = number of isoprene units.
Dolichol and polyprenol are the polyisoprenoid alcohols used as sugar carriers for N-glycosylation in all domains of life. Polyprenol synthesis begins with all-trans-FPP or GGPP, which is elongated by cis-prenyl transferase, using IPP as a substrate and resulting in a trans/cis-Poly-PP (Fig. 5) [21, 46]. This Poly-PP is hydrophobic and, thus, localized within the membrane structure [51]. Although the order of the final steps in this pathway have not been completely elucidated, it is currently believed that Poly-PP is dephosphorylated into polyprenyl phosphate (Poly-P), and further dephosphorylated into polyprenol [52-54]. Polyprenol can then undergo a reduction reaction, catalyzed by polyprenol reductase, in order to form dolichol (Fig. 4) [51, 52, 55, 56]. Dolichol differs from polyprenol in that its α-isoprene unit is saturated. Dolichol can then be phosphorylated by dolichol kinase to form dolichol phosphate (Dol-P) [51, 52, 57, 58]. Sanderman and Strominger discovered a similar kinase for the re-phosphorylation of polyprenol. They report an undecaprenol phosphokinase from Staphylococcus aureus that phosphorylates the 55-carbon polyprenol found in bacteria [21, 59, 60].
These kinases are extremely important because they control the level of phosphorylation of dolichol and polyprenol. Since many of the enzymes involved in the N-glycosylation pathway can only utilize isoprenoids with specific phosphorylation states as substrates, these kinases have the ability to affect the size of the dolichol or polyprenol pool available for use in N-glycosylation[61]. For example, many glycosyltransferases can use Dol-P but not dolichol or Dol-PP as a substrate [62, 63]. The saturation state and length of these polyisoprenoid alcohols are also important for their recognition by the enzymes involved in the N-glycosylation pathway [52, 64-66]. Enzymes such as glycosyltransferases and oligosaccharide transferases in all domains of life have substrate specificity such that they preferentially utilize polyisoprenoids of particular lengths and saturation states [52, 67].
3. Eukaryota
The N-glycosylation pathway in eukaryotes has been extensively researched and characterized (see references [63, 68] and [69] for recent reviews). First described based on experiments done in higher eukaryotes, including plants and animals [63, 70], the basic principles of this pathway were later confirmed to be conserved in lower eukaryotes, such as yeast [63, 71, 72]. Eukaryotes use dolichol pyrophosphate, which has a saturated α-position isoprene unit, as the oligosaccharide carrier in N-linked glycosylation [52, 63, 70, 73]. Dolichol pyrophosphate is biosynthesized on the cytoplasmic face of the endoplasmic reticulum (ER) from FPP and IPP [4, 46, 52], but the biosynthetic pathways that supply the isoprene subunits and the overall length of the polyisoprenoid are species-specific (see subsections 3.1-3.3) [63]. The early steps in the N-glycosylation process, involving the synthesis of a tetradecasaccharide on the Dol-PP carrier seem to be highly conserved across Eucarya and are discussed below. One known exception is in trypanosomatid protozoa, which cannot glucosylate their LLO precursors and therefore do not form the completed tetradecasaccharide [74, 75]. The final processing steps, which occur after the oligosaccharide is transferred onto the nascent polypeptide, vary between organisms, as do the final oligosaccharide structures.
In eukaryotes, N-glycosylation begins on the cytoplasmic face of the ER [76] with the transfer of sugar residues from nucleotide-activated sugar donors, such as uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) and guanosine diphosphate mannose (GDP-Man), onto a dolichol phosphate carrier [62, 77]. In the first reaction, GlcNAc-P is transferred onto Dol-P giving rise to GlcNAc-PP-Dol. Another GlcNAc residue is subsequently added to the structure, followed by 5 mannose groups. These reactions occur with the aid of specific glycosyltransferases, which use sugar nucleotides as substrates and add sugars according to a pre-defined reaction sequence, generating the heptasaccharide Man5GlcNAc2-PP-Dol [4, 77-79]. This heptasaccharide is then flipped across the ER membrane to the luminal face [76]. This translocation is thought to occur with the help of an enzyme “flippase,” [4, 80] which was recently identified as Rft1 in Saccharomycers cerevisiae by Helenius et al. [81]. However, Frank et al. failed to correlate the presence of Rft-1 with flippase activity in fractions from a CM-Sepharose purification [82]. They also saw identical flippase activity in wildtype and Rft-1-depleted yeast ER membrane extracts, leading them to conclude that Rft-1 may not contribute to flippase activity.
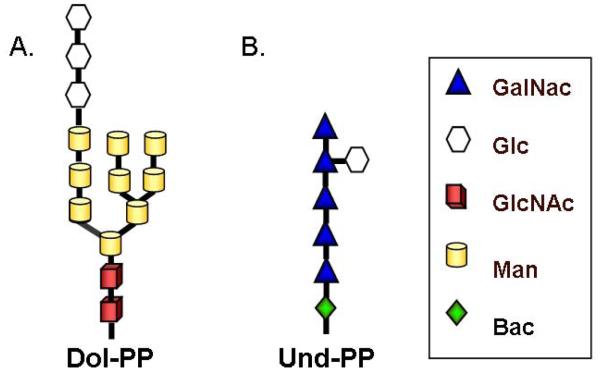
Once on the luminal face of the ER, four additional Man residues, and three glucose (Glc) residues are added to the dolichol-linked oligosaccharide chain. These reactions require glycosyltransferases that utilize lipid activated monosaccharides, such as Dol-P-Man and Dol-P-Glc, as sugar donors [4, 62]. The synthesis of these lipid-linked monosaccharide sugar donors also requires dolichol phosphate, which reacts with UDP-Glc or GDP-Man via additional glycosyltransferases on the cytoplasmic face of the ER to form Dol-P-Man and Dol-P-Glc. These lipid-linked monosaccharides are also translocated across the ER membrane to the lumen through the action of flippases [4, 62]. The final result is a dolichol pyrophosphate-linked tetradecasaccharide with the structure Glc3Man9GlcNAc2-PP-Dol (Fig. 6A) [78, 83, 84]. As was mentioned earlier, trypanosomatid protozoa cannot synthesize this structure because they are unable to synthesize Dol-P-Glc [75]. The genes for certain glucosyltransferases also seem to be absent in the genome of Trypanosoma brucei [74]. Consequently, trypanosomatid protozoa produce LLO with the structure Man9GlcNAc2-PP-Dol [74, 75].
Fig 6.
The sugar structures of the final lipid-linked oligosaccharide in eukaryotes (A.) and bacteria (B.)
The completed oligosaccharides can then be transferred from the dolichol carrier onto specific asparagine residues on a nascent polypeptide located in the ER. This occurs cotranslationally with the help of an oligosaccharide transferrase (OST) enzyme or enzyme complex [73, 84]. After the oligosaccharide is transferred onto the protein, the Dol-PP carrier is de-phosphorylated into Dol-P then diffuses or is flipped back to the cytoplasmic face of the ER [85]. In this way, the dolichol carrier is recycled and can be reutilized for additional rounds of N-glycosylation [52, 85]. For information about the specific enzymes involved in each step of LLO synthesis and oligosaccharide transfer in S. cerevisiae, we refer readers to reviews by Burda and Aebi [63] and Kelleher and Gilmore [69].
3.1 Animalia
Animals require isoprenoids for a wide variety of biological functions. Cholesterol, for example, is involved in maintaining membrane structure [86] and coenzyme Q plays a crucial role in energy synthesis via oxidative phosphorylation [87]. Isoprenoid–derived molecules are also involved in coagulation (vitamin K) and vision (retinol), and can act as signaling hormones (steroids) [7, 86]. Animals utilize only the MVA pathway for isoprenoid synthesis, and therefore make all of their dolichol derivatives from mevalonate via this pathway [35, 88]. Interestingly, animals do not synthesize polyisoprenoid lipids of a single chain length; rather they produce homologous mixtures. These mixtures, or “families”, consist of six to eight different chain lengths with one or two lengths predominating [21, 42]. Most likely, the chain length is determined by the cis-prenyl transferase, which condenses IPP subunits onto FPP and produces dolichols with a distribution of similar lengths [42, 47]. In mammalian cells, dolichols are predominantly 18 – 21 isoprene units in length [52]. Human cells contain dolichol that is primarily 19 isoprene units in length (Dol-19) [21], as do Chinese Hamster Ovary (CHO) cells [42, 89]. Rats contain mostly Dol-18 [21, 90], while Drosophila contain mainly Dol-16 and Dol-17 [91]. Dol-P is required in a number of reactions in the N-glycosylation pathway and the availability of Dol-P is often one of the rate limiting factors in the synthesis of lipid-linked oligosaccharides [63, 92-94].
Though dolichol pyrophosphate is the lipid donor of choice in animals and all eukaryotes, a CHO cell line designated Lec9 was isolated that synthesizes polyprenol, rather than dolichol and utilizes this polyprenol to synthesize its N-glycosylation oligosaccharide donor [21, 95]. The polyprenols in Lec9 cells have the same distribution of chains lengths as dolichols in the parental CHO cells [95]. The Lec9 phenotype is thought to be caused by a deficiency in polyprenol reductase, which reduces polyprenol into dolichol (Fig. 4) [95]. This reduction reaction appears to be a rate limiting step in the synthesis of dolichol [96]. Thus, polyprenol reductase appears to be important in the regulation of N-glycosylation in mammalian cells. This reduction step is also important because many glycosyltransferases involved in the formation of lipid-linked mono- or oligo-saccharides prefer polyisoprenoids with particular saturation states (e.g. Dol-P in animals) as a substrates [42, 67]. For example, in CHO cells, the glycosyltransferase that catalyzes the first step in the N-glycosylation pathway (the formation of GlcNAc-PP-Dol from UDP-GlcNAc and Dol-P) is significantly impaired in its ability to utilize Poly-P compared to Dol-P [64]. Mannosylphosphoryldolichol synthase, which is responsible for the formation of the lipid activated monosaccharide Dol-P-Man, also has a clear preference for the Dol-P substrate [65].
3.2 Fungi
Fungi, represented most commonly in research by the yeast Saccharomyces cerevisiae, also utilize the traditional MVA pathway in their biosynthesis of dolichols [21]. Dolichols in yeast, like mammalian cells, are produced in the ER and exist as a family of different chain length molecules ranging in length from 14-17 isoprene units [52]. The most prevalent dolichols in S. cerevisiae contain 15 – 16 isoprene units, while those in Schizosaccharomyces pombe contain 17 isoprene units [42].
3.3 Plantae
Plants are especially interesting with regard to isoprenoids synthesis, because these photosynthetic organisms are the only ones that regularly utilize both the MVA and DOXP pathways in their production of polyisoprenoids. The isoprenoid metabolic pathways in photosynthetic organisms are involved in the biosynthesis of chlorophyll [97] as well as various plant hormones [98, 99], defensive toxins and antibiotics [100, 101], essential antioxidants [102, 103], and pigments and aromas found in flowers and fruit [104, 105]. Most relevant to this review, however, is the synthesis of dolichol and its derivatives for use in protein N-glycosylation.
Higher plants use both the MVA and DOXP pathways to synthesize isoprenoid compounds, oftentimes in concert. One divergent feature of the two possible routes for IPP and DMAPP synthesis is their different subcellular localizations. The enzymes involved in the MVA pathway are typically located within the cytosol [106, 107], while the DOXP pathway is sequestered within the chloroplast [108-110]. Isoprenoids that are fundamental to photosynthesis, including chlorophyll, carotenoids, and plastoquinone are primarily formed within the chloroplast [106, 111], while other isoprenoids, such as sterols and ubiquinone are mainly formed in the cytosol [106]. The same is true for most diatomaceous algae (i.e. diatoms) [112]. In contrast, the majority of green algae maintain only DOXP isoprenoid metabolism and, as a result, all algal isoprenoids are derived from chloroplastic IPP and DMAPP [113, 114]. The chloroplasts of plants and green algae are believed to represent remnants of prokaryotic photosynthetic cyanobacteria, which sought endosymbiosis within eukaryotic cells [115]. Interestingly, cyanobacteria utilize only the DOXP pathway to produce isoprenoids [113, 116-118], further supporting the idea that chloroplasts, which also contain the DOXP isoprenoid pathway, evolved via endosymbiosis.
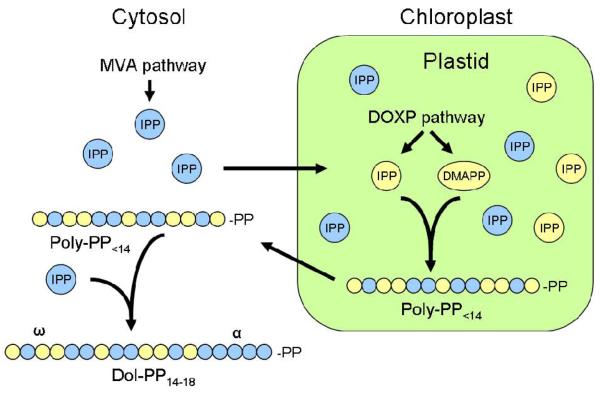
Despite the compartmentalization of the MVA and DOXP pathways, molecular exchange of isoprenoid precursors such as IPP between the cytosol and chloroplast occurs routinely in higher plants [119-124]. Researchers have recently elucidated the extent of this interaction (crosstalk) during the synthesis of dolichol [125]. Skorupinska-Tudek and colleagues investigated the metabolic network responsible for dolichol synthesis in the hairy root of the Coluria geoides plant. Their results showed that dolichol synthesis begins with IPP molecules in the chloroplast. Due to a continuous exchange of intermediates between the MVA and DOXP pathways, the IPP in this reservoir is equally likely to have been produced by the local DOXP pathway or imported from the cytosol (Fig. 7) [124, 125]. Consequently, the carbon found at the ω-terminus of dolichol is not specific to either pathway. The polyisoprenoid chain is then elongated within the chloroplast using IPP from either pathway to form a Poly-PP with less than 14 isoprene units. Finally, this intermediate is exported to the cytosol wherein its conversion to dolichol is completed using IPP derived solely from the MVA pathway [125]. This results in dolichol's α-terminus being comprised solely of carbon of the MVA pathway (Fig. 7). Ultimately, 40-50% of the dolichol molecule was found to originate from the DOXP pathway, indicating significant contributions from both isoprenoid biosynthetic pathways [125].
Fig 7.
Dolichol synthesis in Coluria geoides hairy root. Cytosolic IPP from the MVA pathway can enter the plastid. Dolichol synthesis begins in the plastid where a Poly-PP with less than 14 isoprene units is formed using IPP from both the MVA and DOXP pathways. The Poly-PP is then transported to the cytosol, where synthesis continues using only IPP from the MVA pathway.
Notable diversity has been observed when comparing the families of different length dolichols produced in higher plants and algae. In various model plant systems, including Arabidopsis thaliana, Glycine max (soybean), and Coluria geoides, as well as common plants such as magnolia, spinach, parsley, and perilla, the length of most dolichol molecules falls within the range of 14-18 isoprene units [125-128]. Interestingly, dolichols found in soybean seeds contain one additional isoprene unit compared to those in the roots, shoots, and leaves, which typically have 16 – 17 isoprene units [127]. In the microalga Prototheca zopfii, however, dolichols were found to be considerably longer, ranging from 18 – 21 isoprene units [129]. Conversely, in the green alga Chlamydomonas reinhardtii, dolichol was observed to have the unexpectedly short chain-length of just 11 isoprene units. This structure is similar to the undecaprenyl pyrophosphate used as the oligosaccharide carrier in prokaryotes; however, the molecule does contain the saturated α–isoprene unit that is characteristic of eukaryotes [130].
Since photosynthetic organisms depend on sunlight as both a source of energy and an important environmental cue to elicit metabolic and developmental changes, it is not surprising that sunlight is used to regulate isoprenoid biosynthesis in these organisms. Along the DOXP pathway, the enzyme IspE is responsible for the phosphorylation of the isoprenoid intermediate CDP-ME. In Nicotiana benthamiana, expression of the gene that encodes IspE is induced by light [40]. In Arabidopsis, expression of the gene encoding Dxs is also induced by light [131]. In the MVA pathway, however, expression of the rate-limiting enzyme HMGR appears to be suppressed in Arabidopsis thaliana by prolonged light stimulus [132, 133]. In this case light triggers photoreceptor-mediated signaling cascades which converge to downregulate HMGR expression [133, 134]. Generally, exposure to light causes downregulation of genes in the MVA pathway, while upregulating expression of DOXP pathway genes [133]. The result of this is activation of isoprenoid synthesis in the chloroplast, where many of the key photosynthetic isoprenoids are formed.
4. Bacteria
Although it has been known for decades that polyisoprenoids were involved in the formation of bacterial peptidoglycan [135], Bacteria was the last domain of life in which N-glycosylation was discovered, perhaps because it is such a rare event in these organisms [2, 4, 73]. The DOXP pathway was first discovered in bacteria [9, 12], and while many gram-positive cocci use the traditional MVA pathway for isoprenoid synthesis, many gram-negative bacteria utilize only the DOXP pathway [108, 136]. Bacteria typically employ either the MVA or the DOXP pathway on an exclusive basis [35]. Interestingly, photosynthetic cyanobacteria utilize only the DOXP pathway to isoprenoids [113, 116-118].
In bacteria that utilize the MVA pathway for isoprenoid synthesis, an IPP isomerase is essential for conversion of IPP into DMAPP (Fig. 2). In 2001, Kaneda et al. discovered a novel IPP isomerase in Streptomyces which was not homologous to any previously identified IPP isomerase [137]. Orthologs of this enzyme were subsequently found in the genomes of 35 out of 124 eubacterial families [35]. These novel IPP isomerases were designated type II, while the more traditional IPP isomerases found in eukaryotes and some other bacteria, were designated type I [35, 137]. Bacteria exist that utilize every combination of type I and type II IPP isomerases in combination with the MVA and DOXP pathways. No isomerase at all is detected in about 70% of bacteria that utilize only the DOXP pathway [35]. One study showed that the IPP isomerase gene is not essential in E. coli because they utilize the DOXP pathway and can therefore synthesize both IPP and DMAPP [138]. E. coli do typically express an IPP isomerase, however, which is thought to help adjust the ratio of DMAPP to IPP to optimum levels [35, 50, 138].
Interestingly, the lipid sugar donor involved in N-glycosylation in bacteria is a single polyprenol composed of 11 isoprene units, as opposed to the family of different length isoprenoids that are seen in eukaryotes [21, 42, 139]. The bacterial cis-prenyl transferase, undecaprenyl pyrophosphate synthase (UPPS), is a soluble enzyme that is responsible for adding 8 IPP units to FPP, forming undecaprenyl pyrophosphate (Und-PP) [21, 140]. As opposed to the lipid donor in eukaryotes, the bacterial undecaprenyl pyrophosphate is a polyprenol, in that its α-isoprene unit is unsaturated [52, 84]. Although the 11 isoprene unit undecaprenyl pyrophosphate is the rule, some exceptions exist. In Mycobacterium smegmatis the oligosaccharide is assembled on a Poly-PP either 10 or 7 isoprene units in length [52, 93, 141], while Mycobacterium tuberculosis synthesize only decaprenyl pyrophosphate (10 isoprene units) [93].
Bacterial N-glycosylation was first discovered in C. jejuni and most of our current understanding of the process is based on studies of this organism [2, 142]. Bacteria, like archaea, lack cellular organelles, so glycosylation occurs across the cell membrane [2, 143]. Nucleotide-activated sugars are transferred via glycosyltransferases onto the undecaprenyl pyrophosphate lipid carrier which is located on the cytoplasmic face of the cell membrane [144]. This process is controlled by the pgl gene cluster (for a review see [4]) and results in a heptasaccharide with the following structure: GlcGalNAc5Bac-PP-Und (Bac = bacillosamine, 2,4-diacetamido-2,4,6-trideoxyglucopyranose; GalNAc= N-acetylgalactosamine) ( Fig. 6B) [4, 73, 142, 145]. This lipid-linked oligosaccharide is then flipped across the cell membrane to the periplasm (also by a member of the pgl gene family) and the glycan is transferred en bloc onto select asparagine residues by an oligosaccharide transferase (encoded by pglB in C.jejuni) [4, 73, 142, 146]. There is no evidence that the lipid-linked oligosaccharide is further modified after it is translocated across the membrane [143]. It is still uncertain whether bacterial N-glycosylation occurs co- or post-translationally [4], although it appears to occur independently of the protein translocation machinery since research indicates that the oligosaccharide transferase can transfer N-glycans to completely folded proteins [142, 147].
5. Archaea
The production of isoprenoids in Achaea is typically of interest, not due to its role in glycosylation, but due to the fact that isoprenoids make up a major component of archaeal membranes [88, 148]. In contrast with bacterial and eukaryotic membrane lipids, which consist of consist of fatty acid side chains linked to glycerol via ester linkages [50, 149], archaeal membrane lipids are made up of isoprenoid chains linked to glycerol via ether linkages [88, 150]. Archaea are generally thought to produce isoprenoids through the traditional MVA pathway [35, 88]. This idea was supported by the discovery of homologs to several genes in the MVA pathway, such as HMG-CoA synthase and HMG-CoA reductase, in the Archaeal genome [151-153]. Homologous genes for the last two enzymes in the pathway, however, were not identified [88, 149]. It was thought that these enzymes, phosphomevalonate kinase and mevalonate-5-pyrophosphate decarboxylase, could be nonorthologous enzymes, thus making them more difficult to identify and characterize [88]. In 2005, however, Grochowski et al. proposed an alternative route to produce IPP from acetyl-CoA in Archaea. They discovered an enzyme that catalyzed the formation of IPP from isopentenyl phosphate (IP) called IP kinase and proposed a archaeal variation of the MVA pathway (Fig. 2) [88]. Instead of mevalonate phosphate being phosphorylated, then decarboxylated (as in the typical MVA pathway), they propose a pathway in which mevalonate phosphate is first decarboxylated by a phosphomevalonate decarboxylase into IP and then phosphorylated, via the IP kinase they discovered, into IPP [88].
Until recently the method of DMAPP production in Archaea was a mystery, due to the failure of genomic analysis to identify any archaeal homologs of IPP isomerase [35]. The discovery of the type II IPP isomerase in bacteria [137] resolved this issue, however, and orthologs of this type II enzyme have now been found in all 11 archaeal families [35]. Current evidence suggests that archaea utilize a variation on the traditional MVA pathway, in combination with type II IPP isomerases, in their production of isoprenoids [50].
Unlike eukaryotes and bacteria, archaea do not use a single polyisoprenoid lipid donor in their N-glycosylation pathway. Archaea, like eukaryotes, use dolichol in their synthesis of LLOs but evidence of both Dol-P and Dol-PP being used as oligosaccharide carriers has been reported [73, 143]. In Haloferax volcanii, oligosaccharides are linked to dolichol via a monophosphate bridge [154], while in Halobacterium salinarum, both monophosphate and pyrophosphate linked oligosaccharides are found [84, 155, 156]. The dolichol used by H. volcanii is also unique, in that the ω-terminal isoprene, in addition to α-isoprene unit, is saturated [154]. Archaeal dolichols typically appear to be 10-12 isoprene units in length [84, 154].
Evidence suggests that archaeal N-glycosylation occurs on the external surface of the cell membrane [73, 84, 157]. A process similar to the eukaryotic N-glycosylation system is assumed, in which monosaccharides from nucleotide-activated sugar donors are sequentially added to the dolichol mono- or pyro-phosphate on the cytoplasmic face of the cell membrane [5, 84, 157]. After the oligosaccharide is assembled on the carrier lipid, it is translocated, by an unknown enzyme, across the membrane to the extracellular surface. In a final step, the oligosaccharide is transferred to an asparagine residue of the nascent protein by an oligosaccharide transferase [5, 84, 143]. At present, it is unknown whether this modification occurs post- or co-translationally [5]. An overview of the current understanding of the genes and enzymes involved in this process can be found in an excellent recent review on archaeal N-glycosylation by Yurish-Doutsch et al. [5].
Unlike eukaryotes, archaea display wide diversity in the linking sugars and sugar composition of their glycoproteins [5, 73]. Evidence of glycans containing a variety of saccharides, such as galactofuranose, galactouronic acid, glucose, glucuronic acid, iduronic acid, mannose, N-acetylgalactosamine, N-acetylglucosamine, and rhamnose, has been reported [84, 158-160]. Archaeal N-glycans are typically smaller than their eukaryotic counterparts and thus far, no antennary (branched) structures been detected in archaea [73, 84]. There is also evidence that some LLO are either transiently or permanently chemically modified by sulfation or methylation, and that methylation may be involved in translocation of sulfated oligosacchardies across the membrane [84, 158, 161, 162]. The archaeal N-glycosylation pathway is extremely interesting in its use of aspects from both the eukaryotic (MVA pathway, use of dolichol donor) and bacterial (Type II IPP isomerase) N-glycosylation pathways, but also in the aspects that are unique to the archaeal domain (variety in sugar structure, LLO methylation).
6. Conclusion
Although the eukaryotic isoprenoid and LLO biosynthetic pathways seem well understood, there remain many questions that could be addressed by further research in this area. For example, the final steps in the dolichol recycling mechanism, involving the dephosphorylation of dolichol pyrophosphate and the translocation of the non-glycosylated lipid intermediate back across the ER membrane, remain unclear. The evolving story of the Rft-1 “flippase” illustrates that the identity and specificity of the proteins that catalyze the flipping reactions in N-linked glycosylation remain to be elucidated. The details of polyprenol reductase activity also remain unknown. These basic enzymatic questions remain primarily because of the difficulty in producing substrates as well as the difficulty in isolating and studying the enzymes due to the hydrophobic nature of the membrane proteins themselves.
The bacterial pathway has been studied extensively in C. jejuni, but given the great diversity of this domain, it will be important to identify and fully understand N-glycosylation processes in other representative bacteria. Archaea have the potential to provide a rich source of information on the different pathways possible for generation of variety in both the lipid and oligosaccharide portions of the LLO. Hopefully our understanding of eukaryotic and bacterial N-glycosylation pathways will aid in the elucidation of the process in Archaea and other species. However, the variety and diversity seen in sugar structure and isoprenoid synthesis across the three domains of life show that nature still offers many surprises in the generation and modification of lipid-linked oligosaccharides for protein N-glycosylation.
Acknowledgement
This work was supported by Grant number R01 GM077530 from the National Institute of Health.
List of Abbreviations
- LLO
lipid-linked oligosaccharide
- OST
oligosaccharide transferase
- IPP
isopentenyl pyrophosphate
- DMAPP
dimethylallyl pyrophosphate
- MVA
mevalonate
- DOXP
1-deoxy-D-xylulose-5-phosphate
- HMG-CoA
3-hydroxy-3methylglutaryl-CoA
- HMGS
HMG-CoA synthase
- HMGR
HMG-CoA reductase
- MPPD
MVA-5-pyrophosphate decarboxylase
- Dxs
DOXP synthase
- IspC
DOXP reductase
- MEP
2-C-methyl-D-erythritol,4-phosphate
- CTP
cytidine triphosphate
- CDP-ME
4-pyrophosphocytidyl-2-C-methyl-D-erythritol
- IspD
CDP-ME synthase
- IspE
4-pyrophosphocytidyl-2C-methyl-D-erythritol Kinase
- CDP-MEP
CDP-ME-2-phosphate
- MECPP
2C-Methyl-D-erythritol-2,4-cyclopyrophosphate
- IspF
MECPP synthase
- CMP
cytidine monophosphate
- HMBPP
(E) 1-hydroxy-2-methyl-2--butenyl-4-pyrophosphate
- GPP
geranyl pyrophosphate
- FPP
farnesyl pyrophosphate
- GGPP
geranylgeranyl pyrophosphate
- FGPP
farnesylgeranyl pyrophosphate
- Poly-PP
polyprenyl pyrophosphates
- Poly-P
polyprenyl phosphate
- Dol-P
dolichol phosphate
- Dol-PP
dolichol pyrophosphate
- ER
endoplasmic reticulum
- UDP-GlcNAc
uridine diphosphate N-acetylglucosamine
- GDP-Man
guanosine diphosphate mannose
- Glc
glucose
- Oligo-PP
oligoprenyl pyrophosphate
- UPPS
undecaprenyl pyrophosphate synthase
- Und-PP
undecaprenyl pyrophosphate
- bacillosamine, Bac
2,4-diacetamido-2,4,6-trideoxyglucopyranose
- GalNAc
N-acetylgalactosamine
- IP
isopentenyl phosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mescher MF, Strominger JL. Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 1976;251:2005–14. [PubMed] [Google Scholar]
- 2.Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 1999;32:1022–30. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 3.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- 5.Yurist-Doutsch S, Chaban B, VanDyke DJ, Jarrell KF, Eichler J. Sweet to the extreme: protein glycosylation in Archaea. Mol. Microbiol. 2008;68:1079–84. doi: 10.1111/j.1365-2958.2008.06224.x. [DOI] [PubMed] [Google Scholar]
- 6.Hunter WN. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007;282:21573–7. doi: 10.1074/jbc.R700005200. [DOI] [PubMed] [Google Scholar]
- 7.Kuzuyama T, Seto H. Diversity of the biosynthesis of the isoprene units. Nat. Prod. Rep. 2003;20:171–83. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 8.Withers ST, Keasling JD. Biosynthesis and engineering of isoprenoid small molecules. Appl. Microbiol. Biotechnol. 2007;73:980–90. doi: 10.1007/s00253-006-0593-1. [DOI] [PubMed] [Google Scholar]
- 9.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993;295(Pt 2):517–24. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz W. Eisenreich, M., Arigoni A. Cartayrade, D., Zenk MH, Bacher A. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 1998;5:R221–33. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 11.Flesch G, Rohmer M. Prokaryotic hopanoids: the biosynthesis of the bacteriohopane skeleton. Formation of isoprenic units from two distinct acetate pools and a novel type of carbon/carbon linkage between a triterpene and D-ribose. Eur. J. Biochem. 1988;175:405–11. doi: 10.1111/j.1432-1033.1988.tb14210.x. [DOI] [PubMed] [Google Scholar]
- 12.Broers STJ. Uber die frijhen Stufen der Biosynthese von lsoprenoiden in Escherichia coli. ETH; Zurich: 1994. Thesis. [Google Scholar]
- 13.Schwarz MK. Terpen-Biosynthese in Ginkgo biloba: Eine Uberraschende Geschichte. ETH; Zurich: 1994. Thesis. [Google Scholar]
- 14.Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH. Terpenoid biosynthesis from 1-deoxy-D-xylulose in higher plants by intramolecular skeletal rearrangement. Proc. Natl. Acad. Sci. U S A. 1997;94:10600–5. doi: 10.1073/pnas.94.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohdich F, Hecht S, Gartner K, et al. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. U S A. 2002;99:1158–63. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. Glyceraldehyde 3-Phosphate and Pyruvate as Precursors of Isoprenic Units in an Alternative Non-mevalonate Pathway for Terpenoid Biosynthesis. J. Am. Chem. Soc. 1996;118:2564–2566. [Google Scholar]
- 17.Lynen F. Incorporation of acetate into isoprenoids. In: Wolstenholme GEW, editor. Ciba Foundarion Symposium on the Biosynthesis of Terpenes and Sterols. Churchill, London: 1959. pp. 95–116. [Google Scholar]
- 18.Tavormina PA, Gibbs MH, Huff JW. The utilization of beta-hydroxyl-beta-methylvalerolactone in cholestorol biosynthesis. J. Am. Chem. Soc. 1956;78:4498–4499. [Google Scholar]
- 19.Wright LD, Cresson EL, Skeggs HR, et al. Isolation of a New Acetate-replacing Factor. J. Am. Chem. Soc. 1956;78:5273–5275. [Google Scholar]
- 20.Rudney H. The biosynthesis of Beta-hydroxy-Beta-methylglutaryl coenzyme-A and its conversion to mevalonic acid. In: Wolstenholme GEW, editor. Ciba Foundarion Symposium on the Biosynthesis of Terpenes and Sterols. Churchill, London: 1959. pp. 75–90. [Google Scholar]
- 21.Swiezewska E, Danikiewicz W. Polyisoprenoids: structure, biosynthesis and function. Prog. Lipid Res. 2005;44:235–58. doi: 10.1016/j.plipres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro DJ, Rodwell VW. Regulation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and cholesterol synthesis. J. Biol. Chem. 1971;246:3210–6. [PubMed] [Google Scholar]
- 23.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–30. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 24.Henning U, Moslein EM, Lynen F. Biosynthesis of terpenes. V. Formation of 5-pyrophosphomevalonic acid by phosphomevalonic kinase. Arch. Biochem. Biophys. 1959;83:259–67. doi: 10.1016/0003-9861(59)90031-1. [DOI] [PubMed] [Google Scholar]
- 25.Tchen TT. On the formation of a phosphorylated derivative of mevalonic acid. J. Am. Chem. Soc. 1957;79:6344–6345. [Google Scholar]
- 26.Amdur BH, Rilling H, Bloch K. The enzymatic conversion of mevalonic acid to sqalene. J. Am. Chem. Soc. 1957;79:2646–2647. [Google Scholar]
- 27.Bloch K. Sterol molecule: structure, biosynthesis, and function. Steroids. 1992;57:378–83. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]
- 28.De Waard A, Phillips AH, Bloch K. Mechanism of formation of isopentenyl pyrophosphate. J. Am. Chem. Soc. 1959;81:2913–2914. [Google Scholar]
- 29.Lynen F, Eggerer H, Henning U, Kessel I. Zur Biosynthese der Terpene III. Angew. Chem. 1958;70:738–742. [Google Scholar]
- 30.Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc. Natl. Acad. Sci. U S A. 1998;95:2105–10. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprenger GA, Schorken U, Wiegert T, et al. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. U S A. 1997;94:12857–62. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. U S A. 1998;95:9879–84. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohdich F, Wungsintaweekul J, Fellermeier M, et al. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. U S A. 1999;96:11758–63. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luttgen H, Rohdich F, Herz S, et al. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-Derythritol. Proc. Natl. Acad. Sci. U S A. 2000;97:1062–7. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohdich F, Bacher A, Eisenreich W. Perspectives in anti-infective drug design. The late steps in the biosynthesis of the universal terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. Bioorg. Chem. 2004;32:292–308. doi: 10.1016/j.bioorg.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Herz S, Wungsintaweekul J, Schuhr CA, et al. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. U S A. 2000;97:2486–90. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hecht S, Eisenreich W, Adam P, et al. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. U S A. 2001;98:14837–42. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zepeck F, Grawert T, Kaiser J, et al. Biosynthesis of isoprenoids. purification and properties of IspG protein from Escherichia coli. J. Org. Chem. 2005;70:9168–74. doi: 10.1021/jo0510787. [DOI] [PubMed] [Google Scholar]
- 39.Kemp LE, Alphey MS, Bond CS, et al. The identification of isoprenoids that bind in the intersubunit cavity of Escherichia coli 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase by complementary biophysical methods. Acta. Crystallogr. D Biol. Crystallogr. 2005;61:45–52. doi: 10.1107/S0907444904025971. [DOI] [PubMed] [Google Scholar]
- 40.Ahn CS, Pai H-S. Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis, in organelle biogenesis and cell morphogenesis in Nicotiana benthamiana. Plant Mol. Biol. 2008;66:503–517. doi: 10.1007/s11103-007-9286-0. [DOI] [PubMed] [Google Scholar]
- 41.Carretero-Paulet L, Ahumada I, Cunillera N, et al. Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Plant Physiol. 2002;129:1581–91. doi: 10.1104/pp.003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krag SS. The importance of being dolichol. Biochem. Biophys. Res. Commun. 1998;243:1–5. doi: 10.1006/bbrc.1997.7828. [DOI] [PubMed] [Google Scholar]
- 43.Poulter CD, Rilling HC. The prenyl transfer reaction. Enzymatic and mechanistic studies of the l′-4 coupling reaction in the terpene biosynthetic pathway. Acc. Chem. Res. 1978;11:307–313. [Google Scholar]
- 44.Dorsey JK, Dorsey JA, Porter JW. The purification and properties of pig liver geranyl pyrophosphate synthetase. J. Biol. Chem. 1966;241:5353–60. [PubMed] [Google Scholar]
- 45.Holloway PW, Popjak G. The purification of 3,3-dimethylallyl- and geranyltransferase and of isopentenyl pyrophosphate isomerase from pig liver. Biochem. J. 1967;104:57–70. doi: 10.1042/bj1040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellogg BA, Poulter CD. Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1997;1:570–8. doi: 10.1016/s1367-5931(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 47.Ko TP, Chen YK, Robinson H, et al. Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J. Biol. Chem. 2001;276:47474–82. doi: 10.1074/jbc.M106747200. [DOI] [PubMed] [Google Scholar]
- 48.Liang PH, Ko TP, Wang AH. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002;269:3339–54. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 49.Anderson MS, Yarger JG, Burck CL, Poulter CD. Farnesyl diphosphate synthetase. Molecular cloning, sequence, and expression of an essential gene from Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:19176–84. [PubMed] [Google Scholar]
- 50.Koga Y, Morii H. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones J, Krag SS, Betenbaugh MJ. Controlling N-linked glycan site occupancy. Biochim. Biophys. Acta. 2005;1726:121–37. doi: 10.1016/j.bbagen.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Schenk B, Fernandez F, Waechter CJ. The ins(ide) and out(side) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 2001;11:61R–70R. doi: 10.1093/glycob/11.5.61r. [DOI] [PubMed] [Google Scholar]
- 53.Kato S, Tsuji M, Nakanishi Y, Suzuki S. Enzymatic dephosphorylation of dolichyl pyrophosphate--the bacitracin-sensitive, rate-limiting step for dolichyl mannosyl phosphate synthesis in rat liver microsomes. Biochem. Biophys. Res. Commun. 1980;95:770–6. doi: 10.1016/0006-291x(80)90853-0. [DOI] [PubMed] [Google Scholar]
- 54.Wedgwood JF, Strominger JL. Enzymatic activities in cultured human lymphocytes that dephosphorylate dolichyl pyrophosphate and dolichyl phosphate. J. Biol. Chem. 1980;255:1120–3. [PubMed] [Google Scholar]
- 55.Quellhorst GJ, Jr., Hall CW, Robbins AR, Krag SS. Synthesis of dolichol in a polyprenol reductase mutant is restored by elevation of cis-prenyl transferase activity. Arch. Biochem. Biophys. 1997;343:19–26. doi: 10.1006/abbi.1997.0141. [DOI] [PubMed] [Google Scholar]
- 56.Sagami H, Kurisaki A, Ogura K. Formation of dolichol from dehydrodolichol is catalyzed by NADPH-dependent reductase localized in microsomes of rat liver. J. Biol. Chem. 1993;268:10109–13. [PubMed] [Google Scholar]
- 57.Rossignol DP, Lennarz WJ, Waechter CJ. Induction of phosphorylation of dolichol during embryonic development of the sea urchin. J. Biol. Chem. 1981;256:10538–42. [PubMed] [Google Scholar]
- 58.Volpe JJ, Sakakihara Y, Rust RS. Dolichol kinase and the regulation of dolichyl phosphate levels in developing brain. Brain Res. 1987;428:193–200. doi: 10.1016/0165-3806(87)90117-9. [DOI] [PubMed] [Google Scholar]
- 59.Higashi Y, Siewert G, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. XIX. Isoprenoid alcohol phosphokinase. J. Biol. Chem. 1970;245:3683–90. [PubMed] [Google Scholar]
- 60.Sandermann H, Jr., Strominger JL. C 55 -isoprenoid alcohol phosphokinase: an extremely hydrophobic protein from the bacterial membrane. Proc. Natl. Acad. Sci. U S A. 1971;68:2441–3. doi: 10.1073/pnas.68.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rupar CA, Rip JW, Chaudhary N, Carroll KK. The subcellular localization of enzymes of dolichol metabolism in rat liver. J. Biol. Chem. 1982;257:3090–4. [PubMed] [Google Scholar]
- 62.Taylor ME, Drickamer K. Introduction to Glycobiology. Oxford University Press; New York: 2003. [Google Scholar]
- 63.Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta. 1999;1426:239–57. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 64.McLachlan KR, Krag SS. Substrate specificity of N-acetylglucosamine 1-phosphate transferase activity in Chinese hamster ovary cells. Glycobiology. 1992;2:313–9. doi: 10.1093/glycob/2.4.313. [DOI] [PubMed] [Google Scholar]
- 65.McLachlan KR, Krag SS. Three enzymes involved in oligosaccharide-lipid assembly in Chinese hamster ovary cells differ in lipid substrate preference. J. Lipid Res. 1994;35:1861–8. [PubMed] [Google Scholar]
- 66.Dotson SB, Rush JS, Ricketts AD, Waechter CJ. Mannosylphosphoryldolichol-mediated O-mannosylation of yeast glycoproteins: stereospecificity and recognition of the alpha-isoprene unit by a purified mannosyltransferase. Arch. Biochem. Biophys. 1995;316:773–9. doi: 10.1006/abbi.1995.1103. [DOI] [PubMed] [Google Scholar]
- 67.Palamarczyk G, Lehle L, Mankowski T, Chojnacki T, Tanner W. Specificity of solubilized yeast glycosyl transferases for polyprenyl derivatives. Eur. J. Biochem. 1980;105:517–23. doi: 10.1111/j.1432-1033.1980.tb04527.x. [DOI] [PubMed] [Google Scholar]
- 68.Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 2006;45:6802–18. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- 69.Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 70.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–64. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 71.Kukuruzinska MA, Bergh ML, Jackson BJ. Protein glycosylation in yeast. Annu. Rev. Biochem. 1987;56:915–44. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- 72.Tanner W, Lehle L. Protein glycosylation in yeast. Biochim. Biophys. Acta. 1987;906:81–99. doi: 10.1016/0304-4157(87)90006-2. [DOI] [PubMed] [Google Scholar]
- 73.Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 2008;18:544–550. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 74.Acosta-Serrano A, O'Rear J, Quellhorst G, et al. Defects in the N-linked oligosaccharide biosynthetic pathway in a Trypanosoma brucei glycosylation mutant. Eukaryot. Cell. 2004;3:255–63. doi: 10.1128/EC.3.2.255-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parodi AJ. N-glycosylation in trypanosomatid protozoa. Glycobiology. 1993;3:193–9. doi: 10.1093/glycob/3.3.193. [DOI] [PubMed] [Google Scholar]
- 76.Snider MD, Rogers OC. Transmembrane movement of oligosaccharide-lipids during glycoprotein synthesis. Cell. 1984;36:753–61. doi: 10.1016/0092-8674(84)90355-6. [DOI] [PubMed] [Google Scholar]
- 77.Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 2006;45:6802–18. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- 78.Ko K, Ahn MH, Song M, et al. Glyco-engineering of biotherapeutic proteins in plants. Mol. Cells. 2008;25:494–503. [PubMed] [Google Scholar]
- 79.Rearick JI, Fujimoto K, Kornfeld S. Identification of the mannosyl donors involved in the synthesis of lipid-linked oligosaccharides. J. Biol. Chem. 1981;256:3762–9. [PubMed] [Google Scholar]
- 80.Haselbeck A, Tanner W. Dolichyl phosphate-mediated mannosyl transfer through liposomal membranes. Proc. Natl. Acad. Sci. U S A. 1982;79:1520–4. doi: 10.1073/pnas.79.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Helenius J, Ng DT, Marolda CL, Walter P, Valvano MA, Aebi M. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature. 2002;415:447–50. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- 82.Frank CG, Sanyal S, Rush JS, Waechter CJ, Menon AK. Does Rft1 flip an Nglycan lipid precursor? Nature. 2008;454:E3–4. doi: 10.1038/nature07165. discussion E4-5. [DOI] [PubMed] [Google Scholar]
- 83.Wildt S, Gerngross TU. The humanization of N-glycosylation pathways in yeast. Nat. Rev. Microbiol. 2005;3:119–28. doi: 10.1038/nrmicro1087. [DOI] [PubMed] [Google Scholar]
- 84.Eichler J, Adams MW. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rush JS, Gao N, Lehrman MA, Waechter CJ. Recycling of dolichyl monophosphate to the cytoplasmic leaflet of the endoplasmic reticulum after the cleavage of dolichyl pyrophosphate on the lumenal monolayer. J. Biol. Chem. 2008;283:4087–93. doi: 10.1074/jbc.M707067200. [DOI] [PubMed] [Google Scholar]
- 86.Rip JW, Rupar CA, Ravi K, Carroll KK. Distribution, metabolism and function of dolichol and polyprenols. Prog. Lipid. Res. 1985;24:269–309. doi: 10.1016/0163-7827(85)90008-6. [DOI] [PubMed] [Google Scholar]
- 87.Crane FL. Biochemical functions of coenzyme Q10. J. Am. Coll. Nutr. 2001;20:591–8. doi: 10.1080/07315724.2001.10719063. [DOI] [PubMed] [Google Scholar]
- 88.Grochowski LL, Xu H, White RH. Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 2006;188:3192–8. doi: 10.1128/JB.188.9.3192-3198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stoll J, Rosenwald AG, Krag SS. A Chinese hamster ovary cell mutant F2A8 utilizes polyprenol rather than dolichol for its lipid-dependent asparagine-linked glycosylation reactions. J. Biol. Chem. 1988;263:10774–82. [PubMed] [Google Scholar]
- 90.Keller RK, Jehle E, Adair WL., Jr. The origin of dolichol in the liver of the rat. Determination of the dietary contribution. J. Biol. Chem. 1982;257:8985–9. [PubMed] [Google Scholar]
- 91.Sagami H, Lennarz WJ. Glycoprotein synthesis in Drosophila Kc cells. Biosynthesis of dolichol-linked saccharides. J. Biol. Chem. 1987;262:15610–7. [PubMed] [Google Scholar]
- 92.Crick DC, Rush JS, Waechter CJ. Characterization and localization of a long-chain isoprenyltransferase activity in porcine brain: proposed role in the biosynthesis of dolichyl phosphate. J. Neurochem. 1991;57:1354–62. doi: 10.1111/j.1471-4159.1991.tb08301.x. [DOI] [PubMed] [Google Scholar]
- 93.Crick DC, Schulbach MC, Zink EE, et al. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 2000;182:5771–8. doi: 10.1128/jb.182.20.5771-5778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hubbard SC, Robbins PW. Synthesis and processing of protein-linked oligosaccharides in vivo. J. Biol. Chem. 1979;254:4568–76. [PubMed] [Google Scholar]
- 95.Rosenwald AG, Krag SS. Lec9 CHO glycosylation mutants are defective in the synthesis of dolichol. J. Lipid Res. 1990;31:523–33. [PubMed] [Google Scholar]
- 96.Rosenwald AG, Stanley P, McLachlan KR, Krag SS. Mutants in dolichol synthesis: conversion of polyprenol to dolichol appears to be a rate-limiting step in dolichol synthesis. Glycobiology. 1993;3:481–8. doi: 10.1093/glycob/3.5.481. [DOI] [PubMed] [Google Scholar]
- 97.Nabeta K, Saitoh T, Adachi K, Komuro K. Biosynthesis of phytyl side-chain of chlorophyll a: apparent reutilization of carbon dioxide evolved during acetate assimilation in biosynthesis of chloroplastic isoprenoid. Chem. Commun. 1998:671–672. [Google Scholar]
- 98.Owen SM, Peñuelas J. Oportunistic emissions of volatile isoprenoids. Trends Plant Sci. 2005;10:420–426. doi: 10.1016/j.tplants.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 99.Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–48. doi: 10.1016/s1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- 100.Gibson RW, Pickett JA. Wild potato repels aphids by release of aphid alarm pheromone. Nature. 1983;302:608–609. [Google Scholar]
- 101.Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn SE. Compromised disease resistance in saponin-decifient plants. Proc. Natl. Acad. Sci. USA. 1999;96:12923–12928. doi: 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Demming-Adams B, Adams WW., III Antioxidants in photosynthesis and human nutrition. Science. 2002;298:2149–2153. doi: 10.1126/science.1078002. [DOI] [PubMed] [Google Scholar]
- 103.Peñuelas J, Munné-Bosch S. Isoprenoids: an evolutionary pool for photoprotection. Trends Plant Sci. 2005;10:166–169. doi: 10.1016/j.tplants.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 104.Grotewold E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- 105.Piechulla B, Pott MB. Plant scents – mediators of inter- and intraorganismic communication. Planta. 2003;217:687–689. doi: 10.1007/s00425-003-1047-y. [DOI] [PubMed] [Google Scholar]
- 106.Okada K, Kasahara H, Yamaguchi S, et al. Genetic evidence for the role of isopentenyl diphosphate isomerases in the mevalonate pathway and plant development in Arabidopsis. Plant Cell Physiol. 2008;49:604–616. doi: 10.1093/pcp/pcn032. [DOI] [PubMed] [Google Scholar]
- 107.Disch A, Schwender J, Muller C, Lichtenthaler HK, Rohmer M. Distribution of the mevalonate and glyceraldehyde phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714. Biochem J. 1998;333(Pt 2):381–8. doi: 10.1042/bj3330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eisenreich W, Rohdich F, Bacher A. Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci. 2001;6:78–84. doi: 10.1016/s1360-1385(00)01812-4. [DOI] [PubMed] [Google Scholar]
- 109.Rodríguez-Concepción M, Boronat A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids: a metabolic milestone achieved through genomics. Plant Physiol. 2002;130:1079–1089. doi: 10.1104/pp.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wanke M, Skorupinska-Tudek K, Swiezewska E. Isoprenoid biosynthesis via 1-deoxy-D-xylulose 5-phosphate/2-C-methyl-D-erythritol 4-phosphate (DOXP/MEP) pathway. Acta Biochim. Pol. 2001;48:663–672. [PubMed] [Google Scholar]
- 111.Bouvier F, Rahier A, Camara B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 2005;44:357–429. doi: 10.1016/j.plipres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 112.Massé G, Belt ST, Rowland SJ, Rohmer M. Isoprenoid biosynthesis in the diatoms Rhizosolenia setigera (Brightwell) and Haslea ostrearia (Simonsen) Proc. Natl. Acad. Sci. USA. 2004;101:4413–4418. doi: 10.1073/pnas.0400902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Disch A, Schwender J, Müller C, Lichtenthaler HK, Rohmer M. Distribution of the mevalonate and glyceraldehydes phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714. Biochem. J. 1998;333:381–388. doi: 10.1042/bj3330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schwender J, Gemünden C, Lichtenthaler HK. Chlorophyta exclusively use the 1-deoxyxylulose 5-phosphate/2-C-methylerythritol 4-phosphate pathway for the biosynthesis of isoprenoids. Planta. 2001;212:416–423. doi: 10.1007/s004250000409. [DOI] [PubMed] [Google Scholar]
- 115.Yu J, Ma PJ, Shi DJ, Li SM, Wang CL. Homologous comparisons of photosynthetic system I genes among cyanobacteria and chloroplasts. J. Integr. Plant Biol. 2008;50:929–40. doi: 10.1111/j.1744-7909.2008.00679.x. [DOI] [PubMed] [Google Scholar]
- 116.Cunningham FX, Jr., Lafond TP, Gantt E. Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 2000;182:5841–5848. doi: 10.1128/jb.182.20.5841-5848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ershov YV, Gantt RR, Cunningham FX, Jr., Gantt E. Isoprenoid biosynthesis in Synechocystis sp. strain PCC6803 is stimulated by compounds of the pentose phosphate cycle but not by pyruvate or deoxyxylulose-5-phosphate. J. Bacteriol. 2002;184:5045–5051. doi: 10.1128/JB.184.18.5045-5051.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Okada K, Toshiharu H. Cyanobacterial non-mevalonate pathway: (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase interacts with ferredoxin in Thermosynechococcus elongatus BP-1. J. Biol. Chem. 2005;280:20672–20679. doi: 10.1074/jbc.M500865200. [DOI] [PubMed] [Google Scholar]
- 119.Hampel D, Mosandl A, Wüst M. Biosynthesis of mono- and sesquiterpenes in carrot roots and leaves (Daucus carota L.): metabolic cross talk of cytosolic mevalonate and plastidial methylerythritol phosphate pathways. Phytochem. 2005;66:305–311. doi: 10.1016/j.phytochem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 120.Itoh D, Kawano K, Nabeta K. Biosynthesis of chloroplastidic and extrachloroplastidic terpenoids in liverwort cultured cells: 13C serine as a probe of terpene biosynthesis via mevalonate and non-mevalonate pathways. J. Nat. Prod. 2003;66:332–336. doi: 10.1021/np0204141. [DOI] [PubMed] [Google Scholar]
- 121.Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 2002;277:45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- 122.Yang J-W, Orihara Y. Biosynthesis of abietane diterpenoids in cultured cells of Torreya nucifera var. radicans: biosynthetic inequality of the FPP part and the terminal IPP. Tetrahedron. 2002;58:1265–1270. [Google Scholar]
- 123.Bick JA, Lange BM. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 2003;415:146–154. doi: 10.1016/s0003-9861(03)00233-9. [DOI] [PubMed] [Google Scholar]
- 124.Laule O, Fürholz A, Chang H-S, et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Skorupinska-Tudek K, Poznanski J, Wojcik J, et al. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of dolichols in plants. J. Biol. Chem. 2008;283:21024–21035. doi: 10.1074/jbc.M706069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gutkowska M, Bieńkowski T, Hung VS, et al. Proteins are polyisoprenylated in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2004;322:998–1004. doi: 10.1016/j.bbrc.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 127.Kurisaki A, Sagami H, Ogura K. Distribution of polyprenols and dolichols in soybean plant. Phytochem. 1997;44:45–50. [Google Scholar]
- 128.Ravi K, Rip JW, Carroll KK. Characterization of dolichol and dolichyl phosphate phosphatase from syoa beans (Gylcine max) Biochem. J. 1983;213:513–518. doi: 10.1042/bj2130513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hopp HE, Daleo GR, Romero PA, Lezica RP. Biosynthesis of dolichyl phosphate: characterization and site of synthesis in algae. Plant Physiol. 1978;61:248–251. doi: 10.1104/pp.61.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lang WC. Glycoprotein biosynthesis in Chlamydomonas. Biochem. J. 1984;220:747–754. doi: 10.1042/bj2200747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carretero-Paulet L, Ahumada I, Cunillera N, et al. Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Plant Physiol. 2002;129:1581–91. doi: 10.1104/pp.003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Learned RM. Light suppresses 3-hydroxy-3-methylglutaryl coenzyme A reductase gene expression in Arabidopsis thaliana. Plant Physiol. 1996;110:645–655. doi: 10.1104/pp.110.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodríguez-Concepción M, Forés O, Martínez-García JF, et al. Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell. 2003;16:144–156. doi: 10.1105/tpc.016204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rodríguez-Concepción M. Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem. Rev. 2006;5:1–15. [Google Scholar]
- 135.Anderson RG, Hussey H, Baddiley J. The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane. Biochem. J. 1972;127:11–25. doi: 10.1042/bj1270011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hedl M, Tabernero L, Stauffacher CV, Rodwell VW. Class II 3-hydroxy-3-methylglutaryl coenzyme A reductases. J. Bacteriol. 2004;186:1927–32. doi: 10.1128/JB.186.7.1927-1932.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. U S A. 2001;98:932–7. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hahn FM, Hurlburt AP, Poulter CD. Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 1999;181:4499–504. doi: 10.1128/jb.181.15.4499-4504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wright A, Dankert M, Fennessey P, Robbins PW. Characterization of a polyisoprenoid compound functional in O-antigen biosynthesis. Proc. Natl. Acad. Sci. U S A. 1967;57:1798–803. doi: 10.1073/pnas.57.6.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Allen CM. Purification and characterization of undecaprenylpyrophosphate synthetase. Methods Enzymol. 1985;110:281–99. doi: 10.1016/s0076-6879(85)10085-6. [DOI] [PubMed] [Google Scholar]
- 141.Wolucka BA, McNeil MR, de Hoffmann E, Chojnacki T, Brennan PJ. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 1994;269:23328–35. [PubMed] [Google Scholar]
- 142.Chiba Y, Jigami Y. Production of humanized glycoproteins in bacteria and yeasts. Curr. Opin. Chem. Biol. 2007;11:670–6. doi: 10.1016/j.cbpa.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 143.Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 2006;61:259–68. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- 144.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 2005;3:225–37. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 145.Young NM, Brisson JR, Kelly J, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 2002;277:42530–9. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 146.Wacker M, Linton D, Hitchen PG, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–3. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 147.Kowarik M, Numao S, Feldman MF, et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 2006;314:1148–50. doi: 10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- 148.Langworthy TA. Lipids of Archaebacteria. Archaebacteria, Academic Press; New York: 1985. pp. 459–497. [Google Scholar]
- 149.Boucher Y, Kamekura M, Doolittle WF. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 2004;52:515–27. doi: 10.1111/j.1365-2958.2004.03992.x. [DOI] [PubMed] [Google Scholar]
- 150.Nishimura Y, Eguchi T. Biosynthesis of archaeal membrane lipids: digeranylgeranylglycerophospholipid reductase of the thermoacidophilic archaeon Thermoplasma acidophilum. J. Biochem. 2006;139:1073–81. doi: 10.1093/jb/mvj118. [DOI] [PubMed] [Google Scholar]
- 151.Selkov E, Maltsev N, Olsen GJ, Overbeek R, Whitman WB. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 152.Bult CJ, White O, Olsen GJ, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–73. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 153.Kyrpides NC, Olsen GJ, Klenk HP, White O, Woese CR. Methanococcus jannaschii genome: revisited. Microb. Comp. Genomics. 1996;1:329–38. [PubMed] [Google Scholar]
- 154.Kuntz C, Sonnenbichler J, Sonnenbichler I, Sumper M, Zeitler R. Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii. Glycobiology. 1997;7:897–904. doi: 10.1093/glycob/7.7.897. [DOI] [PubMed] [Google Scholar]
- 155.Eichler J. Facing extremes: archaeal surface-layer (glyco)proteins. Microbiology. 2003;149:3347–51. doi: 10.1099/mic.0.26591-0. [DOI] [PubMed] [Google Scholar]
- 156.Lechner J, Wieland F. Structure and biosynthesis of prokaryotic glycoproteins. Annu. Rev. Biochem. 1989;58:173–94. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- 157.Mescher MF, Strominger JL. Glycosylation of the surface glycoprotein of Halobacterium salinarium via a cyclic pathway of lipid-linked intermediates. FEBS Lett. 1978;89:37–41. doi: 10.1016/0014-5793(78)80517-1. [DOI] [PubMed] [Google Scholar]
- 158.Karcher U, Schroder H, Haslinger E, et al. Primary structure of the heterosaccharide of the surface glycoprotein of Methanothermus fervidus. J. Biol. Chem. 1993;268:26821–6. [PubMed] [Google Scholar]
- 159.Mengele R, Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 1992;267:8182–5. [PubMed] [Google Scholar]
- 160.Paul G, Lottspeich F, Wieland F. Asparaginyl-N-acetylgalactosamine. Linkage unit of halobacterial glycosaminoglycan. J. Biol. Chem. 1986;261:1020–4. [PubMed] [Google Scholar]
- 161.Lechner J, Wieland F, Sumper M. Biosynthesis of sulfated saccharides Nglycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J. Biol. Chem. 1985;260:860–6. [PubMed] [Google Scholar]
- 162.Lechner J, Wieland F, Sumper M. Transient methylation of dolichyl oligosaccharides is an obligatory step in halobacterial sulfated glycoprotein biosynthesis. J. Biol. Chem. 1985;260:8984–9. [PubMed] [Google Scholar]