Abstract
Hsp90 is an ATP-dependent molecular chaperone which assists the maturation of a large set of target proteins. Members of the highly conserved Hsp90 family are found from bacteria to higher eukaryotes, with homologues in different organelles. The core architecture of Hsp90 is defined by the N-terminal ATP binding domain followed by the middle domain and the C-terminal dimerization domain. A long, highly charged linker between the N-terminal domain and the middle domain is a feature characteristic for Hsp90s of eukaryotic organisms. We set out to clarify the function of this linker by studying the effects of deletions in this region in vivo and in vitro. Here we show that increasing deletions in the charged linker region lead to defects ranging from mild temperature sensitivity to a lethal phenotype. The lethal deletion variants investigated in this study still exhibit ATPase activity. Deletion of the charged linker ultimately causes a loss of Hsp90 regulation by co-chaperones, as the sensitivity for Aha1-mediated ATPase acceleration declines, and binding of p23/Sba1 is lost in non-viable deletion constructs. In vivo client assays additionally demonstrated that the deletion of the linker had a pronounced effect on the ability of Hsp90 to facilitate client activation. A partial reconstruction of the linker sequence showed that the supplementation by artificial sequences can rescue the functionality of Hsp90 and restore the conformational flexibility of the protein, required for the processing of client proteins.
Hsp903 is an ATP-dependent molecular chaperone present in the cytosol of eubacteria and eucaryotes. It regulates the maturation and activation of numerous proteins involved in signal transduction, cell cycle control, hormone signaling, and transcription (1–3). Recent crystal structures of HtpG from Escherichia coli and Hsp90 from Saccharomyces cerevisiae show that the overall structural organization of the proteins is highly conserved (4–6). Both prokaryotic and eukaryotic Hsp90 proteins consist of an N-terminal ATP binding domain, a middle domain involved in client protein binding, and a C-terminal dimerization domain. During the ATPase cycle, large conformational changes in the Hsp90 dimer lead to the transient dimerization of the N-terminal domains and their association with the middle domains (7–12). Despite the conservation of the basic molecular architecture and the ATPase mechanism, there are major differences between Hsp90 from prokaryotes and eukaryotes. The most striking difference is the emergence of a large set of co-chaperones in eucaryotes that bind to Hsp90 and seem to modulate and expand its properties (13). Furthermore, in contrast to procaryotes, Hsp90 is an essential protein in eucaryotes (14, 15) and it had been shown in S. cerevisiae that ATP hydrolysis by Hsp90 is required for sustaining its essential function (16–18). Finally, eukaryotic Hsp90 contains additional structural elements compared with prokaryotic Hsp90; that is, a long charged linker between the N terminus and the middle domain and a conserved motif (MEEVD) at the extended C-terminal end of the eucaryotic protein. Although it has been established that the C-terminal motif is the interaction site for TPR domain containing co-chaperones (19–21), the function of the charged linker is less than clear. In general terms, it seems to contribute to the flexibility of the protein as the successful crystallization of Hsp90 from S. cerevisiae required the linker to be shortened and modified (6). Hallmarks of this linker region are its low sequence complexity and the high amount of charged amino acids. In yeast Hsp90, the motif (D/E)(D/E)(D/E)KK is repeated five times in the linker region (22). Various secondary structure prediction programs canonically predict the charged linker region as predominantly in a coiled-coil conformation (23, 24). In a previous complementation study, it was shown that the charged linker is dispensable for the essential function of Hsp90 in S. cerevisiae, as a construct lacking residues 211–259 was still able to support cell proliferation (25). In this study we constructed and analyzed a set of variants of Hsp90 in which the linker region was successively shortened, with a view to determine its contribution to the mechanism of Hsp90 in vivo and in vitro.
MATERIALS AND METHODS
Cloning and Protein Purification
The deletion mutants of yeast Hsp90 were generated using standard molecular biology techniques. The codons of the amino acids to be deleted were omitted by using a primer construct annealing to the 5′- and 3′- flanking regions of the deletion. In the primers an overlap between the N-terminal and the C-terminal fragments was generated, allowing a linker PCR approach to create the final construct. The construct was then inserted into the pRS423 Vector (26) for expression in yeast using the BamHI/NotI restriction sites. For expression in E. coli, the constructs were amplified with NdeI/NotI restriction sites using the respective yeast vector constructs as templates and inserted into the pET28a Vector (Novagen, Madison, WI). All plasmids were sequenced at GATC Biotech (Konstanz, Germany). For the supplementation constructs, Gly-Ser linker sequences were introduced by PCR mutagenesis to bring the length of the linker to the same length as Hsp90 Δ211–259. This resulted in the sequences GSSGSSGIEELNK for Hsp90 Δ211–266SUP and GSSGSSGSSGSSG for Hsp90 Δ211–272SUP. All proteins were expressed and purified according to a published protocol (27) Purified protein was frozen in liquid nitrogen and stored at −80 °C in standard buffer (40 mm HEPES, 150 mm KCl, pH 7.5). The purity of the proteins was assessed by SDS-PAGE and determined to be higher than 98%. The extinction coefficients used for protein concentration calculations were obtained from the ProtParam analysis of the respective primary sequences.
Secondary Structure Measurements
Circular dichroism spectroscopy was performed using a Jasco 715 spectropolarimeter (Jasco, Gross-Umstadt, Germany). Protein samples were diluted to 0.15 mg/ml in 40 mm potassium phosphate, pH 7.5, and measurements were performed in a quartz cuvette with a path length of 0.1 cm. 10 accumulations were performed at a scan speed of 20 nm/min. All spectra were corrected for buffer background.
Analysis of Protein Stability
Protein stability was determined by urea-induced unfolding. Different urea concentrations were added to protein samples of 50 μg/ml and incubated for 24 h. Fluorescence emission spectra were recorded in a Fluoromax-2 instrument (Spex, Edison, NJ) using an excitation wavelength of 280 nm. All spectra were corrected for buffer background. The maximal difference in signal amplitude between the folded and unfolded state of the protein was found to be at 325 nm, and this wavelength was used to monitor the urea-induced unfolding transition. The urea concentration at the midpoint of the transition was derived from the plot of the fluorescence signal versus the urea concentration.
Determination of ATPase Activities
ATPase assays were performed as described earlier using an ATP regenerating system (28). Concentrations of Hsp90 were 2 μm throughout. Assays were measured in 40 mm HEPES, pH 7.5, 150 mm KCl, 5 mm MgCl2, 2 mm ATP, at 30 °C. Data were recorded for 40 min, and the assay was evaluated using the Sigma Plot software package (Systat Software, Erkrath, Germany). For analysis of Aha1-induced activation, the buffer was 40 mm HEPES/KOH, pH 7.5, 20 mm KCl, 5 mm MgCl2, as binding of Aha1 is much tighter at low salt conditions (43).
Nucleotide Binding
ATPase activities were measured at varying ATP concentrations ranging from 25 μm to 2.5 mm. The resulting activities (kcat) were analyzed using least square data analysis and allowed the calculation of the apparent Km of ATP for the different Hsp90 constructs.
SPR Spectroscopy
SPR experiments were performed on a Biacore X Instrument (GE Healthcare). A CM5 chip (GE Healthcare) was loaded with p23/Sba1, yielding 1000 resonance units of immobilized protein. Measurements were performed at 25 °C in 40 mm HEPES, pH 7.5, 150 mm KCl, 5 mm MgCl2 at a flow rate of 20 μl/min. 65-μl injections of Hsp90 were supplemented with 1 mm ATPγS.
Analytical Ultracentrifugation
Analytical ultracentrifugation experiments were performed to assess whether the Hsp90 deletion variants are capable of binding the cochaperone Aha1. Analytical ultracentrifugation was performed in a Beckman ProteomeLab XL-A (Beckman Instruments) equipped with a fluorescence detector (Aviv Biomedical, Lakewood, NJ) (29). Runs were performed at 42,000 rpm with a Ti-50 Rotor (Beckman Instruments).
The formation of larger complexes of Aha1 and Hsp90 was determined from the resulting Svedberg values obtained after the sedimentation runs. For that purpose the raw data were converted to dc/dt figures as described before (30). 1 μm concentrations of labeled Aha1 and 2 μm concentrations of each construct in 40 mm HEPES, 20 mm KCl at pH 7.5 were run at 20 °C.
Fluorescence Resonance Energy Transfer (FRET)
To determine whether the linker variants are capable of N-terminal dimerization, a FRET-based assay was performed (31). Hsp90 was labeled at two engineered cysteine residues (positions 66 and 381) with the dyes Atto 488 and Atto 550 (Atto-tec, Siegen, Germany). 3 μm concentrations of each construct was equilibrated with 0.2 μm concentrations of labeled Hsp90 before the addition of 2 mm ATPγS. Measurements were done in a Jasco fluorescence detector at 15 °C in 40 mm HEPES, 150 mm KCl at pH 7.5. The excitation wavelength was set at 490 nm, whereas the emission wavelength was at 520 nm.
Hsp90 in Vivo Rescue Assay
Hsp90 functionality in vivo was investigated using a plasmid shuffling approach based on the system described previously (32). Hsp90 linker deletion constructs and HtpG constructs were cloned into the vector pRS423 and transformed into a yeast strain devoid of genomic HSP82 and HSC82 that instead carries a plasmid coding for HSP82 under the control of a constitutive glyceraldehyde-3-phosphate dehydrogenase gene promoter. This may result in an overexpression of the respective construct compared with the endogenous situation. The plasmid also contains an URA selection marker, which can be counter-selected by the addition of 5′-FOA to the growth medium (33). Therefore, the loss of this plasmid is required to prevent lethal toxicity of 5′-FOA, and only if the supplied Hsp90 variant on the pRS423 plasmid is functional, will growth of the yeast strain be observed. After repeated 5′-FOA selection, loss of the URA3 plasmid was verified by the inability of the transformants to grow on synthetic dropout medium minus uracil.
In Vivo Client Assays; Glucocorticoid Receptor (GR) Activity, Luciferase Activity, and v-Src Phosphorylation Status
The in vivo activity of heterologous expressed GR depends on functional Hsp90 (34). To test the chaperoning ability of the viable charged linker mutants on GR, we provided the respective strains with a plasmid expressing the GR constitutively (32). Mid-log-phase cells (2 × 106 ml−1) were treated with 10 μm deoxycorticosterone (Sigma) and incubated for 2 h to activate the GR. As transcriptionally active GR binds to a GR responsive element on the plasmid, which in turn controls expression of β-galactosidase, the activity of β-galactosidase serves as an indirect measure of GR activity. β-Galactosidase activity was assayed with the Galacto-Light Plus kit (Applied Biosystems, Foster City, CA), according to a recently published protocol (35).
The in vivo activity of luciferase was monitored using a plasmid encoding firefly luciferase from Photinus pyralis under the control of a Gal promoter (36). Mid-log-phase cells (2 × 106 ml−1) were shifted from glucose via raffinose to galactose containing medium, in which the expression of luciferase was initialized and allowed for 3 h. Then, luminescence detection was carried out as described elsewhere (35).
Hsp90-dependent in vivo maturation of v-Src kinase in S. cerevisiae was monitored with the mouse monoclonal anti-phospho-Tyr-416 antibody (Cell Signaling Technologies, Beverly, MA). Only fully matured and active v-Src is phosphorylated at Tyr-416, thus making this modification an indicator for Hsp90 function. The total amount of expressed v-Src kinase was assayed using the rabbit polyclonal antibody EC10 (Upstate Biotechnology, Lake Placid, NY) which is able to detect both c-Src and v-Src. V-Src expression under the control of a Gal promoter was allowed for 6 h and assayed as described elsewhere (32). To detect the total amount of expressed v-Src or the phosphorylated Tyr-416 in v-Src as a reporter for fully folded and active v-Src, samples of 10 μl of yeast lysate were run on 10% (w/v) SDS-PAGE gels, blotted on a nitrocellulose membrane, and incubated with the either EC10 or anti-phospho-Tyr-416. Further incubation with the corresponding horseradish peroxidase-conjugated secondary antibody (Sigma) was followed by treatment with ECL solution (GE Healthcare). The membrane was exposed to a Kodak X-Omat AR film (Eastman Kodak Co.) for visualization.
RESULTS
The Deletion of the Entire Charged Linker Region of Hsp90 Is Lethal
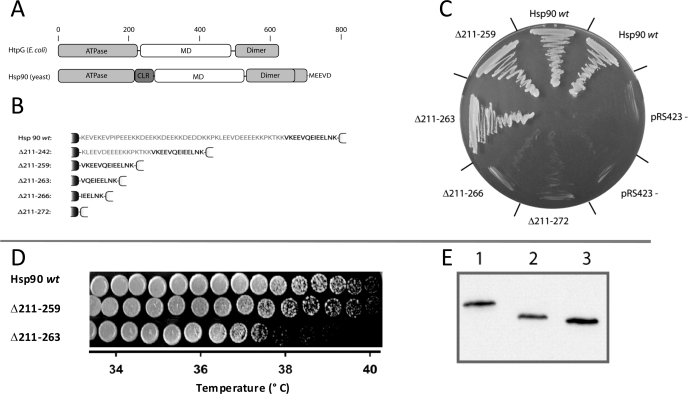
The charged linker region of eucaryotic Hsp90 connects the N-terminal ATP binding domain with the middle domain (Fig. 1A). Although the full-length crystal structure of yeast Hsp90 has been published recently (6), the nature of the charged linker region remains elusive, as the linker had to be shortened and modified to obtain crystals. The crystal structure of yeast Hsp90 together with sequence analysis suggests that the linker ranges from residue 211 up to residue 272. To investigate the function of this highly unusual linker domain, we set out to create and analyze various deletion constructs of Hsp90 ranging from Hsp90 Δ211–259 to Hsp90 Δ211–272 (Fig. 1B). First, we determined whether these constructs confer viability to yeast cells lacking genomic HSP82 and HSC82 by a plasmid shuffling approach (32). For Hsp90 Δ211–266 and for the even larger deletions, no growth on 5′-FOA plates cultivated at 20C, 25, or 30 °C was observed (Fig. 1C). Surprisingly, already the deletion Hsp90 Δ211–263 resulted in a slow growth phenotype, with a doubling time at 25 °C that was 3-fold longer compared with yeast carrying Hsp90 wt and a pronounced temperature sensitivity. Cells carrying the Hsp90 Δ211–263 construct show a growth limit at ∼37.5 °C, whereas the wt strain grows at up to 39.5 °C (Fig. 1D). These results indicate that the presence of a minimal linker region is required for viability in yeast. The threshold for viability seems to be deletions from residues 211 to residues between 263 and 266. Different expression of the Hsp90 variants could not be a reason for the observed effects, as immunoblot analysis showed that the protein levels for all the constructs tested were similar (Fig. 1E).
FIGURE 1.
Deletions of the linker region and their effect on the viability of yeast. A, schematic representation of the domain structure of E. coli HtpG and S. cerevisiae Hsp90. light gray, N-terminal ATP binding domain (ATPase) and C-terminal domain (Dimer); white, middle domain (MD); dark gray, charged linker region (CLR). The domain boundaries are set as described for HtpG in Shiau et al. (5) and for Hsp90 as described in Ali et al. (6). B, Hsp90 linker variants used in this study. A detailed sequence of the charged linker region of the deletion constructs used in this study is shown. The neighboring domains are schematically drawn in black (ATP binding domain) and white (middle domain). Hsp90 wt, wild type Hsp90 of S. cerevisiae; Hsp90 Δ211–259 to Hsp90 Δ211–272, deletions in the charged linker region. C, influence of Hsp90 linker variants on viability of S. cerevisiae. The strains carrying the respective Hsp90 variant were plated on 5′-FOA-HIS agar and incubated for 4 days at 30 °C. All strains used in this study had genomic deletions of HSC82 and HSP82 and carried a rescue plasmid constitutively expressing Hsp90. D, temperature-dependent growth of Hsp90 wt and the viable linker deletion constructs. The growth analysis was carried out on a temperature gradient ranging from 20 to 40 °C. The maximum growth temperature for Hsp90 wt and Hsp90 Δ211–259 was 41 °C, whereas Hsp90 Δ211–263 stops growth above 38 °C. Cells were incubated for 4 days. E, expression levels of the Hsp90 wt and the viable Hsp90 constructs in the HSP82/HSC82 knock-out strain. 2 μl of cell lysate was probed with anti-Hsp90 antibody. Lane 1, Hsp90 wt, lane 2, Hsp90 Δ211–259; lane 3, Hsp90 Δ211–263.
Charged Linker-deleted Hsp90s Have Reduced Chaperone Activity in Vivo
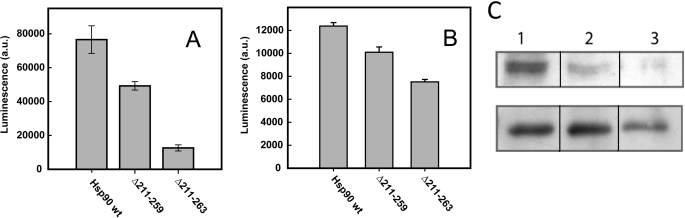
We were interested to test the viable linker deletion constructs Hsp90 Δ211–259 and Hsp90 Δ211–263 together with Hsp90 wt for their effect on client maturation in vivo. A well established in vivo reporter system in yeast is firefly luciferase (36). As Hsp90 influences luciferase folding, the detected luminescence of heterologously expressed firefly luciferase can be used as a direct indicative of in vivo chaperone function of the different Hsp90 constructs on luciferase (35, 37–39). Although lysate from cells harboring the Hsp90 Δ211–259 construct showed a luminescence intensity of around two-thirds that of the signal from lysate of cells expressing Hsp90 wt, the cells carrying the construct Hsp90 Δ211–263 yielded a 6-fold lower luciferase luminescence compared with the Hsp90 wt cells (Fig. 2A).
FIGURE 2.
The charged linker is important for client protein folding in vivo. A, luciferase activity in yeast carrying plasmid-encoded Hsp90 wt, Hsp90 Δ211–259, or Hsp90 Δ211–263 as the only source of Hsp90. Cells were lysed 3 h after galactose-mediated induction of luciferase expression. a.u., arbitrary units. B, in vivo activity of the GR reporter β-galactosidase in yeast carrying plasmid-encoded Hsp90 wt, Hsp90 Δ211–259, or Hsp90 Δ211–263 as the only source of Hsp90. Cells were treated with 10 μm deoxycorticosterone to induce GR transcription activity. After 2 h, β-galactosidase activity was monitored using a luminescence assay. C, levels of active v-Src kinase in the presence of the viable Hsp90 construct strains as the only source of Hsp90. 20 μl of cell lysate was probed with anti-phospho-Tyr-416 antibody. Upper panel, detection of Tyr-416-phosphorylated v-Src; lower lane, detection of total v-Src. Lane 1, Hsp90 wt; lane 2, Hsp90 Δ211–259; lane 3, Hsp90 Δ211–263.
Another widely used system for the characterization of the in vivo chaperone function of Hsp90 is the GR reporter assay (32, 40), in which the activity of the reporter enzyme, β-galactosidase, correlates with the transcriptional activity of the constitutively expressed GR. When expressed in yeast, the GR depends on Hsp90 to reach its active state (40), and therefore, GR activity reflects the chaperoning effect of Hsp90. With yeasts carrying Hsp90 Δ211–259 and Hsp90 Δ211–263, we found a considerable decrease in the activity compared with the strain carrying Hsp90 wt (Fig. 2B), albeit to a smaller extent than in the case of firefly luciferase.
To expand the client range studied, we tested the effect of deletion constructs on Src activity in yeast. The activity of v-Src can be characterized by monitoring the expression and phosphorylation of this Hsp90 client protein. Although the expression level of v-Src does not depend on the Hsp90 system, the active state of v-Src can only be reached with functional Hsp90 (41). Fig. 2C shows that the amount of Tyr-416-phosphorylated v-Src is highest in the yeast strain carrying Hsp90 wt. With the deletion constructs Hsp90 Δ211–259, v-Src levels are significantly reduced, reaching the lowest amount when expressed in the presence of the shortest viable construct of the Hsp90 linker deletion series, Hsp90 Δ211–263. The combined data from the in vivo assays performed with different Hsp90 client proteins suggest that residual activity is maintained in the deletion constructs, which show viability. Interestingly, the temperature sensitivity correlates with reduced client activation for the different constructs studied.
The Structure and Stability of Hsp90 Is Not Affected by Changes in the Charged Linker Region
The temperature sensitivity of mutant proteins, as observed for Hsp90 Δ211–263, is often conferred by a reduced stability of the mutated protein. To assess the impact of the alterations in the charged linker region on the structure of Hsp90, we first analyzed the purified wt and mutant proteins by circular dichroism spectroscopy. Neither in the far-UV nor in the near-UV range could significant changes be observed between wt and mutant proteins (data not shown). Furthermore, we compared the stability of the proteins. The chemical stability was determined by denaturant-induced unfolding transitions. Hsp90 wt and all the variants studied had transition midpoints between 4.7 and 5.1 m urea (Table 1), which is in agreement with previous results (42). These results imply that the deletion of the charged linker has no significant influence on structure and stability of Hsp90 and, therefore, precludes the stability of Hsp90 as the cause for the temperature sensitivity observed in viability assays.
TABLE 1.
Characteristics of Hsp90 linker variants
For viability, see also Fig. 1C. The chemical stability was determined by urea-induced unfolding transitions. kcat and Km values were determined using the regenerative ATPase assay. All methods were used as described under “Materials and Methods.”
| Hsp90 wt | Hsp90 Δ211–242 | Hsp90 Δ211–259 | Hsp90 Δ211–263 | Hsp90 Δ211–266 | Hsp90 Δ211–272 | |
|---|---|---|---|---|---|---|
| Viability (20 °C) | + | + | + | + | - | - |
| Chemical stability (urea) [m] | 4.9 ± 0.1 | 4.9 ± 0.1 | 4.7 ± 0.1 | 4.9 ± 0.1 | 4.8 ± 0.1 | 5.1 ± 0.1 |
| ATPase kcat (min−1) | 0.5 | 0.6 | 0.6 | 0.3 | 0.2 | 0.1 |
| Km (ATP) (μm) | 320 ± 61 | 260 ± 55 | 300 ± 84 | 337 ± 92 | 406 ± 76 | 311 ± 64 |
Lethal-charged Linker Deletion Constructs of Hsp90 Still Have ATPase Activity
The inability of the linker-deletion mutants to process client proteins and their reduced ability to confer viability to yeast cells raised interest in the biochemical properties of these proteins. To date, all known lethal point mutations or deletion constructs of Hsp90 are defective in ATPase activity (16–18). Therefore, we determined the ATPase activity of the different linker variants. For Hsp90 wt, we obtained a kcat of 0.5 min−1 (Table 1), which is in agreement with previously reported values (10, 16). The shortest deletion variant, Hsp90 Δ211–259, had a slightly higher ATPase activity of 0.6 min−1, whereas Hsp90 Δ211–263, Hsp90 Δ211–266, and Hsp90 Δ211–272 showed decreased ATPase activities of 0.3, 0.2, and 0.1 min−1, respectively (Table 1).
A possible cause for a lower ATPase activity could be a decreased affinity for ATP. To test this, we asked whether nucleotide binding is changed in the Hsp90 mutants by determining the apparent Km of ATP for the Hsp90 constructs. All variants had a similar Km for ATP of around 300 to 400 μm (Table 1) showing that deletion of the charged linker region does not affect nucleotide binding to the N-terminal domain of Hsp90 even though ATP hydrolysis is decreased in constructs carrying shortened linker regions.
Charged Linker Deletion Constructs Are Less Sensitive to Co-chaperone Regulation
Deletions in the linker region of Hsp90 apparently affect the ATPase activity without changing ATP binding properties in addition to their reduced ability to activate client proteins. We, therefore, were interested to determine whether the observed effects reflect a modified ability to exert the conformational changes required during the ATPase cycle. To test this we employed the binding of the co-chaperones Aha1, p23/Sba1, and Sti1 as conformational probes of Hsp90. These proteins either stimulate or inhibit the ATPase activity of Hsp90 and bind to specific conformations of Hsp90 (20, 43–45).
The co-chaperone Sti1 is known to bind to Hsp90 mainly via a TPR-mediated interaction, with contributions from a secondary binding site outside the C-terminal domain (45). In the case of Hsp90 wt, Sti1 suppresses the ATPase completely. The same is true for all linker deletions studied (Table 2). In addition, we determined the binding of Sti1 to Hsp90 variants by SPR spectroscopy. Again, the interaction of the linker constructs with Sti1 was similar to the interaction between Sti1 and Hsp90 wt (data not shown).
TABLE 2.
Cochaperone-mediated ATPase regulation of Hsp90 linker constructs
The effects of Sti1 and Aha1 on the ATPase activity of the linker constructs were investigated using the regenerative ATPase assay. The ATPase activity was measured in 40 mm HEPES, 20 mm KCl, 5 mm MgCl2, pH 7.5 at 30 °C. ND, not determined.
| Hsp90 wt | Hsp90 Δ211–242 | Hsp90 Δ211–259 | Hsp90 Δ211–263 | Hsp90 Δ211–266 | Hsp90 Δ211–272 | |
|---|---|---|---|---|---|---|
| Sti1 inhibition | + | + | + | + | + | + |
| Aha1 activation | + | + | + | + | - | - |
| ATPase stimulation factor | 10.5 | 8.3 | 4.4 | 5.1 | 1.8 | 1.4 |
| KD (μm) | 1.6 ± 1.2 | 2.5 ± 2.1 | 2.4 ± 1.3 | 2.4 ± 2.3 | ND | ND |
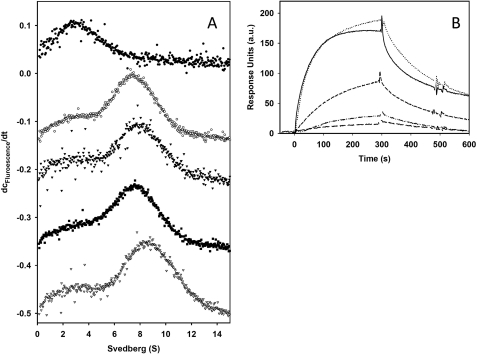
Aha1 was found to be a potent activator of the Hsp90 ATPase (16, 46, 47). Under our conditions, Aha1 stimulates the ATPase of Hsp90 about 10-fold (Table 2), which is in agreement with previously published results (46, 48). The constructs Hsp90 Δ211–242 and Hsp90 Δ211–263 Hsp90 showed stimulation factors comparable with that of the wild type protein, whereas Hsp90 Δ211–266 and Hsp90 Δ211–272 could not be stimulated anymore (Table 2). The inability to stimulate the turnover could principally be because of either an inhibition of binding of Aha1 to Hsp90 or an inhibition of the activation mechanism. We, therefore, subjected the constructs Hsp90 wt, Hsp90 Δ211–259, Hsp90 Δ211–263, and Hsp90 Δ211–266 to analytical ultracentrifugation and fluorescence detection. An AlexaFluor 488-labeled Aha1 was used in the absence and presence of the Hsp90 variants to analyze the amount of Hsp90-complexed Aha1. The uncomplexed Aha1 sediments with an S value of 2.4 (Fig. 3A), whereas the Hsp90-complexed Aha1 sediments with an S value of 6.3–6.6. Therefore, a clear distinction can be made between free and Hsp90-bound Aha1, and information is obtained of whether the respective Hsp90 variant is capable of binding Aha1. Interestingly, all the variants were able to bind Aha1, clearly showing that the lack of stimulation observed in the case of Hsp90 Δ211–266 is because of a defunct activation mechanism.
FIGURE 3.
Influence of the charged linker on co-chaperone interaction. A, binding of the Hsp90 constructs to Aha1 was investigated using analytical ultracentrifugation. 1 μm concentrations of labeled Aha1 and 2 μm concentrations of each construct in 40 mm HEPES, 20 mm KCl at pH 7.5 were subjected to analytical ultracentrifugation at 20 °C. dc/dt profiles are shown for Aha1 alone (●) and the addition of Hsp90 wt (○), Hsp90Δ211–259 (▾), Hsp90Δ211–263 (■), and Hsp90 Δ211–266(▿). B, the interaction of immobilized p23/Sba1 with the Hsp90 constructs was monitored by SPR at 25 °C in 40 mm HEPES, pH 7.5, 150 mm KCl, 5 mm MgCl2, 1 mm ATPγS. Hsp90 wt (—), Hsp90 Δ211–259 (… . .), Hsp90 Δ211–263 (---), Hsp90 Δ211–266 (-..-..-), Hsp90 Δ211–272 (––) to immobilized p23/Sba1 was measured. Injections were 65 μl of a solution of 1 μm Hsp90 construct.
The Hsp90 co-chaperone p23/Sba1 binds exclusively to the ATP-bound, N-terminal closed state of the chaperone and is able to reduce the ATPase activity of Hsp90 (7, 49–52). Therefore, p23/Sba1 is well suited as a conformation-dependent sensor for the Hsp90 ATPase function. We investigated the influence of p23/Sba1 on nucleotide hydrolysis by Hsp90 as well as the binding of Hsp90 constructs to immobilized p23/Sba1 in the presence of ATPγS. Although Hsp90 wt and Hsp90 Δ211–259 show robust binding to p23/Sba1, the last viable construct, Hsp90 Δ211–263, binds only with about half the affinity of the Hsp90 wt-p23/Sba1 interaction. The non-viable constructs Hsp90 Δ211–266 and Hsp90 Δ211–272 no longer interact with Sba1 (Fig. 3B). Thus, the binding of the Hsp90 constructs to p23/Sba1 correlates with their effects on viability.
Based on these results it is reasonable to assume that successive shortening of the linker causes constraints in the Hsp90 protein, which prevent essential conformational changes like the closing of the N-terminal domains to form the ATP hydrolysis competent and p23/Sba1 binding competent state.
Linker Deletion Constructs Are Conformationally Restricted
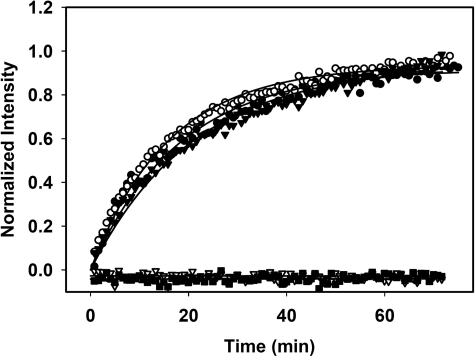
To test the hypothesis of conformational restriction in the linker deletion constructs, we used a FRET-based assay, which uses two labels within one wt subunit and the formation of heterodimers with the linker constructs (31). After formation of the heterodimers, ATPγS was added to induce the conformational changes. All the viable linker constructs formed the N-terminal-dimerized state with similar rates as the wild type protein (Fig. 4). However, the constructs with the shorter linker length Hsp90 Δ211–266 and Hsp90 Δ211–272 were compromised in forming the N-terminal-dimerized conformation. These data explain the reduction in ATPase rate of these constructs compared with the viable Hsp90 variants.
FIGURE 4.
The charged linker influences N-terminal dimerization. The ability of each of the linker deletion constructs to undergo N-terminal dimerization was investigated using a FRET assay (31). 3 μm concentrations each of either Hsp90 wt (●), Hsp90 Δ211–242 Hsp90 (○), Hsp90 Δ211–259 (▾), Hsp90Δ211–263 (■), or Hsp90 Δ211–266 (▿) were equilibrated with 0.2 μm concentrations each of doubly labeled Hsp90 wt in 40 mm HEPES, 150 mm KCl, 5 mm MgCl2 at pH 7.5 until subunit exchange was complete. The addition of ATPγS was used to start of the assay.
Supplementation of the Linker Region Rescues Viability
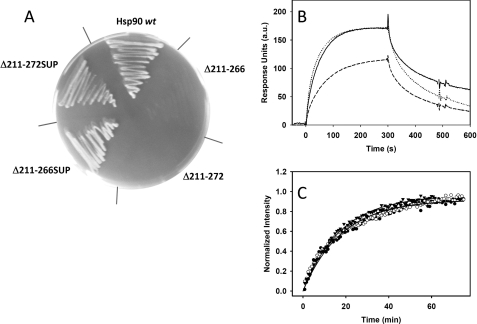
To analyze whether the length of the charged linker or its characteristics are essential for viability, we tested two different replacement constructs, Hsp90 Δ211–266SUP and Hsp90 Δ211–272SUP. In Hsp90 Δ211–266SUP and Hsp90 Δ211–272SUP, positions 259–266 or 259–272, respectively, were replaced by a Gly-Ser spacer of the corresponding length, resulting in proteins which shared the linker length with the viable Hsp90 Δ211–259 but contained Gly-Ser stretches either from amino acid 259 to 266 (Hsp90 Δ211–266SUP) or from amino acid 259 to 272 (Hsp90 Δ211–272SUP). The analysis of these constructs resulted in a viable phenotype for Hsp90 Δ211–266SUP and even for Hsp90 Δ211–272SUP (Fig. 5A).
FIGURE 5.
Shortened linker variants can be rescued by artificial linker sequences. A, influence of Hsp90 supplemented linker variants on viability of S. cerevisiae. The strains carrying the respective Hsp90 variant were plated on 5′-FOA-HIS agar and incubated for 4 days at 30 °C. B, the interaction of p23/Sba1 with the Hsp90 constructs was monitored by SPR at 25 °C in 40 mm Hepes, pH 7.5, 150 mm KCl, 5 mm MgCl2, 1 mm AMP-PNP. Binding of 1 μm of Hsp90 wt (—), Hsp90 Δ211–266SUP (… . .), or Hsp90 Δ211–272SUP (---) to immobilized p23/Sba1 was measured with injections of 65 μl. C, the ability of Hsp90 wt (●), Hsp90 Δ211–266SUP (○), or Hsp90 Δ211–272SUP (▾) to undergo N-terminal dimerization was investigated using the described FRET assay in 40 mm HEPES, 150 mm KCl, 5 mm MgCl2 at pH 7.5. The addition of ATPγS was used to start the assay.
We aimed to understand the response of the two reporter constructs and, therefore, analyzed the ATPase activity of the two purified replacement constructs. Hsp90 Δ211–272SUP showed a kcat of 0.3 min−1, and the longer construct showed a wt-like ATPase activity of 0.7 min−1. Both constructs showed wt-like Km values (Table 3).
TABLE 3.
Characteristics of supplemented linker constructs
The non-viable linker deletions Hsp90 Δ211–266 and Hsp90 Δ211–272 were characterized with the respect to the ATPase rate, Km value, and stimulation by the co-factor Aha1 using the regenerative ATPase assay.
| Hsp90 Δ211–266SUP | Hsp90 Δ211–272SUP | |
|---|---|---|
| ATPase | ||
| kcat (min−1) | 0.7 | 0.3 |
| Km (μm) | 293 ± 120 | 313 ± 98 |
| Aha1 activation | + | + |
| ATPase stimulation factor | 14.5 | 5.2 |
| KD (μm) | 2.0 ± 1.5 | 1.8 ± 1.2 |
Although both linker deletions did not show any detectable activation in the presence of Aha1, the constructs carrying the Gly-Ser repeat supplement were stimulated in the presence of Aha1 by a factor of 5 (Hsp90 Δ211–272SUP) and 16 (Hsp90 Δ211–266SUP) (Table 3), implying again that the supplementation of linker length by a variable sequence results in a protein which is better capable to fulfill its enzymatic functions. Hsp90 Δ211–266SUP did not show any deviation from the Hsp90 wt protein with respect to this assay (Fig. 5B). The same is true for p23/Sba1 interaction, where the longer deletions had been unable to bind (Fig. 3B). Here, Hsp90 Δ211–266SUP showed the same binding capability as Hsp90 wt, whereas Hsp90 Δ211–272SUP is to some extent restrained (Fig. 5B). Furthermore, the replacement constructs were capable of forming the N-terminal dimers as evidenced in the FRET assay (Fig. 5C). These data confirm the importance of the charged linker for Hsp90 to perform conformational rearrangements.
DISCUSSION
The molecular chaperone Hsp90 is an ATP-driven machine whose conformational changes are required to perform a complex reaction cycle (31). Comparing prokaryotic and eukaryotic Hsp90 homologues, two specific regions obviously differ strongly. One is a flexible linker region connecting the N-terminal (ATPase) and middle domains, and the other one is a C-terminal extension, which serves as the primary binding site for TPR domain-containing co-chaperones. Both regions are dispensable in yeast (25), which raised questions about their function. Especially, the function of the linker sequence remained enigmatic. We deleted this sequence to a different extent, including one version that did not have any linker sequence and two versions which had artificial amino acid stretches to connect the two domains. Our results highlight a minimal linker length required to retain viability. Interestingly, the full deletion of the linker, although leaving Hsp90 structurally intact and preserving ATP binding, does reduce ATP hydrolysis to values similar to those known from the isolated N-terminal domain/middle domain construct (53). Hsp90 activity can be rescued when an artificial linker is introduced, implying that one function of the linker region is to contribute conformational freedom for connection of the N and M domains. It might well be that conformational changes at the N-M interface require this conformational space to rearrange productively during the ATPase cycle. This alone appears to be not sufficient given the observation that the amino acids 266–272 are required for purposes, which cannot be fully supplemented fully by a Gly-Ser stretch as judged by the lower ATP turnover of 211–272SUP compared with 211–266SUP and differences in Aha1 interaction. Analysis of x-ray structures of HtpG and Grp94 (4, 5) suggests that these amino acid participate in the formation of a β-sheet-like structure with the N-terminal domain. However, this is difficult to decide based on the crystal structures, as both proteins were strongly mutated within the linker region to allow crystallization. Nevertheless, the participation of this sequence in a β-sheet structure could explain the requirement for certain amino acids within this region.
The analysis of the co-chaperone interaction with the Hsp90 linker deletion variants allowed us to further scrutinize the function of the linker region. Some co-chaperones can be used as conformational sensors as they associate with specific states of Hsp90 during the ATPase cycle (45, 46, 51). The interaction with Sti1 was not significantly influenced by the linker deletions. This may be explained with the relative flexibility of the adaptor protein Sti1, which could compensate for a changed domain orientation in the charged linker constructs of Hsp90. The data for the p23/Sba1 interaction on the other hand suggest that the conformational changes leading to the N-terminal-dimerized state of Hsp90 seem to be inhibited by the linker deletion. This conformation is induced upon ATP binding (7). The conformational changes leading to this state are rate-limiting during the hydrolysis cycle (9). Apparently, the deletion of the linker slows the repositioning of the N and the M domain at the N-M interface. Interestingly, also the ability of Aha1 to stimulate the ATP turnover is strongly reduced in the linker variants or even absent in the maximum deletion construct. Our data indicate that the Aha1-induced ATPase stimulation is only possible with unconstrained N-M interactions. It is reasonable to assume that if this interaction is sterically inhibited, Aha1 can no longer exert stimulatory effects even though Aha1 binding itself is not affected.
To date, the ability to hydrolyze ATP was believed to be an essential prerequisite for in vivo functionality of Hsp90 wt as ATPase-defective mutants are not able to replace Hsp90 in an Hsp90-deficient background (16, 17). Our results indicate that ATP hydrolysis alone is not sufficient for viability. Also, conformationally restricted but ATPase-active Hsp90 versions are unable to replace Hsp90 wt. The unrestricted motility of the N-M interface seems required for in vivo functions even more than for the ATP turnover. This is true even for the smallest deletions used in this study, which are not affected in their ATP turnover yet, but their chaperone effect on known in vivo clients of Hsp90, like the GR, luciferase, and v-Src kinase, is diminished. The severely compromised yeast strains carrying the Hsp90 linker constructs show a reduced ability to interact with Hsp90 clients, consistent with the view that the charged linker region is involved in Hsp90 chaperone function (54). The recent structure of the kinase Cdk4 in complex with Hsp90-Cdc37 implies client interactions, which span the N-terminal part of Hsp90 and the middle domain (55). It might very well be that the conformational flexibility required at the N-M interface is even more pronounced in the case of client binding as compared with ATP hydrolysis. The strongly extended linker region, which can be found in all eukaryotic Hsp90 proteins, might therefore result from the need of a highly flexible contact surface that is able to integrate various client proteins into the Hsp90 dimer structure.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie (to J. B. and K. R.).
This work was supported by grants from the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie (to J. B. and K. R.).
- Hsp90
- heat shock protein 90
- HSP82
- the S. cerevisiae gene for Hsp90
- Hsp90 ΔLinker
- yeast Hsp90 after deletion of the charged region (amino acids 211–259)
- HtpG
- Hsp90 from E. coli
- 5′-FOA
- 5′-fluoroorotic acid
- FRET
- fluorescence resonance energy transfer
- GR
- glucocorticoid receptor
- SPR
- surface plasmon resonance
- wt
- wild type
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- MP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- TPR
- tetratricopeptide repeat.
REFERENCES
- 1.Pearl L. H., Prodromou C. (2006) Annu. Rev. Biochem. 75,271–294 [DOI] [PubMed] [Google Scholar]
- 2.Picard D. (2002) Cell. Mol. Life Sci. 59,1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pratt W. B., Morishima Y., Murphy M., Harrell M. (2006) Handb. Exp. Pharmacol. 172,111–138 [DOI] [PubMed] [Google Scholar]
- 4.Dollins D. E., Warren J. J., Immormino R. M., Gewirth D. T. (2007) Mol. Cell 28,41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiau A. K., Harris S. F., Southworth D. R., Agard D. A. (2006) Cell 127,329–340 [DOI] [PubMed] [Google Scholar]
- 6.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Nature 440,1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prodromou C., Panaretou B., Chohan S., Siligardi G., O'Brien R., Ladbury J. E., Roe S. M., Piper P. W., Pearl L. H. (2000) EMBO J. 19,4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weikl T., Muschler P., Richter K., Veit T., Reinstein J., Buchner J. (2000) J. Mol. Biol. 303,583–592 [DOI] [PubMed] [Google Scholar]
- 9.Richter K., Reinstein J., Buchner J. (2002) J. Biol. Chem. 277,44905–44910 [DOI] [PubMed] [Google Scholar]
- 10.Richter K., Soroka J., Skalniak L., Leskovar A., Hessling M., Reinstein J., Buchner J. (2008) J. Biol. Chem. 283,17757–17765 [DOI] [PubMed] [Google Scholar]
- 11.Leskovar A., Wegele H., Werbeck N. D., Buchner J., Reinstein J. (2008) J. Biol. Chem. 283,11677–11688 [DOI] [PubMed] [Google Scholar]
- 12.Frey S., Leskovar A., Reinstein J., Buchner J. (2007) J. Biol. Chem. 282,35612–35620 [DOI] [PubMed] [Google Scholar]
- 13.Wandinger S. K., Richter K., Buchner J. (2008) J. Biol. Chem. 283,18473–18477 [DOI] [PubMed] [Google Scholar]
- 14.Bardwell J. C., Craig E. A. (1988) J. Bacteriol. 170,2977–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. (1989) Mol. Cell. Biol. 9,3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obermann W. M., Sondermann H., Russo A. A., Pavletich N. P., Hartl F. U. (1998) J. Cell Biol. 143,901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panaretou B., Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1998) EMBO J. 17,4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter K., Moser S., Hagn F., Friedrich R., Hainzl O., Heller M., Schlee S., Kessler H., Reinstein J., Buchner J. (2006) J. Biol. Chem. 281,11301–11311 [DOI] [PubMed] [Google Scholar]
- 19.Chen S., Sullivan W. P., Toft D. O., Smith D. F. (1998) Cell Stress Chaperones. 3,118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prodromou C., Siligardi G., O'Brien R., Woolfson D. N., Regan L., Panaretou B., Ladbury J. E., Piper P. W., Pearl L. H. (1999) EMBO J. 18,754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101,199–210 [DOI] [PubMed] [Google Scholar]
- 22.Farrelly F. W., Finkelstein D. B. (1984) J. Biol. Chem. 259,5745–5751 [PubMed] [Google Scholar]
- 23.Combet C., Blanchet C., Geourjon C., Deléage G. (2000) Trends Biochem. Sci. 25,147–150 [DOI] [PubMed] [Google Scholar]
- 24.Pollastri G., McLysaght A. (2005) Bioinformatics 21,1719–1720 [DOI] [PubMed] [Google Scholar]
- 25.Louvion J. F., Warth R., Picard D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93,13937–13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110,119–122 [DOI] [PubMed] [Google Scholar]
- 27.Richter K., Muschler P., Hainzl O., Buchner J. (2001) J. Biol. Chem. 276,33689–33696 [DOI] [PubMed] [Google Scholar]
- 28.Tamura J. K., Gellert M. (1990) J. Biol. Chem. 265,21342–21349 [PubMed] [Google Scholar]
- 29.Kroe R. R., Laue T. M. (2009) Anal. Biochem 390,1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stafford W. F., 3rd (1992) Anal. Biochem. 203,295–301 [DOI] [PubMed] [Google Scholar]
- 31.Hessling M., Richter K., Buchner J. (2009) Nat. Struct. Mol. Biol. 16,287–293 [DOI] [PubMed] [Google Scholar]
- 32.Nathan D. F., Lindquist S. (1995) Mol. Cell. Biol. 15,3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boeke J. D., LaCroute F., Fink G. R. (1984) Mol. Gen. Genet. 197,345–346 [DOI] [PubMed] [Google Scholar]
- 34.Picard D., Khursheed B., Garabedian M. J., Fortin M. G., Lindquist S., Yamamoto K. R. (1990) Nature 348,166–168 [DOI] [PubMed] [Google Scholar]
- 35.Wandinger S. K., Suhre M. H., Wegele H., Buchner J. (2006) EMBO J. 25,367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodsky J. L., Lawrence J. G., Caplan A. J. (1998) Biochemistry 37,18045–18055 [DOI] [PubMed] [Google Scholar]
- 37.Ahner A., Whyte F. M., Brodsky J. L. (2005) Arch. Biochem. Biophys. 435,32–41 [DOI] [PubMed] [Google Scholar]
- 38.Kimura Y., Rutherford S. L., Miyata Y., Yahara I., Freeman B. C., Yue L., Morimoto R. I., Lindquist S. (1997) Genes Dev. 11,1775–1785 [DOI] [PubMed] [Google Scholar]
- 39.Wegele H., Wandinger S. K., Schmid A. B., Reinstein J., Buchner J. (2006) J. Mol. Biol. 356,802–811 [DOI] [PubMed] [Google Scholar]
- 40.Nathan D. F., Vos M. H., Lindquist S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94,12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y., Singer M. A., Lindquist S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakob U., Meyer I., Bügl H., André S., Bardwell J. C., Buchner J. (1995) J. Biol. Chem. 270,14412–14419 [DOI] [PubMed] [Google Scholar]
- 43.Siligardi G., Hu B., Panaretou B., Piper P. W., Pearl L. H., Prodromou C. (2004) J. Biol. Chem. 279,51989–51998 [DOI] [PubMed] [Google Scholar]
- 44.Siligardi G., Panaretou B., Meyer P., Singh S., Woolfson D. N., Piper P. W., Pearl L. H., Prodromou C. (2002) J. Biol. Chem. 277,20151–20159 [DOI] [PubMed] [Google Scholar]
- 45.Richter K., Muschler P., Hainzl O., Reinstein J., Buchner J. (2003) J. Biol. Chem. 278,10328–10333 [DOI] [PubMed] [Google Scholar]
- 46.Meyer P., Prodromou C., Liao C., Hu B., Mark R. S., Vaughan C. K., Vlasic I., Panaretou B., Piper P. W., Pearl L. H. (2004) EMBO J. 23,511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. (2003) Mol. Cell 11,647–658 [DOI] [PubMed] [Google Scholar]
- 48.Lotz G. P., Lin H., Harst A., Obermann W. M. (2003) J. Biol. Chem. 278,17228–17235 [DOI] [PubMed] [Google Scholar]
- 49.Fang Y., Fliss A. E., Rao J., Caplan A. J. (1998) Mol. Cell. Biol. 18,3727–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chadli A., Bouhouche I., Sullivan W., Stensgard B., McMahon N., Catelli M. G., Toft D. O. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,12524–12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richter K., Walter S., Buchner J. (2004) J. Mol. Biol. 342,1403–1413 [DOI] [PubMed] [Google Scholar]
- 52.McLaughlin S. H., Sobott F., Yao Z. P., Zhang W., Nielsen P. R., Grossmann J. G., Laue E. D., Robinson C. V., Jackson S. E. (2006) J. Mol. Biol. 356,746–758 [DOI] [PubMed] [Google Scholar]
- 53.Wegele H., Muschler P., Bunck M., Reinstein J., Buchner J. (2003) J. Biol. Chem. 278,39303–39310 [DOI] [PubMed] [Google Scholar]
- 54.Scheibel T., Siegmund H. I., Jaenicke R., Ganz P., Lilie H., Buchner J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan C. K., Gohlke U., Sobott F., Good V. M., Ali M. M., Prodromou C., Robinson C. V., Saibil H. R., Pearl L. H. (2006) Mol. Cell 23,697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]