Abstract
Breast cancer susceptibility gene BRCA1 is implicated in the control of mitotic progression, although the underlying mechanism(s) remains to be further defined. Deficiency of BRCA1 function leads to disrupted mitotic machinery and genomic instability. Here, we show that BRCA1 physically interacts and colocalizes with Nlp, an important molecule involved in centrosome maturation and spindle formation. Interestingly, Nlp centrosomal localization and its protein stability are regulated by normal cellular BRCA1 function because cells containing BRCA1 mutations or silenced for endogenous BRCA1 exhibit disrupted Nlp colocalization to centrosomes and enhanced Nlp degradation. Its is likely that the BRCA1 regulation of Nlp stability involves Plk1 suppression. Inhibition of endogenous Nlp via the small interfering RNA approach results in aberrant spindle formation, aborted chromosomal segregation, and aneuploidy, which mimic the phenotypes of disrupted BRCA1. Thus, BRCA1 interaction of Nlp might be required for the successful mitotic progression, and abnormalities of Nlp lead to genomic instability.
The successful mitosis requires the assembly of a strictly bipolar mitotic apparatus that will ensure that chromosomes equally distribute to the daughter cells. This process is controlled by the centrosomes that are required for spindle formation and function (1). Abnormalities of centrosome have been demonstrated to cause chromosomal missegregation and generation of aneuploidy, consequently leading to cell malignant transformation and tumorigenesis (2, 3). The machinery that controls centrosome stability involves multiple important cellular proteins, including p53 (4), BRCA1 (5), Gadd45 (6, 7), p21 (8), and Cdk2/cyclin E (9). The precise coordination among those regulators maintains centrosome duplication and stability. Prior to mitosis, centrosomes undergo maturation (10), which is characterized by centrosome enlargement, recruitment of γ-tubulin, and an increased microtubule nucleation activity (11, 12). Centrosome maturation is regulated by several mitotic kinases (13), such as Plk1 (Polo-like kinase 1) (14), Aurora-A (15), and Nek2, a member of NIMA (never in mitosis gene A)-related kinase (16). Recently, a Plk1-regulated ninein-like protein, termed Nlp, has been characterized as an important molecule involved in centrosome maturation (17). Nlp interacts with γ-tubulin ring complex and stimulates microtubule nucleation in the interphase. Upon the G2/M transition, Nlp is subjected to phosphorylation by Plk1 and Nek2 (17, 18) and departs from the centrosome. It is thus suggested that the delicate association of Nlp with the centrosome is required for proper centrosome maturation and spindle assembly (17).
BRCA1, a breast cancer susceptibility gene that accounts for more than 70% of hereditary breast cancer cases, is a critical regulator in the control of cell cycle progression (19, 20). BRCA1 interacts with multiple important cellular proteins, including RAD51 (21), BRCA2 (22), p53 (23), c-Myc (24), and p300 proteins (25). It is speculated that the BRCA1 protein may exert its control over cellular functions by acting as a platform for these proteins to converge and interact and may, therefore, create interactive modes for regulating their respective functions. BRCA1 is linked to the control of centrosome stability (26). Mouse embryonic fibroblasts (MEFs)3 carrying targeted deletion of exon 11 of the Brca1 gene exhibit centrosome amplification and abnormalities of spindle formation (5). BRCA1 may regulate centrosome duplication, probably through its interacting proteins such as p53 (23), BRCA2 (27), Cdk2 (28), and γ-tubulin (29–31), or its downstream genes such as p21 (32) and Gadd45a (33, 34). Most recently, BRCA1 was reported to be required for mitotic spindle assembly through its interaction with three spindle pole proteins, TPX2, NuMA, nuclear mitotic apparatus protein; and XRHAMM, Xenopus homolog to human RHAXX (35). These findings strongly suggest that BRCA1 is involved in the mitotic machinery. However, the importance of BRCA1 in the control of mitotic progression still remains to be further defined.
In this report, we demonstrate that BRCA1 physically interacts and colocalizes with Nlp. Nlp centrosomal localization and its protein stability are likely dependent on normal cellular BRCA1 function. Suppression of Nlp using the siRNA approach disturbs the process of chromosomal segregation and results in aberrant spindle formation, failure of chromosomal segregation, and aneuploidy.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Cell lines used were human cervical cancer line HeLa, human osteosarcoma line U2OS, human breast carcinoma line HCC1937 that harbors homozygous mutant BRCA1, primary human normal fibroblast line GM00380, and Chinese hamster ovary cell line. Both HCC1937/BRCA1 and HCC1937/GFP cells, which are isogenic lines of HCC1937, were kindly provided by D. M. Livingston of Harvard Medical School and maintained in ACL4 medium. HCC1937/BRCA1 is a HCC1937 cell line that stably expresses GFP-BRCA1, but HCC1937/control is used as a control for the HCC1937/BRCA1 cell line (36). Cell transfection was carried out as described previously (34).
Plasmid Clones
For construction of pEGFP-Nlp, the KIAA0980 fragment (75–4848) was inserted into the sites between SalI and SmaI of the pEGFP-C3 plasmid. GST-Nlp plasmid was constructed by ligating KIAA0980 fragment (75–4848) into the BamHI and NotI sites of pGEX-5X-1 vector. Additionally, Myc-tagged BRCA1 was made by inserting the open reading frame region of BRCA1 into the pCS2-MT vector. GST-BRCA1 was made by cloning BRCA1 cDNA into the pGEX-5X-1 vector. The Myc-Nlp plasmid was made by cloning the cDNA fragment of KIAA0980 into the NcoI and XhoI sites of the pCS2+MT vector.
Cellular Protein, Western Blotting Analysis, and Antibodies
Cellular proteins were prepared and subjected to immunoblotting analysis as described previously. For immunoprecipitation, cellular lysates were incubated with 10 μl of antibody and 20 μl of protein A/G agarose beads at 4 °C for 4 h. Immunocomplexes were washed with lysis buffer and examined by immunoblotting analysis (34). KIAA0980/Nlp polyclonal antibody was produced using Ac-CQEKVDKLKEQFEKNTKSD-amide short peptide (putative KIAA0980 protein 1310–1327 amino acids) from Invitrogen. In addition, the following antibodies were also used in the experiments; BRCA1, green fluorescent protein (GFP), c-Myc, p53, and actin were commercially provided from Santa Cruz Biotechnology.
GST Protein Preparations
Escherichia coli were collected by centrifugation and resuspended in cold sodium chloride-Tris-EDTA buffer. After incubation with lysozyme (100 μg/ml), bacteria were treated with DTT: Dithiothreitol (10 mm) and Sarkosyl (0.7%) and subjected to sonication. Following centrifugation, supernatants were mixed with glutathione-agarose beads at 4 °C overnight. After washing with PBS, glutathione-agarose bead-conjugated GST fusion proteins were ready for use in the designed experiments.
siRNA Treatment
The NLP siRNA sequence was designed as 5′-(GUG AGU CUU GAG GAA UUC C)d(TT)-3′ and 5′-(C AUG UAG AUU UGA GAG AGA)d(TT)-3′. The nonspecific siRNA (control) sequence was designed as 5′-(AUU GUA UGC GAU CGC AGA C)d(TT)-3′. These sequences do not match any other sequences in the GenBankTM. Transfection assays were conducted with 5 μl of 20 μm siRNA and 1 μl of Lipofectamine. Cells were incubated with siRNA for 24–96 h until they were ready to assay for gene knockdown analysis.
Immunofluorescent Staining of Centrosome
Cells were placed in 100-mm dishes. 16 h later, cells were fixed with 2% paraformaldehyde in PBS for 10 min at room temperature and washed with PBS. Cells were treated with cold methanol at −20 °C and followed by 0.5% Triton X-100 and 0.5% bovine serum albumin in PBS for 30 min. After incubation with antibodies to γ-tubulin and Nlp, the plates were washed and incubated with Cy3- or Cy4-conjugated goat anti mouse IgG. Following washing with PBS, Cells were visualized using an Olympus fluorescent microscope, and photographs were generated using a Kodak digital camera.
RESULTS
Nlp Is Characterized as a BRCA1-associated Protein and Colocalized with BRCA1
To gain molecular insights into the biological function of BRCA1, a yeast two-hybrid screen approach was employed to identify BRCA1-interacting proteins. Four different baits that harbor the C-terminal region of BRCA1 protein from 1293 to 1863 amino acids were used to screen a HeLa cDNA library. Following two-round screenings, one partial cDNA clone that interacts with the BRCA1 bait was identified as the KIAA0980. Recently, this gene product was characterized as a Plk1-regulated substrate and named Nlp (ninein-like protein) due to its significant similarity to ninein (17).
An initial effort was made to determine the physical interaction between BRCA1 and Nlp proteins. GST-BRCA1 or GST alone proteins were incubated with the lysates isolated from HeLa cells transfected with Myc-tagged Nlp vector. Following GST pulldown and immunoblotting analysis, Myc-tagged Nlp was detected in GST-BRCA1 pulldown complexes but not in GST pulldown complexes (Fig. 1A). Similarly, GST-Nlp, GST-p53, and GST-actin were incubated with cell lysates containing Myc-tagged BRCA1. Both GST-Nlp and GST-p53 (positive control) were shown to bring down Myc-tagged BRCA1 but not GST-actin or GST alone protein (Fig. 1B). The physical interactions between endogenous Nlp and BRCA1 proteins were also examined. Cellular lysates were prepared from HeLa cells and incubated with anti-BRCA1 or anti-Nlp antibodies. Following immunoprecipitation, Nlp protein was observed in the immunocomplex prepared with antibody to BRCA1, and BRCA1 was also detected in Nlp immunoprecipitates (Fig. 1C). In addition, the in vitro translated Nlp and the purified FLAG-Nlp were pulled down by GST-BRCA1, further supporting a direct interaction of Nlp with BRCA1 (Fig. 1D and supplemental Fig. 1A). Moreover, anti-Nlp antibody was incubated with cell lysates extracted from HCC1937 cells (BRCA1-deficient line) that were transfected with wild-type (WT) or mutant BRCA1 (P1749R and Y1853insA) expression vectors. Interestingly, only WT BRCA1 was detected in the Nlp immunocomplexes, but both P1749R and Y1853insA were not found in the immunoprecipitates by Nlp (supplemental Fig. 1B).
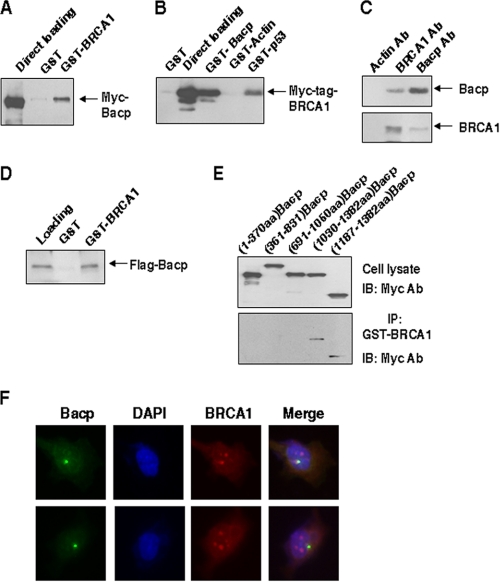
FIGURE 1.
Interaction of Nlp with BRCA1. A, Myc-tagged Nlp vector was expressed in HeLa cells. Cellular extracts were incubated with GST-BRCA1, and GST pulldown complexes were analyzed with anti-Myc antibody. B, cell lysates were made from HeLa cells transfected with Myc-tagged BRCA1 vector and incubated with GST proteins. The GST pulldown complexes were examined with anti-Myc antibody. C, HeLa cell lysates were immunoprecipitated with anti-actin, anti-BRCA1, and anti-Nlp antibodies (Ab). The immunocomplexes were analyzed with antibodies against BRCA1 and Nlp, respectively. D, purified FLAG-tagged Nlp from lysates of HeLa cells transfected with p3XFlag-Nlp was incubated, pulled down with GST-BRCA1, and analyzed by immunoblotting assay. E, a series of Myc-tagged Nlp deletion mutants were transiently introduced into HeLa cells. Whole cell lysates were prepared and incubated with GST-BRCA1. The GST pulldown complexes were analyzed with anti-Myc antibody. IB, immunoblot. F, U2OS cells were fixed and stained with antibodies to BRCA1 and Nlp and examined for BRCA1 (green spots) and Nlp (red spots). DAPI, 4′,6-diamidino-2-phenylindole.
To identify the regions required for Nlp interaction with BRCA1 protein, a series of Myc-tagged Nlp deletion protein expression vectors were introduced into HeLa cells, and cell lysates were incubated with GST-BRCA1. Following pulldown and immunoblotting analysis, the full-length Nlp, Nlp-(1030–1382), and Nlp-(1187–1382) were detected in the GST-BRCA1 complexes, but Nlp N-terminal fragments including Nlp-(1–370), Nlp-(361–831), and Nlp-(691–1050) were undetectable (Fig. 1E). Along with the observations that both Nlp-(1030–1382) and Nlp-(1187–1382) were able to immunoprecipitate endogenous BRCA1 (results not shown), these results indicate that the C-terminal region harboring 1187–1382 amino acids is required for Nlp association with BRCA1.
We also performed immunofluorescent staining with antibodies to BRCA1 and Nlp. The staining patterns of Nlp (green spots) were found to substantially overlap with BRCA1 protein (red spots) (Fig. 1F). Additionally, we transfected pEGFP-Nlp into U2OS cells and carried out immunostaining with BRCA1 antibody. The GFP-Nlp fusion protein was seen to localize with BRCA1 (results not shown). Collectively, the results in Fig. 1 demonstrate a specifically physical association of Nlp with BRCA1.
Functional Cellular BRCA1 Is Required for Nlp Localization to Centrosome
The centrosomal localizations of Nlp were further confirmed by double staining experiments using polyclonal anti-Nlp (green spots) and monoclonal anti-γ-tubulin (red spots) antibodies in human HeLa or U2OS cells (supplemental Fig. 2A) or introduction of pEGFP-Nlp expression vectors into Chinese hamster ovary or HeLa cells (supplemental Fig. 2B). Next, the multiple deletion mutants of GFP-Nlp fusion protein vectors were used to map the regions required for centrosome localization of Nlp (Fig. 2A). Full-length Nlp-(1–1382) revealed normal centrosomal localization. Similarly, two C-terminal fragments, Nlp-(1030–1382) and Nlp-(1187–1382), also showed normal centrosomal localization. In contrast, three N-terminal fragments, Nlp-(1–380), Nlp-(361–831), and Nlp-(651–1050), displayed aberrant centrosomal localization, as reflected by the abnormal aggregations of Nlp fusion proteins (Fig. 2A). These observations suggest that the C terminus of Nlp might be required for its localization at centrosomes.
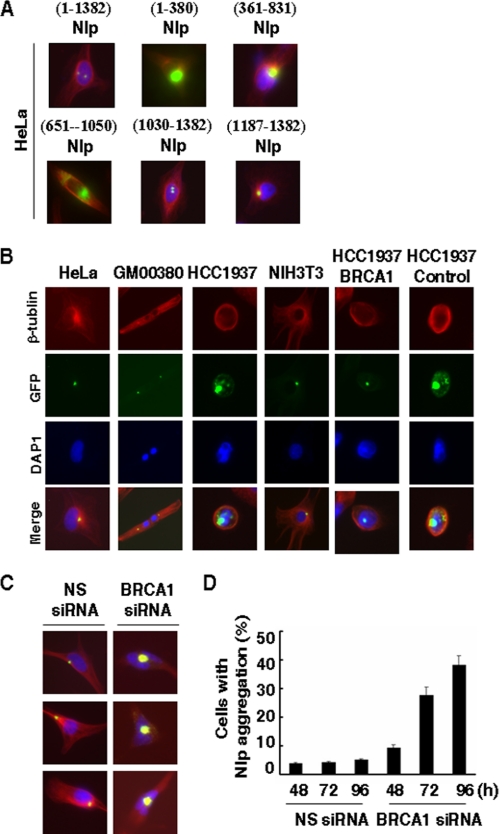
FIGURE 2.
BRCA1 dependence of Nlp colocalization to centrosome. A, requirement of Nlp C terminus for its colocalization to the centrosome. A series of pEGFP-Nlp expression vectors were introduced into HeLa cells. 48 h later, cells were subjected to fixation and staining with 4′,6-diamidino-2-phenylindole and β-tubulin. Cells were examined with a fluorescent microscope. B, a variety of human and mouse cells with known BRCA1 status were transiently transfected with pEGFP-Nlp. 48 h later, cells were fixed and examined for Nlp colocalization to the centrosome (green spots). DAP1, 4′,6-diamidino-2-phenylindole. C, HeLa cells were co-transfected with pEGFP-Nlp expression vector and 21-nucleotide siRNA duplexes that target BRCA1 mRNA transcript. As a control, nonspecific 21-nucleotide siRNA duplexes (NS siRNA) were included in the experiments. Cells were fixed and subjected to examine Nlp colocalization to the centrosomes. D, the quantitative results of Nlp protein aggregation in HeLa cells treated with BRCA1 siRNA were obtained from C. The experiments were repeated three times, and at least 500 cells were examined in each single assay.
Considering that the C terminus of Nlp is also critical for its interaction with BRCA1 (Fig. 1E), we examined whether Nlp centrosomal localization is correlated to cellular BRCA1 status. Following the introduction of pEGFP-Nlp into HeLa, U2OS, RKO, GM00380, NIH3T3, and Chinese hamster ovary cells, which contain wild-type BRCA1, GFP-Nlp was found to localize at centrosomes (Fig. 2B). Interestingly, in HCC1937 cells that harbor homozygous BRCA1 mutations, GFP-Nlp displayed abnormal patterns of subcellular localization as manifested by protein aggregation (Fig. 2B). In addition, two isogenic cell lines (HCC1973/BRCA1 and HCC1973/control) (36) derived from HCC1973 were employed. In HCC1973/BRCA1 cells, which are stably transfected with wild-type BRCA1 expression vector and express functional BRCA1 protein, GFP-Nlp resumed normal centrosomal localization. However, in HCC1973/control cells, GFP-Nlp still revealed abnormal centrosomal localization, as seen in parental HCC1973 cells (Fig. 2B). Similarly, WT BRCA1 or mutant BRCA1 (P1749R and Y1853insA) expression vectors were transiently introduced into HCC1973 cells and followed by fluorescent examination. We found that Nlp displayed centrosomal localization in cells expressing WT BRCA1 but not in cells containing mutant forms (results not shown).
Next, HeLa cells were co-transfected with pEGFP-Nlp vector and BRCA1 siRNA. Following the knockdown of endogenous BRCA1 (Fig. 3, C and D), there were substantial increases in abnormal aggregations of Nlp fusion protein (Fig. 2, C and D). In contrast, nonspecific siRNA had no significant effect on the Nlp-centrosomal localization. Together with the observations in Fig. 2B, these data suggest that functional cellular BRCA1 might be required for Nlp colocalization to centrosomes.
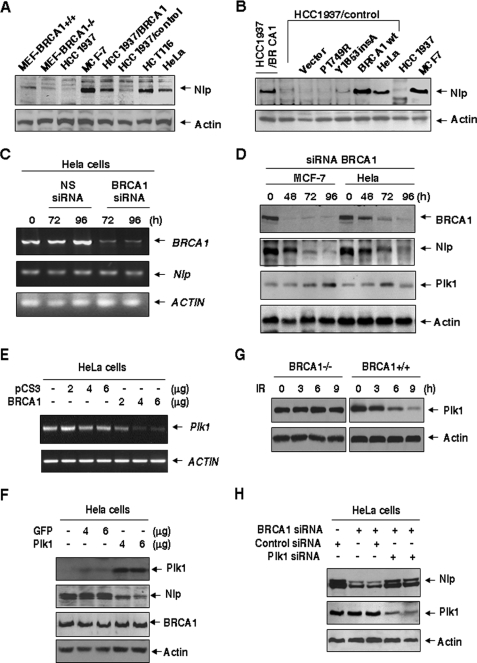
FIGURE 3.
BRCA1 dependence of Nlp protein stability. A, a variety of human and mouse cells with known BRCA1 status were collected for immunoblotting analysis with the Nlp antibody. B, HCC1937/control cells were transiently transfected with either WT BRCA1 or mutated BRCA1 (P1749R and Y1853insA) expression vectors and followed by immunoblotting assay with Nlp. C, HeLa cells were treated with BRCA1 siRNA or nonspecific (NS) siRNA, and BRCA1 mRNA was examined at the indicated time points. D, cell lysates were collected from HeLa cells treated with BRCA1 siRNA and subjected to immunoblotting analyses with antibodies to Nlp and Plk1. E, different amounts of BRCA1 expression vector were introduced into HeLa cells, and the levels of PLK1 mRNA were measured using the reverse transcription-PCR approach. F, HeLa cells were transfected with the indicated amounts of either GFP-Plk1 or pGFP vectors and subjected to analysis with antibodies to Nlp and BRCA1. G, both WT BRCA1 and BRCA1-deficient cells were treated with 10 grays of ionizing radiation (IR), and Plk1 expression was examined 8 h later. H, disruption of Plk1 rescues Nlp down-regulation caused by suppression of BRCA1. Plk1 siRNA were co-introduced with BRCA1 siRNA into HeLa cells. Nlp and Plk1 expression was analyzed by immunoblotting assay with antibodies to Nlp and Plk1.
Functional BRCA1 Maintains Nlp Stability
It has been proposed that Nlp/Nlp protein stability might be associated with its centrosomal localization (17). Nlp/Nlp expression is cell cycle-dependent and regulated by anaphase-promoting complex (APC) (37). Considering that the centrosomal localization of Nlp is dependent on BRCA1, we examined Nlp expression in cells with known BRCA1 status. In the human cell lines (MCF-7, HCT116, HeLa, and HCC1937/BRCA1) with functional BRCA1, Nlp protein was evidently detected, although its levels varied among different lines. In contrast, there were extremely low levels of Nlp in HCC1937 and HCC1937/control cells. The similar patterns of Nlp expression were observed in the normal MEFs and the MEFs derived from BRCA1 knockouts (Fig. 3A). Next, several vectors expressing either WT BRCA1 or mutated BRCA1 (P1749R and Y1853insA) were transfected into HCC1937/control cells. Nlp expression was clearly rescued following the introduction of WT BRCA1, but no evident changes in the cells expressing BRCA1 mutants were seen (Fig. 3B). We employed the siRNA approach to inhibit endogenous BRCA1 (Fig. 3C) and found that suppression of BRCA1 was coupled with the reduction of Nlp protein expression (Fig. 3D) but not NLP mRNA.
Because Plk1 regulates Nlp centrosomal localization (17) and BRCA1 suppresses Plk1 (38), we examined Plk1 expression after employment of BRCA1 siRNA. Interestingly, expression of Plk1 was elevated following BRCA1 inhibition (Fig. 3D), suggesting that BRCA1 regulation of Nlp stability might involve inhibition of Plk1. These results were further supported by the observation that introduction of BRCA1 expression vector into HeLa cells significantly suppressed mRNA levels of Plk1 (Fig. 3E) and that overexpression of Plk1 substantially down-regulated endogenous Nlp but had no effect on BRCA1 expression (Fig. 3F). Additionally, BRCA1 is likely required for suppression of Plk1 after ionizing radiation (Fig. 3G), which is similar to the report by others (38). Consistently, employment of Plk1 siRNA was shown to partially rescue the Nlp degradation caused by BRCA1 siRNA in HeLa cells (Fig. 3H). Furthermore, introduction of C-terminal Nlp that could compete for the binding of endogenous Nlp to BRCA1 resulted in reduction of Nlp expression (data not shown). Taken together, these findings indicate that BRCA1 regulates Nlp expression, probably through its inhibitory role in regulating Plk1.
Suppression of Endogenous Nlp Abrogates Chromosomal Segregation
Much effort was then taken to examine whether depletion of endogenous Nlp would interfere with chromosomal segregation and mitotic progression. Following treatment with Nlp siRNA, both HeLa and GM00380 cells silenced for Nlp revealed dramatic binuclear or multinuclear cells, coupled with bigger sizes (Fig. 4A). In contrast, nonspecific siRNA treatment had no effect on those cells. Quantitative analysis showed that after treatment with Nlp siRNA for 96 h, more than 80% of HeLa cells and 20% of GM00380 cells displayed such abnormal nuclear patterns (Fig. 4B), indicating that disruption of Nlp causes chromosomal instability. Similarly, suppression of endogenous BRCA1 by siRNA also resulted in multiple nuclei in both cell lines (Fig. 4A). To confirm the specific effect of Nlp siRNA, the rescuing experiments were included, and results are shown in Fig. 4C. Multinuclear and centrosome amplification caused by Nlp siRNA were only rescued by co-introduction of Nlp expression vector but not by BRCA1 vector. Vice versa, BRCA1 siRNA-induced multinuclear and centrosome amplification were only rescued by BRCA1 expression vector and not by Nlp expression vector. Therefore, disruption of endogenous Nlp results in chromosomal instability, which mimics the phenotypes of disrupted BRCA1.
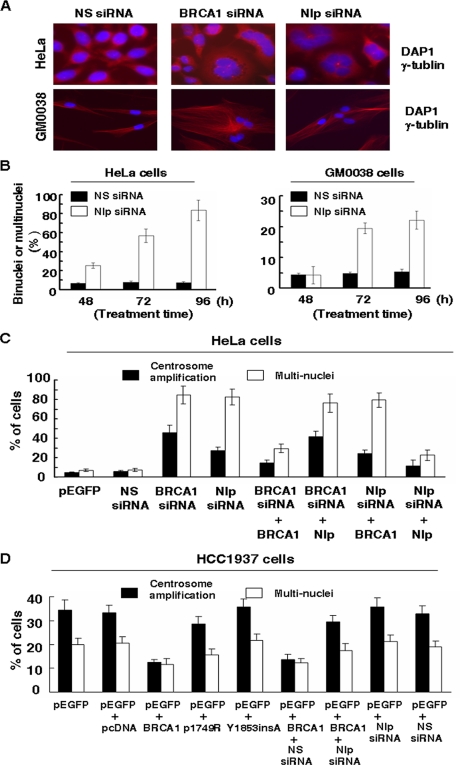
FIGURE 4.
Knockdown of Nlp expression results in multiple nuclei in human cells. A, HeLa and GM00380 cells were treated with Nlp siRNA or BRCA1 siRNA. 96 h later, cells were incubated with γ-tubulin, stained with 4′,6-diamidino-2-phenylindole (DAP1), and subjected to examination of nuclei with a fluorescent microscope. NS siRNA, nonspecific siRNA. B, the quantitative results of multiple nuclei in HeLa and GM00380 cells treated with Nlp siRNA. Cells with two or more nuclei were scored, and at least 500 cells were counted in each determination. C, HeLa cells were transfected with the indicated siRNA (BRCA1, Nlp, or nonspecific siRNA) in the presence of either BRCA1 or Nlp expression vectors. 96 h later, cells were subjected to analyses for centrosome amplifications and multiple nuclei. D, HCC1937 cells were transfected with pEGFP and BRCA1 (WT or mutants) expression vectors. In some cases, nonspecific siRNA or Nlp siRNA were included. 96 h after transfection, GFP-positive cells were subjected to analyses for centrosome amplifications and multiple nuclei. In each assay, 500 GFP-positive cells were counted, and the experiments were repeated three times.
To confirm whether Nlp is an important effector required for BRCA1 function in regulating genomic stability, WT BRCA1 or mutant BRCA1 expression vectors were transfected into HCC1937 cells, which have significant centrosome amplification and multiple nuclei due to BRCA1 deficiency. Expression of exogenous WT BRCA1 substantially reduced centrosome amplifications and multiple nuclei, but two BRCA1 mutants (P1749R and Y1853insA) had no improving effect on centrosomal abnormalities of HCC1937 cells. When Nlp siRNA were co-introduced into cells to suppress endogenous Nlp, the rescuing role of WT BRCA1 in HCC1937 was greatly eliminated. However, introduction of either Nlp siRNA or nonspecific siRNA into HCC1973 cells did not show exacerbated defect (Fig. 4D). These observations indicate that Nlp is a critical molecule for BRCA1 in regulating centrosome stability.
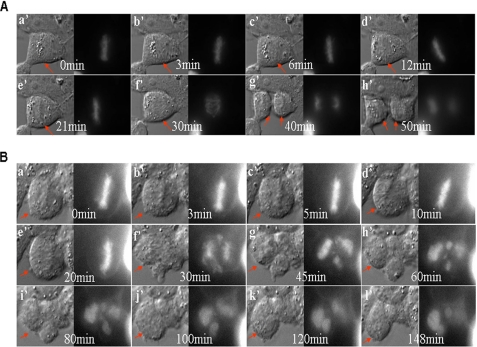
The time-lapse imaging analyses were employed to examine chromosomal segregation in HEK293T cells silenced for Nlp. Following treatment with control (nonspecific) siRNA, cells exhibited a normal mitotic process (Fig. 5A).
FIGURE 5.
Time-lapse images of human cells treated with Nlp siRNA. A, HEK293T cells were co-transfected with GFP-histone 2B expression vector and control siRNA and examined for chromosomal segregation using the time-lapse images approach. Cells exhibited normal chromosomal segregation and successful completion of cell division (a′–h′). Arrows point to a successful completion of mitosis. B, the GFP-histone 2B expression vector and Nlp siRNA were introduced into HEK293T cells, and time-lapse image analyses were carried out to examine chromosomal segregation. Apparently, cells silenced for endogenous Nlp displayed abrogated chromosomal segregation and mitotic failure (a′–l′). Arrows point to the unsuccessful completion of cell division.
In contrast, cells silenced for Nlp failed to complete the mitotic process. As shown in Fig. 5B, cells with disrupted Nlp were likely capable of entering metaphase, as reflected by the alignment of chromosomes in the equatorial plate (panel a). It appeared that cells could also move into anaphase and start chromosomal segregation. However, chromosomes did not separate evenly and migrated toward no specific direction instead of toward the two opposite poles (panel f–h). Cells also exhibited unsuccessful cytokinesis and ended up with the pattern of multiple nuclei (panels g–l).
Additionally, HeLa cells expressing GFP-α-tubulin were treated with control siRNA or Nlp siRNA and subjected to time-lapse imaging analyses. We found that cells treated with nonspecific siRNA revealed an intact bipolar spindle and performed a normal mitotic process (supplemental Fig. 3A), but cells silenced for Nlp exhibited disorganized mitotic spindles after metaphase and failed to complete mitotic process (supplemental Fig. 3B). These findings go along with the observations that cells treated with Nlp siRNA revealed multiple nuclei (Fig. 4), indicating that Nlp plays a critical role in spindle formation, chromosomal segregation, and cytokinesis and that disruption of Nlp results in abrogation of mitotic progression and genomic instability (multiple nuclei).
DISCUSSION
The physiological importance of BRCA1 is greatly determined by its interactions with many important cellular proteins (21–25). It is thought that BRCA1 may serve as a platform for some important cellular proteins to interact, and therefore, that it plays its controlling roles over cellular functions. In the current study, a series of direct biochemical data strongly demonstrate that Nlp is a BRCA1-interacting protein and that its C terminus is required for Nlp interaction with BRCA1 (Fig. 1), which suggests that the interaction between Nlp and BRCA1 might bridge the roles of BRCA1 in the control of mitotic progression. Strikingly, the findings in this report indicate that BRCA1 is likely required for Nlp centrosomal localization. In addition to the observations that normal Nlp centrosomal localization correlates to functional BRCA1 status, disruption of endogenous BRCA1 using the siRNA approach or deletion of the BRCA1-binding region of Nlp abrogate the property of Nlp localizing to the centrosome (Fig. 2). However, BRCA1 is also able to participate in the mitotic machinery through its E3 ubiquitin ligase activity. BRCA1 forms complexes with spindle pole proteins including TPX2, NuMA, and XRHAMM and specifically attenuates XRHAMM function, thereby ensuring the normal concentration of TPX2 on spindle poles and proper spindle pole assembly. However, this function of BRCA1 is independent of centrosome (35).
It has been proposed that Nlp stability might be associated with its centrosomal localization. When it departs from the centrosome, Nlp would undergo protein degradation (17). If this is the case, the BRCA1 regulation of Nlp centrosome localization would be important for the maintenance of Nlp stability. In support of this hypothesis, we have found that Nlp expression is dependent on normal cellular BRCA1 function. In cells with abnormal BRCA1 status, Nlp expression displays extremely low or undetectable levels. Suppression of endogenous BRCA1 via the siRNA approach or disruption of Nlp association with BRCA1 using C-terminal fragment results in enhanced degradation of Nlp (Fig. 3).
We have recently shown that Nlp is a fast turnover protein and expressed in a cell cycle-dependent manner. Its degradation is regulated by APC/c (37). It is thus speculated that disruption of the interaction between BRCA1 and Nlp might cause abnormal or unphysiological displacement of Nlp from the centrosome, and in turn, leads to fast degradation of Nlp by APC-mediated ubiquitination. Taken together, BRCA1 interaction of Nlp is critical for the maintenance of Nlp centrosome localization and protein stability. Because the Nlp centrosomal localization is important for centrosome maturation and spindle formation, Nlp interaction with BRCA1 appears to be a vital event in the regulation of mitotic progression.
Our findings are consistent with the idea that BRCA1 is a key molecule involved in the control of mitotic events. Despite its association with γ-tubulin (29–31) and spindle pole proteins including TPX2, NuMA, and XRHAMM (35), the BRCA1 interaction with Nlp is of great importance. Given the facts that Nlp centrosome localization and stability are controlled by both BRCA1 and Plk1 (Fig. 2), the dynamic interactions among BRCA1, Plk1, and Nlp are critical for mitotic machinery. This can be emphasized by the following facts. First of all, Nlp is important for spindle formation (17), chromosomal segregation, and cytokinesis (Fig. 5). Its is likely that the fine levels of Nlp appear to be required for its physiological function because suppression of Nlp causes aberrant mitotic spindle structure and leads to failures of chromosomal segregation (Figs. 4 and 5). Secondly, BRCA1 interaction of Nlp regulates Nlp centrosomal localization and Nlp stability. Disruption of BRCA1 abrogates Nlp localization at the centrosome (Fig. 2) and promotes Nlp degradation (Fig. 3). Thirdly, Plk1 phosphorylation of Nlp causes Nlp displacement from the centrosome, and in turn, regulates Nlp protein stability (17). Finally, the data presented in this study and by others (12) suggest that BRCA1 can serves as a negative regulator for Plk1 expression (Fig. 3).
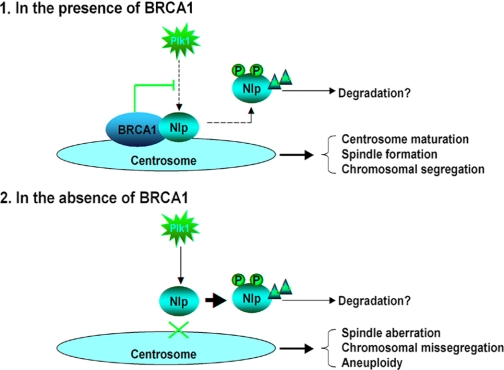
Thus, it can be reasonably interpreted (Fig. 6) that in mammalian cells, BRCA1 and Plk1 might work together to build up a delicate machinery to maintain a fine level of Nlp protein and precisely regulate Nlp dynamics of centrosomal localization to ensure successful spindle formation and chromosomal segregation. During the normal cell cycle progression, BRCA1 functions in balancing Plk1 activity and enabling Nlp to play a role in mitotic machinery. However, loss of BRCA1 may result in unbalanced and overpowered Plk1 activity, which causes “abnormal Nlp displacement” from the centrosome and Nlp degradation, and in turn, abrogates spindle formation and chromosomal segregation. This is in line with the observations that disruption of endogenous Nlp exhibited similar phenotypes (such as chromosomal abnormalities) to that seen with abrogated BRCA1 (Fig. 4). Therefore, the proper “switch on/off” of Nlp centrosomal localization is apparently vital machinery in the process of mitotic progression and mainly controlled by BRCA1 and Plk1. Abnormalities of this machinery via either loss of BRCA1 or amplified Plk1 activity will lead to aberrant mitotic spindle and chromosomal instability, and subsequently, result in tumorigenesis.
FIGURE 6.
The model for the role of the Nlp interaction with BRCA1 in mitosis. 1, in the presence of BRCA1, Nlp interacts with BRCA1 and thus localizes at the centrosome, and in turn, promotes centrosome maturation. This process is required for spindle formation and chromosomal segregation. It is likely that BRCA1 and Plk, as well as other mitotic kinases, act together to establish a fine balance to precisely regulate Nlp displacement and degradation. 2, in the absence of BRCA1, Nlp is unable to localize to the centrosome and likely suffers rapid degradation, which might be enhanced by Plk1-mediated phosphorylation and phosphorylation mediated by other mitotic kinases. Disruption of Nlp localization to the centrosome in turn leads to abnormal spindle formation and chromosomal missegregation, which greatly contribute to tumorigenesis.
In conclusion, we have demonstrated that BRCA1 interacts with Nlp. Several lines of evidence in our study indicate that Nlp interaction with BRCA1 is required for the maintenance of Nlp centrosomal localization and protein stability and is essential for mitotic progression. Therefore, these findings provide a novel link that connects BRCA1 with mitotic machinery, further supporting the notion that BRCA1 is an important regulator of cell cycle progression.
Supplementary Material
Acknowledgments
We greatly thank D. M. Livingston and R. Scully of Harvard Medical School for both HCC1937/BRCA1 and HCC1937/GFP cells, C. Deng of the National Institutes of Health for brca1 knockouts MEFs, and T. Nagase of Kazusa DNA Research Institute, Chiba, Japan for KIAA0980 (NLP) cDNA clone.
This work was supported by funding from the 973 National Key Fundamental Research Program of China (Grant 2009CB521801) and the National Natural Science Foundation of China (Grants 30730046 and 30721001).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- MEF
- mouse embryonic fibroblast
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- WT
- wild type
- APC
- anaphase-promoting complex
- NuMA
- Nuclear mitotic apparatus protein
- XRHAMM
- Xenopus homolog to human RHAXX.
REFERENCES
- 1.Doxsey S. J. (2001) Nat. Cell Biol 3,E105–E108 [DOI] [PubMed] [Google Scholar]
- 2.Salisbury J. L., Whitehead C. M., Lingle W. L., Barrett S. L. (1999) Biol. Cell 91,451–460 [PubMed] [Google Scholar]
- 3.Marx J. (2001) Science 292,426–429 [DOI] [PubMed] [Google Scholar]
- 4.Fukasawa K., Choi T., Kuriyama R., Rulong S., Vande Woude G. F. (1996) Science 271,1744–1747 [DOI] [PubMed] [Google Scholar]
- 5.Xu X., Weaver Z., Linke S. P., Li C., Gotay J., Wang X. W., Harris C. C., Ried T., Deng C. X. (1999) Mol. Cell 3,389–395 [DOI] [PubMed] [Google Scholar]
- 6.Hollander M. C., Sheikh M. S., Bulavin D. V., Lundgren K., Augeri-Henmueller L., Shehee R., Molinaro T. A., Kim K. E., Tolosa E., Ashwell J. D., Rosenberg M. P., Zhan Q., Fernández-Salguero P. M., Morgan W. F., Deng C. X., Fornace A. J., Jr. (1999) Nat. Genet. 23,176–184 [DOI] [PubMed] [Google Scholar]
- 7.Shao S., Wang Y., Jin S., Song Y., Wang X., Fan W., Zhao Z., Fu M., Tong T., Dong L., Fan F., Xu N., Zhan Q. (2006) J. Biol. Chem. 281,28943–28950 [DOI] [PubMed] [Google Scholar]
- 8.Tarapore P., Horn H. F., Tokuyama Y., Fukasawa K. (2001) Oncogene 20,3173–3184 [DOI] [PubMed] [Google Scholar]
- 9.Spruck C. H., Won K. A., Reed S. I. (1999) Nature 401,297–300 [DOI] [PubMed] [Google Scholar]
- 10.Rieder C. L., Faruki S., Khodjakov A. (2001) Trends Cell Biol. 11,413–419 [DOI] [PubMed] [Google Scholar]
- 11.Khodjakov A., Rieder C. L. (1999) J. Cell Biol. 146,585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blagden S. P., Glover D. M. (2003) Nat Cell Biol. 5,505–511 [DOI] [PubMed] [Google Scholar]
- 13.Donaldson M. M., Tavares A. A., Hagan I. M., Nigg E. A., Glover D. M. (2001) J. Cell Sci. 114,2357–2358 [DOI] [PubMed] [Google Scholar]
- 14.Nigg E. A. (1998) Curr. Opin. Cell Biol. 10,776–783 [DOI] [PubMed] [Google Scholar]
- 15.Goepfert T. M., Brinkley B. R. (2000) Curr. Top. Dev. Biol. 49,331–342 [DOI] [PubMed] [Google Scholar]
- 16.Fry A. M., Meraldi P., Nigg E. A. (1998) EMBO J. 17,470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casenghi M., Meraldi P., Weinhart U., Duncan P. I., Körner R., Nigg E. A. (2003) Dev. Cell 5,113–125 [DOI] [PubMed] [Google Scholar]
- 18.Rapley J., Baxter J. E., Blot J., Wattam S. L., Casenghi M., Meraldi P., Nigg E. A., Fry A. M. (2005) Mol. Cell. Biol. 25,1309–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L. M., Ding W. (1994) Science 266,66–71 [DOI] [PubMed] [Google Scholar]
- 20.Easton D. F., Ford D., Bishop D. T. (1995) Am. J. Hum. Genet. 56,265–271 [PMC free article] [PubMed] [Google Scholar]
- 21.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M. (1997) Cell 88,265–275 [DOI] [PubMed] [Google Scholar]
- 22.Rajan J. V., Wang M., Marquis S. T., Chodosh L. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93,13078–13083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., Somasundaram K., Peng Y., Tian H., Zhang H., Bi D., Weber B. L., El-Deiry W. S. (1998) Oncogene 16,1713–1721 [DOI] [PubMed] [Google Scholar]
- 24.Wang Q., Zhang H., Kajino K., Greene M. I. (1998) Oncogene 17,1939–1948 [DOI] [PubMed] [Google Scholar]
- 25.Pao G. M., Janknecht R., Ruffner H., Hunter T., Verma I. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,1020–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng C. X. (2002) Oncogene 21,6222–6227 [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Tombline G., Weber B. L. (1998) Cell 92,433–436 [DOI] [PubMed] [Google Scholar]
- 28.Okada S., Ouchi T. (2003) J. Biol. Chem. 278,2015–2020 [DOI] [PubMed] [Google Scholar]
- 29.Hsu L. C., Doan T. P., White R. L. (2001) Cancer Res. 61,7713–7718 [PubMed] [Google Scholar]
- 30.Sankaran S., Starita L. M., Groen A. C., Ko M. J., Parvin J. D. (2005) Mol. Cell. Biol. 25,8656–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starita L. M., Machida Y., Sankaran S., Elias J. E., Griffin K., Schlegel B. P., Gygi S. P., Parvin J. D. (2004) Mol. Cell. Biol. 24,8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somasundaram K., El-Deiry W. S. (1997) Oncogene 14,1047–1057 [DOI] [PubMed] [Google Scholar]
- 33.Harkin D. P., Bean J. M., Miklos D., Song Y. H., Truong V. B., Englert C., Christians F. C., Ellisen L. W., Maheswaran S., Oliner J. D., Haber D. A. (1999) Cell 97,575–586 [DOI] [PubMed] [Google Scholar]
- 34.Fan W., Jin S., Tong T., Zhao H., Fan F., Antinore M. J., Rajasekaran B., Wu M., Zhan Q. (2002) J. Biol. Chem. 277,8061–8067 [DOI] [PubMed] [Google Scholar]
- 35.Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., Livingston D. M. (2006) Cell 127,539–552 [DOI] [PubMed] [Google Scholar]
- 36.Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D. M. (1999) Mol. Cell 4,1093–1099 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Zhan Q. (2007) J. Biol. Chem. 282,17712–17719 [DOI] [PubMed] [Google Scholar]
- 38.Ree A. H., Bratland A., Nome R. V., Stokke T., Fodstad Ø. (2003) Oncogene 22,8952–8955 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.