Abstract
Comparative global proteome analyses were performed on Leptospira interrogans serovar Copenhageni grown under conventional in vitro conditions and those mimicking in vivo conditions (iron limitation and serum presence). Proteomic analyses were conducted using iTRAQ and LC-ESI-tandem mass spectrometry complemented with two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry. A total of 563 proteins were identified in this study. Altered expression of 65 proteins, including upregulation of the L. interrogans virulence factor Loa22 and 5 novel proteins with homology to virulence factors found in other pathogens, was observed between the comparative conditions. Immunoblot analyses confirmed upregulation of 5 of the known or putative virulence factors in L. interrogans exposed to the in vivo-like environmental conditions. Further, ELISA analyses using serum from patients with leptospirosis and immunofluorescence studies performed on liver sections derived from L. interrogans-infected hamsters verified expression of all but one of the identified proteins during infection. These studies, which represent the first documented comparative global proteome analysis of Leptospira, demonstrated proteome alterations under conditions that mimic in vivo infection and allowed for the identification of novel putative L. interrogans virulence factors.
Keywords: comparative proteomics, iTRAQ, two-dimensional gel electrophoresis, mass spectrometry, Leptospira, virulence factors, pathogenesis
Short abstract
The L. interrogans proteome was analyzed using iTRAQ and 2DGE. These analyses identified 563 proteins and altered expression of 65 proteins upon growth of L. interrogans under in vivo-like conditions, including upregulation of the L. interrogans virulence factor Loa22, a putative lipoprotein with primary amino acid sequence similarity to the outer surface protein ErpY of B. burgdorferi, and 4 additional proteins with homology to virulence factors found in other pathogens.
Introduction
Leptospirosis is a widespread zoonotic disease caused by various pathogenic serovars of the spirochete bacterium Leptospira.1,2 Leptospirosis has an average annual global prevalence of approximately 500 000 severe human cases.3,4 The total number of worldwide infections is expected to be grossly underestimated, due to inefficient diagnosis resulting from extensive serological diversity among pathogenic Leptospira species and an array of disease symptoms ranging from mild jaundice to severe liver and/or kidney failure.4−7Leptospira are transmitted from infected animals to humans either by direct contact with bodily fluids or indirectly via water contaminated with urine from an infected animal.(8) Pathogenic Leptospira enter the host via intact mucosal linings or through abrasions in the epidermis.4,8 Although leptospirosis can be effectively treated with tetracyclines and β-lactam/cephalosporin antibiotics in the early stages of the disease,(4) accurate disease diagnosis at this stage of infection is rarely achieved. Leptospirosis control is further hindered by the lack of an effective vaccine. Research focusing on the molecular components utilized by Leptospira to establish infection will permit the identification of novel virulence factors that may facilitate vaccine design and the development of novel diagnostics.
Many pathogenic bacteria establish infection by altering the expression of their proteome (defined as the entire complement of proteins expressed by a genome, cell, tissue or organism(9)) in response to environmental cues such as iron availability and the presence of host factors. Pathogenic bacteria use various outer membrane proteins (OMPs) to detect environmental changes, which in turn leads to the activation of signal transduction pathways, alteration of the proteome, and induction of conditions that favor pathogen survival.(10) Paradoxically, the same OMPs that facilitate pathogen survival can also lead to pathogen demise if detected by the host immune system.(11) Therefore, the regulation of bacterial OMPs and subcellular proteins involved in OMP regulation is crucial for the survival of the invading bacteria and for their ability to establish infection.
Multiple reports have established that Leptospira respond to altered environmental conditions at both the transcriptome12,13 and proteome levels.14−25 Genome-wide transcriptional analyses have previously been conducted on Leptospira interrogans serovar Lai exposed to temperature shifts from environmental/laboratory conditions to physiological conditions using DNA microarrays and real-time reverse transcription-PCR. These studies demonstrated regulation of genes whose encoded products can be classified based upon various functional groups, including energy production, protein export, heat shock protection, and chaperone activity.12,13 At the protein level, previous gel-based proteomic analyses conducted on L. interrogans have collectively (1) identified and determined cellular location for a diverse repertoire of L. interrogans proteins,15−18 (2) demonstrated the immunoreactive potential of some of these identified proteins,17,19,21 and (3) revealed that L. interrogans display unique protein expression profiles within different microenvironments.14,20,22−25 In particular, immunoblot analysis of outer membrane protein extracts of L. interrogans cultured under conditions of iron limitation, using sera collected from patients with leptospirosis, revealed expression of four immunoreactive OMPs of approximate molecular weights of 50, 55, 60, and 64 kDa.(14) Additionally, comparative proteomic analysis of L. interrogans cultured at temperatures above 30 °C in media containing 10% fetal bovine serum (FBS) and depleted of iron (−Fe) revealed altered regulation of various OMPs and cleavage of the lipoprotein LipL32 into various isoforms.(20)
In the present study, we have extended the above-mentioned gel-based proteomic investigations by performing both nongel-based and gel-based proteomic analyses on an organismal level for L. interrogans serovar Copenhageni strain Fiocruz L1-130 (henceforth referred to as L. interrogans). Comparative analyses were conducted on total protein extracted from bacteria that had been grown under conventional in vitro conditions and those mimicking in vivo conditions (iron limitation and serum presence, defined as in vivo-like conditions). These comparative culture conditions selected for identification of proteins that were differentially expressed upon exposure to host components (serum) and iron limitation independent of proteomic changes induced by a temperature shift. These studies provide the first comprehensive, comparative, and quantitative analysis of Leptospira total proteome alterations in response to environmental conditions that mimic those found within the host.
In the nongel-based proteomic approach, isobaric tags for relative and absolute quantitation (iTRAQ, for a review see ref (26)) and liquid chromatography-electrospray ionization (LC-ESI) tandem mass spectrometry were utilized. This approach combines the ability of iTRAQ to facilitate simultaneous protein abundance determination between different experimental conditions with the specificity of mass spectrometry for protein identification. Briefly, in iTRAQ proteins derived from a complex mixture are first digested by trypsin into peptides and an isobaric tag is covalently added onto primary amines within the peptides at N-termini and lysine side chains.(27) Tandem mass spectrometry analysis of the labeled peptides allows for quantitation of the isobaric tags and, accordingly, the peptide to which the tag is coupled. The isobaric nature of the tags ensures that the tagged peptides originating from the comparative conditions are treated as equal for separation purposes but can be accurately quantitated during tandem mass spectrometry analysis. This approach offers several advantages over traditional proteomic methodologies. First, the reduced spectral complexity associated with the isobaric tags used in iTRAQ allows for quantitation determination via comparison of reporter ion peak intensities during collision induced dissociation.(27) Second, iTRAQ/LC/ESI-tandem mass spectrometry allows accurate identification and quantitation of all proteins within a complex sample, irrespective of molecular weight or isoelectric point,26,28 thus facilitating analysis of a diverse set of proteins with various functional roles, divergent cellular locations, and differing abundance.(29) Finally, iTRAQ permits simultaneous proteomic analysis of multiple samples,(27) thereby facilitating statistical verification of changes in protein expression between different environmental conditions.(30) This methodology has been successfully used to analyze the proteomic profiles of Halobacterium salinarum NRC-1(31) and Cyanobacteria(32) in relation to salt tolerance, Nostoc punctiforme ATCC 29133 during different nitrogen fixation stages,(33)Mycobacterium ulcerans during mycolactone biosynthesis,(34) and Escherichia coli grown in either Lauria Broth media or fresh milk,(35) thus, displaying the effectiveness of this methodology for analysis of bacterial proteomes.
To complement and confirm our proteomic findings obtained via the nongel-based proteomic approach, we conducted simultaneous gel-based proteomic analyses using two-dimensional gel electrophoresis (2DGE) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry.20,36,37 The combined iTRAQ and 2DGE experiments identified a total of 563 expressed leptospiral proteins. Altered expression levels were observed for 65 of these proteins, including 6 proteins that have been previously shown to play a direct role in the virulence of either L. interrogans or other pathogenic bacteria.
Materials and Methods
Leptospira and Culture Conditions
L. interrogans serovar Copenhageni strain Fiocruz L1-130 is a clinical isolate originating from Salvador, Brazil(2) that was kindly provided by Dr. David A. Haake (Veterans Affairs Greater Los Angeles Health Care System, Los Angeles, California). Prior to protein extraction for proteomic analysis, bacteria were passaged in vitro a maximum of six times via cultivation in Ellinghausen and McCullough(38) as modified by Johnson and Harris(39) (EMJH) media at 29.5 °C. Bacteria were diluted in phosphate buffered saline (PBS) containing 5 mM MgCl2 to allow for accurate enumeration using a Petroff Hauser counting chamber (Fisher Scientific, Ottawa, Ontario) and a darkfield microscope (Nikon ECLIPSE 50i, Mississauga, Ontario). For media shift experiments, cultures were grown to a density of 1 × 108 cells/mL, centrifuged at 8500g, and resuspended at the same concentration in either (1) fresh EMJH media preincubated at 37 °C or (2) EMJH media depleted of iron by overnight incubation with 0.4 mM 2,2-dipyridyl(20) (Sigma, Oakville, Ontario, Canada) and supplemented with FBS (Sigma) to a final concentration of 10% (−Fe/FBS media) at 37 °C. Cultures were then incubated for an additional 72 h at 37 °C prior to harvest.
Protein Extraction and Sample Preparation
Subsequent to harvest, bacteria were washed twice with repeated steps of centrifugation at 9500g and resuspended in PBS supplemented with 5 mM MgCl2.(20) Pellets obtained from the comparative conditions were lyophilized, weighed, and resuspended to equal dry mass per volume. For iTRAQ and immunoblotting analyses, total protein was prepared according to the following protocol: pellets were resuspended in 0.1% SDS, lysed via sonication (Sonicator 3000, Masonix, Farmindale, NY) using 4 W of continuous power for a total of 3 × 30 s on ice, centrifuged at 16 000g to remove debris, and subjected to overnight acetone precipitation at −20 °C. Protein precipitates were centrifuged at 10 000g (30 min at 4 °C), excess acetone was removed, and the resulting pellets were resuspended in 1 mL of 100 mM triethylammonium bicarbonate/4 M urea/0.2% sodium dodecyl sulfate and subjected to a second acetone precipitation. For 2DGE experiments, pellets were resuspended to a concentration of 2.5 mg/mL of dry weight in IEF buffer (5 M urea, 2 M thiourea, 2 mM tributyl phosphine (Sigma), 2% CHAPS (Sigma), 1% carrier ampholytes (Bio-Rad Laboratories, Mississauga, Ontario), 2% 3-(Decyldimethylammonio)propanesulfonate inner salt (Sigma), 40 mM TRIS-HCl, and 0.002% bromphenol blue). Lysis was achieved via sonication followed by the addition of 150 U of benzonase (Sigma) per mL of lysate and incubation at room temperature for 20 min. The resulting lysate was centrifuged at 16 000g to remove debris and the isolated supernatant was used in IEF experiments.
iTRAQ Analysis
Total protein samples were submitted to the University of Victoria-Genome BC Proteomics Centre for iTRAQ analysis. Trypsin digestion and isobaric tag labeling were performed according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA).
iTRAQ Experimental Design
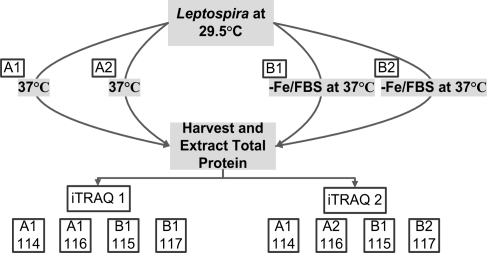
To control for biological, technical and experimental variations, two independent iTRAQ experiments were performed using the experimental strategy developed by Gan et al.(30) Figure 1 shows an overview of the iTRAQ experimental design used to analyze the L. interrogans protein samples. Biological variation was controlled for at the culture level using pooled cultures, where A1 and B1 each represent 75 independent cultures and A2 and B2 each represent 25 independent cultures. Following tryptic digestion of all four samples (A1, A2, B1, B2), samples A1 and B1 were split into three equal volumes, and the resulting 8 samples were labeled with appropriate isobaric tags as depicted in Figure 1. Replicate samples (2 × A1 and 2 × B1) in the first iTRAQ experiment (iTRAQ1) controlled for technical variation within the same iTRAQ experiment and analysis of A1 and B1 in the second iTRAQ experiment (iTRAQ2) controlled for experimental variation between independent iTRAQ experiments. Finally, comparison of the experimental replicates A1 to A2 and B1 to B2 in iTRAQ2 allowed evaluation of variation introduced via the protein extraction steps.
Figure 1.
iTRAQ experimental design. Pooled biological samples were used to control for biological variation by combining 75 independent cultures in samples A1 (shifted to 37 °C) and B1 (shifted to 37 °C in −Fe/FBS media) and 25 independent cultures in samples A2 (shifted to 37 °C) and B2 (shifted to 37 °C in −Fe/FBS media). To control for technical variation within a single iTRAQ experiment and experimental variation between two iTRAQ experiments, samples A1 and B1 were split into 3 equal volumes subsequent to tryptic digestion but prior to isobaric tag labeling. Comparison of A1 to A1 and B1 to B1 in iTRAQ1 and A1 to A2 and B1 to B2 within iTRAQ2 controlled for technical variation within the same iTRAQ experiment, while comparison of A1 and B1 between iTRAQ1 and iTRAQ2 controlled for experimental variation between separate iTRAQ experiments. Numbers 114, 115, 116, and 117 correspond to the isobaric tags used for labeling the samples. The overall experimental strategy was adapted from Gan et al.(30)
Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry
Mass spectrometry was performed at the University of Victoria-Genome BC Proteomics Centre.(40) Briefly, LC-MS/MS analysis was performed using an integrated Famos autosampler, SwitchosII switching pump, and UltiMate micro pump (LC Packings, Amsterdam, The Netherlands) system with an Hybrid Quadrupole-TOF LC-MS/MS Mass Spectrometer (QStar Pulsar i) equipped with a nanoelectrospray ionization source (Proxeon, Odense, Denmark) and fitted with a 10 μm fused-silica emitter tip (New Objective, Woburn, Massachusetts). Chromatographic separation was achieved on a 75 μm × 15 cm C18 PepMap Nano LC column (3 μm, 100 Å, LC Packings) and a 300 μm × 5 mm C18 PepMap guard column (5 μm, 100 Å, LC Packings) was in place before switching inline with the analytical column and the MS. The mobile phase (solvent A) consisted of water/acetonitrile (ACN) (98:2 (v/v)) with 0.05% formic acid (FA) for sample injection and equilibration on the guard column at a flow rate of 100 μL/min. A linear gradient was created upon switching the trapping column inline by mixing with solvent B which consisted of ACN/water (98:2 (v/v)) with 0.05% FA and the flow rate was reduced to 200 nL/min for high resolution chromatography and introduction into the mass spectrometer.
Samples were brought up to 20 μL with 5% ACN and 3% FA and transferred to autosampler vials (LC Packings). Ten microliters of sample was injected in 95% solvent A (10 mM KPO4 (pH 2.7), 25% ACN) and allowed to equilibrate on the trapping column for 10 min to wash away any contaminants. Upon switching inline with the MS, a linear gradient from 95% to 40% solvent A developed for 40 min, and in the following 5 min, the composition of mobile phase was increased to 20% A before decreasing to 95% A for a 15 min equilibration before the next sample injection. MS data was acquired automatically using Analyst QS 1.0 software Service Pack 8 (ABI MDS SCIEX, Concord, Canada). An information-dependent acquisition method consisting of a 1 s TOFMS survey scan of mass range 400−1200 amu and two 2.5 s product ion scans of mass range 100−1500 amu was performed. The two most intense peaks over 20 counts with charge state 2−5 were selected for fragmentation and a 6 amu window was used to prevent the peaks from the same isotopic cluster from being fragmented again. Once an ion was selected for MS/MS fragmentation, it was put on an exclude list for 180 s. Curtain gas was set at 23, nitrogen was used as the collision gas, and the ionization tip voltage used was 2700 V.
If the observed A215 was greater than 0.1 for any fraction collected during the strong cation exchange 2.5 h gradient, then 95−50% solvent A was used to compensate for the higher peptide concentration in that fraction. Data files were processed using the Protein Pilot software (Version 2.0.1, Applied Biosystems).
iTRAQ Data Analysis
Data analysis was first conducted by exporting all data sets from Protein Pilot to Microsoft Excel (Office 2007, Microsoft, Seattle, WA). These were then converted to text files to be further analyzed using Statistical Analysis System (SAS) software Version 9.1 of the SAS System for Windows (SAS Institute, Inc., Cary, NC).
To be considered as significantly altered in expression, a protein and it's representative peptides must have met the following criteria: (1) the protein had to demonstrate a database match with a leptospiral protein; (2) the protein had to be represented by at least two peptides of differing amino acid sequence;26,41,42 (3) the test to determine if the ratio of protein expression equals 1 had to have a p-value less than 0.05 for at least 2 of the 4 possible comparisons (e.g., for iTRAQ1; 115:114, 115:116, 117:114 and 117:116); and (4) experimental and nonexperimental 95% confidence intervals for a given protein ratio must not have overlapped. Confidence intervals for the average protein ratios were calculated on the log10 scale and then back-transformed to the linear scale (the same scale as the original ratio).
To obtain an estimate of the average peptide ratio for a protein within a replicate experiment (replicate i), Protein Pilot calculates a weighted average of the log10 peptide ratios as
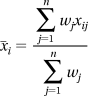 |
where n is the number of peptides used to identify a protein; xij is the log10 ratio of peptide j for replicate i; i = 1, 2, ..., N,j = 1, 2, ..., n; wj is the weight given to peptide j where wj = 1/%Errorj; and the percent error for each peptide ratio is formulated by Protein Pilot as
where AreaA and AreaB are the areas for each peak in the ratio of experimental to nonexperimental peptides. ErrorA and ErrorB are the mass spectrometry measurement errors for AreaA and AreaB, respectively.
As previously discussed, iTRAQ experiments were designed to control for biological, experimental, and technical variation (Figure 1) resulting in four iTRAQ replicate experiments. This provides up to four estimates of the average peptide ratio for each protein. These estimates were averaged to obtain the grand mean of the log10 peptide ratio which is calculated as x̅. = ∑Ni=1(x̅i/N), where, for the experimental ratios, N is the number of replicate experiments that satisfied criteria 1, 2, and 3 above. For the nonexperimental ratios, N is the number of replicate experiments that satisfied criteria 1 and 2 above. Thus, N varied between 2 and 4 depending on the protein.
As each replicate experiment provides an estimate of the average peptide ratio, the variation among these estimates (the empirical variance) is used to obtain the standard error of the grand mean.(43) The estimate of the empirical variance of the grand mean log10 peptide ratio is calculated as
Finally, a 95% confidence interval for the average log10 peptide ratio for a protein is obtained using x̅. ± tN−1,0.025(Vâr(x̅.))1/2, where tN−1,0.025 is the critical value of the Student t distribution with N − 1 degrees of freedom that corresponds with a 95% confidence level. The upper and lower levels of the confidence interval were back-transformed from log10 space into linear space to obtain 95% confidence intervals for the average peptide ratio. All calculations were automated by writing custom scripts in SAS available from the authors.
2DGE Experiments
Supernatants from protein extraction experiments were split into three equal volumes and run in triplicate for a total of 6 IEF and SDS-PAGE experiments (3 replicates for L. interrogans in unmodified culture media and 3 replicates for L. interrogans in −Fe/FBS media) conducted concurrently to limit experimental and technical variation. Isoelectric focusing was conducted using the Ettan IPGphor isolectric focusing system (Amersham Bioscience, Piscatway, NJ) and nonlinear pH 3−11 immobilized pH gradient isoelectric focusing strips (Bio-Rad Laboratories). The run parameters included overnight hydration of the strips with sample in IEF buffer at room temperature under a gradient voltage to a maximum of 100 V. Isoelectric focusing run parameters included gradient voltage to 3500 V over 12 h and a hold at 3500 V for 18 h. The strips were then incubated at room temperature for 20 min in equilibration buffer (6 M urea, 2% SDS, 0.36 M Tris-HCl, 20% glycerol, 5 mM tributyl phosphine, and 2.5% acrylamide) and subsequently used in SDS-PAGE experiments. SDS-PAGE was conducted on an Etta Dalt Twelve system (Amersham) using 8−16% gradient acrylamide gels. Gels were subsequently stained with colloidal Coomassie Blue G-250, and proteins that reproducibly exhibited differential protein expression levels between the experimental conditions were cored for in-gel tryptic digest.(20) Core digests were C8 ZipTip purified (Bio-Rad Laboratories), manually spotted on MALDI plates, and overlaid with α-cyano-4-hydroxycinnamic acid (Sigma) at a concentration of 10 mg/mL in 50% ACN/0.1% trifluoroacetic acid for MALDI-TOF mass spectrometry analyses.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry
MALDI-TOF MS (Voyager-DE STR; Applied Biosystems) was conducted in reflector mode under high voltage settings. Laser intensities ranged from 2500 to 2800 Hz depending on the observed signal-to-noise ratios in the mass spectra. A three point calibration was performed under reflector mode using an angiotensin tryptic digest calibration mix (Sigma). Prior to database analysis, mass spectra were noise filtered, baseline corrected, two point internally calibrated using trypsin m/z peaks 841.51 and 2210.1, and, depending on signal-to-noise ratios, the detection level was adjusted between 2 and 15%. Peptide mass fingerprint searches were conducted using the Mascot peptide mass fingerprinting database (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF) and search parameters were set as follows: taxonomy was set to “other bacteria” or “all entries” for identification of leptospiral proteins and protein contaminants, respectively; trypsin was selected as the enzyme; the number of missed cleavage sites was set to one; variable modifications were set to carbamidomethyl and methionine oxidation; mass tolerance was set between 10 and 50 ppm; and mass values were set to monoisotopic. Proteins with significant scores and at least four representative peptides were considered for further computer database analyses.
Computer Database Analysis
Proteins were annotated and accession numbers were assigned using the ExPASy proteomics server (http://ca.expasy.org/). Proteins deemed as significantly altered in expression were further confirmed using an annotated leptospiral genome database operated and maintained by Dr. T. Seemann in collaboration with the Victorian Bioinformatics Consortium at Monash University, Clayton, Victoria, Australia. The cellular localization of proteins was predicted with PSORTb v.2.0 (http://www.psort.org/psortb/index.html) using the Gram-negative algorithm, while LipoP 1.0 (http://www.cbs.dtu.dk/services/LipoP/), SpLip (http://jcslab.vbi.vt.edu/splip/) and SignalP (http://www.cbs.dtu.dk/services/SignalP/) bioinformatic tools were utilized to predict lipoproteins and signal peptides.
Recombinant Protein Expression and Purification
All recombinant proteins were prepared as follows. Open reading frames (ORFs) corresponding to LIC12032, LIC13166, LIC11966, and LIC12575 were PCR amplified from L. interrogans serovar Copenhageni strain Fiocruz L1-130 genomic DNA, while LIC13050 was amplified from L. interrogans serovar Copenhageni strain RJ15958 genomic DNA. Primer pairs used in the amplifications are listed in Table 1. The LIC13050 PCR product was ligated first into the donor vector pDONR201 followed by the expression vector pDEST17 (Gateway Technology, Invitrogen, Carlsbad, CA). Amplicons corresponding to LIC12032, LIC13166, LIC11966, and LIC12575 were ligated first into the cloning vector pJET1 (CloneJet, Fermentas, Ontario, Canada), digested with NdeI and either HindIII (LIC12032), EcoRI (LIC13166), or XhoI (LIC11966 and LIC12575) followed by ligation into a similarly digested pET28a expression vector (Novagen, Gibbstown, NJ). The sequence and reading frame of the expression constructs were verified by DNA sequencing with vector-specific primers. The LIC13050/pDEST17 construct was transformed into the E. coli expression strain BL21-AI and all other constructs were transformed into the E. coli expression strain BL21 Star (DE3) (both from Invitrogen). Expression of the constructs and purification of the resulting six-histidine-tagged recombinant proteins were performed as follows. Recombinant expression was induced during log-phase growth using final concentrations of 1 mM isopropyl β-d-thiogalactopyranoside for LIC11966, LIC12575, LIC12032, and LIC13166 and 0.2% l-arabinose for LIC13050. Bacteria were harvested by centrifugation and lysed via sonication (3 × 20 s) in the presence of a protease inhibitor cocktail (Catalogue number 539134; Calbiochem, San Diego, CA). For LIC11966, LIC12575, and LIC13050, insoluble recombinant protein preparations were purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (QIAGEN, Mississauga, Ontario, Canada) as previously described.(44) For LIC12032 and LIC13166, soluble recombinant protein preparations were purified using Ni-NTA agarose beads according to the ProBond native purification protocol (Invitrogen). Briefly, bacterial lysates (see above) were centrifuged at 10 000g at 4 °C for 15 min and the supernatant applied to Ni-NTA agarose beads (QIAGEN) in a 50 mL conical tube. The supernantant-Ni-NTA agarose bead suspension was incubated overnight at 4 °C on a hematology/chemistry mixer 346 (Fisher Scientific) followed by centrifugation at 200g at 4 °C and removal of the supernatant. Beads were washed twice with wash buffer (60 mM NaH2PO4, 500 mM NaCl, and 20 mM imidazole, pH 8.0) and the protein was eluted with elution buffer (identical to the wash buffer except containing 250 mM imidazole). The expressed recombinant proteins were dialyzed against PBS and quantitated using the BCA Protein Assay kit (Pierce, Rockford, IL).
Table 1. ORF-Specific Primers Used to Amplify Fragments for Recombinant Expression.
| ORF | sense primera | antisense primera | size (bp) | portion of ORF (bp) |
|---|---|---|---|---|
| LIC13050 | 5′-CAGGAAGATCTGGATGAAAG | 5′-TTATTTTTTTGTAGGTTGAGTAGTAGT | 1114 | 70−1114 |
| LIC12032 | 5′-CTAGACCATATGATGAGTAGAAAAACCCTTACTAC | 5′-GTCAGAAGCTTTTAAACTCGACTCAGAGCAAG | 1470 | 1−1470 |
| LIC13166 | 5′-CTAGACCATATGAACAATCAGGGCGGTAATCAG | 5′-GTCAGGAATTCTTAAGGTCTAACCGAAATCAC | 864 | 82−864 |
| LIC11966 | 5′-CTAGACCATATGATGAAAAAACATTCTATCAGTAAAATC | 5′-GTCAGCTCGAGTTATTGAGAAGCGTATTCTTTCGC | 504 | 1−504 |
| LIC12575 | 5′-CTAGACCATATGTTAGAATATGCTTATAAACATAGAG | 5′-GTCAGCTCGAGTCATTTAGCGAGATAATAGCGCAAC | 604 | 943−1518 |
Underlined nucleotides represent incorporated restriction sites.
Antibodies
Polyclonal rabbit antiserum was prepared against each of LIC12032, LIC13166, LIC11966, and LIC12575 by ImmunoPrecise Antibodies (Victoria, British Columbia, Canada). Monoclonal antibody 1H8 against FlaA1 has been described previously.(45) Mouse antiserum specific for Loa22 (LIC10191) was kindly provided by Drs. Nobuo Koizumi and Haruo Watanabe (National Institute of Infectious Diseases, Tokyo, Japan). LIC13050-specific rabbit antiserum was a gift from Dr. David A. Haake (Veterans Affairs Greater Los Angeles Health Care System, Los Angeles, CA).
Immunoblot Analyses
Bacteria were grown and shifted as described in the section. Cultures were harvested at 10 000g,, washed twice with wash buffer (PBS and 5 mM MgCl2), split into duplicate samples, and lyophilized. Equal dry weights were resuspended to a concentration of 7.5 mg/mL with sample buffer (100 mM Tris pH 6.8, 8 M urea, 2% SDS (w/v), 10% glycerol (v/v), and 100 mM DTT) and sonicated (Sonicator 3000) at 6 W for 30 s continuous bursts on ice until a homogeneous mixture was obtained. Samples were centrifuged at 16 000g and the supernatant was removed and heated at 40 °C for 30 min. Ten microliters of protein sample for LIC13166, LIC12032, LIC11966, and LIC10191 and 30 μL of protein sample for LIC12575 and LIC13050 were separated on 15% sodium dodecyl sulfate-polyacrylamide gels. High-range molecular mass markers (Spectra Broad Range Protein Ladder; Fermentas, Burlington, Ontario, Canada) were used as standards. Proteins were electrophoretically transferred to Immobilon-polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA). Immunoblots were blocked with 2.5% milk powder in Tris-buffered saline, pH 7.4, with 0.05% Tween 20 (TBST) for 1 h at room temperature. Membranes were washed 2 × 5 min with TBST followed by incubation for 90 min at room temperature with a 1:2000 dilution of LIC10191-specific mouse antiserum; a 1:2000 dilution of rabbit antiserum specific for LIC13050, LIC12032, LIC13166, and LIC11966; or a 1:1000 dilution of rabbit antiserum specific for LIC12575, all diluted in 2.5% milk powder/TBST. To normalize the samples, a 1:2000 dilution of anti-FlaA1 1H8 monoclonal antibody was included within each of these incubations. FlaA1 was demonstrated through iTRAQ analyses to remain unchanged between the comparative conditions and has been used elsewhere as a normalization control. Membranes were washed with TBST 4 × 5 min, incubated with a 1:10 000 dilution of anti-mouse IgG (H+L) IRDye700 conjugate for LIC13166; anti-mouse IgG (H+L) IRDye800 conjugate for LIC13050, LIC12032, LIC11966, LIC12575, and LIC10191; and anti-rabbit IgG (H+L) IRDye800 conjugate for LIC13166, LIC13050, LIC12032, LIC11966, and LIC12575 (IRDye conjugates were purchased from Rockland Immunochemicals for Research, Gilbertsville, PA) in 2.5% milk powder/TBST for 60 min at room temperature followed by washing 4 × 5 min with TBST. Image analyses were performed with an Odyssey infrared imaging system equipped with Version 2.1 software (LI-COR Bioscience, Lincoln, NE).
Serum Samples
Serum samples collected from patients with severe leptospirosis were a kind gift from Dr. Albert I. Ko (Centro de Pesquisa Gonçalo Moniz, Fundação Oswaldo Cruz, Salvador, Bahia, Brazil). Samples were obtained from the collection of the Oswaldo Cruz Foundation (Salvador, Bahia, Brazil) and have been described elsewhere.(46) A pooled sample was prepared by combining 5 μL of serum collected from each of 15 individuals with laboratory-confirmed leptospirosis. Normal human serum was obtained from Lonza (Basel, Switzerland). Approval was obtained from the Human Subjects Institutional Review Board of the University of Victoria, as well as the Oswaldo Cruz Foundation.
Enzyme-Linked Immunosorbent Assay (ELISA)
Unless otherwise stated, all incubations were performed at 37 °C in a humidified chamber. Ninety-six-well plates (Nunc-Immuno Maxisorp; Sigma) were coated in triplicate overnight with 100 ng per well of rLIC13050, rLIC12032, rLIC13166, rLIC11966, or rLIC12575 in 50 mM sodium carbonate-bicarbonate buffer (pH 9.6). Plates were washed with PBS, pH 7.4, with 0.05% Tween 20 (PBST) and blocked with 200 μL per well of 3% bovine serum albumin (BSA; Sigma)/PBST for 60 min. Plates were washed with PBST and 100 μL of a 1:500 dilution of pooled laboratory-confirmed leptospirosis sera or normal human sera prepared in 3% BSA/PBST was added to wells containing recombinant proteins and to wells lacking recombinant protein (for background control) and incubated for 60 min. Plates were washed with PBST and 100 μL of a 1:3000 dilution of goat anti-human IgG (Fab specific) peroxidase conjugate (Sigma) prepared in 3% BSA/PBST was added. Following a final incubation for 60 min, plates were washed with PBST and developed for 10 min at room temperature with 100 μL of tetra-methylbenzidine-H2O2 substrate (TMB; Kirkegaard & Perry Laboratories, Gaithersburg, MD). Absorbance was measured at 600 nm on a Synergy HT microplate reader (BioTek, VT). Values were background subtracted and statistical analyses were performed using the Student’s t test.
Immunofluorescence Microscopy
Paraffin-embedded, formalin-fixed (PEFF) tissue samples of golden Syrian hamsters (Mesocricetus auratus) infected with L. interrogans serovar Pomona strain 11000-74A were analyzed by indirect immunofluorescence microscopy as described previously.47,48 Briefly, paraffin was removed from 4 μm-thick sections of PEFF samples adhered to glass slides using standard procedures, then blocked with 10% normal horse sera. Sections were incubated overnight at 4 °C with rabbit antibodies prepared to either LipL21(49) or LIC13050. Slides were washed with PBS and then incubated for 1 h at room temperature in the dark with goat anti-rabbit Alexa 488-conjugated secondary antibody (Molecular Probes, Eugene, OR). After washing the slides with PBS, the samples were counterstained with DAPI (4′, 6-diamidino-2-phenylindole, 1.5 mg/mL), and coverslips mounted using SlowFade Light antifade (Molecular Probes). Stained tissue sections were examined with a Nikon Eclipse E800 microscope (Nikon Instruments, Inc., Melville, NY) using a 100× Pan Fluor objective. Separate images captured with a Spot RT color CCD camera (Diagnostic Instruments, Inc., Sterling Heights, MI) using B-2A and UV-2E/C filters were merged using Spot Advanced Software (Diagnostic Instruments, Inc.). Use of serial tissue sections enabled analysis of adjacent sections incubated with different primary antibodies.
Results and Discussion
Overview of Global Proteome Analyses
In this study, global proteome analyses were conducted to directly investigate the differential proteomic profiles that result upon exposure of L. interrogans to environmental conditions that mimic those found within the host. Comparative conditions consisted of growth of L. interrogans at 37 °C in either unmodified culture media or culture media that had been depleted of iron and supplemented with 10% FBS (−Fe/FBS media representative of in vivo-like conditions). Although it should be stressed that these conditions are entirely in vitro in nature, these analyses revealed two findings. First, they provided a novel global analysis of the L. interrogans proteome. And second, the comparative conditions allowed for identification of the complete repertoire of proteins that are differentially expressed in specific response to limited iron availability and the presence of serum factors, two conditions that represent, at least in part, the environment present within a host.
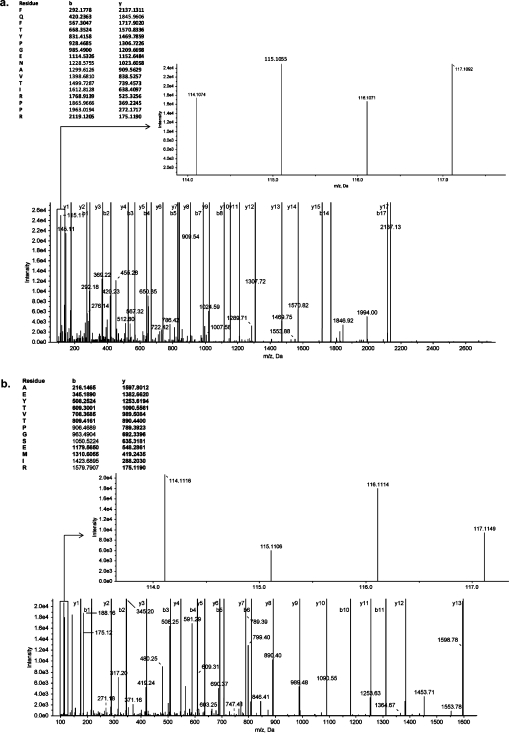
Two independent yet complementary quantitative global proteomic analytical techniques were used to investigate the proteomes isolated from the comparative growth conditions. Proteomes were first analyzed using iTRAQ combined with LC-ESI tandem mass spectrometry. This approach allows direct determination of relative protein abundance levels between comparative conditions. Representative MS/MS spectra for peptides identified from a endoflagellar filament core protein (LIC11890) and a probable aconitate hydratase (LB327) are shown in Figure 2, representing proteins that are up- and downregulated in L. interrogans grown in −Fe/FBS culture media, respectively. The peaks of iTRAQ signature ions (114 and 116 for proteins isolated from L. interrogans grown in unmodified culture media and 115 and 117 for proteins isolated from L. interrogans grown in −Fe/FBS culture media) are shown as insets. To reduce the possibility of both false positive protein identification and variation in protein quantitation, biological, technical and experimental variations were controlled for by designing and analyzing iTRAQ experiments as outlined in Figure 1.26,27,30,41,42 Briefly, biological variation was controlled for by extracting protein from pooled independent culture conditions, while technical variation in our sample processing was controlled for by conducting analyses on two independent pooled samples for each culture condition. Variability within the same iTRAQ experiment and between separate iTRAQ experiments was controlled for by conducting analyses on samples A1 and B1 twice in iTRAQ1 and once in iTRAQ2. Statistical analyses were subsequently performed to determine proteins that were altered in expression levels under −Fe/FBS culture conditions within a 95% confidence interval. This method of analysis allowed for determination of statistically significant altered expression levels from “control” conditions, regardless of the observed fold change. Collectively, this strategy for experimental design and statistical analysis allowed for stringent and sensitive determination of altered protein expression levels in response to the in vivo-like environmental conditions. To further increase confidence in assigning protein expression levels, we also performed gel-based proteomics via 2DGE combined with MALDI-TOF mass spectrometry.
Figure 2.
Mass spectra of iTRAQ reporter ion m/z peak intensities used for peptide quantitation and of m/z peaks used for peptide sequence identification. Upon collision induced dissociation of gated peptides of a given precursor mass, reporter ions are released and their relative intensities used for quantitation. Representative MS/MS spectra for peptides identified from (a) an endoflagellar filament core protein (LIC11890) and (b) a probable aconitate hydratase (LB327) suggest up- and downregulation, respectively, in response to growth in −Fe/FBS media.
Together iTRAQ and 2DGE analyses identified 563 L. interrogans proteins (see Tables 2 and 3 and Supporting Information Tables S1−S3 and Figures S1−S3). This corresponds to approximately 15% of the total protein-expressing ORFs of L. interrogans serovar Copenhageni strain Fiocruz L1-130 (563/3728 total)(50) and, to our knowledge, represents the highest comparative global proteomic coverage achieved to date in Leptospira. Of the identified proteins, 65 demonstrated statistically significant alterations in expression levels between the two experimental conditions tested (see Tables 2 and 3). The highly sensitive iTRAQ analyses identified 62 proteins that were altered in expression (Table 3), while the 2DGE analyses identified a narrower range of 6 proteins altered in expression levels (Table 2 and Supporting Information Figure S3), a finding consistent with the reduced experimental sensitivity associated with 2DGE technology. The average fold change in expression levels as determined by iTRAQ analyses ranged from −5.863 to 2.731 (Table 3), which compares with other investigations focused on comparative global proteome analyses.51−53
Table 2. Proteins Exhibiting Altered Expression Levels in L. interrogans in Response to in vivo-like Conditions (Exposure to −Fe/FBS) As Determined by 2DGE.
| accession no. | protein | ORF | protein expression level |
|---|---|---|---|
| Energy Production and Metabolism (1) | |||
| Q72VD7 | Electron transfer flavoprotein beta-subunit EtfB | LIC10361 | Downregulated |
| Confirmed/Potential Virulence Factors (2) | |||
| Q72MY9 | Putative glycosyl hydrolase | LIC13050 | Upregulated |
| Q72MM7 | Putative coagulase | LIC13166 | Upregulated |
| Chaperones and Post-Translational Regulation (2) | |||
| CH10_LEPIC | GroES | LIC11336 | Downregulated |
| Q72QA2 | IbpA-1 | LIC12210 | Downregulated |
| Chemotaxis and Motility (1) | |||
| Q72R58 | Periplasmic flagellin FlaB1 | LIC11890 | Upregulated |
Table 3. Proteins Exhibiting Altered Expression Levels in L. interrogans in Response to in vivo-like Conditions (Exposure to −Fe/FBS) As Determined by iTRAQa.
| iTRAQ1 |
iTRAQ2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| protein | ORF | controlb | experimentalb | fold changec | controlb | experimentalc | fold changec | overall fold change |
| Energy Production and Metabolism (18) | ||||||||
| Bifunctional dihydrolipoyllysine-residue acetyltransferase/dihydrolipoyllysine-residue succinyltransferase | LIC1_ SPN3184 | NS | NS | NS | 0.905−1.239 | 1.507−1.74 | 1.384 | 1.384 |
| Cytochrome C oxidase subunit II | LIC10208 | 0.971−1.123 | 1.221−1.395 | 1.214 | NS | NS | NS | 1.214 |
| Transketolase, C-terminal subunit | LIC11355 | NS | NS | NS | 0.872−1.275 | 1.369−1.562 | 1.209 | 1.209 |
| Cytochrome C oxidase, subunit I | LIC10209 | 0.996−1.064 | 1.185−1.29 | 1.196 | NS | NS | NS | 1.196 |
| Nucleoside-diphosphate kinase | LIC13326 | 0.977−1.042 | 0.863−0.945 | −1.072 | NS | NS | NS | −1.072 |
| Acyl carrier protein | LIC20065 | 0.922−1.39 | 0.629−0.911 | −1.076 | NS | NS | NS | −1.076 |
| Methionine adenosyl transferase | LIC11354 | 1.106−1.107 | 0.708−0.942 | −1.107 | NS | NS | NS | −1.107 |
| Ketol-acid reductoisomerase | LIC13393 | 0.718−1.557 | 0.6−0.709 | −1.041 | 0.942−1.236 | 0.649−0.804 | −1.209 | −1.125 |
| Sugar pyridoxal-phosphate-dependent aminotransferase | LIC12198 | 1.011−1.155 | 0.707−0.859 | −1.187 | NS | NS | NS | −1.187 |
| Thiosulfate sulfurtransferase | LIC11115 | 0.712−1.524 | 0.52−0.595 | −1.229 | 0.923−1.332 | 0.553−0.758 | −1.286 | −1.257 |
| Succinyl-CoA synthetase, alpha subunit | LIC12574 | 1.027−1.095 | 0.685−0.805 | −1.270 | NS | NS | NS | −1.270 |
| Acyl dehydratase | LIC12739 | NS | NS | NS | 0.762−1.936 | 0.362−0.634 | −1.310 | −1.310 |
| NADH dehydrogenase (ubiquinone), E chain | LIC12745 | 0.838−1.36 | 0.413−0.759 | −1.402 | NS | NS | NS | −1.402 |
| Isopropylmalate Isomerase | LIC11821 | NS | NS | NS | 0.606−1.823 | 0.376−0.43 | −1.434 | −1.434 |
| Nitrogen regulatory protein PII | LIC10440 | 0.848−1.279 | 0.501−0.568 | −1.526 | NS | NS | NS | −1.526 |
| Electron transfer flavoprotein, alpha subunit | LIC10360 | 0.949−1.093 | 0.47−0.502 | −1.918 | 0.956−1.245 | 0.541−0.695 | −1.429 | −1.674 |
| Electron transfer flavoprotein, beta subunit | LIC10361 | 1.015−1.065 | 0.446−0.497 | −2.043 | NS | NS | NS | −2.043 |
| Aconitate hydratase | LIC20249 | 0.98−1.043 | 0.415−0.423 | −2.314 | 0.85−1.317 | 0.376−0.455 | −1.942 | −2.128 |
| Protein Synthesis (17) | ||||||||
| 50S Ribosomal protein L7/L12 | LIC10752 | 0.81−1.355 | 0.69−0.778 | −1.055 | 0.932−1.223 | 0.733−0.899 | −1.075 | −1.065 |
| 50S Ribosomal protein L4 | LIC12872 | 0.941−1.15 | 0.751−0.84 | −1.139 | NS | NS | NS | −1.139 |
| 50S Ribosomal protein L2 | LIC12870 | 0.962−1.149 | 0.751−0.858 | −1.140 | NS | NS | NS | −1.140 |
| 50S Ribosomal protein L3 | LIC12873 | NS | NS | NS | 0.883−1.446 | 0.561−0.821 | −1.151 | −1.151 |
| Ribosomal protein S12 | LIC10755 | NS | NS | NS | 0.913−1.169 | 0.702−0.802 | −1.178 | −1.178 |
| 50S Ribosomal protein L15 | LIC12854 | NS | NS | NS | 0.87−1.573 | 0.474−0.731 | −1.263 | −1.263 |
| 30S Ribosomal Protein S13 | LIC12849 | 0.943−1.098 | 0.706−0.73 | −1.291 | 1.077−1.317 | 0.536−0.858 | −1.238 | −1.265 |
| Protein-synthesizing GTPase complex, EF-Tu component | LIC12875 | 0.952−1.08 | 0.717−0.746 | −1.284 | NS | NS | NS | −1.284 |
| 50S Ribosomal protein L10 | LIC10751 | 0.834−1.423 | 0.499−0.635 | −1.360 | 0.911−1.48 | 0.519−0.787 | −1.228 | −1.294 |
| 30S Ribosomal protein S17 | LIC12864 | 1.009−1.027 | 0.713−0.755 | −1.339 | NS | NS | NS | −1.339 |
| 30S Ribosomal protein S1 | LIC12447 | NS | NS | NS | 0.68−1.91 | 0.362−0.536 | −1.355 | −1.355 |
| 50S Ribosomal protein L16 | LIC12866 | 0.836−1.246 | 0.539−0.57 | −1.478 | 0.953−1.12 | 0.698−0.765 | −1.262 | −1.370 |
| 50S Ribosomal protein L11 | LIC10749 | 0.776−1.387 | 0.5−0.568 | −1.404 | NS | NS | NS | −1.404 |
| 50S Ribosomal protein L22 | LIC12868 | 0.833−1.243 | 0.552−0.595 | −1.428 | NS | NS | NS | −1.428 |
| 30S Ribosomal protein S3 | LIC12867 | 1.011−1.034 | 0.667−0.699 | −1.432 | NS | NS | NS | −1.432 |
| 50S Ribosomal protein L1 | LIC10750 | 1.024−1.039 | 0.65−0.7 | −1.437 | NS | NS | NS | −1.437 |
| 50S Ribosomal protein L29 | LIC12865 | NS | NS | NS | 1.093−1.404 | 0.296−0.508 | −2.082 | −2.082 |
| Unknown Function (9) | ||||||||
| Protein of unknown function | LIC10314 | NS | NS | NS | 1.035−1.039 | 1.116−1.341 | 1.180 | 1.180 |
| Protein of unknown function | LIC13123 | 0.975−1.066 | 1.198−1.247 | 1.169 | NS | NS | NS | 1.169 |
| Protein of unknown function | LIC10672 | NS | NS | NS | 0.858−1.323 | 1.341−1.557 | 1.164 | 1.164 |
| Protein of unknown function | LIC10027 | 0.895−1.138 | 1.297−1.315 | 1.158 | NS | NS | NS | 1.158 |
| Protein of unknown function | LIC13314 | NS | NS | NS | 1.044−1.063 | 1.077−1.265 | 1.108 | 1.108 |
| Protein of unknown function | LA0268 | NS | NS | NS | 0.796−1.329 | 0.678−0.767 | −1.073 | −1.073 |
| Protein of unknown function | LIC11784 | NS | NS | NS | 0.803−1.483 | 0.405−0.542 | −1.571 | −1.571 |
| Protein of unknown function | LIC13183 | NS | NS | NS | 1.085−1.317 | 0.308−0.461 | −2.220 | −2.220 |
| Conserved protein of unknown function | LIC11059 | NS | NS | NS | 1.205−1.304 | 0.099−0.187 | −5.863 | −5.863 |
| Confirmed/ Potential Virulence Factors (5) | ||||||||
| Putative glycosyl hydrolase | LIC13050 | NS | NS | NS | 0.538−2.069 | 5.271−5.441 | 2.731 | 2.731 |
| Putative Erp Y-like lipoprotein | LIC11966 | NS | NS | NS | 0.889−1.176 | 1.975−2.063 | 1.755 | 1.755 |
| OmpA-family lipoprotein (Loa22) | LIC10191 | 0.908−1.141 | 1.267−1.317 | 1.152 | NS | NS | NS | 1.152 |
| Catalase | LIC12032 | 0.956−1.114 | 1.197−1.29 | 1.151 | NS | NS | NS | 1.151 |
| TolC-like protein | LIC12575 | 0.8−1.325 | 1.378−1.512 | 1.122 | NS | NS | NS | 1.122 |
| Chaperones and Post-Translational Regulation (3) | ||||||||
| Sensor histidine kinase and response regulator of a two component complex | LIC11709 | NS | NS | NS | 0.786−1.366 | 0.471−0.688 | −1.333 | −1.333 |
| Endopeptidase Clp, ATP-dependent proteolytic subunit | LIC12017 | 0.905−1.136 | 0.379−0.393 | −2.313 | NS | NS | NS | −2.313 |
| Preprotein translocase, SecA subunit | LIC11944 | 0.931−1.127 | 0.328−0.408 | −2.485 | NS | NS | NS | −2.485 |
| Transcription (4) | ||||||||
| Transcription termination factor Rho | LIC12636 | NS | NS | NS | 0.845−1.254 | 0.757−0.809 | −1.049 | −1.049 |
| Transcription elongation factor | LIC12706 | NS | NS | NS | 0.906−1.157 | 0.498−0.591 | −1.631 | −1.631 |
| Two-Component Response Regulator Transcriptional Regulator Protein | LIC13088 | 0.942−1.119 | 0.49−0.572 | −1.733 | NS | NS | NS | −1.733 |
| Response Regulator | LIC20254 | 1.055−1.061 | 0.478−0.561 | −1.825 | NS | NS | NS | −1.825 |
| Chemotaxis and Motility (4) | ||||||||
| Methyl-accepting chemotaxis protein | LIC11523 | 0.858−1.343 | 1.593−1.952 | 1.409 | NS | NS | NS | 1.409 |
| Endoflagellar filament core protein | LIC11890 | 0.996−1.029 | 1.293−1.356 | 1.303 | NS | NS | NS | 1.303 |
| Methyl-accepting chemotaxis protein | LIC12921 | 0.954−1.115 | 1.162−1.242 | 1.111 | 1−1.018 | 1.064−1.181 | 1.111 | 1.111 |
| Response regulator receiver domain | LIC11526 | 0.724−1.557 | 0.51−0.659 | −1.176 | NS | NS | NS | −1.176 |
| Cell Shape (1) | ||||||||
| Actin-like ATPase involved in cell morphogenesis | LIC11258 | 0.938−1.078 | 0.512−0.532 | −1.787 | 1.049−1.137 | 0.547−0.712 | −1.467 | −1.627 |
| DNA Regulation (1) | ||||||||
| RecA recombinase | LIC11745 | NS | NS | NS | 0.725−1.51 | 0.592−0.683 | −1.090 | −1.090 |
NS: Not significant. Protein quantitation was deemed not significant if any of the following conditions were found: proteins represented by less than two peptides; proteins displaying a p-value greater than 0.05; or proteins having an error factor of greater than two or overlapping 95% confidence values for their respective average ratios.
A 95% confidence interval for a given protein ratio.
Value was obtained by averaging absolute high and low control and experimental protein ratios in log space. Values were then transformed into linear space and experimental values divided by control values.
Assignment of Functional Categories
To evaluate the global protein expression changes in L. interrogans in response to a shift to −Fe/FBS media, proteins altered in expression were assigned to 9 distinct categories based upon their database-assigned functionality (Tables 2 and 3). L. interrogans shifted to −Fe/FBS media altered the expression of proteins with predicted functional roles corresponding to energy production and metabolism (18), protein synthesis (17), unknown function (9), confirmed/potential virulence factors (6), chaperones and post-translational regulation (5), transcription (4), chemotaxis and motility (4), cell shape (1) and DNA repair (1). The broad changes in observed protein expression levels are consistent with a previous genome-wide transcriptional analysis performed on L. interrogans serovar Lai strain Lai in response to a temperature shift, which revealed altered expression of 106 genes.(12) However, whereas overall upregulation was observed in transcript levels of chaperones and protein synthesis machinery in response to a temperature shift, in our studies, we observed a general trend toward downregulation of proteins belonging to the same functional categories in response to a shift to −Fe/FBS media.
The following sections describe in detail the proteins identified via our proteomic analyses to be altered in expression levels in response to the in vivo-like growth conditions compared to control in vitro conditions. Proteins are discussed in the context of leptospiral pathogenesis within the selected functional categories of energy production and metabolism, chaperones and protein synthesis, motility and chemotaxis, and confirmed/potential virulence factors.
Energy Production and Metabolism
Within our studies, numerous proteins mediating key steps of oxidative phosphorylation, amino acid biosynthesis, and nucleotide metabolism were downregulated in response to a shift to −Fe/FBS media (Supporting Information Figure S4). Downregulation of proteins involved in energy production and metabolism was in accordance with the downregulation of 17 proteins involved in protein synthesis. Experimentally, L. interrogans shifted to −Fe/FBS media displayed slower growth rates and generation times. It follows that the observed downregulation of energy production and metabolic processes correlates well with the slower generation time in −Fe/FBS media. As iron is an essential cofactor for many enzymes that are in turn required for cell viability,(54) bacteria have evolved various mechanisms for sensing changes in iron concentration within their environment and for iron acquisition via siderophore expression. Hence, a rationale for the reduced energetic and metabolic processes may have been the inability to acquire necessary iron for optimal functioning of these processes, thereby resulting in downregulation of the proteins involved in these pathways when the L. interrogans were shifted to the −Fe/FBS media.
Chaperones and Protein Synthesis
The stress response pathway includes the alteration of expression of chaperones, a group of proteins with broad molecular functions ranging from protein repair and regulation to protein export (for a review see ref (55)). In our studies, shifting L. interrogans to −Fe/FBS media resulted in downregulation of 3 chaperones, namely, small heat shock protein IbpA-1, GroES, and ClpA-1 (Tables 2 and 3). Downregulation of chaperone expression fits with the overall trend toward decreased protein synthesis as there would be fewer proteins requiring repair, regulation and export to noncytoplasmic locations.
As previously stated, we also observed downregulation of 17 proteins involved in protein synthesis in −Fe/FBS media (Table 3). This number is likely an underestimation of the actual number of downregulated proteins mediating protein synthesis, due to technical limitations associated with conducting proteome analyses on a complex sample. In L. interrogans, genes encoding ribosomal proteins appear to be organized in operons of varying sizes, and in select regions of the genome these operons are tandemly organized. As expression generally occurs at the operon level rather than the individual gene level, detection of downregulation of individual protein products in our studies suggests downregulation of the operon as a whole. The downregulated proteins we detected comprised 16 ribosomal proteins and the GTPase complex of the EF-Tu component TufB, which facilitates aminoacyl-tRNA binding to the A-site of ribosomes during translation. As translational machinery was downregulated in −Fe/FBS media, it is likely that downregulation of proteins involved in energy production, metabolism, export and chaperones was occurring, at least partially, at the translational level. Combined, these results correlate well with the slower growth rate observed in the −Fe/FBS media.
Flagellar and Chemotaxis Proteins
An important step in the infection process is the use of chemotactile and flagellar proteins for traversal of host barriers resulting in dissemination to deeper tissues.56,57 Directional changes and swimming behavior of motile bacteria is a complex process involving the regulation of an extensive number of proteins at both the translational and post-translational levels in response to altered environmental conditions (for a review see ref (58)). Traversal of host barriers is not a random event, as previously shown by motile Treponema denticola mutants that do not express the chemotaxis protein histidine kinase CheA and/or the chemoreceptors DmcA or DmcB. These mutants show only 2−30% penetration rates of epithelial cell layers when compared to the wild type.(59) It has also previously been observed that the swimming behavior of motile pathogens is a specific result of differential concentration gradients of attractants and repellents. In particular virulent, but not avirulent, Leptospira demonstrate strong chemoattractant behavior toward hemoglobin, as shown by movement of virulent Leptospira toward hemoglobin but not distilled water in a chemotaxis assay utilizing semisolid media-filled U-shaped polypropylene tubes.(60)
In contrast to the overall trend toward downregulation observed for the majority of the aforementioned leptospiral proteins, proteins involved in motility were generally observed to be upregulated in L. interrogans shifted to −Fe/FBS media (Table 3). Directed motility and chemotaxis are central to spirochete virulence,(61) and thus, the observed upregulation of flagellar proteins in −Fe/FBS media parallels the experimental conditions that mimic an in vivo environment. The sole chemotaxis protein that was downregulated corresponded to the two-component response regulator CheY which is involved in facilitating directional changes in motile bacteria in response to chemotactile stimuli.62,63
Confirmed/Potential Virulence Factors
Through our comparative proteomic experimental approach, we observed upregulation of 6 proteins that fit within the confirmed/potential virulence factor category (Tables 2 and 3). One of these proteins (Loa22) has been previously identified in L. interrogans as a virulence factor. The remaining 5 upregulated proteins within this category share significant sequence similarity with proteins shown to play key roles in the pathogenesis of other organisms, including coagulase, catalase, TolC, glycosyl hydrolase, and the outer surface lipoprotein ErpY. The following sections describe each of these upregulated leptospiral proteins within the functional context of pathogenesis of either L. interrogans for previously characterized proteins or other pathogens for previously uncharacterized proteins. In addition, the results of experiments conducted to independently confirm upregulation of protein expression and verify protein expression during the course of infection are included. Discussion of the putative ErpY-like lipoprotein is excluded as the immunoblot analysis did not confirm upregulation of this protein under the tested in vivo-like environmental conditions (see below).
1. Loa22
Previous gel-based analyses investigating the outer membrane proteome of L. interrogans serovar Copenhageni strain RJ16441 during infection of Hartley male guinea pigs revealed that the L. interrogans surface lipoprotein Loa22 is expressed during infection.(64) Furthermore, Himar1 transposon mutagenesis of the gene encoding Loa22 rendered L. interrogans serovar Lai strain Lai avirulent in Hartley male guinea pig and Golden Syrian male hamster models of leptospirosis and Loa22 genetic complementation in these mutants restored virulence.(65) Combined, these studies definitively show that Loa22 is essential for L. interrogans virulence. In our studies, we detected a 1.152-fold upregulation of Loa22 upon a shift to −Fe/FBS media (Table 3). To our knowledge, this is the first documented report of regulation of Loa22 expression in response to defined stimuli that mimic infection, namely iron limitation and the presence of serum.
2. Putative Coagulase
A noteworthy observation arising from the 2DGE analyses was the upregulation of a protein in response to the −Fe/FBS media that shared amino acid sequence similarity with a region of the Staphylococcus aureus surface protein coagulase (Table 2; 53% similarity/26% identity over 128 amino acids). In S. aureus, this protein has been shown to contribute to bacterial pathogenesis in two ways. First, it plays a pivotal role in the development of blood-borne infection through binding of the blood factor prothrombin to the bacterial surface, which upon activation to thrombin results in subsequent conversion of fibrinogen to fibrin. This proteolytic cascade results in the formation of fibrin clots on the bacterial surface,(66) thus, causing the bacteria to become refractory to neutrophil phagocytosis(67) and promoting evasion of the host immune response. Second, in S. aureus, the coagulase protein further contributes to pathogenesis by aiding invasion of deeper tissues via degradation of the host fibrinogen barriers to fibrin. In support of this proposed functional role, expression of the S. aureus coagulase has been associated with hematogenous pulmonary infection in mice.(68)
3. Putative Catalase
An additional obstacle that pathogenic bacteria must overcome to establish infection is host-induced oxidative stress and the release of reactive oxygen species. Catalase and peroxidase activity and H2O2 detoxification have previously been established in Leptospira.69,70 In our studies, we have detected 1.151-fold upregulation of a catalase (KatE) in response to growth of L. interrogans in −Fe/FBS media (Table 3). In L. interrogans, the gene encoding this protein resides in a putative two-component operon with a gene encoding the ankyrin-like protein AnkB, a genetic organization that is mirrored in the pathogen Pseudomonas aeruginosa. In P. aeruginosa, KatB and AnkB, which localize to the periplasmic space and inner membrane, respectively, physically interact and form an antioxidant scaffolding that functions as a protective lattice network for optimal H2O2 degradation and protection from reactive oxygen species, thus, promoting bacterial survival.
4. TolC-like Outer Membrane Protein
During the infection process, Leptospira specifically target the kidneys and liver and can attain bacterial loads as high as 109 bacteria/g of guinea pig liver.(71) To ensure bacterial survival, Leptospira must possess efficient efflux systems for removal of potentially toxic compounds. A canonical example of an efflux system is the E. coli AcrAB-TolC system belonging to the resistance-nodulation-cell division superfamily of efflux systems.72,73 The TolC protein of this complex is an OM porin that many Gram-negative transport systems require for a functional efflux system.(74) Studies utilizing TolC mutants have revealed increased sensitivity to bile salts in Salmonella enterica(75) and Vibrio cholera.(76) The critical importance of TolC in spirochete pathogenesis has recently been established in Borrelia burgdorferi where a TolC− mutant was unable to establish infection in mice.(77) In the present study, a TolC-like protein was observed to be upregulated 1.122-fold in L. interrogans grown in −Fe/FBS media (Table 3). Interestingly, previous investigations conducted on the outer membrane proteome of Leptospira have failed to detect this protein.16,18,22,78
5. Putative Glycosyl Hydrolase
To reach their target organs, Leptospira must be capable of traversal and invasion of host tissue barriers. Indeed, L. interrogans have been shown to efficiently traverse epithelial cell barriers in vitro using a transwell assay procedure.(79) One of the preliminary steps in traversal of host barriers is the degradation of mucin barriers. In the mammalian reovirus pathogen, the σ1 coat protein has been shown to possess a glycosyl hydrolase activity that functions to degrade mucin, thus, facilitating infection of Madin Darby canine kidney (MDCK) cells.(80) The requirement for glycosyl hydrolases in establishing infection has also been shown in Streptococcus pneumoniae where a SP2159 (coding for a family 98 glycosyl hydrolase(81)) mutant displayed attenuated virulence in a murine model of infection.(82) In our studies, we have detected 2.731-fold upregulation of a putative glycosyl hydrolase predicted to contain a bacterial fibronectin type III (Fn3) domain (38% similarity/20% identity over 320 amino acids) in response to our in vivo-like conditions (Table 3). In bacteria, extracellular glycosyl hydrolases have been observed to contain fibronectin type III domains.(83) Indeed, in L. interrogans, this protein has been shown to localize to the bacterial surface through a previous study examining the subset of outer membrane proteins.(16)
6. Verification of Protein Expression During Infection
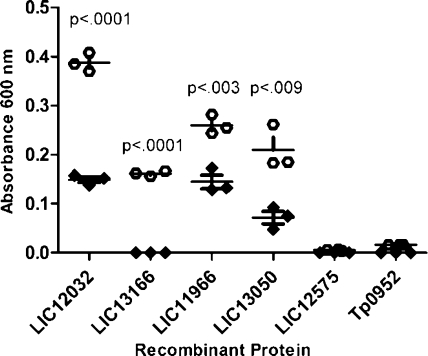
To verify expression of the confirmed/potential virulence factors during infection, two independent experimental approaches were utilized. In the first approach, the antibody reactivity of serum samples collected from patients with leptospirosis was tested against the panel of proteins identified through the proteomic analyses (excluding Loa22 which has previously been well-established to be expressed during infection).(65) For these studies, recombinant versions of each of the virulence factors were heterologously produced and subsequently analyzed via ELISA for specific reactivity with pooled serum from patients with leptospirosis (n = 15). As shown in Figure 3, the pooled serum samples from patients with leptospirosis demonstrated significantly higher reactivity against 4 of the 5 recombinant L. interrogans proteins (p < 0.009) compared to normal human serum, while no significant reactivity was observed against an irrelevant spirochete recombinant protein (Tp0952). The presence of antibodies against these leptospiral proteins in serum samples collected from natural leptospirosis infections is indicative of expression of the proteins during the course of infection. No reactivity was observed against the TolC-like protein (LIC12575), a finding which indicates either lack of expression in vivo or lack of reactivity against the portion of the recombinant protein that was produced (amino acids 315−506 out of 525 total). Antibody reactivity against the putative catalase (LIC12032), the ErpY-like lipoprotein (LIC11966), and the putative coagulase (LIC13166) was also detected in line blot experiments using sera from rabbits infected with virulent L. interrogans serovar Pomona (data not shown), suggesting in vivo expression of these proteins during infection in rabbits.
Figure 3.
Reactivity of human serum to recombinant L. interrogans proteins as determined by ELISA. A 1:500 dilution of pooled serum samples collected from each of 15 individuals with laboratory-confirmed leptospirosis (open circles) and normal human serum (closed diamonds) were tested against the indicated recombinant L. interrogans proteins as well as a negative control recombinant Treponema pallidum protein (Tp0952) in triplicate. Reactivity was measured via absorbance at 600 nm and background subtracted values (no recombinant protein) were used in two-tailed t test calculations to determine statistical significance.
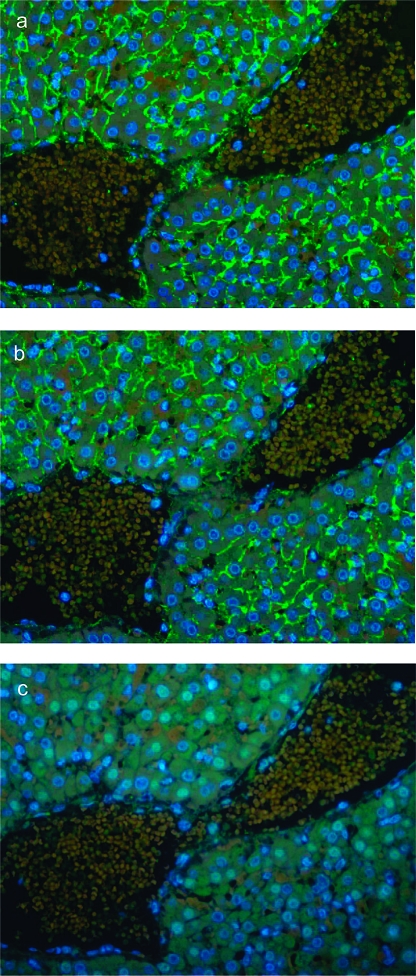
The second approach utilized immunofluorescence microscopy performed on liver sections prepared from L. interrogans serovar Pomona-infected hamsters. Fixed tissue sections were probed with primary rabbit polyclonal antibody against the panel of proteins identified (with the exception of Loa22, as the ORF coding for this protein has not been found in Pomona) and LipL21 (Figure 4a) as a positive control. Preimmune rabbit serum was used as a negative control in these studies (Figure 4c). Fluorescence reactivity indicated expression of the putative glycosyl hydrolase (LIC13050) in liver sections of Pomona-infected hamsters (Figure 4b). Lack of reactivity against the other four proteins may have been due to a lack of expression of these proteins in serovar Pomona.
Figure 4.
In situ expression of L. interrogans proteins in golden Syrian hamster liver tissue. Immunofluorescence microscopy was used to evaluate expression of selected L. interrogans proteins during infection. Shown are PEFF tissue sections prepared from hamsters infected with L. interrogans serovar Pomona and probed with primary polyclonal antisera raised against LipL21 (a) and LIC13050 (b). A negative control using preimmune rabbit serum (c) is shown. In the liver, L. interrogans are found predominantly in the intracellular junctions (a and b), consistent with previous findings. Fluorescence reactivity was detected for the positive control LipL21 (a) and the putative glycosyl hydrolase (LIC13050) (b), while no fluorescence was observed for the negative control (c) or the 4 other proteins tested (LIC12032, LIC13166, LIC11966, and LIC12575-data not shown).
7. Confirmation of Upregulation of Protein Expression in L. interrogans Grown in −Fe/FBS Media
To confirm our iTRAQ quantitation results, we performed immunoblot analysis to determine the level of expression of each of the identified proteins within the virulence factor category in response to growth within −Fe/FBS media (Table 4). In concordance with our iTRAQ results, immunoblot analyses verified upregulation of expression for 5 of the 6 proteins within the virulence factor category upon growth of L. interrogans in −Fe/FBS media. Specifically, after normalizing the results against FlaA1 expression, fold changes of 1.576, 1.350, 1.270, 1.020, and 1.005 were obtained for the TolC-like outer membrane protein, the putative glycosyl hydrolase, Loa22, the putative catalase, and the putative coagulase, respectively. Overall, the increased expression levels observed by immunoblot analyses for these proteins were similar to the results obtained via the 2DGE and iTRAQ experiments (refer to Tables 2 and 3, respectively), although the fold changes obtained from each of the approaches differed slightly. The only protein for which the immunoblot analyses did not confirm the upregulation observed in the iTRAQ analyses was the putative ErpY-like lipoprotein, which showed a 1.309-fold downregulation upon growth of L. interrogans in −Fe/FBS media.
Table 4. Proteins Exhibiting Altered Expression Levels in L. interrogans in Response to in vivo-like Conditions (Exposure to −Fe/FBS) As Determined by Immunoblot Analyses.
| accession no. | protein | ORF | controla | experimentala | fold change |
|---|---|---|---|---|---|
| Q72PA0 | TolC-like protein | LIC12575 | 9.9 | 15.6 | 1.576 |
| Q72MY9 | Putative glycosyl hydrolase | LIC13050 | 3.1 | 4.2 | 1.350 |
| Q72VV5 | OmpA-family lipoprotein (Loa22) | LIC10191 | 18.3 | 23.2 | 1.270 |
| Q72QS7 | Catalase | LIC12032 | 6.3 | 6.4 | 1.020 |
| Q72MM7 | Putative coagulase | LIC13166 | 41.5 | 41.7 | 1.005 |
| Q72QY9 | Putative ErpY-like lipoprotein | LIC11966 | 12.3 | 9.4 | −1.309 |
Average values (n = 2).
Conclusions
In this study, we have identified leptospiral proteins with potential roles in the infection process by analyzing L. interrogans global proteome expression using an in vitro environment designed to mimic in vivo infection. Our study revealed that L. interrogans undergo a complex change in protein expression profiles in response to a shift to the in vivo-like environment of −Fe/FBS media. Specifically, proteins involved in the process of protein synthesis were downregulated, reflecting the general trend toward downregulation of proteins involved in energy production, metabolism, and regulation. A shift to −Fe/FBS media also caused upregulation of the majority of proteins involved in motility, an expected response since motility is required for infection in all pathogenic spirochetes. Finally, our proteomic studies also revealed that, in response to iron limitation and the presence of serum factors, L. interrogans upregulate a number of known and potential virulence factors, as determined by iTRAQ and 2DGE analyses. These include the previously identified outer membrane lipoprotein Loa22 and 5 novel potential virulence factors that exhibit sequence similarity to proteins shown to play key functional roles in other pathogens, including the outer surface lipoprotein ErpY, coagulase, catalase, TolC, and glycosyl hydrolase. Immunoblot analyses verified upregulation for 5 of these 6 proteins between the comparative conditions, and immunological experiments verified in vivo expression of all but one of the identified protein. Future characterization of these leptospiral proteins will significantly advance our understanding of this important pathogen, and future proteomic analyses can expand on this study by investigating the global L. interrogans proteome response to additional environmental conditions that are representative of the environment found within a host. Moreover, the described “infection proteomics” approach can be applied to study the molecular mechanisms of infection in other pathogens. The advantage of this approach is that it examines the organism at the phenotypic level, confirming expression of proteins previously designated as hypothetical and incorporating the myriad complexity that is encoded at both the post-transcriptional and post-translational levels.
Acknowledgments
The authors would like to thank Ami Frank and Richard Hornsby for excellent technical assistance, Dr. David Haake for the generous gifts of L. interrogans serovar Copenhageni strain Fiocruz L1-130 and anti-LIC13050 serum, Drs. Albert Ko and Wesley C. Van Voorhis for providing Leptospira-positive serum samples, Dr. Torsten Seemann at the Victorian Bioinformatics Consortium for providing access to the spirochete database, Dr. Martin Boulanger for his gift of the pET28a vector and assistance with construct design, Anna von Rossum and Marcus Barron for assistance with recombinant protein production, Dr. Christoph Borchers, Derek Smith, Monica Elliott, Darryl Hardie, and the members of the University of Victoria-Genome BC Proteomics Centre for their support and guidance with the proteomic analyses, Paul Hauer, Mary Rasmusson, and Sue Whitaker for supplying tissue from infected hamsters, and Judy Stasko for histology support. This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (327186; C.E.C.), the British Columbia Proteomics Network (C.E.C.), the Canada Foundation for Innovation (C.E.C.), and the British Columbia Knowledge Development Fund (C.E.C.). P.A.C. is a NHMRC CJ Martin Fellow and C.E.C. is a Canada Research Chair in Molecular Pathogenesis and a Michael Smith Foundation for Health Research Scholar.
Supporting Information Available
Figures of annotated MS/MS spectra for single peptide represented proteins; annotated mass spectra used for identification of proteins in 2DGE experiments; and protein regulation in Leptospira in response to culture shift to 37 °C and to media containing −Fe/FBS media as determined by 2DGE. Tables listing Leptospira proteins identified through iTRAQ analyses; peptide evidence for all identified proteins; peptide mass fingerprints and search parameters for proteins identified in 2DGE experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Faine S.; Stallman N. D. Amended descriptions of the genus Leptospira Noguchi 1917 and the species L. interrogans (Stimson 1907) Wenyon 1926 and L. biflexa (Wolbach and Binger 1914) Noguchi 1918. Int. J. Syst. Bacteriol. 1982, 32, 461–463. [Google Scholar]
- Ko A. I.; Galvao Reis M.; Ribeiro Dourado C. M.; Johnson W. D. Jr.; Riley L. W. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 1999, 354, 820–825. [DOI] [PubMed] [Google Scholar]
- Vinetz J. M. Leptospirosis. Curr. Opin. Infect. Dis. 2001, 14, 527–538. [DOI] [PubMed] [Google Scholar]
- Bharti A. R.; Nally J. E.; Ricaldi J. N.; Matthias M. A.; Diaz M. M.; Lovett M. A.; Levett P. N.; Gilman R. H.; Willig M. R.; Gotuzzo E.; Vinetz J. M. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [DOI] [PubMed] [Google Scholar]
- O’Neil K. M.; Rickman L. S.; Lazarus A. A. Pulmonary manifestations of leptospirosis. Rev. Infect. Dis. 1991, 13, 705–709. [DOI] [PubMed] [Google Scholar]
- Heron L. G.; Reiss-Levy E. A.; Jacques T. C.; Dickeson D. J.; Smythe L. D.; Sorrell T. C. Leptospirosis presenting as a haemorrhagic fever in a traveller from Africa. Med. J. Aust. 1997, 167, 477–479. [DOI] [PubMed] [Google Scholar]
- Monsuez J. J.; Kidouche R.; Le Gueno B.; Postic D. Leptospirosis presenting as haemorrhagic fever in visitor to Africa. Lancet 1997, 349, 254–255. [DOI] [PubMed] [Google Scholar]
- Farr R. W. Leptospirosis. Clin. Infect. Dis. 1995, 21, 1–6. [DOI] [PubMed] [Google Scholar]
- Wasinger V. C.; Cordwell S. J.; Cerpa-Poljak A.; Yan J. X.; Gooley A. A.; Wilkins M. R.; Duncan M. W.; Harris R.; Williams K. L.; Humphery-Smith I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [DOI] [PubMed] [Google Scholar]
- Braun V.; Braun M. Iron transport and signaling in Escherichia coli. FEBS Lett. 2002, 529, 78–85. [DOI] [PubMed] [Google Scholar]
- Nart P.; Holden N.; McAteer S. P.; Wang D.; Flockhart A. F.; Naylor S. W.; Low J. C.; Gally D. L.; Huntley J. F. Mucosal antibody responses of colonized cattle to Escherichia coli O157-secreted proteins, flagellin, outer membrane proteins and lipopolysaccharide. FEMS Immunol. Med. Microbiol. 2007, 52, 59–68. [DOI] [PubMed] [Google Scholar]
- Qin J. H.; Sheng Y. Y.; Zhang Z. M.; Shi Y. Z.; He P.; Hu B. Y.; Yang Y.; Liu S. G.; Zhao G. P.; Guo X. K. Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar lai strain 56601. BMC Microbiol. 2006, 6, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.; Bulach D. M.; Powell D. R.; Haake D. A.; Matsunaga J.; Paustian M. L.; Zuerner R. L.; Adler B. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 2006, 74, 5848–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velineni S.; Asuthkar S.; Sritharan M. Iron limitation and expression of immunoreactive outer membrane proteins in Leptospira interrogans serovar icterohaemorrhagiae strain lai. Indian J. Med. Microbiol. 2006, 24, 339–342. [DOI] [PubMed] [Google Scholar]
- Haake D. A.; Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect. Immun. 2002, 70, 4936–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. A.; Xu X.; Matsunaga J.; Sanchez Y.; Ko A. I.; Haake D. A.; Adler B. Surfaceome of Leptospira spp. Infect. Immun. 2005, 73, 4853–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakolvaree Y.; Maneewatch S.; Jiemsup S.; Klaysing B.; Tongtawe P.; Srimanote P.; Saengjaruk P.; Banyen S.; Tapchaisri P.; Chonsa-nguan M.; Chaicumpa W. Proteome and immunome of pathogenic Leptospira spp. revealed by 2DE and 2DE-immunoblotting with immune serum. Asian Pac. J. Allergy Immunol. 2007, 25, 53–73. [PubMed] [Google Scholar]
- Monahan A. M.; Callanan J. J.; Nally J. E. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 2008, 76, 4952–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro H.; Croda J.; Flannery B.; Mazel M.; Matsunaga J.; Galvao Reis M.; Levett P. N.; Ko A. I.; Haake D. A. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 2001, 69, 4958–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. A.; Cordwell S. J.; Bulach D. M.; Haake D. A.; Adler B. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 2002, 70, 2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin S.; Timoney J. F.; Nally J.; Verma A. Host-inducible immunogenic sphingomyelinase-like protein, Lk73.5, of Leptospira interrogans. Infect. Immun. 2004, 72, 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E.; Whitelegge J. P.; Bassilian S.; Blanco D. R.; Lovett M. A. Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infect. Immun. 2007, 75, 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E.; Timoney J. F.; Stevenson B. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 2001, 69, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E.; Artiushin S.; Timoney J. F. Molecular characterization of thermoinduced immunogenic proteins Q1p42 and Hsp15 of Leptospira interrogans. Infect. Immun. 2001, 69, 7616–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J.; Lo M.; Bulach D. M.; Zuerner R. L.; Adler B.; Haake D. A. Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect. Immun. 2007, 75, 2864–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal K.; Choe L. H.; Lee K. H. Shotgun proteomics using the iTRAQ isobaric tags. Briefings Funct. Genomics Proteomics 2006, 5, 112–120. [DOI] [PubMed] [Google Scholar]
- Ross P. L.; Huang Y. N.; Marchese J. N.; Williamson B.; Parker K.; Hattan S.; Khainovski N.; Pillai S.; Dey S.; Daniels S.; Purkayastha S.; Juhasz P.; Martin S.; Bartlet-Jones M.; He F.; Jacobson A.; Pappin D. J. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. [DOI] [PubMed] [Google Scholar]
- Washburn M. P.; Wolters D.; Yates J. R. 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001, 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Cong Y. S.; Fan E.; Wang E. Simultaneous proteomic profiling of four different growth states of human fibroblasts, using amine-reactive isobaric tagging reagents and tandem mass spectrometry. Mech. Ageing Dev. 2006, 127, 332–343. [DOI] [PubMed] [Google Scholar]
- Gan C. S.; Chong P. K.; Pham T. K.; Wright P. C. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 2007, 6, 821–827. [DOI] [PubMed] [Google Scholar]
- Leuko S.; Raftery M.; Burns B.; Walter M.; Neilan B. Global protein-level responses of Halobacterium salinarum NRC-1 to prolonged changes in external sodium chloride concentrations. J. Proteome Res. 2009, 8, 2218–2225. [DOI] [PubMed] [Google Scholar]
- Pandhal J.; Ow S. Y.; Wright P. C.; Biggs C. A. Comparative proteomics study of salt tolerance between a nonsequenced extremely halotolerant Cyanobacterium and its mildly halotolerant relative using in vivo metabolic labeling and in vitro isobaric labeling. J. Proteome Res. 2009, 8, 818–828. [DOI] [PubMed] [Google Scholar]
- Ow S. Y.; Noire J.; Cardona T.; Taton A.; Lindblad P.; Stensjo K.; Wright P. C. Quantitative overview of N2 fixation in Nostoc punctiforme ATCC 29133 through cellular enrichments and iTRAQ shotgun proteomics. J. Proteome Res. 2009, 8, 187–198. [DOI] [PubMed] [Google Scholar]
- Tafelmeyer P.; Laurent C.; Lenormand P.; Rousselle J. C.; Marsollier L.; Reysset G.; Zhang R.; Sickmann A.; Stinear T. P.; Namane A.; Cole S. T. Comprehensive proteome analysis of Mycobacterium ulcerans and quantitative comparison of mycolactone biosynthesis. Proteomics 2008, 8, 3124–3138. [DOI] [PubMed] [Google Scholar]
- Lippolis J. D.; Bayles D. O.; Reinhardt T. A. Proteomic changes in Escherichia coli when grown in fresh milk versus laboratory media. J. Proteome Res. 2009, 8, 149–158. [DOI] [PubMed] [Google Scholar]
- Drummelsmith J.; Winstall E.; Bergeron M. G.; Poirier G. G.; Ouellette M. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J. Proteome Res. 2007, 6, 4690–4702. [DOI] [PubMed] [Google Scholar]
- Zeaiter Z.; Cohen D.; Musch A.; Bagnoli F.; Covacci A.; Stein M. Analysis of detergent-resistant membranes of Helicobacter pylori infected gastric adenocarcinoma cells reveals a role for MARK2/Par1b in CagA-mediated disruption of cellular polarity. Cell. Microbiol. 2008, 10, 781–794. [DOI] [PubMed] [Google Scholar]
- Ellinghausen H. C. Jr.; McCullough W. G. Nutrition of Leptospira pomona and growth of 13 other serotypes: Fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 1965, 26, 45–51. [PubMed] [Google Scholar]
- Johnson R. C.; Harris V. G. Differentiation of pathogenic and saprophytic letospires. Growth at low temperatures. J. Bacteriol. 1967, 94, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. A.; Overall C. M.; Smith D. Proteomic identification of cellular protease substrates using isobaric tags for relative and absolute quantification. Curr. Protoc. Protein Sci. 2007, 21, 1–21. [DOI] [PubMed] [Google Scholar]
- Wiese S.; Reidegeld K. A.; Meyer H. E.; Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [DOI] [PubMed] [Google Scholar]
- Chong P. K.; Gan C. S.; Pham T. K.; Wright P. C. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: Implication of multiple injections. J. Proteome Res. 2006, 5, 1232–1240. [DOI] [PubMed] [Google Scholar]
- Burnham K. P.; Anderson D. R..; White G. C. B., C.; Pollock K. H.. American Fisheries Society; American Fisheries Society: Bethesda, MD, 1987; pp 242−243. [Google Scholar]
- Cameron C. E.; Lukehart S. A.; Castro C.; Molini B.; Godornes C.; Van Voorhis W. C. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J. Infect. Dis. 2000, 181, 1401–1413. [DOI] [PubMed] [Google Scholar]
- Trueba G. A.; Bolin C. A.; Zuerner R. L. Characterization of the periplasmic flagellum proteins of Leptospira interrogans. J. Bacteriol. 1992, 174, 4761–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhis W. C.; Barrett L. K.; Lukehart S. A.; Schmidt B.; Schriefer M.; Cameron C. E. Serodiagnosis of syphilis: antibodies to recombinant Tp0453, Tp92, and Gpd proteins are sensitive and specific indicators of infection by Treponema pallidum. J. Clin. Microbiol. 2003, 41, 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J.; Werneid K.; Zuerner R. L.; Frank A.; Haake D. A. LipL46 is a novel surface-exposed lipoprotein expressed during leptospiral dissemination in the mammalian host. Microbiology 2006, 152, 3777–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner R. L.; Cameron C. E.; Raverty S.; Robinson J.; Colegrove K. M.; Norman S. A.; Lambourn D.; Jeffries S.; Alt D. P.; Gulland F. Geographical dissemination of Leptospira interrogans serovar Pomona during seasonal migration of California sea lions. Vet. Microbiol. 2008, 137, 105–110. [DOI] [PubMed] [Google Scholar]
- Cullen P. A.; Haake D. A.; Bulach D. M.; Zuerner R. L.; Adler B. LipL21 is a novel surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 2003, 71, 2414–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento A. L.; Ko A. I.; Martins E. A.; Monteiro-Vitorello C. B.; Ho P. L.; Haake D. A.; Verjovski-Almeida S.; Hartskeerl R. A.; Marques M. V.; Oliveira M. C.; Menck C. F.; Leite L. C.; Carrer H.; Coutinho L. L.; Degrave W. M.; Dellagostin O. A.; El-Dorry H.; Ferro E. S.; Ferro M. I.; Furlan L. R.; Gamberini M.; Giglioti E. A.; Goes-Neto A.; Goldman G. H.; Goldman M. H.; Harakava R.; Jeronimo S. M.; Junqueira-de-Azevedo I. L.; Kimura E. T.; Kuramae E. E.; Lemos E. G.; Lemos M. V.; Marino C. L.; Nunes L. R.; de Oliveira R. C.; Pereira G. G.; Reis M. S.; Schriefer A.; Siqueira W. J.; Sommer P.; Tsai S. M.; Simpson A. J.; Ferro J. A.; Camargo L. E.; Kitajima J. P.; Setubal J. C.; Van Sluys M. A. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 2004, 186, 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y. S.; Fan E.; Wang E. Simultaneous proteomic profiling of four different growth states of human fibroblasts, using amine-reactive isobaric tagging reagents and tandem mass spectrometry. Mech. Ageing Dev. 2006, 127, 332–343. [DOI] [PubMed] [Google Scholar]
- Liu T.; Donahue K. C.; Hu J.; Kurnellas M. P.; Grant J. E.; Li H.; Elkabes S. Identification of differentially expressed proteins in experimental autoimmune encephalomyelitis (EAE) by proteomic analysis of the spinal cord. J. Proteome Res. 2007, 6, 2565–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glen A.; Gan C. S.; Hamdy F. C.; Eaton C. L.; Cross S. S.; Catto J. W.; Wright P. C.; Rehman I. iTRAQ-facilitated proteomic analysis of human prostate cancer cells identifies proteins associated with progression. J. Proteome Res. 2008, 7, 897–907. [DOI] [PubMed] [Google Scholar]
- Louvel H.; Bommezzadri S.; Zidane N.; Boursaux-Eude C.; Creno S.; Magnier A.; Rouy Z.; Medigue C.; Saint Girons I.; Bouchier C.; Picardeau M. Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp. J. Bacteriol. 2006, 188, 7893–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Vollmer A.; Van Dyk T. K. Stress responsive bacteria: biosensors as environmental monitors. Adv. Microb. Physiol. 2004, 49, 131–174. [DOI] [PubMed] [Google Scholar]
- Dons L.; Eriksson E.; Jin Y.; Rottenberg M. E.; Kristensson K.; Larsen C. N.; Bresciani J.; Olsen J. E. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 2004, 72, 3237–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. M.; Chen Y. T.; Andermann T. M.; Carter J. E.; McGee D. J.; Ottemann K. M. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 2007, 75, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon N. W.; Goldstein S. F. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 2002, 36, 47–73. [DOI] [PubMed] [Google Scholar]
- Lux R.; Miller J. N.; Park N. H.; Shi W. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect. Immun. 2001, 69, 6276–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuri K.; Takamoto Y.; Okada M.; Hiramune T.; Kikuchi N.; Yanagawa R. Chemotaxis of leptospires to hemoglobin in relation to virulence. Infect. Immun. 1993, 61, 2270–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R.; Moter A.; Shi W. Chemotaxis in pathogenic spirochetes: directed movement toward targeting tissues. J. Mol. Microbiol. Biotechnol. 2000, 2, 355–364. [PubMed] [Google Scholar]
- Park S. Y.; Chao X.; Gonzalez-Bonet G.; Beel B. D.; Bilwes A. M.; Crane B. R. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol. Cell 2004, 16, 563–574. [DOI] [PubMed] [Google Scholar]
- Motaleb M. A.; Miller M. R.; Li C.; Bakker R. G.; Goldstein S. F.; Silversmith R. E.; Bourret R. B.; Charon N. W. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J. Bacteriol. 2005, 187, 7963–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N.; Watanabe H. Molecular cloning and characterization of a novel leptospiral lipoprotein with OmpA domain. FEMS Microbiol. Lett. 2003, 226, 215–219. [DOI] [PubMed] [Google Scholar]
- Ristow P.; Bourhy P.; McBride F. W.; Figueira C. P.; Huerre M.; Ave P.; Girons I. S.; Ko A. I.; Picardeau M. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 2007, 3, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz C.; McDevitt D.; Foster T. J.; Cheung A. L. Influence of Agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 1996, 64, 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarestrup F. M.; Scott N. L.; Sordillo L. M. Ability of Staphylococcus aureus coagulase genotypes to resist neutrophil bactericidal activity and phagocytosis. Infect. Immun. 1994, 62, 5679–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai T.; Tomono K.; Yanagihara K.; Yamamoto Y.; Kaku M.; Hirakata Y.; Koga H.; Tashiro T.; Kohno S. Role of coagulase in a murine model of hematogenous pulmonary infection induced by intravenous injection of Staphylococcus aureus enmeshed in agar beads. Infect. Immun. 1997, 65, 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corin R. E.; Boggs E.; Cox C. D. Enzymatic degradation of H2O2 by Leptospira. Infect. Immun. 1978, 22, 672–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P. J.; Larson A. D.; Cox C. D. Catalase activity in Leptospira. J. Bacteriol. 1964, 88, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faine S.; Adler B.; Bolin C.; Perolat P.. Leptospira and Leptospirosis, 2nd ed.; MediSci; Melbourne, Australia, 1999; pp 30−32. [Google Scholar]
- Fralick J. A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 1996, 178, 5803–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H. Jr.; Tam R.; Reizer A.; Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol. Microbiol. 1994, 11, 841–847. [DOI] [PubMed] [Google Scholar]
- Paulsen I. T.; Park J. H.; Choi P. S.; Saier M. H. Jr. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 1997, 156, 1–8. [DOI] [PubMed] [Google Scholar]
- Ramos-Morales F.; Prieto A. I.; Beuzon C. R.; Holden D. W.; Casadesus J. Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 2003, 185, 5328–5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina J. E.; Mekalanos J. J. Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 2001, 69, 4681–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis I.; Denker K.; Ostberg Y.; Andersen C.; Benz R.; Bergstrom S. An RND-type efflux system in Borrelia burgdorferi is involved in virulence and resistance to antimicrobial compounds. PLoS Pathog. 2008, 4, e1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally J. E.; Whitelegge J. P.; Aguilera R.; Pereira M. M.; Blanco D. R.; Lovett M. A. Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of Leptospira interrogans serovar Copenhageni. Proteomics 2005, 5, 144–152. [DOI] [PubMed] [Google Scholar]
- Barocchi M. A.; Ko A. I.; Reis M. G.; McDonald K. L.; Riley L. W. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect. Immun. 2002, 70, 6926–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaillon M.; Senechal S.; Bernier L.; Lemay G. A glycosyl hydrolase activity of mammalian reovirus sigma1 protein can contribute to viral infection through a mucus layer. J. Mol. Biol. 1999, 286, 759–773. [DOI] [PubMed] [Google Scholar]
- Higgins M. A.; Abbott D. W.; Boulanger M. J.; Boraston A. B. Blood group antigen recognition by a solute-binding protein from a serotype 3 strain of Streptococcus pneumoniae. J. Mol. Biol. 2009, 388, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava D. L.; Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 2002, 45, 1389–1406. [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R.; Henrissat B.; Kilburn D. G.; Miller R. C. Jr.; Warren R. A. Domains in microbial beta-1, 4-glycanases: Sequence conservation, function, and enzyme families. Microbiol. Rev. 1991, 55, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.