Abstract
Cell surface proteins are excellent targets for diagnostic and therapeutic interventions. By using bioinformatics tools, we generated a catalog of 3,702 transmembrane proteins located at the surface of human cells (human cell surfaceome). We explored the genetic diversity of the human cell surfaceome at different levels, including the distribution of polymorphisms, conservation among eukaryotic species, and patterns of gene expression. By integrating expression information from a variety of sources, we were able to identify surfaceome genes with a restricted expression in normal tissues and/or differential expression in tumors, important characteristics for putative tumor targets. A high-throughput and efficient quantitative real-time PCR approach was used to validate 593 surfaceome genes selected on the basis of their expression pattern in normal and tumor samples. A number of candidates were identified as potential diagnostic and therapeutic targets for colorectal tumors and glioblastoma. Several candidate genes were also identified as coding for cell surface cancer/testis antigens. The human cell surfaceome will serve as a reference for further studies aimed at characterizing tumor targets at the surface of human cells.
Keywords: colorectal tumors, CT antigens, glioblastoma, transmembrane, tumor cell surface antigens
With the availability of the human genome sequence, an important goal of current biological research is a more specific and accurate annotation of human genes. One critical property is the subcellular localization of gene products, because this affects their use as potential diagnostic and therapeutic targets. In this respect, the identification of cell surface proteins is of particular interest (1–3) because these proteins represent ideal therapeutic targets. Indeed, cell surface proteins have proved to be relevant to many areas of medicine, and a number of monoclonal antibodies against them are approved for therapeutic applications by the Food and Drug Administration, particularly in cancer therapy. Furthermore, cell surface proteins are also excellent targets for diagnostic assays, especially in biological fluids. On the other hand, there are several issues that make cell surface proteins difficult to manipulate biochemically. First, their hydrophobic transmembrane (TM) domain makes them insoluble. Second, several posttranslational modifications are not executed in commonly used expression systems. Finally, interactions involving cell surface proteins usually have an extremely short half-life (on the order of milliseconds), which has an effect on the development of purification protocols. Despite these limitations, decades of intensive research of cell surface proteins have generated a significant information base. Ideally, this information should be analyzed in a genome-wide context.
We generated here a catalog of more than 3,700 genes believed to encode proteins located at the surface of human cells. For the sake of simplicity, we will call this catalog the “human cell surfaceome.” An integrated database with both public and original information was produced. In this report, we explore the diversity of the cell surfaceome at four different levels: xenogenetic, allogenetic, clonogenetic, and epigenetic. In addition, the expression pattern of the surfaceome in human tumors was evaluated. In our search for new therapeutic and diagnostic targets, a large-scale quantitative real-time PCR (qPCR) was carried out for 593 gene transcripts in tumor samples from colorectal tumors and glioblastomas (GBMs), as well as in a panel of normal tissues. We were thus able to identify sets of genes with either a restricted expression in normal tissues and/or genes that are differentially expressed in tumors.
Results and Discussion
Definition of the Human Cell Surfaceome.
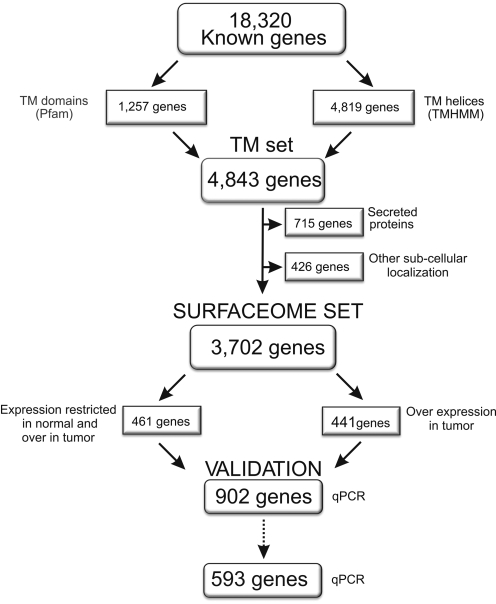
Fig. 1 shows a schematic view of our strategy to identify human gene coding for cell surface proteins. First, we searched the whole set of known human genes (the Reference Sequence collection) for an annotated TM domain as reported by Pfam (4). This search returned 1,257 genes. In parallel, we submitted all of the translated sequences from the Reference Sequence collection to TMHMM (5), an algorithm that identifies TM domains by using a hidden Markov model-based strategy. TMHMM is considered the best algorithm for the prediction of TM domains, with specificity and sensitivity above 99% (5). TMHMM functions ab initio, and therefore it returned a larger set of TM proteins (4,819 genes). As expected, the great majority of the candidates identified by the Pfam analysis were within the set identified by TMHMM, resulting in a merged, nonredundant set of 4,843 gene candidates. Because TMHMM also identifies signal peptides of secreted proteins, we next excluded from our dataset all cases in which the region identified as TM by TMHMM was unique and within the first 50 aa, thus characterizing a signal peptide. This left us with 4,128 gene candidates. Because TM domains are not restricted to cell surface proteins, those candidates having a TM domain but already known to be exclusive to other cell membranes were excluded: Gene Ontology (GO) was used to filter out those cases, which left us with our final set of genes (3,702) believed to encode cell surface proteins. A list of all surfaceome genes is available in Table S1. We expect that our final list contains false-positive candidates because of a lack of information regarding their subcellular localization. One way to evaluate the rate of false-positives is to search for proteins in our list without an indication of cell surface localization and assume that all of them are false-positives. This is very stringent, because many proteins classified as false-positives will be classified as cell surface in the future, but give us an upper limit for false-positives. Manual inspection of annotation data from most of the candidates suggests that the rate of false-positives does not correspond to more than 15% of our final list of genes.
Fig. 1.
Overall representation of our bioinformatics strategy to identify human genes coding for cell surface proteins and to select a subset of these genes for experimental validation. A total of 18,320 known human genes were submitted to Pfam and TMHMM. A nonredundant set of 4,843 genes were classified as having a TM domain. After excluding genes coding for proteins secreted or located in other subcellular compartments, we defined 3,702 genes as belonging to the surfaceome set. A subset of 902 genes were selected for experimental validation.
Because the surfaceome set is restricted to those proteins that have a TM domain, proteins present at the cell surface but anchored or associated with the external side of the plasma membrane by means other than TM domains (e.g., phosphatidylinositol-anchored) are not present in our dataset. The absence of a reliable sequence feature that characterizes these proteins prevented us from identifying ab initio these types of proteins, which will have to be later incorporated in our dataset.
To evaluate whether our dataset reliably represents the collection of cell surface proteins, a GO analysis was performed to evaluate whether it was enriched with GO categories clearly associated with plasma membrane. We found that categories such as “cell adhesion,” “transport,” and “cell–cell signaling” were significantly enriched in our dataset (Fig. S1). As another approach to test the robustness of our dataset, the representation of known families of cell surface proteins, like G-protein-coupled receptors (GPCRs), solute carrier (SLC) proteins, and cluster of differentiation (CD) antigens, was measured in the human cell surfaceome. Table 1 shows that it was possible to identify the great majority of these proteins: 85% of all 921 GPCRs, 81% of all 354 SLCs, and 82% of all 372 CDs. Those that were not identified either do not have a TM domain or were annotated as located in other subcellular compartments (Table 1). For most of these cases, further functional studies will be needed to confirm their subcellular localization. The Ig family of cell surface receptors was not analyzed because of the lack of a well-curated dataset. Because many members of this family are also members of the CD family, we believe that our human cell surfaceome well represents members of the Ig family as well.
Table 1.
Representation of known cell surface gene categories within the human surfaceome
| Category | Number of genes |
||
|---|---|---|---|
| GPCRs | SLCs | CDs | |
| Total | 921 | 354 | 372 |
| Present in surfaceome set, no. (%) | 779 (85) | 287 (81) | 304 (82) |
| Absent from surfaceome set, no. | |||
| Secreted proteins | 12 | 5 | 15 |
| Other subcellular localization | 11 | 28 | 6 |
| Without TM domain | 119 | 34 | 47 |
The total number of genes (first row) coding for GPCR, SLC, and CD were obtained by querying several databases (see SI Methods for more details). The number of genes classified in these three categories and absent in our surfaceome database is identified.
The 3,702 surfaceome set corresponds to ≈20% of all known human genes. This number is in accordance with other similar estimates for human and mouse (3, 6). The chromosomal distribution of the surfaceome set resembled the distribution of human genes in general except for a higher density on chromosome 11, due to a large cluster of olfactory receptors that map to this chromosome (Table S1).
Genetic Diversity Within the Human Surfaceome.
Twenty years ago, Rettig and Old (1) classified the genetic diversity at the cell surface level into four categories: (i) xenogenetic diversity, defined by the pattern of conservation and novelty between species; (ii) allogenetic diversity, defined by genetic polymorphisms; (iii) clonogenetic diversity, defined by somatic changes in specific cell populations within an individual; and (iv) epigenetic diversity, defined by differences in gene expression between cell types.
With the availability of many genome-wide types of data, we decided to explore these categories of diversity at the cell surface in more detail. Clonogenetic diversity (e.g., somatic mutations) will be explored in the context of the cancer surfaceome and published elsewhere.
Xenogenetic Diversity.
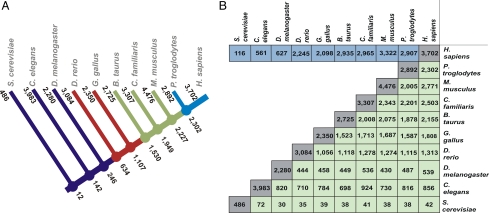
The availability of genome sequences for a large variety of species covering the major groups in the tree of life allowed us to compare the human surfaceome with other species. First, the same bioinformatics strategy used for humans was used to identify the surfaceome of nine other species (Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Gallus gallus, Bos taurus, Canis familiaris, Mus musculus, and Pan troglodytes). Fig. 2A is a schematic view of our findings. The major feature observed in our analysis is the increase in surfaceome complexity with the emergence of multicellularity. Although in Metazoa the surfaceome size is, on average, 20% of all genes in a given species, the same number is less than 10% for yeast. This could be largely accounted for by complexity involved in the construction of organs and their constituent cell types, as well as the emergence of an extracellular matrix (and their receptors). We then compared the surfaceome of all 10 species by using HomoloGene (www.ncbi.nlm.nih.gov/HomoloGene). The number of conserved genes in all pairwise comparisons is found in Fig. 2B. Although only 116 surfaceome genes (3% of the human cell surfaceome) are conserved between human and yeast, we observed a higher degree of conservation among mammals (1,530 genes are shared in all mammals—41% of the human cell surfaceome). A more detailed comparative analysis of the surfaceome in these species will be published elsewhere.
Fig. 2.
Overall pattern of conservation in the surfaceomes of 10 species. (A) Numbers at branching nodes represent the number of surfaceome genes conserved between the species that diverged at that node. To be classified as conserved, a gene must be present in the surfaceome of all descendent species. Numbers at the end of the branches indicate the size of the surfaceome set in the respective species. (B) Pairwise comparisons of the surfaceome for all 10 species. The row in blue represents the level of conservation of the human surfaceome in all other nine species, whether they are surfaceome genes in the respective species. The lower half in green represents the level of conservation when the surfaceomes of both species are taken into account. For example, there are 2,005 genes conserved and classified as surfaceome in both M. musculus and P. troglodytes.
Allogenetic Diversity.
To explore the distribution of genetic polymorphisms in the human cell surfaceome, information available in the Single Nucleotide Polymorphism database (dbSNP) was integrated into our database. There are two ways in which genetic polymorphisms can affect the cell surface: (i) SNPs occurring in the enzymes involved in posttranslational modifications, such as carbohydrate synthesis, which may have an indirect effect on the surfaceome, and (ii) SNPs occurring directly in the genes belonging to the surfaceome set. Nevertheless, we only evaluated here the distribution of both coding and noncoding SNPs occurring directly in the surfaceome set. Although the surfaceome genes were slightly enriched for both coding and noncoding SNPs compared with all remaining human genes, the difference was not statistically significant (Fig. S2a). As expected, HLA coding genes, belonging to the MHC, are the most polymorphic genes in the surfaceome set (Table S1). Interestingly, genes classified as CD have a higher density of coding SNPs than GPCRs and SLCs (Fig. S2b). As can be seen in Fig. S2b, this effect is not due to the higher density of SNPs restricted to HLA genes, but rather a higher density in many CD genes throughout the SNP density distribution.
Detailed studies of polymorphisms affecting the surfaceome may be critical for the effective use of personalized medicine, especially for issues related to drug-response phenotypes.
Epigenetic Diversity.
We capitalized on Massively Parallel Signature Sequencing (MPSS) expression data (7) from a panel of normal tissues to define the expression profile of the human cell surfaceome in normal cells/tissues. This large-scale profiling of the surfaceome in normal tissues has great potential value in facilitating the further development of therapeutic and diagnostic protocols. A limitation of this tissue analysis is that it does not provide cell type-specific expression information, but rather a composite of all of the different cell types in a tissue. The same limitation also exists for the analysis presented in the next section related to the cancer surfaceome. The characterization of the surfaceome specific to different cell types will require transcriptional profiling of single cells or immunohistochemistry analysis of cellular expression patterns as in the Protein Atlas initiative (www.proteinatlas.org).
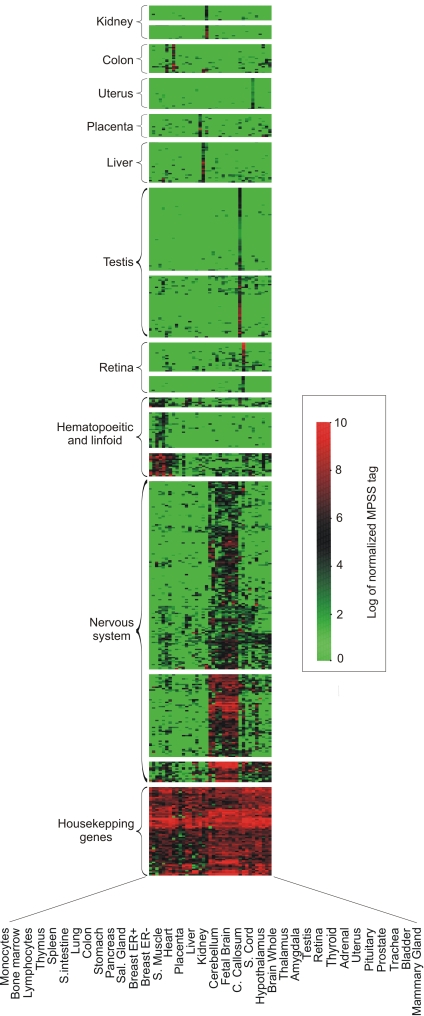
Fig. 3 depicts the complex pattern of the surfaceome expression in normal tissues. Almost half of the surfaceome set (42%) presents a very broad expression pattern, being expressed in more than 20 normal tissues. A smaller fraction (≈85 genes) is expressed in all normal MPSS libraries, a typical feature of housekeeping genes. On the other hand, a significant number of surfaceome genes (13%) have a restricted expression pattern among normal tissues. These gene products, if expressed in cancer, would be candidate tumor targets. We have also clearly identified a group of genes with preferential expression in certain tissues, such as brain and testis, as shown in Fig. 3. Genes having an expression restricted to testis (see below) are of interest as candidate genes coding for cancer/testis (CT) antigens (8).
Fig. 3.
MPSS expression profile of a subset of surfaceome genes in normal tissues. Surfaceome genes were arbitrarily chosen based on their expression pattern. Genes showing a tissue-biased expression were emphasized, as were genes showing a broad expression pattern (genes classified as “Housekeeping” at the bottom of the heatmap). The heatmap was generated by a log transformation of the normalized frequency of an MPSS tag (tags per million) specific for each gene. Each row represents a single gene, and each column represents a different tissue. Color reflects the expression of a gene in a given tissue, based on the frequency of an MPSS tag specific for that gene.
The Cancer Surfaceome.
In exploring the human cell surfaceome with respect to differential expression in tumors, emphasis was placed on finding genes exhibiting a restricted expression in normal tissues and a differential expression in tumor samples, as well as identifying genes showing differential higher expression in tumors regardless of their expression in normal tissues.
The analysis was restricted to two types of tumors—GBMs and colorectal tumors—based on the abundance of publicly available gene expression analyses as well as our access to samples and clinical information. The surfaceome database was used to select genes for further experimental validation based on their expression in 305 SAGE (Serial Analysis of Gene Expression) libraries (97 and 208 derived from normal and tumor samples, respectively) extracted from SAGE Genie (9) and 37 normal libraries of MPSS (7). We selected 461 genes with restricted expression in normal tissues and expression in any tumor type, and 441 genes showing a significantly higher expression in GBMs or colorectal tumors (Fig. 1). This analysis generated a subset of 902 genes that were submitted to experimental validation by using a platform for large-scale measurement of cDNA levels (Biomark–Fluidigm). All primers and probes used in this study are listed in Table S2. Sixty-five RNA samples were used: 21 normal tissues (available commercially), 15 tumor cell lines of diverse origins, 11 colorectal tumors with two pools of normal colon, and 11 GBMs with five pools of normal brain. Details on all of the samples can be found in Table S3. After one qPCR measurement, all genes (593 genes) that were expressed in at least one sample were selected for subsequent analyses.
Genes with a Restricted Expression in Normal Tissues and Differential Expression in GBMs or Colorectal Tumors.
Among the dataset of genes showing a restricted expression in normal tissues (Table S4), genes showing a differential expression in GBMs and/or colorectal tumors were identified. We found five genes with a restricted expression in normal tissues and a differential expression in colorectal tumors: MUC17, UNC93A, TMEM211, SLCO1B3, and FAT (Fig. S3B). Among these, SLCO1B3 (also known as LST-2) seems the most interesting, for several reasons. First, overexpression of this gene has been clearly observed in a variety of tumors, including breast, prostate, and gastrointestinal cancers (10, 11). Second, this gene has been reported to code for the key methotrexate transporter in colorectal cancer (12). Finally, SLCO1B3 has been seen to be frequently mutated in colorectal cancer (Table S1) (13).
Fourteen genes have been identified that show a restricted expression in normal tissues and a differential expression in GBMs: CDH10, GSG1L, IL1RAP, TRPM8, CXorf1, TMTC3, SMO, CNGA3, CSF1R, ADORA3, ATP6V0E1, CD99, LTBR, and RARRES3 (Fig. S3A). CDH10, a type II classical cadherin, shows restricted expression in normal brain and overexpression in GBM. Another interesting gene, SMO, is a known oncogene for basal cell carcinoma (14) that activates the Hedgehog pathway, leading to an increase in angiogenic factors (15), cyclins D1 and B1 (16), and antiapoptotic genes, and a decrease in apoptotic genes (FAS) (17). In addition, ADORA3, an adenosine receptor, has been observed to be highly expressed in a variety of tumors and has been suggested as a potential therapeutic target (18).
Genes Differentially Expressed in Colorectal Tumors and GBMs.
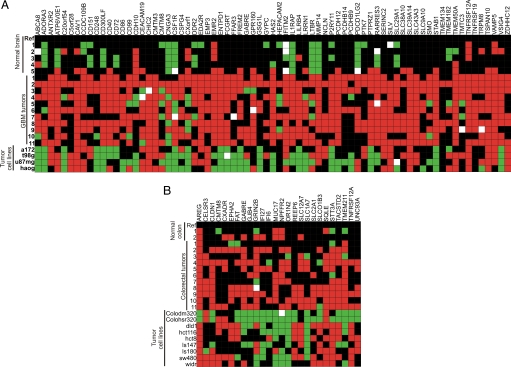
We next looked at genes differentially expressed in GBM and colorectal tumors regardless of their expression pattern in normal tissues other than their tissue of origin (Table S1). Fig. 4 shows the expression profile of 73 (Fig. 4A) and 26 (Fig. 4B) genes for GBMs and colorectal tumors, respectively. Among the genes identified as differentially expressed in colorectal tumors is TACSTD2 (also known as TROP2), a recently described oncogene (19). This gene has also been identified in genetic translocations involving cyclin D1 in ovarian and breast tumors (20). Also for colorectal tumors, we were able to identify EPHA2, an ephrin receptor, as a potential tumor target.
Fig. 4.
Expression profile of cell surface-encoding genes differentially expressed in GBMs (A) and colorectal (B) tumors as evaluated by qPCR in 65 RNA samples of normal tissues, GBMs, and colorectal tumors and cell lines derived from these tumor types. Heatmap was generated by averaging three qPCR experiments presented as fold change values. Each row represents a single gene, and each column represents a sample. Noninformative reactions are represented by white spots. Red squares represent genes overexpressed (fold change three times higher than standard deviation) in relation to the reference. Green squares represent genes down-regulated in relation to the reference. Black squares represent genes equally expressed between sample and reference. Differential expression is shown for GBMs (73 genes) and colorectal tumors (26 genes).
Among the genes differentially expressed in GBM, CD248 (endosialin or TEM1) is one of the most interesting because it was originally characterized as the target of a monoclonal antibody (FB5) that recognizes vasculature in several tumor types (21), and it was subsequently identified as highly expressed transcript in endothelial cells of colorectal cancer (22). CD248 is strongly overexpressed in GBM (23) but, as expected from its localization in tumor vasculature but not tumor cells, GBM cell lines show no CD248 expression (Fig. 4A).
CMTM8, GABRE, GPR172A, SLC12A7, and TNFRSF12A (see below) are other genes that are overexpressed both in GBM and colorectal samples. SLC12A7, a potassium/chloride transporter, is involved in the invasiveness and proliferation of cervical cancer and ovarian cancer cells when activated by insulin-like growth factor 1 (24).
A Pathway-Based View of the Cancer Surfaceome.
The qPCR expression data, together with the remaining genomics data assembled in this study, provide the basis for constructing a rudimentary map of the genetic/epigenetic alterations related to the surfaceome in both types of tumors (Fig. S4). The approach taken here was based on searches in protein–protein interaction (PPI) networks looking for common interaction partners among differentially expressed surfaceome genes. In addition, we have also looked at the KEGG pathways (www.genome.ad.jp/kegg/pathway.html) identified for these genes, as well as the corresponding literature. For example, a number of overexpressed cell surface proteins in GBM are involved in RAS signaling, a pathway known to be important in this tumor type (25). However, one of the most provocative findings from this analysis was the overexpression in GBM of a number of cell surface proteins, including CAV1, TNFRSF12A, TNFRSF19, EDA2R, CD40, and LTBR, that signal through the TNF receptor-associated factor (TRAF) family. TRAF proteins, known to activate JUN and NF-κB, are important in mediating cell survival/death (26, 27). Recently, Zheng et al. (28) reported that the silencing of TRAF2 suppressed growth of a GBM cell line and sensitized the cells to radiation. Tumors exploit inflammatory responses to enhance their own growth and invasiveness, and TNF, a proinflammatory cytokine, appears to act as an endogenous tumor promoter in some systems (29). Production of TNF is elevated in various types of human cancer, has positive correlation with tumor grade, and associates with poor prognosis (30, 31). Taken together, our results suggested that these cell surface TNF receptors may be important for the growth and invasion of GBM.
Regarding colorectal tumors, the same type of analysis points to the importance of the ERK/MAPK pathway because overexpression of several cell surface-coding genes that activate this pathway were identified, including EPHA2, AREG, GRIN2B, CELSR3, SLC12A7, and MC1R. Amphiregulin (AREG), a cell surface member of the EGF family (32), is released after cleavage by ADAM17, and it can then bind to EGFR, leading to activation of the ERK/MAPK pathway.
CT Genes Coding for Cell Surface Antigens.
More than 90 genes coding for CT antigens have been identified in the last decades (8). The striking feature of a substantial number of CT genes is their restricted expression in normal tissues—testis—and their anomalous expression in a wide array of human malignancies (8). Because of their high degree of cancer specificity, CT antigens, such as MAGE-3 and NY-ESO-1, have been prime cancer vaccine targets (33, 34). CT antigens characteristically have an intracellular location, nuclear and/or cytoplasmic. Clearly, cell surface antigens with CT features would be of great interest for antibody-based therapies, but to date no validated CT cell surface antigens have been demonstrated. Based on that, and as a first step, we screened the MPSS and qPCR dataset for all surface coding genes with predominant or exclusive expression in testis. More than 119 candidate genes were identified (Table S5). To further enrich our set of candidates with bona fide CT characteristics, we evaluated the expression pattern of all 119 candidates in UniGene (www.ncbi.nlm.nih.gov/unigene) and selected those annotated with restricted expression in testis. A total of 22 candidates of the 119 were selected, as shown in Table S5.
Among these, the anion transporter SLCO6A1 was identified previously by Lee et al. (35) as a CT antigen by serological screening of recombinant expression libraries of human cancer with human serum (SEREX), providing validation for our approach to defining putative cell surface CT antigens. Another cell surface coding gene, FMR1NB (also known as NY-SAR-35), was also identified by SEREX as having a characteristic CT pattern of expression (36). However, the subcellular localization of FMR1NB is still uncertain.
Because the great majority of testis-restricted CT antigens are coded for by the X chromosome, we examined the surfaceome encoded by this chromosome. There are 134 TM-containing genes mapped to the X chromosome (Table S1). By using the MPSS analysis shown in Fig. 3, we observed that 6 of 134 genes have an expression pattern biased toward testis: TMEM31, FMR1NB, FATE1, IL13R2, GPA34, and ACE2. In addition to FMR1NB (CT37), two other antigens, FATE1 (CT43) and IL13R2 (CT13), have been identified previously as CT antigens, although FaTe1 also appears to be localized to the endoplasmic reticulum, according to ontology databases. TMEM31, a poorly characterized gene found to be differentially expressed in melanoma (37), represents a promising candidate cell surface CT coding gene. Interestingly, we have also identified GPA34, a cell surface antigen already identified as a candidate for immunotherapy in stomach and ovarian tumors (38). However, GPA34 is also expressed in normal stomach and a limited number of other normal tissues in addition to testis, and so it would be considered a testis-selective, not testis-restricted, CT antigen. Similarly, ACE-2 would also be considered a testis-selective CT antigen because of its predominant expression in testis and its expression pattern in normal tissues. Candidate CT cell surface genes listed in Table S5 were also analyzed in Oncomine (www.oncomine.org) by looking for genes overexpressed in tumors. A total of 14 of the 22 genes showed overexpression in at least one tumor type (P < 0.01).
A surprising outcome of this search for cell surface antigens with CT characteristics is the paucity of candidates, despite the extensive list of highly cancer-specific intracellular CT antigens.
Final Remarks.
We have used bioinformatics tools to define the human cell surfaceome, the set of putative cell surface proteins encoded by the human genome. Our methods agree with other estimates that at least 20% of all human genes code for proteins that are located at the cell surface. By integrating data from several large-scale platforms, we extensively annotate the human surfaceome, giving a special attention to xenogenetic, allogenetic, and epigenetic diversity.
By using gene expression data, we identified a subset of genes for experimental validation by using a large-scale qPCR platform. We identified dozens of genes as candidate tumor markers for GBM and colorectal tumors. Although there was public evidence for a tumor-specific expression for some candidates, we were able to identify many previously uncharacterized markers that considerably expand the list of potentially useful drug targets. Because we have evaluated the expression of the surfaceome in a panel of normal tissues, our analyses are also useful to avoid possible clinical side effects based on the expression of a potential target in any given normal tissue. We envisage that the present dataset will be of general value to the cancer research community.
Materials and Methods
Samples were obtained after explicit informed consent and with local ethics committee approval. Total RNA was prepared from cultured cells and from tissues by using TRIzol (Invitrogen). The qPCRs were performed in 96.96 dynamic array chips (Fluidigm) following the fabricant instructions. Bioinformatics analyses were performed as described previously (39, 40). Detailed information for materials and methods is described in SI Methods.
Supplementary Material
Acknowledgments.
We are indebted to Douglas Cancherini, Paula Asprino, Gladis Wilner, Bruna Quevedo, Ana Paula Medeiros Silva, Ken Livak, and Min Lin for technical assistance. This work was supported by intramural funds from the Ludwig Institute for Cancer Research and National Institutes of Health Fogarty International Center Grant 5D43TW007015-02.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907939106/DCSupplemental.
References
- 1.Rettig WJ, Old LJ. Immunogenetics of human cell surface differentiation. Annu Rev Immunol. 1989;7:481–511. doi: 10.1146/annurev.iy.07.040189.002405. [DOI] [PubMed] [Google Scholar]
- 2.Clark HF, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diehn M, Bhattacharya R, Botstein D, Brown PO. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 6.Davis MJ, et al. Differential use of signal peptides and membrane domains is a common occurrence in the protein output of transcriptional units. PLoS Genet. 2006;2:e46. doi: 10.1371/journal.pgen.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jongeneel CV, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson AJ, et al. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 9.Boon K, et al. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muto M, et al. Human liver-specific organic anion transporter-2 is a potent prognostic factor for human breast carcinoma. Cancer Sci. 2007;98:1570–1576. doi: 10.1111/j.1349-7006.2007.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada A, et al. Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res. 2008;14:3312–3318. doi: 10.1158/1078-0432.CCR-07-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe T, et al. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology. 2001;120:1689–1699. doi: 10.1053/gast.2001.24804. [DOI] [PubMed] [Google Scholar]
- 13.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 14.Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 15.Lee SW, Moskowitz MA, Sims JR. Sonic hedgehog inversely regulates the expression of angiopoietin-1 and angiopoietin-2 in fibroblasts. Int J Mol Med. 2007;19:445–451. [PubMed] [Google Scholar]
- 16.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66:2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 17.Athar M, et al. Inhibition of smoothened signaling prevents ultraviolet B-induced basal cell carcinomas through regulation of Fas expression and apoptosis. Cancer Res. 2004;64:7545–7552. doi: 10.1158/0008-5472.CAN-04-1393. [DOI] [PubMed] [Google Scholar]
- 18.Madi L, et al. The A3 adenosine receptor is highly expressed in tumor versus normal cells: Potential target for tumor growth inhibition. Clin Cancer Res. 2004;10:4472–4479. doi: 10.1158/1078-0432.CCR-03-0651. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, et al. Identification of Trop-2 as an oncogene and an attractive therapeutic target in colon cancers. Mol Cancer Ther. 2008;7:280–285. doi: 10.1158/1535-7163.MCT-07-2003. [DOI] [PubMed] [Google Scholar]
- 20.Guerra E, et al. A bicistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res. 2008;68:8113–8121. doi: 10.1158/0008-5472.CAN-07-6135. [DOI] [PubMed] [Google Scholar]
- 21.Rettig WJ, et al. Identification of endosialin, a cell surface glycoprotein of vascular endothelial cells in human cancer. Proc Natl Acad Sci USA. 1992;89:10832–10836. doi: 10.1073/pnas.89.22.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.St CB, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 23.Simonavicius N, et al. Endosialin (CD248) is a marker of tumor-associated pericytes in high-grade glioma. Mod Pathol. 2008;21:308–315. doi: 10.1038/modpathol.3801006. [DOI] [PubMed] [Google Scholar]
- 24.Shen MR, et al. Insulin-like growth factor 1 stimulates KCl cotransport, which is necessary for invasion and proliferation of cervical cancer and ovarian cancer cells. J Biol Chem. 2004;279:40017–40025. doi: 10.1074/jbc.M406706200. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arron JR, Walsh MC, Choi Y. TRAF-mediated TNFR-family signaling. Curr Protoc Immunol. 2002 doi: 10.1002/0471142735.im1109ds51. Chapter 11:Unit 11.9D. [DOI] [PubMed] [Google Scholar]
- 27.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp Med. 2000;191:1233–1240. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng M, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570–7578. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng SM, et al. Interferon-gamma regulation of TNFalpha-induced matrix metalloproteinase 3 expression and migration of human glioma T98G cells. Int J Cancer. 2007;121:1190–1196. doi: 10.1002/ijc.22729. [DOI] [PubMed] [Google Scholar]
- 30.Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 31.Mantovani G, et al. Serum levels of leptin and proinflammatory cytokines in patients with advanced-stage cancer at different sites. J Mol Med. 2000;78:554–561. doi: 10.1007/s001090000137. [DOI] [PubMed] [Google Scholar]
- 32.Gschwind A, Hart S, Fischer OM, Ullrich A. TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. EMBO J. 2003;22:2411–2421. doi: 10.1093/emboj/cdg231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coulie PG, et al. A monoclonal cytolytic T-lymphocyte response observed in a melanoma patient vaccinated with a tumor-specific antigenic peptide encoded by gene MAGE-3. Proc Natl Acad Sci USA. 2001;98:10290–10295. doi: 10.1073/pnas.161260098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gnjatic S, et al. NY-ESO-1: Review of an immunogenic tumor antigen. Adv Cancer Res. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]
- 35.Lee SY, et al. Identification of the gonad-specific anion transporter SLCO6A1 as a cancer/testis (CT) antigen expressed in human lung cancer. Cancer Immun. 2004;4:13. [PubMed] [Google Scholar]
- 36.Lee SY, et al. Immunomic analysis of human sarcoma. Proc Natl Acad Sci USA. 2003;100:2651–2656. doi: 10.1073/pnas.0437972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha C, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scanlan MJ, et al. Glycoprotein A34, a novel target for antibody-based cancer immunotherapy. Cancer Immun. 2006;6:2. [PubMed] [Google Scholar]
- 39.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galante PA, et al. Sense-antisense pairs in mammals: Functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.