Abstract
Background
Artificial selection results in phenotypic evolution. Maize (Zea mays L. ssp. mays) was domesticated from its wild progenitor teosinte (Zea mays subspecies parviglumis) through a single domestication event in southern Mexico between 6000 and 9000 years ago. This domestication event resulted in the original maize landrace varieties. The landraces provided the genetic material for modern plant breeders to select improved varieties and inbred lines by enhancing traits controlling agricultural productivity and performance. Artificial selection during domestication and crop improvement involved selection of specific alleles at genes controlling key morphological and agronomic traits, resulting in reduced genetic diversity relative to unselected genes.
Scope
This review is a summary of research on the identification and characterization by population genetics approaches of genes affected by artificial selection in maize.
Conclusions
Analysis of DNA sequence diversity at a large number of genes in a sample of teosintes and maize inbred lines indicated that approx. 2 % of maize genes exhibit evidence of artificial selection. The remaining genes give evidence of a population bottleneck associated with domestication and crop improvement. In a second study to efficiently identify selected genes, the genes with zero sequence diversity in maize inbreds were chosen as potential targets of selection and sequenced in diverse maize landraces and teosintes, resulting in about half of candidate genes exhibiting evidence for artificial selection. Extended gene sequencing demonstrated a low false-positive rate in the approach. The selected genes have functions consistent with agronomic selection for plant growth, nutritional quality and maturity. Large-scale screening for artificial selection allows identification of genes of potential agronomic importance even when gene function and the phenotype of interest are unknown. These approaches should also be applicable to other domesticated species if specific demographic conditions during domestication exist.
Key words: Maize, Zea mays L. ssp. mays, teosinte, Zea mays subspecies parviglumis, agronomic traits, artificial selection, domestication, plant breeding, DNA sequence, genetic diversity, HKA (Hudson–Kreitman–Aguadé) test, bottleneck, coalescent simulation
INTRODUCTION
Artificial selection is the process of intentional or unintentional modification of individuals in a population through human action, resulting in phenotypic evolution of plants and animals. For plants, key events in the advancement of human civilization were the domestication of crop plants from wild progenitor populations and improvement through crop breeding. Crop varieties consequently experienced strong selection at genes controlling agronomically important traits. Therefore genes that are identified as targets of artificial selection can be assumed to be important genetic factors controlling agronomic traits (Vigouroux et al., 2002b; Yamasaki et al., 2005).
An essential goal for plant geneticists is to identify and characterize the genes responsible for phenotypic variation. Using this knowledge, plant breeders can improve crop varieties for key agronomic traits. To determine genomic regions contributing to agronomic traits, gene and quantitative trait locus (QTL) mapping has been conducted, but this approach has rarely led to candidate gene isolation. In domesticated plants, only a limited number of such genes have been isolated and characterized based on information from gene/QTL analysis [examples include: fw2·2 in tomato (Frary et al., 2000), Hd1 in rice (Yano et al., 2000), tga1 in maize (Wang et al., 2005) and Gn1a in rice (Ashikari et al., 2005)]. Another promising approach – association analysis (Thornsberry et al., 2001) – searches for a significant correlation between genotype and phenotype and helps identify the genes that control phenotypic variation. Both gene/QTL and association approaches depend on segregating phenotypic and molecular genetic variation, and multiple functional alleles may be segregating among modern maize lines. In contrast, genes that have experienced strong artificial selection may have retained only a single functional allele. Therefore, gene/QTL and association methods among cultivars may miss this interesting class of genes, i.e. those genes that lack genetic diversity because of their history of selection.
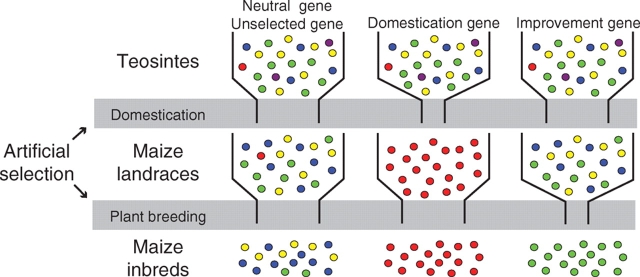
The loss of genetic diversity in crop varieties involves not only directional selection but also population bottleneck effects in a species specific manner. Bottlenecks affect all genes in the genome and modify the distribution of genetic variation among loci. In particular, recent population bottlenecks reduce genetic diversity relative to an ancestral population, cause an excess of high frequency polymorphisms (Tajima, 1989), and increase levels of linkage disequilibrium (LD) among polymorphic sites (Pritchard and Przeworski, 2001). The magnitude and variance of the reduction in genetic diversity reflect the demographic history of the species. To be considered a selected gene, a candidate gene must exhibit a loss of genetic diversity greater than can be expected from bottleneck effects alone (Fig. 1). On the other hand, genetic diversity in neutral and unselected genes is expected to be reduced only by bottleneck effects, thereby retaining more diversity than selected genes (Fig. 1). These facts lead directly to the prediction that genes strongly impacted by domestication and plant breeding are enriched in the subset of genes that exhibit low genetic diversity in modern improved varieties. Note that this model in Fig. 1 can be proposed for species such as maize that had a single domestication event (Matsuoka et al., 2002). If a crop species has experienced multiple independent domestications such as rice (Londo et al., 2006), one should propose multiple models similar to Fig. 1. We acknowledge that the model in Fig. 1 represents a gross oversimplification of the second bottleneck for crop improvement which occurred multiple times on different subsets of landrace germplasm.
Fig. 1.
Effect of artificial selection on the genetic diversity of maize genes (Yamasaki et al., 2005). Artificial selection in maize can be divided into two stages: domestication and improvement (plant breeding). The coloured circles represent different alleles. The shaded areas indicate bottleneck effects placed on all genes by the processes of domestication and improvement. The model assumes that there will be three types of genes; neutral (unselected) genes that show reduction of diversity by the general bottleneck effects, domestication genes in which diversity is greatly reduced by selection between the teosintes and landraces, and improvement genes in which diversity is greatly reduced by selection between the landraces and inbreds.
How can one conduct a comprehensive search for selected genes? This review is a report on new methodologies to identify this selected class of genes that contribute to agronomic traits in maize (Zea mays L. ssp. mays).
MAIZE DOMESTICATION
Maize was domesticated from its wild progenitor teosinte. Teosinte is the common name for annual and perennial species of the genus Zea native to Mexico and Central America: Z. diploperennis Iltis, Doebley & Guzman, Z. perennis (Hitchc.) Reeves & Mangelsdorf, Z. luxurians (Durieu & Ascherson) Bird, Z. nicaraguensis Iltis & Benz, Z. mays ssp. mexicana (Schrader) Iltis, Z. mays ssp. parviglumis Iltis & Doebley and Z. mays ssp. huehuetenangensis (Iltis & Doebley) Doebley (reviews in Doebley, 2004; Matsuoka, 2005). By using microsatellite data of diverse maize landraces and the three wild taxa of Z. mays, Matsuoka et al. (2002) revealed that Zea mays ssp. parviglumis is the ancestor of maize and supported a single domestication event in southern Mexico. For this review, teosinte hereafter refers to Zea mays ssp. parviglumis. By these molecular data and archaeological records, the maize domestication event is estimated to be between 6000 and 9000 years ago (Piperno and Flannery, 2001; Matsuoka et al., 2002).
Maize and teosinte differ in many aspects of plant morphology and productivity. For example, long lateral branches are observed in teosinte. These lateral branches are tipped with tassels (male inflorescence) in teosinte whereas maize reveals short ear (female inflorescence)-tipped lateral branches. Teosinte has a small number of kernels per ear (about 5–12) and the kernels were enveloped by a stony casing. Maize can produce 500 or more kernels per ear, which are naked without the hard seed coat; making it easy to harvest and consume. At maturity, the teosinte ear disarticulates to disperse the seeds whereas maize kernels remain firmly attached to the cob (non-shattering).
The maize domestication event resulted in the original maize landrace varieties, which were spread throughout the Americas by Native Americans and adapted to a wide range of environmental conditions (Smith, 1998). Starting with landraces, 20th century plant breeders selected maize inbred lines for use in hybrid maize production (Walden, 1979). They improved yield, resistance to biotic and abiotic stresses and seed nutritional quality. However, despite its selection history, most maize genes retain high levels of nucleotide diversity (Tenaillon et al., 2001; Wright et al. 2005; Yamasaki et al., 2005).
Several maize genes affected by artificial selection have already been identified: c1 (Hanson et al., 1996), tb1 (Wang et al., 1999), su1 (Whitt et al., 2002), Y1 (Palaisa et al., 2003), ba1 (Gallavotti et al., 2004) and tga1 (Wang et al., 2005). These genes were isolated by transposon tagging or QTL mapping and map-based cloning in crosses between maize and teosinte, and subsequently confirmed for selection in DNA sequence diversity using diverse maize lines and teosinte accessions. tb1 (teosinte branched1) controls lateral branch morphology (Doebley et al., 1997). Wang et al. (1999) examined sequence polymorphisms in tb1 in maize landraces and teosintes, and defined the gene region selected by domestication. It is the 5' regulatory, not the protein-coding region that exhibits a severe loss of diversity in maize landraces relative to teosintes. In the limited examples (tb1 and tga1) examined in maize to date, the region of reduced polymorphism is short, presumably reflecting high effective rates of recombination as maize is an outcrossing species. This results in high resolution to delimit the region under selection and aids in confirmation of a single gene as the target of selection. Another domestication gene tga1 (teosinte glume architecture1) controls the hardened and protective casing that envelopes the kernel in teosinte (Wang et al., 2005). The candidate region was narrowed within about 1 kb by a map-based cloning strategy. Seven fixed single nucleotide polymorphisms (SNPs) were identified between maize landraces and teosintes: one nonsynonymous substitution (i.e. amino acid substitution) and six SNPs in the 5' side of the promoter region.
TEST FOR ARTIFICIAL SELECTION
The neutral theory of Kimura (1968) proposed that most of polymorphisms are selectively neutral and their evolutionary fate is determined by genetic drift. The neutral equilibrium model was established based on this theory and assumes random mating and constant long-term population size. This model provides a basis for null hypotheses. The search for selection using patterns of DNA sequence diversity involves detecting inconsistency in the pattern of genetic polymorphism compared with the expectation of the neutral equilibrium model. There are three types of natural selection: positive selection (sometimes referred to as directional selection), negative selection (referred to as purifying selection) and balancing selection. Positive selection favours a mutation and therefore the allele frequency shifts in one direction. Artificial selection is a form of positive selection. Positive selection results in a reduction of DNA sequence diversity at the target of selection and in a region neighbouring the target. Negative selection is the selective removal of deleterious mutations or alleles from a population, leading to a reduction in genetic diversity. Balancing selection represents the long-term selective maintenance of multiple alleles, and this process elevates sequence diversity.
There are several molecular population genetics tests to infer positive selection in plants (Wright and Gaut, 2004). Tajima's D (Tajima, 1989) measures the frequency spectrum for sequence polymorphism: the difference between θ derived from number of segregating sites (Watterson, 1975) and π from the average number of pairwise nucleotide differences (Tajima, 1983). Under the neutral equilibrium model, the Tajima's D statistic is expected to be zero. However, because π incorporates frequency information while θ does not, departures from the standard neutral model affecting the frequencies of mutations will lead to non-zero values. A negative value of Tajima's D statistic indicates an excess of rare sequence variants and thus the possibility of recent positive selection as new polymorphisms accumulate as rare variants post-selection. In contrast, a positive value of Tajima's D suggests balancing selection because the long-term maintenance of distinct haplotypes can lead to an excess of intermediate frequency variants.
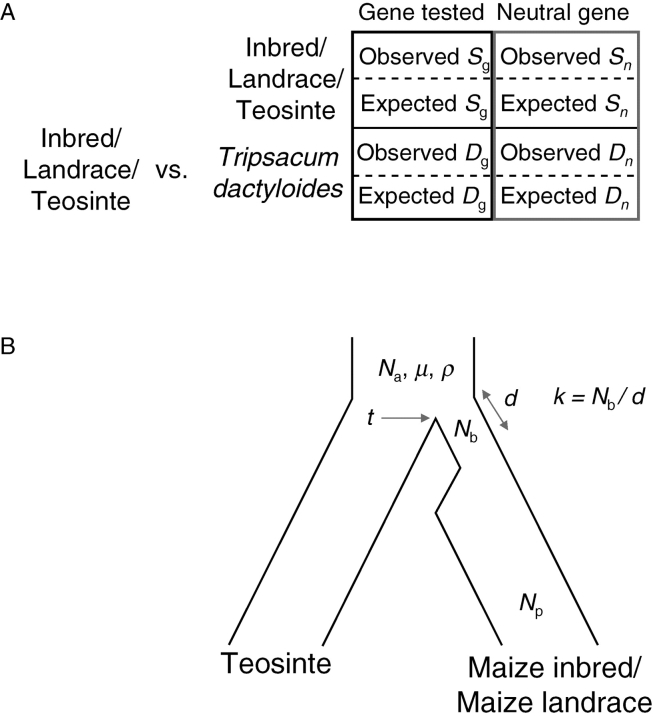
Another test for selection, the HKA (Hudson–Kreitman–Aguadé) test requires an outgroup sequence to compare rates of divergence between species to levels of polymorphism within species (Hudson et al., 1987), compared with neutral genes. Under the neutral equilibrium model, all loci should have an equivalent ratio of polymorphism to divergence, since both depend on the underlying mutation rate. The number of intraspecific polymorphisms is compared between a putatively selected gene and the reference neutral gene by the χ2 test (Fig. 2A). If a gene has significantly reduced intraspecific polymorphisms relative to divergence (i.e. observed Sg of gene tested compared with expected Sg in Fig. 2A), it is evidence that the polymorphism level has been diminished by directional selection. Recently, multilocus data have been generally used to detect a significant difference among loci because it is difficult to reject the neutral equilibrium model at a single locus, and the results of pairwise tests can be dependent on the choice of putative ‘neutral’ locus (Wang and Hey, 1996; Wright and Charlesworth, 2004). Simulation studies have demonstrated that the HKA test is an appropriate and powerful tool to find genes affected by artificial selection (Innan and Kim, 2004).
Fig. 2.
Hudson–Kreitman–Aguadé (HKA) test (A) and coalescent simulation test (B) (Wright et al., 2005; Yamasaki et al., 2005). (A) The HKA test considers whether a gene of interest is selected using a maize neutral gene as reference and Tripsacum dactyloides as outgroup species. S, The number of segregation sites; D, divergence from T. dactyloides. Subscripts ‘g’ and ‘n’ exhibit belonging to gene tested and neutral genes, respectively. (B) Coalescent model with a population bottleneck. Na, The ancestral population size; μ, the mutation rate; ρ, the population-recombination parameter (Hudson, 2001); t, the time of the split between maize and teosinte; d, the duration of the maize population bottleneck; k, the severity of the bottleneck; Nb, the population size of maize during bottleneck; Np, the present size of maize.
The neutral equilibrium model assumes a constant long-term population size. However, crop species have experienced population bottlenecks during domestication. Bottleneck effects can increase the variance in sequence diversity across loci and skew the frequency spectrum. To address this problem, coalescent simulations (CS) construct and analyse random genealogies incorporating mutation, recombination and demographic history of the species under study. Coalescence in genealogical trees means to merge ancestral lineages going backwards in time (Kingman, 1982; Hudson, 1983a, b; Tajima, 1983).
Coalescent simulations were used to fit the best model of a population bottleneck leading to maize, and this model was then used as the ‘null’ model in which to test for artificial selection on a subset of genes. The coalescent model incorporates information about maize demographic history, such as the domestication time approx. 7500 years ago (Eyre-Walker et al., 1998; Tenaillon et al., 2004) and the inference of a single domestication event (Matsuoka et al., 2002) (Fig. 2B). The model assumes a simple population split at time t = 7500 generations, with one population undergoing a bottleneck of size Nb and duration d. Genetic diversity data from teosinte is used to control for variation in stochastic effects, mutation rates and recombination rates among loci. The primary parameter of interest is bottleneck severity k, which is the ratio of the size of bottlenecked population Nb to duration of the bottleneck d in generations (Fig. 2B). By varying this parameter, the likelihood of a given bottleneck model was estimated by calculating the proportion of simulations which fit with the observed maize diversity data for each locus. By rejecting simulations which did not show a good fit with the observed teosinte data, the analysis incorporated the ancestral diversity data into the model. Maximum likelihood methods estimated k at 2·45 for the full dataset (Wright et al., 2005). The simulated data under the estimate of 2·45 are generally consistent with the observed data on genetic diversity, recombination and the SNP frequency spectrum (Wright et al., 2005), suggesting that the demographic model explains most features of the data. If the maximum estimate of d for the domestication of maize is <2800 years (Eyre-Walker et al., 1998), assuming one generation per year for an annual plant, Nb is 6860 chromosomes. This implies fewer than 3500 individuals, or <10 % of the teosinte population (Hilton and Gaut, 1998; Vigouroux et al., 2002a) was involved in the domestication event. Simulations can then be used to test whether the loss of diversity in maize inbreds vs. teosintes and maize landraces vs. teosintes at a particular gene is too great to be explained by demographic effects alone. One can identify a candidate gene that experienced artificial selection during domestication and/or plant breeding if inbreds vs. teosintes were significant for selection. A candidate ‘domestication’ gene was identified if the gene was significant for selection in both landraces vs. teosintes and inbreds vs. teosintes whereas a candidate ‘improvement’ gene was inferred if inbreds vs. teosintes, but not landraces vs. teosintes, were significant (Fig. 1; Wright et al., 2005; Yamasaki et al., 2005). For identification of an improvement gene, the CS test cannot be applied directly between landraces and inbreds because the ancestral population in our CS test follows the neutral equilibrium model: maize landraces are unreasonable to fit the neutral equilibrium model because they have recently experienced a population bottleneck.
GENOMIC SCREENING FOR ARTIFICIAL SELECTION IN MAIZE GENOME
Two large-scale screens were conducted to discover genes responsible for artificial selection in maize (Wright et al., 2005; Yamasaki et al., 2005). Wright et al. (2005) sequenced regions of 774 randomly selected maize genes in a sample of 14 maize inbred lines and 16 inbred teosintes. Simple sequence repeat (SSR) polymorphism data was used to choose 14 maize inbreds lines with maximum allelic diversity (Liu et al., 2003). The 16 teosinte accessions were chosen based on geographic criteria to represent all areas where Z. mays ssp. parviglumis is found. This sample of maize inbreds has 57 % of SNP variability found in the teosinte sample, consistent with a population bottleneck during domestication and maize improvement. Also, the distribution of Tajima's D statistic (Tajima, 1989) in maize inbreds and teosintes revealed the expected shift towards a higher frequency of rare variants as expected after a recent bottleneck. Using a novel CS approach, Wright et al. (2005) determined by a likelihood ratio test that the multilocus data are best explained by the presence of two classes of genes: non-selected genes that have experienced the neutral bottleneck and selected genes that have a more severe bottleneck. The class of selected genes has undergone a severe bottleneck of more than ten times the intensity of non-selected genes. Under this model, the likelihood analysis estimates that 2–4 % of maize genes have been selected during domestication and maize breeding. If it is assumed that the sample of genes is representative of the maize genome and if maize contains 59 000 genes (Messing et al., 2004), a minimum of 1200 genes (≈59 000 × 2 %) throughout the genome have been targets of selection. A number of candidate genes selected in this study (Wright et al., 2005) map near QTLs for phenotypic differences identified between maize and teosinte (Doebley et al., 1990; Doebley and Stec, 1991, 1993). The combination of QTL mapping from crosses between domesticated species and the wild ancestor with ‘selective sweep mapping’ thus provides a powerful approach for narrowing down genomic regions that have been targets of artificial selection.
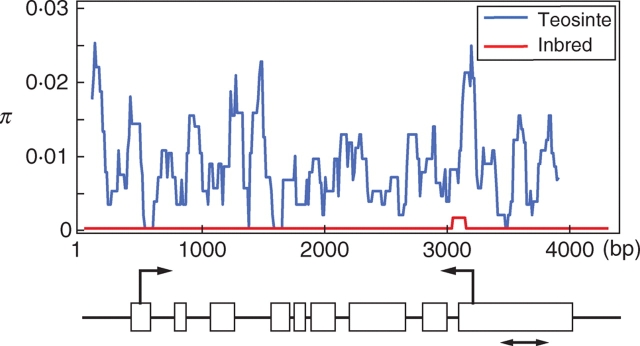
Because the first genomic screening indicated only 2–4 % were selected, Yamasaki et al. (2005) proposed a more efficient screen for the identification of genes affected by artificial selection. Yamasaki et al. (2005) obtained sequence alignments at 1095 randomly selected maize genes using a sample of 14 maize inbred lines and identified 35 genes with zero SNP diversity as potential targets of selection. From Fig. 1, note that genes strongly impacted by selection are enriched in the subset of genes that exhibit very low genetic diversity in modern inbreds. Although the lack of nucleotide diversity in these genes could reflect a history of selection during domestication or improvement, the low diversity in maize could also reflect low diversity in teosinte and/or the demographic effects of domestication, plant breeding and/or chance events (e.g. genetic drift). In order to distinguish between selection and other effects and also to determine if selection occurred primarily during domestication or crop improvement, Yamasaki et al. (2005) sequenced the same region of these 35 genes in the 16 teosinte accessions and 16 diverse maize landraces. The 16 maize landraces represent all areas in which maize was grown at the time of the discovery of the New World (Tenaillon et al., 2001). They identified 17 of 35 genes as candidates for selection by either CS or HKA analysis. There were four domestication and four improvement genes identified in common between the CS and HKA analyses. One limitation to these approaches for identifying selected genes is that the short length of the alignments (generally 300–500 bp) restricts the power of the approach. Longer sequences would increase the power to identify selection. Yamasaki et al. (2005) performed extended sequence analysis of the eight genes positive for selection by both CS and HKA to demonstrate that the false-positive rate was low. In Fig. 3, the sliding-window graph of sequence diversity in an auxin response factor gene is shown as an example of evidence for strong selection throughout the entire gene. The relative ratio of sequence diversity measure π (Tajima, 1983) at all nucleotide sites in inbred/teosinte was 0·005, indicating that the inbreds have lost 99·5 % of genetic diversity present in the teosinte sample. The extended sequencing clearly confirmed this candidate as a selected gene. Since longer sequencing could not necessarily increase the accuracy of identifying selected genes (Yamasaki et al., 2005; Teshima et al., 2006), the remaining genes with low sequence diversity in maize inbreds are potential selected genes.
Fig. 3.
Sliding-window analysis of the genetic diversity π of auxin response factor 1-like gene (AY107195) in maize inbreds and teosintes. This gene experienced selection throughout the entire gene sequence. For sliding-window analysis, π was calculated for segments of 100 bp at 10-bp intervals. Horizontal and vertical axes on the graphs indicate DNA sequence position and genetic diversity π, respectively. Red and blue lines indicate genetic diversity in inbreds and teosinte, respectively. For gene structure under the sliding-window graphs, white bars indicate the predicted exons and black lines indicate introns or genomic regions. Left-pointing arrows and right-pointing arrows indicate the positions for predicted start codons and stop codons, respectively. The lines with a double arrow under the gene structure indicate the sequencing regions in initial short screening.
FUNCTIONS OF SELECTED GENES IN MAIZE
From the candidate selected genes identified in maize (Wright et al., 2005; Yamasaki et al., 2005), a homology search provided clues to the functions and traits under selection. Several candidate genes are involved in plant growth, e.g. auxin response factor and GTP binding protein, suggesting a contribution to the morphological difference between maize and teosinte. Other candidates are associated with amino acid biosynthesis and protein catabolism. Amino acid composition is different among maize inbreds, landraces and teosinte accessions (S. A. Flint-Garcia and M. McMullen, unpubl. res.). Thus, nutritional quality may have been a major target of human selection. This result complements the finding that many genes in the starch synthesis pathway in maize also show evidence of human selection (Whitt et al., 2002). One candidate gene shows homology to circadian clock genes and may have been selected for an aspect of maize maturity. Another candidate gene encodes a putative methyl binding domain protein. DNA methylation is a common factor in epigenetic gene regulation in plants (Bender, 2004). Several candidates have no homology to known genes and protein. One of the advantages of an unbiased genomic scan is that genes of unknown function can be identified as selection candidates. The results suggest that genes controlling a wide range of traits have been targets of selection. Selective sweep mapping thus functions to further delimit regions identified by QTL approaches, and also provides new testable hypotheses about the physiological and biochemical targets of artificial selection.
APPLICATION OF THESE APPROACHES TO OTHER CROP SPECIES
These approaches may be applied to other crop species but specific demographic conditions must exist. First, there must be a high level of genetic diversity in the progenitor species. Also there must be a relatively relaxed bottleneck in domestication permitting a high level of diversity in neutral genes before the loss of diversity by selection can be detected. A second species-specific consideration is the degree of LD surrounding selected genes. In neutral genes in maize, even among diverse inbred lines, LD is generally less than a gene unit in length (Remington et al., 2001; Tenaillon et al., 2001). Artificial selection is expected to increase LD at target genes relative to neutral gene. It is therefore striking that the selective sweep at tb1 extended only 60–90 kb upstream of the gene and does not contain other genes (Clark et al., 2004). However, for the Y1 locus, a gene under recent and strong selection in maize breeding programmes (an improvement selection), the effects of selection were evident up to 600 kb downstream of the target gene in the yellow endosperm subset of maize lines (Palaisa et al., 2004). If LD among cultivars is extensive in a genome-wide manner, it is impossible to tell targets of selection from neighbouring unselected genes.
The most rigorous search for signatures of selection in a crop other than maize has been conducted in sorghum (Sorghum bicolor). Despite scanning a total of 445 genes by SSR diversity (Casa et al., 2005, 2006) or resequencing (Hamblin et al., 2006), unequivocal evidence of artificial selection was not demonstrated. The authors attribute this result to lower levels of sequence diversity in neutral genes in sorghum compared with maize and to genome wide departures from neutral frequency distributions indicative of a complex demographic history for cultivated sorghum. The search for selected genes is expected to be even more difficult in a species such as soybean (Glycine max) were the sequence diversity is extremely low in cultivars (Zhu et al., 2003) and LD is extensive, even among landrace accessions (Hyten et al. 2007). In comparing sorghum and soybean to maize, these differences in outcome are often attributed to sorghum and soybean being inbreeding rather than outcrossing. Inbreeding reduces effective recombination rates and therefore is expected to lead to reduced diversity and extensive LD. A study of wild barley (Hordeum vulgare ssp. spontaneum) accessions found almost identical sequence diversity and limited LD as in the maize progenitor teosinte (Morrell et al., 2005). The similarity of barley to maize contrasts with extensive LD established in landrace and cultivars after domestication and crop improvement (Caldwell et al., 2006). Possibly more promising as an acceptable model for screens for selection is sunflower (Helianthus annuus) where the levels of diversity and LD in wild and cultivated accessions are similar to maize (Liu and Burke, 2006). Clearly, the applicability of genomic scans for selection depends on the mating system and demographic history of the species.
USE OF SELECTED GENES IN MAIZE IMPROVEMENT
Teosintes and maize landraces are potential genetic resources to improve key agronomic traits. Even though there is not yet an example for using alleles from teosintes to improve maize, beneficial alleles for agronomic traits for cultivated rice (Oryza sativa) were identified from a wild rice relative Oryza rufipogon (Xiao et al., 1998). There are two approaches to reintroducing variation at selected genes into maize breeding. The alleles from teosintes at domestication genes and from landraces for improvement genes could be introduced into maize breeding programmes. It is specifically these genes that need to be added to maize breeding from exotic sources to broaden the genetic base of maize. Although the selected alleles were favoured within the populations under selection, there can still be promising unknown alleles that may improve agronomic traits. Alternatively, transgenic alteration of the expression patterns of selected genes can be tested for desired effects on the relevant agronomic traits.
The studies reported in this review depended on resequencing methods to determine polymorphism levels within taxa. With the advances in massively parallel sequencing methods we may soon be approaching the potential for complete gene content comparisons among cultivars and progenitors of crop plants, enabling a true genome search for adaptive genes. The ultimate value of these studies awaits demonstration that crop improvement can be advanced by manipulation of this interesting class of genes discovered by genomic screens for selection.
ACKNOWLEDGEMENTS
We thank Brandon Gaut for helpful discussions and comments on the manuscript. This research was supported by National Science Foundation Plant Genome Awards DBI0321467 and by research funds provided by USDA-ARS and the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad in 2004. We thank the American Society of Plant Biologists as copyright holder for permission to reuse Fig. 1. Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Bender J. DNA methylation and epigenetics. Annual Review of Plant Biology. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Caldwell KS, Russell J, Langridge P, Powell W. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species. Hordeum vulgare. Genetics. 2006;172:557–567. doi: 10.1534/genetics.104.038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Mitchell SE, Hamblin MT, Sun H, Bowers JE, Paterson AH, et al. Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theoretical and Applied Genetics. 2005;111:23–30. doi: 10.1007/s00122-005-1952-5. [DOI] [PubMed] [Google Scholar]
- Casa AM, Mitchell SE, Jensen JD, Hamblin MT, Paterson AH, Aquadro CF, et al. Evidence for a selective sweep on chromosome 1 of cultivated sorghum. Plant Genome. 2006;46:S27–S40. [Google Scholar]
- Clark RM, Linton E, Messing J, Doebley JF. Pattern of diversity in the genomic region near the maize domestication gene tb1. Proceedings of the National Academy of Science of the USA. 2004;101:700–707. doi: 10.1073/pnas.2237049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. The genetics of maize evolution. Annual Review of Genetics. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A. Genetic analysis of the morphological differences between maize and teosinte. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Wendel J, Edwards M. Genetic and morphological analysis of a maize-teosinte F2 population: implications for the origin of maize. Proceedings of the National Academy of Science of the USA. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proceedings of the National Academy of Science of the USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Frary A, Grandillo S, van der Knaap E, Cong B, et al. fw2·2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, et al. The role of barren stalk1 in the architecture of maize. Nature. 2004;432:630–635. doi: 10.1038/nature03148. [DOI] [PubMed] [Google Scholar]
- Hamblin MT, Casa AM, Sun H, Murray SC, Paterson AH, Aquadro CF, et al. Challenges of detecting directional selection after a bottleneck: lessons from. Sorghum bicolor. Genetics. 2006;173:953–964. doi: 10.1534/genetics.105.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Gaut BS, Stec AO, Fuerstenberg SI, Goodman MM, Coe EH, et al. Evolution of anthocyanin biosynthesis in maize kernels: the role of regulatory and enzymatic loci. Genetics. 1996;143:1395–1407. doi: 10.1093/genetics/143.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton H, Gaut BS. Speciation and domestication in maize and its wild relatives: evidence from the Globulin-1 gene. Genetics. 1998;150:863–872. doi: 10.1093/genetics/150.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Properties of a neutral allele model with intragenic recombination. Theoretical Population Biology. (a) 1983;23:183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Testing the constant-rate neutral allele model with protein sequence data. Evolution. (b) 1983;37:203–217. doi: 10.1111/j.1558-5646.1983.tb05528.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Choi IY, Song Q, Shoemaker RC, Nelson RL, Costa JM, et al. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics. 2007;175:1937–1944. doi: 10.1534/genetics.106.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proceedings of the National Academy of Science of the USA; 2004. pp. 10667–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kingman JFC. On the genealogy of large populations. Journal of Applied Probability. 1982;19A:27–43. [Google Scholar]
- Liu A, Burke JM. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics. 2006;173:321–330. doi: 10.1534/genetics.105.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics. 2003;165:2117–2128. doi: 10.1093/genetics/165.4.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveal multiple independent domestications of cultivated rice, Oryza sativa. Proceedings of the National Academy of Science of the USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y. Origin matters: lessons from the search for the wild ancestor of maize. Breeding Science. 2005;55:383–390. [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Jesus Sanchez G, Buckler E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proceedings of the National Academy of Science of the USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, et al. Sequence composition and genome organization of maize. Proceedings of the National Academy of Science of the USA. 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell PL, Toleno DM, Lundy KE, Clegg MT. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proceedings of the National Academy of Science of the USA. 2005;102:2442–2447. doi: 10.1073/pnas.0409804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa K, Morgante M, Tingey S, Rafalski A. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proceedings of the National Academy of Science of the USA. 2004;101:9885–9890. doi: 10.1073/pnas.0307839101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa KA, Morgante M, Williams M, Rafalski A. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. The Plant Cell. 2003;15:1795–1806. doi: 10.1105/tpc.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno DR, Flannery KV. The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications. Proceedings of the National Academy of Science of the USA. 2001;98:2101–2103. doi: 10.1073/pnas.98.4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. American Journal of Human Genetics. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Science of the USA; 2001. pp. 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BD. The emergence of agriculture. New York, NY: W. H. Freeman & Co; 1998. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays L. ssp. mays; Proceedings of the National Academy of Science of the USA; 2001. pp. 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, U'Ren J, Tenaillon O, Gaut BS. Selection versus demography: a multilocus investigation of the domestication process in maize. Molecular Biology and Evolution. 2004;21:1214–1225. doi: 10.1093/molbev/msh102. [DOI] [PubMed] [Google Scholar]
- Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Reseach. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES. Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Jaqueth JS, Matsuoka Y, Smith OS, Beavis WD, Smith JSC, et al. Rate and pattern of mutation at microsatellite loci in maize. Molecular Biology and Evolution. 2002;19:1251–1260. doi: 10.1093/oxfordjournals.molbev.a004186. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, McMullen M, Hittinger CT, Houchins K, Schulz L, Kresovich S, et al. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication; Proceedings of the National Academy of Science of the USA; 2002. pp. 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden BD. Maize breeding and genetics. Chichester: John Wiley & Sons; 1979. [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, et al. The origin of naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Hey J. The speciation history of Drosophila pseudoobscura and close relatives: inferences from DNA sequence variation at the period locus. Genetics. 1996;144:1113–1126. doi: 10.1093/genetics/144.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology. 1975;7:188–193. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ES. Genetic diversity and selection in the maize starch pathway. Proceedings of the National Academy of Science of the USA; 2002. pp. 12959–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Charlesworth B. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution. 2004;22:506–519. doi: 10.1093/molbev/msi035. [DOI] [PubMed] [Google Scholar]
- Wright SI, Vroh Bi I, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, et al. Identification of trait-improving quantitative trait loci alleles from a wild rice relative. Oryza rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Tenaillon MI, Vroh Bi I, Schroeder SG, Sanchez-Villeda H, Doebley JF, et al. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. The Plant Cell. 2005;17:2859–2872. doi: 10.1105/tpc.105.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, Van Tassel CP, Matukumalli LK, Grimm DR, et al. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]