Nanomaterials modified with biological recognition motifs acquire a myriad of functions that can be exploited for the diagnosis and treatment of cancer. Nevertheless, while bioactive domains can be used to target nanoparticles to cell receptors, shuttle them across cell membranes, and activate cell signaling, such modifications typically include cationic or hydrophobic regions that lead to rapid reticuloendothelial system (RES) clearance of particles from the blood, ultimately reducing particle accumulation in tumors.[1,2] Further functionalization with hydrophilic polymers like poly(ethylene glycol) (PEG) can improve blood half-lives and tumor accumulation, but often at the expense of efficient ligandmediated nanoparticle binding.[3-5] To address this tradeoff between improved biodistribution and optimal functionality on nanoparticles, we present a general strategy for reversibly veiling bioactive domains on nanoparticles using sterically protective polymers. We demonstrate that these materials effectively accumulate via the hyperpermeable vasculature of tumors and can be activated by cancer-secreted proteases to unveil hidden functional domains.
Several cleavable polymers relying on reducible,[6] pH-sensitive,[7-15] and protease-cleavable linkages[16,17] have been described in the literature. While all of these systems enable rapid polymer release in the presence of appropriate physiological triggers, nanoparticles employing reducible or pH-sensitive linkages require transport through cell membranes or diffusion to acidic environments that may be hard to reach. Tumor-associated proteases, however, are expressed at the invasive front of tumors and at sites of angiogenesis, areas that are more easily accessed via passive particle accumulation, thus making nanoparticles sensitive to such enzymes excellent candidates for cancer therapy. Previously, we demonstrated that veiling particles with protease-cleavable polymers effectively suppresses the binding of complementary small molecules and larger proteins on nanoparticles.[17,18] In this work we extend the utility of this technique by demonstrating that these coatings can be optimized to deter nanoparticle-cell interactions and to confer favorable circulation and accumulation properties in vivo. In contrast to the reported use of protease-cleavable PEGs to destabilize and fuse liposomes[19,20] or the use of cleavable polyanionic peptides to electrostatically neutralize cationic domains,[21,22] this strategy exploits the entropic penalty imparted by hydrophilic polymers on approaching surfaces to veil and unveil the bioactivity of surface ligands. Consequently, this technique may be used to veil bioactive domains that need not be cationic or lipid-like, thereby enabling the regulation of functions besides fusion or internalization, such as cell binding or cell signaling.
Using fluorescence imaging and MRI, we demonstrate that protease-removable polymer coatings effectively suppress cell uptake of nanoparticles bearing cell internalization domains, while proteolytic cleavage by MMP-2, a protease upregulated in angiogenesis, invasion, and metastasis,[23-26] restores internalization function. In vivo, reversible polymer veiling greatly extends nanoparticle circulation in the blood over unveiled particles, and enhances tumor accumulation. We confirm that cleavable coatings on extravasated nanoparticles are removed in the tumor, thus establishing the potential of this design for unveiling bioactive ligands in response to disease-associated triggers on a variety of nanoparticle platforms.
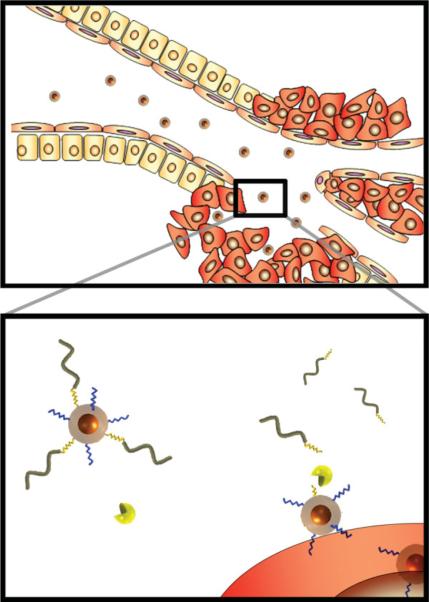
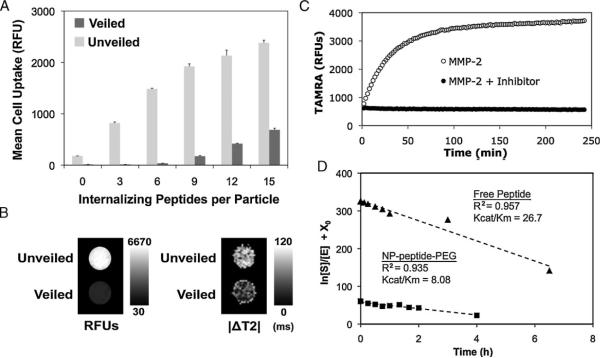
Figure 1 shows a schematic model of nanoparticles bearing protease-removable polymer coatings that veil the function of bioactive surface ligands. Two species, a cell internalization domain and a removable hydrophilic polymer, consisting of a linear PEG tethered by an MMP-2 cleavable substrate, are conjugated onto the surface of a magnetofluorescent dextrancoated iron oxide nanoparticle. Prior to activation, the hydrophilic polymer prevents both RES-mediated clearance of the particles and systemic action of the bioactive ligand, in this case an internalizing domain. Previously we identified a removable polymer coating that was optimal for veiling and unveiling ligands mediating interparticle interactions.[17] We hypothesized that this approach could be extended to veil particle-cell interactions. To test this, we conjugated particles with the removable polymer coating and varying densities of cell internalization domains and then measured the uptake of veiled and protease-activated (unveiled) particles by HT-1080 cells using flow cytometry. Particles with lower domain densities were taken up minimally by cells in both the veiled and unveiled state, while particles with higher domain densities were taken up by cells even with the intact polymer coating. An optimized particle design was selected based on a high level of internalization of unveiled particles and a low level of internalization of veiled particles, with the optimum ratio of internalization domains resulting in a 40-fold increase in cell accumulation (Figure 2A). This particle had, on average, 6 internalization domains per nanoparticle and 60 cleavable PEGs. Polymer coatings increased the hydrodynamic diameter of nanoparticles from 63 nm to 85 nm (Supporting Information, Figure 1). Once activated by MMPs, unveiled particles did not aggregate, but remained colloidally stable in solution (Supporting Information, Figure 1).
Figure 1.
Schematic depiction of removable polymer coatings that veil and unveil bioactive ligands on a nanoparticle surface. A hydrophilic polymer (wavy-gray) linked via MMP cleavable substrates (jagged-yellow) veils the activity of a cell-internalizing domain (jagged-blue) on the surface of a magnetofluorescent nanoparticle. Veiled particles have extended circulation times that enable their passive accumulation in tumors. Extravasated particles are activated by MMP-2 in the micro-environment to unveil internalizing domains, which associate with the cell membrane and shuttle nanoparticles into cells.
Figure 2.
Optimization and characterization of nanoparticle veiling, activation, and internalization. A) A library of nanoparticles with removable polymer coatings and a varying density of internalization ligands was screened for relative uptake by HT-1080 cancer cells before (veiled, dark gray) and after (unveiled, light gray) MMP cleavage. A density of 6 cell-internalizing peptides per particle demonstrated optimal veiling and internalization. Error bars are standard deviations from three separate experiments. B) Cells incubated with veiled and unveiled nanoparticles for 5 h were imaged by (left) a fluorescence scanner or (right) MRI, demonstrating the dual contrast properties of the nanoparticles and the correlated fluorescent and magnetic domain uptake of unveiled particles. C) MMP-mediated removal of polymer coatings relieves TAMRA-iron quenching interactions, thereby enabling remote monitoring of protease activation. D) The Kcat/Km for peptide-polymer NPs (squares) and free peptide (triangles) was determined to be 8.42 and 26.7 μM-1h-1, respectively, by measuring the cleavage of the substrate by MMP-2 over time. Polymer veiling and immobilization of the cleavable peptide substrate reduces its associated MMP-2 Kcat/Km within a practical range, 3.2 fold.
To verify that internalization function is indeed restored after removal of the polymer coating, epifluorescence microscopy was used to monitor the trafficking of unveiled nanoparticles as they traveled from the cell membrane toward the nucleus through intracellular organelles, a punctate pattern greatly reduced with veiled particles (Supporting Information, Figure 2). Flow cytometry and microscopy studies using other cell lines confirmed that this effect is not specific to HT-1080 cells (Supporting Information, Figure 3). The magnetic properties of the iron oxide core particles used in these studies can also be used to confirm cell uptake of the particles with MRI. A T2 mapping sequence was used to detect T2 changes in cells that had been incubated with veiled and unveiled nanoparticles for 5 h and imaged by a 4.7T MRI (Bruker). The internalization of nanocrystal cores leads to a measurable decrease in T2 signal which was significantly greater with unveiled particles and well correlated with signal changes detected in a planar fluorescence scan (Licor - Figure 2B).
Given that the orientation of the cleavable substrate in this system is constrained between the nanoparticle and 10 kDa PEG, we sought to confirm that proteases could access the peptide backbone by comparing the catalytic efficiency (Kcat/Km) of MMP-2 for its substrate on the particle with the substrate free in solution. Since peptide-PEG domains were labeled with TAMRA in a position removed upon cleavage, activation of nanoparticles in solution and determination of the Kcat/Km can be monitored by the release of the TAMRA dye (Figures 2C, 2D).[18] Using this approach, it was determined that proteases effectively accessed the peptide backbone for cleavage and that PEG shielding and particle immobilization resulted in only a 3.2-fold decrease in the Kcat/Km, a favorable reduction considering the order of magnitude decreases that have been reported with MMP-2 substrates on other immobilized polymers.[27]
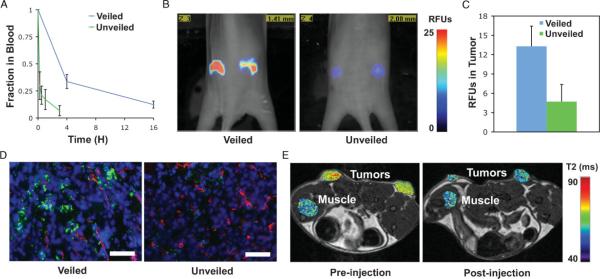
After completing these proof-of-principle experiments in vitro, we sought to characterize how the different surface properties of veiled and unveiled particles affect blood circulation times in vivo. To this end, we systemically administered to mice (via intravenous tail injections) veiled nanoparticles consisting of cleavable L-isomer amino acid peptide substrates (veiled L-AA), in addition to non-cleavable particles with D-isomer substrate analogs (veiled D-AA) and unveiled controls. The circulation time of cleavable particles was not significantly altered from that of non-cleavable controls, suggesting that the cleavable coating withstands degradation by proteases in the blood (Supporting Figure 4A). This is supported by in vitro experiments that show that this MMP-responsive polymer is not activated by serum or serum enzymes.[18] Particles with unveiled bioactive domains had significantly reduced blood half-lives, clearing from the blood approximately 8 times faster than veiled particles, with more than 25% of PEG-veiled nanoparticles still in the blood at 4 h compared to unveiled particles that had 25% remaining after only 30 min (Figure 3A).
Figure 3.
Effects of removable polymer coatings on the blood-clearance and tumor accumulation of nanoparticles. A) Nanoparticles bearing MMP cleavable polymer coatings (veiled) have greater blood clearance times compared with particles that have had the coating removed by MMPs (unveiled). Error bars indicate standard deviation of three animals. B) Fluorescence molecular tomography (FMT) of regions of interest (ROIs) selected around bilateral flank tumors from two representative animals shows intravenous injections of veiled nanoparticles yield greater accumulation in tumors after 48 h as compared to unveiled controls. C) Analysis of nanoparticle accumulation in the tumor at 48 h by FMT demonstrates superior accumulation of veiled particles as compared to unveiled controls. Error bars represent standard deviation of three animals. D) Representative histological sections confirm the increased accumulation of veiled nanoparticles versus unveiled controls after 48 h; nanoparticles (green), blood vessels (red), nuclei (blue). Scale bar is 250 μm. E) T2 map of tumor and muscle ROIs after intravenous injection show enhanced contrast from veiled nanoparticles in the tumor versus normal tissue (muscle) at 24 h post-injection.
Because the rate-limiting step of nanoparticle transport into the tumor is extravasation, size and circulation time are the most important factors governing their accumulation.[28]
Empirically, 80-100nm nanoparticles bearing targeting ligands that bind tumor cells in the extravascular space do not show improved accumulation over untargeted controls, though these strategies do improve therapeutic efficacy.[29,30] In accordance with these results, the activation and subsequent internalization of cleavable particles should not enhance their accumulation over non-cleavable particles that remain in the extracellular space. Using fluorescence molecular tomography (FMT) we show that cleavable particles do not accumulate significantly more than non-cleavable controls in tumor regions of interest over 48 h (Supporting Information, Figure 4B), but that they do accumulate significantly more than nanoparticles that have their bioactive domains unveiled (Figures 3B, 3C). Histological analysis of tumors confirms increased accumulation of veiled particles as compared to unveiled particles and shows that particles have moved beyond vascular borders (Figure 3D). Ultimately, these results translated to post-injection changes in T2 relaxation times in tumors, but not normal muscle, by veiled nanoparticles administered to xenograft mice (Figure 3E).
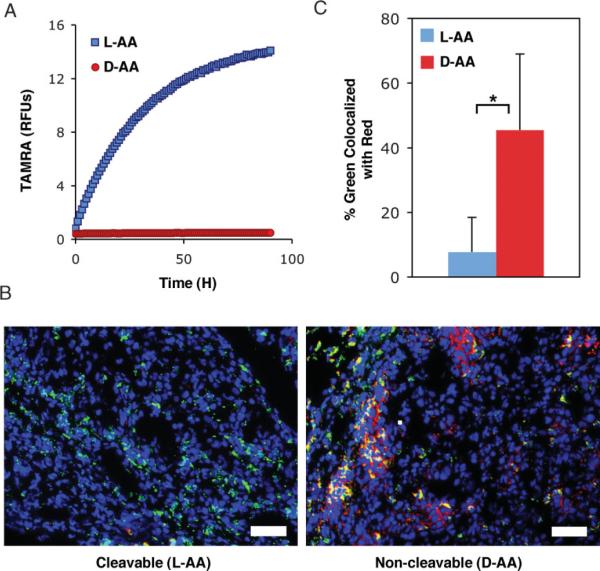
While these results generally exhibit the ability of veiled nanoparticles to passively accumulate in tumors and enable both fluorescent and magnetic-resonance tumor imaging, we aimed to demonstrate that cleavable particles are unveiled by endogenous MMP expression in tumor xenografts. MMP activation of cleavable (L-AA) particles by MMP-2 occurs rapidly with the majority of the polymer removed within an hour when incubated with enzyme in solution at a concentration even lower than what is estimated to be in xenograft tumors (63 μM vs. 1 μM;[19] Supplementary Figure 5). Monitoring the removal and subsequent dequenching of TAMRA fluorophores attached to the leaving peptide-PEG shows cleavable (L-AA) particles to be activated by MMPs, while non-cleavable (D-AA) particles are not (Figure 4A). This removal of TAMRA-labeled peptide-PEG from the particle by MMP activation can be used as a sensor of protease unveiling in vivo. To measure this activation we performed colocalization analysis on fluorescent micrographs of peptide-PEG (TAMRA-labeled) and nanoparticles (VT-680-labeled) from histological sections of tumors harvested 48 h after injection (Figure 4B). A pixel-by-pixel analysis of histological sections from tumors measuring the percentage of particles (green pixels) that colocalized with the polymer coating (red pixels) shows that the association of particles with their removable coating is reduced nearly sixfold for cleavable particles, indicating the cleavable particle is in fact activated in the tumor (Figure 4C).
Figure 4.
Removable polymer coatings veil nanoparticles in the blood, but are effectively released in tumors. A) Monitoring the release of TAMRA-iron quenching interactions shows that particles with cleavable L-isomer peptides (L-AA) are activated by MMPs, while particles with non-cleavable D-isomer peptides (D-AA) remain intact. B) Representative RGB merge of nanoparticles (green), removable polymer (red), and nuclei (blue) in tumor sections harvested 48 h after injection shows a loss in signal from the removable polymer with cleavable peptides, but not non-cleavable controls. Scale bar is 250 μm. C) The percentage of particles colocalized with the removable polymer in tumor sections harvested 48 h after injection is significantly reduced for the cleavable (L-AA) particle.
The removal of the polymer in the tumor highlights a key feature of this system, the implicitly engineered spatial regulation of ligand function: particles remain in a functionally latent state in the vascular space (enabling improved circulation times and tissue accumulation) until they extravasate and are activated by cancer-associated proteases in the tumor microenvironment (enabling improved site-specific functionality). In this paper we have built on previous work in which the entropic penalty of PEG coatings was used to veil and unveil ligands mediating particleparticle interactions by extending this strategy to veil and unveil ligands mediating particle-cell interactions. Additionally we have shown that removable polymer coatings confer favorable tumor targeting properties in vivo. In the future, the incorporation of core particles carrying drug or nucleic acid cargo and bioactive domains mediating cell signaling or cellspecific binding in this strategy could provide a cytotoxic response to protease unveiling and ultimately lead to a multifunctional platform for improved imaging and therapy.
Experimental Section
Synthesis of nanoparticles
The nanoparticles used in these experiments were synthesized, cross-linked, aminated, and labeled with a near-infrared fluorophore (VivoTag 680) according to published protocols.[31] To conjugate species onto nanoparticles, surface amines were functionalized with SIA (Nsuccinimidyl iodoacetate) to make them thiol reactive. A fluoroscein isothiocyanate (FITC)-labeled polyarginine cell internalizing peptide, NH2-RRRRGRRRRK(FITC)GC, and a TAMRAlabeled protease-cleavable PEG, prepared by coupling the amine terminus of an MMP-2 cleavable peptide substrate, NH2-GK(TAMRA)GPLGVRGC, to 10 kDa NHS-PEG[18], were then linked to nanoparticles via thiol groups on the cysteine residues at the carboxyl termini. A more detailed protocol is available in the Supporting Information.
Flow cytometry
HT-1080 human fibrosarcoma cells (ATCC) were cultured in 24-well plates and grown to 80% confluency using ATCC recommended media. Veiled and MMP pre-cleaved nanoparticles (100 μL at 0.1 mg/mL Fe) were added to 400 μL cell culture media with 25 μM Galardin and incubated over cells for 1 h. Adherent cells were detached from the tissue culture plate with 0.25% trypsin, washed in phosphate buffered saline (PBS), and analyzed on a Becton Dickin LSR II using a 633 nm excitation source and a 690/40 band pass filter to detect VT-680 labeled nanoparticles in cells.
MMP activation
Unless otherwise stated, pre-cleaved (unveiled) particles were prepared by incubating nanoparticles with 20 μg/mL collagenase (Clostridiopeptidase A) in 0.1 M HEPES 0.15 M NaCl pH 7.2 (HEPES buffer) with 5 mM CaCl2 for 3 h at room temperature. Activation was monitored by the decrease of TAMRA quenching at an excitation of 515 nm and emission of 580 nm. Addition of 25 μM of the broad-spectrum MMP inhibitor (Galardin) prevented cleavage of peptide-PEGs as monitored by dequenching (Supplementary Figure 4).
Kcat/Km determination
Cloaked nanoparticles (0.05 mg/mL Fe) coated with 2.9 μmM of peptide-PEG substrate in HEPES buffer with 5 mM CaCl2 were incubated with recombinant MMP-2 (0.724 μmg/mL) at room temperature and monitored fluorometrically to assess activation. The Vmax of fluorescence release of particles at this concentration was linearly related to that of particles at concentrations half and twofold as much, indicating that the substrate concentration [S] was much less than the binding constant Km in this experimental setup. Activation experiments were quenched by the addition of 0.1 M ethylenediaminetetraacetic acid (EDTA) at 1:9 v/v. Particles were ultracentrifuged and the supernatant collected to measure product formation. Similarly free peptide ([S] = 15.45 μM) in HEPES buffer +5mM CaCl2 was incubated with recombinant MMP-2 (0.3367 μg/mL). Activation was quenched by the addition of 0.1 M EDTA at a 1:9 v/v and cleavage was monitored using a fluorescamine assay. The Vmax of substrate cleavage during the first 30 min for substrate concentrations of 15.45 μM and 7.75 μM were linearly related confirming that the experiment was operating in a range of [S] much less than Km. The reaction was driven to completion over 24 h and the change in fluorescamine signal at various time points was used to determine the substrate concentration.
Multimodal imaging in agarose wells
A 5% agarose solution in water was boiled and then cooled in a cell culture dish containing well molds from centrifuge tubes. Each well was filled with 8 million cells from a 40% confluent T-150 flask. HT-1080 cells in these flasks were incubated with nanoparticles (1 μg/mL Fe) in Dulbecco's Modified Eagle Medium (DMEM) with serum media for various times. Particles were removed after incubation and cells were trypsinized, washed in PBS, fixed overnight in 50 μl of PBS with 4% paraformaldehyde, and transferred to agarose wells for imaging. MRI images were taken on a Bruker 4.7 T magnet, 7 cm core. A series of 32 images with multiples of 15 ms echo times and a TR of 3000 ms were acquired. T2 maps were obtained for each well using the T2 fit map plug-in in OsiriX imaging software. A fluorescence scan through the wells was acquired on an Odyssey Infrared System (Licor) using the 700-emission channel to detect VT-680 labeled particles.
Xenograft animals
Nude mice were injected s.c. bilaterally in the hind flank with 2 × 106 HT-1080 cells. After 1-2 weeks animals were anaesthetized with isoflurane and injected through the tail vein with nanoparticles (4-10 mg/kg Fe). Animals were imaged before and 24 h after intravenous injection of nanoparticles (10 mg/Kg Fe) on a 4.7 T Bruker magnet. A series of 16 images with multiples of 8.6 ms echo times and a TR of 2133.3 ms was acquired. T2 maps were obtained for regions of interest using the T2 fit map plug-in in OsiriX. At 48 h animals were imaged by a fluorescence molecular tomography (FMT) imaging system (Visen Medical). Quantitative analysis of relative nanoparticle uptake in tumors by FMT was assessed by selecting regions of interest around tumor masses 4-6 mm in diameter. Quantitative measurements on dye concentrations were normalized by the total injected dose in each animal to yield relative fluorescent units (RFUs). Blood half-lives were determined by the decrease in particle fluorescence (VT-680) of 25 μL blood samples withdrawn periodically into heparinized capillary tubes from the sub-orbital space. Withdrawn blood was diluted with 50 μL of PBS containing 10 mM EDTA, centrifuged for 1 min at 1000 rpm to remove RBCs, and read on an near-IR scanner (LICOR, 700 channel). Animals were euthanized by cervical dislocation under anesthesia and tumors were harvested, embedded in OCT, and stored at -80°C for cryosectioning. Samples were cut into 5 mm sections using a cryotome and fixed in cold acetone for staining and imaging.
Colocalization analysis
Histological sections were labeled with an anti-TAMRA primary antibody (AbD Serotec) and an Alexa-750 secondary antibody (Invitrogen) to confirm the presence of TAMRA-labeled polymer on nanoparticles in the tumor tissue. Twelve image fields from three different tumor specimens were acquired for animals injected with cleavable (L-AA) and noncleavable (D-AA) nanoparticles. To cancel the background signal from noise and non-specific antibody binding, the cumulative distribution of pixel intensity data from all analyzed fields was generated for VT-680 and TAMRA antibody channels, and a value determined from the inflection point was subtracted from all images. The number of non-zero pixels in the particle fluorescence channel that colocalized with non-zero signal from the TAMRA channel were counted and divided by the total number of particle pixels to obtain a percentage of colocalized particles for each image from 23 images.
Supplementary Material
Acknowledgments
This work was supported by NIH (BRP: R01CA124427-01), NIH/NCI (U54 CA119349, U54 CA119335), Packard Fellowship (1999-1453). T. J. H. acknowledges support from the NIH-NIBIB (EB 006324). G.v.M. acknowledges support from the Whitaker Foundation and the National Science Foundation. M.E.L. acknowledges support from the Paul E. Gray Award Fund. We thank Dr. Ralph Weissleder and the CMIR for assistance with MRI and FMT.
Footnotes
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
References
- [1].Moghimi SM, Hunter AC, Murray JC. Pharmacol. Rev. 2001;53:283. [PubMed] [Google Scholar]
- [2].Weissleder R, Bogdanov A, Neuwelt EA, Papisov M. Adv. Drug Delivery Rev. 1995;16:321. [Google Scholar]
- [3].Alexander S. J. Phys. 1977;38:983. [Google Scholar]
- [4].Degennes PG. Macromolecules. 1980;13:1069. [Google Scholar]
- [5].Storm G, Belliot SO, Daemen T, Lasic DD. Adv. Drug Delivery Rev. 1995;17:31. [Google Scholar]
- [6].Zalipsky S, Qazen M, Walker JA, Quinn YP, Huang SK. Bioconjugate Chem. 1999;10:703. doi: 10.1021/bc990031n. [DOI] [PubMed] [Google Scholar]
- [7].Kale AA, Torchilin VP. J. Liposome Res. 2007;17:197. doi: 10.1080/08982100701525035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee ES, Na K, Bae YH. Nano Lett. 2005;5:325. doi: 10.1021/nl0479987. [DOI] [PubMed] [Google Scholar]
- [9].Torchilin VP. Adv. Drug Delivery Rev. 2006;58:1532. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- [10].Boomer JA, Thompson DH. Chem. Phys. Lipids. 1999;99:145. doi: 10.1016/s0009-3084(99)00033-x. [DOI] [PubMed] [Google Scholar]
- [11].Guo X, Szoka FC. Bioconjugate Chem. 2001;12:291. doi: 10.1021/bc000110v. [DOI] [PubMed] [Google Scholar]
- [12].Kratz F, Beyer U, Schutte MT. Crit. Rev. Ther. Drug Carrier Syst. 1999;16:245. doi: 10.1615/critrevtherdrugcarriersyst.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- [13].Roux E, Passirani C, Scheffold S, Benoit JP, Leroux JC. J. Controlled Release. 2004;94:447. doi: 10.1016/j.jconrel.2003.10.024. [DOI] [PubMed] [Google Scholar]
- [14].Simoes S, Moreira JN, Fonseca C, Duzgunes N, Lima M. C. P. de. Adv. Drug Delivery Rev. 2004;56:947. doi: 10.1016/j.addr.2003.10.038. [DOI] [PubMed] [Google Scholar]
- [15].Turk MJ, Reddy JA, Chmielewski JA, Low PS. Biochim. Biophys. Acta-Biomembranes. 2002;1559:56. doi: 10.1016/s0005-2736(01)00441-2. [DOI] [PubMed] [Google Scholar]
- [16].Bremer C, Tung CH, Weissleder R. Nat. Medicine. 2001;7:743. doi: 10.1038/89126. [DOI] [PubMed] [Google Scholar]
- [17].Harris TJ, Maltzahn G. von, Derfus AM, Ruoslahti E, Bhatia SN. Angew. Chem. Int. 2006;45:3161. doi: 10.1002/anie.200600259. [DOI] [PubMed] [Google Scholar]
- [18].Maltzahn G. von, Harris TJ, Park J-H, Min DH, Schmidt AJ, Sailor MJ, Bhatia SN. J. Am. Chem. Soc. 2007;129:6064. doi: 10.1021/ja070461l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hatakeyama H, Akita H, Kogure K, Oishi M, Nagasaki Y, Kihira Y, Ueno M, Kobayashi H, Kikuchi H, Harashima H. Gene Therapy. 2007;14:68. doi: 10.1038/sj.gt.3302843. [DOI] [PubMed] [Google Scholar]
- [20].Zhang JX, Zalipsky S, Mullah N, Pechar M, Allen TM. Pharmacol. Res. 2004;49:185. doi: 10.1016/j.phrs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- [21].Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc. Nat. Acad. Sci. U. S. A. 2004;101:17867. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang Y, So MK, Rao JH. Nano Lett. 2006;6:1988. doi: 10.1021/nl0611586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davidson B, Goldberg I, Kopolovic J, Lerner-Geva L, Gotlieb WH, Ben-Baruch G, Reich R. Gynecol. Oncol. 1999;73:372. doi: 10.1006/gyno.1999.5381. [DOI] [PubMed] [Google Scholar]
- [24].Fang JM, Shing Y, Wiederschain D, Yan L, Butterfield C, Jackson G, Harper J, Tamvakopoulos G, Moses MA. Proc. Nat. Acad. Sci. U. S. A. 2000;97:3884. doi: 10.1073/pnas.97.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Giannelli G, Marzillier J. Falk, Schiraldi O, Stetler Stevenson WG, Quaranta V. Science. 1997;277:225. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- [26].Stearns ME, Wang M. Cancer Res. 1993;53:878. [PubMed] [Google Scholar]
- [27].Chau Y, Tan FE, Langer R. Bioconjugate Chemistry. 2004;15:931. doi: 10.1021/bc0499174. [DOI] [PubMed] [Google Scholar]
- [28].Gabizon A, Shmeeda H, Horowitz AT, Zalipsky S. Adv. Drug Delivery Rev. 2004;56:1177. doi: 10.1016/j.addr.2004.01.011. [DOI] [PubMed] [Google Scholar]
- [29].Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Proc. Nat. Acad. Sci. U. S. A. 2007;104:15549. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kirpotin DB, Drummond DC, Shalaby MR, Hong KL, Nielsen UB, Marks JD, Benz CC, Park JW. Cancer Res. 2006;66:6732. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- [31].Josephson L, Tung CH, Moore A, Weissleder R. Bioconjugate Chem. 1999;10:186. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.