Abstract
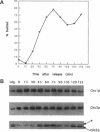
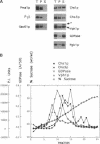
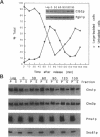
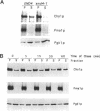
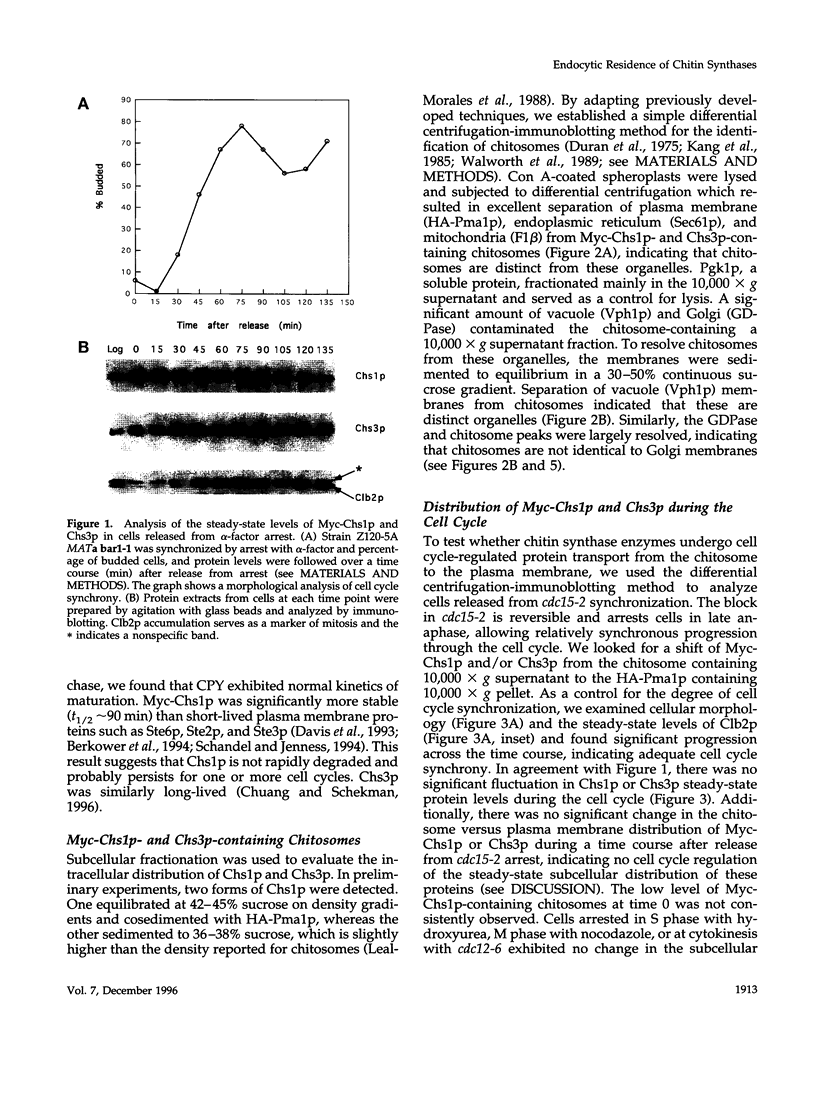
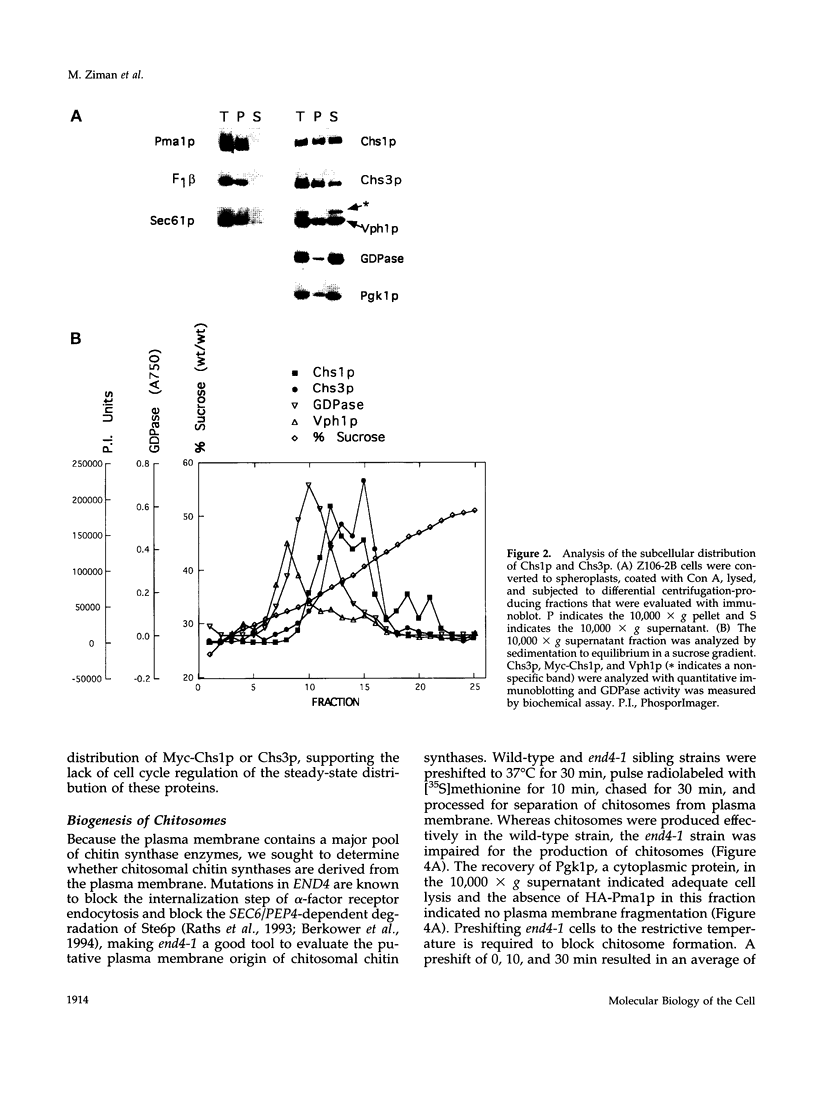
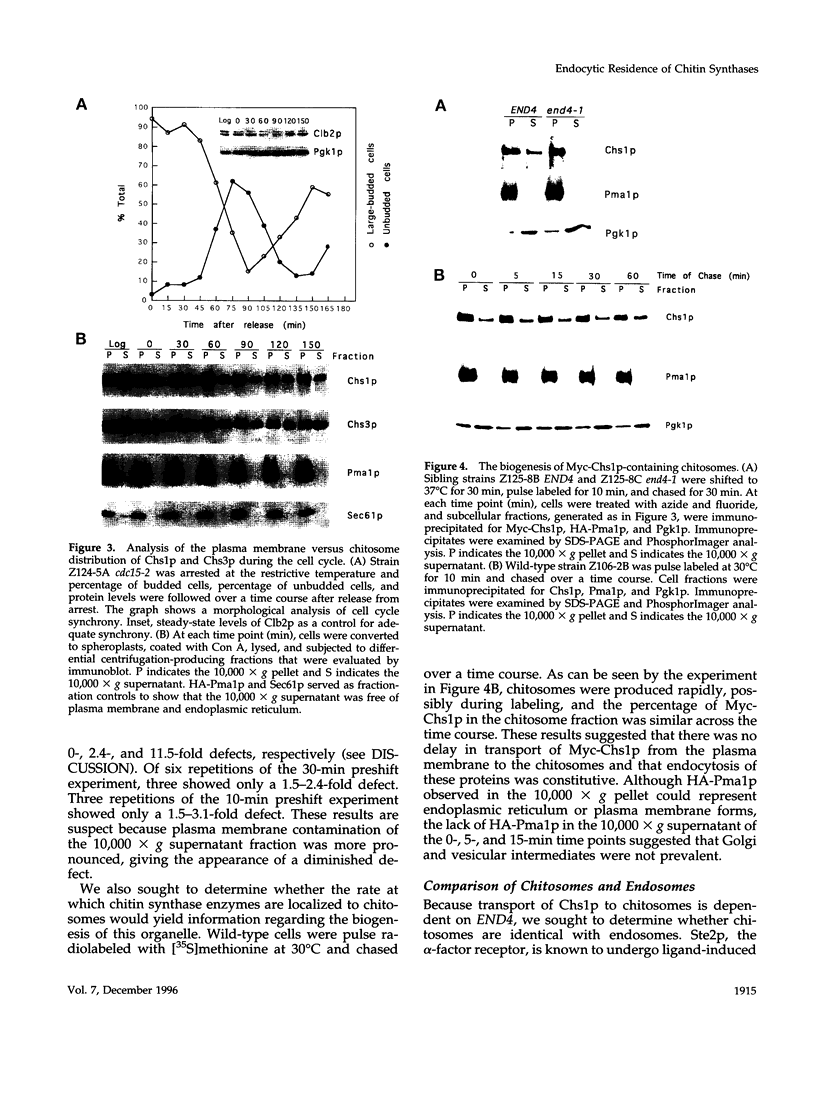
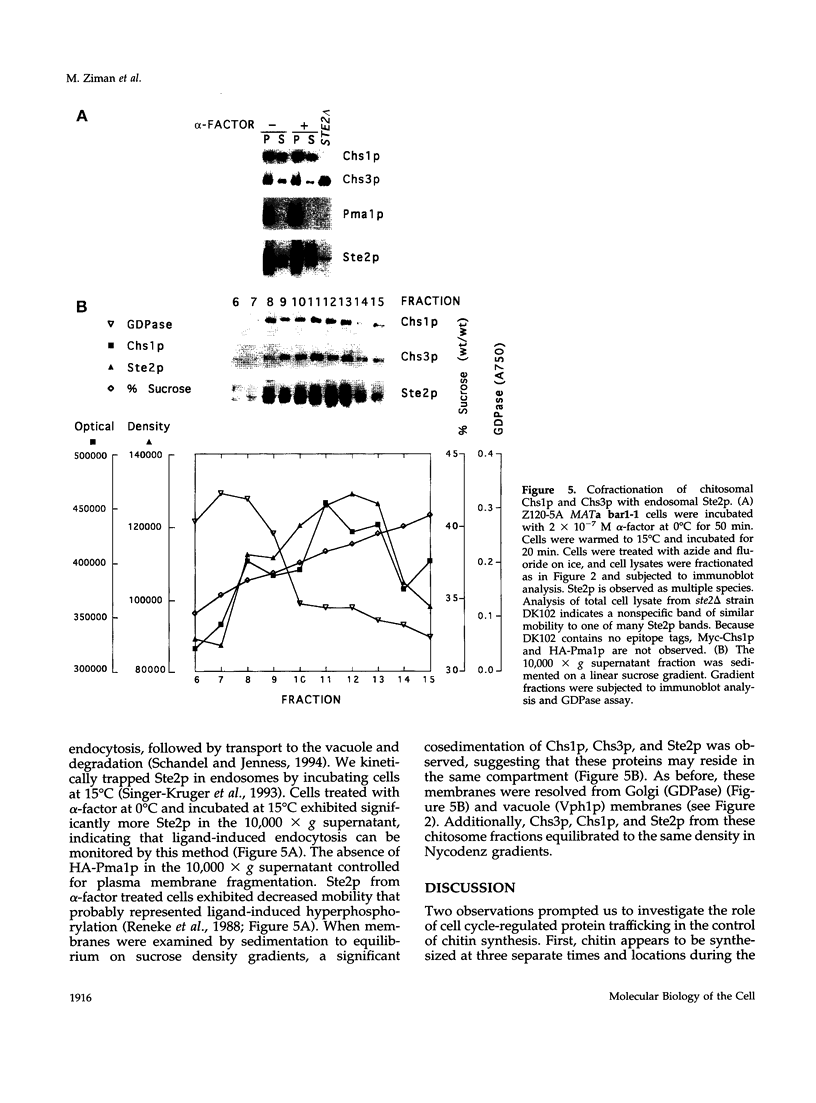
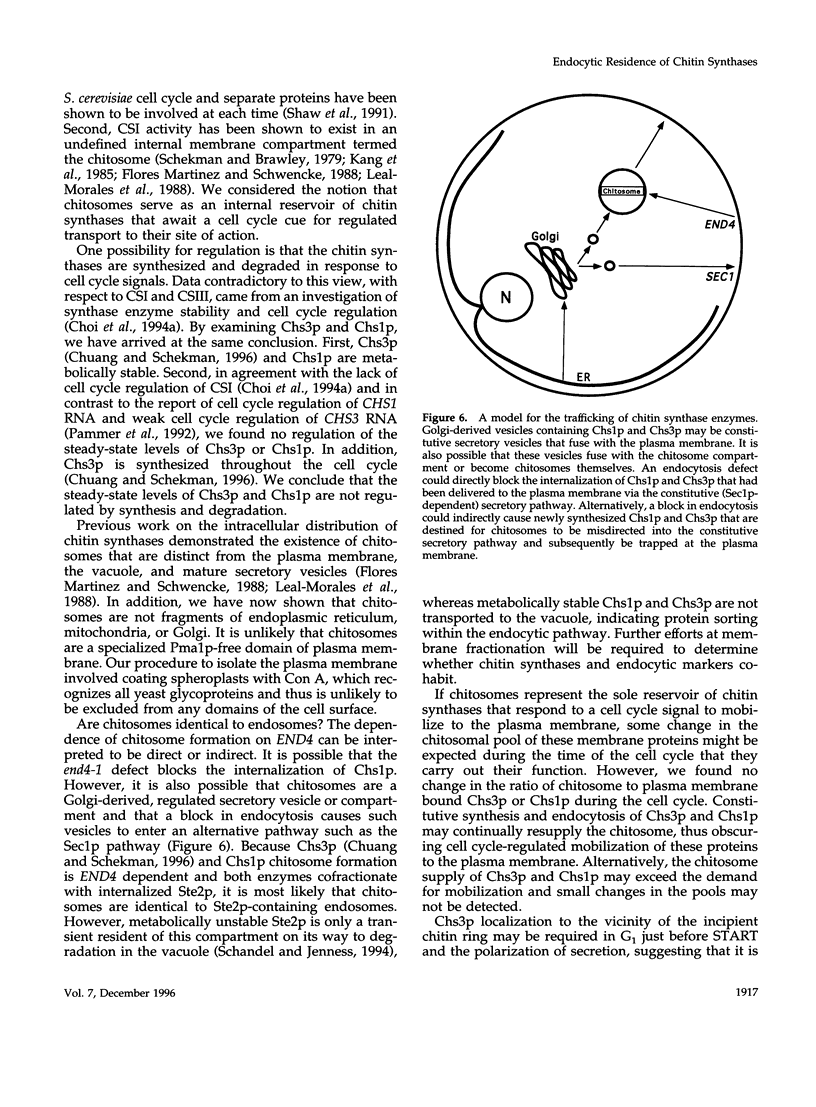
In Saccharomyces cerevisiae, the synthesis of chitin, a cell-wall polysaccharide, is temporally and spatially regulated with respect to the cell cycle and morphogenesis. Using immunological reagents, we found that steady-state levels of Chs1p and Chs3p, two chitin synthase enzymes, did not fluctuate during the cell cycle, indicating that they are not simply regulated by synthesis and degradation. Previous cell fractionation studies demonstrated that chitin synthase I activity (CSI) exists in a plasma membrane form and in intracellular membrane-bound particles called chitosomes. Chitosomes were proposed to act as a reservoir for regulated transport of chitin synthase enzymes to the division septum. We found that Chs1p and Chs3p resided partly in chitosomes and that this distribution was not cell cycle regulated. Pulse-chase cell fractionation experiments showed that chitosome production was blocked in an endocytosis mutant (end4-1), indicating that endocytosis is required for the formation or maintenance of chitosomes. Additionally, Ste2p, internalized by ligand-induced endocytosis, cofractionated with chitosomes, suggesting that these membrane proteins populate the same endosomal compartment. However, in contrast to Ste2p, Chs1p and Chs3p were not rapidly degraded, thus raising the possibility that the temporal and spatial regulation of chitin synthesis is mediated by the mobilization of an endosomal pool of chitin synthase enzymes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeijon C., Orlean P., Robbins P. W., Hirschberg C. B. Topography of glycosylation in yeast: characterization of GDPmannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appeltauer U., Achstetter T. Hormone-induced expression of the CHS1 gene from Saccharomyces cerevisiae. Eur J Biochem. 1989 Apr 15;181(1):243–247. doi: 10.1111/j.1432-1033.1989.tb14718.x. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- Baum P., Thorner J., Honig L. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4962–4966. doi: 10.1073/pnas.75.10.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C., Loayza D., Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994 Nov;5(11):1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol. 1992 Apr;12(4):1764–1776. doi: 10.1128/mcb.12.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- Bulawa C. E., Osmond B. C. Chitin synthase I and chitin synthase II are not required for chitin synthesis in vivo in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7424–7428. doi: 10.1073/pnas.87.19.7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., Slater M., Cabib E., Au-Young J., Sburlati A., Adair W. L., Jr, Robbins P. W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986 Jul 18;46(2):213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Cabib E., Bowers B. Chitin and yeast budding. Localization of chitin in yeast bud scars. J Biol Chem. 1971 Jan 10;246(1):152–159. [PubMed] [Google Scholar]
- Cabib E., Sburlati A., Bowers B., Silverman S. J. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1989 May;108(5):1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Silverman S. J., Shaw J. A. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J Gen Microbiol. 1992 Jan;138(1):97–102. doi: 10.1099/00221287-138-1-97. [DOI] [PubMed] [Google Scholar]
- Choi W. J., Cabib E. The use of divalent cations and pH for the determination of specific yeast chitin synthetases. Anal Biochem. 1994 Jun;219(2):368–372. doi: 10.1006/abio.1994.1278. [DOI] [PubMed] [Google Scholar]
- Choi W. J., Santos B., Durán A., Cabib E. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol Cell Biol. 1994 Dec;14(12):7685–7694. doi: 10.1128/mcb.14.12.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. J., Sburlati A., Cabib E. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4727–4730. doi: 10.1073/pnas.91.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid V. J., Durán A., del Rey F., Snyder M. P., Nombela C., Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995 Sep;59(3):345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N. G., Horecka J. L., Sprague G. F., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993 Jul;122(1):53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán A., Bowers B., Cabib E. Chitin synthetase zymogen is attached to the yeast plasma membrane. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3952–3955. doi: 10.1073/pnas.72.10.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M. P., Correa J. U., Cabib E. Activation of chitin synthetase in permeabilized cells of a Saccharomyces cerevisiae mutant lacking proteinase B. J Bacteriol. 1982 Dec;152(3):1255–1264. doi: 10.1128/jb.152.3.1255-1264.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores Martinez A., Schwencke J. Chitin synthetase activity is bound to chitosomes and to the plasma membrane in protoplasts of Saccharomyces cerevisiae. Biochim Biophys Acta. 1988 Dec 22;946(2):328–336. doi: 10.1016/0005-2736(88)90408-7. [DOI] [PubMed] [Google Scholar]
- Grandin N., Reed S. I. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993 Apr;13(4):2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. L., Na S., Zhu X., Seto-Young D., Perlin D. S., Teem J. H., Haber J. E. Dominant lethal mutations in the plasma membrane H(+)-ATPase gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10531–10535. doi: 10.1073/pnas.91.22.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. J., Stevens T. H. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol Biol Cell. 1994 Sep;5(9):1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. S., Au-Young J., Cabib E. Modification of yeast plasma membrane density by concanavalin A attachment. Application to study of chitin synthetase distribution. J Biol Chem. 1985 Oct 15;260(23):12680–12684. [PubMed] [Google Scholar]
- Leal-Morales C. A., Bracker C. E., Bartnicki-Garcia S. Localization of chitin synthetase in cell-free homogenates of Saccharomyces cerevisiae: chitosomes and plasma membrane. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8516–8520. doi: 10.1073/pnas.85.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993 Mar;120(6):1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccecchini M. L., Rudin Y., Blobel G., Schatz G. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc Natl Acad Sci U S A. 1979 Jan;76(1):343–347. doi: 10.1073/pnas.76.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammer M., Briza P., Ellinger A., Schuster T., Stucka R., Feldmann H., Breitenbach M. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast. 1992 Dec;8(12):1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- Raths S., Rohrer J., Crausaz F., Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993 Jan;120(1):55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneke J. E., Blumer K. J., Courchesne W. E., Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988 Oct 21;55(2):221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Bartnicki-Garcia S. Synthesis of cell wall microfibrils in vitro by a "soluble" chitin synthetase from Mucor rouxii. Science. 1974 Oct 25;186(4161):357–359. doi: 10.1126/science.186.4161.357. [DOI] [PubMed] [Google Scholar]
- Sburlati A., Cabib E. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J Biol Chem. 1986 Nov 15;261(32):15147–15152. [PubMed] [Google Scholar]
- Schandel K. A., Jenness D. D. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994 Nov;14(11):7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Brawley V. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc Natl Acad Sci U S A. 1979 Feb;76(2):645–649. doi: 10.1073/pnas.76.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Durán A., Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991 Jul;114(1):111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S. H., Chrétien P., Bastien L., Slilaty S. N. Primary sequence of the glucanase gene from Oerskovia xanthineolytica. Expression and purification of the enzyme from Escherichia coli. J Biol Chem. 1991 Jan 15;266(2):1058–1063. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J., Sburlati A., Slater M. L., Cabib E. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4735–4739. doi: 10.1073/pnas.85.13.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J., Shaw J. A., Cabib E. Proteinase B is, indeed, not required for chitin synthetase 1 function in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991 Jan 15;174(1):204–210. doi: 10.1016/0006-291x(91)90506-3. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger B., Frank R., Crausaz F., Riezman H. Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J Biol Chem. 1993 Jul 5;268(19):14376–14386. [PubMed] [Google Scholar]
- Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992 Feb;3(2):129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso M. H., Mol P. C., Shaw J. A., Cabib E., Durán A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J Cell Biol. 1991 Jul;114(1):101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N. C., Goud B., Ruohola H., Novick P. J. Fractionation of yeast organelles. Methods Cell Biol. 1989;31:335–356. doi: 10.1016/s0091-679x(08)61618-0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Wuestehube L. J., Schekman R. W. Reconstitution of transport from endoplasmic reticulum to Golgi complex using endoplasmic reticulum-enriched membrane fraction from yeast. Methods Enzymol. 1992;219:124–136. doi: 10.1016/0076-6879(92)19015-x. [DOI] [PubMed] [Google Scholar]