Abstract
The basic helix-loop-helix transcription factors are present in animals, plants and fungi and play important roles in the control of cellular proliferation, tissue differentiation, development and detoxification. Although insect genomes contain more than 50 Helix-Loop-Helix transcription factors, the functions of only a few are known. RNAi has become a widely used tool to knockdown the expression to analyze the function of genes. As RNAi works well in Tribolium castaneum, we utilized this insect and RNAi to determine functions of 19 bHLH transcription factors belonging to PAS and HES families during the larval stages of the red flour beetle, T. castaneum. We searched the genome sequence of T. castaneum and identified 53 bHLH genes. Phylogenetic analyses classified these 53 genes into ten families; PAS, HES, Myc/USF, Hand, Mesp, Shout, p48, NeuroD/Neurogenin, Atonal and AS-C. In RNAi studies, knocking-down the expression of seven members of the PAS and HES families affected the growth and development of T. castaneum. An inability to grow to reach critical weight to undergo metamorphosis, failure to complete larval-pupal or pupal-adult ecdysis and abnormal wing development are among the most common phenotypes observed in RNAi insects. Among the bHLH transcription factors studied, the steroid receptor coactivator (SRC) showed the most severe phenotypes. Knock-down in the expression of the gene coding for SRC caused growth arrest by affecting the regulation of lipid metabolism. These studies demonstrate the power of RNAi for functional characterization of members of the multigene families in this model insect.
Keywords: Red flour beetle, bHLH transcription factors, RNA interference, Steroid receptor coactivator
1. Introduction
A superfamily of transcription factors containing basic Helix-Loop-Helix (bHLH) domain play important roles in the control of cell proliferation, determination and tissue differentiation during the development of animals and plants (Jan and Jan, 1993; Weintraub, 1993; Hassen and Bellen, 2000). The bHLH motif was first identified by Murre and colleagues (Murre et al., 1989a) in two murine transcription factors E12 and E47. The bHLH domain is approximately 60 amino acids in length and comprises a DNA- binding basic region of 15 amino acids followed by two α- helices separated by a variable loop region (Ferre-D’ Amare et al., 1993). Besides binding to DNA, the bHLH domain also promotes dimerization, allowing the formation of homodimer or heterodimer complexes (Murre et al., 1989b; Kadesh, 1993).
Several bHLH genes have been identified in various organisms, whose genome sequences are available. These include eight bHLH genes in yeast, 39 in Caenorhabditis elegans, 39 in Gallus gallus, 39 in Brachydanio rerio, 46 in Ciona intestinalis, 47 in Xenopus laevis, 59 in Drosophila melanogaster, 50 in Tribolium castaneum, 87 in Lagocephalus lagocephalus, 102 in Mus musculus, 118 in Homo sapiens, 167 in Oryza sativa and 147 in Arabidopsis (Ledent et al., 2002; Satou et al., 2003; Simionato et al., 2007; Li et al., 2006; Toledo-ortiz et al., 2002). Recently, in Silk worm, Bombyx mori 52 bHLH genes have been identified (Wang et al., 2007). The members of the bHLH superfamily have been classified into various families as defined by bHLH- Per, Arnt, Sim (PAS) domain, Hairy-Enhancer of split (HES), Myc/Upstream Transcription Factor (USF), Atonal, Mesp, Hand, p48, Shout and Achaete- scute (AS-C) (Moore et al., 2000). Previous studies had identified bHLH genes from different organisms including both vertebrates and invertebrates. A phylogenetic analysis based on a sample of 122 bHLH sequences from animals, plants and fungi has lead to classifying them into four monophyletic groups of proteins named A, B, C, and D (Atchley and Fitch, 1997). Phylogenetic analysis has classified 52 bHLH genes in B. mori into 39 bHLH families and six groups (Wang et al., 2007). We searched recently completed red flour beetle, T. castaneum genome sequence deposited into the National Center for Biotechnology Information (NCBI) and Human Genome Sequencing Center (HGSC) databases (Tribolium Genome Sequencing Consortium, 2008) and identified 53 bHLH genes. Phylogenetic analysis classified these 53 proteins into ten families.
RNA-aided gene silencing is a highly conserved cellular mechanism in eukaryotic organisms (Fire et al., 1998; Meister and Tuschl, 2004; Mello and Conte, 2004). It has become a widely used tool to knock-down and analyze the function of genes. Systemic RNAi has been first described in plants as a type of post-transcriptional gene silencing (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998). There are many reports on identification and classification of bHLH superfamily members, but there are very few reports on functional characterization of members of this family. Since the systemic RNAi works very well in T. castaneum (Tomoyasu and Denell, 2004; Bucher et al., 2002), the functional characterization of the members of two important families, PAS and HES, was performed in this model insect. RNAi analyses showed that knocking-down the expression of seven genes coding for bHLH-transcription factors belonging to PAS and HES families affected the growth and development during larval and pupal stages of T. castaneum. Growth arrest, failure to undergo ecdysis from larval to pupal or pupal to adult stages and abnormal wing phenotypes are among the most common phenotypes observed in RNAi insects.
2. Materials and methods
2.1. Tree building and sequence alignment
Full length bHLH protein sequences of the T. castaneum and one closest homologue from Drosophila melanogaster, Apis mellifera or Gallus gallus were aligned in MAFFT (Ahola et al., 2006) with the L-INS-i iterative refinement option on the MAFFT server (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/). MAFFT with the L-INS-i option has shown to be the most accurate and consistent method for aligning protein sequences (Atlekar et al., 2004; Golubchik et al., 2007). Additional alignment parameters included the scoring matrix set to BLOSUM62, gap opening penalty to 1.53, and the offset value to 0.00. The aligned protein sequences were converted to a nexus file in BioEdit version 7.0.9.0 (Hall, 1999). The nexus file was analyzed under Bayesian optimality criterion using MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001; Katoh et al., 2005) run in parallel (Ronquist and Huelsenbeck, 2003) on a quad core Mac Pro running Mac OS X version 10.4.10. Two independent runs, each containing four chains, were executed using a mixture of amino acid models (aamodelpr=mixed) over 10,000,000 generations sampling every 100 generations with the outgroup set to gene HLH106. Upon completion of the analysis, probabilities were checked for stationarity across all parameters using Tracer version 1.4 (http://tree.bio.ed.ac.uk/software/tracer/). Burnin was set at 2,500,000 generations and probabilities and trees were summarized.
2.2. Rearing and staging of larvae
GA-1 strain of T. castaneum was used for all the experiments. Beetles were reared in organic whole wheat flour containing 10% yeast at 30°C. Newly ecdysed final instar larvae were staged based on the length and untanned white cuticle. Fast moving, smaller and slender larvae were used for the penultimate instar injections.
2.3. RNA isolation and cDNA synthesis
Total RNA was isolated from 5-6 larvae for each sample at each time point using the TRI reagent (Molecular Research Center Inc., Cincinnati, OH). The RNA was treated with DNase l (Ambion Inc., Austin, TX) and cDNA was synthesized using cDNA synthesis kit (Bio- Rad Laboratories, Hercules, CA) in a 20μl reaction volume.
2.4. Quantitative real-time reverse transcriptase PCR (qRT-PCR)
The relative mRNA expression of bHLH-PAS and HES family genes and knock-down levels of these genes after dsRNA injections were determined using MyiQ single color real-time polymerase chain reaction (PCR) detection system (Bio-Rad Laboratories, Hercules, CA). Primers designed based on bHLH-PAS and HES family genes (Supplementary, Table 1) were used to quantify the mRNA levels. Knock-down levels of the genes involved in the insulin signaling and Target of Rapamycin (TOR) pathways after the TcSRC dsRNA injection was also determined by using qRT-PCR. qRT-PCR reactions were performed using a common program as follows: initial incubation of 95°C for 3 min followed by 40 cycles of 95°C for 10s, 60°C for 20s, 72°C for 30s. Standard curves were obtained using a ten-fold serial dilution of pooled cDNA. Quantitative mRNA measurements were performed in triplicate and normalized to an internal control of T. castaneum ribosomal protein rp49 mRNA.
2.5. Double-stranded RNA synthesis
Genomic DNA was extracted from the T. castaneum adults using DNeasy Tissue Kit (Qiagen). Two μl of genomic DNA was used as a template in a 50 μl PCR. Primers containing the bHLH gene sequences and T7 Polymerase Promoter at the 5’ end of both the forward primer and reverse primer were used to amplify the 150-550 bp regions (Supplementary, Table 2). Eight microliters of PCR product was used as a template to synthesize the dsRNA using MEGA script RNAi kit (Ambion Inc., Austin, TX) as per the instruction manual. After the DNase and RNase treatment purification of dsRNA was done by using Phenol/ chloroform extraction followed by ethanol precipitation and finally the pellet was dried and dissolved in 15 μl of Nuclease- free water. The quality of dsRNA was checked by running an aliquot on an agarose gel. The concentration of dsRNA was determined using Nanodrop (NanoDrop Products, Wilmington, DE).
2.6. Microinjection of larvae
Larvae were anaesthetized using ether vapors for 6 min, and then placed in a line on double-sided sticky tape over a glass slide. The bHLH gene dsRNA was diluted to a concentration of 5 μg/ μl and 0.2 μl of the dsRNA was injected into each larva on the dorsal side near the first or second abdominal segments. Needles used for microinjections were pulled using a needle puller (Model P-2000 Sutter Instruments co.) from a 3.5 inch glass capillary tube (Drummond). DsRNA prepared using 800 bp bacterial malE gene was used as a negative control. The injected larvae were allowed to recover for 1 hr and transferred to the diet and reared under standard conditions.
2.7. Recording of Larval weights
TcSRC and malE dsRNAs were injected into the newly ecdysed final instar larvae. Larvae were weighed from third day after the injection of dsRNA. Larval weights were recorded 3rd, 4th and 5th day after dsRNA injection. Most of the malE dsRNA injected larvae became pupae by 6th day after dsRNA injection.
2.8. Measurement of nutrient levels
The amount of total proteins was estimated using the Bradford assay. A 1 μg/μl solution of bovine serum albumin (BSA) was used as the standard, from which 0-20 μg calibration series were prepared. One final instar larvae was placed in each centrifuge tube and crushed with a homogenizer in 200 μl of phosphate buffered saline (PBS) solution. Each sample was sonicated for 30 sec and 2 μl of each sample, 800 μl of water and 200 μl of Bradford reagent were added and mixed in a new tube. The optical densities (OD) were read at 625 nm.
Total amount of carbohydrates was determined using an Anthrone based method (Van Handel, 1985). A 1 μg/μl solution of glycogen was used as the standard, from which 0-200 μg calibration series were prepared. One final instar larvae was placed in each tube and crushed with a homogenizer in 1 ml of Anthrone reagent. Standards and samples were heated at 92 ° C for 17 min. The samples were allowed to cool down to room temperature and OD was measured at 625 nm.
The amount of total lipids was estimated using Vanillin reagent (Van Handel, 1985) method. A 1 μg/μl solution of commercial vegetable oil was used as a standard by preparing 0-400 μg calibration series. One larva was placed in each tube and crushed with a homogenizer in 500 μl of Chloroform-methanol. Samples were kept in a heating block to evaporate the chloroform-methanol. After evaporating the solvent, 200 μl of sulfuric acid was added and samples were heated in a heating block at 99° C for 10 min. The samples were cooled to room temperature and 800 μl of vanillin reagent was added to each tube and mixed well. Standards and samples were incubated for 30 min. OD of samples were read at 490 nm.
2.9. Statistical analysis
Analysis of variance was performed using SAS version 9.0 software (SAS Institute Inc., Cary, NC, USA) to test for statistical differences among treatments (α = 0.05). Pair-wise comparisons were made using the protected least squares difference (LSD) method.
3. Results
3.1. Identification of bHLH genes in T. castaneum
The blastP program was used to search T. castaneum genome sequences deposited in NCBI (http://www.ncbi.nlm.nih.gov) and the HGSC databases (http://www.hgsc.bcm.tmc.edu/projects/tribolium) and identified 53 bHLH family members (Tables 1a and 1b). Amino acid sequences of the bHLH domains of several known bHLH proteins and the predicted degenerate sequence based on multiple bHLH domains were used as queries (Atchley et al., 1999). NCBI conserved domain program (http://www.ncbi.nlm.nih.gov) was used to confirm the presence of the HLH domain in all the identified T. castaneum bHLH genes. The identified genes were named with the letters Tc for T. castaneum followed by the name of the previously identified member of the bHLH superfamily that showed the highest amino acid identity with the T. castaneum deduced amino acid sequence over the entire length of the protein. The deduced amino acid sequences of the 53 bHLH genes identified in T. castaneum genome and one closest homologue for each gene were subjected to phylogenetic analysis. This analysis classified 53 bHLH proteins into ten families; PAS, HES, Myc/USF, Hand, Mesp, Shout, p48, NeuroD/ Neurogenin, Atonal and AS-C (Fig. 1). We identified two additional members of the PAS family (TcSim1, TcSim2) and one additional member (Hey2) of the HES family when compared to the previous study which identified 50 bHLH transcription factors in T. castaneum (Simionato et al., 2007).
Table 1.
| a. Accession and GLEAN numbers of PAS and HES family genes identified in T. castaneum | ||
|---|---|---|
| Name of Gene | Accession Number | GLEAN Number |
| Hairy | XP_971935 | GLEAN_12851 |
| Side2 | XP_973515 | GLEAN_11804 |
| Deadpan | XP_967694 | GLEAN_05224 |
| E(spl)mgamma2 | XP_972493 | GLEAN_05541 |
| E(spl)mgamma1 | XP_972685 | GLEAN_06580 |
| Side1 | XP_975187 | GLEAN_13080 |
| Hey1 | XP_968161 | GLEAN_01593 |
| Hey2 | XP_001812240 | GLEAN_12119 |
| SRC | XP_967666 | GLEAN_14256 |
| Tango | XP_970422 | GLEAN_13793 |
| Cycle | NP_001107795 | GLEAN_02494 |
| Clock | XP_967106 | GLEAN_00088 |
| Trachealess | XP_967112 | GLEAN_01448 |
| Sim1 | XP_967930 | GLEAN_16205 |
| Sim2 | - | GLEAN_16204 |
| Hypoxia | XP_967427 | GLEAN_13241 |
| Spineless | XP_967876 | GLEAN_11105 |
| Dysfusion | - | GLEAN_13566 |
| Met/GCE | XP_966542 | GLEAN_03908 |
|
b. Accession and GLEAN numbers of Myc/USF, Hand, Shout, p48, NeuroD/Neurogenin, Atonal, AS-C and miscellaneous genes identified in T. castaneum | ||
| Name of gene | Accession Number | GLEAN Number |
| TcHLH106 | XP_974195 | GLEAN_07163 |
| TcBigmax | XP_967151 | GLEAN_00954 |
| TcAP-4 | XP_967737 | GLEAN_07652 |
| TcMax | XP_975370 | GLEAn_09976 |
| TcMyc | - | GLEAN_00123 |
| TcMnt | XP_967000 | GLEAN_15361 |
| TcFlocculin like | XP_001811168 | GLEAN_12658 |
| TcUSF2 | XP_974456 | GLEAN_07937 |
| TcHLH3B | XP_969923 | GLEAN_09117 |
| TcTwist | CAH25640 | GLEAN_14598 |
| TcHand | XP_972310 | GLEAN_04726 |
| TcTF21 | XP_968572 | GLEAN_02349 |
| TcParaxis | XP_001808472 | GLEAN_15754 |
| TcDelilah2 | XP_973102 | GLEAN_07598 |
| TcDelilah1 | XP_973186 | GLEAN_07594 |
| TcShout | XP_967920 | GLEAN_05579 |
| TcFer3 | XP_969923 | GLEAN_01473 |
| TcFer2 | XP_971276 | GLEAN_13785 |
| TcFer1 | XP_969845 | GLEAN_14105 |
| TcDimmed | XP_971229 | GLEAN_06222 |
| TcTap | XP_970244 | GLEAN_13793 |
| TcNeuroD | - | GLEAN_01576 |
| TcBeta3 | XP_974793 | GLEAN_14712 |
| TcHLH4C | XP_971274 | GLEAN_07725 |
| TcNautilus | XP_972025 | GLEAN_15855 |
| TcCato | XP_974243 | GLEAN_03171 |
| TcAtonal | XP_970709 | GLEAN_11336 |
| TcAmos | XP_974297 | GLEAN_03170 |
| TcScute | - | GLEAN_07826 |
| TcAsense | NP_001034533 | GLEAN_08437 |
| TcAsh | NP_001034537 | GLEAN_08433 |
| TcDaughterless | XP_973272 | GLEAN_09743 |
| TcEmc | XP_970015 | GLEAN_00024 |
| TcMitf | XP_975837 | GLEAN_14225 |
Fig.1.
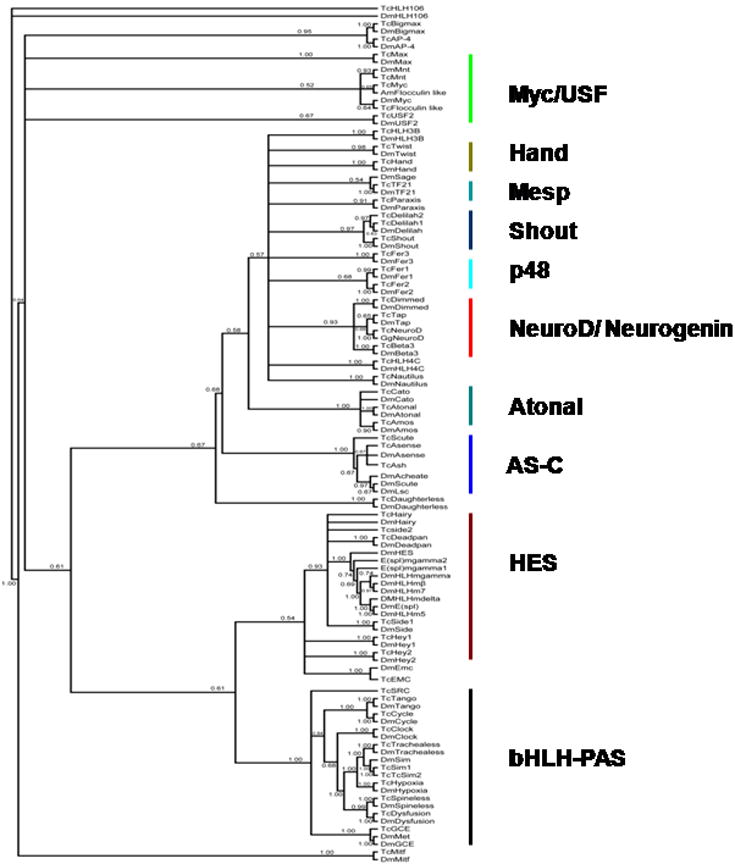
Phylogentic tree of 53 bHLH superfamily members and one of their closest homologues. The amino acid sequences were aligned in MAFFT and converted to a nexus file in BioEdit. The nexus file was analyzed under Bayesian optimality criterion using MrBayes version 3.1.2. The summary tree obtained by keeping burnin at 2,500,000 generations and probabilities is shown. Phylogenetic analysis classified 53 bHLH genes into ten families; PAS, HES, Myc/USF, Hand, Mesp, Shout, p48, NeuroD/ Neurogenin, Atonal and AS-C. Ten proteins such as TcHLH106, TcBigmax, TcAP-4, TcHLH3B, TcHLH4C, TcNautilus, TcExtramacrochaete (Emc), TcDaughterless, TcParaxis and TcMitf did not fall into any of these ten families and are grouped under miscellaneous family.
3.1.1. PAS family
The PAS domain is named after three proteins, D. melanogaster period (per), the human aryl hydrocarbon receptor nuclear translocator (ARNT) and D. melanogaster single-minded (Sim) (Zelzer et al., 1997). The members of this family are characterized by the presence of a 260-310 residue long PAS domain that is mainly involved in dimerization (Kewley et al., 2004). The PAS proteins are known to heterodimerize with other transcription factors and regulate various developmental processes. Eleven members of the PAS family have been identified in the red flour beetle (Fig. 1). In the red flour beetle, only one protein related to both germ cell expressed (GCE) and Methoprene-tolerant protein (Met) of D. melanogaster is present (Fig. 1). In mosquitoes also a single ortholog of DmMet and GCE is present (Wang et al., 2007). Two proteins related to D. melanogaster Sim are present in the T. castaneum genome (TcSim1 and TcSim2, Table 1a). The TcSim1 protein showed 48% identity over the entire length of the protein and 93% identity in the HLH region with the DmSim (Table 2a). TcSim2 showed 33% identity over the entire length of protein and 93% identity in the HLH region with the DmSim (Table 2a). T. castaneum Steroid Receptor Coactivator (TcSRC) showed 14% amino acid identity in the overall protein and 48% in the bHLH region with DmTaiman (Table 2a). In contrast, TcSRC showed 17% amino acid identity in the overall protein and 71% in the bHLH region with human SRC (Table 2a) suggesting that this bHLH protein is closer to human SRC than to DmTaiman; therefore, this protein was named TcSRC.
Table 2.
| a. Amino acid identity of bHLH-PAS and HES genes between beetle and fruitfly; beetle and vertebrates | ||||||
|---|---|---|---|---|---|---|
| bHLH gene | Fly homologue | Vertebrate homologue | % Overall identify | % HLH identity | % Overall identity | % HLH identity |
| TcHairy | DmHairy | HsHES-1 | 45 | 89 | 39 | 83 |
| TcSide2 | DmSide | HsHES1 | 24 | 62 | 20 | 55 |
| TcDeadpan | DmDeadpan | HsHES-1 | 32 | 90 | 29 | 77 |
| TcE(spl)mgamma2 | DmE(spl)mgamma | HsHES-4 | 42 | 81 | 26 | 62 |
| TcE(spl)mgamma1 | DmE(spl)mgamma | HsHES-1 | 45 | 90 | 30 | 62 |
| TcSide1 | DmSide | HsHES-1 | 35 | 83 | 29 | 65 |
| TcHey1 | DmHey1 | HsHey1 | 16 | 88 | 38 | 88 |
| TcHey2 | DmHey2 | HsHey2 | 32 | 87 | 38 | 88 |
| TcSRC | DmTaiman | HsSRC | 14 | 48 | 17 | 71 |
| TcTango | DmTango | HsARNT | 58 | 100 | 49 | 93 |
| TcCycle | DmCycle | HsCycle | 41 | 91 | 30 | 74 |
| TcClock | DmClock | HsClock | 45 | 81 | 33 | 62 |
| TcTrachealess | DmTrachealess | HsNPAS | 42 | 98 | 24 | 93 |
| TcSim1 | DnSim | HsSim | 48 | 93 | 41 | 91 |
| TcSim2 | DmSim | HsSim | 33 | 93 | 21 | 91 |
| TcHypoxia | DmHypoxia | HsHIF | 25 | 68 | 26 | 70 |
| TcSpineless | DmSpineless | HsAhr | 57 | 67 | 26 | 65 |
| TcDysfusion | DmDysfusion | HsNXF | 68 | 78 | 28 | 31 |
| TcMet/GCE | DmGCE/Met | - | 24/25 | 53/51 | - | - |
|
b. Amino acid identity of Myc/USF, Hand, Shout, p48, NeuroD/Neurogenin, Atonal, AS-C and miscellaneous genes between beetle and fruitfly; beetle and vertebrates | ||||||
| bHLH Name | Fly homologue | Vertebrate homologue | % Overall identity | % HLH identity | % Overall identity | % HLH identity |
| TcHLH106 | DmHLH106 | HsSREBP2 | 29 | 77 | 29 | 69 |
| TcBigmax | DmBigmax | HsBigmax | 43 | 90 | 52 | 82 |
| TcAP-4 | DmAP-4 | HsAP-4 | 30 | 86 | 31 | 89 |
| TcMax | DmMax | HsMax | 47 | 56 | 51 | 87 |
| TcMyc | DmMyc | HsMyc | 17 | 64 | 13 | 51 |
| TcMnt | DmMnt | HsMnt | 18 | 65 | 21 | 78 |
| TcFlocculin like | - | - | - | - | - | - |
| TcUSF2 | DmUSF2 | HsUSF | 17 | 65 | 21 | 78 |
| TcHLH3B | DmHLH3B | HsSCL | 21 | 82 | 21 | 75 |
| TcTwist | DmTwist | HsTwist | 31 | 79 | 40 | 92 |
| TcHand | DmHand | HsHand | 42 | 75 | 38 | 79 |
| TcTF21 | DmHLH54F | HsTF21 | 39 | 55 | 41 | 89 |
| TcParaxis | DmParaxis | HsParaxis | 31 | 60 | 38 | 82 |
| TcDelilah2 | DmDelilah | HsNDF6 | 33 | 79 | 22 | 56 |
| TcDelilah1 | DmDelilah | HsNEUROD2 | 33 | 76 | 22 | 61 |
| TcShout | DmNet | HsATOH8 | 33 | 58 | 25 | 56 |
| TcFer3 | DmFer3 | HsFer3 | 43 | 92 | 53 | 90 |
| TcFer2 | DmFer2 | MdFer2 | 70 | 86 | 52 | 91 |
| TcFer1 | DmFer1 | HsPTF1 | 40 | 54 | 35 | 56 |
| TcDimmed | DmDimmed | HsMIST1 | 37 | 67 | 29 | 62 |
| TcTap | DmTap | HsNEUROG2 | 29 | 76 | 27 | 79 |
| TcNeuroD | - | HsNeuroD1 | 31 | 79 | ||
| TcBeta3 | DmBeta3 | HsBeta3 | 46 | 96 | 35 | 98 |
| TcHLH4C | DmNHLH1 | HsNHLH1 | 46 | 94 | 54 | 95 |
| TcNautilus | DmNautilus | HsMyf5 | 40 | 62 | 35 | 55 |
| TcCato | DmCato | HsATOH1 | 35 | 72 | 34 | 77 |
| TcAtonal | DmAtonal | HsATOH1 | 53 | 85 | 40 | 67 |
| TcAmos | DmAmos | HsATOH1 | 38 | 77 | 33 | 74 |
| TcScute | DmScute | HsMash-3 | 26 | 50 | 28 | 53 |
| TcAsense | DmAsense | HsMash-3 | 32 | 82 | 15 | 46 |
| TcAsh | DmAcheate | HsASH1 | 32 | 71 | 26 | 72 |
| TcDaughterless | DmDaughterless | HsTCF12 | 33 | 87 | 22 | 69 |
| TcEmc | DmEmc | - | 37 | 76 | ||
| TcMitf | DmMitf | HsMitf | 39 | 96 | 29 | 80 |
3.1.2. HES family
HES family members play key roles during embryogenesis, cell proliferation and tissue differentiation. These genes display an oscillatory expression pattern and control the timing of biological events such as somite segmentation. Most of these genes participate in Notch signaling pathway which mediates cell-cell communication (Muskavitch, 1994). The members of this family contain additional domains such as orange and WRPW. These domains allow these proteins to repress transcription by interacting with proteins such as Groucho (Fischer and Caudy, 1998). In D. melanogaster twelve members of the HES family have been identified. In T. castaneum we could identify only eight members of the HES family (Fig. 1). The homologs of DmHLHmβ, DmHLHmdelta, DmHLHm5, DmHLHm7 are not present in the T. castaneum genome. In T. castaneum two genes closely related to DmSide have been identified and named as TcSide1 and TcSide2. TcSide1 showed 35% amino acid identity in overall protein and 83% identity in the bHLH region with DmSide. TcSide2 showed 24% amino acid identity in overall protein and 62% identity in the bHLH region with DmSide (Table 2a). TcExtramacrochaete (Emc) was not considered as a member of the HES family (Fig. 1) because it lacks orange domain, a characteristic feature of all the HES family members.
3.1.3. Myc/USF family
Myc/USF family of bHLH proteins are widely expressed in many different cell types. Myc family genes are among the most frequently affected genes in tumors. These proteins contain a leucine-zipper domain at carboxy-terminal to the bHLH region (Luscher, 2001). Myc proteins regulate translation initiation (Schmidt, 2004) and also function as transcriptional activators when they form heterodimers with Max proteins (Schmidt, 2004). Max is known to form homodimers and heterodimers with other bHLH proteins including Mad (Nair and Burley, 2003).
Phylogenetic analyses did not satisfactorily resolve TcMyc, dMyc, TcFlocculin-like and Apis mellifera Flocculin-like; the support values are very low (Fig. 1). This could be due to insignificant differences in their sequences. AmFlocculin-like gene identified in a T. castaneum genome is not present in vertebrates but is present in the genomes of Anopheles gambiae, Aedes aegypti, A. mellifera, Culex pipiens, and a parasitic wasp Nasonia vitripennis.
3.1.4. Hand family
In the Hand family two genes have been identified in both D. melanogaster and T. castaneum.
3.1.5. Mesp family
In this family only one gene was identified, TcTF21, whereas in D. melanogaster two genes (DmTF21 and DmSage) have been identified in this family. The homolog for DmSage is absent in T. castaneum. Mesp family proteins are necessary for mesoderm segmentation initiation. Sage protein expression during the zygotic stage starts at stage 10 and persists until stage 15 (Moore et al., 2000).
3.1.6. Shout family
In this family we identified three genes in T. castaneum, whereas in D. melanogaster only two genes have been identified (Fig. 1). Only one Delilah gene has been identified in D. melanogaster. Two genes closely related to DmDelilah have been identified in T. castaneum (TcDelilah1 and TcDelilah2).
3.1.7. p48 family
In both D. melanogaster and T. castaneum three members of this family have been identified (Fig. 1). DmFer1, DmFer2 and DmFer3 are most closely related to the bHLH domain of the p48 subunit of the pancreatic, exocrine cell-specific transcription factor in the mouse (Krapp et al., 1996).
3.1.8. NeuroD/ Neurogenin family
We identified four genes in this family in T. castaneum (Fig. 1), whereas in D. melanogaster only three members of this family have been identified. TcNeuroD identified in the T. castaneum genome codes for a member of NeuroD/ Neurogenin family. None of the six insect genomes sequenced showed a protein closely related to TcNeuroD (Fig. 1). The human homologue for TcNeuroD is HsNeuroD1 (31% overall identity and 79% identity in the bHLH region) (Table 2b).
3.1.9. Atonal family
In this family, we identified three genes which are similar to the three members of this family identified in D. melanogaster. The members of this family are mainly involved in nervous system development.
3.1.10. AS-C family
AS-C family contains four genes in D. melanogaster whereas in the red flour beetle, we could identify only three genes coding for members of this family. Lethal of scute (L’sc) is absent in the red flour beetle genome (Fig. 1). Previous studies reported that the number of AS-C genes increased during insect evolution, particularly in the Diptera lineage to provide the flexibility that is required for more complex transcriptional regulation (Skaer et al., 2002).
Members of the AS-C family are known to be involved in neurogenesis. These proteins include D. melanogaster proneural proteins, achaete, scute, L’sc and asense (Ghysen et al., 1993). In vertebrates, Mash-1, Math-1 and the neurogenins are important in the initial determination of neurons, whereas Neuro-D, NeuroD2 and MATH-2 act as differentiation factors (Lee, 1997). The AS-C proteins function at several stages of neurogenesis in D. melanogaster.
3.1.11. Miscellaneous genes
TcHLH106, TcBigmax, TcAP-4, TcHLH3B, TcHLH4C, TcNautilus, TcEmc, TcDaughterless, TcParaxis and TcMitf did not fall into any of the ten families; therefore, these genes are classified in a miscellaneous gene family.
3.2. Functional characterization of PAS and HES family transcription factors
As a first attempt at determining the role of all the members belonging to two important families, PAS and HES, in growth and development of the T. castaneum, dsRNA for 19 genes were prepared and injected into newly molted final instar or in some cases into penultimate instar. To determine the efficiency of knock-down in expression of these genes in the larvae injected with respective dsRNA, the mRNA levels of these genes were determined at 72 hr after injection of dsRNA. As shown in Fig. 2, the mRNA levels of all the genes tested were reduced between 50-90% in bHLH dsRNA injected larvae when compared to their levels in control insects injected with malE dsRNA. Among PAS family genes, TcSim1 dsRNA injection resulted in 50% reduction in the mRNA levels of this gene in RNAi insects when compared to its expression in control insects injected with malE dsRNA. The other ten genes showed around 80% reduction in their respective mRNA levels in RNAi insects (Fig. 2A). Among HES family members, TcE(spl)mgamma2, Deadpan, and Hey2 dsRNA showed around 50% knock-down efficiency while others showed between 80-90% knock-down efficiency (Fig. 2B). We knocked-down the expression of about 400 genes in T. castaneum and learned that about 50% knock-down efficiency is sufficient to observe phenotypes in RNAi insects. For example, TcMet is one of the most difficult genes to achieve more than 50% knock-down efficiency. Yet, Met RNAi insects always showed severe phenotypes including disruption of larval-pupal ecdysis and induction of precocious adult development upon partial ecdysis (Parthasarathy et al., 2008). To avoid non-target effects in RNAi insects, the most variable region of each gene was selected for dsRNA synthesis. The effects of knock-down in the expression of bHLH transcription factor genes on the growth and development of larvae, pupae and adults are discussed below.
Fig.2.
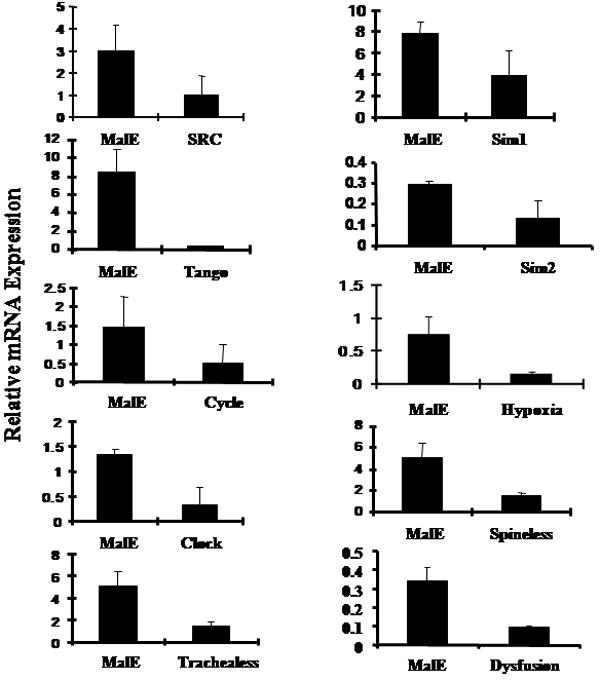
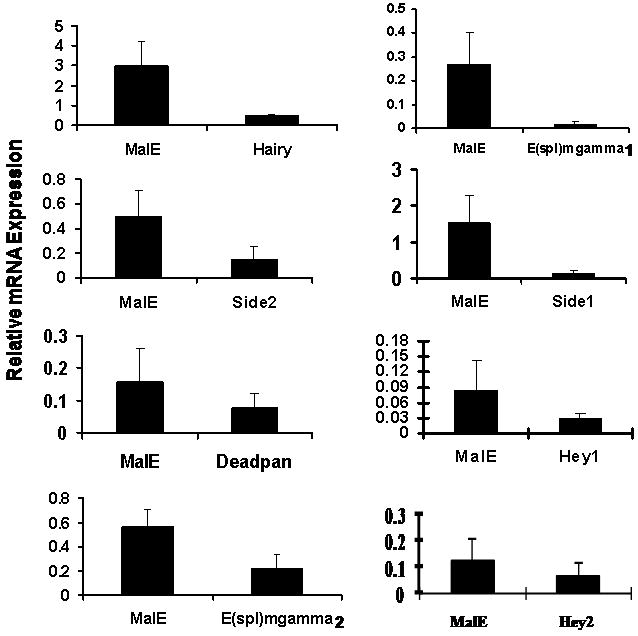
A. bHLH-PAS or malE dsRNA was injected into one-day-old final instar. qRT-PCR was used to quantify mRNA levels in larvae injected with bHLH-PAS gene or malE dsRNA. Mean + SD (n=3) are shown. The mRNA levels were normalized using the levels of rp49 as an internal control. mRNA levels of all the bHLH-PAS genes were reduced in the range of 50-90% when compared to their levels in the control larvae injected with malE dsRNA.
B. HES or malE dsRNA was injected into one-day-old final instar. qRT-PCR was used to quantify mRNA levels in larvae injected with HES gene or malE dsRNA. Mean+SD (n=3) are shown. The mRNA levels were normalized using the levels of rp49 as an internal control. mRNA levels of all the HES genes were reduced in the range of 50-90% when compared to their levels in the control larvae injected with malE dsRNA.
3.2.1. PAS family
Knocking-down in the expression of six of the eleven members of this family affected growth and development of T. castaneum. TcMet is one of the most studied members of this family. Injection of TcMet dsRNA into final instar larvae derailed the development of all the injected insects; none of them were able to develop into adults (Table 3). Most of the dsRNA injected insects died as larvae and a few developed to become pupae and died during the pupal stage (Table 3). The larvae injected with TcMet dsRNA during the penultimate or final larval instar showed phenotypes similar to the allatectomized lepidopteran final instar larvae that were attempting to undergo larval-pupal metamorphosis. Recently, knocking-down of Met during third or fourth larval instars has caused the precocious metamorphosis two instars later (fifth or sixth) instead of seventh or eighth instars (Konopova and Jindra, 2007). Knocking-down in the expression of TcMet disrupted larval-pupal ecdysis and induced precocious adult development upon partial ecdysis (Parthasarathy et al., 2008). These studies clearly established an important role for TcMet in juvenile hormone action. Since JH regulates almost every aspect of an insect’s life, it is likely that TcMet plays a key role in almost every stage of an insect’s life. Further studies are required to elucidate the mechanism of TcMet action in juvenile hormone signal transduction.
Table 3.
Effect of injection of dsRNA of bHLH-PAS or HES family genes into final instar larvae
| Name of gene | # injected larvae | # dead larvae | # dead pupae | # abnormal adults | # normal adults | % insects affected |
|---|---|---|---|---|---|---|
| Mal E | 40 | 4 | 0 | 0 | 36 | 10 |
| Hairy | 28 | 3 | 1 | 0 | 24 | 11 |
| Side2 | 31 | 4 | 0 | 0 | 26 | 13 |
| Deadpan | 30 | 1 | 2 | 0 | 27 | 11 |
| E(spl)mgamma2 | 25 | 1 | 1 | 0 | 23 | 8 |
| E(spl)mgamma1 | 38 | 14 | 20 | 0 | 4 | 89 |
| Side1 | 28 | 4 | 1 | 0 | 23 | 18 |
| Hey1 | 22 | 0 | 1 | 0 | 20 | 5 |
| Hey2 | 29 | 0 | 3 | 0 | 26 | 10 |
| SRC | 40 | 40 | 0 | 0 | 0 | 100 |
| Tango | 40 | 40 | 0 | 0 | 0 | 100 |
| Cycle | 40 | 4 | 0 | 0 | 36 | 10 |
| Clock | 40 | 2 | 0 | 0 | 38 | 5 |
| Trachealess | 40 | 38 | 0 | 0 | 2 | 95 |
| Sim1 | 40 | 3 | 0 | 0 | 37 | 7.5 |
| Sim2 | 20 | 3 | 0 | 0 | 18 | 15 |
| Hypoxia | 40 | 15 | 8 | 0 | 17 | 58 |
| Dysfusion | 20 | 3 | 0 | 0 | 17 | 15 |
| Spineless | 40 | 2 | 38 | 0 | 0 | 100 |
| Met/GCE | 69 | 58 | 11 | 0 | 0 | 100 |
Most of the larvae injected with four other members of the PAS family, TcSRC, TcTango, TcSpineless and TcTrachealess dsRNA died during larval or pupal stages (Table 3). In addition, about 58% of insects injected with TcHypoxia dsRNA died during the larval or pupal stages (Table 3). The larvae injected with TcCycle, TcClock, TcSim1, TcSim2 and Tcdysfusion dsRNA were able to develop into normal adults similar to the control malE injected larvae (Table 3).
The TcSRC dsRNA injected larvae remained small and did not reach critical weight to undergo metamorphosis. Larvae looked healthy similar to the larvae injected with malE dsRNA until seven days after injection. The feeding of these larvae was similar to the control larvae until ten days after injection when the feeding rate was decreased and larval size was reduced when compared to control larvae injected with malE dsRNA (Fig. 3C). The TcSRC dsRNA injected larvae became sluggish and eventually died (Fig. 3D). The TcSRC dsRNA and the control malE injected larvae were monitored for their growth from the third day till five days after the injection of dsRNA. In contrast to the malE injected final instar larvae, the TcSRC dsRNA injected larvae did not increase in weight and did not reach critical weight to undergo metamorphosis into pupae (Fig. 4A). TcSRC dsRNA injected larvae weighed less when compared to the control malE injected larvae (Fig. 4A).
Fig.3.

Phenotypes observed after injection of bHLH-PAS and HES family genes dsRNAs. Control malE or bHLH dsRNA was injected into one-day-old final instar. The pictures shown are various phenotypes observed during the larval and pupal stages. (A, B) final instar larvae injected with malE dsRNA and the pupae formed from the final instar larvae injected with malE dsRNA; (C, D) SRC dsRNA injected larvae died ten to fifteen days after injection; (E, F) tango and trachealess dsRNA injected insects died during the quiescent stage; (G) pupae formed with problems in wing development after the injection of Hypoxia dsRNA; (H) abnormal pupae with defects in wing development after the injection of E(spl)mgamma1 dsRNA.
Fig. 4.
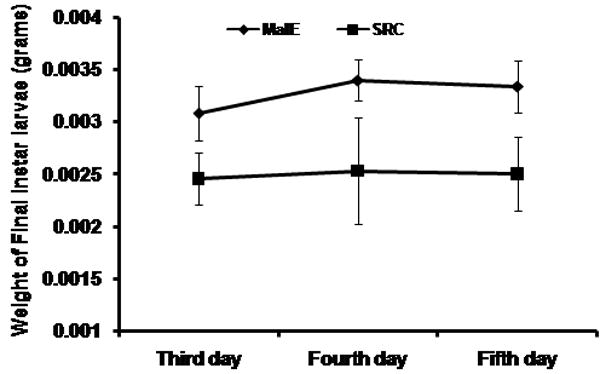
(A) Comparison of larval weights between malE and TcSRC dsRNA injected larvae. Larvae were injected with dsRNA at 24 h after ecdysis into final instar. Larvae were weighed from third day till fifth day after the injection of dsRNA. Mean +SD (n=16) are shown.
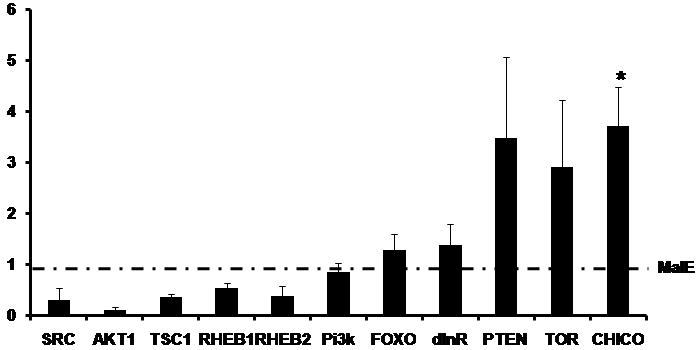
(B) The expression ratio of mRNA levels of SRC, AKT1, TSC1, RHEB1, RHEB2, Pi3k, FOXO, dInR, PTEN, TOR and CHICO in insects injected with TcSRC dsRNA using qRT-PCR. Injections were done at 24 h after ecdysis into final instar. Total RNA was extracted from pools of three larvae for each treatment at six days after injection. The mRNA levels were normalized using the levels of rp49 as an internal control. The expression levels of each gene in the corresponding control insects injected with malE dsRNA was set as 1. Means+SE for three biological replicates are shown. Paired T-test with unequal mean of variances was performed at P ≤ 0.05.
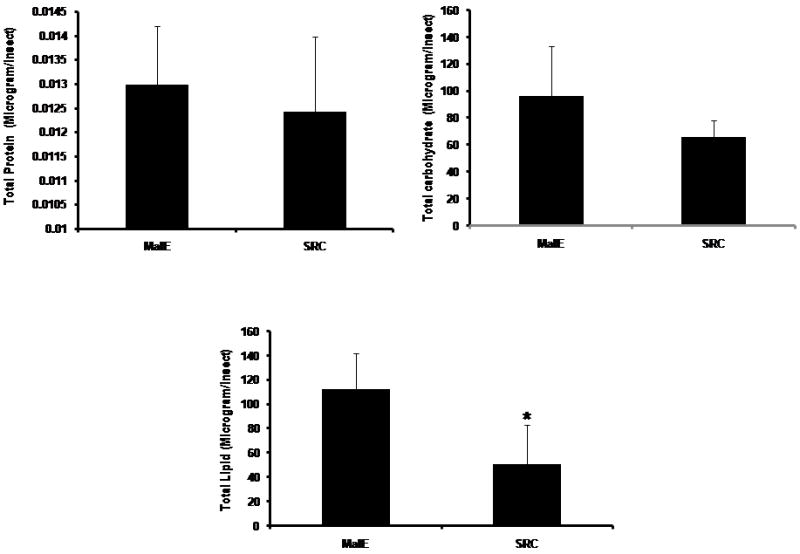
(C) Estimation of Total protein, carbohydrate and lipids between malE and TcSRC dsRNA injected final instar larvae. Larvae were injected with dsRNA at 24 h AEFI. Total protein, carbohydrate and lipids were estimated using Bradford, Anthrone-based and Vanillin-reagent methods at five days after the injection of dsRNA. Mean +SD (n=7) are shown. Paired T-test with unequal mean of variances was performed at P ≤ 0.05.
Insulin-like peptides (ILPs) and the insulin receptor signaling pathways are known to play a very important role in regulating the nutrition-dependent growth rates in insects. To confirm whether the expression of any of the genes involved in the insulin signaling and TOR pathways was affected in the TcSRC RNAi insects, the mRNA levels of select genes coding for proteins known to be involved in these pathways were quantified by qRT-PCR. The mRNA levels of these select genes were compared between the malE and TcSRC RNAi insects at six days after the injection of dsRNA. As expected in TcSRC dsRNA injected insects the level of SRC was 3-fold lower than in malE injected insects (Fig. 4B). The mRNA levels of genes Akt1, Tuberous Sclerosis dimer 1 (TSC1) and Ras Homolog Enhanced Brain 1, 2 (RHEB1, RHEB2) were 3 to 4-fold lower in TcSRC dsRNA injected insects when compared to their expression in control malE dsRNA injected insects (Fig. 4B). The expression of three genes, phosphatase and tensin homolog (PTEN), tor and chico was 3 to 4-fold higher in TcSRC dsRNA injected insects when compared to malE injected insects. The expression of three genes Forkhead box, Class O (FOXO), Insulin Receptor homologue (dInR) and Phosphatidylinositide 3-kinase (Pi3k) is similar in TcSRC dsRNA injected insects when compared to the malE injected insects (Fig. 4B). These studies suggest that TcSRC directly or indirectly regulates the expression of genes involved in insulin-signaling and TOR pathways.
To determine the role of TcSRC in nutrient metabolism, the amount of total proteins, carbohydrates and lipids were estimated in the final instar larvae at five days after the injection of malE or TcSRC dsRNA. Total carbohydrates, proteins and lipids were estimated using Bradford, Anthrone-based, and Vanillin reagent methods. As shown in Fig. 4C, there was no significant change in the amount of carbohydrate and protein levels between the malE and TcSRC dsRNA injected insects. Total carbohydrate content was decreased by about 30% in TcSRC dsRNA injected insects but was not significantly different from the control malE injected larvae (Fig. 4C). The amount of total lipids was reduced by about 50-60% in TcSRC dsRNA injected insects and showed a significant decrease from the malE injected final instar larvae (Fig. 4C). These studies suggest that TcSRC plays a critical role in lipid metabolism.
Dissection of RNAi larvae at seven days after injection of dsRNA did not show significant abnormalities in the midgut, central nervous system and other tissues (data not shown). In the larvae that had survived up to twenty days, the food was accumulated in the hindgut of the alimentary canal. 100% of the dsRNA injected larvae were dead by twenty days after injection. When TcSRC dsRNA was injected into penultimate instars, all the larvae stopped growing and did not reach critical weight to undergo the next molt to final instar; these larvae remained at this stage and eventually died (Table 4).
Table 4.
Effect of silencing of bHLH-PAS genes on penultimate instar larvae
| Name of gene | # injected larave | # dead larvae | # dead pupae | # adnormal adults | # normal adults | % insects affected |
|---|---|---|---|---|---|---|
| MalE | 20 | 2 | 0 | 0 | 18 | 10 |
| Tango | 20 | 20 | 0 | 0 | 0 | 100 |
| Trachealess | 20 | 20 | 0 | 0 | 0 | 100 |
| Hypoxia | 20 | 5 | 0 | 0 | 13 | 25 |
| Spineless | 20 | 6 | 14 | 0 | 0 | 100 |
| SRC | 20 | 20 | 0 | 0 | 0 | 100 |
Injection of TcTango dsRNA into the final and penultimate instars immediately after molting caused the death of the larvae within one week after injection (Tables 3, 4). Death of the larvae was initiated after the third day after injection and by the seventh day all the insects were dead. Major external symptoms observed were reduction in total body size, shrinkage in the thoracic region and change in color to dark brown (Figure 3E). ARNT is the vertebrate homologue of D. melanogaster Tango and is involved in toxin metabolism (Crews and Fan, 1999). Knock-down in the expression of TcTango appears to affect growth and metabolism because most of the Tango dsRNA injected insects died during the quiescent stage prior to pupation.
Most of the final instars injected with TcTrachealess dsRNA died within seven days after injection (Table 3). TcTrachealess dsRNA injected larvae were healthy during initial stages, fed, grew and entered the quiescent stage. Then they became sluggish and died during the quiescent stage (Fig. 3F). In the dead larvae, the abdominal region resembled the normal insects in the quiescent stage, but the head and the thoracic regions were elongated and turned abnormal with crinkled abdomen and legs (Fig. 3F). TcTrachealess dsRNA injected penultimate instar could not molt into the final instar and died as the penultimate instar (Table 4). Similar to TcTango knockdown, TcTrachealess knock-down also affected growth and metabolism, because TcTrachealess dsRNA injected insects died prior to larval-pupal metamorphosis. DmTango protein is known to heterodimerize with several other bHLH-PAS proteins including DmTrachealess and DmSim to regulate transcription in the trachea and central midline, respectively (Sonnenfeld et al., 1997). It is also known to heterodimerize with DmDysfusion to regulate the tracheal tip fusion in D. melanogaster (Jiang and Crews, 2003).
Injection of TcHypoxia dsRNA into the final instar led to the death of 38% of the insects during the larval stage. About 20% of the injected larvae died during the pupal stage and some of the pupae and adults developed from dsRNA injected larvae showed abnormal wings with forewings shorter than the hind wings (Fig. 3G). Injection of TcHypoxia dsRNA into penultimate instar did not show much effect and 75% of larvae were able to develop into normal adults (Table 4). D. melanogaster Hypoxia is the closest homolog of mammalian Hypoxia-inducible factor (HIF) – α protein (Bacon et al., 1998). In both vertebrates (Bruick and Knight, 2001) and D. melanogaster (Lavista-Llanos et al., 2002) Hypoxia plays a major role in regulating responses to decreased oxygen concentrations.
Almost all the final and penultimate instars injected with TcSpineless dsRNA progressed through development and ecdysed into the pupal stage and died during the pupal stage. The phenotypes of the pupae developed from TcSpineless dsRNA injected larvae were similar to the pupae developed from control larvae injected with malE dsRNA. However, the pupae developed from TcSpineless dsRNA injected larvae were not able to develop into adults and died during the pupal stage. Most (66%) of penultimate instars injected with TcSpineless dsRNA progressed until pupal stage and died during the pupal stage. These data suggest that TcSpineless is required for adult development but not larval or pupal growth and development. DmSpineless encodes the closest known homolog of the mammalian Aryl hydrocarbon receptor (Ahr), a transcription factor of the bHLH-PAS family. Loss-of-function alleles of spineless in D. melanogaster have led to the transformation of distal antenna to leg, deletion of distal leg structures and reduction in the size of bristles in adults (Duncan et al., 1998). Recent RNAi studies in T. castaneum showed that the antennal identity in the flagellum is specified by spineless (Angelini et al., 2008). The effect of spineless knock-down in antennal structure depended on the timing of dsRNA injection, insects injected shortly before pupation showed fusions in the flagellum, but not transformation, suggesting that spineless has distinct roles at different developmental stages. We were unable to monitor the adult phenotypes in TcSpineless RNAi insects as 100% of the dsRNA injected final and penultimate instars died during the pupal stage (Tables 3, 4).
The final instars injected with TcClock, TcCycle, TcDysfusion, TcSim1 or TcSim2 dsRNA did not show significant effects and more than 60% of the injected larvae were able to develop into normal adults. Although TcCycle and TcClock were shown to be involved in circadian rhythm (Dunlap, 1999) knocking-down the expression of these genes in T. castaneum did not block growth or development. One possible reason could be that the low levels of TcCycle and TcClock proteins present in RNAi animals are sufficient to regulate circadian rhythm during the larval and pupal stages. Knock-down efficiency of the expression of TcCycle and TcClock genes was quantified using qRT-PCR and more than 90% efficiency in the knock-down in gene expression was observed (Fig. 2A).
It is interesting that the PAS domain containing proteins makes up about 20% of members of the bHLH superfamily and more than 50% of the genes belonging to the bHLH-PAS family showed an effect on growth and development after knock-down in their expression. These data suggest that bHLH proteins that contain PAS domains are more critical to growth and development of T. castaneum and other insects.
3.2.2. HES family
DsRNA of HES family members or malE were injected into newly molted final instar larvae and their development was monitored. Except for TcE(spl)mgamma1, knock-down in the expression of all other seven bHLH family members showed no significant effect on growth and development as most of the larvae injected with dsRNA of these genes progressed through larval, pupal and adult stages similar to the larvae injected with malE dsRNA (Table 3). Out of the 38 larvae injected with TcE(spl)mgamma1 dsRNA, 14 died during the larval stage, 20 died during the pupal stage and only four of them developed into adults (Table 3). The pupae developed from dsRNA injected larvae showed defects in wing development (Fig. 3H) and Table 3). The E(spl) complex of genes in D. melanogaster shows a distinct pattern of expression in the wing imaginal discs (Jennings et al., 1995) and the absence of E(spl) genes had resulted in the formation of thickened veins. Also, clones of cells lacking all E(spl) genes that abut the wing margin did not produce wing nicks (de celis et al., 1996). Some of the dsRNA injected larvae were able develop into pupae, and the pupae showed abnormal wing development Wings were not completely formed, and the abdomen was exposed resembling a naked pupa. The development of forewings was more affected than the development of hind wings, as a result, the wings were short and devoid of venation.
3.3. Developmental expression profiles of bHLH-PAS and HES family genes
As described in the previous section, seven genes coding for members of PAS and HES families affected growth and development in RNAi analysis. To determine the expression patterns of these genes during final instar and pupal stages, the mRNA levels of these genes were determined using qRT-PCR in the whole body samples collected at 12 h intervals during the final instar, quiescent and pupal stages. TcMet/GCE expression profiles have been reported recently (Parthasarathy et al., 2008). The mRNAs of the rest of the six PAS and HES family genes that affected growth and development of T. castaneum larvae were detected during the final instar, quiescent and pupal stages (Fig. 5). TcTrachealess mRNA was detected during the final instar, quiescent and pupal stages and one small peak of expression was observed at 60 h after ecdysis into the final instar (AEFI). TcSRC mRNA was detected during the final instar, quiescent and pupal stages and showed increases at the end of larval and pupal stages. TcHypoxia mRNA was detected during the final instar, quiescent and pupal stages and showed an increase at the end of pupal stage. TcTango mRNA levels were lower at the beginning of final instar but increased beginning at 48 h AEFI, the mRNA levels reached the maximum by 60 h AEFI. The mRNA levels then decreased and remained low throughout quiescent stage. The mRNA levels increased again after ecdysis into the pupal stage and remained high until end of pupal stage. TcE(spl)mgamma1 and TcSpineless mRNA levels were detected during the final instar, quiescent and pupal stages and showed two peaks of expression at the beginning of final instar and at the end of pupal stage (Fig. 5). These qRT-PCR studies showed that the six genes coding for PAS and HES family members and play important roles in growth and development of T. castaneum are expressed during final instar, quiescent and pupal stages.
Fig. 5.
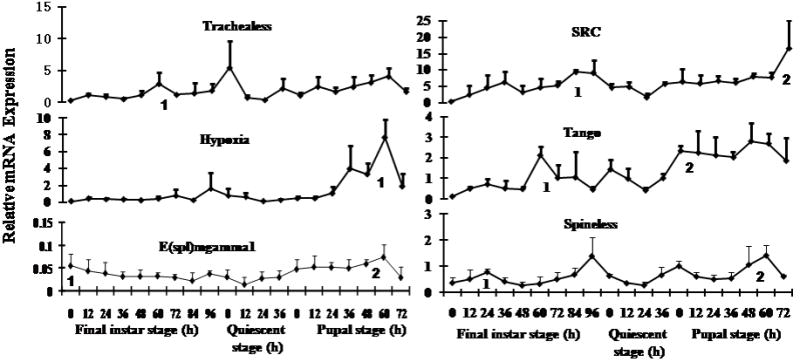
Expression profiles of six bHLH PAS and HES family genes in the whole body of T. castaneum determined by quantitative real-time PCR (qRT-PCR). Samples were collected at 12 h interval during the final instar and pupal stages. Total RNA was extracted from pools of three larvae for each treatment. The mRNA levels were normalized using the levels of rp49 as an internal control. Mean+SD of two independent experiments with three replicates each are shown. Statistical analysis was performed as described in the Materials and methods section and peaks of expression are marked with numbers.
4. Discussion
The main finding of this study is the identification of six members of the PAS and one member of the HES family transcription factors that play key roles in growth and development of the red flour beetle, T. castaneum. Since the main goal of this study is functional characterization of the members of this superfamily identified in T. castaneum and the functions of bHLH proteins are often influenced by the presence of various functional domains, the phylogenetic analyses were performed using the whole protein sequence. The phylogenetic analyses classified the 53 members into 10 families and ten members that did not group with any one of these 10 families were placed in the miscellaneous family. Similar analysis in the previous studies classified 46 members of the bHLH superfamily in D. melanogaster into eight subfamilies (bHLH-PAS, HES, bHLH-zip, Atonal-related, Mesp-related, Shout, Hand, and p48-related) (Moore et al., 2000). As shown in Table 5, based on phylogenetic analysis the Atonal family was further classified into two families Atonal and NeuroD/ Neurogenin families (Ledent and Vervoort, 2001). AS-C family genes were not included in the data set that was used for phylogenetic analysis in D. melanogaster study (Moore et al., 2000) but in our analysis we have included AS-C family genes.
Table 5.
Comparison of bHLH genes present in Tribolium castaneum, Drosophila melanogaster, and Bombyx mori
| Family Name | T. castaneum | D. melanogaster | B. mori |
|---|---|---|---|
| bHLH-PAS | 11 | 11 | 12 |
| HES | 8 | 12 | 7 |
| Myc/ USF | 5 | 4 | 5 |
| Hand | 2 | 2 | 2 |
| Mesp | 1 | 2 | 2 |
| Shout | 3 | 2 | 2 |
| p48 | 3 | 3 | 2 |
| NeuroD / Neurogenin | 4 | 3 | 3 |
| Atonal | 3 | 3 | 1 |
| AS- C | 3 | 4 | 4 |
| Miscellaneous | 10 | 10 | 10 |
| Total | 53 | 56 | 52 |
Data for T. castaneum and D. melanogaster were from this study and data for B. mori was from a Bombyx study (Wang et al., 2007).
As summarized in Table 6, seven out of the 19 bHLH transcription factors that showed phenotypes during the metamorphosis in dsRNA injected T. castaneum showed more or less similar functions described in D. melanogaster. Discovering the function of the remaining bHLH transcription factor, SRC homologue in growth of T. castaneum is one the major findings of this study. In vertebrates there are three isoforms of SRC, SRC-1, SRC-2 and SRC-3, whereas in T. castaneum only one SRC protein has been identified; therefore, the red flour beetle may provide a simpler model system to study the mechanism of SRC action.
Table 6.
Comparison of Phenotypes observed in T. castaneum and D. melanogaster
| Gene | Tribolium castaneum | Drosophila melanogaster |
|---|---|---|
| Trachealess | Final instar larvae were healthy, fed and grew until four days after injection and entered quiescent stage. After that they became sluggish and died during the quiescent stage. | Is required for tubulogenesis during embryonic stage (Isaac and Andrew, 1996). Trachealess mutants display no trachea and requires for tracheal and salivary gland development (Parrish et al., 2000). Silencing the gene using RNAi caused defects in muscle development and dendrite morphogenesis during the embryonic stage (Parrish et al., 2000). |
| Tango | Tango injected larvae were dead during quiescent stage similar to the trachealess injected larvae. Tango and trachealess injected larvae would have been dead due to the improper functioning of major systems such as tracheal and nervous system. | Tango mutants show central nervous system midline and tracheal defects during embryonic stage (Jiang and Crews, 2003). RNAi of tango resulted in reduced arborization of dendritic neurons, defects in muscle and dendrite morphogenesis during embryonic stage (Parrish et al., 2000). |
| Spineless | Most of the dsRNA injected larvae died during the pupal stage. Spineless seems to play a key role during adult development in Tribolium similar to its role in Drosophila as spineless mutants showed severe abnormalities in the adult flies compared to the other stages. | Spineless mutants display transformation of distal antennae to leg, reduction in size of bristles and deletion of distal leg structures in adults (Duncan et al., 1998). Severe loss of function mutation resulted in amplification of sex combs on first leg in adult flies (Kuzin et al., 1997). Plays a key role for establishing retinal mosaic required for color vision (Wernet et al., 2006). Necessary for diversification and dendrite morphology of Drosophila dendritic arborization neurons during all stages of fly (Kim et al., 2006). |
| SRC | Larval growth is blocked and the larvae did not reach critical weight to undergo molt to the next stage. The phenotypes observed is similar to the SRC-3 knock-out mice (Wang et al., 2006; Xu et al., 2000) and the HLH domain of TcSRC is more closely related to vertebrate SRC rather than to DmTaiman. | Mutations in taiman caused defects in migration of specific follicle cells and border cells in ovary (Bai et al., 2000). |
| Met | Single homologue of Met is present. Silencing of Met during early stage beetle larvae has lead to precocious metamorphosis (Konopova and Jindra, 2007). Pre-mature development of adult structures was observed during larval-pupal metamorphosis in Met RNAi insects (Parthasarathy et al., 2008). |
Two homologues of Met are present (Met & GCE). Met mutants showed increase resistance to JH or its analogue, methoprene (Wilson and Fabian, 1986). Met mutant flies are viable (Wilson and Ashok, 1998). |
| Hypoxia | Most of the dsRNA injected insects died during the larval stage and a few died during the pupal stage. Some of the larvae that were able to develop into pupae showed problems in wing development and the wings were shorter and did not cover the abdomen. | Regulates responses to decreased oxygen concentrations. Plays an important role in tracheal outgrowth during oxygen deprived conditions (Lavista-Llanos, 2002). |
| E(spl)mga- mma1 | Some of the injected larvae died during quiescent stage. Pupae developed from E(spl)mgamma1 dsRNA injected larvae showed defects in wing development and development was arrested during pupal stage. Wings were shorter and did not fold properly. As this gene is expressed in wing imaginal discs, silencing of this gene would have lead to the formation of improper wings during the pupal stage. | Plays an important role in Notch signaling pathway (Lecourtois and Schweisguth, 1995; Bailey and Posakony, 1995). Loss of function results in neural hyperplasia and cell death (Knust et al., 1987). Is required for the segregation of a single sensory organ precursor in wing morphogenesis (de celis et al, 1996). E(spl) complex of genes show a distinct pattern of expression in wing imaginal discs (Jenning et al., 1995). |
TcSRC dsRNA injected final instar larvae showed reduced weight gain when compared to the malE injected final instar larvae and the larvae were unable to grow or increase in size to reach critical weight for metamorphosis and eventually died (Fig. 3D and Fig. 4A). In the penultimate instar of both Manduca sexta and Drosophila melanogaster, it has been shown that the larva must surpass the threshold size for metamorphosis to occur (Nijhout, 1975; Zhou et al., 2004). In TcSRC dsRNA injected larvae the amount of total lipids is reduced by about 50-60% when compared to the malE injected larvae (Fig. 4C). Earlier studies have shown that failure to surpass the minimum viable weight (minimal weight at which the amount of fat body storage is sufficient for survival through metamorphosis) in the last larval instar of M. sexta impaired the metamorphosis to occur (Nijhout, 1975). The decrease in the amount of total lipids in TcSRC dsRNA injected larvae would have directly affected the fat body metabolism and the larvae failed to reach the minimum viable weight to undergo metamorphosis. In the previous studies, SRC-2 knock-out mice have shown reduced weight gain and more hyperactive brown adipose tissue and a less white adipose tissue (Picard et al., 2002). In SRC-3 knock-out mice adipocyte differentiation assessed in mouse embryonic fibroblasts (MEFs) was severely impaired compared to the wild type animals (Louet et al., 2006). These studies suggest that SRC regulation of lipid metabolism is conserved between insects and vertebrate. In addition, some of the genes involved in the insulin signaling and TOR pathways were down regulated in TcSRC dsRNA injected insects (Fig. 4B). Recent studies in the D. melanogaster have highlighted the role of insulin like peptides (DILP’s) and the insulin receptor-signaling pathway in regulating the nutrition dependent growth rates (Oldham and Hafen, 2003). Therefore, it is likely that SRC either directly or indirectly regulates the expression of genes involved in insulin and TOR signaling pathways. Further studies are required to uncover mechanism of action on SRC in regulation of genes involved in insulin and TOR signaling pathways.
The D. melanogaster homolog for SRC is Taiman, a nuclear receptor coactivator. D. melanogaster Taiman is shown to be involved in follicular cell migration (Bai et al., 2000). Even though T. castaneum is more closely related to D. melanogaster than to the vertebrates, the % overall and % HLH identity of TcSRC with HsSRC is higher (17 and 71 respectively, Table 2a) when compared to its amino acid identity with DmTaiman (14% overall identity and 48% HLH identity). More details about the phenotypes observed in T. castaneum and comparisons with D. melanogaster mutants are summarized in Table 6. The mosquito homologue of the vertebrate SRC is FISC, a p160 coactivator of the ecdysone receptor (EcR) complex. Protein-protein interactions between the nuclear receptor BetaFTZ-F1 and FISC are required for the stage specific expression of 20E effector genes during mosquito reproduction (Zhu et al., 2006). DsRNA studies in T. castaneum showed that TcSRC is required for growth and development of this insect. The current studies on T. castaenum and the previous studies on related proteins in the fruit fly and mosquito suggest that T. castaneum SRC plays multiple roles in growth, development and reproduction by acting as a coactivator of nuclear receptors as reported in D. melanogaster (Bai et. al., 200) and A. aegypti (Zhu et al., 2006). However, whether SRC functions as only nuclear receptor coactivator in all these regulatory roles is not clear from these studies and requires further investigation.
In conclusion, Tribolium genome sequence was searched and identified 53 members of the bHLH superfamily. Phylogenetic analysis classified these 53 genes into ten families. Functional analysis using RNAi showed that seven members belonging to bHLH-PAS and HES families play a major role in the growth and metamorphosis of T. castaneum. More than 90% of larvae injected with TcTango, TcTrachealess, TcSRC, TcMet, died during the larval stage (Fig. 7). More than 90% of larvae injected with TcSpineless died during pupal stage (Fig. 7). These studies demonstrate the power of RNAi for functional characterization of members of the multigene families in this model insect. This first study on bHLH transcription factors in T. castaneum identified seven bHLH proteins that play key roles in growth and metamorphosis. The precise mechanism of action of these seven transcription factors in regulation of growth and metamorphosis of T. castaneum require further studies. The current contribution lays a solid foundation to begin studies on mechanism of action of individual bHLH transcription factors in regulation of growth and metamorphosis in T. castaneum. Since seven out of the 53 bHLH proteins are required for growth and metamorphosis of T. castaneum, these could serve as target sites to develop management tools to control this important storage pest. Some of the bHLH proteins such as TcSRC show well conserved function between insects and humans and these proteins are known to play key roles in causing human diseases such as cancer. T. castaneum could provide a simpler model system to study the function of these proteins.
Supplementary Material
Fig.6.
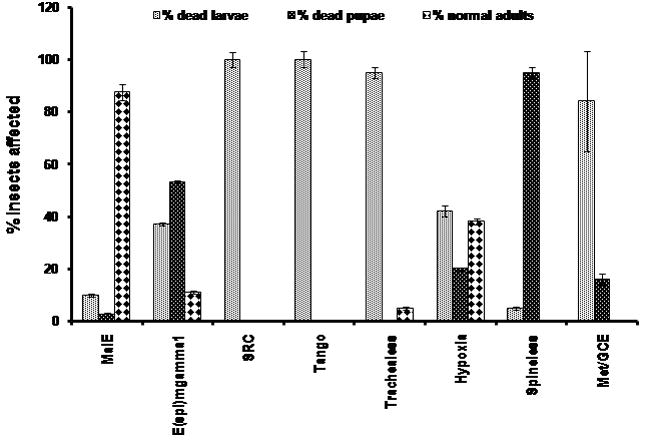
The effect of knock-down expression of bHLH genes on the development of T. castaneum. The proportion (percent) of dsRNA injected insects died during larval, pupal stages and unaffected insects (healthy adults) for each bHLH gene are shown. Mean+SD of two or three independent experiments are shown.
Acknowledgments
This work was supported by National Science Foundation (IBN-0421856) and National Institutes of Health (GM070559-05). This is contribution number 07-09-075 from the Kentucky Agricultural Experimental Station.
Abbreviations
- bHLH
basic helix-loop-helix
- PAS
Per, Arnt, Sim
- HES
Hairy-enhancer of split
- USF
Upstream transcription factor
- AS-C
Achaete-scute
- NCBI
National center for biotechnology information
- HGSC
Human genome sequencing center
- dsRNA
Double stranded RNA
- qRT-PCR
Quantitative real-time reverse transcriptase PCR
- PCR
Polymerase chain reaction
- TOR
Target of Rapamycin
- BSA
Bovine serum albumin
- PBS
Phosphate buffered saline
- OD
Optical density
- ARNT
Aryl hydrocarbon receptor nuclear translocator
- Met
Methoprene-tolerant protein
- GCE
Germ cell expressed
- SRC
Steroid receptor coactivator
- Emc
Extra macrochaete
- L’sc
Lethal of scute
- AEFI
After ecdysis into final instar
- ILP’s
Insulin-like peptides
- TSC1
Tuberous Sclerosis dimer 1
- RHEB1, RHEB2
Ras Homolog Enhanced Brain 1, 2
- Pi3k
Phosphatidylinositide 3-kinase
- PTEN
Phosphatase and tensin homolog
- FOXO
Forkhead box, Class O
- dInR
Drosophila Insulin Receptor
- HIF
Hypoxia-inducible factor
- Ahr
Aryl hydrocarbon receptor
- MEFs
Mouse embryonic fibroblasts
- DILPs
Drosophila insulin-like peptides
- EcR
Ecdysone receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahola V, Aittokallio T, Vihinen M, Uusipaikka E. A statistical score for assessing the quality of multiple sequence alignments. BMC Bioinformatics. 2006;7:484. doi: 10.1186/1471-2105-7-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altekar GS, Dwarkadas JP, Huelsenbeck P, Ronquist F. Parallel Metropolis- coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kikuchi M, Jockusch EL. Genetic patterning in the adult capitate antenna of the Tribolium castaneum. Dev Biol. 2008;327:240–251. doi: 10.1016/j.ydbio.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A. Positional Dependence, Cliques, and predictive Motifs in the bHLH Protein Domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon NC, Wappner P, O’ Rourke JF, Bartlett SM, Shilo B, Pugh CW, Ratcliffe PJ. Regulation of the Drosophila bHLH- PAS protein Sima by hypoxia: functional evidence for homology with HIF-1 alpha. Biochem Biophys Res Commun. 1998;3:811–816. doi: 10.1006/bbrc.1998.9234. [DOI] [PubMed] [Google Scholar]
- Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a D. melanogaster protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;5545:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Bucher G, Scholten J, Kingler M. Parental RNAi in Tribolium (coleopteran) Curr Biol. 2002;12:85–86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- de Celis JF, de Celis J, Ligoxygakis P, Preiss A, Delidakis C, Bray S. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development. 1996;122:2719–2728. doi: 10.1242/dev.122.9.2719. [DOI] [PubMed] [Google Scholar]
- Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by Spineless - aristapedia, a homolog the mammalian dioxin receptor. Genes Dev. 1998;9:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Ferre- D’Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric B/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischer A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C, Jan LY, Jan YN. Cell interactions and gene interactions in peripheral neurogenesis. Genes Dev. 1993;7:723–733. doi: 10.1101/gad.7.5.723. [DOI] [PubMed] [Google Scholar]
- Golubchik T, Wise MJ, Easteal S, Jermiin LS. Mind the Gaps: Evidence of Bias in Estimates of Multiple Sequence Alignments. Mol Bio Evol. 2007;24:2433–2442. doi: 10.1093/molbev/msm176. [DOI] [PubMed] [Google Scholar]
- Hall TA BioEdit. A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Handel EV. Rapid determination of Glycogen and Sugars in Mosquitoes. J AM MOSQ CONTROL ASSOC. 1985;1:299–301. [PubMed] [Google Scholar]
- Handel EV. Rapid determination of Total Lipids in Mosquitoes. J AM MOSQ CONTROL ASSOC. 1985;1:302–304. [PubMed] [Google Scholar]
- Hassen BA, Bellen HJ. Doing the MATH: Is the mouse a good model for fly development. Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- Jennings B, de Celis J, Delidakis C, Preis A, Bray S. Role of Notch and achaete-scute complex in the expression of Enhancer of split bHLH proteins. Development. 1995;120:3745–3752. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Jiang L, Crews ST. The D. melanogaster dysfusion basic helix- loop- helix (bHLH)- PAS gene control tracheal fusion and levels of trachealess bHLH-PAS protein. Mol Cell Biol. 2003;23:5625–5637. doi: 10.1128/MCB.23.16.5625-5637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesh T. Consequences of heterodimeric interactions among helix-loop-helix proteins. Cell Growth Differ. 1993;4:49–55. [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acid Res. 2005;32:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-smith A. The mammalian basic helix-loop-helix PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Kim MD, Jan YN, Jan YN. The bHLH-PAS protein spineless is necessary for the Diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20:2773–2778. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E, Bremer KA, Vassin H, Ziemer A, Tepass U, Campos-ortega JA. The enhancer of split locus and neurogenesis in Drosophila melanogaster. Dev Biol. 1987;122:262–273. doi: 10.1016/0012-1606(87)90351-4. [DOI] [PubMed] [Google Scholar]
- Konopova B, Jindra M. Juvenile hormone resistance gene methoprene tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc Natl Acad Sci U S A. 2007;104:10488–10493. doi: 10.1073/pnas.0703719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA- binding subunit of transcription factor PTF1 is a new exocrine pancreas- specific basic helix-loop-helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Kuzin B, Doszhanov K, Mazo A. Interaction between spineless- aristapedia Gene and genes from antennapedia and bithorax complexes of Drosophila melanogaster. Int J Dev Biol. 1997;41:867–875. [PubMed] [Google Scholar]
- Lavista- Llanos S, Centanin L, Irisarri M, Russo DM, Gleadle JM, Bocca SN, Muzzopappa M, Ratcliffe PJ, Wappner P. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol Cell Biol. 2002;19:6842–6853. doi: 10.1128/MCB.22.19.6842-6853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA- binding protein mediates the transcriptional activation of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the basic helix- loop-helix poteins. Genome Biol. 2002;3:research0030.1–research0030.18. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The Baisc Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome- wide analysis of basic/helix- loop- helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet JF, Coste A, Amazit L, Tannour-Louet M, Wu RC, Tsai MJ, Auwerx J, O’Malley BW. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci USA. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix- loop-helix factors in D. melanogaster. Proc Natl Acad Sci U S A. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, Mc Caw PS, Baltimore D. A new DNA binding and dimerizing motif in immunoglobulin enhancer binding, Daughterless, MyoD and Myc proteins. Cell. 1989a;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C, Mc Caw PS, Vaessin H, Caudy M, Jan LY, Cabrera CV, Buskin JN, Hauschka SD, Lasssar AB, Weintraub H. Interactions between heterologous helix- loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989b;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Muskavitch MAT. Delta-Notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Nair SK, Burley S. X-ray structures of Myc-Max and Mad-Max recognizing DNA: molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. A threshold size for metamorphosis in the tobacco hornworm, Manduca sexta (L) Biol Bull. 1975;149:214–225. doi: 10.2307/1540491. [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgene- specific post-transcriptional silencing is transmitted grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for morphogenesis of Drosophila sensory dendrites. Genes Dev. 2000;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Tan A, Palli SR. bHLH- PAS family transcriptior factor methoprene-tolerant plays a key role in JH action in preventing the premature development od adult structures during larval-pupal metamorphosis. Mech Dev. 2008;125:601–616. doi: 10.1016/j.mod.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O’Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Levine M, Kohara Y, Rokhsar D, Satoh N. A genomewide survey of developmentally genes in Ciona intestinalis I Genes for bHLH transcription factors. Dev Genes Evol. 2003;213:213–221. doi: 10.1007/s00427-003-0319-7. [DOI] [PubMed] [Google Scholar]
- Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix- loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaer N, Pistillo D, Gilbert JM, Lio P, Wulbeck C, Simpson P. Gene duplication at the achaete-scute complex and morphological complexity of the peripheral nervous system in Diptera. Trends Genet. 2002;18:399–405. doi: 10.1016/s0168-9525(02)02747-6. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crew S. The D. melanogaster tango gene encodes a bHLH-pAS protein that is orthologous to mammalian Arnt and control CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Toledo-ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop- helix transcription factor family. Plant Cell. 2002;5:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;7190:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC. Systemic signaling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen K, Yao Q, Wang W. The basic helix-loop-helix transcription factor family in Bombyx mori. Dev Genes Evol. 2007;217:715–723. doi: 10.1007/s00427-007-0184-x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Rose DW, Hermanson O, Liu F, Herman T, Wu W, Szeto D, Gleiberma A, Krones A, Pratt K, Rosenfeld R, Glass CK, Rosenfeld MG. Regulation of Somatic growth by the p160 coactivator p/CIP. Proc Natl Acad Sci USA. 2000;97:13549–13554. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Baumann A, Wilson TG. D. melanogaster melanogaster Methoprene-tolerant (Met) gene homologs from three mosquito species: members of PAS transcriptional factor family. J Insect Physiol. 2007;53:246–253. doi: 10.1016/j.jinsphys.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The MyoD family and myogenesis: Redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for color vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TG, Fabian J. A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev Biol. 1986;118:190–201. doi: 10.1016/0012-1606(86)90087-4. [DOI] [PubMed] [Google Scholar]
- Wilson TG, Ashok M. Insecticide resistance resulting from an absence of target- site gene product. Proc Natl Acad Sci U S A. 1998;95:14040–14044. doi: 10.1073/pnas.95.24.14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-komiya H, Deng C, O’Malley BW. The Steroid Receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ ACTR/ TRAM-1) is required for normal growth, puberty, female reproductive function and mammary gland development. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E, Wappner P, Shilo B. The PAS domain confers target gene specificity of D. melanogaster bHLH/PAS proteins. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhou B, Truman JW, Riddiford LM. Overexpression broad: a new insight into its role in the Drosophila prothoracic gland cells. J Exp Biol. 2004;207:1151–1161. doi: 10.1242/jeb.00855. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen L, Sun G, Raikhel AS. The competence factor beta Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol. 2006;26:9402–9412. doi: 10.1128/MCB.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


