Abstract
In DNA methylation microarray analysis, quantitative assessment of intermediate methylation levels in samples with various global methylation levels is still difficult. Here, specifically for methylated DNA immunoprecipitation-CpG island (CGI) microarray analysis, we developed a new output value. The signal log ratio reflected the global methylation levels, but had only moderate linear correlation (r = 0.72) with the fraction of DNA molecules immunoprecipitated. By multiplying the signal log ratio using a coefficient obtained from the probability value that took account of signals in neighbouring probes, its linearity was markedly improved (r = 0.94). The new output value, Me value, reflected the global methylation level, had a strong correlation also with the fraction of methylated CpG sites obtained by bisulphite sequencing (r = 0.88), and had an accuracy of 71.8 and 83.8% in detecting completely methylated and unmethylated CGIs. Analysis of gastric cancer cell lines using the Me value showed that methylation of CGIs in promoters and gene bodies was associated with low and high, respectively, gene expression. The degree of demethylation of promoter CGIs after 5-aza-2'-deoxycytidine treatment had no association with that of induction of gene expression. The Me value was considered to be useful for analysis of intermediate methylation levels of CGIs.
Keywords: epigenetics, CpG island microarray, 5-aza-2′-deoxycytidine, methylation silencing, gastric cancer
1. Introduction
DNA methylation plays a critical role during mammalian development and differentiation. Methylation of a CpG island (CGI) in a gene promoter region has been known to repress transcription of its downstream gene.1 At the same time, DNA methylation statuses of CGIs are faithfully inherited upon cell replication,2 and are considered to work as a stable switch of gene transcription.1 Once a promoter CGI is aberrantly methylated, it leads to permanent aberrant silencing of its downstream gene. Aberrant DNA methylation is deeply involved in human cancers,3 and is also likely to be involved in other human-acquired disorders.4
There is a great interest in genome-wide analysis of DNA methylation, and new technologies involving microarrays and next-generation sequencers are being developed,5 replacing traditional techniques.6 In comparison with techniques using next-generation sequencers, microarray techniques are cost-effective and do not need complex bioinformatics. Their hybridization probes can be prepared by bisulphite modification of unmethylated cytosines, use of methylation-sensitive restriction enzymes, and affinity purification. The affinity purification can be performed by an antibody against 5-methylcytidine or by methylated DNA binding domains (MBDs). It has an advantage over the use of restriction enzymes, since genomic regions analysed by affinity purification are not limited to restriction sites of methylation-sensitive enzymes.
Methylated DNA immunoprecipitation (MeDIP)-microarray analysis has been used to obtain a high-resolution whole-genome DNA methylation profile of various genomes.7–17 However, quantitative assessment of intermediate methylation levels has been hampered by the difficulty in appropriate normalization. Methylation levels have a unique distribution pattern that is essentially different from gene transcription and is likely to be bimodal.17,18 Also, global methylation levels in different samples are highly variable, and there are few reference genes that have consistent methylation levels across various samples. To overcome these issues, two methods (Batman and MEDME) were recently developed.14,16
Batman (Bayesian tool for methylation analysis) transforms a signal log ratio of an individual probe to a value of methylation level taking account of the methylation levels of nearby CpG sites using standard Bayesian techniques. It is capable of processing data obtained by microarray and by next-generation sequencers. The method was validated by bisulphite sequencing of sperm samples,14 and its validity in samples with different global methylation levels remains to be established. MEDME (modelling experimental data with MeDIP enrichment) weighs signal log ratios of individual probes using a logistic model and signals obtained by neighbouring probes and by using completely methylated DNA samples.16 Both Batman and MEDME had good correlation with methylation levels obtained by bisulphite sequencing (R2 = 0.82 and 0.75, respectively). Also, both are capable of processing data from both CpG-rich and -poor regions, and this made their conversion algorithm complex as a trade-off.
In this study, we developed a novel output value, the ‘Me value’, that can be calculated from raw output values, and confirmed that the value had a linear correlation with methylation levels of genomic regions using samples with various global methylation levels. The Me value was used to clarify how methylation of CGIs in various positions against transcription start sites (TSSs) is associated with gene expression, and how demethylation of CGIs is associated with re-expression of genes.
2. Materials and methods
2.1. Cell lines and tissue samples
AGS and KATOIII gastric cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA) and the Japanese Collection of Research Bioresources (Tokyo, Japan), respectively. HSC39 and HSC57 gastric cancer cell lines were gifted by Dr K. Yanagihara, National Cancer Center Research Institute, Tokyo, Japan. Treatment with a demethylating agent, 5-aza-2'-deoxycytidine (5-aza-dC, Sigma, St Louis, MO, USA), was performed as in our previous study (AGS, 3 days, 1 µM).19 A normal gastric tissue sample was prepared by pooling endoscopic biopsy specimens from three healthy volunteers with informed consents. High-molecular-weight DNA was extracted by the phenol/chloroform method with RNase A treatment.
2.2. MeDIP and quantification of the number of immunoprecipitated DNA molecules
Five micrograms of genomic DNA were sonicated by a VP-5s homogenizer (TAITEC, Saitama, Japan) to fragment lengths between 200 and 800 bp. The mode of fragment length was about 300 bp. After heat denaturation at 95°C for 10 min, DNA was incubated with 5 µg antibody against 5-methylcytidine (Diagnode, Liége, Belgium) in 1× IP buffer [10 mM Na-phosphate, pH 7.0, 140 mM NaCl, 0.05% (w/v) Triton X-100] at 4°C overnight. Immune complexes were collected with Dynabeads Protein A (Invitrogen Dynal AS, Oslo, Norway), washed with 1× IP buffer four times, treated with Proteinase K, and purified by phenol and chloroform extraction and isopropanol precipitation.
To assess the fraction of immunoprecipitated (IP) DNA molecules among that of the total DNA (whole cell extract DNA, WCE) molecules, the number of IP and WCE molecules was quantified by real-time PCR using SYBR® Green I (BioWhittaker Molecular Applications, Rockland, ME, USA) and an iCycler Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) as described previously.20 All primers used in this study are listed in Supplementary Table S1.
2.3. CGI microarray analysis
Methylation microarray analysis was carried out using a human CGI oligonucleotide microarray (Agilent Technologies, Santa Clara, CA, USA) that contained 237 220 probes in or within 95 bp either side of a CGI and covered 27 800 CGIs with an average probe spacing of 100 bp. IP from 4.5 µg of sonicated DNA and 1.0 µg of WCE, without any amplification, were labelled with Cy5 and Cy3, respectively, using an Agilent Genomic DNA Labeling Kit PLUS (Agilent Technologies). Labelled DNA was hybridized to the microarray at 67°C for 40 h with constant rotation (20 rpm), and then scanned with an Agilent G2565BA microarray scanner (Agilent Technologies).
From the scanned data, signal values of the IP and WCE were obtained using Feature Extraction Ver.9.1 (Agilent Technologies). These two signal values were normalized using background subtraction, and signal ratio (IP/WCE), signal log ratio [log2(IP/WCE)], P[X], and P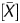 were obtained using Agilent G4477AA ChIP Analytics 1.3 software (Agilent Technologies). The P[X] and P
were obtained using Agilent G4477AA ChIP Analytics 1.3 software (Agilent Technologies). The P[X] and P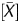 values, which are used in chromatin immunoprecipitation (ChIP)-on-chip analysis to obtain a binding call,21–25 were defined as the probability how the X (
values, which are used in chromatin immunoprecipitation (ChIP)-on-chip analysis to obtain a binding call,21–25 were defined as the probability how the X (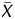 ) value deviates from Gaussian distribution of X (
) value deviates from Gaussian distribution of X (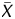 ) values of the entire genome of a sample. Here, the X value for a probe was obtained as the difference between the IP and the WCE signals after adjusting the symmetry of its distribution. The
) values of the entire genome of a sample. Here, the X value for a probe was obtained as the difference between the IP and the WCE signals after adjusting the symmetry of its distribution. The 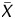 value for a probe was calculated as an average X, taking account of signals for neighbouring probes (within 1 kb of the probe). In addition, to calculate signal log ratio in experiments specifically referred to, the two signal values were also normalized by the Median and the Lowess normalization methods. The microarray results were submitted to the GEO database (GSE15291).
value for a probe was calculated as an average X, taking account of signals for neighbouring probes (within 1 kb of the probe). In addition, to calculate signal log ratio in experiments specifically referred to, the two signal values were also normalized by the Median and the Lowess normalization methods. The microarray results were submitted to the GEO database (GSE15291).
2.4. Calculation of the Me value
The Me value of each probe (site Me value) was calculated as Me value = [signal log ratio × (1 − P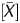 ) − k]/l + 0.5. The P
) − k]/l + 0.5. The P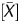 value and signal log ratio normalized using background subtraction were used for this formula. The [signal log ratio × (1 − P
value and signal log ratio normalized using background subtraction were used for this formula. The [signal log ratio × (1 − P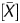 )] value mostly ranged from 0 to 2.6 in this study, and in general, the distribution depends on the microarray platform. Accordingly, the constant l was fixed at 2.6 in this study, so that the Me value would be within a range between 0 and 1. Me values larger than 1 and those smaller than 0, which were occasionally produced after calculation, were corrected to 1 and 0, respectively. The constant k was calculated as [the signal log ratio of CGIs that had a 50% fraction of DNA molecules IP (1.7 in this study) – 0.4], which equalled to 1.3 in this study. The signal log ratio of CGIs with 50% methylation depends on the microarray platform, labelling method, and mixture rate of IP and WCE, but does not need to be changed once established to suit a protocol.
)] value mostly ranged from 0 to 2.6 in this study, and in general, the distribution depends on the microarray platform. Accordingly, the constant l was fixed at 2.6 in this study, so that the Me value would be within a range between 0 and 1. Me values larger than 1 and those smaller than 0, which were occasionally produced after calculation, were corrected to 1 and 0, respectively. The constant k was calculated as [the signal log ratio of CGIs that had a 50% fraction of DNA molecules IP (1.7 in this study) – 0.4], which equalled to 1.3 in this study. The signal log ratio of CGIs with 50% methylation depends on the microarray platform, labelling method, and mixture rate of IP and WCE, but does not need to be changed once established to suit a protocol.
The Me value was calculated only for probes with high reliability. To select such probes, first, probes that yielded extremely high signal intensities (5-fold higher than average) for the WCE (Cy-3) were excluded. Since the signals obtained for the WCE should be the same theoretically for all the probes, extremely high signals were considered to be due to cross-hybridization. Then, continuity of signal log ratios of neighbouring probes was enforced. If the value of a probe was higher than those of neighbouring probes on both sides, it was corrected to their average because the value was likely to be an error. In addition, efficiency in labelling and hybridization in each microarray analysis was monitored by the signal log ratio and the fraction of DNA molecules IP by MeDIP at 10 probe loci. The data processing for the Me value was performed by Excel 2007 (Microsoft, Redmond, WA, USA), and the templates are available upon request.
2.5. Definitions of genomic regions
The position of each probe against a TSS was determined using UCSC hg18 (NCBI Build 36.1, March 2006). A single CGI was defined as an assembly of probes in the CGI microarray with intervals <500 bp. CGIs were classified into four categories: upstream CGIs (within 10 kb upstream of the TSS), divergent CGIs (within 10 kb upstream of the TSSs of two genes that are transcribed in opposite directions), gene body CGIs, and downstream CGIs (within 10 kb downstream of genes). A CGI spanning both an upstream region and a gene body was split into an upstream CGI and a gene body CGI. A putative promoter region (promoter) was defined as a region between a TSS, determined by UCSC hg18 (NCBI Build 36.1, March 2006) and its 200 bp upstream. According to these definitions, 34 697 assemblies of probes were defined as CGIs, and 9624 assemblies were defined as promoters. Genes with multiple promoters were analysed as different genes because of their multiple TSSs. CGIs that could not be classified by these criteria (4164 CGIs) were omitted from the following analysis. An average number of probes that covered a single CGI (or a single promoter) was 6.8 (2.0), and the distribution of the numbers is shown in Supplementary Fig. S1.
2.6. Expression microarray analysis
Microarray analysis of gene expression was performed using GeneChip (Affymetrix) as described previously,19,26 and the signal intensities were normalized, so that the average intensity of all the genes on a microarray would be 500. The average signal intensity of all the probes for a gene was used as its expression level. Genes with signal intensities of 1000 or more and of 250 or less were defined as those with high and low expression, respectively.
2.7. Bisulphite treatment, methylation-specific PCR, and bisulphite sequencing
Bisulphite modification was performed using BamHI-digested genomic DNA as descried previously.26 Methylation-specific PCR (MSP) was performed using bisulphite-treated DNA as descried previously.26 Bisulphite sequencing was performed after cloning the PCR product (10 clones or more for each sample). We used the data of methylation status previously analysed.19,26–30
2.8. Determination of methylation levels of a genomic region and CGI (or promoter) using the microarray data
A methylation level of a genomic region analysed by quantitative PCR of IP DNA molecules was assessed by an output value of a probe within the PCR product and closest to the forward primer, or a probe closest to the PCR product when no probes were present within the PCR product. A methylation level of a region analysed by bisulphite sequencing (200 bp) was assessed by an output value of a probe in the centre of the region. This was possible because bisulphite sequencing was performed for a region larger than 100 bp upstream and downstream of a probe, and methylation statuses of CpG sites within the 200 bp region were scored. A methylation level of a CGI (or promoter) was assessed by an average of site Me values of the probes located within the CGI (or promoter), which was defined as the CGI (or promoter) Me value.
3. Results and discussion
3.1. Assessment of current output values for methylation levels
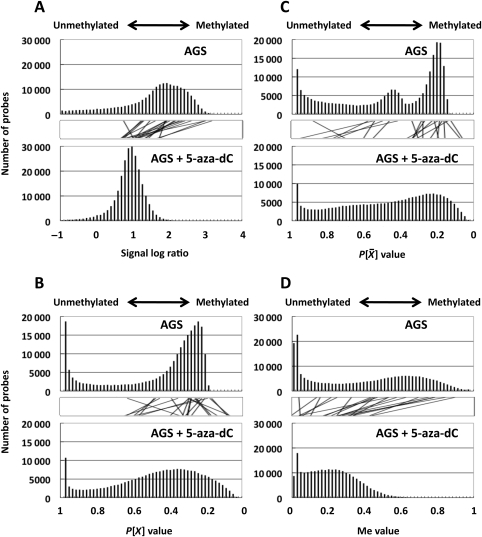
We first examined whether or not distribution of the available output values reflected the global methylation levels. The output values analysed were: (i) the signal log ratio, which is most frequently used in microarray analysis, (ii) the P[X] value, and (iii) P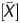 value, which is often used in ChIP-on-chip analysis. Their distribution was analysed in two samples, a cell line (AGS) with frequent CGI methylation26 and the same cell line after treatment with 5-aza-dC. Our previous study showed that AGS after 5-aza-dC treatment has demethylation of at least 421 promoter CGIs,19 and its global methylation level was expected to shift towards unmethylated ranges. Among the three values, the signal log ratio showed such a shift (Fig. 1A). On the other hand, the P[X] and P
value, which is often used in ChIP-on-chip analysis. Their distribution was analysed in two samples, a cell line (AGS) with frequent CGI methylation26 and the same cell line after treatment with 5-aza-dC. Our previous study showed that AGS after 5-aza-dC treatment has demethylation of at least 421 promoter CGIs,19 and its global methylation level was expected to shift towards unmethylated ranges. Among the three values, the signal log ratio showed such a shift (Fig. 1A). On the other hand, the P[X] and P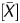 values did not show such a shift (Fig. 1B and C).
values did not show such a shift (Fig. 1B and C).
Figure 1.
Distribution of methylation levels assessed by the available output values and the Me value in a gastric cancer cell line, AGS, and the same cell line after 5-aza-dC treatment. The analysis was performed for the signal log ratio with background subtraction normalization (A), the P[X] value (B), the P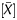 value (C), and the Me value (D). Lines between the two bar graphs indicate values of individual probes (randomly selected representative 20 values). Distributions of the signal log ratio and the Me value reflected the global methylation level, as shown by their shift towards smaller values in cells with 5-aza-dC treatment. In contrast, those of the P[X] and P
value (C), and the Me value (D). Lines between the two bar graphs indicate values of individual probes (randomly selected representative 20 values). Distributions of the signal log ratio and the Me value reflected the global methylation level, as shown by their shift towards smaller values in cells with 5-aza-dC treatment. In contrast, those of the P[X] and P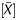 values did not show the shift.
values did not show the shift.
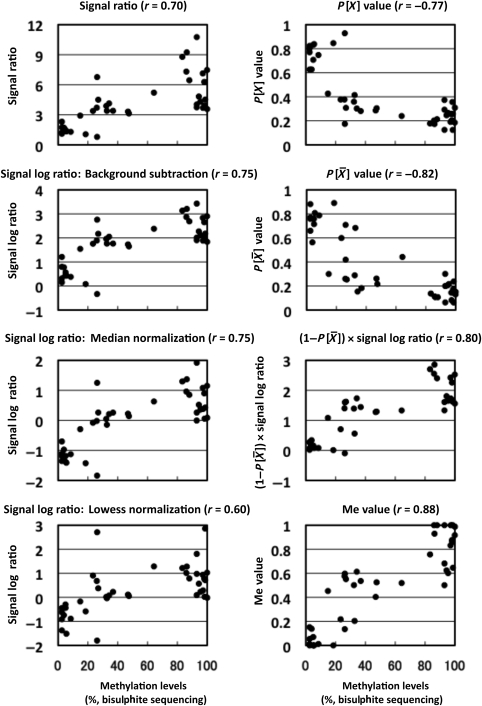
Next, a linear correlation with methylation level, represented here by the fraction of DNA molecules IP by the anti-5-methylcytidine antibody, was examined for individual output values using 31 genomic regions located within various CGIs. In addition to the three output values, (iv) the signal ratio (background subtraction normalization), (v) the signal log ratio with Median normalization, and (vi) the signal log ratio with the Lowess normalization were analysed. Although P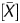 gave a correlation coefficient of −0.91 (r, Pearson's), the absolute values of correlation coefficients using other output values were smaller than 0.72 (Supplementary Fig. S2). Median and Lowess normalization did not improve the linear correlation.
gave a correlation coefficient of −0.91 (r, Pearson's), the absolute values of correlation coefficients using other output values were smaller than 0.72 (Supplementary Fig. S2). Median and Lowess normalization did not improve the linear correlation.
3.2. Development of a novel output value ‘Me value’
To improve the linearity of the signal log ratio, which reflected the global methylation levels, P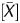 , which had a strong linear correlation, was used as a coefficient to multiply it. Because P
, which had a strong linear correlation, was used as a coefficient to multiply it. Because P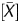 showed an inverse correlation, (1 − P
showed an inverse correlation, (1 − P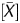 ) was used as a coefficient to multiply the signal log ratio ‘=(1 − P
) was used as a coefficient to multiply the signal log ratio ‘=(1 − P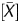 ) × signal log ratio’. This value showed a higher correlation coefficient (0.93) than the other output values (Supplementary Fig. S2). This value was scaled to a value with a minimum value of 0 and a maximum value of 1 using the constants in Section 2, and the scaled output value was designated as the ‘Me value’. The Me value had the largest correlation coefficient (0.94) among all of the output values analysed. The distribution of the Me value reflected the global methylation levels by showing a shift towards smaller values after demethylation (Fig. 1D).
) × signal log ratio’. This value showed a higher correlation coefficient (0.93) than the other output values (Supplementary Fig. S2). This value was scaled to a value with a minimum value of 0 and a maximum value of 1 using the constants in Section 2, and the scaled output value was designated as the ‘Me value’. The Me value had the largest correlation coefficient (0.94) among all of the output values analysed. The distribution of the Me value reflected the global methylation levels by showing a shift towards smaller values after demethylation (Fig. 1D).
The Me value was generated taking advantage of the signal log ratio and P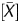 . The signal log ratio was not quantitative but reflected the global methylation levels. On the other hand, P
. The signal log ratio was not quantitative but reflected the global methylation levels. On the other hand, P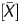 had a high linear correlation with the fraction of DNA molecules IP by the anti-5-methylcytidine antibody. The P
had a high linear correlation with the fraction of DNA molecules IP by the anti-5-methylcytidine antibody. The P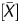 value is obtained as a probability value that takes account of neighbouring probes, and reflects the methylation level of a small local region. Since the vast majority of CpG sites within a CGI are (un)methylated when the CGI is (un)methylated,31 the P
value is obtained as a probability value that takes account of neighbouring probes, and reflects the methylation level of a small local region. Since the vast majority of CpG sites within a CGI are (un)methylated when the CGI is (un)methylated,31 the P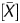 value was considered to have an advantage in faithful reflection of the local methylation status.
value was considered to have an advantage in faithful reflection of the local methylation status.
3.3. High accuracy of MeDIP-CGI microarray with Me value
In addition to the linear correlation between the Me value and the fraction of DNA molecules IP, a linear correlation between the Me value and the fraction of methylated CpG sites was analysed. Fractions of methylated CpG sites of 11 genomic regions (each 200 bp) with a variety of methylation levels were obtained by bisulphite sequencing in four different cell lines with different global methylation levels26 (44 values in total). The Me value was obtained for a probe (site Me value) in the centre of the genomic region analysed. A strong correlation (r = 0.88, Fig. 2) was observed. The correlation was stronger than any of those obtained by the other output values. When the analysis was limited to genomic regions with intermediate Me values, the correlation coefficients were 0.75 (Me value: 0.1–0.9); 0.53 (0.2–0.8); 0.47 (0.3–0.7); and 0.19 (0.4–0.6). These data further supported that a site Me value reflects a methylation level of a small genomic region (200 bp), even if it is within an intermediate range, and that the Me value is useful for the analysis of various biological samples, such as tissue samples.
Figure 2.
Correlation between an output value and the fraction of methylated CpG sites by bisulphite sequencing. Forty samples with variable methylation levels (11 loci in four cell lines) were analysed by bisulphite sequencing, and the fraction of methylated CpG sites among all the CpG sites within 200 bp of the microarray probes was obtained. The numbers of CpG sites in this region ranged from 7 to 21. The Me value had a linear correlation with the fraction (r = 0.88, Pearson's correlation coefficient).
Cancer tissues sometimes show mixtures of methylated and unmethylated CpG sites on the same DNA molecule (mosaic pattern). Even in this case, multiple CpG sites are usually located within the average size of shearing DNA (300 bp) because there are 9–53 CpG sites in 300 bp regions of promoter CGIs.20 If two or more CpG sites are methylated, such DNA molecules are reported to be efficiently IP.9 Therefore, it is expected that, for most CGIs, the Me value will work even in samples with mosaic pattern methylation, such as cancer tissues.
Next, detection of completely methylated and unmethylated statuses of CGIs was attempted. Using output values other than the Me value, this has been achieved by optimizing cut-off values depending upon samples because their global methylation levels were highly variable. Since the Me value well reflected the global methylation levels, we tried to use cut-off CGI Me values common to different samples. Methylation statuses of 113 CGIs in four cell lines with different global methylation levels26 (452 values in total) were scored using various cut-off CGI Me values. A high specificity with little compromise of sensitivity was achieved with the cut-off values of 0.6 and 0.4 for highly methylated and unmethylated CGIs, respectively (Supplementary Fig. S3). Accuracies for methylated and unmethylated CGIs with these cut-off values were 71.8 and 83.8%, respectively (Table 1). Since these cut-off CGI Me values worked in the four samples with different global methylation levels, Me values of 0.6 and 0.4 were considered to be suitable to identify highly methylated and unmethylated CGIs, respectively, with reasonable accuracies.
Table 1.
Comparison between methylation statuses determined using the CGI Me value and those by MSP
| MSP | MeDIP-CGI microarray | AGS | HSC39 | HSC57 | KATOIII | Total |
|---|---|---|---|---|---|---|
| Methylated | Highly methylated | 64 | 17 | 12 | 36 | 129 |
| Moderately methylated | 29 | 13 | 6 | 23 | 71 | |
| Unmethylated | 4 | 3 | 6 | 8 | 21 | |
| Unmethylated | Highly methylated | 1 | 6 | 7 | 4 | 18 |
| Moderately methylated | 2 | 8 | 11 | 4 | 25 | |
| Unmethylated | 11 | 56 | 61 | 21 | 149 | |
| Methylated/unmethylated | Highly methylated | 0 | 4 | 5 | 8 | 17 |
| Moderately methylated | 1 | 4 | 2 | 5 | 12 | |
| Unmethylated | 1 | 2 | 2 | 4 | 9 | |
| Not amplified by MSP | 0 | 0 | 1 | 0 | 1 | |
| Methylated | Sensitivity | 0.660 | 0.515 | 0.500 | 0.537 | 0.584 |
| Specificity | 0.938 | 0.875 | 0.864 | 0.739 | 0.848 | |
| Accuracy | 0.699 | 0.770 | 0.786 | 0.619 | 0.718 | |
| Unmethylated | Sensitivity | 0.786 | 0.800 | 0.772 | 0.724 | 0.776 |
| Specificity | 0.949 | 0.884 | 0.758 | 0.857 | 0.884 | |
| Accuracy | 0.929 | 0.832 | 0.768 | 0.823 | 0.838 |
Using 113 CGIs, we compared methylation statuses determined using the Me value and those by MSP.
Methylated/unmethylated, both methylated and unmethylated DNA molecules were detected by MSP.
These data showed that, for quantification of methylation levels, the Me value had linearity similar to Batman and MEDME.14,16 Although samples analysed were different, the coefficient of determination (R2, Pearson's) obtained by the Me value was 0.77, being in the same range as Batman (0.82) and MEDME (0.75). The linearity of the Me value was validated also using four samples with different global methylation levels, which has not been done with Batman (one sample) and MEDME (two samples). Further, the CGI Me value enabled us to score completely methylated and unmethylated statuses of CGIs in samples with variable global methylation levels using common cut-off values. It is also of note that the Me value can be conveniently calculated using a spreadsheet and a commercially available software, such as Excel (Microsoft).
3.4. Application of the Me value to analysis of the association between CGI methylation and gene expression
Using the Me value, methylation profiles of CGIs and promoters were analysed in four gastric cancer cell lines, AGS cells after 5-aza-dC treatment, and one normal gastric tissue sample (a pool of four samples from four individuals). CGIs (promoters) with CGI (promoter) Me values more than 0.6 were classified as ‘methylated’. Of the 30 533 CGIs analysed, 3768–7310 CGIs were methylated in the cancer cell lines, and 3393 CGIs were methylated in the normal sample (Table 2). However, methylation was infrequent in promoters (Table 2, Supplementary Fig. S4A and B). The ratio of methylated promoters in cancer cell lines (6.7–12.5%) was in accordance with that in a previous report (10.9% in lung cancer cell lines).32 The difference between overall CGIs and promoters was clearer in the normal gastric tissue than in the human gastric cancer cell lines. Interestingly, CGIs in the vicinity of the LINE and SINE repetitive elements had lower Me values than those further away from them in cancer cell lines (Supplementary Fig. S4C and D). A small peak was observed at 300 bp from SINE, and CGIs closer to SINE and LINE were less methylated than those further apart from SINE and LINE. There is a possibility that a boundary function of these repetitive elements prevents methylation beyond the boundary spread across into CGIs. Actually, SINE is reported to have binding sites for the TFIIIC transcription factor, and to protect promoter CGIs from repressive chromatin modifications.33
Table 2.
DNA methylation profiles of CGIs according to their positions against TSSs obtained by MeDIP-CGI microarray
| Total number analysed | Number of methylated CpG islands and promoters |
||||||
|---|---|---|---|---|---|---|---|
| AGS | HSC39 | HSC57 | KATOIII | AGS + 5-aza-dC (1 µM) | Normal gastric tissue | ||
| CpG islands | |||||||
| All CGI | 30 533 | 7310 (23.9%) | 3768 (12.3%) | 4663 (15.3%) | 6460 (21.2%) | 4 (0.0%) | 3393 (11.1%) |
| Upstream | 10 709 | 1911 (17.8%) | 937 (8.7%) | 794 (7.4%) | 1556 (14.5%) | 1 (0.0%) | 260 (2.4%) |
| Gene body (+1 to +1 k) | 10 654 | 1607 (15.1%) | 867 (8.1%) | 665 (6.2%) | 1328 (12.5%) | 0 (0.0%) | 194 (1.8%) |
| Gene body (+1 to +5 k) | 2050 | 694 (47.0%) | 508 (24.8%) | 598 (29.2%) | 809 (39.5%) | 1 (0.0%) | 353 (17.2%) |
| Gene body (more than +5 k) | 4431 | 2438 (55.0%) | 1099 (24.8%) | 2171 (49.0%) | 2155 (48.6%) | 2 (0.0%) | 2360 (53.3%) |
| Downstream | 1186 | 561 (47.3%) | 310 (26.1%) | 376 (31.7%) | 509 (42.9%) | 0 (0.0%) | 216 (18.2%) |
| Divergent | 1503 | 99 (6.6%) | 47 (3.1%) | 59 (3.9%) | 103 (6.9%) | 0 (0.0%) | 10 (0.7%) |
| Promoters | 9624 | 1205 (12.5%) | 792 (8.2%) | 641 (6.7%) | 1142 (11.9%) | 3 (0.0%) | 113 (1.2%) |
| Probes | 237 202 | 70 027 (29.5%) | 43 825 (18.5%) | 48 292 (20.4%) | 65 192 (27.5%) | 723 (0.3%) | 27 017 (11.4%) |
Definitions of the CGI positions are described in Section 2.
The numbers of methylated CGIs were larger in AGS and KATOIII than in HSC39 and HSC57, which was in accordance with our previous findings.26 The difference among the four cell lines was observed not only at the overall genome level but also on individual chromosomes (Supplementary Fig. S5). In contrast, the numbers of methylated promoters had the same difference at the overall genome level, but some distortions were present on specific chromosomes. For example, HSC39 had the largest numbers of methylated promoters on chromosomes 15, 18, and 22, and HSC57 had the largest number on chromosome 21. This result suggested that promoters have unique mechanisms to be methylated in cancer cells, such as functional selection of a cell with methylation of specific promoters.
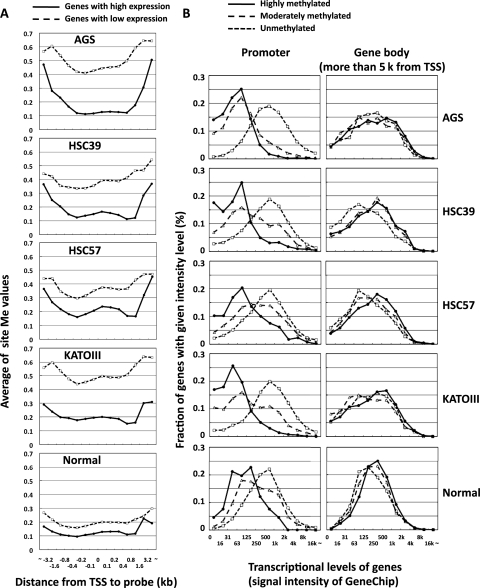
Gene expression profiles were also obtained by expression microarray analysis of the same four cancer cell lines and the normal gastric tissue sample. When genes were classified by their expression levels, genes with high expression had lower methylation levels in their promoters than those with low expression in all cell lines (Fig. 3A). When genes were classified by methylation levels (unmethylated, moderately methylated, and highly methylated), genes with high methylation in their promoters had lower expression than those with low methylation (left panel in Fig. 3B). In contrast, genes with high methylation in their gene bodies (5 kb or more downstream of TSSs) had slightly, but significantly, higher expression levels (right panel). These results clearly showed that methylation of gene body CGIs was associated with increased gene expression, as in previous reports.34–37 Finally, genes with low expression in normal gastric tissue had higher methylation levels of promoters in cancer cell lines (Supplementary Fig. S6). This result was in line with the fact that genes with low expression are susceptible to DNA methylation.20,38,39
Figure 3.
Association between DNA methylation level and gene expression. (A) Average methylation levels (site Me values) of genes with high and low expression. Genes with low expression (dashed line) showed significantly higher methylation levels than those with high expression (solid line) in positions close to TSSs. (B) Gene expression levels according to methylation statuses of the promoters (left panel) and gene bodies (right panel). Genes were classified into unmethylated (CGI Me value <0.4, dotted line), moderately methylated (0.4 ≤ CGI Me value ≤ 0.6, dashed line), and highly methylated (CGI Me value >0.6, solid line). Methylation of promoters was strongly associated with low expression (P = 8 × 10−203 in AGS, t-test of expression of methylated genes and unmethylated genes), and methylation of CGIs in gene bodies was associated with higher expression (P = 6 × 10−4 in AGS).
3.5. Application of the Me value to analyse the effect of a demethylating agent
Treatment of cells with 5-aza-dC induces demethylation of various CGIs, but the degree depending upon their positions against TSSs has not been clarified. The relationship between the degree of demethylation of promoters and that of expression induction has not been clarified, either. To address these two issues, we analysed methylation and gene expression levels in AGS before and after 5-aza-dC treatment.
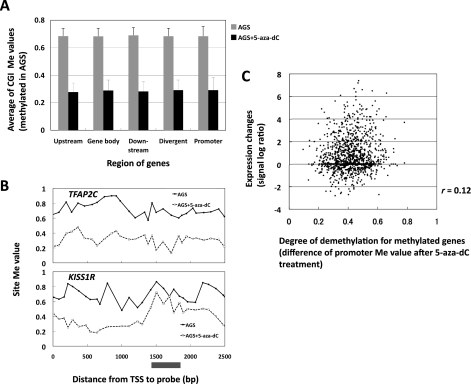
Demethylation of the genes with methylated CGIs or promoters (Me value > 0.6) before 5-aza-dC treatment was analysed. The average degree of demethylation was not influenced by the positions of CGIs against their TSS (Fig. 4A), whereas the degree of demethylation of individual CGIs was highly variable (representative genes in Fig. 4B). The average degree was not influenced by the distance between a CGI and repetitive elements (LINE, SINE), either (data not shown). There was no correlation (r = 0.12, Fig. 4C) between the degree of demethylation of promoters (decrease of Me value in MeDIP-CGI microarray) and that of induction of gene expression (fold increase of signal log ratio in expression microarray analysis). The majority of genes with methylation of their promoters showed little or no increase of expression after 5-aza-dC treatment. The number of methylated promoters identified by MeDIP-microarray analysis was much larger than that of genes identified as silenced by expression microarray analysis after 5-aza-dC treatment (Table 3).
Figure 4.
Degree of demethylation according to the positions against TSSs, and the lack of association between the degree of demethylation and that of re-expression. (A) Degree of demethylation according to the positions against TSSs. Methylation levels were analysed for CGIs methylated in AGS without 5-aza-dC treatment. 5-Aza-dC treatment induced demethylation of upstream CGIs, gene body CGIs, downstream CGIs, divergent CGIs and promoters to similar degrees. (B) Site Me value of TFAP2C and KISS1R in AGS and AGS treated with 5-aza-dC. In 1400–1800 bp against TSS, the degree of demethylation was high in TFAP2C and low in KISS1R, which representatively showed variation among individual genes. (C) Degree of demethylation of promoters and degree of re-expression in AGS. The degree of demethylation of promoters was not associated with that of re-expression of methylated genes after 5-aza-dC treatment.
Table 3.
The number of methylated promoters by MeDIP-microarray analysis and silenced genes by expression microarray after 5-aza-dC treatment
| Number of methylated promoter CGIs |
||||
|---|---|---|---|---|
| AGS | HSC39 | HSC57 | KATOIII | |
| MeDIP-CGI microarraya | 1205 | 792 | 641 | 1142 |
| Expression microarray after 5-aza-dC treatmentb (Moriguchi et al.26) | 25 | 4 | 1 | 41 |
aPromoters identified as methylated by MeDIP-microarray analysis, among 9624 genes arrayed on CpG island microarray (Agilent).
bThe genes in which a 16-fold increase of expression after treatment of 5-aza-dC (optimized concentrations in each cell line) was observed by expression microarray, which confirmed its methylation in promoter by MSP, among 19 421 genes arrayed on GeneChip Human Genome 133 Plus 2.0 (Affymetrix).
These results showed that expression cannot be induced for the majority of genes with methylation of their promoters even with a demethylating agent, possibly due to the lack of transcriptional factors or the presence of inactive histone modifications. Genes with low transcription tend to become methylated,20,38,39 and such genes are unlikely to be expressed even if the methylation is removed. Caution is necessary when the relationship between methylation of the promoter and the expression of a gene is interpreted.
3.6. Application of the Me value in future studies
The differential role of DNA methylation in promoters and gene bodies and the lack of association between the degree of demethylation of promoters and that of induction of gene expression were clearly shown due to the accuracy of the Me value. We here focused on CGIs using a CGI microarray. Roles of methylation of CpG-poor genomic regions in genome regulation are not as clear as those of CGIs,10,40 and focusing on CpG-rich regions is advantageous to isolate genes of interest. Also, the collection efficiency of methylated DNA by MeDIP or MBD is low in CpG-poor genomic regions,7,9,41 and analysis of both CpG-poor and -rich regions leads to a complicated algorithm for normalization. The selective analysis of CGIs enabled us to develop a simple, but reliable output value.
The use of MeDIP allowed us to analyse methylation levels of any genomic regions regardless of the presence of restriction sites, and we were able to focus on promoters in some part of this study. We did not amplify the IP DNA during preparation of hybridization probes to avoid any amplification bias, and this was enabled by the use of a microarray platform with a high sensitivity and low signal/noise ratio. Under these optimal conditions, we achieved a high reproducibility (r = 0.98) and simplicity for the quantitative methylation microarray analysis.
In conclusion, we developed a new output value for microarray analysis suitable for quantitative assessment of DNA methylation levels. The Me value will be useful in genome-wide screening using heterogeneous samples.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by a Grant-in-Aid for the Third-term Cancer Control Strategy Program from the Ministry of Health, Labour and Welfare, Japan. K.G. and H.T. are recipients of a Research Resident Fellowship from the Foundation for Promotion of Cancer Research.
Acknowledgements
We thank Dr Toshiki Taya (Agilent Technologies) for his knowledgeable and devoted technical assistance.
Footnotes
Edited by Minoru Yoshida
References
- 1.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Ushijima T., Watanabe N., Okochi E., Kaneda A., Sugimura T., Miyamoto K. Fidelity of the methylation pattern and its variation in the genome. Genome Res. 2003;13:868–74. doi: 10.1101/gr.969603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 6.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat. Rev. Cancer. 2005;5:223–31. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 7.Weber M., Davies J.J., Wittig D., et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X., Yazaki J., Sundaresan A., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Keshet I., Schlesinger Y., Farkash S., et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–53. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 10.Weber M., Hellmann I., Stadler M.B., et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 11.Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi H., Nagae G., Tsutsumi S., et al. High-resolution mapping of DNA methylation in human genome using oligonucleotide tiling array. Hum. Genet. 2007;120:701–11. doi: 10.1007/s00439-006-0254-6. [DOI] [PubMed] [Google Scholar]
- 13.Jacinto F.V., Ballestar E., Ropero S., Esteller M. Discovery of epigenetically silenced genes by methylated DNA immunoprecipitation in colon cancer cells. Cancer Res. 2007;67:11481–6. doi: 10.1158/0008-5472.CAN-07-2687. [DOI] [PubMed] [Google Scholar]
- 14.Down T.A., Rakyan V.K., Turner D.J., et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng A.S., Culhane A.C., Chan M.W., et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–96. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelizzola M., Koga Y., Urban A.E., et al. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–9. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straussman R., Nejman D., Roberts D., et al. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16:564–71. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 18.Buck M.J., Lieb J.D. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–60. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita S., Tsujino Y., Moriguchi K., Tatematsu M., Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakajima T., Yamashita S., Maekita T., Niwa T., Nakazawa K., Ushijima T. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int. J. Cancer. 2009;124:905–10. doi: 10.1002/ijc.24018. [DOI] [PubMed] [Google Scholar]
- 21.Lee T.I., Rinaldi N.J., Robert F., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 22.Pokholok D.K., Harbison C.T., Levine S., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Hahn M.A., Hahn T., Lee D.H., et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–9. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong H., Yauk C.L., Rowan-Carroll A., et al. Identification of thyroid hormone receptor binding sites and target genes using ChIP-on-chip in developing mouse cerebellum. PLoS One. 2009;4:e4610. doi: 10.1371/journal.pone.0004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke X.S., Qu Y., Rostad K., et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriguchi K., Yamashita S., Tsujino Y., Tatematsu M., Ushijima T. Larger numbers of silenced genes in cancer cell lines with increased de novo methylation of scattered CpG sites. Cancer Lett. 2007;249:178–87. doi: 10.1016/j.canlet.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Kaneda A., Kaminishi M., Yanagihara K., Sugimura T., Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645–50. [PubMed] [Google Scholar]
- 28.Miyamoto K., Asada K., Fukutomi T., et al. Methylation-associated silencing of heparan sulfate d-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene. 2003;22:274–80. doi: 10.1038/sj.onc.1206146. [DOI] [PubMed] [Google Scholar]
- 29.Abe M., Watanabe N., McDonell N., et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology. 2008;74:50–60. doi: 10.1159/000139124. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita S., Takahashi S., McDonell N., et al. Methylation silencing of transforming growth factor-beta receptor type II in rat prostate cancers. Cancer Res. 2008;68:2112–21. doi: 10.1158/0008-5472.CAN-07-5282. [DOI] [PubMed] [Google Scholar]
- 31.Eckhardt F., Lewin J., Cortese R., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukasawa M., Kimura M., Morita S., et al. Microarray analysis of promoter methylation in lung cancers. J. Hum. Genet. 2006;51:368–74. doi: 10.1007/s10038-005-0355-4. [DOI] [PubMed] [Google Scholar]
- 33.Tomilin N.V. Regulation of mammalian gene expression by retroelements and non-coding tandem repeats. Bioessays. 2008;30:338–48. doi: 10.1002/bies.20741. [DOI] [PubMed] [Google Scholar]
- 34.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 35.Smith J.F., Mahmood S., Song F., et al. Identification of DNA methylation in 3′ genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2:161–72. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
- 36.Ball M.P., Li J.B., Gao Y., et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch T.A., Wu X., Zhong X., Riggs A.D., Pfeifer G.P. A human B cell methylome at 100-base pair resolution. Proc. Natl Acad. Sci. USA. 2009;106:671–8. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J.Z., Stirzaker C., Harrison J., Melki J.R., Clark S.J. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–61. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- 39.de Smet C., Loriot A., Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol. Cell Biol. 2004;24:4781–90. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irizarry R.A., Ladd-Acosta C., Wen B., et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rauch T., Li H., Wu X., Pfeifer G.P. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–47. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.