Abstract
An analysis of Group IVA (GIVA) phospholipase A2 (PLA2) inhibitor binding was conducted using a combination of deuterium exchange mass spectrometry (DXMS) and molecular dynamics (MD). Models of the GIVA PLA2 inhibitors pyrrophenone and the 2-oxoamide AX007 docked into the protein were designed based on deuterium exchange results, and extensive molecular dynamics simulations were run to determine protein-inhibitor contacts. The models show that both inhibitors interact with key residues that also exhibit changes in deuterium exchange upon inhibitor binding. Pyrrophenone is bound to the protein through numerous hydrophobic residues located distal from the active site, while the oxoamide is bound mainly through contacts near the active site. We also show differences in protein dynamics around the active site between the two inhibitor-bound complexes. This combination of computational and experimental methods is useful in defining more accurate inhibitor binding sites, and can be used in the generation of better inhibitors against GIVA PLA2.
INTRODUCTION
The Group IVA phospholipase A2 (GIVA PLA2), also know as cPLA2 for cytosolic PLA2, is a member of the superfamily of phospholipase A2 enzymes that cleave a fatty acid from the sn-2 position of phospholipids.1,2 The products of this reaction, a free fatty acid and a lysophospholipid play important roles as lipid second messengers. GIVA PLA2 was isolated in 1990 from U937 cells,3 and was discovered to be composed of a C2 domain, and an α/β hydrolase domain containing the active site.4 The GIVA PLA2 is specific for phospholipids with arachidonic acid in the sn-2 position, and the release of arachidonic acid is the first step in the production of eicosanoids and leukotrienes which play important roles in many inflammatory diseases.5 Experiments performed using mice deficient in the GIVA PLA2 enzyme have proven that GIVA PLA2 is the critical PLA2 enzyme for eicosanoid generation in many inflammatory disease models.6-8
The enzyme was shown through site directed mutagenesis to contain an active site dyad composed of Ser-228 and Asp-549,9 and this was later confirmed through x-ray crystallography of the enzyme.10 The enzyme contains an amphipathic lid region from 415-432 that prevents accession of phospholipid into the active site.10 The lid region has two disordered regions from 408-412, and 433-457 that may act as hinges that allow the lid region to open. It has been shown that this lid is in the open conformation when the enzyme is in the presence of lipid vesicles (its natural substrate) or when inhibitor is bound in the active site.11
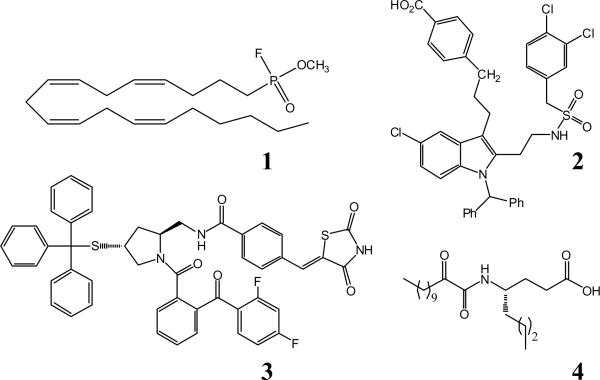
The knowledge that GIVA PLA2 plays an important functional role in many inflammatory diseases has sparked an interest in the production of specific inhibitors against this enzyme. The first inhibitors of this enzyme were based around the specificity of the enzyme for phospholipids with arachidonic acid in the sn-2 position, and as such arachidonyl trifluoromethyl ketones (ATK) and methyl arachidonyl fluorophosphonate (MAFP) (1) were synthesized and found to inhibit the enzyme in platelet models of eicosanoid generation.12-14 In recent years many different strategies have been pursued to create effective and specific GIVA PLA2 inhibitors. These have included indole derivatives developed by Wyeth Pharmaceuticals (2),15-18 pyrrolidine based inhibitors by Shionogi Pharmaceuticals (3),19-22 substituted propan-2-ones by Astra Zeneca and the Lehr group,23-26 as well as 2-oxoamide compounds by the Kokotos and Dennis groups (4) as shown in Figure 1.27-30 Of these inhibitors, there exist two docked structures in the GIVA PLA2 active site, generated through computer modeling,15,31 but there are no in depth examinations of the binding pocket contacts between inhibitor and enzyme.
Figure 1. Inhibitors of GIVA PLA2.
1. MAFP. 2. Efipladib. 3. Pyrrophenone. 4. AX007
The pyrrolidine derived inhibitor pyrrophenone displays some of the best inhibition but (due to chemical properties) is not useful as a drug.18 We have previously shown that the 2-oxoamide compounds show an antihyperalgesic effect in rat models.32 The invention of better 2-oxoamide inhibitors is a promising drug strategy, and to such end, we set out to model the 2-oxoamide inhibitor AX007, as well as the pyrrolidine derived inhibitor pyrrophenone, bound in the active site. This required a technique to monitor changes in protein structure upon inhibitor binding.
Peptide amide hydrogen deuterium exchange analyzed via liquid chromatography/mass spectrometry has been widely used to analyze protein-protein interactions,33,34 protein conformational changes,35,36 and protein dynamics.37 We have previously used this technique to explore changes in lipid binding with the GIVA PLA2 and discovered changes in exchange profiles in the presence of the irreversible inhibitor MAFP.11 The DXMS technique, in conjunction with site-directed mutagenesis, has recently been used to identify regions interacting with different inhibitors.38,39
Coupled with these experimental techniques, computational methods can be employed to study the atomic-level details in the GIVA PLA2-Inhibitor complex. Extensive simulations of the phospholipase A2`s have been carried out. Most notably, Wee et al. recently conducted a coarse-grained simulation of the pancreatic phospholipase A2, in which they demonstrate how the enzyme adheres to the lipid bilayer.40 Quantum mechanical methodologies have also been applied to the phospholipase system.41 This work has proven vital to the understanding of phospholipase A2 chemistry and dynamics.
In turn, by running molecular dynamics (MD) simulations of GIVA PLA2 with inhibitor, one can observe how the latter docks into and stabilizes itself in the enzyme. Contacts between the inhibitor and specific residues of GIVA PLA2 can also be identified. This information augments the results from the deuterium exchange technique which at this time lacks the resolution to achieve single-residue data. In lieu of known crystal structures depicting the enzyme-inhibitor complex, this computational work affords working models of the complexes and characterizes key enzyme-inhibitor interactions. The MD simulations and subsequent analysis aid in drawing comparisons between the oxoamide and pyrrophenone complexes.
The study of these two very different inhibitors provides an excellent model for generalized GIVA PLA2 inhibition. The dual techniques of deuterium exchange mass spectrometry and MD simulation are excellent methods to probe the dynamical changes induced by binding of inhibitors to any enzyme. This study also represents a continuation of our deuterium exchange studies on the PLA2 family of enzymes.11,42,43 We have identified specific regions of the protein that interact with the oxoamide and pyrrophenone inhibitors, and we have carried out extensive computer simulations to create a model of inhibitor binding in the active site. We have also identified significant differences in the way pyrrophenone and oxoamide bind GIVA PLA2. This work leads to the possibilities of enhanced rational drug design through the powerful combination of experimental and computational work.
MATERIALS AND METHODS
All reagents were analytical reagent grade or better. Pyrrophenone was the generous gift from Shionogi, and the 2-oxoamide AX007 was synthesized as described.28
Protein Expression and Purification
C-terminal His6-tagged GIVA PLA2 was expressed using recombinant baculovirus in a suspension culture of Sf9 insect cells. The cell pellet was lysed in 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM β-mercaptoethanol, and 2 mM EGTA and than the insoluble portion was removed by centrifugation at 12,000 x g for 30 min. The supernatant was passed through a column comprised of 6 ml nickel-nitrilotriacetic acid agarose (Qiagen, Valencia, CA). The protein in the native state was eluted in the “protein buffer” (25 mM Tris-HCl pH 8.0, 100 mM NaCl, 125 mM imidazole and 2 mM dithiothreitol). The protein concentration was measured using the Bradford assay (Bio-rad) to manufacturer's standards, and the activity was assayed using mixed micelles in a modified Dole assay.44 Purified GIVA PLA2 (2 mg/ml) was stored in the protein buffer on ice for DXMS experiments. Experiments were performed immediately after elution from the nickel column.
Preparation of Deuterated samples for On-exchange Experiments
D2O buffer contained 10 mM Tris (pD 7.5), 50 mM NaCl in 98% D20. Hydrogen/deuterium exchange experiments were initiated by mixing 20 μl of GIVA PLA2 (containing 40 μg) in protein buffer with 60 μl of D2O buffer to a final concentration of 73% D2O at pH 7.5. In inhibitor binding experiments, the GIVA PLA2 in protein buffer was pre-incubated in the presence of 40 μM pyrrophenone, 40 μM oxoamide AX007, or DMSO control. The inhibitors were added from 600 μM stock dissolved in DMSO. The final concentration of DMSO was 1.5% for all experiments. The inhibitors were allowed to preincubate with the enzyme for 10 minutes at 23°C before addition of D2O buffer. The D2O buffer was added and the samples were incubated at 23°C for an additional 10, 30, 100, 300, 1000, 3000 or 10000 seconds. The dilution of inhibitors with D2O buffer gave a final inhibitor concentration of 10 μM. The deuterium exchange was quenched by adding 120 μl of ice-cold quench solution (0.96% formic acid, 1.66 M guanidine hydrochloride (GdHCl)) that acidified the sample to a final pH = 2.5 and concentrations of formic acid of 0.58%, and 1 M GdHCl. The samples were placed on ice for 10 min to partially denature the protein and obtain optimal peptide maps. Vials with frozen samples were stored at -80 °C until analysis, usually within three days.
Proteolysis-liquid-chromatography-mass spectrometry analysis of samples
All steps were performed at 0°C as previously described.11, 33,35
Data processing
SEQUEST software (Thermo Finnigan Inc.) was used to identify the sequence of the peptide ions as previously described.11 DXMS Explorer (Sierra Analytics Inc, Modesto CA) was used for the analysis of the mass spectra, and all data processing was the same as previously described. 11,33,35 The deuteration level of each peptide was calculated by the ratio of the incorporated deuteron number to the maximum possible deuteration number. Peptide deuteration levels in replicate samples, measured by our DXMS methods, have been found to vary by less than 10%, and we therefore regard changes greater than 10% as significant.33 All experiments were performed at least twice, and representative data is shown. Trends in the data were similar from experiment to experiment, but total deuterium content varied by roughly 5-10% in similar experiments carried out weeks apart. For all peptides shown in the Figures, different peptides that cover the same region are included in the supplementary information as Supplemental Figure 1.
Molecular Dynamics Simulations
In total, three systems were simulated: the apo form of GIVA PLA2; GIVA PLA2 with pyrrophenone bound; and GIVA PLA2 with oxoamide bound. The structure of GIVA PLA2 was obtained from the Protein Data Bank (code 1CJY).10 In this reported structure, several segments were missing including residues 407-414, 431-462, 498-538, and 626-632. These apparently flexible regions were modeled into chain B of 1CJY using SWISS MODEL (Supplemental Figure 2).45 The resulting complete structure of GIVA PLA2 was optimized by 500 steps each of steepest decent and conjugate gradient energy minimizations and used in all three systems.
The inhibitors pyrrophenone and oxoamide were constructed using the Accelrys® Discovery Studio package, in which they were built and energy minimized to obtain their initial conformations. Each inhibitor was manually placed in the active site of GIVA PLA2 using the aforementioned knowledge of the active site and residues implicated in binding. This was carried out using the Visual Molecular Dynamics (VMD) package,46 which was also used for further construction of all 3 systems. The protein (or the protein-inhibitor complex) was placed in a simulation box of dimensions 127 × 73 × 91 Å, solvated with approximately 22750 TIP3P water molecules, and neutralized with 28 sodium counterions (~80,000 atoms total for each system).
The minimization steps and all subsequent simulations were carried out using the NAMD molecular dynamics package.47 For the apo structure, an energy minimization was performed on the system first with the protein backbone atoms fixed—for 25000 steps, to allow the water/ions to conform to the shape of the protein—and then with no such constraint—for another 25000 steps, to relieve any unfavorable contacts in the entire system. For the inhibitor-bound systems, a similar energy minimization scheme was performed, with the addition of an extra minimization holding the inhibitor coordinates fixed, to relieve unfavorable contacts between the inhibitor and protein. For all three systems, a heating step was performed, in which the system was gradually heated (~50 ps) to a temperature of 300 K. During this heating and then the equilibration phase, the protein backbone and inhibitor atoms were restrained with a force constant of 5 kcal mol-1 Å-2. In the equilibration phase, pressure coupling was added and a series of restrained MD simulations were conducted. The restraints were gradually relieved and the free, un-restrained system was equilibrated for 1 ns further. The production runs for each system commenced; each protein (or protein-inhibitor complex) was simulated for 50 ns.
For each of the production runs, the temperature was maintained at 300K using the Langevin thermostat, with a coupling constant of 2 ps-1. Pressure was maintained at 1.01325 kPa using the Langevin piston method, with the `GroupPressure' and `FlexibleCell' parameters turned on. Bonds to hydrogen atoms were held fixed while using a 2 fs timestep. Non-bonded interactions and full electrostatics was calculated every 1 and 2 steps, respectively. Non-bonded interactions were smoothly switched off between 8.5 and 10 Å, while the cut-off for pairlist distance was set to 12 Å. Long-range electrostatic forces were evaluated using the particle mesh Ewald (PME) method.48 The CHARMM22/27 all-hydrogen parameter files were used for protein and inhibitors in all three simulations.49 The standard protein/amino acid parameters were used for GIVA PLA2 and pyrrophenone (by analogy to various amino acid side chain structures). The oxoamide was parameterized according to the lipid parameters provided in CHARMM27.
RESULTS AND DISCUSSION
GIVA PLA2 Digestion Map
The protein digestion procedure was optimized to produce a peptide map that yielded the best coverage of GIVA PLA2 as described previously.11,43 The optimized condition identified 185 distinct peptides that gave 94% coverage of the GIVA PLA2 sequence (Supplemental Figure 3). From this group, the 71 peptides with the best signal to noise ratio with the least amount of redundant data were used to generate the Figures in the manuscript as described previously.11,43 All peptides were analyzed for deuterium content as a comparison, but only the ones with non-redundant data were used in the generation of figures. Additional peptides that overlap regions shown in the figures are shown in supplemental figure 1.
Modeling and Simulation
The overall structure of the protein remained close to the crystal structure in all three simulations (Supplemental Figure 4). The regions missing in the X-ray structure, but model-built in the simulations, exhibited large fluctuations throughout the simulations, which is consistent with their being disordered in the crystals. However, the majority of the modeled regions remained solvated, without making significant contact with the rest of the protein. As a result, their motion did not affect the active site region of the enzyme. In the apo form, GIVA PLA2 shows no significant conformational changes, as expected. In the inhibitor-bound forms, both pyrrophenone and the oxoamide show considerable movement in the first half of simulation but settle to a converged conformation and location in the last 25 ns, as judged by the root mean square (RMSD) of the inhibitors (see Supplemental Figure 4). Thus, the last 25 ns of each simulation were used in all subsequent simulation analysis.
GIVA PLA2 Pyrrophenone Binding Experiments
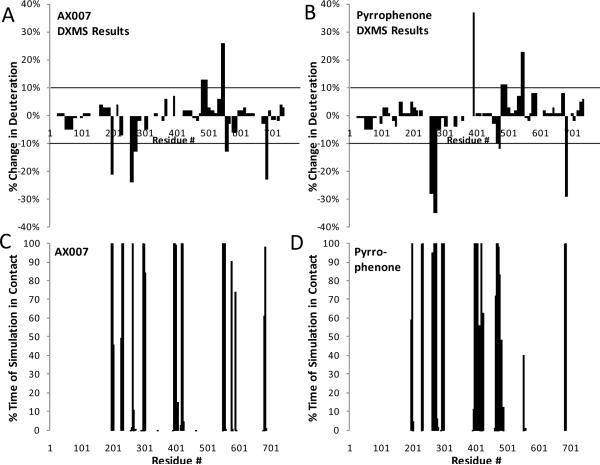
We examined both the pyrrolidine derived inhibitor pyrrophenone as well as the 2-oxoamide derived inhibitor AX007. These compounds are structurally quite different and target different functionalities of the GIVA PLA2. Therefore, determining exactly how these inhibitors bind is an important goal, because it allows the possibility of combining the best parts of each inhibitor to form new, more effective inhibitors to allow for further structure-activity studies. On-exchange experiments were performed on the intact GIVA PLA2 enzyme in the presence of both 10 μM pyrrophenone and 10 μM of the 2-oxoamide inhibitor AX007 to determine if inhibitor binding caused changes in deuterium exchange rates. The experiments were carried out at relatively low ratios of protein to inhibitor (1:2) in 1.5% DMSO to prevent possible complications from inhibitor aggregation and misleading deuterium exchange results. Experiments were carried out at seven time points varying from 10 to 10000 seconds. Both inhibitors showed multiple regions of the protein with greater than 10% change in the on-exchange rates between the inhibitor-bound and apo forms at all time points. These percent increases and decreases in on-exchange rates showed a strong correlation with computational data mapping percent chance of contact per residue number (Figure 2). The residues from 292-298 and 401-417 predicted to be in contact with the inhibitor (within 5 Å) from modeling and that show no changes in deuterium exchange are all located in regions with either extremely slow or rapid exchange, and hence there is no significant difference in exchange between apo- and halo- forms (see Supplemental Figure 5). The difference in on-exchange at the 100 second time point captured all of the major changes, and was used to generate the data shown in Figure 2, 3, and 5.
Figure 2. Deuterium exchange information compared to computer simulation results.
Panels A and B: The percent change in deuterium exchange between inhibitor free GIVA PLA2 and oxoamide-bound (panel A) or pyrrophenone-bound (panel B) GIVA PLA2 at 100 seconds of on-exchange is shown. Each bar represents a region in which deuterium exchange was quantified. All changes greater than 10% are considered significant. Panels C and D: The percent chance of specific residues that were within 5 Å of the docked inhibitor in the molecular dynamics simulation are plotted for oxoamide-bound (panel C) and pyrrophenone-bound (panel D) PLA2.
Figure 3. Deuterium exchange upon binding of 10 μM pyrrophenone.
The number of incorporated deuterons at seven time points in eight different regions, 256-265, 268-279, 393-397, 466-470, 473-478, 481-495, 543-553, and 684-689 in GIVA PLA2 are plotted onto the docked model of pyrrophenone binding at 50 ns of simulation time (the inhibitor is shown in space filled form). Decreases or increases in deuteration greater than 10% at the 100 second time point are represented by the color scheme in the legend.
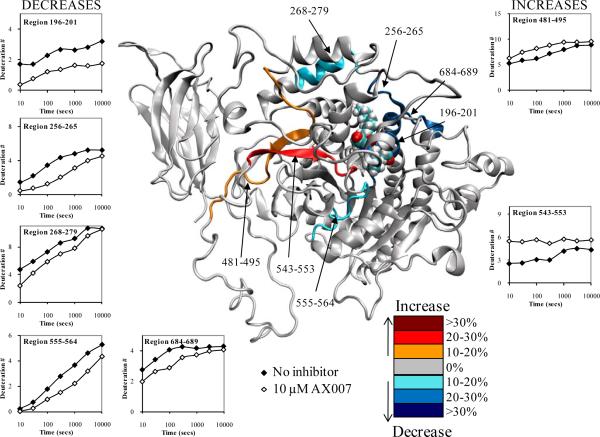
Figure 5. Deuterium exchange upon binding of 10 μM AX007.
The number of incorporated deuterons at seven time points in seven different regions, 196-201, 256-265, 268-279, 555-564, 481-495, 543-553, and 684-689 in GIVA PLA2 are plotted onto the docked model of the oxoamide AX007 binding at 50 ns of simulation time (the inhibitor is shown in space filled form) Decreases or increases in deuteration greater than 10% at the 100 second time point are represented by the color scheme in the legend.
The inhibitor pyrrophenone was synthesized in 2001;20 and contains a thiazoloidinedione ring postulated to target Arg-200 and a carbonyl group bridging the two benzoyl groups that is expected to target the active site Ser-228. This class of inhibitors was also shown, through structure-activity work, to have large increases in inhibitory potency with the addition of large bulky lipophilic substituents, suggesting the presence of a hydrophobic binding pocket in the enzyme.19 Using deuterium exchange and modeling we planned to test this hypothesis based on structure-activity work.
Eight regions of the GIVA PLA2 exhibited significant changes in deuterium exchange in the presence of pyrrophenone. Figure 3 shows these results both quantitatively and visually imposed on snapshots from the MD simulations. Three regions of the protein, residues 393-397, 481-495, and 543-553 exhibited increased rates of exchange in the presence of pyrrophenone. Regions 393-397 and 543-553 had greater differences in on-exchange rates (between apo- and pyrrophenone-bound enzymes) at early time points of roughly 20-30%, with the difference going to zero at later time points. Region 393-397 contains Ala-396 and Phe-397 in contact with pyrrophenone. Region 481-495 had a constant 10-15% increase in exchange at all time points. These are the exact regions that we have previously shown had increases in on-exchange rates in the presence of the potent irreversible GIVA PLA2 inhibitor MAFP, as well as natural phospholipid substrate vesicles.11 We have hypothesized that these regions show an increase in exchange due to the opening of the lid region from 415-432, and that pyrrophenone also causes an opening of the lid region upon binding in the active site. In turn, this opening event induces an increase in the solvent accessibility and results in higher on-exchange rates. This lid opening was not seen in our simulations, and this is most likely a time dependent process that is too slow to view with molecular dynamics.
Five regions of the protein, residues 256-265, 268-279, 466-470, 473-478, and 684-689 demonstrated decreases in exchange between apo- and pyrrophenone-bound enzymes (Figure 3). Region 268-279 exhibits greater than 30% decreases in exchange at all time points. Correlating with the MD simulation, this region harbors multiple residues that are in constant contact with the pyrrophenone. Regions 256-265 and 684-689 exhibit 20-30% decreases at early time points and drop to less than 10% at later time points. These regions also contain multiple hydrophobic residues demonstrated by simulation to be in contact with pyrrophenone; yet, this is to a lesser extent (and thus less of a decrease in exchange) as compared to region 268-279. Regions 466-470 and 473-478 showed 10-15% decreases in on-exchange rates from 30 to 300 seconds but no differences in exchange at earlier or later time points. We assume that the deuterium exchange rates are a combination of the individual rates of numerous amide hydrogens. In region 466-470 the rate profile suggests there is a ser of amide hydrogens that are not exchanged in either the apo or inhibitor bound structure at 30 seconds, and hence no difference in exchange is seen. At 100 and 300 seconds these amide hydrogens are more deuterated in the apo structure and have lower levels of exchange in the inhibitor bound structure due to decreases in solvent accessibility, which gives a 10-15% decrease. At 1000 seconds, however, these amides are fully exchanged in both the apo form and inhibitor bound form, which gives similar deuteration levels at this time point.
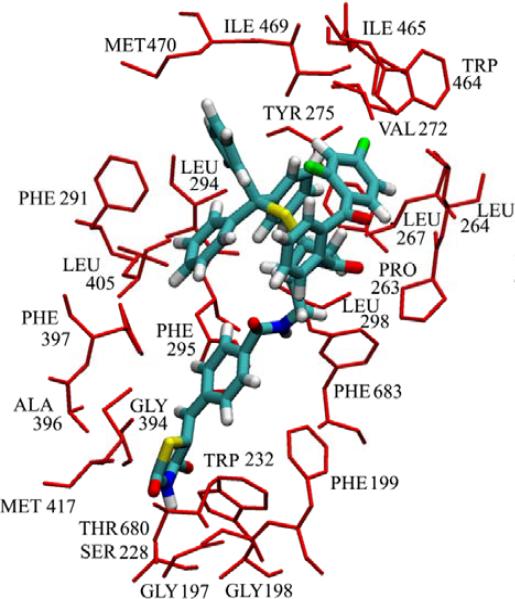
Our deuterium exchange results show decreases in exchange in numerous regions containing hydrophobic regions and this matches our modeling work as shown in Figure 4, where Pro-263, Leu-264, Leu -267, Val-272, Tyr-275, Trp-464, Ile-465, Ile-469, Met-470, and Phe-683 all make contact with the numerous phenyl groups in pyrrophenone. In addition region 282-291, which includes Phe-291 shows a constant 7% decrease across all time points, and has been included in supplemental figure 1. These residues most likely are acting as the hydrophobic pocket postulated through structure activity work with pyrrophenone, and our results correlate with recent data from Wyeth showing that GIVA PLA2 inhibitors become more potent when the steric bulk of the inhibitor is increased in functionally allowed regions.18 However we find that the thiazoloidinedione functionality targets Ser-228, rather than targeting Arg-200, as originally suggested by Shionogi,19,20 with the carbonyl bridging the two phenyl groups in pyrrophenone being located at a large distance from the active site serine.
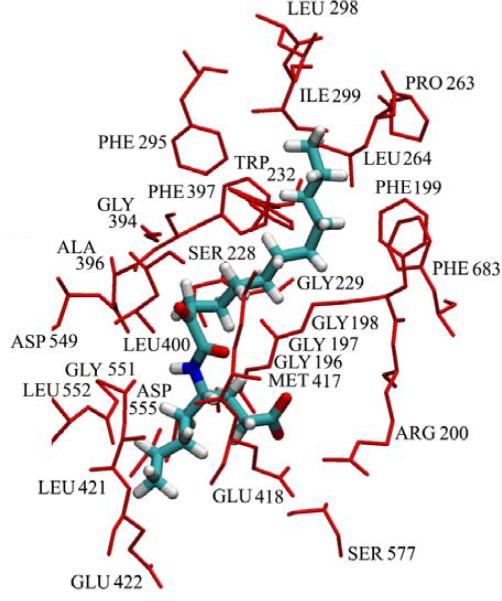
Figure 4. Residues involved in binding pyrrophenone.
The residues that have contact with pyrrophenone greater than 90% of the time in the molecular dynamics simulation are represented as red sticks and labeled on the figure. The inhibitor is shown in the licorice representation, with carbon, hydrogen, oxygen, nitrogen and phosphorus atoms colored cyan, white, red, blue and yellow, respectively.
GIVA PLA2 Oxoamide Binding Experiments
The 2-oxoamide inhibitor AX007 was originally synthesized and shown to be an effective GIVA PLA inhibitor in 2002.27 It was postulatedto target GIVA PLA2 via an interaction between its 2-oxo amide functionality and the active site Ser-228. Also, the carboxylic acid moiety of the oxoamide was designed to target Arg-200, while the inhibitor's long fatty acyl tail positions itself in the hydrophobic binding pocket.28 Using deuterium exchange and modeling we planned to test these hypothesis based on structure-activity work.
Seven regions of the GIVA PLA2 exhibited significant changes in deuterium exchange in the presence of the oxoamide (Figure 5). Two regions of the protein, residues 481-495, and 543-553 exhibit an increase in exchange in the presence of the oxoamide. These regions show the exact same deuterium on-exchange profile as the pyrrophenone-bound enzyme. However, the region 393-397 does not show any difference in exchange in the presence of the oxoamide. This result allows for multiple interpretations; perhaps the lid region is opened in a different way (as compared to the enzyme-pyrrophenone complex), thus only increasing solvent accessibility for regions 481-495 and 543-553, but not for region 393-397. Or, the lid region may be opened in the same way, but increased contacts between region 393-397 and the oxoamide cause a comparative decrease in exchange rates between the two inhibitor-bound structures.
Five regions of the protein, residues 196-201, 256-265, 268-279, 555-564, and 684-689 exhibit decreases in exchange in the presence of the oxoamide. Regions 256-265 and 684-689 reveal the same on-exchange pattern in the presence of both the oxoamide and pyrrophenone. Simulation shows that along these regions, both inhibitors make similar hydrophobic contacts with Pro-263, Leu-264, and Phe-683 as shown in Figures 4 and 6. Region 268-279 has a 10-15% decrease in exchange at all time points, which is much lower than the corresponding differences in this region in the pyrrophenone-enzyme complex. This much smaller decrease in exchange in the oxoamide-enzyme complex correlates well with the simulation, which shows no residues in 268-279 making contacts with the inhibitor. Region 555-564 displays a 10-20% decrease in exchange at all time points in the presence of the oxoamide. This region contains Asp-555 and neighbors Gly-551/Leu-552, all of which are in constant contact with the oxoamide during the simulation. Recent work by us has shown that the short, nonpolar, aliphatic R-group substituent on the oxoamide AX007 (residing on the linker between the 2-oxo and carboxylic acid) increased potency. We postulated that this is facilitated by a hydrophobic pocket in the enzyme that can accommodate this particular group. 28,30 The residues in and around 555-564 appear to constitute this pocket, as shown here by both on-exchange results and simulation. Region 196-201 exhibits a 20-30% decrease in exchange at all time points in the presence of the oxoamide. This region contains the proposed oxy-anion hole—residues Gly-196, Gly-197, Gly-198, Gly-229, and Arg-200— required for catalytic activity, as well as Phe-199, which is part of the hydrophobic pocket for the substrate. The modeling data shows an interaction between the carboxylic acid of the oxoamide and Arg-200, as well as the carbonyl of the 2-oxoamide in contact with the oxyanion hole composed of the numerous glycine residues.
Figure 6. Residues involved in the binding of oxoamide AX007.
The residues that have contact with the oxoamide greater than 90% of the time in the molecular dynamics simulation are represented as red sticks and labeled on the figure. The inhibitor is shown in the licorice representation, with carbon, hydrogen, oxygen, nitrogen and phosphorus atoms colored cyan, white, red, blue and yellow, respectively.
Differences in Oxoamide and Pyrrophenone Binding
Numerous regions of the protein show the same decreases or increases in exchange with both pyrrophenone and the oxoamide AX007. These regions include 256-265, 481-495, 543-553, and 684-689, and they all show similar contacts in these regions between both AX007 and pyrrophenone as shown in Figures 4 and 6. There are also regions such as 466-470, 473-478, and 555-564 which show changes only in the presence of one or the other inhibitor, and this is explained by specific contacts only seen between pyrrophenone or the oxoamide and the protein.
However region 196-201 acts as an interesting example of the differences between pyrrophenone and oxoamide inhibitor binding. There are four different regions in the enzyme 196-201, 225-232, 577-591, and 670-682 that show a greater than 10% change in exchange between the pyrrophenone bound structure and the oxoamide bound structure (Figure 7). Many of these peptides do not show a change in deuterium exchange greater than 10% as compared to the apo structure. However, the comparison between pyrrophenone and the oxoamide does show a greater than 10% change in exchange. These peptide regions are all in or near the active site of the enzyme. Region 225-232, which contains the active site residue Ser-228, never exchanges greater than 25% at any time point. Yet, there is a greater than 10% change in exchange between the oxoamide and pyrrophenone-bound studies, with the oxoamide-enzyme complex showing less exchange than the pyrrophenone-bound sample.
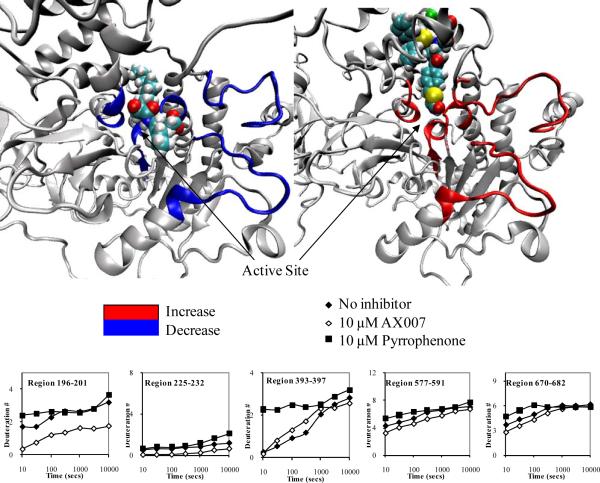
Figure 7. Different inhibitors cause different rates of exchange of the active site residues of GIVA PLA2.
The number of incorporated deuterons at seven time points in five different regions, 196-201, 225-232, 393-397, 577-591, and 670-682 in GIVA PLA2 are plotted onto the docked models. Areas that show both a greater than 10% change in exchange between oxoamide and pyrrophenone in at least 3 consecutive time points as well as less exchange with the oxoamide compared to the apo structure (left), and more exchange with pyrrophenone compared to the apo structure (right) are shown in color in the respective structures.
For regions 577-591 and 670-682, the main effects are most likely localized to 577-580 and 680-682, respectively, which are located within 5 Å of the active site. These results show that the 2-oxoamide inhibitor AX007 decreases the solvent accessibility of the active site while pyrrophenone has the opposite effect (an increase). These results suggest an increase in flexibility of the active site in the presence of pyrrophenone with a concurrent decrease in flexibility in the presence of the oxoamide. This result also explains why there are numerous residues shown in contact with pyrrophenone in figure 4 that do not have a decrease in exchange, due to increases in exchange caused by increased flexibility in the active site that counters decreases in exchange caused by inhibitor bindinig decreaseing solvent accessibility. These results also help to explain why region 393-397 has an increase in exchange with pyrrophenone and not with the oxoamide. This region is located near the active site, and increases in exchange are seen with the presence of pyrrophenone, MAFP, and natural phospholipid substrate,11 but not the oxoamide. The decreased solvent accessibility and lack of flexibility of the active site in the presence of the oxoamide would explain the lack of exchange increases in region 393-397. From viewing Figure 7, it is also apparent that the oxoamide mainly occupies the active site area, while pyrrophenone is mainly bound in the cap region near the interfacial binding surface of the enzyme.
CONCLUSIONS
These results have greatly enhanced our knowledge of how these two different inhibitors bind GIVA PLA2 and have allowed us to model all the residues contacting both inhibitors. This will allow us to create new inhibitors combining the 2-oxoamide functionality with a bulky lipophilic substituent in place of the acyl fatty acid tail to mimic how pyrrophenone binds GIVA PLA2 through multiple hydrophobic contacts located on or near the cap region. This study is the first to combine deuterium exchange mass spectrometry with molecular dynamics simulations for the determination of inhibitor binding. This methodology provides an exciting new tool to develop better inhibitors, and we plan to continue this work through the synthesis and testing of new GIVA PLA2 inhibitors based on our results.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Howard Hsu for constructive review of this manuscript. We also thank the following agencies for funding: NIH GM20501 (E.A.D.), and CA099835, CA118595, GM037684, AI0220221, AI022160 (V.L.W); a Discovery Grant (UC10591) from the University of California Industry-University Cooperative Research Program (V.L.W); NIH GM31749 and NSF Grants MCB-0506593 and MCA93S013 (J.A.M.). This work was also supported by the Howard Hughes Medical Institute, San Diego Supercomputing Center, the W.M. Keck Foundation, Accelrys, Inc., the National Biomedical Computational Resource, and the Center for Theoretical Biological Physics.
Footnotes
SUPPORTING INFORMATION Additional experimental information on peptide maps, computational modeling, and supporting data are found in supplemental data available online. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFEERENCES
- (1).Schaloske RH, Dennis EA. Biochim Biophys Acta. 2006;1761:1246–59. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- (2).Six DA, Dennis EA. Biochim.Biophys.Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- (3).Clark JD, Milona N, Knopf JL. Proc Natl Acad Sci U S A. 1990;87:7708–12. doi: 10.1073/pnas.87.19.7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Clark JD, Lin LL, Kriz RW, Ramesha CS, Sultzman LA, Lin AY, Milona N, Knopf JL. Cell. 1991;65:1043–51. doi: 10.1016/0092-8674(91)90556-e. [DOI] [PubMed] [Google Scholar]
- (5).Funk CD. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- (6).Bonventre JV, Huang Z, Taheri MR, O'Leary E, Li E, Moskowitz MA, Sapirstein A. Nature. 1997;390:622–625. doi: 10.1038/37635. [DOI] [PubMed] [Google Scholar]
- (7).Nagase T, Uozumi N, Ishii S, Kume K, Izumi T, Ouchi Y, Shimizu T. Nat.Immunol. 2000;1:42–46. doi: 10.1038/76897. [DOI] [PubMed] [Google Scholar]
- (8).Uozumi N, Kume K, Nagase T, Nakatani N, Ishii S, Tashiro F, Komagata Y, Maki K, Ikuta K, Ouchi Y, Miyazaki J, Shimizu T. Nature. 1997;390:618–622. doi: 10.1038/37622. [DOI] [PubMed] [Google Scholar]
- (9).Pickard RT, Chiou XG, Strifler BA, DeFelippis MR, Hyslop PA, Tebbe AL, Yee YK, Reynolds LJ, Dennis EA, Kramer RM, Sharp JD. The Journal of Biological Chemistry. 1996;271:19225–19231. doi: 10.1074/jbc.271.32.19225. [DOI] [PubMed] [Google Scholar]
- (10).Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS. Cell. 1999;97:349–60. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- (11).Burke JE, Hsu YH, Deems RA, Li S, Woods VL, Jr., Dennis EA. J Biol Chem. 2008;283:31227–36. doi: 10.1074/jbc.M804492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Liu S, Laliberte F, Falgueyret JP, Street I, Huang Z. Faseb Journal. 1995;9:A1310–A1310. [Google Scholar]
- (13).Riendeau D, Guay J, Weech PK, Laliberte F, Yergey J, Li C, Desmarais S, Perrier H, Liu S, Nicoll-Griffith D. J.Biol.Chem. 1994;269:15619–15624. [PubMed] [Google Scholar]
- (14).Trimble LA, Street IP, Perrier H, Tremblay NM, Weech PK, Bernstein MA. Biochemistry. 1993;32:12560–12565. doi: 10.1021/bi00210a002. [DOI] [PubMed] [Google Scholar]
- (15).McKew JC, Lovering F, Clark JD, Bemis J, Xiang Y, Shen M, Zhang W, Alvarez JC, Joseph-McCarthy D. Bioorg Med Chem Lett. 2003;13:4501–4. doi: 10.1016/j.bmcl.2003.08.070. [DOI] [PubMed] [Google Scholar]
- (16).McKew JC, Foley MA, Thakker P, Behnke ML, Lovering FE, Sum FW, Tam S, Wu K, Shen MW, Zhang W, Gonzalez M, Liu S, Mahadevan A, Sard H, Khor SP, Clark JD. J Med Chem. 2006;49:135–58. doi: 10.1021/jm0507882. [DOI] [PubMed] [Google Scholar]
- (17).Lee KL, Foley MA, Chen L, Behnke ML, Lovering FE, Kirincich SJ, Wang W, Shim J, Tam S, Shen MW, Khor S, Xu X, Goodwin DG, Ramarao MK, Nickerson-Nutter C, Donahue F, Ku MS, Clark JD, McKew JC. J Med Chem. 2007;50:1380–400. doi: 10.1021/jm061131z. [DOI] [PubMed] [Google Scholar]
- (18).McKew JC, Lee KL, Shen MW, Thakker P, Foley MA, Behnke ML, Hu B, Sum FW, Tam S, Hu Y, Chen L, Kirincich SJ, Michalak R, Thomason J, Ipek M, Wu K, Wooder L, Ramarao MK, Murphy EA, Goodwin DG, Albert L, Xu X, Donahue F, Ku MS, Keith J, Nickerson-Nutter CL, Abraham WM, Williams C, Hegen M, Clark JD. J Med Chem. 2008;51:3388–413. doi: 10.1021/jm701467e. [DOI] [PubMed] [Google Scholar]
- (19).Seno K, Okuno T, Nishi K, Murakami Y, Watanabe F, Matsuura T, Wada M, Fujii Y, Yamada M, Ogawa T, Okada T, Hashizume H, Kii M, Hara S, Hagishita S, Nakamoto S, Yamada K, Chikazawa Y, Ueno M, Teshirogi I, Ono T, Ohtani M. J.Med.Chem. 2000;43:1041–1044. doi: 10.1021/jm9905155. [DOI] [PubMed] [Google Scholar]
- (20).Seno K, Okuno T, Nishi K, Murakami Y, Yamada K, Nakamoto S, Ono T. Bioorganic & Medicinal Chemistry Letters. 2001;11:587–590. doi: 10.1016/s0960-894x(01)00003-8. [DOI] [PubMed] [Google Scholar]
- (21).Ono T, Yamada K, Chikazawa Y, Ueno M, Nakamoto S, Okuno T, Seno K. Biochemical Journal. 2002;363:727–735. doi: 10.1042/0264-6021:3630727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Flamand N, Picard S, Lemieux L, Pouliot M, Bourgoin SG, Borgeat P. British Journal of Pharmacology. 2006;149:385–392. doi: 10.1038/sj.bjp.0706879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Connolly S, Bennion C, Botterell S, Croshaw PJ, Hallam C, Hardy K, Hartopp P, Jackson CG, King SJ, Lawrence L, Mete A, Murray D, Robinson DH, Smith GM, Stein L, Walters I, Wells E, Withnall WJ. Journal of Medicinal Chemistry. 2002;45:1348–1362. doi: 10.1021/jm011050x. [DOI] [PubMed] [Google Scholar]
- (24).Ludwig J, Bovens S, Brauch C, Elfringhoff AS, Lehr M. Journal of Medicinal Chemistry. 2006;49:2611–2620. doi: 10.1021/jm051243a. [DOI] [PubMed] [Google Scholar]
- (25).Hess M, Elfringhoff AS, Lehr M. Bioorganic & Medicinal Chemistry. 2007;15:2883–2891. doi: 10.1016/j.bmc.2007.02.016. [DOI] [PubMed] [Google Scholar]
- (26).Fritsche A, Elfringhoff AS, Fabian J, Lehr M. Bioorganic & Medicinal Chemistry. 2008;16:3489–3500. doi: 10.1016/j.bmc.2008.02.019. [DOI] [PubMed] [Google Scholar]
- (27).Kokotos G, Kotsovolou S, Six DA, Constantinou-Kokotou V, Beltzner CC, Dennis EA. J Med Chem. 2002;45:2891–2893. doi: 10.1021/jm025538p. [DOI] [PubMed] [Google Scholar]
- (28).Kokotos G, Six DA, Loukas V, Smith T, Constantinou-Kokotou V, Hadjipavlou-Litina D, Kotsovolou S, Chiou A, Beltzner CC, Dennis EA. J Med Chem. 2004;47:3615–3628. doi: 10.1021/jm030485c. [DOI] [PubMed] [Google Scholar]
- (29).Stephens D, Barbayianni E, Constantinou-Kokotou V, Peristeraki A, Six DA, Cooper J, Harkewicz R, Deems RA, Dennis EA, Kokotos G. Journal of Medicinal Chemistry. 2006;49:2821–2828. doi: 10.1021/jm050993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Six DA, Barbayianni E, Loukas V, Constantinou-Kokotou V, Hadjipavlou-Litina D, Stephens D, Wong AC, Magrioti V, Moutevelis-Minakakis P, Baker SF, Dennis EA, Kokotos G. Journal of Medicinal Chemistry. 2007;50:4222–4235. doi: 10.1021/jm0613673. [DOI] [PubMed] [Google Scholar]
- (31).Gopalsamy A, Yang H, Ellingboe JW, McKew JC, Tam S, Joseph-McCarthy D, Zhang W, Shen M, Clark JD. Bioorg Med Chem Lett. 2006;16:2978–81. doi: 10.1016/j.bmcl.2006.02.067. [DOI] [PubMed] [Google Scholar]
- (32).Yaksh TL, Kokotos G, Svensson CI, Stephens D, Kokotos CG, Fitzsimmons B, Hadjipavlou-Litina D, Hua XY, Dennis EA. Journal of Pharmacology and Experimental Therapeutics. 2006;319:466–475. doi: 10.1124/jpet.105.091686. [DOI] [PubMed] [Google Scholar]
- (33).Hamuro Y, Anand GS, Kim JS, Juliano C, Stranz DD, Taylor SS, Woods VL., Jr. J Mol Biol. 2004;340:1185–96. doi: 10.1016/j.jmb.2004.05.042. [DOI] [PubMed] [Google Scholar]
- (34).Mandell JG, Falick AM, Komives EA. Proc Natl Acad Sci U S A. 1998;95:14705–10. doi: 10.1073/pnas.95.25.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Brudler R, Gessner CR, Li S, Tyndall S, Getzoff ED, Woods VL., Jr. J.Mol.Biol. 2006;363:148–160. doi: 10.1016/j.jmb.2006.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hoofnagle AN, Resing KA, Goldsmith EJ, Ahn NG. Proc Natl Acad Sci U S A. 2001;25:956–61. doi: 10.1073/pnas.98.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wales TE, Engen JR. Mass Spectrom.Rev. 2006;360:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- (38).Brier S, Lemaire D, DeBonis S, Forest E, Kozielski F. Journal of Molecular Biology. 2006;20:360–376. doi: 10.1016/j.jmb.2006.04.062. [DOI] [PubMed] [Google Scholar]
- (39).Brier S, Lemaire D, DeBonis S, Kozielski F, Forest E. Rapid Communications in Mass Spectrometry. 2006;95:456–462. doi: 10.1002/rcm.2329. [DOI] [PubMed] [Google Scholar]
- (40).Wee CL, Balali-Mood K, Gavaghan D, Sansom MSP. Biophys. J. 2008;95:1649–1657. doi: 10.1529/biophysj.107.123190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bala P, Grochowski P, Nowinski K, Lesyng B, McCammon JA. Biophys. J. 2000;79:1253–1262. doi: 10.1016/S0006-3495(00)76379-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Burke JE, Karbarz MJ, Deems RA, Li S, Woods VL, Jr., Dennis EA. Biochemistry. 2008;47:6451–9. doi: 10.1021/bi8000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hsu YH, Burke JE, Stephens DL, Deems RA, Li S, Asmus KM, Woods VL, Jr., Dennis EA. J Biol Chem. 2008;283:9820–7. doi: 10.1074/jbc.M708143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lucas KK, Dennis EA. Prostaglandins Other Lipid Mediat. 2005;77:235–48. doi: 10.1016/j.prostaglandins.2005.02.004. [DOI] [PubMed] [Google Scholar]
- (45).Peitsch MC. Biochemical Society Transactions. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- (46).Humphrey W, Dalke A, Schulten K. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (47).Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. The Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- (49).MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.