Abstract
The amino acid sequences of transmembrane regions of helical membrane proteins are highly constrained, diverging at slower rates than their extramembrane regions and than water-soluble proteins. Moreover, helical membrane proteins seem to fall into fewer families than water-soluble proteins. The reason for the differential restrictions on sequence remains unexplained. Here, we show that the evolution of transmembrane regions is slowed by a previously unrecognized structural constraint: Transmembrane regions bury more residues than extramembrane regions and soluble proteins. This fundamental feature of membrane protein structure is an important contributor to the differences in evolutionary rate and to an increased susceptibility of the transmembrane regions to disease-causing single-nucleotide polymorphisms.
Keywords: disease mutation, potassium channel, protein folding, protein stability, single nucleotide polymorphisms
Evolutionary rates vary considerably in different cellular compartments (1). Membrane proteins have been found to diverge faster overall than soluble proteins (2, 3), but this increased rate is confined entirely to the rapidly evolving extramembrane regions. Transmembrane regions, on average, diverge much more slowly than the extramembrane regions more slowly than soluble proteins (1, 4–6).
A major factor controlling protein sequence divergence is the need to preserve protein function by maintaining a folded structure (7). Because the physical forces that drive folding can change with environment, proteins in different cellular locations can be subject to distinct evolutionary constraints. Membrane proteins, in particular, must accommodate to a dramatically varied environment, ranging from hydrocarbon chains in the bilayer core to water as they emerge from the membrane (8, 9). It therefore seems possible that distinct structural imperatives found in different environments could be an important contributor to evolutionary rates. An obvious sequence adaptation is the hydrophobic matching of the protein exterior, reflected in an apolar transmembrane amino acid composition. Although amino acid diversity is more limited in the transmembrane segments, simple compositional differences do not explain the slower divergence rates of transmembrane regions (1, 4, 5).
Here, we find that the transmembrane regions of membrane proteins bury more residues on average than soluble proteins and much more than extramembrane regions, a possible mechanism for increasing stabilization in the absence of the hydrophobic effect. Because buried residues evolve at slower rates than surface residues (10–12), the higher level of residue burial in the transmembrane regions leads to slower sequence divergence. Moreover, we find that higher residue burial may explain a higher prevalence of disease-causing mutations in the transmembrane region of membrane proteins compared with the extramembrane regions.
Results and Discussion
Transmembrane Regions Bury More Residues.
Fig. 1A shows plots of the fractional surface area buried per residue versus oligomer size for transmembrane regions, extramembrane regions, and soluble proteins. Transmembrane segments clearly bury more of their surface on average than soluble proteins and much more than the extramembrane regions. When transmembrane segments are compared only with α-helices of extramembrane regions or α-helices of helical soluble proteins, the difference, albeit less pronounced, still remains (Fig. 1B). We note that the average surface area buried per residue is similar in membrane and soluble α-helices, because transmembrane segments bury smaller residues on average (13, 14) (supporting information (SI) Fig. S1). Transmembrane helices, however, bury more of their available surface and thus, in effect, use more residues for structure maintenance than soluble protein helices and much more than extramembrane helices.
Fig. 1.
Transmembrane segments bury a larger fraction of their surface area on average than soluble proteins or membrane protein extramembrane segments. The plot shows the average fraction of surface area buried per residue as a function of the number of residues in the native oligomer for (A) transmembrane segments (red circles), all residues of water-soluble proteins (open green squares), and all residues of extramembrane segments (blue diamonds) and for (B) transmembrane segments (red circles), helices of helical soluble proteins (open green squares), and helices of extramembrane segments (blue diamonds).
The reason for the higher burial rate for transmembrane helices is unclear. It is possible that increased burial is driven by a need to maximize van der Waals packing. Alternatively, the use of small residues that can facilitate polar backbone interactions and reduce entropy costs simply may necessitate a closer apposition of the transmembrane helices, increasing the rate of burial (15).
Is Residue Burial an Important Factor Controlling Evolutionary Rates?
The higher level of residue burial in transmembrane helices could impose a greater structural constraint on the rate of sequence divergence compared with environments that demand less residue burial. Nevertheless, many factors influence sequence divergence rates, so how important is simple residue burial in explaining the slower divergence rates in the transmembrane segments?
To assess the impact of residue burial on conservation differences in the transmembrane versus the extramembrane environments, we compared the divergence rates of residues grouped according to their extent of burial. In the extreme scenario, in which residue burial is the only factor controlling the disparity in evolutionary rates, buried residues in the transmembrane segments showed essentially the same variability as buried residues in the extramembrane segments. The same finding was true for exposed residues.
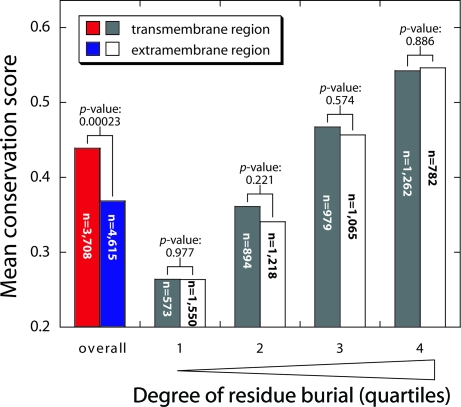
From 19 distinct helical membrane proteins of known structure, we collected all 21 unique polytopic chains and prepared sequence alignments of family members. Conservation scores were calculated for each position using the trident scoring method (16), with the conservation scores adjusted to account for trivial composition effects as described in SI Materials (results are given in Table S1). Fig. 2 shows the distribution of conservation scores for the transmembrane regions and the extramembrane regions. As expected, the extramembrane segments had lower conservation scores overall than the transmembrane regions (P = 2.3 × 10−4), corroborating the slower divergence of the transmembrane regions compared with the extramembrane regions. When residues were divided according to their degree of burial, however, the scores were very similar on average (Fig. 2). For both the extramembrane regions and transmembrane regions, the conservation scores increase with increasing burial, as expected, but for residues with a similar level of burial the conservation scores are statistically indistinguishable for the 2 regions. Thus, within a given membrane protein family, the rate of residue burial seems to have a significant influence on divergence rates.
Fig. 2.
Comparison of the divergence rates as a function of burial for the residues in transmembrane and extramembrane regions of integral membrane proteins. The average conservation scores in different categories are shown. The pair of histogram bars on the left reports the average normalized conservation scores for all residues in the transmembrane region (red bar) and the extramembrane region (blue bar). The next 3 pairs of histogram bars represent the average conservation scores at different levels of burial for the transmembrane region (gray bars) and extramembrane region (white bars). The residues in the combined transmembrane and extramembrane regions were divided according to their level of burial into quartiles of equal numbers: 1 (0–52% buried), 2 (52.1–81.3% buried), 3 (81.4–97.3% buried), and 4 (97.4–100% buried). The p-values shown were calculated using a paired t test, in which each pair is the conservation score for the extramembrane and transmembrane region within a particular family (Table S1). The number of residues in each category, n, is shown on the histogram bars.
Effect on Deleterious SNPs.
The high level of residue burial also could increase the susceptibility of transmembrane regions to deleterious substitutions, such as those that occur in genetic diseases. In water-soluble proteins, 80% of disease-causing mutations were found to destabilize structure (17). For membrane proteins, however, it remains unclear how important structural factors are compared with the many other mechanisms that can compromise protein viability, such as the impairment of membrane insertion or the alteration of functional sites. If transmembrane residue burial is an important factor in genetic disease etiology, we would expect disease-causing mutations would be (i) more probable in the transmembrane domains and (ii) targeted to buried residues. If another factor is the primary cause of disease, there might be little or no correlation with structural parameters. A strong bias for disease-causing SNPs to occur in the transmembrane regions of G protein-coupled receptors (6, 18) and potassium channels (19) has been observed, although the structural basis of this observation has not been investigated. We therefore collected a set of disease-causing variants as described in Methods and listed in Table S2.
To assess the relative preference for disease mutations in different structural categories, we define a disease bias ratio (DBR) as follows:
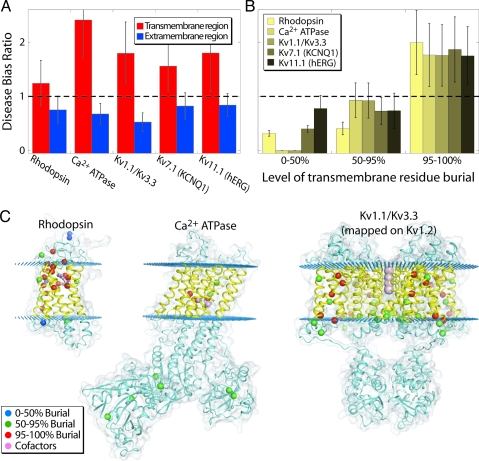
where F(D,i) is the fraction of all disease-causing mutations in category i and F(i) is the fraction of all residues in category i. Thus, if the DBR is >1, disease mutants are more prevalent than expected by chance in category i. Consistent with our hypothesis and prior observations, there is a clear preference for disease-causing mutations to reside in the transmembrane regions for each of the protein families (Fig. 3A). Moreover, in the transmembrane regions, the DBR increases dramatically as residues become more buried (Fig. 3B). The strong bias for transmembrane disease mutations to occur in buried residues suggests that transmembrane segments are more structurally sensitive than extramembrane segments because of the higher level of residue burial.
Fig. 3.
There is a bias for disease-causing mutations to occur in buried positions of the transmembrane helices. (A) Disease-causing mutations are more likely in the transmembrane regions. The plot shows the DBR observed for the transmembrane (red) and extramembrane (blue) regions. For KCNQ1 and hERG, the transmembrane/extramembrane assignment was taken from Jackson and Acili (19); for the other entities it was taken directly from the mapping onto the structures. The transmembrane region of rhodopsin has 167 residues and 17 disease mutations, whereas the soluble region has 162 residues and 9 disease mutations; the transmembrane region of Ca2+ ATPase has 180 residues and 5 disease mutations, whereas the soluble region has 814 residues and 6 disease mutations; the transmembrane region of the voltage-gated K+ channel (Kv1.1/Kv1.3) has 144 residues and 8 disease mutations, whereas the soluble region has 242 residues and 4 disease mutations; the transmembrane region of KCNQ1 (Kv7.1) has 146 residues and 49 disease mutations, whereas the soluble region has 458 residues and 81 disease mutations; the transmembrane region of hERG (Kv11.1) has 151 residues and 40 disease mutations, whereas the soluble region has 753 residues and 93 disease mutations. (B) Disease-causing mutations in the transmembrane region are biased toward buried residues. The DBR for the residues in the transmembrane regions are grouped by the degree of side-chain burial for the residues that were mappable onto the structures (see Table S2). (C) The position of disease variants on the protein structures. The sequences were aligned to 3 proteins of known structure: rhodopsin (1GZM) (34), Ca2+ ATPase (1SU4) (35), and voltage-gated K+ channel Kv 1.2 (2R9R) (36). Kv1.1/Kv3.3 were aligned to the Kv1.2 structure. The α-carbons of the residues associated with disease-causing mutations are shown as spheres color-coded according to the degree of side-chain burial for the membrane proteins used in A and B. Blue corresponds to 0–50% buried, green to 50–95% buried, and red to 95–100% buried. The transmembrane and extramembrane regions are highlighted in yellow and blue, respectively, and are separated by planes of blue dots. Cofactors and substrates for each protein (retinal for rhodopsin, Ca2+ for Ca2+ ATPase, and K+ for Kv 1.2) are highlighted in violet. KCNQ1 and hERG are not included in this figure because only partial alignments to the known Kv1.2 structure are possible.
Conclusion
Our results indicate how environmental influences on the ability to fold may limit membrane protein evolution. The hydrophobic effect is a dominant contributor to the structure stabilization of soluble proteins and the extramembrane regions of membrane proteins (7, 20), but water is essentially absent in the hydrocarbon core of the bilayer, where membrane proteins must operate. Consequently, the relative importance of other forces, such as van der Waals packing and hydrogen bonds, must increase in the apolar environment of the membrane core (13). To make good use of dispersion forces and polar interactions, membrane proteins therefore may need to pack a larger fraction of their surface area to maintain a stable structure. Regardless of the reason for additional packing, this physical constraint seems to be important for disease etiology and could be a factor in the smaller number of integral-membrane protein families that seem to exist compared with water-soluble protein families (21, 22). It has been suggested that water-soluble proteins evolved from the extramembrane segments of primordial membrane proteins (23). If so, the ability to break out of the folding constraints imposed by the membrane may have been a key factor in the early evolution of life.
Materials and Methods
Protein Structure Database Analysis.
A set of 31 helical membrane proteins (Table S3) of known structure were selected using the membrane proteins of known 3D structure database (24) and 533 water soluble proteins of known structure were selected from the ACT database (27), so that none had >30% sequence identity with any others in the respective sets. Quaternary structures of membrane proteins were obtained from the PQS (25) and OPM (26) databases, and quaternary structures of the soluble proteins were determined using the ACT database (27). The transmembrane domain boundaries were taken from the OPM database (26). Solvent accessibilities were determined using the method of Le Grand and Merz (28) as implemented in EZPROT (27). The atomic radii and free residue areas were taken from ref. 29. Cofactors were included in the solvent accessibility calculations, but substrates and other bound molecules were removed.
Helical soluble proteins were defined as water-soluble proteins whose total residue content constitutes at least 50% helix-structured residues. A set of 137 helical water-soluble proteins was obtained from the 533 water-soluble protein structures.
Sequence Alignments.
Proteins from our membrane protein list were compared with sequences from the UniProt sequence database using Blast 2.0 (37). Hits with a minimum sequence identity threshold of 30% and a minimum overlap in length of 70% were extracted and aligned using ClustalW (30). A final set of 21 protein families that have highly informative conservation scores (> 90% diversity of scores) was selected for further analysis.
Conservation Scoring.
The trident scoring method in SCORECONS was used to obtain conservation scores (16). Trident is an entropy-based method that also utilizes amino acid physico-chemical properties and is weighted by the sequence similarity of family members. Scores were considered only for positions in the alignment with <80% gaps; the remainder were considered noninformative and were removed.
There is an inherent bias in conservation scores between the transmembrane and extramembrane regions because of the lower residue diversity in the apolar membrane regions. To remove this bias, we determined the average conservation score obtained for random sequences with compositions of either the extramembrane or transmembrane regions. We determined this score by creating 2 random pseudofamilies 200 residues long with 200 family members. The pseudofamilies then were scored as a multiple alignment using SCORECONS. The random score for the transmembrane region was 0.136 and for the extramembrane region was 0.055. Because for both regions the maximum score is 1.0, the expected range of values is ≈10% smaller for the transmembrane region sequences because of composition alone. To correct for this small difference, a normalization was applied to both transmembrane and extramembrane residue scores according to the formula:
where NS is the normalized score, OS is the original score, and RS is the randomized score.
Identification and Structure Mapping of Disease-Causing Variants.
The membrane proteins for which an experimental structure from a mammalian species is available were used to identify disease-causing variant alleles of genes from the Online Mendelian Inheritance In Man (OMIM) database (31), which contains data on human monogenic disorders. We were able to find homologues with disease-causing variant data for 3 proteins of known structure (requiring >30% sequence identity). Only those nonsynonymous SNP disease-causing variants that result in an amino acid change were used; others, such as those resulting in a termination or those from deletions, were removed from the set. To map the residues on the known structures, the sequences corresponding to the disease-causing genes were aligned to the sequence of the protein with an experimental structure using BLAST. The proteins used were rhodopsin (aligned to 1GZM), calcium ATPases (aligned to 1SU4, 1T5S, 1WPE, 1WPG, and 2AGV), and the voltage-gated potassium channels Kv1.1 and Kv3.3 (aligned to 2R9R). In addition, we made use of hand-curated alignments of KCNQ1 (32) and hERG (33) to portions of the 2R9R (Kv1.2) sequence and the disease-mutation database for these proteins compiled by Jackson and Accili (19) (see Table S2).
Supplementary Material
Acknowledgments.
We thank Yungok Ihm for help with the membrane protein structure database and members of the laboratory for helpful comments on the manuscript. The work was funded by National Institutes of Health Grants R01 GM063919 and R01 GM081783.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906390106/DCSupplemental.
References
- 1.Julenius K, Pedersen AG. Protein evolution is faster outside the cell. Mol Biol Evol. 2006;23:2039–2048. doi: 10.1093/molbev/msl081. [DOI] [PubMed] [Google Scholar]
- 2.Plotkin JB, Dushoff J, Fraser HB. Detecting selection using a single genome sequence of M. tuberculosis and P. falciparum. Nature. 2004;428:942–945. doi: 10.1038/nature02458. [DOI] [PubMed] [Google Scholar]
- 3.Volkman SK, et al. Excess polymorphisms in genes for membrane proteins in Plasmodium falciparum. Science. 2002;298:216–218. doi: 10.1126/science.1075642. [DOI] [PubMed] [Google Scholar]
- 4.Tourasse NJ, Li WH. Selective constraints, amino acid composition, and the rate of protein evolution. Mol Biol Evol. 2000;17:656–664. doi: 10.1093/oxfordjournals.molbev.a026344. [DOI] [PubMed] [Google Scholar]
- 5.Leabman MK, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee A, et al. Distribution analysis of nonsynonymous polymorphisms within the G-protein-coupled receptor gene family. Genomics. 2003;81:245–248. doi: 10.1016/s0888-7543(03)00009-0. [DOI] [PubMed] [Google Scholar]
- 7.Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: Tolerance to amino acid substitutions. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 8.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 9.White SH, Ladokhin AS, Jayasinghe S, Hristova K. How membranes shape protein structure. J Biol Chem. 2001;276:32395–32398. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- 10.Eyre TA, Partridge L, Thornton JM. Computational analysis of alpha-helical membrane protein structure: Implications for the prediction of 3D structural models. Protein Engineering, Design and Selection. 2004;17:613–624. doi: 10.1093/protein/gzh072. [DOI] [PubMed] [Google Scholar]
- 11.Goldman N, Thorne JL, Jones DT. Assessing the impact of secondary structure and solvent accessibility on protein evolution. Genetics. 1998;149:445–458. doi: 10.1093/genetics/149.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lio P, Goldman N, Thorne JL, Jones DT. PASSML: Combining evolutionary inference and protein secondary structure prediction. Bioinformatics (Oxford, UK) 1998;14:726–733. doi: 10.1093/bioinformatics/14.8.726. [DOI] [PubMed] [Google Scholar]
- 13.Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S, Vakser IA. Side chains in transmembrane helices are shorter at helix-helix interfaces. Proteins. 2000;40:429–435. doi: 10.1002/1097-0134(20000815)40:3<429::aid-prot80>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 15.MacKenzie KR, Engelman DM. Structure-based prediction of the stability of transmembrane helix-helix interactions: The sequence dependence of glycophorin A dimerization. Proc Natl Acad Sci USA. 1998;95:3583–3590. doi: 10.1073/pnas.95.7.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 17.Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Balasubramanian S, Xia Y, Freinkman E, Gerstein M. Sequence variation in G-protein-coupled receptors: Analysis of single nucleotide polymorphisms. Nucleic Acids Res. 2005;33:1710–1721. doi: 10.1093/nar/gki311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson HA, Accili EA. Evolutionary analyses of KCNQ1 and HERG voltage-gated potassium channel sequences reveal location-specific susceptibility and augmented chemical severities of arrhythmogenic mutations. BMC Evolutionary Biology. 2008;8:188. doi: 10.1186/1471-2148-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Gerstein M, Engelman DM. Transmembrane protein domains rarely use covalent domain recombination as an evolutionary mechanism. Proc Natl Acad Sci USA. 2004;101:3495–3497. doi: 10.1073/pnas.0307330101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberai A, Ihm Y, Kim S, Bowie JU. A limited universe of membrane protein families and folds. Protein Sci. 2006;15:1723–1734. doi: 10.1110/ps.062109706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi N, Yanagawa H. Origins of globular structure in proteins. FEBS Lett. 1998;430:150–153. doi: 10.1016/s0014-5793(98)00674-7. [DOI] [PubMed] [Google Scholar]
- 24.White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 25.Henrick K, Thornton JM. PQS: A protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 26.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of proteins in membranes database. Bioinformatics (Oxford, UK) 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 27.Pettit FK, Bare E, Tsai A, Bowie JU. HotPatch: A statistical approach to finding biologically relevant features on protein surfaces. J Mol Biol. 2007;369:863–879. doi: 10.1016/j.jmb.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Grand S, Merz K. Rapid approximation to molecular surface area via the use of Boolean logic and look-up tables. J Comput Chem. 1993;14:349–352. [Google Scholar]
- 29.Richmond TJ, Richards FM. Packing of alpha-helices: Geometrical constraints and contact areas. J Mol Biol. 1978;119:537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith JA, Vanoye CG, George AL, Jr, Meiler J, Sanders CR. Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry. 2007;46:14141–14152. doi: 10.1021/bi701597s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wynia-Smith SL, Gillian-Daniel AL, Satyshur KA, Robertson GA. hERG gating microdomains defined by S6 mutagenesis and molecular modeling. J Gen Physiol. 2008;132:507–520. doi: 10.1085/jgp.200810083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 35.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 36.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.