Abstract
AGAMOUS-like6 (AGL6) genes encode MIKC-type MADS box transcription factors and are closely related to SEPALLATA and AP1/FUL-like genes. Here, we focus on the molecular evolution and expression of the AGL6-like genes in grasses. We have found that AGL6-like genes are expressed in ovules, lodicules (second whorl floral organs), paleas (putative first whorl floral organs), and floral meristems. Each of these expression domains was acquired at a different time in evolution, indicating that each represents a distinct function of the gene product and that the AGL6-like genes are pleiotropic. Expression in the inner integument of the ovule appears to be an ancient expression pattern corresponding to the expression of the gene in the megasporangium and integument in gymnosperms. Expression in floral meristems appears to have been acquired in the angiosperms and expression in second whorl organs in monocots. Early in grass evolution, AGL6-like orthologs acquired a new expression domain in the palea. Stamen expression is variable. Most grasses have a single AGL6-like gene (orthologous to the rice [Oryza sativa] gene MADS6). However, rice and other species of Oryza have a second copy (orthologous to rice MADS17) that appears to be the result of an ancient duplication.
INTRODUCTION
The identity of floral organs in most angiosperms is specified by a combination of transcription factors in the MADS box family (Ng and Yanofsky, 2001). These transcription factors fall into large clades, often named for the characteristic protein in Arabidopsis thaliana, such as AGAMOUS, PISTILLATA, APETALA3, APETALA1, and SEPALLATA (Becker and Theissen, 2003). Whereas many of these clades of proteins have been studied extensively, others are much more poorly known. One of these less well-studied clades includes proteins that are coorthologous to the Arabidopsis protein AGL6.
Phylogenetic analysis of MADS box genes and/or proteins has shown that AGL6-like genes are sister to the SEPALLATA-like genes (E-function) (Purugganan et al., 1995; Theissen et al., 2000; Zahn et al., 2005). AGL6-like genes have been found in both in angiosperms and gymnosperms but not in ferns (Theissen et al., 2000). All members of the AGL6 and SEPALLATA clades share a conserved hydrophobic motif in the C-terminal domain, suggesting that they might potentially interact (Moon et al., 1999; Litt and Irish, 2003; Vandenbussche et al., 2003; Zahn et al., 2005).
AGL6-like genes have been characterized as floral specific in several Angiosperms; however, some studies reported expression in vegetative tissues (Ma et al., 1991; Rounsley et al., 1995). AGL6 function has been examined by Hsu et al. (2003) and Fan et al. (2007), who overexpressed AGL6 in Arabidopsis. In both cases, the inflorescences of the transgenic plants were determinate and the flowers showed homeotic transformations, where sepals are transformed into carpel-like organs bearing ovules and petals are transformed into staminoids. Expression patterns of AGL6-like genes in the flower vary from one lineage to the next, further complicating efforts to understand gene function (see Supplemental Tables 1 and 2 online). Some of these differences may be due to the different methods used (i.e., RNA gel blots, RT-PCR, microarrays, and in situ hybridization).
Grasses are morphologically unique in having highly modified flowers (florets) collected into novel structures called spikelets (Clifford, 1987). Each spikelet is subtended by two outer bracts (glumes) and contains, depending on the species, from 1 to 40 florets. Each floret has a central gynoecium and three to six stamens but does not have obvious petals and sepals. Instead, the fertile organs are surrounded by two to three fleshy structures known as lodicules and two sterile bracts, the lemma and the palea. Lodicules are in the position of second whorl organs in more conventional flowers, and like such organs, their identity is specified by homologs of APETALA3 and PISTILLATA (Ambrose et al., 2000; Whipple et al., 2004; Whipple and Schmidt, 2006). However, homology of the lemma and palea is controversial. Stebbins (1956) comments in passing that the palea is derived from two fused tepals, and some genetic evidence supports this point of view (Ambrose et al., 2000). However, the position and morphological characteristics of the lemma and palea are so different from conventional tepals that other authors have instead concluded that they are not perianth at all (Clifford, 1987).
AGL6-like genes in grasses might be involved in lodicule, stamen, carpel, and seed development. For instance, rice (Oryza sativa) has two AGL6-like genes, MADS6 and MADS17. Based on RNA gel blots, Moon et al. (1999) showed that MADS6 is expressed strongly in lodicules and carpels and weakly in lemma. However, in situ hybridization showed that MADS6 is expressed in floral meristems and carpels, but no expression was reported for lemma, palea, and stamens (Pelucchi et al., 2002). Besides the microarrays that reported MADS17 expression during floral and seed development (Arora et al., 2007), no other expression studies have been reported for MADS17. In maize (Zea mays) two AGL6-like genes, zag3 and zag5, have been characterized (Mena et al., 1995). zag3 mRNA in situ hybridization showed that it is expressed in both upper and lower floral meristems but not in the lemma and stamens (Thompson et al., 2009). Later in development, zag3 is expressed in developing lodicules, palea, carpels, and the inner integument. Characterization of a zag3 mutant, the only loss-of-function mutant available for an AGL6-like gene, demonstrates a role in floral meristem and organ development (Thompson et al., 2009). No data are available on the specific floral organs where zag5 is expressed.
The function of AGL6 genes during flower development in grasses as well in other lineages is therefore not clear, although they may play a role in perianth and gynoecium development. Given the characterization of a loss-of-function mutant for AGL6 in maize (Thompson et al., 2009), we set out to determine the generality of the maize expression patterns and molecular evolution of this gene subfamily in the grasses. Accordingly, we have reconstructed the evolutionary history of AGL6-like genes in grasses and have analyzed gene expression patterns during spikelet and floret development. We have found that the rice duplication is restricted to Oryza, and the zag3/zag5 duplication in maize is characteristic only of maize and its sister genus Tripsacum. More importantly, we find that the AGL6-like genes in the grasses have discrete expression domains that have been acquired sequentially in evolutionary time. Expression of AGL6-like genes in the ovule is ancient, apparently shared with gymnosperms, whereas expression in floral meristems is shared among flowering plants. Expression in floral organs of the second whorl is less widespread, and expression in the palea (presumptive first whorl) is a novel trait that correlates with the origin of the grass spikelet.
RESULTS
The AGL6-like Duplicate Genes Evolve at Different Rates
To reconstruct the evolutionary history of AGL6-like genes in grasses, we PCR amplified AGL6-like genes from disparate grass species to supplement data available in GenBank (see Supplemental Table 3 online). Sequences spanned the three regions of the gene, the I (intervening, important for specification of dimerization), K (keratin-like, facilitates dimerization), and C (C terminus, functions as a transactivation region and contributes to the formation of multimeric MADS box protein complexes), regions but excluded the highly conserved MADS box and sometimes 25 bp at the 3′ end of the C terminus. Most of the divergence among the sequences appears in the C-terminal region. All sequences had a single open reading frame and no premature stop codons. All amino acid sequences shared a conserved motif at the end of the C terminus (MLGWVL).
The Oryza species investigated had two copies of AGL6-like genes, corresponding to MADS6 and MADS17, as previously described. We used two methods to assess the timing of the MADS6/MADS17 duplication and arrived at contradictory results. First, we determined the presence/absence of the two paralogs in grasses outside the genus Oryza. No homolog of MADS17 was found in the maize, sorghum (Sorghum bicolor), or Brachypodium genomes or in any other genome database (see Methods). In addition, multiple efforts at PCR and sequencing of multiple clones failed to detect MADS17-like sequences in other grasses or nongrass monocots; therefore, we conclude that MADS17 is truly absent in all grasses except Oryza. Within Oryza, the MADS17-like sequences (254 amino acids long) differ by 71 amino acids from the paralogous MADS6-like sequences (250 amino acids long) (see Supplemental Figure 1 online), with most substitutions located at the C terminus. Among the Oryza MADS17-like sequences, however, both coding regions and intron sequences were conserved. Taken together, these data suggest that MADS17 is the product of a duplication event at the origin of the genus Oryza.
The second method we used to assess the timing of the MADS6/MADS17 duplication was to analyze the sequences phylogenetically; for this analysis, we retrieved similar trees for all analytical methods. Phylogenetic analysis offers a different estimate of the timing of the MADS6 and MADS17 duplication, which appears after the divergence of Restionaceae and before the divergence of Joinvilleaceae. However, we cannot reject the possibility that the duplication occurred at the same time as the whole-genome duplication of the grasses, after the divergence of Joinvilleaceae but before the origin of the family (Figure 1). The S-H test rejects all other possible placements of this duplication at P values < 0.05; statistical phylogenetic evidence thus argues against the possibility that the duplication was specific only to Oryza.
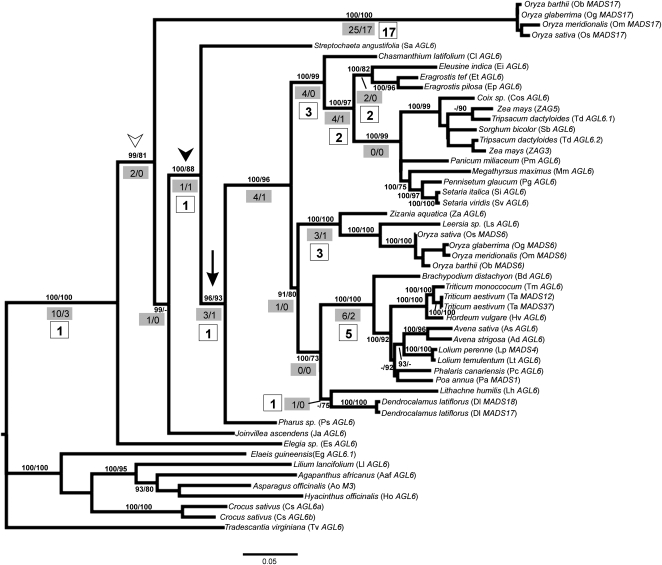
Figure 1.
Maximum Likelihood Tree of AGL6-like Genes in Grasses.
Numbers on branches, Bayesian posterior probabilities (>90) and likelihood bootstrap support values (>70). White arrowhead, inferred Os MADS6/Os MADS17 duplication event. Black arrowhead, an alternative hypothesis of the duplication event that cannot be rejected by Shimodaira-Hasegawa test. Black arrow indicates grass spikelet origin. Numbers in gray rectangles indicate total number of nonsynonymous substitutions and number of unique nonsynonymous substitutions. Numbers in white boxes indicate the number of unreversed nonsynonymous substitutions. Only amino acid changes of branches up to the grass subfamily levels are presented.
The branch leading to the Os MADS17 clade is the longest branch in the tree (Figure 1), and relative rates tests show that MADS17 has evolved significantly faster than MADS6 (P = 0.01963). Twenty-five amino acid substitutions characterized the MADS17 clade, and 17 are unique substitutions that do not change elsewhere in the clade (Figure 1, number in white box). Most of these mutations are located at the K-domain and C terminus. Nine substitutions change the chemical properties of the residues (i.e., 83 D-G, 94 T-H, 139 C-Y, 205 A-R, and 260 V-P) with the remainder being conservative substitutions (see Supplemental Figure 1 online). MADS17 orthologs also differ in the highly conserved motif that characterizes the MADS6-like sequences in the grasses (256-261 MLGWVL versus VMGWPL). In addition, MADS17 has an extra five amino acids at the N terminus of the MADS box (MDRSE) that are not found in other known AGL6-like genes or MADS box genes.
The duplication of zag3 and zag5 (both genes of maize) may have occurred before the common ancestor of Zea plus Tripsacum. However, the duplication could have been before the common ancestor of Zea, Tripsacum, Sorghum, and Coix (the zag3/Td AGL6.2 + Sb AGL6 + Cos AGL6 clade). Neither relationship is well supported and cannot be distinguished statistically. There is no evidence that zag5/zag3 are evolving at different rates.
Within the Os MADS6 clade, the phylogeny largely agrees with that proposed for the grass family by the Grass Phylogeny Working Group (2001), except that our data show the Ehrhartoideae diverging before the clade comprised of Bambusoideae and Pooideae (Figure 1). The position of Chasmanthium latifolium, sister to Panicoideae plus Chloridoideae, is likely to be a sampling artifact, reflecting the lack of sequences for other taxa in the PACCMAD clade. Only one amino acid substitution characterizes the AGL6/Os MADS6 clade (before the divergence of Streptochaeta) and has remained unchanged since then. A single amino acid in the K-domain (S → T; black arrow in Supplemental Figure 2 online) changed before the divergence of Pharus and has been unchanged ever since; this correlates with the origin of the grass spikelet (Figure 1).
All AGL6-like genes are under purifying selection, although sites do not evolve homogeneously across the gene (see Supplemental Statistical Tests online; codons showed ω values from 0.01390 [purifying selection] to 0.72797 [relaxed selection]). We found no evidence of positive selection; none of the AGL6-like sequences have overall ω values greater than one. In addition, we found no evidence for a change in selective pressure along the branches leading to Os MADS17, AGL6/Os MADS6, or the spikelet-bearing grasses where there is a marked change in gene expression pattern.
AGL6 mRNA Expression Patterns
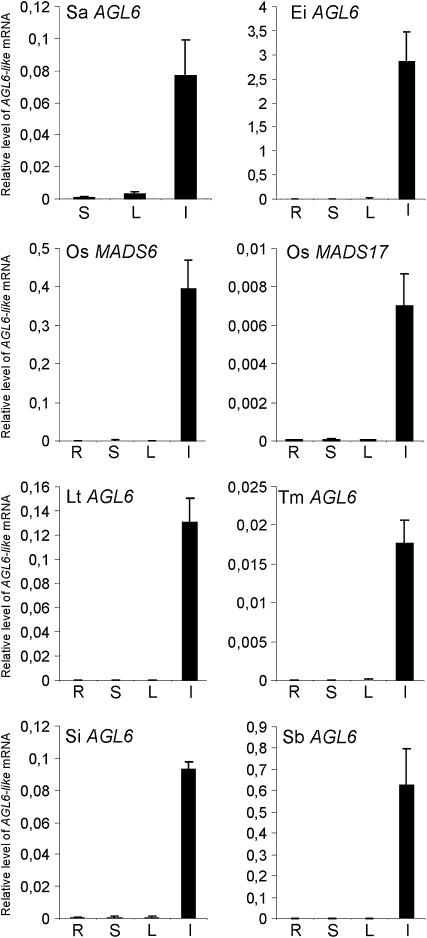
To determine organ-level patterns of AGL6-like gene expression in disparate grasses, we used quantitative PCR (Q-PCR). Q-PCR with AGL6-specific primers showed that gene expression is restricted largely to inflorescence tissue (Figure 2). No significant expression was detected in the leaf, culm, and root. In the nongrass monocot Agapanthus africanus, AGL6-mRNA was detected in the inner tepals as well as in the carpels (see Supplemental Figure 3 online).
Figure 2.
AGL6-like Q-PCR Products from RNA of Roots, Stems, Leaves, and Inflorescence in Grasses.
Relative expression to ACTIN is presented, with sd for three biological replicates and three Q-PCR replicates. Sa, Streptochaeta angustifolia; Ei, Eleusine indica; Os, Oryza sativa; Lt, Lolium temulentum; Tm, Triticum monococcum; Si, Setaria italica; Sb, Sorghum bicolor. R, roots; S, stems, L, leaves, I, inflorescence.
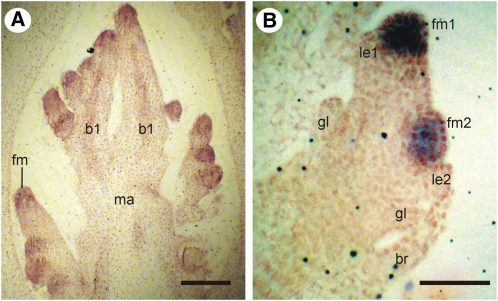
To determine tissue-specific patterns of AGL6 mRNA expression, we conducted in situ hybridizations on inflorescences of several grass species. The specificity of the probe was verified by DNA gel blot hybridization; in all cases, the probe detected a single band. In all species examined, AGL6 mRNA is first detected in floral meristems during early inflorescence development (Figures 3A and 3B; O. sativa and Setaria italica are representative). No expression was detected in the main axis of the inflorescence and branches nor was there expression in glumes.
Figure 3.
AGL6-like mRNA Expression in Floral Meristems during Early Inflorescence and Spikelet Development.
(A) Os MADS17 in floral meristems, young O. sativa inflorescence.
(B) Si AGL6 is expressed in upper and lower floral meristems, young S. italica spikelet, but not in glumes and lemma. b1, primary branch; br, bristle; fm, floral meristem; fm1, floral meristem of the upper floret; fm2, floral meristem of the lower floret; gl, glume; le1, lemma of the upper floret; le2, lemma of the lower floret; ma, main axis of the inflorescences.
Bars = 100μm.
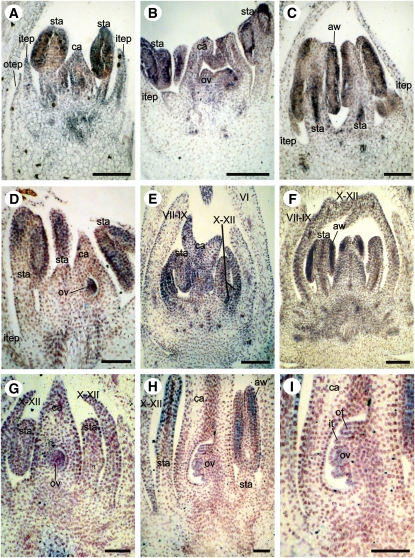
Joinvillea ascendens (Joinvilleaceae) has a typical monocot floral plan with three outer tepals, three inner tepals, six stamens and three carpels. Ja AGL6 was first detected in the inner tepal primordia as well as in the young anthers and gynoecium (carpels and ovule) (Figures 4A to 4C). Later, expression in the inner tepals and carpels decreased, but expression was detectable in the stamens; gynoecial expression was restricted to the ovule (Figure 4D). Very early developmental stages were unavailable, so we could not check floral meristem expression.
Figure 4.
AGL6-like Expression in J. ascendens (Joinvilleaceae) and S. angustifolia (Anomochlooideae).
(A) to (D) Ja AGL6 in J. ascendens.
(A) and (B) Expression in inner tepal primordia, young stamens, carpels, and ovules.
(C) Expression is lost in inner tepals.
(D) Late expression only in anther wall and ovule.
(E) to (I) Sa AGL6 in S. angustifolia at successive stages of development.
(E) and (F) Early expression in bracts X to XII, in young anthers, and carpels.
(G) Late expression reduced in bracts X to XII and carpels but maintained in anthers and restricted to ovule.
(H) Expression in anther wall and ovule.
(I) Higher magnification of (H), showing expression in inner and outer integuments and embryo sac.
VI, S. angustifolia bract; VII-IX, S. angustifolia bracts corresponding to the outer whorl; X-XII, S. angustifolia bracts corresponding to the inner whorl; aw, anther wall; ca, carpel primordia; it, inner integument of the ovule; itep, inner tepals; ot, outer integument of the ovule; otep, outer tepals; ov, ovule; sta, stamens. Bars = 100 μm.
In Streptochaeta angustifolia (Anomochlooideae), the spikelet equivalent has been described as a complex of 11 or 12 bracts (I to XII) that initiate before the stamens and carpels. Previous studies suggested that bracts VII to VIII (or IX) represent the outer whorl and bracts X to XII are modifications of the inner tepals or second whorl of a typical monocot flower (Whipple et al., 2007). Bracts X to XII initiate as a whorl outside the stamens and inside the VII-VIII-(IX) whorl. Sa AGL6 was first detected in the floral meristem (see Supplemental Figure 4 online) and subsequently in bracts X to XII as well as in young anthers and gynoecium (Figures 4E and 4F). Later, expression in bracts X to XII and in the carpels is reduced, but Sa AGL6 is still expressed in the anthers; gynoecial expression is restricted to the ovule (Figures 4G to 4I). We found no expression in bracts VII and VIII.
Lithachne humilis (Bambusoideae) bears female and male spikelets in separate inflorescences. We observed the female inflorescences, which contain one spikelet with a female floret plus a basal sterile spikelet characterized by a single scale or a tiny vestige of a male spikelet. Lh AGL6 expression was detected in the female spikelet, but no expression was observed in the vestigial male basal spikelet (Figure 5A). In the fertile spikelet, Lh AGL6 was detected in palea as well as in the carpel primordia (Figure 5B). Expression was not detected in what we assume are reduced lodicules. Very early developmental stages were unavailable, so we could not check floral meristem expression.
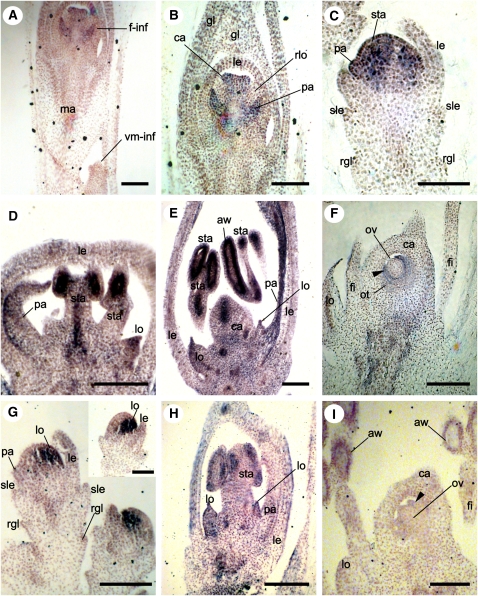
Figure 5.
AGL6-like Expression in L. humilis (Bambusoideae) and O. sativa (Ehrhartoideae).
(A) and (B) L. humilis, female inflorescence.
(A) Lh AGL6 in female but not vestigial male spikelet.
(B) Lh AGL6 in palea and carpels.
(C) to (F) O. sativa. Os MADS6 at successively older stages of development.
(C) Expression in palea and young stamen primordia.
(D) Expression in palea, lodicules, and anthers of young stamens.
(E) Expression in palea, lodicules, anther wall of young stamens, and carpel primordia.
(F) Expression restricted to inner integument of ovule (arrowhead).
(G) to (I) Os MADS17 at successively older stages of development.
(G) Expression in lodicule primordia.
(H) Expression in lodicules and anther walls.
(I) Expression in anther wall but not in carpels or the inner integument (arrowhead).
aw, anther wall; ca, carpel primordia; fi, stamen filament; f-inf, female inflorescence; gl, glume; it, inner integument of the ovule; le, lemma; lo, lodicules; ma, main inflorescence axis; ot, outer integument of the ovule; ov, ovule; pa, palea; rgl, rudimentary glume; rlo, reduced lodicule; sle, sterile lemma; sta, stamens; vm-inf, vestigial male inflorescence. Bars = 100 μm.
The O. sativa (Ehrhartoideae) spikelet contains three florets, one bisexual and fertile and two reduced to sterile lemmas. The glumes are tiny flaps of tissue below the sterile lemmas and are often known as rudimentary glumes. Os MADS6 mRNA was detected in young palea and stamen primordia (Figure 5C). Later in development, Os MADS6 mRNA was also detected in lodicules, gynoecium, and anther walls (Figures 5D and 5E). Still later, expression was only observed in the inner integument of the ovule (Figure 5F).
The expression pattern of Os MADS17 is distinct from that of Os MADS6 starting at an early stage of development. Os MADS17 mRNA is initially detected in the floral meristem (Figure 3A) and later becomes restricted to a group of cells next to the developing lemma that we interpret as young lodicule primordia. No expression was detected in the young palea primordium or in any other region of the floral meristem (Figure 5G). Later, expression was also observed in lodicules and in the anther wall, but no significant expression was detected in the carpels and inner integument (Figures 5H and 5I).
Lolium temulentum and Triticum monococcum (Pooideae) have inflorescences comprised of sessile spikelets with each spikelet containing four to 10 and two to eight florets, respectively, subtended by one (Lolium) or two glumes (Triticum). Lt AGL6 and Tm AGL6 show the same pattern of expression within spikelets. Both genes are expressed in palea, lodicules, and carpels (Figures 6A to 6C; only in situs of T. monoccocum are shown). In addition, AGL6-like genes are expressed in floral meristems in both species (see Supplemental Figure 4 online).
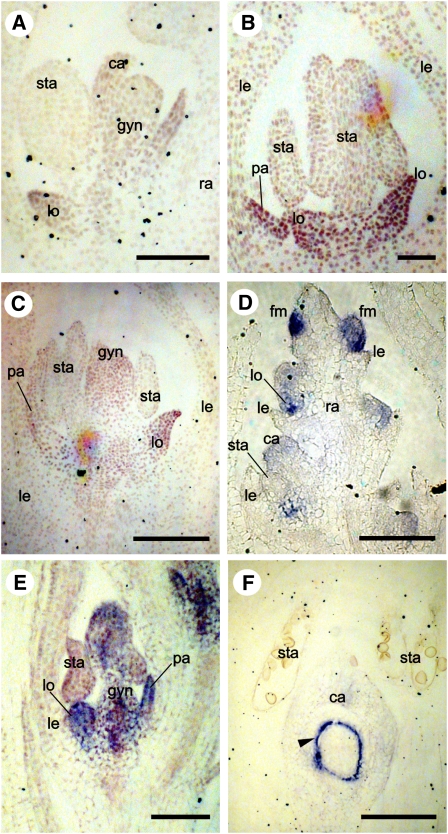
Figure 6.
AGL6-like Expression in T. monoccocum (Pooideae) and E. indica (Chloridoideae) at Successively Older Stages of Development.
(A) to (C) Tm AGL6 in T. monococcum.
(A) and (B) Expression in lodicules and carpels (A) and palea (B).
(C) Later expression in palea, lodicules, and gynoecium.
(D) to (F) Ei AGL6 in E. indica.
(D) Expression in floral meristems, lodicules, and carpel primordia.
(E) Expression in lodicules, palea and gynoecium.
(F) Late expression in the inner integument (arrowhead).
ca, carpel primordia; fm, floral meristem; gl, glume; gyn, gynoecium; le, lemma; lo, lodicules; ra, rachilla; pa, palea; sta, stamens. Bars = 100 μm.
Eleusine indica (Chloridoideae) has sessile spikelets containing three to 15 florets subtended by two glumes. Within a spikelet, Ei AGL6 mRNA is detected in the distal floral meristems as well as in floral organs of the proximal florets, which are more advanced in development (Figure 6D). Ei AGL6 is expressed in the palea, lodicules, and carpel primordia (Figures 6E). Later, Ei AGL6 mRNA was only detected in the inner integument of the ovule (Figure 6F).
S. bicolor (Panicoideae) has an inflorescence comprised of pedicellate and sessile spikelets, each of which includes two florets. Florets in the spikelet mature basipetally. The upper floret of the sessile spikelet is bisexual, whereas the upper floret of the pedicellate spikelet is staminate or sterile, with the pistil aborting early in development. In upper florets, the lemma and palea wrap around the flower with the margins of the palea inserted inside the margins of the lemma; this means that sections will often capture the palea and lemma on both sides of the floral organs. The lower floret in both the sessile and pedicellate spikelets includes only a floral meristem and a sterile lemma. Sb AGL6 is expressed throughout the floral meristem (delimited by the corresponding lemma) of both florets in the sessile and pedicellate spikelet (Figure 7A) and later is detected in the palea and carpel primordia (Figure 7B). Later, Sb AGL6 is still expressed in palea and gynoecium and is also detectable in lodicules (Figures 7C to 7E). Finally, expression is only detectable in the inner integument of the ovule (Figure 7F).
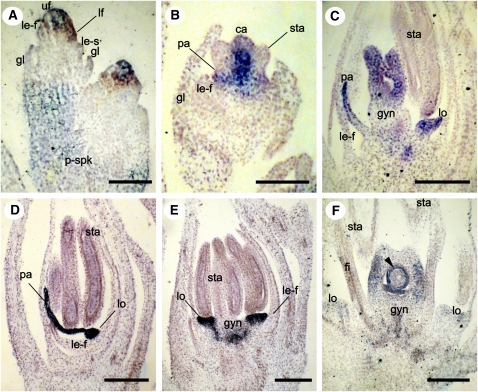
Figure 7.
AGL6-like Expression in Young Spikelets of S. bicolor (Panicoideae) at Successively Older Stages of Development.
(A) Sb AGL6 in the floral meristem of both florets of sessile and pedicellate spikelets.
(B) Expression in palea and carpel primordia.
(C) Expression in palea, lodicules, and gynoecium.
(D) and (E) Two successive sections of the same spikelet.
(D) Expression in developing palea and lodicules.
(E) Expression in lodicules and gynoecium.
(F) Expression in the integument (arrowhead).
ca, carpel primordia; fi, filament of the stamen; gl, glume; gyn, gynoecium; le-f, fertile lemma; le-s, sterile lemma; lf, lower floral meristem; lo, lodicules; ra, rachilla; p-spk, paired spikelet; pa, palea; sta, stamens; uf, upper floral meristem. Bars = 100 μm.
S. italica (Panicoideae) inflorescences bear pedicellate spikelets, each subtended by one to several undifferentiated branches (bristles; Doust and Kellogg, 2002). Each spikelet has an upper complete floret and a lower reduced floret. Si AGL6 was first detected in both floral meristems (Figure 8A). Later, the gene is expressed in palea, lodicules, and carpel primordia (Figures 8B to 8F). Si AGL6 is on in the lower floral meristem even late in development (when all floral organs of the upper floret were initiated) even though no floral organs will form (Figure 8E). Finally, as in other species, late expression is restricted to the inner integument of the ovule of the upper floret (Figure 8F).
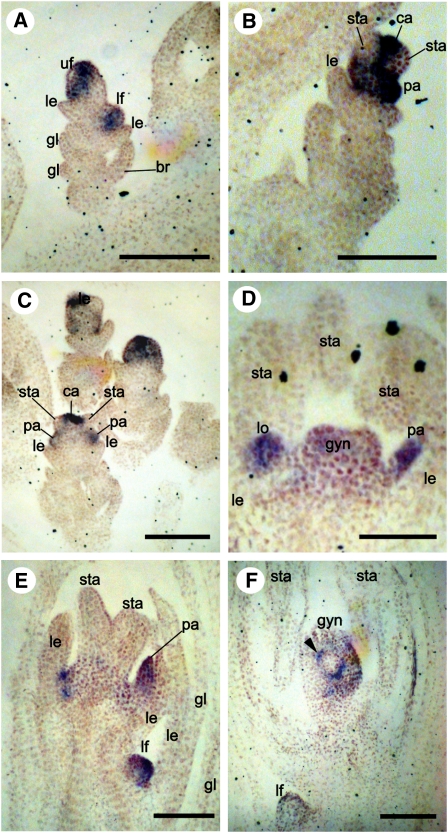
Figure 8.
AGL6-like Expression in Young Spikelets of S. italica (Panicoideae) at Successively Older Stages of Development.
(A) Expression in both floral meristems
(B) and (C) Expression in palea and carpel primordia.
(D) Expression in palea, lodicules, and gynoecium.
(E) Expression in palea of upper floret as well in lower floret meristem.
(F) Expression restricted to the inner integument (arrowhead).
br, bristle; ca, carpel primordia; gl, glume; gyn, gynoecium; le, lemma; lf, lower floral meristem; lo, lodicules; pa, palea; sta, stamens; uf, upper floral meristem. Bars = 100 μm.
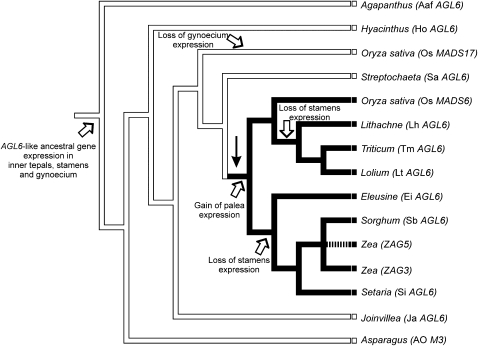
Reconstruction of the ancestral expression pattern estimates that the ancestral AGL6-like gene was expressed in the lodicules (inner tepals), stamens, and carpels but not in the glumes, lemma, or palea (Figure 9). This ancestral condition was maintained in S. angustifolia after which the expression in palea was gained in the rest of the grasses. Also, expression in the gynoecium was lost once in Os MADS17, and expression in stamens was independently lost twice (in the Bambusoideae + Pooideae and in the PACCMAD clades).
Figure 9.
Parsimony Ancestral State Reconstruction of AGL6-like Gene Expression Patterns.
Solid branches indicate gain of palea expression. Dashed branch indicates missing data. Black arrow indicates spikelet origin.
DISCUSSION
Sequence Evolution of AGL6-like Genes in Poales
Our data on the timing of the rice MADS6/MADS17 duplication are contradictory. Phylogenetic analysis of AGL6-like genes in Poales indicates an ancient duplication event, giving rise to the paralogous clades of genes. Placement of the MADS17 clade after the divergence of Restionaceae and before that of Joinvilleaceae suggests that the duplication occurred before the common ancestor of the grasses and Joinvilleaceae, but we cannot reject the hypothesis of a duplication event just after the divergence of Joinvilleaceae, in the common ancestor of the grasses. The latter placement of the duplication is consistent with the ample evidence indicating a whole-genome duplication that characterizes all grasses, but likely not Joinvillea (Kellogg and Bennetzen, 2004; Kellogg, 2006).
If the AGL6/MADS6 and MADS17 duplication reflects the whole-genome duplication of the grasses, we would expect to find orthologs of MADS17 in all grasses. We were therefore surprised that we could not find sequences homologous to MADS17 outside the genus Oryza. Our phylogeny implies that, after the duplication, MADS17 was lost multiple times and has been retained only in Oryza.
In contrast with the phylogenetic data, presence/absence data of MADS17 suggest that the duplication occurred before the diversification of Oryza. The hypothesis is also supported by the positions of MADS6 and MADS17 on rice chromosomes 2 and 4, respectively (coordinates 27870242-27877829 and 29123231-29128740; http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice), in duplicated segments of the genome. This genomic duplication event may be unique to Oryza (http://www.tigr.org/tdb/e2k1/osa1/segmental_dup/500kb/segdup_500kb.shtml; see also Arora et al., 2007). If this hypothesis is correct, then the position of MADS17-like sequences in the phylogeny is an artifact caused by high rates of molecular evolution.
Sequences from zag3 and zag5 duplicate genes formed two clades within Zea-Tripsacum, supporting the idea that zag3/zag5 were duplicates in the tetraploidy event preceding the Zea-Tripsacum divergence (Gaut and Doebley, 1997). A detailed characterization of zag3 is presented by Thompson et al. (2009).
Although selection among AGL6-like genes varies, we found no evidence for positive selection among sites. Our data are consistent with the structure of the MIKC MADS box genes, in which different regions of the genes are under different selective pressures (Becker and Theissen, 2003). MADS box proteins all share a highly conserved DNA binding domain (the MADS domain), a less-conserved intervening (I) domain, a keratin-like (K) domain, and a highly variable C terminus (Riechmann et al., 1996a, 1996b). The MADS box domains are generally under purifying selection, while selection is less intense in the C terminus.
Expression of AGL6-like Genes in Floral Meristems May Indicate a Role in Floral Meristem Identity
In grasses, AGL6-like gene expression is floral specific. This contrasts with the situation in gymnosperms and some eudicots where expression is also observed in vegetative tissues (Ma et al., 1991; Rounsley et al., 1995; Tandre et al., 1995; Mouradov et al., 1998).
AGL6 mRNA was first detected in floral meristems in all grasses examined, even in floral meristems that never form floral organs, such as those in the sterile florets of Setaria and Sorghum. This result agrees with the loss-of-function mutant reported in maize (Thompson et al., 2009). Interestingly, the zag3 mutant in maize, bearded-ear, produces extra organs from the upper floral meristem and axillary meristems from the lower floral meristem of the tassel and ear. Thus, zag3 in maize affects the upper and lower meristem differentially, being required for meristem determinacy as well as floral organ number and development (Thompson et al., 2009).
In non-Poales monocots and in eudicots, mRNA of AGL6 orthologs is also detected in floral meristems prior to floral organ formation (Boss et al., 2002; Hsu et al., 2003; Losa et al., 2004; Fan et al., 2007). All these results together indicate that expression of AGL6 in floral meristems has been conserved at least since the origin of the angiosperms. Because it is not clear what the homolog of the flower is in gymnosperms, we have no particular expectation of the early expression pattern in gymnosperms.
AGL6-like Genes Are Markers for the Palea in Grasses
The lemma and palea are organs unique to the grass floret. The palea is located outside the lodicules on the floral axis, whereas the lemma appears to be borne on the rachilla and subtends the other organs of the floret. The palea is generally morphologically distinct from the lemma and in many grasses is two keeled, although in others it has a single keel like the lemma. There are two opposing interpretations of the homology of the palea: (1) The palea has been interpreted as a prophyll rather than a floral organ, based on its two keels and adaxial position, characteristics that it has in common with a standard monocot prophyll (Linder, 1987; Clayton, 1990; Stapleton, 1997; Soreng and Davis, 1998; Judziewicz et al., 1999; Rudall and Bateman, 2004). However, very few monocots have a prophyll adaxial to the flower; thus, a prophyll in this position would be highly unusual and would represent a novelty in the grasses. (2) The palea has been interpreted as two fused adaxial tepals from the outer perianth whorl (Stebbins, 1956). Most monocots have tepals in this position, and fusion is not uncommon (e.g., in some orchids). Genetic evidence also supports this interpretation; when APETALA3-like genes are mutated in grasses, lodicules are converted to palea/lemma-like organs, as would be expected if lodicules were derived from inner tepals and the palea from the outer ones (Irish, 1998; Schmidt and Ambrose, 1998; Ambrose et al., 2000).
Most genes expressed in the palea are also expressed in the lemma, giving no clue how the two organs might be differentiated (Jeon et al., 2000; Prasad et al., 2001; Malcomber and Kellogg, 2004; Preston and Kellogg, 2006, 2007). However, the CYCLOIDEA (CYC)-like homolog RETARDED PALEA1 (REP1) in rice is a palea-specific gene that plays essential roles in palea initiation by regulating cell proliferation and expansion but does not affect lemma development (Yuan et al., 2009). In the rep1 mutant, palea development is delayed and the palea morphology is also altered. The palea of the rep1 mutant showed asymmetrical differentiation of its cells as well as an increased number of vascular bundles resembling a lemma-like organ (Yuan et al., 2009).
Our data show that AGL6-like genes could be another player in establishing palea development because expression in the developing palea is conserved among all spikelet-bearing grasses. Interestingly, no expression was identified in the bracts of S. angustifolia that have been thought to be homologous to the palea (Sajo et al., 2008). Members of the genus Streptochaeta, which is in a clade sister to all other grasses, do not have a true lemma and palea, nor do they have spikelets. Instead, the flowers are borne at the ends of bracteate inflorescence branches. Thus, true spikelets evolved after the divergence of Streptochaeta from the spikelet-bearing grasses, at the same time that AGL6-like genes develop their novel expression domain in the palea. It would be of interest to assess AGL6-like expression in rep mutants of rice to determine if AGL6-like in the palea is affected by the loss of function of CYC genes.
In addition to AGL6-like genes, LHS1 and FUL1/2 are also expressed in the palea (Malcomber and Kellogg, 2004; Reinheimer et al., 2006; Preston and Kellogg, 2007), and these proteins can interact (Moon et al., 1999). Dimerization of MADS box proteins is facilitated in part by formation of an amphipathic helix in the K-domain, made up of conserved hydrophobic residues (Ma et al., 1991; Shore and Sharrocks, 1995; Davies et al., 1996; Fan et al., 1997). We identified three nonsynonymous substitutions in the K domain coincident with the gain of palea expression of AGL6-like genes, one of which has remained unchanged throughout subsequent grass evolution. It would be interesting to explore whether the changes in codon sites on AGL6-like sequences on the branch given rise to the spikelet-bearing grasses may be involved in changes in dimerization patterns, although it is equally possible that the novel expression pattern of the AGL6-like gene in the palea is the result of cis-regulatory changes. Palea development may thus be under the control of a complex network of protein interactions and regulations that is distinct from that of the lemma.
AGL6-like Genes May Have Been Involved in Inner Perianth Whorl Evolution during Angiosperm Diversification
Expression of AGL6-like genes in the perianth differs among angiosperm taxa (see Supplemental Tables 1 and 2 online). AGL6-like genes are expressed exclusively in the second whorl in species that have flowers with novel structures such as the cap in Vitis vinifera (Boss et al., 2002) or the lip in Oncidium (Hsu et al., 2003); otherwise, AGL6 expression was detected in both whorl 1 and whorl 2 (Ma et al., 1991; Tsuchimoto et al., 2000; Losa et al., 2004; Kim et al., 2005; Chanderbali et al., 2006; Fan et al., 2007). In grasses, previous studies suggest that AGL6 orthologs are expressed in lodicules as well as in the palea (Mena et al., 1995; Moon et al., 1999; Aiguo and Griffin, 2002; Petersen et al., 2004; Thompson et al., 2009). In addition, the loss-of-function zag3 mutant in maize has organ number and developmental defects in the position of the lemma/palea and lodicules (Thompson et al., 2009).
AGL6-like gene expression is conserved in the second whorl in grasses as well as in close relatives. Aaf AGL6 (A. africanus) and Ja AGL6 (J. ascendens) mRNA was detected in the inner tepals. In S. angustifolia, Sa AGL6 mRNA was detected in the bracts representing the inner whorl. In the rest of the grasses examined, AGL6-like mRNA was detected in lodicules. These results, together with functional data and previous expression analysis showing heterogeneous expression patterns in perianth whorls, suggest that perhaps AGL6-like genes may control some aspects of inner perianth formation and evolution.
AGL6-like Expression Is Conserved in Carpel Development and Ovule Formation but Not in Stamens
We found conserved expression of AGL6 orthologs during carpel initiation and in the ovule in the grasses (in particular in the developing integument of the ovule) and also in its close relatives. Similar results have been obtained in zag3 in situ hybridization and, moreover, zag3 mutants produce extra carpels in the ear (Thompson et al., 2009). These data are consistent with many other studies that have detected expression in carpels and ovules in angiosperms (Ma et al., 1991; Mena et al., 1995; Rounsley et al., 1995; Moon et al.,1999; Tsuchimoto et al., 2000; Boss et al., 2002; Pelucchi et al., 2002; Hsu et al., 2003; Losa et al., 2004; Petersen et al., 2004; Chanderbali et al., 2006; Fan et al., 2007), except in Persea and Magnolia where RT-PCR failed to detect AGL6 expression in the carpel (Kim et al., 2005). Curiously, yeast two-hybrid data fail to show any interaction of AGL6-like proteins with B and C class MADS box proteins in rice (Moon et al., 1999), whereas the maize AGL6 homolog, ZAG3, does interact with the AG homolog, ZAG1 (Thompson et al., 2009).
The expression and function of AGL6-like genes in carpel and ovule development can be traced back to the gymnosperms. In situ hybridization in Pinus radiata has shown that the AGL6 homologs Pr MADS2 and Pr MADS3 are expressed strongly in developing ovuliferous scale primordia and in the ovule (Mouradov et al., 1998), and similar data for Gnetum parviflorum show expression of Gp MADS3 in developing ovules and the three envelopes that surround them (Shindo et al., 1999). RNA gel blots of reproductive tissues in Gnetum gnemon show that the AGL6 homologs GGM9 and GGM11 are expressed in the female cones (Winter et al., 1999). This expression pattern is consistent with the homology between the megasporangium in gymnosperms and angiosperms. Moreover, it suggests that AGL6-like gene expression might be a marker that could be used to explore the presumed homology between the single integument of conifers, the inner integument of angiosperms, and the three envelopes surrounding the ovule in gnetophytes. As pointed out by Shindo et al. (1999), the outer two ovular envelopes in gnetophytes are unlikely to be homologous to the angiosperm perianth and are more likely to be homologous to the ovuliferous scale of conifers.
By contrast, expression of AGL6-like genes is not conserved during stamen development. Expression in microsporangia was detected in gymnosperms as well as in several angiosperms but not in all (Ma et al., 1991; Mena et al., 1995; Rounsley et al., 1995; Tandre et al., 1995; Liu and Podila, 1997; Mouradov et al., 1998; Moon et al.,1999; Winter et al., 1999; Tsuchimoto et al., 2000; Boss et al., 2002; Pelucchi et al., 2002; Becker et al., 2003; Hsu et al., 2003; Losa et al., 2004; Petersen et al., 2004; Chanderbali et al., 2006; Fan et al., 2007). Our in situ hybridizations showed that AGL6-like genes are expressed in stamens only in J. ascendens, the grass S. angustifolia in Anomochlooideae, and in Ehrhartoideae. Interestingly, all these species have more than three stamens. When stamen expression occurs at all, it appears strongly during early organ initiation as well as later during anther and possibly tapetum development. In the zag3 mutant of maize, extra stamens, as well as fused or transformed stamens, are formed (Thompson et al., 2009). zag3 expression in stamens was not detected, suggesting that all abnormalities in stamen formation may be due to floral meristem defects.
Divergent Expression Patterns of Rice MADS6 and MADS17
In rice, MADS6 and MADS17 have different expression patterns. Both genes are expressed in the floral meristem and are later expressed in lodicules and stamens; we hypothesize that they are functionally redundant in these tissues. These expression domains are shared with the outgroups of the grasses and are presumed to be ancestral. MADS6 has also retained the ancestral expression pattern (and function; cf. Thompson et al., 2009) in the gynoecium, whereas MADS17 has lost this expression domain and function. In addition, MADS6 has acquired a new expression domain, in the palea. Such changes in expression domains are common among duplicated genes (for examples, see Conant and Wolfe, 2008) and reflect complex patterns of mutation and selection following the duplication event.
We hypothesize that MADS17 may have different interacting partners than MADS6 because of the substantial differences in the C terminus. The C terminus of MADS box proteins is known to be essential for protein–protein interaction and transcriptional activation (Egea-Cortines et al., 1999; Honma and Goto, 2001). In several cases, C-terminal evolution has been linked to changes in gene interactions and consequently in gene function (Vandenbussche et al., 2003).
In summary, our comparative data show that the AGL6-like genes have multiple expression patterns that have originated at different evolutionary times; the fact that these expression domains originated millions of years apart suggests (but does not prove) that they each represent distinct developmental roles for the proteins. Gene expression during carpel development is likely as old as the seed plants themselves, whereas the expression in meristems and apparent functional role in meristem determinacy may be angiosperm specific. The proteins are presumably deployed in floral organ identity in different ways and at different times in angiosperm evolution, as indicated by variable expression patterns in stamens and outer and inner perianth whorls. Deployment of AGL6-like in the inner perianth whorl is shared among the grasses and their immediate relatives, whereas only the grasses express AGL6-like in the outer whorl. In the latter case, the novel gene expression pattern correlates with a highly modified and novel floral organ, the grass palea.
METHODS
Sequencing and Phylogenetic Analyses
Selection of species was based on phylogenetic position and variation in spikelet and flower morphology. Plants were grown in the greenhouse at the University of Missouri–St. Louis or the Missouri Botanical Garden.
A pool of MADS box cDNAs was produced following methods of Malcomber and Kellogg (2004). AGL6 homologs were PCR amplified from pooled MADS box cDNAs using a degenerate forward primer (see Supplemental Table 4 online), designed to bind at the 5′ end of the I box, and a degenerate reverse primer at the 3′ end of the C-terminal domain (AGL6_POAC_805R, 5′-TTCATGCTGGGRTGGGTT-3′). PCR products were gel purified and subcloned before sequencing. Gene names of new AGL6-like sequences begin with the initials of the genus and species. Of the sequences included in these analyses, 36 are new and 16 were obtained from GenBank; the sequence of Elegia sp was generated in the lab of S. Malcomber. Accession numbers for the sequences used are provided in Supplemental Table 3 online.
Sequences were aligned using MUSCLE (Edgar, 2004a, 2004b), and the alignment corrected manually in MacClade 4 (Maddison and Maddison, 2003). Phylogenetic trees were generated with maximum parsimony and maximum likelihood algorithms, implemented in PAUP* version 4.0b10 (Swofford, 2001); tree support was assessed with bootstrap analysis. Bayesian phylogenetic estimates were run for 5 million generations in MrBayes 3.2 (Huelsenbeck and Ronquist, 2001). Maximum likelihood and Bayesian analyses implemented the GTR+I+G model of evolution, based on results from MrModelTest 2.2 (Nylander, 2004). To identify amino acid sites distinguishing the major clades, codon transitions were mapped onto the AGL6-like gene tree (Maddison and Maddison, 2003).
Os MADS17-like Sequences
We were unable to isolate MADS17 sequences in any genus other than Oryza sativa by the PCR strategy mentioned above. To test whether we had simply missed the MADS17-like sequences, we performed RT-PCR in Joinvillea ascendens and 21 grass species using a wide variety of primers (see Supplemental Table 4 online); PCR products were cloned and sequenced, but only MADS6-like sequences were retrieved outside Oryza. We also searched for MADS17-like sequences in the nucleotide collection and genomic survey sequence databases at NCBI (http://blast.ncbi.nlm.nih.gov), the nonredundant protein sequence database at NCBI (http://blast.ncbi.nlm.nih.gov), Gramene (http://www.gramene.org), the Plant Genome DataBase (http://www.plantgdb.org/), the rice genome at The Institute for Genomic Research (http://www.tigr.org/), the maize genome sequence (http://maizesequence.org), the Brachypodium genome (http://www.brachypodium.org/), and the sorghum genome (http://www.phytozome.net/cgi-bin/gbrowse/sorghum/). Searches excluded the MADS box to avoid retrieving all members of the gene family. For greater specificity, additional searches used only the C terminus.
To investigate syntenic regions in the rice and maize (Zea mays) genomes, we used the coordinates of MADS17 (Chr4:29082088..29102087) in the rice genome displayed at Gramene (http://www.gramene.org) and the maize genome displayed in (1) Gramene, (2) the maize genome sequence browser (http://maizesequence.org), and (3) Synteny (http://synteny.cnr.berkeley.edu/CoGe/.Read:Lyons).
Statistical Tests of Sequences
To test whether we could statistically reject alternative hypotheses for the timing of the AGL6/MADS6/MADS17 duplication and the zag3/zag5 duplication, we performed a Shimodaira-Hasegawa test (Goldman et al., 2000). We also tested whether there was any evidence for positive selection on different parts of the protein or on particular branches of the gene tree, using the program codeml from the PAML package, version 4 (Yang, 2007). To test whether MADS17/MADS6 as well as zag5/zag3 duplicates show differences in evolutionary rates, we used the relative rates test proposed by Tajima (1993) as implemented in MEGA4 (Tamura et al., 2007). Details of these tests and tabulations of the results are in the Supplemental Statistical Test online.
Characterization of Expression
For Q-PCR, RNA was extracted from roots, stems, leaves, and inflorescences tissues using RNAwiz solution (Ambion) according to the manufacturer's instructions. About 1 μg total RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). Q-PCR was done on the MyIQ single-color real-time detection system (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) and specific primers designed to bind the 3′ untranslated region (UTR; see Supplemental Table 4 online). Data presented are the summary of three biological replicates and three PCR replicates. Data were normalized against ACTIN expression.
We also performed RT-PCR using specific primers designed to bind the 5′ C terminus of the coding region and the 3′ UTR (see Supplemental Table 4 online), except for Phalaris canariensis, Eragrostis tef, and Megathyrsus maximus, in which primers bind the 5′ end of the I box and the 3′ end of the C terminus. The RT reaction was performed using Superscript One-Step RT-PCR with Platinum Taq (Invitrogen) as per the manufacturer's instructions. ACTIN was used as a positive control.
For in situ hybridization, young inflorescences in different stages of development were dissected, fixed, and dehydrated as described by Malcomber and Kellogg (2004). AGL6-specific cDNA probes were generated using a nested PCR approach (Preston and Kellogg, 2007). The first round of PCR used specific primers complementary to the 5′ end of the C terminus and the 3′ end of the 3′ UTR using a polyT-adaptor. The second round used primers complementary to the 5′ end of the C terminus and the 3′ of the C terminus or the 3′ end of the 3′ UTR (see Supplemental Table 4 online). Probes were 300 to 410 bp long. Probe hydrolysis followed Jackson (1991). Probe hybridization, posthybridization washing, immunolocalization, and colorimetric detection were performed as described by Jackson et al. (1994) and Malcomber and Kellogg (2004). Photos were taken after 1 d of staining and were imported into Photoshop and adjusted for contrast, brightness, and color balance. Hybridizations with antisense probes were repeated in separate experiments on material from at least two different plants for Lithachne humilis, Hordeum vulgare, Triticum monoccocum, and Lolium temulentum. For J. ascendens, Streptochaeta angustifolia, O. sativa, Leersia sp, Eleusine indica, Setaria italica, and Sorghum bicolor, the experiment was repeated three or four times using different probes, different hydrolysis times, and on independently fixed and embedded material. No signal was detected in any of the control hybridizations with sense probes, except for occasional faint, nonspecific background.
DNA Gel Blot Hybridization
Total DNA was extracted from 300 mg of leaf tissue using a modified CTAB protocol (Doyle and Doyle, 1987). Approximately 10 μg of total DNA was digested with the enzyme BamHI, EcoRI, or HindIII, and digested DNA was run on a 0.8% agarose gel overnight. Digested DNA was blotted and hybridized at moderate stringency following the protocol of Laurie et al. (1993).
Maximum Parsimony Ancestral State Reconstructions
Ancestral expression patterns were estimated using maximum parsimony ancestral state reconstruction in MacClade 4 (Maddison and Maddison, 2003). We supplemented our own data with published sequence and expression data for Hyacinthus orientalis (Hyacinthaceae) (Fan et al., 2007), Asparagus officinalis (Asparagaceae) (Losa et al., 2004), and Z. mays (Thompson et al., 2009).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ496625 to GQ496659 as presented in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Differences in Amino Acid Composition between the Paralogous Copies MADS17 and MADS6.
Supplemental Figure 2. Alignment of the Predicted Amino Acid Sequences of AGL6-like Genes Included in This Study.
Supplemental Figure 3. AGL6-like RT-PCR in Agapanthus africanus and Disparate Grasses.
Supplemental Figure 4. AGL6-like Expression in Floral Meristems of Streptochaeta angustifolia (Anomochlooideae) and Triticum monoccocum (Pooideae).
Supplemental Table 1. Expression Patterns Recovered for AGL6-like Homologs in Gymnosperm Species.
Supplemental Table 2. AGL6-like Gene Expression Patterns Recovered in Eudicots and Monocots.
Supplemental Table 3. AGL6 Homologs Included in This Study.
Supplemental Table 4. Primers Used for Isolation of Os MADS6-Like Sequences and Os MADS17-Like Sequences, and Expression Studies in Poaceae and Other Monocots.
Supplemental Data Set 1. Text File of Alignment Corresponding to Figure 1 and Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank the Missouri Botanical Garden and the USDA for access to plant materials; Kenneth Olsen, Briana Gross, Pu Huang, and Barbara Schaal for access to Oryza material; Sarah Youngstrom for help with subcloning; Jinshun Zhong for help with RNA extractions; Simon Malcomber for assistance with in situ hybridizations; Xuemin (Sam) Wang, Liang Guo, and Geliang Wang for assistance with real time Q-PCR; and Simon Malcomber and Felipe Zapata for assistance and helpful discussions. We are also indebted to Simon Malcomber and Ashley Christensen for the unpublished sequence of Elegia and Sarah Hake, Beth Thompson, and members of the Kellogg lab for critical reading of the manuscript. This work was funded by National Science Foundation Grant DBI-0604923 to E.A.K.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Renata Reinheimer (reinheimerr@umsl.edu) and Elizabeth A. Kellogg (kellogge@umsl.edu).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Aiguo, L., and Griffin, J.D. (2002). Cloning and analysis of a MADS-box gene expressed in Poa annua md-2 apices during floral meristem formation. Int. J. Plant Sci. 163 43–50. [Google Scholar]
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. [DOI] [PubMed] [Google Scholar]
- Arora, R.P., Agarwal, R., Swatismita, S., Ashok, K.S., Vijay, P.K., Akhilesh, K.T., and Sanjay, K. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8 242–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, A., Saedler, H., and Theissen, G. (2003). Distinct MADS-box gene expression patterns in the reproductive cones of the gymnosperm Gnetum gnemon. Dev. Genes Evol. 213 567–572. [DOI] [PubMed] [Google Scholar]
- Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29 464–489. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Sensi, E., Hua, C., Davies, C., and Thomas, M.R. (2002). Cloning and characterization of grapevine (Vitis vinifera L.). MADS-box genes expressed during inflorescence and berry development. Plant Sci. 162 887–895. [Google Scholar]
- Chanderbali, A.S., Kim, S., Buzgo, M., Zheng, Z., Oppenheimer, D.G., Soltis, D.E., and Soltis, P.S. (2006). Genetic footprints of stamen ancestors guide perianth evolution in Persea (Lauraceae). Int. J. Plant Sci. 167 1075–1089. [Google Scholar]
- Clayton, W.D. (1990). The spikelet. In Reproductive Versatility in the Grasses, G.P. Chapman, ed (Cambridge, UK: Cambridge University Press.), pp. 32–51.
- Clifford, H.T. (1987). Spikelet and floral morphology. In Grass Systematics and Evolution, T.R. Soderstrom, K.W. Hilu, C.S. Campbell, and M.E. Barkworth, eds (Washington, DC: Smithsonian Institute), pp. 21–30.
- Conant, G.C., and Wolfe, K.H. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 9 938–950. [DOI] [PubMed] [Google Scholar]
- Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996). Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Doust, A.N., and Kellogg, E.A. (2002). Inflorescence diversification in the panicoid “bristle grass” clade (Paniceae, Poaceae): evidence from molecular phylogenies and developmental morphology. Am. J. Bot. 89 1203–1222. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, J.L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem. Bull. 19 11–15. [Google Scholar]
- Edgar, R.C. (2004. a). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004. b). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, H.Y., Hu, Y., Tudor, M., and Ma, H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12 999–1010. [DOI] [PubMed] [Google Scholar]
- Fan, J.H., Li, W.Q., Dong, X.C., Guo, W., and Shu, H.R. (2007). Ectopic expression of a hyacinth AGL6 homolog caused earlier flowering and homeotic conversion in Arabidopsis. Sci. China C Life Sci. 50 676–689. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N., Anderson, J.P., and Rodrigo, A.G. (2000). Likelihood based tests of topology in phylogenetics. Syst. Biol. 49 652–670. [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001). Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 88 373–457. [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Hsu, H.F., Huang, C.H., Chou, L.T., and Yang, C.H. (2003). Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 44 783–794. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., and Ronquist, F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Irish, E.E. (1998). Grass spikelets: A thorny problem. Bioessays 20 789–793. [Google Scholar]
- Jackson, D. (1991). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M. McPherson, eds. (London: Oxford University Press), pp. 163–174.
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Jeon, J.S., Lee, S., Jung, K.H., Yang, W.S., Yi, G.H., Oh, B.G., and An, G. (2000). Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol. Breed. 6 581–592. [Google Scholar]
- Judziewicz, E.J., Clark, L.G., Londoño, X., and Stern, M.J. (1999). American Bamboos (Washington, DC: Smithsonian Institution Press).
- Kellogg, E.A. (2006). Progress and challenges in studies of the evolution of development. J. Exp. Biol. 57 3505–3516. [DOI] [PubMed] [Google Scholar]
- Kellogg, E.A., and Bennetzen, J.L. (2004). The evolution of nuclear genome structure in seed plants. Am. J. Bot. 91 1709–1725. [DOI] [PubMed] [Google Scholar]
- Kim, S., Koh, J., Yoo, M.J., Kong, H., Hu, Y., Ma, H., Soltis, P.S., and Soltis, D.E. (2005). Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. 43 724–744. [DOI] [PubMed] [Google Scholar]
- Laurie, D.A., Pratchett, N., Devos, K.M., Leitch, I.J., and Gale, M.D. (1993). The distribution of RFLP markers on chromosome 2 (2H) of barley in relation to the physical and genetic location of 5S rDNA. Theor. Appl. Genet. 87 177–183. [DOI] [PubMed] [Google Scholar]
- Linder, H.P. (1987). The evolutionary history of the Poales/Restionales – A hypothesis. Kew Bull. 42 297–318. [Google Scholar]
- Litt, A., and Irish, V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.J., and Podila, G.K. (1997). Characterization of a MADS-box gene (accession no. Y09611) from immature female cones of red pine (PGR 97-032). Plant Physiol. 117 55–62. [Google Scholar]
- Losa, A., Caporali, E., Spada, A., Martinelli, S., and Marziani, G. (2004). AOM3 and AOM4: Two MADS box genes expressed in reproductive structures of Asparagus officinalis. Sex. Plant Reprod. 16 215–221. [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5 484–495. [DOI] [PubMed] [Google Scholar]
- Maddison, D.R., and Maddison, W.P. (2003). MacClade: Analysis of Phylogeny and Character Evolution. (Sunderland, MA: Sinauer Associates).
- Malcomber, S.T., and Kellogg, E.A. (2004). Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16 1692–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena, M., Mandel, M.A., Lerner, D.R., Yanofsky, M.F., and Schmidt, R.J. (1995). A characterization of the MADS-box gene family in maize. Plant J. 8 845–854. [DOI] [PubMed] [Google Scholar]
- Moon, Y.H., Kang, H.G., Jung, J.Y., Joen, J.S., Sung, S.K., and An, G. (1999). Determination of the motif responsible for interaction between the rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiol. 120 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., Glassick, T.V., Hamdorf, B.A., Murphy, L.C., Marla, S.S., Yang, Y., and Teasdale, R.D. (1998). Family of MADS-box genes expressed early in male and female reproductive structures of Monterey Pine. Plant Physiol. 117 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M., and Yanofsky, M.F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2 186–195. [DOI] [PubMed] [Google Scholar]
- Nylander, J.A.A. (2004). MrModeltest v2. (Uppsala, Sweden: Evolutionary Biology Centre).
- Pelucchi, N., Fornara, F., Favalli, C., Masiero, S., Lago, C., Pe, M.E., Colombo, L., and Kater, M.M. (2002). Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 15 113–122. [Google Scholar]
- Petersen, K., Didion, T., Andersen, C.H., and Nielsen, K.K. (2004). MADS-box genes from perennial ryegrass differentially expressed during transition from vegetative to reproductive growth. J. Plant Physiol. 161 439–447. [DOI] [PubMed] [Google Scholar]
- Prasad, K., Sriram, P., Kumar, C.S., Kushalappa, K., and Vijayraghavan, U. (2001). Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 211 281–290. [DOI] [PubMed] [Google Scholar]
- Preston, J.C., and Kellogg, E.A. (2006). Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, J.C., and Kellogg, E.A. (2007). Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses. Plant J. 52 69–81. [DOI] [PubMed] [Google Scholar]
- Purugganan, M.D., Rounsley, S.D., Schmidt, R.J., and Yanofsky, M.F. (1995). Molecular evolution of flower development: Diversification of the plant MADS-box regulatory gene family. Genetics 140 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheimer, R., Malcomber, S.T., and Kellogg, E.A. (2006). Evidence for distinct roles of the SEPALLATA gene LEAFY HULL STERILE1 in Eleusine indica and Megathyrsus maximus (Poaceae). Evol. Dev. 8 293–303. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Krizek, B.A., and Meyerowitz, E.M. (1996. a). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., Wang, M., and Meyerowitz, E.M. (1996. b). DNA binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall, R.J., and Bateman, R.M. (2004). Evolution of zygomorphy in monocot flowers: Iterative patterns and developmental constraints. New Phytol. 162 25–44. [Google Scholar]
- Sajo, M.G., Longhi-Wagner, H.M., and Rudall, P.J. (2008). Reproductive morphology of the early-divergent grass Streptochaeta and its bearing on the homologies of the grass spikelet. Plant Syst. Evol. 275 245–255. [Google Scholar]
- Schmidt, R.J., and Ambrose, B.A. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1 60–67. [DOI] [PubMed] [Google Scholar]
- Shindo, S., Ito, M., Ueda, K., Kato, M., and Hasebe, M. (1999). Characterization of MADS genes in the gymnosperm Gnetum parvifolium and its implication on the evolution of reproductive organs in seed plants. Evol. Dev. 1 180–190. [DOI] [PubMed] [Google Scholar]
- Shore, P., and Sharrocks, A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229 1–13. [DOI] [PubMed] [Google Scholar]
- Stapleton, C.M.A. (1997). The morphology of woody bamboos. In The Bamboos, G.P. Chapman, ed (London: Academy Press), pp. 251–267.
- Stebbins, G.L. (1956). Cytogenetics and evolution of the grass family. Am. J. Bot. 43 890–905. [Google Scholar]
- Soreng, R.J., and Davis, J.I. (1998). Phylogenetic and character evolution in the grass family (Poaceae): Simultaneous analysis of morphological and chloroplast DNA restriction site character sets. Bot. Rev. 64 1–85. [Google Scholar]
- Swofford, D.L. (2001). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). (Sunderland, MA: Sinauer Associates).
- Tajima, F. (1993). Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tandre, K.A., Sundas, V.A., and Engström, A.P. (1995). Conifer homologues to genes that control floral development in angiosperms. Plant Mol. Biol. 27 69–87. [DOI] [PubMed] [Google Scholar]
- Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J.T., Münster, T., Winter, K.U., and Saedler, H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42 115–149. [PubMed] [Google Scholar]
- Thompson, B.E., Bartling, L., Whipple, C., Hall, D.H., Sakai, H., Schmidt, R., and Hake, S. (2009). bearded-ear encodes a MADS-box transcription factor critical for maize floral development. Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- Tsuchimoto, S., Mayama, T., van der Krol, A., and Ohtsubo, E. (2000). The whorl-specific action of a petunia class B floral homeotic gene. Genes Cells 5 89–99. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, M., Theissen, G., Van de Peer, Y., and Gerats, T. (2003). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31 4401–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, K.U., Becker, A., Münster, T., Kim, J.T., Saedler, H., and Theissen, G. (1999). MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc. Natl. Acad. Sci. USA 96 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple, C.J., Ciceri, P., Padilla, C.M., Ambrose, B.A., Bandong, S.L., and Schmidt, R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091. [DOI] [PubMed] [Google Scholar]
- Whipple, C.J., and Schmidt, R.J. (2006). Genetics of grass flower development. Adv. Bot. Res. 44 385–424. [Google Scholar]
- Whipple, C.J., Zanis, M., Kellogg, E.A., and Schmidt, R.J. (2007). Conservation of B class MADS-box gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. USA 104 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yuan, Z., Gao, S., Xue, D.W., Luo, D., Li, L.T., Ding, S.Y., Yao, X., Wilson, Z.A., Qian, Q., and Zhang, D.B. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn, L.M., Kong, H., Leebens-Mack, J.H., Kim, S., Soltis, P.S., Landherr, L.L., Soltis, D.E., Depamphilis, C.W., and Ma, H. (2005). The evolution of the SEPALLATA subfamily of MADS-Box genes: A preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.