Abstract
Inherited predisposition to disease is often linked to reduced activity of a disease associated gene product. Thus, quantitation of the influence of inherited variants on gene function can potentially be used to predict the disease relevance of these variants. While many disease genes have been extensively characterized at the functional level, few assays based on functional properties of the encoded proteins have been established for the purpose of predicting the contribution of rare inherited variants to disease. Much of the difficulty in establishing predictive functional assays stems from the technical complexity of the assays. However, perhaps the most challenging aspect of functional assay development for clinical testing purposes is the absolute requirement for validation of the sensitivity and specificity of the assays and the determination of positive predictive and negative predictive values of the assays relative to a “gold standard” measure of disease predisposition. In this commentary we provide examples of some of the functional assays under development for several cancer predisposition genes [BRCA1, BRCA2, CDKN2A, and mismatch repair (MMR) genes MLH1, MSH2, MSH6, and PMS2] and present a detailed review of the issues associated with functional assay development. We conclude that validation is paramount for all assays that will be used for clinical interpretation of inherited variants of any gene, but note that in certain circumstances information derived from incompletely validated assays may be valuable for classification of variants for clinical purposes when used to supplement data derived from other sources.
Keywords: unclassified variant, functional assay, BRCA1, BRCA2, MMR, MLH1, MSH2, MSH6, PMS2, CDKN2A
The problem of unclassified variants
Genetic testing for cancer predisposition has become widespread, and is particularly sought after by individuals with a family history of cancer primarily because of the potential for improved risk assessment. Genetic testing of predisposition genes can establish the presence of cancer predisposing alterations or the absence of any specific changes. However, the finding of a variant of uncertain significance (often called VUS), is another possible result that can complicate rather than improve the risk assessment process. In this case, a change in the nucleotide sequence is found for which there is not enough information to decide whether it affects the function of the gene product and by consequence influences cancer risk. Especially in the case of genes that strongly affect risk, such non-informative results can be a source of anxiety to individuals and their offspring because they will not be able to use the information from genetic testing to modify behavior or lifestyle, or to make important clinical decisions that may, in many cases, involve prophylactic surgery. In addition, all first-degree relatives including non-carriers are considered at risk as long as the contribution of the variant to disease cannot be assessed, resulting in frequently unnecessary lifelong anxiety and prophylactic screening.
In the last few years, this problem has received the attention of a large number of investigators from different disciplines, as illustrated in this special issue. In this article we focus on the use of laboratory tests (functional assays) that assess the impact of genetic variants on the activity of the gene product. The principle behind the use of functional assays is that because cancer predisposition seems to be due to the inheritance of alterations that result in loss of function of tumor suppressor genes, detection of a decrease in activity of a tumor suppressor likely represents increased cancer predisposition. Thus, functional assays that can establish and possibly quantify alterations in the activity of a tumor suppressor variant can potentially be used to predict whether the variant predisposes to disease or alternatively has no significant influence on cancer risk.
In order to provide some insight into the use of functional assays for evaluation of cancer predisposition we review the basis of specific functional assays for BRCA1 (NM_007294.2) and BRCA2 (NM_000059.3), mismatch repair (MMR) genes and CDKN2A (NM_058197.3) and address their strengths and limitations. This is not a complete overview of all potential functional assays that have relevance to cancer predisposition testing. However, because the assays described here have been studied in some detail they are particularly useful for illustrating common problems associated with performance of functional assays and interpretation of results. In summary, we aim to a) provide a guide that can be used by the non-specialist to judge the validity and reliability of certain assays; b) call attention to common misconceptions about functional assays; and c) foster the development of better assays for a wide variety of genes involved in cancer predisposition.
BRCA1 and BRCA2
BRCA1 and BRCA2 variants and cancer
Mutations that truncate and inactivate BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) and predispose to breast and ovarian cancer have been identified throughout the genes. The lifetime risk of breast cancer for Caucasians with BRCA1 and BRCA2 truncating mutations is between 55% and 85% and the lifetime risk of ovarian cancer is between 20% and 40% [Antoniou, et al., 2005].
While thousands of BRCA1 and BRCA2 truncating mutations have been associated with increased risk of cancer in carriers, the contribution of other BRCA1 and BRCA2 variants to cancer risk remains largely undefined. VUS are mainly missense mutations but also include a number of intronic variants and in-frame deletions and insertions [Szabo, et al., 2004]. To date over 2,000 unique BRCA1 and BRCA2 missense variants have been identified, a subset of which are listed in the open access, on-line Breast Cancer Information Core (BIC) Database (http://research.nhgri.nih.gov/bic/) that functions as a repository of sequence alterations in BRCA1 and BRCA2. The uncertainty in establishing the disease relevance of BRCA1 and BRCA2 variants is an important obstacle to identifying individuals at risk of cancer and to providing appropriate health care options and counseling.
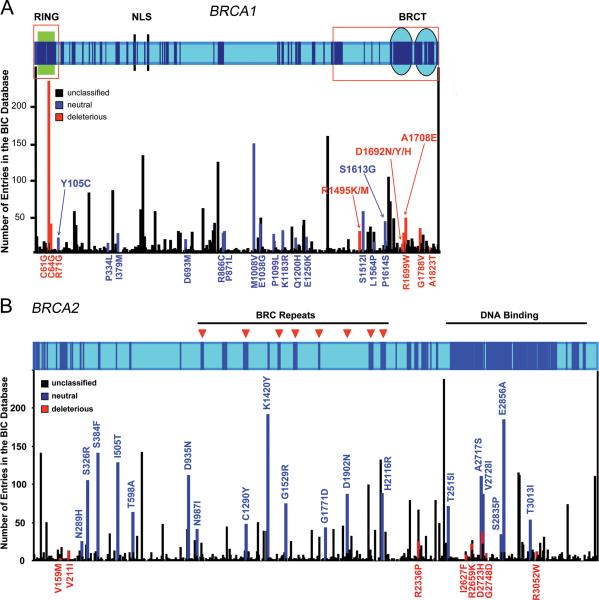
At present, the clinical relevance of relatively few of these variants has been established. These include 133 common variants that were recently classified as neutral/ no clinical significance [Easton, et al., 2007] and a small number of variants located in evolutionarily conserved residues [Tavtigian et al., 2008a] or splicing consensus sites that were classified as deleterious using a statistical genetic combined likelihood model [Easton, et al., 2007; Goldgar et al., 2008], or in vivo splicing/exon skipping analysis [Spurdle et al., 2008]. Classification of other BRCA1 and BRCA2 variants as cancer predisposing or neutral has proven problematic because it is not known whether these subtle changes alter the function of the proteins sufficiently to predispose to cancer and there is insufficient information from family studies of these rare variants to allow classification using the likelihood model. In Figure 1 we illustrate the problem of missense mutations by showing the location and frequency of all missense variants in BRCA1 and BRCA2 listed in the BIC database and by indicating the small number of variants that have been definitively classified as either deleterious or neutral. Because of this problem, functional assays that assess the impact of amino acid changes on BRCA1 and BRCA2 protein function have been proposed as an alternative approach to the classification of the cancer relevance of BRCA1 and BRCA2 variants.
Figure 1.
A. Diagram of human BRCA1 depicting defined domains and motifs (Green box, RING domain; Blue circles, BRCT domains; Black bars, nuclear localization signals). Regions showing conservation above 0.75 in a multiple sequence alignment (from human to puffer fish) are shown as dark blue boxes. Regions analyzed by domain-specific functional assays (ubiquitin ligase for the RING region; transcription assay for the C-terminal region) are depicted as red empty boxes. Graph aligned to the top diagram shows the number of entries in the BIC database for each missense variant recorded to date. Variants are shown as black (unclassified), red (pathogenic), and blue (neutral) bars. All pathogenic and neutral variants as classified in the BIC database are shown. B. Diagram for BRCA2 depicting the BRC repeats (arrows) and DNA binding domain. Regions showing conservation above 0.75 in a multiple sequence alignment (from human to puffer fish) are shown as dark blue boxes. Aligned graph shows position and frequency of all missense mutations in the BIC database. Variants are shown as black (unclassified), red (pathogenic), and blue (neutral) bars. All pathogenic and neutral variants as classified in the BIC database are shown.
BRCA1 and BRCA2 function
Functional assays that can measure the influence of variants on protein activity can only be established following development of a detailed understanding of how a protein functions in a cell and contributes to cancer predisposition when mutated. Here we outline the features of BRCA1 and BRCA2 that have been utilized for development of functional assays.
BRCA1 is composed of 22 coding exons, and encodes a large protein of 1863 amino acids. Disruption of BRCA1 has been associated with centrosome amplification, aneuploidy, chromosomal rearrangements and loss of maintenance of DNA integrity. These effects reflect the role of BRCA1 in fundamental cellular processes including transcriptional regulation, DNA repair and regulation of centrosome function and cell cycle checkpoint activity. The BRCA1 C-terminal BRCT domains activate transcription in yeast and mammalian cells [Monteiro, et al., 1996] and bind to several transcription factors including p53, c-myc and CtIP [Ouchi, et al., 1998; Pao, et al., 2000; Wang, et al., 1998; Yu, et al., 1998]. BRCA1 mutant cells from humans and mice are deficient in homologous recombination repair of DNA double strand breaks [Moynahan, et al., 1999]. BRCA1 also functions as an E3 ubiquitin ligase through the interaction of the N-terminal RING domain with BARD1 [Baer and Ludwig, 2002], and is associated with mono-ubiquitination of γ-tubulin in the centrosome [Starita, et al., 2004]. Similarly, the BRCA2 gene is comprised of 10.5 kb of coding sequence in 26 exons and encodes a very large 384 kD protein. Like BRCA1, BRCA2 has a role in the fundamental cellular processes of DNA repair, centrosome regulation and maintenance of chromosomal stability. BRCA2 interacts with the Rad51 DNA recombination repair protein and binds to single and double stranded DNA through a large C-terminal DNA binding domain. Through these interactions BRCA2 is thought to recognize sites of double strand DNA breaks and mediate homologous recombination repair [Davies, et al., 2001; Yang, et al., 2002]. BRCA2 has been identified as the FANC-D1 Fanconi anemia gene [Howlett, et al., 2002] and BRCA2 deficient or mutant cell lines are highly sensitive to DNA damaging and crosslinking agents that induce double strand DNA breaks [Kraakmanvan der Zwet, et al., 2002; Moynahan, et al., 2001]. BRCA2 controls mitotic checkpoint activity [Tutt, et al., 1999; Yu, et al., 2000], is also localized in the centrosome where it maintains normal centrosome number and function and is involved in the final stages of cell division (cytokinesis) through undefined mechanisms [Daniels, et al., 2004]. Disruption of these functions of BRCA2 by truncating mutations likely accounts for the aneuploidy and centrosome amplification observed in BRCA2 deficient or mutant cells.
Functional assays
Since the exact biochemical functions of the multifunctional BRCA1 and BRCA2 proteins are not fully defined, it is not clear which in vitro assays measure functions important in carcinogenesis and which are useful in defining whether a variant is pathogenic or not. It has been necessary to develop multiple assays that evaluate specific functions in cells that are known to be dependent on BRCA1 or BRCA2. In some cases, the resulting assays may be limited to specific domains of these proteins. Candidate functional assays must also be fully evaluated for the ability to discriminate between a series of well-defined positive and negative controls [Phelan, et al., 2005; Vallon-Christersson, et al., 2001; Wu, et al., 2005]. Currently, two assays for BRCA1 (transcription activation and combined BARD1 binding/ubiquitin ligase activity) and two assays for BRCA2 (homologous recombination repair and centrosome amplification) meet these criteria.
Transcription Activation (TA) assay
Early studies of BRCA1 noted that the carboxy-terminal region of BRCA1 functions as a transactivation domain when expressed as a fusion to a heterologous DNA binding domain (DBD) in yeast and mammalian cells [Monteiro, 2000; Monteiro, et al., 1996]. Cancer-associated nonsense, frameshift, and missense mutations, but not benign variants, in this region of BRCA1 were found to impair this transcriptional activity [Hayes, et al., 2000]. This led to the development of a transcription assay based on the notion that it is a reliable monitor of the integrity of the BRCT domain [Williams, et al., 2004] and that it can reliably predict the functional effects of mutations.
The assay can be performed in yeast and mammalian cells using expression vector constructs (pLex9 and pcDNA3, respectively) encoding a DNA binding domain (DBD) fusion to residues 1396−1863 of BRCA1. In yeast, the pLex9-BRCA1 construct is transformed into cells with a lacZ reporter construct driven by a lexA binding sites. Transcriptional activity is measured by a quantitative liquid ß-galactosidase assay [Vallon-Christersson, et al., 2001]. In human cells (HEK 293T), the variants in a GAL4 DBD fusion are co-transfected with a luciferase reporter driven by five GAL4 binding sites (pG5luc) [Monteiro, et al., 1997]. An internal control of Renilla luciferase normalizes the luciferase activity from the reporter. Protein levels measured by western blot allow adjustment for transfection efficiency and expression levels. In this assay wild type BRCA1 and the S1613G neutral variant can serve as positive controls and the M1775R, Y1853X and A1708E cancer-associated mutations can serve as negative controls in both yeast and mammalian cells [Hayes, et al., 2000] [Vallon-Christersson, et al., 2001].
This assay has been used to evaluate over 50 different VUS from the BRCA1 BRCT domains. Only one example of discordance between the yeast and mammalian assay results has been observed. In this case, variant R1699W was shown to be a temperature-sensitive mutant of BRCA1 [Worley, et al., 2002]. Efforts to validate the assay have focused on the correlation between the assay results and the results from the likelihood model that is based on family data. While this approach has been limited by the relatively small number of variants from the BRCA1 BRCT domains that have been classified by the likelihood model, a strong correlation appears to exist [Carvalho, et al., 2007]. Specifically, the TA assay correctly classified 24 of 24 known deleterious and neutral variants giving sensitivity of between 69% and 100% and specificity of between 77% and 100%. Further studies aimed at correlation with sequence conservation algorithms [Tavtigian et al., 2008b], and overall posterior probabilities of cancer causality derived from the combination of sequence analysis, pathology and family data [Goldgar et al., 2008] are needed to better define the sensitivity, specificity of the assay and its predictive value for all C-terminal BRCA1 variants. Overall, while this assay may be an excellent method for evaluating the effects of variants on BRCA1 function, it is presently limited to the C-terminus of BRCA1 (aa 1396−1863) and it is not likely that other regions of the protein can be interrogated using this assay.
BARD1 binding/Ubiquitin ligase activity
BRCA1 displays E3 ubiquitin ligase activity mediated by an interaction with BARD1 through the N-terminal RING-finger domain [Baer and Ludwig, 2002; Brzovic, et al., 2003; Chen, et al., 2002; Hashizume, et al., 2001; Lorick, et al., 1999; Ruffner, et al., 2001]. Cancer-predisposing mutations within the RING domain of BRCA1 have been correlated with loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity [Ruffner, et al., 2001]. Based on these observations, a functional assay that measures the influence of BRCA1 N-terminal variants on the interaction between BRCA1 and BARD1 and between BRCA1 and UbcH5a (E2 ubiquitin conjugating enzyme) was proposed [Morris, et al., 2006]. Initially, yeast 2-hybrid (Y2H) screenings of mutagenized BRCA1 N-terminal region using BARD1 or UbcH5a as baits were used to identify changes in residues found as germline changes in patients that resulted in loss of interaction. Subsequently, the influence of all variants in the first 100 amino acids of BRCA1 on the interaction with BARD1 and UbcH5a were tested. The interaction with UbcH5a was sensitive to variants throughout N-terminus of BRCA1, whereas the BARD1 interaction was sensitive to a subset of BRCA1 variants that also inhibited UbcH5a binding. Importantly, variants classified as deleterious disrupted the interaction with BARD1 and UbcH5a and also showed loss of ubiquitin ligase activity [Morris, et al., 2006]. These positive controls established the potential utility of this assay for classification of BRCA1 variants. However, further studies aimed at defining the correlation with the family based likelihood model and posterior probability of causality are needed in order to establish the sensitivity and specificity of the assay. Similarly to the TA assay, the BARD1 binding/ubiquitin ligase assay is domain-specific and may not be used to interrogate variants in other regions of BRCA1.
Homologous recombination repair assay
The ability of BRCA2 to associate with Rad51 and influence homologous recombination repair of DNA double strand breaks can be measured using a green fluorescent protein (GFP)-dependent homology directed repair assay [Moynahan, et al., 2001]. This assay involves generation of a stable cell line with a single copy of a repair reporter plasmid (DR-GFP) composed of two differentially mutated GFP genes, one of which contains a unique I-SceI restriction endonuclease recognition site. A double strand break is introduced into the plasmid at the I-Sce1 site and the break is repaired by homologous recombination involving the downstream inactivated GFP gene as a template. This results in GFP expression that can be measured by flow cytometry. This homologous recombination assay has been established in several DNA repair deficient cell lines including the BRCA2-deficient V-C8 cell line [Sakai, et al., 2008; Wu, et al., 2005; Xia, et al., 2006]. V-C8-DR-GFP cells can be either transiently or stably transfected with the I-Sce1 construct and either wildtype or mutant forms of epitope-tagged full-length BRCA2 and the number of GFP positive cells enumerated. In the case of transient transfection, the efficiency of transfection can be defined by immunofluorescence analysis of epitope-tagged BRCA2 constructs and used to normalize the results. While it is possible to conduct this assay using partial BRCA2 proteins, studies performed with full-length BRCA2 are more credible since this approach accounts for the influence of BRCA2 structure and the other domains of this multifunctional protein on homologous recombination repair activity. In this assay wild type BRCA2 and the Y42C neutral variant can serve as positive controls and the 6174delT and D2723H cancer-associated mutations can serve as negative controls.
Approximately 30 BRCA2 variants have been studied using this approach, including 25 from the DNA binding domain [Farrugia, et al., 2008; Wu, et al., 2005]. Efforts to validate the assay have focused on the correlation between the assay results and the results from the family based likelihood model. As with the TA assay for BRCA1 this approach has been limited by the relatively small number of variants that have been classified by the likelihood model. However, a very strong correlation appears to exist [Farrugia, et al., 2008] suggesting high sensitivity, specificity, and predictive value for this assay. Further studies aimed at establishing the correlation with sequence conservation measures [Tavtigian et al., 2008b], and overall posterior probabilities of cancer causality derived from the combination of sequence analysis and family data [Goldgar et al., 2008] are needed to better define the predictive value of the assay. Similarly to the BRCA1 assays, the homologous recombination assay may be domain-specific. The assay may also apply to other BRCA2 variants, since other domains of BRCA2, such as the Rad51 binding BRC repeats, are involved in DNA repair, and variants in the N-terminal PALB2 interaction domain of BRCA2 influence the homologous recombination activity of full-length BRCA2 [Xia, et al., 2006]. However, it is currently unclear whether the correlation between the assay and cancer risk will hold for these other functionally important domains because no missense substitutions in the domains other than the DNA binding domain have yet been shown to confer high risk via the integrated model.
Centrosome amplification
BRCA2 deficient cancer cell lines and mouse embryo fibroblasts grown in culture have significant aneuploidy and centrosome amplification and all BRCA2 mutant tumors in both humans and mice have substantial centrosome amplification [Tutt, et al., 1999]. In addition, mutant forms of BRCA2 can function as dominant negatives, when overexpressed, leading to centrosome amplification and aneuploidy. It is unclear how mutant forms of BRCA2 lead to centrosome amplification, but it has been suggested that the phenotype is a combination of inappropriate duplication of centrosomes in S-phase and failure of cytokinesis in M-phase of the cell cycle [Daniels, et al., 2004]. However, these observations suggest that measurement of the ability of BRCA2 variants to induce centrosome amplification is a useful method for identifying mutations that disrupt the role of BRCA2 in mitotic regulation.
To measure the influence of mutant forms of BRCA2 on centrosome and centriole amplification, GFP-tagged mutant or wildtype forms of BRCA2 are transiently expressed in cells with a wildtype complement of centrosomes (n≤2) or centrioles (n≤4). Cells are subjected to direct immunofluorescence using α-centrin-2 antibodies to detect centrioles and centrosomes [Farrugia, et al., 2008; Wu, et al., 2005] and the numbers of centrosomes in GFP positive BRCA2 expressing cells are subsequently enumerated using fluorescence or confocal microscopy. Cells with ≥2 centrosomes or ≥4 centrioles are considered to have centrosome amplification. As with the homologous recombination assay the wildtype BRCA2 and Y42C variant can serve as positive controls and the 6174delT truncation mutant and the D2723H deleterious missense mutant can serve as negative controls.
Approximately 30 BRCA2 variants have been studied using this approach, including 25 from the DNA binding domain [Farrugia, et al., 2008; Wu, et al., 2005]. While studies have focused on this region, there is no specific evidence indicating that the influence of BRCA2 on the centrosome is mediated by the DNA binding domain. Thus, this assay may also be useful for evaluation of variants in other parts of BRCA2. Results from the assay have been compared with the results from the family based likelihood model for the purpose of assay validation. Although a strong correlation appears to exist [Farrugia, et al., 2008], two mutations that alter homologous recombination activity of BRCA2 do not influence centrosome number, indicating that the Centrosome Amplification assay may have lower sensitivity [and lower negative predictive (NPV) value] than the homologous recombination assay but equally high specificity and positive predictive value (PPV). These findings suggest that the BRCA2 protein may have completely independent functions. Some variants may influence homologous recombination repair of DNA and centrosome amplification and other variants may cause centrosome amplification but have no effect on homologous recombination activity. As a result, both assays should probably be performed for each variant to minimize the risk that a specific functional effect is overlooked. However, one caveat to this approach is that it is not yet clearly established that induction of centrosome amplification alone predisposes to cancer. Further studies aimed at correlating these assays with sequence conservation measures [Tavtigian et al., 2008a], and overall posterior probabilities of cancer causality derived from the combination of sequence analysis, pathology and family data [Goldgar et al., 2008] are needed to better define the sensitivity, specificity, and both NPV and PPV of the assay.
Other assays for BRCA1
Several other domain specific assays have been used to determine the functional impact of changes in BRCA1. While, in some cases these assays may provide important contributions to classification and to our understanding of the function of BRCA1 they have not yet been systematically analyzed against a set of genetically-defined variants to determine their predictive value. They include the small colony phenotype Assay [Coyne, et al., 2004; Humphrey, et al., 1997; Monteiro and Humphrey, 1998], the protease sensitivity assay [Williams, et al., 2003; Williams and Glover, 2003], and the phosphopetide binding assay [Botuyan, et al., 2004; Clapperton, et al., 2004; Glover, et al., 2004; Rodriguez, et al., 2003; Williams, et al., 2004]. All of them are specific for the BRCT domains.
The small colony phenotype assay measures the ability of GAL4 DNA binding domain fusions to induce growth arrest in yeast. Deleterious variants abrogate this activity [Coyne, et al., 2004; Humphrey, et al., 1997] providing an easy scoring method to classify variants. It is not known the mechanism by which BRCA1 BRCT domains induce growth arrest. One possibility is that the overexpression of DNA damage response proteins forcibly targeted to DNA (by the GAL4DBD moiety) triggers the activation of cellular DNA damage response in the absence of DNA damage as was recently demonstrated [Soutoglou and Misteli, 2008]. Importantly, at least for some proteins, this activity seems to be dependent on the presence of BRCT domains [Soutoglou and Misteli, 2008].
The protease sensitivity assay is based on the idea that variants that induce conformational changes will expose regions of the BRCT domain susceptible to protease digestion that are not exposed in the context of the wild type protein [Williams, et al., 2003; Williams and Glover, 2003]. It is still unclear whether more subtle deleterious changes in surface residues would be correctly identified in this assay. Nevertheless, the simplicity of the assay makes it attractive to large scale studies.
Finally, the phosphopeptide binding assay emerged from recent findings that identified a groove in BRCT domains from several proteins that recognized peptide motifs that are phosphorylated by PI3K-like enzymes such as ATM and ATR [Botuyan, et al., 2004; Clapperton, et al., 2004; Glover, et al., 2004; Rodriguez, et al., 2003; Williams, et al., 2004]. These findings suggested that BRCT domains may function as signal transduction recognition modules for phosphoserine and phosphothreonines analogous to SH2 domains and phosphotyrosine. It has been shown that several deleterious variants disrupt phosphopeptide recognition, correlating this activity with tumor suppression. It will be interesting to see how these different assays compare with the others discussed in the above in terms of predictive value and coverage.
Protein and mRNA stability and splicing
Several variants in the BRCA1 and BRCA2 genes alter mRNA stability, potentially through a mechanism related to nonsense mediated mRNA decay. Similarly certain variants may influence protein degradation. Thus, a subset of variants may inactivate BRCA1 or BRCA2 function and predispose to cancer by influencing protein and mRNA stability rather than through a specific structural or functional change in the proteins. Since functional assays that rely on overexpression of variants may fail to detect these effects, a comprehensive analysis of mRNA and protein half lives should probably be considered in association with any functional assays [Lovelock, et al., 2007]. Similarly, a number of variants that have been implicated in cancer predisposition using the family based likelihood model have been shown to cause splicing aberrations and exon skipping. These findings suggest that functional testing of BRCA1 and BRCA2 variants should include an evaluation of splicing through a combination of in silico prediction algorithms and RT-PCR analysis of RNA extracted from blood or tissue specimens of patients [Spurdle et al., 2008].
Lynch syndrome and Mismatch Repair
MMR Genes and cancer predisposition
Lynch syndrome (often named hereditary nonpolyposis colorectal cancer, HNPCC; MIM# 120435) is an autosomal dominant hereditary disease caused by germline mutations in mismatch repair (MMR) genes. Patients suffering from this syndrome inherit mutations in one allele of any of four MMR genes, most frequently MSH2 (NM_000251.1) or MLH1 (NM_000249.2) and, more rarely, in MSH6 (NM_000179.2) or PMS2 (NM_000535) [Li, 2008; Peltomaki, 2001; Peltomaki, 2003]. Sporadic somatic mutation or loss of heterozygosity of the wild-type allele, leaving the cell with a defective MMR system, results in genetic instability and a spontaneous mutator phenotype. This permits the accumulation of mutations that are thought to drive or contribute to the neoplastic transformation process. The majority of MMR gene mutations in Lynch syndrome patients cause truncations and loss of function of the affected polypeptide. However, missense variants comprise a significant proportion of the mutations in patients that fulfill the criteria for Lynch syndrome (∼10% of MSH2, ∼30% of MLH1, and ∼37% of MSH6) [Nystrom-Lahti, et al., 2002] and the functional consequences of many of these variants are not clear.
The canonical MMR pathway
MMR is a post-replicative DNA repair system that corrects errors by the replicative DNA polymerases δ or ε, including DNA base-base mispairing and insertion/deletion loops (IDL). The system originated in prokaryotes and is well conserved through all eukaryotes. IDL arise during slippage of the polymerase during replication of simple sequence repeats (‘microsatellites’) like the mononucleotide repeat (A)n or the dinucleotide repeat (CA)n [Kunkel and Erie, 2005; Li, 2008]. MMR thereby protects the cells from spontaneous mutations, including the lengthening and shortening of short oligonucleotides repeats known as microsatellites (a phenomenon known as microsatellite instability, MSI). The major MMR pathway is initiated by the recognition of a mismatch by the MutSα heterodimer consisting of the Msh2 and Msh6 proteins. MutSα is responsible for the recognition of base-base mismatches and IDLs in mono- to tetranucleotide repeats. A minor MMR pathway, triggered by MutSβ (MSH2-MSH3), is responsible for the redundant repair of, preferentially dinucleotide and larger, IDLs. Upon DNA mismatch recognition, MutSα (or MutSβ) recruits MutLα, another heterodimer that consists of MLH1-PMS2, that acts as a matchmaker protein as well as an endonuclease [Kunkel and Erie, 2005; Li, 2008]. The resulting nick serves as entry point for exonucleases such as hEXO1 that carry out DNA excision. The MMR excision intermediate is protected from nuclease degradation by Replication Protein A (RPA). The excised DNA strand is resynthesized by Polδ or ε in the presence of the polymerase PCNA, the clamp loader RFC and RPA [Kunkel and Erie, 2005; Li, 2008].
Biochemistry of mismatch repair
MutSα is able to recognize most base-base mismatches and short IDLs. The crystal structures of MutSα and its prokaryotic homodimeric homolog MutS show that the protein senses perturbed base stacking rather than loss of base pairing, by probing the deformability of the helix. This common recognition mode implies a rather low discriminatory power, which lies in the order of 20−50 fold. To validate mismatch binding, MutS has a kinetic ‘proofreading’ activity that is mediated by the exchange of ADP for ATP in a Walker-type ATPase domain. In case of inadvertent homoduplex binding ATP is rapidly hydrolyzed accompanied by release of MutSα from the DNA [Kunkel and Erie, 2005; Li, 2008]. In case of mismatch binding the ATP-bound form of MutSα is stabilized, forming a DNA-embracing clamp that allows binding of MutLα (or MutL). MutLα also has an ATPase domain, and an ADP-ATP cycle [Li, 2008; Sacho, et al., 2008]. In its ATP-bound form it coordinates downstream events in mismatch repair, like hEXO1 binding and endonucleolysis. The requirement of primary mismatch binding followed by a double ATP-driven verification mechanism implies that amino acid changes in MutSα or MutLα may affect any of a number of biochemical processes, including mismatch recognition, intramolecular signaling, ATPase cycling and PCNA or hEXO1 binding. This variety of defined activities of the mismatch repair proteins make pinpointing the biochemical defect of a variant difficult.
Classification of MMR gene variants
Variants in MMR genes may affect splicing by influencing consensus splice sites or forming cryptic splice sites, or altering binding sites for splicing enhancer proteins. Thus, as with BRCA1 and BRCA2, the influence of variants on splicing should be assessed where possible using web-based algorithms that predict effects of variants on splice sites, and/or RT-PCR analysis [reviewed in Spurdle et al., 2008].
If splice alterations are not suspected and the MMR gene variant is not silent, a dysfunctional protein may or may not result. However, some single amino acid substitutions can give rise to partly active, dominant negative, unstable, or nonfunctional proteins depending on the nature of the individual mutation. Defective MMR caused by missense mutations can be a result of (1) inactivation of enzymatic activity (ATP binding/hydrolysis), (2) defective protein-protein interaction (complex formation), (3) defective protein-DNA binding (mismatch recognition), (4) defective subcellular localization of MMR proteins, (5) altered expression of MMR proteins (stochiometry of MMR complexes), and (6) altered stability of MMR proteins.
Functional assays for MMR gene variants
A multitude of assays in different experimental systems have been developed to investigate whether variants found in possible Lynch syndrome patients cause functional defects resulting in cancer predisposition. These assays are the subject of a recent review [Ou, et al., 2007] and therefore are only summarized here. Roughly, the assays can be subdivided into three categories, (1) assays based on MMR in vivo, (2) assays that measure MMR in vitro and (3) assays that measure specific biochemical and cell biological properties of MMR variants.
In vivo assays
These assays exploit the functional conservation of MMR throughout evolution, enabling investigation of the functional consequences of variants in both autologous and heterologous systems. In particular, the amenability of the yeast Saccharomyces cerevisiae to mutation assays can be exploited to assess the activity of human MMR gene variants. This can be done by expressing the corresponding variant in the yeast MMR gene ortholog [Drotschmann, et al., 1999a; Drotschmann, et al., 1999b; Polaczek, et al., 1998; Shcherbakova and Kunkel, 1999]. The loss of dominant-negative interference after expression of variant MLH1 protein within yeast with intact MMR may indicate its loss-of function [Takahashi, et al., 2007]. Unfortunately, yeast in vivo assays may be of limited value for routine testing since they are limited to variants in conserved regions of the protein, the dominant-negative effect may partially be dependent on the reporter gene [Takahashi, et al., 2007] and may be prone to artifacts due to aberrant expression of the introduced gene [Shcherbakova and Kunkel, 1999].
Cell based assays using human lymphoblastoid cell lines have also been used to evaluate MMR variants. Specifically, cell lines from MSH2 heterozygous, but not from MLH1 heterozygous patients, demonstrate acquired tolerance to methylating agents [Marra, et al., 2001], reflecting haploinsufficiency of the wild type MSH2 allele. However, the sensitivity and specificity of this assay is poor.
Cell-free MMR assays
The canonical MMR reaction can be fully performed in vitro using cell extracts [Longley, et al., 1997]. These extracts are added to a plasmid substrate carrying a defined mismatch within a restriction endonuclease site. Repair of the mismatch results in restoration of the restriction site, which allows simple assessment of the repair efficiency by analysis of the cleaved product by gel electrophoresis. The inclusion of an MMR variant in this assay allows the assessment of its activity; variants that have lost activity are considered pathogenic.
Several forms of this assay have been developed: (1) introduction of a variant-encoding gene in cells with an endogenous deficiency of the gene followed by performance of the MMR assay on extracts from these complemented cells [Marra, et al., 2001; Takahashi, et al., 2007; Trojan, et al., 2002], (2) Production of the variant protein (and its heterodimeric partner) in a heterologous expression system, like insect cells, followed by complementation of a cell extract with a deficiency in the gene in a MMR assay [Ollila, et al., 2006; Raevaara, et al., 2005].
In these studies, assays showing loss of MMR activity in vitro were considered the gold standard for pathogenicity, despite the absence of complete validation of this method. In vitro results were supported by clinicopathologic and evolutionary sequence alignment-based predictions of pathogenicity where possible, and by loss of other biochemical characteristics of the variants such as in vivo stability, subcellular localization and heterodimerization. However, clinicopathological data alone were often insufficient to establish that the kindreds with the relevant variants had Lynch syndrome, so the predictive values of each of these methods could not be determined.
While the in vitro MMR assays appear to be highly predictive of strongly pathogenic mutations in MLH1 and MSH2, it is not known how best to use the results from the assays in the clinic. Specifically, the assays are technically complex and have not been formally validated relative to a gold standard such as clinicopathologic data or disease co-segregation data from patients and families. Overall, although the assays show great promise, it remains to be determined what the standards should be before such assays can be considered sufficient for clinical interpretation.
MMR protein-specific assays
Other biochemical consequences of variants can also be assessed. These assays focus on individual aspects of protein function like (for MSH2/MSH6) mismatch binding and the ADP→ATP cycle [Heinen, et al., 2002] and (for both MSH2/MSH6 and MLH1/PMS2) protein stability and intracellular localization and protein-protein interaction [Jager, et al., 2001; Ollila, et al., 2006; Raevaara, et al., 2005]. Although these assays can pinpoint a specific functional defect conferred by an MMR gene variant, confirming and extending loss of function found in in vitro MMR assays, they are not amenable to routine screening of variants.
A proposed decision tree for the analysis of MMR gene variants
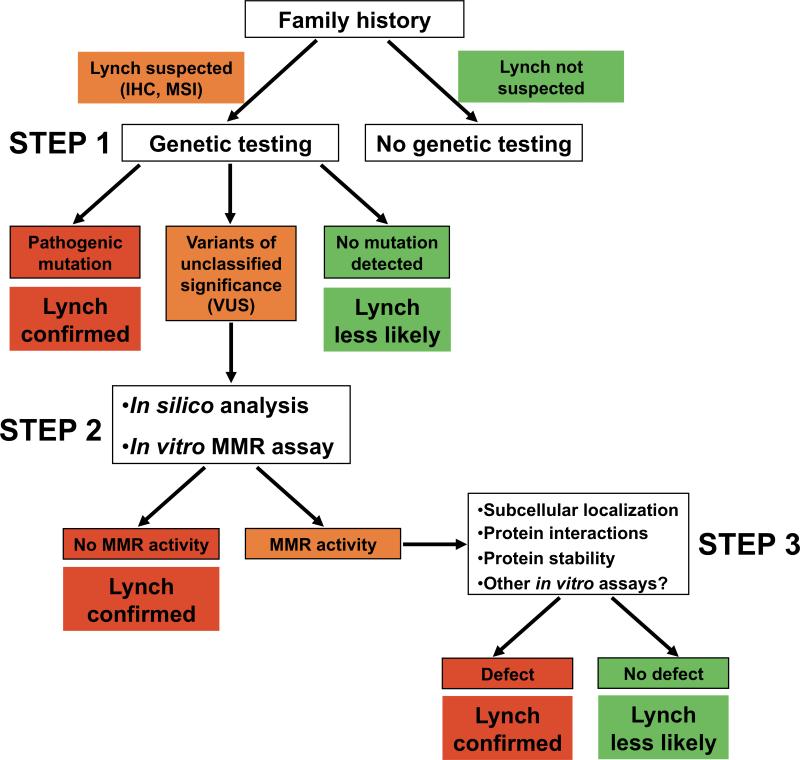
Often, the first step in “biochemical diagnosis” of HNPCC is tumor testing looking for MMR protein loss by immunohistochemistry and for MSI. This is followed by sequencing relevant MMR genes. These procedures can be performed by most diagnostic laboratories (Figure 2). If a missense variant is identified, the second step can be analysis of splice aberrations [Spurdle et al., 2008] and of protein folding in silico [Tavtigian et al., 2008b], followed by an in vitro test of variant function. New in vitro MMR assays [de Wind, unpublished data] that can be optimized for use in most diagnostic laboratories equipped for genetic testing, allowing their independent validation, may help this process.
Figure 2.
Proposed decision tree for the in vitro analysis of variants in MMR genes in suspected Lynch syndrome cases.
Lack of MMR activity in in vitro assays likely indicates clinical HNPCC. Based on the wealth of evidence linking MMR defects to clinical Lynch syndrome, results indicating that a variant is deficient in MMR can be considered strong qualitative evidence that the variant is pathogenic. Clear loss of function detected in technically verified MMR assays, when considered in conjunction with other lines of evidence, may be used to help classify a variant as Pathogenic (Class 5) or Likely Pathogenic (Class 4) [Plon et al., 2008].
Importantly, a number of variants may display normal activity in MMR in vitro assays. These variants may be neutral with no influence on cancer or may have defects in protein localization, expression or stability that are not assessed by the in vitro assays. In this case, the variant will continue to Step 3 for detailed studies of MMR protein stability, protein-protein interactions and translocations in the cell. These analyses are complex and must be carried out in specialized laboratories.
The Working Group encourages studies that will validate MMR assays relative to a gold standard such as segregation with cancer in families or clincopathologic measures, and will quantify the PPV and NPV of each assay. Because of the established link between MMR defects and clinical Lynch syndrome, we anticipate that predictive values of validated MMR assays will be high. A variant exhibiting loss of MMR activity (Figure 2, step 2), or loss of more specific properties (Figure 2, step 3) in an assay with known NPV and PPV should not need further testing by other methods. However, it may prove difficult to optimally validate functional assays due to limited availability of “gold standard genetic data”. Therefore, a single functional assay may not be definitive, and optimal classification may require integration of data from different functional assays, in silico sequence conservation analyses of the variants, segregation with disease in families, and clinicopathological features to determine a posterior probability of pathogenicity [Goldgar et al., 2008].
CDKN2A (p16, Ink4a)
CDKN2A variants and cancer
Variants in the CDKN2A gene (also known as p16 and Ink4a; MIM# 600160) are associated with 20−38% of cases of Familial Melanoma (FM; MIM# 155601) [Goldstein, 2004; Goldstein, et al., 2006]. Because most variants are found in small kindreds with few cases of cancer, it is often difficult to establish whether variants predispose to cancer or not based solely on assessment of family history. As a result, many of these variants remain unclassified, and some classifications of pathogenic mutations are based on only one or two kindreds, with only moderate statistical certainty. In an effort to improve this situation, multiple assays based on the known activity of the CDKN2A protein have been developed to classify variants as deleterious (inactivate CDKN2A function) or not (no influence on function).
In this context, CDKN2A illustrates several of the issues important to the interpretation of functional assays in all cancer susceptibility genes: choice of assay, establishing standards that will be used to decide clinical effect (e.g., deciding on a threshold cutoff when gradations of decreased function are observed), and assessing the predictive value of a functional assay (what is the likelihood that a single result is correct or incorrect).
Functional results for over 100 CDKN2A variants (88 missense and 16 non-missense) have been reported in over 30 papers. Because many variants have been tested multiple times, often in different assays, a total of around 200 functional conclusions have been reported. At least 2−3% of reported conclusions must be incorrect, since different studies come to conflicting conclusions (wild type vs. loss of function) for five variants (c.24_47dup24, c.212A>G p.N71S, c.298G>C p.A100P, c.334C>G p.R112G, c.352G>A p.A118T).
CDKN2A Function
In order to interpret functional assays of CDKN2A (wild type and variants), it is important to understand some details of CDKN2A structure and function. Wild type CDKN2A induces cell cycle arrest in G1 phase and cell senescence through the retinoblastoma (RB) tumor suppressor pathway. It is a small protein (156 AA) that consists of four “ankyrin repeats” that align to form a compact, tightly folded “V-shaped” convex structure [Greenblatt, et al., 2003; Sharpless, 2005]. The convex surface binds to cyclin dependent kinases CDK4 and CDK6, inducing a conformational change in the kinase that inhibits phosphorylation of the RB protein. Unphosphorylated RB releases transcriptional factor E2F, resulting in transcription of genes that affect cell growth and senescence [Ruas and Peters, 1998]. Mutations can inactivate CDKN2A by either interfering with CDK binding or causing unfolding of the compact CDKN2A structure.
Functional Assays
Two main CDKN2A functions can be easily measured in vitro: 1) Binding of full-length, wild type or variant CDKN2A protein to CDK4 and CDK6. This can be measured in cell-free assays with labeled proteins, in two-hybrid assays, or by co-immunoprecipitation (28 reports, 121 variants). 2) Cell cycle arrest and cell growth inhibition. In vitro cell growth assays can measure changes in cell proliferation, either by cell count or more commonly by Flow Cytometry measuring the fraction of cells in each phase of the cell cycle. Tumor cell lines that lack CDKN2A protein but express RB are transfected with plasmids coding for wild type or variants of CDKN2A and cell cycle inhibition is determined. All cell types with these features seem to behave similarly. The advantage of cell growth or cell cycle assays is that they measure a phenotype that is clearly relevant to tumorigenesis. Their disadvantage is that cell culture conditions may influence cell growth and complicate interpretation of cell inhibition. Results must be compared to controls using wild type CDKN2A (positive cell cycle arrest) and vector only (negative cell cycle arrest). In both of these types of assays, some variants are temperature sensitive; they function normally at 20−34°C but are impaired at 40°C [Parry and Peters, 1996].
Other binding partners of CDKN2A have been identified: RelA, in the NFkB pathway [Becker, et al., 2005], and ISOC2, a novel protein of unknown function [Huang, et al., 2007]. However, the functional consequences and clinical relevance of CDKN2A binding to these proteins is unclear.
Interpretation of Functional Assay Results
Although the major CDKN2A functions that are associated with cancer development seem clear, there are still unresolved issues regarding how to use in vitro functional assays to help establish pathogenicity. Some variants are deficient in all in vitro functional assays, but other variants show discordant results among assays. There is no consensus regarding which assay, or set of assays, is most appropriate for determining the clinical effect of a CDKN2A variant.
Impaired binding to CDK4 and CDK6 usually but not always indicates impaired cell cycle inhibition. Of variants that failed to bind CDK4 and CDK6, 18/22 (81.8%) and 10/10 variants, respectively, failed to cause cell cycle arrest. For two variants, binding to both CDK4 and CDK6 was reported to be impaired, but cell cycle arrest was intact [Rizos, et al., 2001]. Four variants (c.71G>C p.R24P, c.212A>G p.N71S, c.242C>T p.P81L, c.260G>C p.R87P) bind normally to CDK6 but fail to bind CDK4; only R24P clearly loses cell cycle inhibitory activity, whereas the others show intermediate results.
However, binding assays for both CDK4 and CDK6 often give misleading results suggesting lack of pathogenicity. Approximately half of variants tested for both functions show normal CDK4 or CDK6 binding but still fail to cause cell cycle arrest. Despite these inconsistencies, CDK binding is the most common method for assessing function (23 reports, 121 conclusions, many variants tested multiple times).
Mixed results have been described in different assays for 40 variants within the same report. In half of these cases (19% of all variants tested), a report concluded that the variant was deficient in tumor suppressor function despite normal results in at least one assay. In half of the cases (19% of all variants tested) the conclusion was that the pathogenicity of the variant was uncertain. For seven of the uncertain variants, other reports concluded that the variant was deficient in function, and in two cases other reports concluded that the variant retained wild type function.
Thus, a single assay is unlikely to be definitive in determining whether a CDKN2A variant is deleterious or not, and CDK binding assays alone are clearly inadequate. If CDK binding assays are used, they must also be supplemented by cell cycle assays, especially if normal binding is found. Of 31 missense variants associated with FM, 17 have been tested by either CDK binding or cell inhibition assays. All are clearly deficient in at least one assay (deficient binding n=13, deficient cell inhibition n=13). Two missense variants (c.106G>C p.A36P, c.142C>A p.P48T) and an 8-amino acid duplication [Monzon, et al., 1998; Walker, et al., 1999] have shown normal CDK binding but loss of growth arrest [Becker, et al., 2001; Della Torre, et al., 2001]. Only one FM-associated variant, c.199G>A p.G67S, has shown normal cell cycle inhibition in a cellular assay despite abnormal CDK binding [Rizos, et al., 2001]. The explanation for this is unclear. The common neutral variant, c.442G>A p.A148T, has shown wild type activity in both assays, multiple times [Ruas and Peters, 1998]. Since some variants are temperature sensitive, assays should be done at higher temperatures if wild type results are found at lower temperatures.
Other assays that have been used less frequently include inhibition of kinase activity (10 reports) and measurement of protein structure changes [Byeon, et al., 1998; Zhang and Peng, 1996]. These assays are more technically demanding than CDK binding or cell cycle inhibition assays. Only one FM-associated variant, c.247C>A p.H83N, has not been tested in vitro for binding or cell inhibitory activity, but has been shown to be deficient in other assays (decreased kinase inhibition, structural abnormality by NMR) [Byeon, et al., 1998].
Standards for Reporting Results
Both binding and cell cycle inhibition assays show a continuum of deficient activity, from complete abrogation to small decrements in activity (20% or less compared to wild type). Early reports were reluctant to declare a variant deleterious in the setting of partial activity [Ranade, et al., 1995]. Subsequent experience has clearly demonstrated that some pathogenic variants completely abrogate function, but others (e.g., c.159G>C p.M53I c.301G>T p.G101W) may show intermediate results on binding, kinase, and growth assays [Becker, et al., 2001; Koh, et al., 1995; Yang, et al., 1995]. A consensus is required regarding the degree of functional loss that is required in order to classify a variant as pathogenic.
Predictive Values
Establishing a predictive value for a test requires that a set of known deleterious and known neutral variants be studied to establish sensitivity, specificity, and positive (PPV, predicting deleterious) and negative (NPV, predicting neutral) predictive values. For CDKN2A, to date, the PPV of binding assays to both CDK proteins for predicting phenotype appears to be about 83%. The PPV of cell cycle inhibition assays compared with clinical phenotype is currently 93%. However, this is based on a review of multiple studies using heterogeneous methods. Only six clearly neutral missense variants are known; three of these have been tested and found to have wild type function. A systematic study of these methods, in a standardized format, is needed to formally establish how well the results correlate with other predictors of risk, such as segregation of cancer in families, and how the results can be translated to the cancer genetics clinic.
Discussion
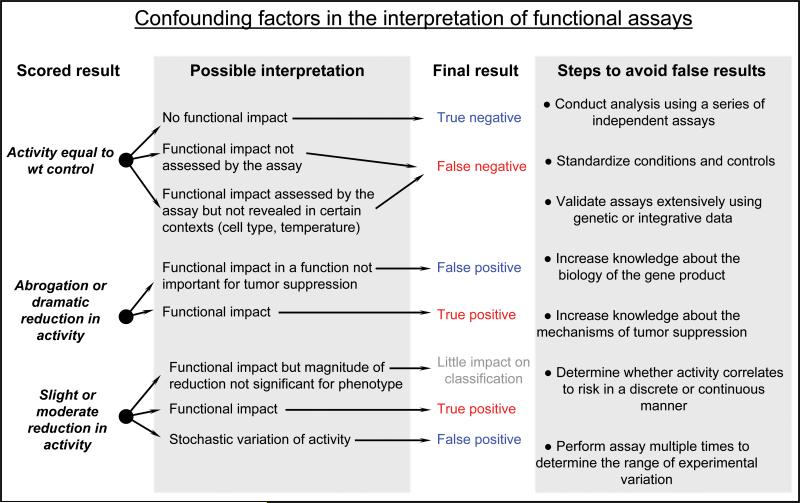
The examples described above provide some insight into the complexity associated with functional assay development and interpretation. It is clear that assays for different genes have their own particular issues but there are also issues that are common for all of the assays discussed here. We briefly discuss some of the central issues associated with the use of functional assays for classification of VUS (Figure 3).
Figure 3.
Confounding factors in the interpretation of functional assays.
Do specific functional assays account for all functional effects?
The answer to this question depends on the nature of the assay and also on the function of the gene product. The protein binding and activity assays for MMR proteins are well-defined. In contrast, in the setting of the multifunctional BRCA1 and BRCA2 proteins, no single assay will account for the functional effects of all variants. There is also uncertainty in the best assays to use for CDKN2A variants, even though the main phenotypic abnormality contributing to cancer is well understood. In addition, variants that induce aberrant splicing, exon skipping or mRNA/protein stability will be overlooked in the setting of most functional assays. As a result it seems unlikely that the influence of every variant can be accounted for using functional assays.
Can variants be definitively shown to have no effect?
There are a variety of considerations for variants that show wild type activity in functional assays. The NPV for each assay should be determined on a panel of known mutations and known neutral variants in order to quantify the interpretation of a wild type result [Goldgar et al., 2008]. Aberrant splicing, exon skipping or mRNA/protein stability also must be considered using in silico and in vitro approaches when interpreting negative functional results [Spurdle et al., 2008].
Negative results may also occur if a variant is in a different domain, one that does not influence the function being assayed. A variant may disrupt a specific binding site for a protein interaction that while important for tumor suppression is not assessed in the assay. In this setting the variant would be inappropriately viewed as “neutral”. Results from assays that do not apply to the domains in which variants are located must be excluded for the classification process. Defining variants as neutral may involve a laborious and hierarchical approach to functional studies, and knowing the NPV of each assay will be important in reaching conclusions regarding protein function. Final classification of neutral will likely require integration of multiple lines of evidence [Goldgar et al., 2008].
How should differences in results among assays be interpreted?
Discordant results may be very informative. One interpretation is that any in vitro result of functional loss should lead to a classification of pathogenic, as has been applied to MMR genes and CDKN2A variants. Alternatively, it is possible that some assays assess a bona fide function of a protein that is not necessarily required for tumor suppression. Validation of the PPV of all assays is therefore critical. Each biochemical pathway has specific features that demand sophistication in interpreting the results of assays. Therefore, the process of validation may require ongoing evaluation by panels with expertise in all aspects of the pathway and disease. Periodic consensus statements may be required, similar to what has been done for classification of carcinogens. Such statements should attempt to be comprehensive in evaluating data and transparent in discussing what conclusions were reached and why, as has been done for classifying BRCA mutations [Greenblatt et al., this issue]. The Working Group encourages this unified view of functional assays, which can be applied to all attempts to use in vitro data to classify genetic variants.
Are there specific thresholds for function?
Results from functional assays tend to cover the entire spectrum of responses within the dynamic range of each assay. Given the variation in results this raises the question of whether to use a specific cutoff point or a continuous measure of activity for distinguishing between inactivating and neutral effects. While validation of the sensitivity, specificity, and PV of an assay may result in the definition of a specific cutoff point, a continuous measure may be most useful in the absence of comprehensive validation. In the case of a continuous measure, one could assign variants whose values are comparable to known damaging mutants to a class called “damaging” and variants whose values are comparable to wild type or known neutral variants as “neutral”. The class that falls in between could be considered unknown. Alternatively, somewhat arbitrary “favor damaging” or “favor neutral” categories could be used. The most appropriate system may be different for different genes. Panels of experts for each gene should agree on classification schemes and their rationales before the results are used clinically.
How can functional effect translate to risk?
Establishing a predictive value for a test requires that a set of known deleterious and known neutral variants be studied to establish sensitivity, specificity, PPV and NPV. This method establishes the error rate of a given assay. This can subsequently be converted into a likelihood ratio that can be used in a Bayesian integrative analysis, in which data from family studies and sequence conservation analysis can be combined with functional assay data to generate an overall likelihood or odds that a variant is pathogenic or neutral. This step has not yet been accomplished for any of the assays described above, although advances in the area are expected in the near future. In the absence of these data, the functional assays are currently only used to qualitatively cross-validate and improve confidence in results from other models.
How should the sensitivity, specificity, and PV of the assays be determined?
Where possible, assays should be validated against a “gold standard” measure of the disease phenotype, such as genetic and clinicopathologic data from families (co-segregation of variants with disease) using known pathogenic and known neutral variants. Initially, few such known variants may be available for a given gene. An extension of this approach is the integrated Bayesean likelihood model that has been applied to BRCA1 and BRCA2 variants. This model accounts for co-segregation data, family history of cancer, co-occurrence of variants with known pathogenic mutations and tumor pathology data and appears to be the best developed method for this purpose. However, at this time this model is limited to genes with established estimates of penetrance [Goldgar et al., 2008] and by the availability of family data for a large number of mutations. Indeed, it has even been difficult to establish the sensitivity and specificity of BRCA1 and BRCA2 assays because of limited availability of family data. An attractive feature of the Bayesean likelihood model is that new classification techniques such as in vitro assays, when validated and quantified against a panel of known variants, can be added to the baseline statistical genetic assessment for a variant.
The greatest hindrance of assay validation today is the absence of “gold standard” data for established pathogenic and neutral variants. As a result, validation studies tend to be underpowered and yield wide confidence intervals. The certainty with which a variant can be classified by functional assays is limited, and results remain qualitative and not quantitative.
Should qualitative functional assay data be used clinically?
The ultimate goal of the use of functional tests is to provide information for the individual to make informed clinical decisions. In principle, only the most rigorously evaluated information should be used so that when a classification is made it carries a high level of certainty. It can be argued however, that such an approach, while scientifically sound, denies the patients and their families information that may not have achieved the “gold standard”. The patient and clinicians may prefer to make decisions based on reasonable estimates of moderately high or low risk from incomplete and less-than-optimal data rather than using no data at all [Plon et al., 2008]. If this argument is accepted then several of the functional assays described above are already useful or will shortly be useful for clinical classification of variants, in a qualitative manner.
For these reasons it might be useful to use a hierarchical validation scheme that mixes quantitative and qualitative elements. Statistical genetic data are currently the most powerful because they are quantitative. Some in silico sequence conservation methods have also been quantified, to a much less rigorous degree at this time [Tavtigian et al., 2008a,b]. Although predictive values have not been determined for in vitro functional assays, some do appear to be robust enough to contribute qualitatively to classification. This is particularly the case for in vitro MMR activity assays because the MMR pathway is well defined, both biochemically and genetically and it is established that defective MMR is causative for Lynch syndrome. If we find a complete defect of a variant in the in vitro MMR assay, and knowing that an MMR activity defect causes Lynch syndrome, at this stage we may diagnose the patient as such and dismiss the non-variant carrying relatives from screening (Figure 2, step 2). Thus, an in vitro functional defect identified by a technically validated assay for MMR activity should be considered strong qualitative evidence.
In addition, newly developed assays that are quickly validated against a smaller panel of variants might also contribute to such a scheme. Since many variants will be rare, accumulating enough genetic data may take many years and in some cases never be achieved, so qualitative functional conclusions may provide an important contribution. One caveat for this approach is that since less-than-optimal data can be misinterpreted and misconstrued, it is crucial to ensure that the data from functional assays that is released is either of highest certainty or, if not, then clearly specified as tentative or uncertain.
Will functional assays ever be used for clinical classification?
Assay results as they stand now can be one of several tools to help with clinical decisions. At this time, results from most assays must be considered qualitative evidence. Once assays are validated against standards and Type I and Type II errors are evaluated, assays can likely be used for classification of variants. Whether they should be used for clinical decisions is a question that needs to be addressed together by laboratory scientists, clinicians, clinical geneticists, and patient advocates. Decisions will probably depend on whether individuals are comfortable using the results for clinical decisions knowing that they may include flaws. If that is the case, one needs to make sure that the target audience understands what data were used, all the limitations in the assays, all possible pitfalls, and also understands the significance of each class of variant used in the classification. Results can be disseminated to the biomedical community through publications and through locus sprecific databases [Greenblatt et al., this issue] so that recommendations are uniform. In addition, genetic counselors and clinicians may need specific training in interpretation of results and/or teaching aids to help with delivery of the information to patients.
Summary
This working group believes that some in vitro functional data (e.g., mismatch repair, cell cycle inhibition, DNA repair and transcription activation and splicing assays) can be used to help classify variants at this time. However, standards for performing each assay must be agreed upon by the research and clinical community.
Acknowledgments
The authors would like to acknowledge support from the NIH Breast Cancer Specialized Program in Research Excellence grant P50 CA116201 (FJC), from NIH grants CA116167 (FJC), CA92309 (ANAM) and CA96536 (MSG), from the Lake Champlain Cancer Research Organization (MSG), American Cancer Society award RSG-04-220-01-CCE (FJC), Florida Breast Cancer Coalition Foundation (ANAM), Danish Cancer Society (LJR), Danish Research Council (LJR) and EU grant LSHC-CT-2005-18754 (LJR, RH, NW).
Appendix
Members of the IARC Working Group on Unclassified Genetic Variants:
Paolo Boffetta, IARC, France; Fergus Couch, Mayo Clinic, USA; Niels de Wind, Leiden University, the Netherlands; Douglas Easton, Cambridge University, UK; Diana Eccles, University of Southampton, UK; William Foulkes, McGill University, Canada; Maurizio Genuardi, University of Florence, Italy; David Goldgar, University of Utah, USA; Marc Greenblatt, University of Vermont, USA; Robert Hofstra, University Medical Center Groningen, the Netherlands; Frans Hogervorst, Netherlands Cancer Institute, the Netherlands; Nicoline Hoogerbrugge, University Medical Center Neimejen, the Netherlands; Sharon Plon, Baylor University, USA; Paolo Radice, Istituto Nazionale Tumori, Italy; Lene Rasmussen, Roskilde University, Denmark; Olga Sinilnikova, Hospices Civils de Lyon, France; Amanda Spurdle, Queensland Institute of Medical Research, Australia; Sean Tavtigian, IARC, France.
References
- Antoniou AC, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Olsson H, Johannsson O, Borg A, Pasini B, others Breast and ovarian cancer risks to carriers of the BRCA1 5382insC and 185delAG and BRCA2 6174delT mutations: a combined analysis of 22 population based studies. J Med Genet. 2005;42(7):602–3. doi: 10.1136/jmg.2004.024133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12(1):86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Becker TM, Rizos H, de la Pena A, Leclercq IA, Woodruff S, Kefford RF, Mann GJ. Impaired inhibition of NF-kappaB activity by melanoma-associated p16INK4a mutations. Biochem Biophys Res Commun. 2005;332(3):873–9. doi: 10.1016/j.bbrc.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Becker TM, Rizos H, Kefford RF, Mann GJ. Functional impairment of melanoma-associated p16(INK4a) mutants in melanoma cells despite retention of cyclin-dependent kinase 4 binding. Clin Cancer Res. 2001;7(10):3282–8. [PubMed] [Google Scholar]
- Botuyan MV, Nomine Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12(7):1137–46. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci U S A. 2003;100(10):5646–51. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon IJ, Li J, Ericson K, Selby TL, Tevelev A, Kim HJ, O'Maille P, Tsai MD. Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1(3):421–31. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- Carvalho MA, Marsillac SM, Karchin R, Manoukian S, Grist S, Swaby RF, Urmenyi TP, Rondinelli E, Silva R, Gayol L, others Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 2007;67(4):1494–501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277(24):22085–92. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11(6):512–8. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- Coyne RS, McDonald HB, Edgemon K, Brody LC. Functional characterization of BRCA1 sequence variants using a yeast small colony phenotype assay. Cancer Biol Ther. 2004;3(5):453–7. doi: 10.4161/cbt.3.5.809. [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306(5697):876–9. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7(2):273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Della Torre G, Pasini B, Frigerio S, Donghi R, Rovini D, Delia D, Peters G, Huot TJ, Bianchi-Scarra G, Lantieri F, others CDKN2A and CDK4 mutation analysis in Italian melanoma-prone families: functional characterization of a novel CDKN2A germ line mutation. Br J Cancer. 2001;85(6):836–44. doi: 10.1054/bjoc.2001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drotschmann K, Clark AB, Kunkel TA. Mutator phenotypes of common polymorphisms and missense mutations in MSH2. Curr Biol. 1999a;9(16):907–10. doi: 10.1016/s0960-9822(99)80396-0. [DOI] [PubMed] [Google Scholar]
- Drotschmann K, Clark AB, Tran HT, Resnick MA, Gordenin DA, Kunkel TA. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci U S A. 1999b;96(6):2970–5. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, others A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81(5):873–83. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia DJ, Agarwal MK, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Wadum L, Johnson K, Mentlick J, Tavtigian SV, others Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Research. 2008 doi: 10.1158/0008-5472.CAN-07-1587. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29(11):579–85. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Iversen ES, Greenblatt MS, IARC Unclassified Genetic Variants Working Group Integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29 doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM. Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum Mutat. 2004;23(6):630. doi: 10.1002/humu.9247. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, Azizi E, Bianchi-Scarra G, Bishop DT, Bressac-de Paillerets B, others High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66(20):9818–28. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Beaudet JG, Gump JR, Godin KS, Trombley L, Koh J, Bond JP. Detailed computational study of p53 and p16: using evolutionary sequence analysis and disease-associated mutations to predict the functional consequences of allelic variants. Oncogene. 2003;22(8):1150–63. doi: 10.1038/sj.onc.1206101. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Brody LC, Foulkes W, Genuardi M, Hofstra R, Plon S, Sijmons RH, Sinilnikova OM, Spurdle AB, IARC Unclassified Genetic Variants Working Group Locus-Specific Databases (LSDBs) and the Classification of Variants in Cancer Susceptibility Genes. Hum Mutat. 2008;29 [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276(18):14537–40. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Hayes F, Cayanan C, Barilla D, Monteiro AN. Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 2000;60(9):2411–8. [PMC free article] [PubMed] [Google Scholar]
- Heinen CD, Wilson T, Mazurek A, Berardini M, Butz C, Fishel R. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1(5):469–78. doi: 10.1016/s1535-6108(02)00073-9. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, others Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297(5581):606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Huang X, Shi Z, Wang W, Bai J, Chen Z, Xu J, Zhang D, Fu S. Identification and characterization of a novel protein ISOC2 that interacts with p16INK4a. Biochem Biophys Res Commun. 2007;361(2):287–93. doi: 10.1016/j.bbrc.2007.06.181. [DOI] [PubMed] [Google Scholar]
- Humphrey JS, Salim A, Erdos MR, Collins FS, Brody LC, Klausner RD. Human BRCA1 inhibits growth in yeast: potential use in diagnostic testing. Proc Natl Acad Sci U S A. 1997;94(11):5820–5. doi: 10.1073/pnas.94.11.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager AC, Rasmussen M, Bisgaard HC, Singh KK, Nielsen FC, Rasmussen LJ. HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene. 2001;20(27):3590–5. doi: 10.1038/sj.onc.1204467. [DOI] [PubMed] [Google Scholar]
- Koh J, Enders GH, Dynlacht BD, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375(6531):506–10. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, others Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol Cell Biol. 2002;22(2):669–79. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272(16):10917–21. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96(20):11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock PK, Spurdle AB, Mok MT, Farrugia DJ, Lakhani SR, Healey S, Arnold S, Buchanan D, Couch FJ, Henderson BR, others Identification of BRCA1 missense substitutions that confer partial functional activity: potential moderate risk variants? Breast Cancer Res. 2007;9(6):R82. doi: 10.1186/bcr1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra G, D'Atri S, Yan H, Perrera C, Cannavo E, Vogelstein B, Jiricny J. Phenotypic analysis of hMSH2 mutations in mouse cells carrying human chromosomes. Cancer Res. 2001;61(21):7719–21. [PubMed] [Google Scholar]
- Monteiro AN. BRCA1: exploring the links to transcription. Trends Biochem Sci. 2000;25(10):469–74. doi: 10.1016/s0968-0004(00)01632-7. [DOI] [PubMed] [Google Scholar]
- Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci U S A. 1996;93(24):13595–9. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro AN, August A, Hanafusa H. Common BRCA1 variants and transcriptional activation. Am J Hum Genet. 1997;61(3):761–2. doi: 10.1086/515515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro AN, Humphrey JS. Yeast-based assays for detection and characterization of mutations in BRCA1. Breast Dis. 1998;10(1−2):61–70. doi: 10.3233/bd-1998-101-208. [DOI] [PubMed] [Google Scholar]
- Monzon J, Liu L, Brill H, Goldstein AM, Tucker MA, From L, McLaughlin J, Hogg D, Lassam NJ. CDKN2A mutations in multiple primary melanomas. N Engl J Med. 1998;338(13):879–87. doi: 10.1056/NEJM199803263381305. [DOI] [PubMed] [Google Scholar]
- Morris JR, Pangon L, Boutell C, Katagiri T, Keep NH, Solomon E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum Mol Genet. 2006;15(4):599–606. doi: 10.1093/hmg/ddi476. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–8. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7(2):263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Perrera C, Raschle M, Panyushkina-Seiler E, Marra G, Curci A, Quaresima B, Costanzo F, D'Urso M, Venuta S, others Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer. 2002;33(2):160–7. [PubMed] [Google Scholar]
- Ollila S, Sarantaus L, Kariola R, Chan P, Hampel H, Holinski-Feder E, Macrae F, Kohonen-Corish M, Gerdes AM, Peltomaki P, others Pathogenicity of MSH2 missense mutations is typically associated with impaired repair capability of the mutated protein. Gastroenterology. 2006;131(5):1408–17. doi: 10.1053/j.gastro.2006.08.044. [DOI] [PubMed] [Google Scholar]
- Ou J, Niessen RC, Lutzen A, Sijmons RH, Kleibeuker JH, de Wind N, Rasmussen LJ, Hofstra RM. Functional analysis helps to clarify the clinical importance of unclassified variants in DNA mismatch repair genes. Hum Mutat. 2007;28(11):1047–54. doi: 10.1002/humu.20580. [DOI] [PubMed] [Google Scholar]
- Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci U S A. 1998;95(5):2302–6. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao GM, Janknecht R, Ruffner H, Hunter T, Verma IM. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci U S A. 2000;97(3):1020–5. doi: 10.1073/pnas.97.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D, Peters G. Temperature-sensitive mutants of p16CDKN2 associated with familial melanoma. Mol Cell Biol. 1996;16(7):3844–52. doi: 10.1128/mcb.16.7.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaki P. DNA mismatch repair and cancer. Mutat Res. 2001;488(1):77–85. doi: 10.1016/s1383-5742(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21(6):1174–9. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Phelan CM, Dapic V, Tice B, Favis R, Kwan E, Barany F, Manoukian S, Radice P, van der Luijt RB, van Nesselrooij BP, others Classification of BRCA1 missense variants of unknown clinical significance. J Med Genet. 2005;42(2):138–46. doi: 10.1136/jmg.2004.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon SE, Eccles DM, Easton DF, Foulkes W, Genuardi M, Greenblatt MS, Hogervorst FBL, Hoogerbrugge N, Spurdle AB, Tavtigian S, IARC Unclassified Genetic Variants Working Group Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29 doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaczek P, Putzke AP, Leong K, Bitter GA. Functional genetic tests of DNA mismatch repair protein activity in Saccharomyces cerevisiae. Gene. 1998;213(1−2):159–67. doi: 10.1016/s0378-1119(98)00150-4. [DOI] [PubMed] [Google Scholar]
- Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, McKinnon W, Duraisamy S, others Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129(2):537–49. doi: 10.1016/j.gastro.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Ranade K, Hussussian CJ, Sikorski RS, Varmus HE, Goldstein AM, Tucker MA, Serrano M, Hannon GJ, Beach D, Dracopoli NC. Mutations associated with familial melanoma impair p16INK4 function. Nat Genet. 1995;10(1):114–6. doi: 10.1038/ng0595-114. [DOI] [PubMed] [Google Scholar]
- Rizos H, Darmanian AP, Holland EA, Mann GJ, Kefford RF. Mutations in the INK4a/ARF melanoma susceptibility locus functionally impair p14ARF. J Biol Chem. 2001;276(44):41424–34. doi: 10.1074/jbc.M105299200. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278(52):52914–8. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378(2):F115–77. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci U S A. 2001;98(9):5134–9. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol Cell. 2008;29(1):112–21. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, others Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451(7182):1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576(1−2):22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Kunkel TA. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19(4):3177–83. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320(5882):1507–10. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Couch FJ, Hogervorst FBL, Radice P, Sinilnikova OM, IARC Unclassified Genetic Variants Working Group Prediction and Assessment of Splicing Alterations. Hum Mutat. 2008;29 [Google Scholar]
- Starita LM, Machida Y, Sankaran S, Elias JE, Griffin K, Schlegel BP, Gygi SP, Parvin JD. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol Cell Biol. 2004;24(19):8457–66. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo CI, Worley T, Monteiro AN. Understanding germ-line mutations in BRCA1. Cancer Biol Ther. 2004;3(6):515–20. doi: 10.4161/cbt.3.6.841. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67(10):4595–604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- Tavtigian S, Byrnes GB, Goldgar DE, Thomas A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum Mutat. 2008a;29 doi: 10.1002/humu.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB, IARC Unclassified Genetic Variants Working Group In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat. 2008b;29 doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122(1):211–9. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9(19):1107–10. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondum-Nielsen K, Gerdes AM, Moller P, Kristoffersson U, Olsson H, others Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet. 2001;10(4):353–60. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GJ, Gabrielli BG, Castellano M, Hayward NK. Functional reassessment of P16 variants using a transfection-based assay. Int J Cancer. 1999;82(2):305–12. doi: 10.1002/(sici)1097-0215(19990719)82:2<305::aid-ijc24>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang H, Kajino K, Greene MI. BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene. 1998;17(15):1939–48. doi: 10.1038/sj.onc.1202403. [DOI] [PubMed] [Google Scholar]
- Williams RS, Chasman DI, Hau DD, Hui B, Lau AY, Glover JN. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J Biol Chem. 2003;278(52):53007–16. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- Williams RS, Glover JN. Structural consequences of a cancer-causing BRCA1-BRCT missense mutation. J Biol Chem. 2003;278(4):2630–5. doi: 10.1074/jbc.M210019200. [DOI] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11(6):519–25. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Worley T, Vallon-Christersson J, Billack B, Borg A, Monteiro AN. A naturally occurring allele of BRCA1 coding for a temperature-sensitive mutant protein. Cancer Biol Ther. 2002;1(5):497–501. doi: 10.4161/cbt.1.5.164. [DOI] [PubMed] [Google Scholar]
- Wu K, Hinson SR, Ohashi A, Farrugia D, Wendt P, Tavtigian SV, Deffenbaugh A, Goldgar D, Couch FJ. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65(2):417–26. [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297(5588):1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yang R, Gombart AF, Serrano M, Koeffler HP. Mutational effects on the p16INK4a tumor suppressor protein. Cancer Res. 1995;55(12):2503–6. [PubMed] [Google Scholar]
- Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14(11):1400–6. [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273(39):25388–92. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Zhang B, Peng Z. Defective folding of mutant p16(INK4) proteins encoded by tumor-derived alleles. J Biol Chem. 1996;271(46):28734–7. [PubMed] [Google Scholar]