Abstract
The leader protein of cardioviruses, Theiler's murine encephalomyelitis virus (TMEV) and encephalomyocarditis virus (EMCV), is a multifunctional protein known to antagonize type I interferon expression and to interfere with nucleocytoplasmic trafficking of host proteins and mRNA. This protein plays an important role in the capacity of TMEV to establish persistent infection of the central nervous system. Mutant forms of the TMEV leader protein were generated by random mutagenesis and selected after retroviral transduction on the basis of the loss of the highly toxic nature of this protein. Selected mutations define a short C-terminal domain of the leader conserved in TMEV and Saffold virus but lacking in the EMCV leader and thus called the Theilo domain. Mutations in this domain had a dramatic impact on TMEV L protein activity. Like the zinc finger mutation, Theilo domain mutations affected all of the activities of the L protein tested: interferon gene transcription and IRF-3 dimerization antagonism, alteration of nucleocytoplasmic trafficking, nucleoporin 98 hyperphosphorylation, and viral persistence in vivo. This suggests that the Zn finger and the Theilo domain of the protein cooperate for function. Moreover, the fact that all of the activities tested were affected by these mutations suggests that the various leader protein functions are somehow coupled.
Theiler's murine encephalomyelitis virus (TMEV) is a neurotropic picornavirus that belongs to the Cardiovirus genus (reviewed by Brahic et al. [5]). The leader (L) protein of TMEV is a short (76 amino acids), very acidic protein. This protein helps the establishment of persistent TMEV infections in the central nervous system by antagonizing innate host defenses. It inhibits the transcription of type I interferon (IFN) and selected cytokine and chemokine genes, likely through inhibition of IRF-3 dimerization (19, 25, 28, 33, 34). It also interferes with nucleocytoplasmic trafficking of cellular proteins and blocks mRNA export from the nucleus (11, 28). These activities correlate with the phosphorylation of nucleoporin 98 (Nup98) (28).
The sequence of the L protein contains three domains: a zinc finger domain that was shown to bind divalent cations (7), an acidic central domain, and a Ser/Thr-rich domain (see Fig. 2). The L protein encoded by encephalomyocarditis virus (EMCV) shows 35% amino acid identity with the TMEV L protein. In the EMCV L protein, the zinc finger and the acidic domain are conserved but the C-terminal region encompassing the Ser/Thr-rich domain is lacking. In spite of this difference, L proteins of cardioviruses share the abilities to antagonize IFN production, to affect nucleocytoplasmic trafficking of mRNA and proteins, and to promote nucleoporin hyperphosphorylation (3, 16, 21, 25-27, 36).
FIG. 2.
Alignment of Cardiovirus L protein sequences and positions of selected mutations. Position of the domains defined in the L sequences are indicated above the alignment. Mutations of the L sequence of the DA1 TMEV strain are indicated: mutations selected after random mutagenesis for the loss of L toxicity are encircled in light gray. The 14 residues forming the Theilo domain are shown in bold characters in the DA1 sequence and framed. Among these, the Thr corresponding to a putative phosphorylation site is indicated. Residues mutated in the Zn, 3D-3A, and 3S-3A mutants are outlined. The extent of the Δ6-67 deletion is shown under the sequence.
Likely as a consequence of mRNA nuclear export inhibition, the TMEV L protein mediates shutoff of host protein synthesis and is very toxic when expressed in cells (2, 11, 28). In this work, we took advantage of this toxicity to select L mutants that lost the ability to shut off host protein synthesis in order to identify critical domains of the L protein and to test whether the multiple activities of the L protein can be uncoupled.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells were cultured as previously described (34). BALB/3T3, L929, and Phoenix-Eco cells were cultured in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (MP Biologicals), 100 IU of penicillin/ml, and 100 μg of streptomycin/ml. Phoenix-Eco cells were kindly provided by G. Nolan via the ATCC (SD-3444).
TMEV derivatives were produced by electroporation of BHK-21 cells (23) with genomic RNA transcribed in vitro from plasmids carrying the corresponding cDNAs. Virus DA1 was produced from plasmid pTMDA1 (10, 22, 23). Virus TM598 is a DA1 derivative carrying mutations that disrupt the zinc finger motif of the L protein (Zn mutation, previously referred to as an Lcys mutation) (34). FB05, FB06, and FB07 are DA1 derivatives bearing the M60V, W62R, and T63A mutations in the L coding region, respectively. Plasmids pFB05, pFB06, and pFB07, carrying the corresponding viral cDNA, were obtained as follows. A synthetic sequence made of annealed primers (TM896-TM897 for M60V, TM898-TM899 for W62R, and TM900-TM901 for T63A) was used to replace the segment of the L coding region between the unique XhoI and BsiWI restriction sites of pTM410, a subclone carrying the 5′ 1,730 nucleotides of the DA1 virus (31). Mutated L coding regions were then transferred into pTMDA1 derivates as BbrPI-BsiWI fragments. KJ6 is a DA1 derivative containing capsid mutations that adapt the virus to infect L929 cells with high efficiency (17). The corresponding zinc finger mutant is called TM659 (34). SB3, another derivative of KJ6, carries a deletion encompassing codons 6 to 67 of the L region (Δ6-67) (28). FB09, FB10, and FB11 are KJ6 derivatives bearing the M60V, W62R, and T63A mutations, respectively. These viruses were obtained as described above for the DA1 derivatives. KJ6 derivatives used at a multiplicity of infection of 2 PFU per cell yielded nearly 100% L929 cell infection, as assessed by VP1 immunostaining at 8 h postinfection (hpi) (data not shown).
Plasmid construction.
Plasmids were constructed in the pcDNA3 vector backbone (Invitrogen), in which gene transcription is under the control of the cytomegalovirus (CMV) immediate-early promoter.
A series of plasmids was designed to express fusion proteins constructed between the L variants and enhanced green fluorescent protein (eGFP). Plasmids pCER01, pCER02, and pCER03, encoding LWT-, LZn-, and LΔ-eGFP fusions, respectively, were described previously (28). Plasmids encoding the mutant LM60V-, LW62R-, LT63A-, and LP66S-eGFP fusions were obtained by cloning, into pCER01, the BamHI-BstEII mutated fragment of L amplified by PCR from genomic DNA of the clones selected after random mutagenesis (see below for the mutagenesis and mutant selection procedures used). To construct the T63D mutant encoded by pCER58, the L region was PCR amplified from pCER01 derivatives with primers TM247 and TM904 and the BamHI-BsiWI segment of the PCR product was cloned into pCER01. The 3D-3A mutant (D44A, D45A, D46A) encoded by pCER31 was obtained by cloning, into pCER01, the BamHI-XhoI fragment of the PCR product obtained after amplification of L with primers TM245 and TM831. The 3S-3A mutant (S51A, S53A, S54A) encoded by pCER35 was obtained by cloning, into pCER01, the BamHI-BsiWI fragment of the PCR product obtained after amplification of L with primers TM245 and TM832 (Table 1).
TABLE 1.
Plasmids carrying the viral cDNA of constructed mutants and plasmids expressing L protein variants
| Mutation | Plasmid carrying viral cDNA |
Plasmid expressing L protein variant |
|||
|---|---|---|---|---|---|
| WT capsid | Capsid adapted to L929 cells | PCMV-L | PCMV-L-eGFP | PCMV-L-IRES-eGFP | |
| None (WT) | pTMDA1 | pKJ6 | pTM553 | pCER01 | pTM625 |
| Zn/Lcysa | pTM598 | pTM659 | pTM592 | pCER02 | pTM626 |
| Δ (or Δ6-67)b | pSB3 (Δ6-67)b | pTM641 (Δ6-67)b | pCER03 | pTM624 | |
| M60V | pFB05 | pFB09 | pCER44c | pCER40c | pCER48c |
| W62R | pFB06 | pFB10 | pCER56c | pCER50c | pCER57c |
| T63A | pFB07 | pFB11 | pCER45c | pCER41c | pCER49c |
| P66S | pCER62c | pCER61c | pCER63c | ||
| T63D | pCER59c | pCER58c | pCER60c | ||
| 3D-3A | pCER30 | pCER31 | pCER70 | ||
| 3S-3A | pCER34 | pCER35 | pCER71 | ||
The formerly named Lcys mutation was renamed the Zn mutation for clarity.
Δ6-67 (where indicated), deletion of codons 6 to 67 of L. Δ, complete deletion of L.
Plasmid in which the first two AUG codons (codons 1 and 5) of the alternative L* open reading frame were changed to ACG without modification of the L amino acid sequence.
A second series of plasmids was designed to express the L variants alone: the LWT, LZn, and LΔ6-67 proteins are encoded by pTM553, pTM592, and pTM641, respectively (28). The plasmids expressing the other mutant proteins were obtained by replacing, in pTM533, the BamHI-BsiWI fragment of L with the mutated fragment taken from the first series of plasmids (Table 1). A third series of plasmids was derived from pTM625 (L-internal ribosome entry site [IRES]-eGFP). These plasmids coexpress the L variants and eGFP (28). Plasmids pTM626 and pTM624 express the LZn and LΔ proteins, respectively (28). The other L-IRES-eGFP constructs were obtained by cloning the mutated sequences from the first plasmid series, as BamHI-BsiWI fragments, into pTM625. Table 1 summarizes the properties of the various L-expressing plasmids used in this study. For cloning, PCR amplification was done with Pfu polymerase (Fermentas). The primers used for PCR and mutagenesis are presented in Table 2.
TABLE 2.
Primers used in this study
| Name | Sequence (5′→3′) | Usea |
|---|---|---|
| TM245 (s) | CTG GCT AAC TAG AGA ACC CA | Mutagenesis |
| TM247 (s) | TAA TAC GAC TCA CTA TAG GGA | Mutagenesis |
| TM281 (s) | ACC CGG GTC GAG GTA GGC GT | amplification |
| TM595 (as) | GGA CAC GCT GAA CTT GTG G | Mutagenesis |
| TM687 (as) | CGT CCA GCT CGA CCA GGA TG | Amplification |
| TM831 (as) | AAA GCT CGA GTT CGA AGG CCA GAA GAC GGC AGC GGC CAG GTC CAC ACA AAG AAG | Mutagenesis |
| TM832 (as) | AAA GCG TAC GAG CGG TAA GTC AGT CCA TTC CAT TGT TTC AGA TTG ATT GGC CGC GTT CGC AGG CCA GAA GAC GTC ATC | Mutagenesis |
| TM896 (s) | TCG AGC AAT CAA TCT GAA ACA GTG GAA TGG ACT GAC TTA CCG CTC | Mutagenesis |
| TM897 (as) | GTA CGA GCG GTA AGT CAG TCC ATT CCA CTG TTT CAG ATT GAT TGC | Mutagenesis |
| TM898 (s) | TCG AGC AAT CAA TCT GAA ACA ATG GAA CGG ACT GAC TTA CCG CTC | Mutagenesis |
| TM899 (as) | GTA CGA GCG GTA AGT CAG TCC GTT CCA TTG TTT CAG ATT GAT TGC | Mutagenesis |
| TM900 (s) | TCG AGC AAT CAA TCT GAA ACA ATG GAA TGG GCT GAC TTA CCG CTC | Mutagenesis |
| TM901 (as) | GTA CGA GCG GTA AGT CAG CCC ATT CCA TTG TTT CAG ATT GAT TGC | Mutagenesis |
| TM904 (as) | TAT CGC GTA CGA GCG GTA AGT CAT CCC ATT CCA TTG T | Mutagenesis |
| TM346 (s) | GCC GCT CTT CAC ACC CAT | qPCR virus |
| TM347 (as) | AGC AGG GCA GAA AGC ATC AC | qPCR virus |
| TM425 (s) | ACT CCC TGC TGC TTT GCC TA | qPCR RANTES |
| TM426 (as) | CCC ACT TCT TCT CTG GGT TGG | qPCR RANTES |
| TM642 (s) | ATG AAC AAC AGG TGG ATC CTC C | qPCR IFN-β |
| TM643 (as) | AGG AGC TCC TGA CAT TTC CGA A | qPCR IFN-β |
qPCR, quantitative PCR.
Plasmid pCS41 is a pcDNA3 derivative expressing the firefly luciferase gene (30). Plasmid pCER07 was derived from the retroviral vector pQCXIN (Clontech). In this vector, the CMV promoter was replaced with a tetracycline-inducible promoter (seven tet operators followed by a minimal CMV promoter) taken from plasmid pUHD10-3 (14). Transcription from this promoter is activated by the artificial rtTA transcription factor upon doxycycline addition (15). The BstEII restriction site present in the GAG region of the vector was eliminated by site-directed mutagenesis. The BamHI-XbaI fragment of pCER01 that encodes the L-eGFP fusion was then cloned into the modified vector to yield pCER07.
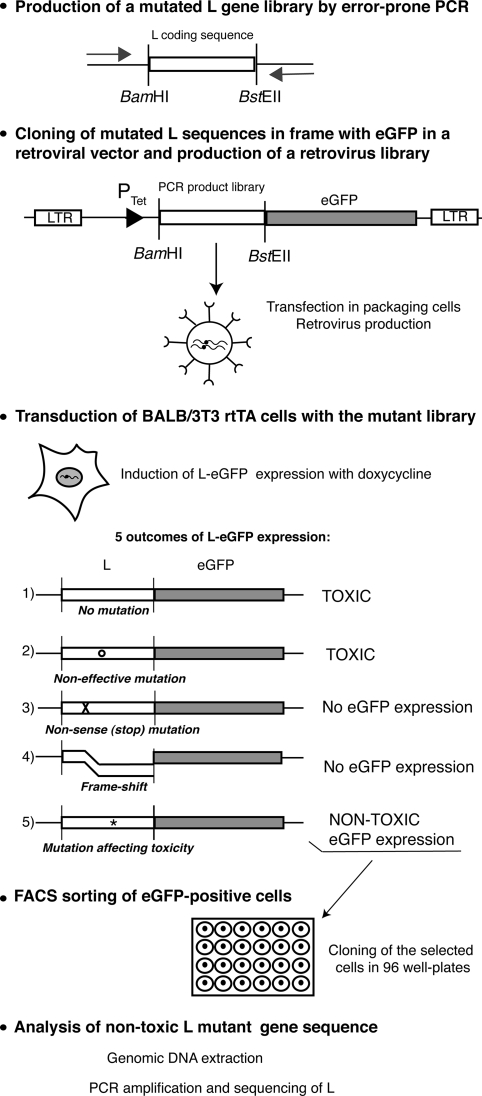
Random mutagenesis and mutant selection (see Fig. 1).
FIG. 1.
Strategy used to select L protein mutants lacking toxicity. Random mutagenesis was performed by error-prone PCR amplification of the L protein coding region. The PCR product was cloned into a retroviral vector as a BamHI-BstEII fragment to generate a library of mutant L sequences cloned in frame with the eGFP coding sequence. In these constructs, transcription of the L-eGFP fusion is driven by a tetracycline-inducible promoter. BALB/3T3-rtTA cells transduced with the constructs were treated with doxycycline to induce expression of the L-eGFP fusion. Only cells expressing a nontoxic mutant L-eGFP fusion are expected to be eGFP positive and able to grow. Thus, transduced cells were sorted for green fluorescence by flow cytometry and cloned. After growth, genomic DNA was extracted from cellular clones and the L coding sequence was PCR amplified and sequenced to identify mutations that abrogated L protein toxicity. LTR, long terminal repeat.
Mutagenic PCR was performed with primers TM247 and TM595 on pCER24, a variant of pCER01 in which the L* putative initiation codons were replaced with ACG codons. Error-prone PCR conditions included an imbalanced proportion of deoxynucleoside triphosphates (dCTP and dTTP at 0.5 mM and dGTP and dATP at 0.2 mM), the use of a Taq polymerase lacking 3′-5′ exonuclease activity (aTaq; Promega), and a high number (n = 35) of amplification cycles. The BamHI-BstEII fragment of the PCR product pool was cloned into the pCER07 retroviral vector, in frame with eGFP and under the control of the tetracycline-inducible promoter. A mutated L-eGFP retrovirus library was then produced by transfecting the pool of constructs into Phoenix-Eco cells by calcium phosphate transfection. Retroviruses were collected from the supernatant of the packaging cells 24 h posttransfection and used to transduce BALB/3T3-rtTA cells which stably express the tetracycline-activated rtTA transcription factor. Two days after transduction, expression of the L-eGFP constructs was induced with 5 μg/ml doxycycline. Three days after the addition of doxycycline, eGFP-expressing cells were sorted by flow cytometry (FACS Vantage; Becton Dickinson) and cloned in 96-well plates. Cells were sorted as FL1-positive and FL2-negative cells to minimize contamination by autofluorescent cells. The sorted cell population represented about 0.02% of the total cells. After 10 to 21 days of growth, genomic DNA of growing cell clones was extracted by boiling the cells at 100°C for 20 min. The L coding region was PCR amplified from these samples (Taq polymerase from Biotools) with primers TM687 and TM281, and PCR products were sequenced.
DNA transfection of BALB/3T3 cells and functional L testing.
Plasmid DNA was transfected with TransIT-LT1 transfection reagent (Mirus) according to the manufacturer's recommendations. We used 1.5 to 3 μl of transfection reagent with 0.5 to 1 μg of DNA to transfect about 300,000 cells grown in a well of a 24-well plate.
Lactate dehydrogenase (LDH) assay (Roche) was performed on the supernatant of BALB/3T3 cells transfected for 48 h with pcDNA3 derivatives expressing the L protein variants alone.
Inhibition of firefly luciferase expression (Luciferase Reporter Gene Assay; Roche) by L protein was assessed 7 h after cotransfection of BALB/3T3 cells with 100 ng of pCS41 and 400 ng of the pcDNA3 derivatives expressing the L protein variants.
Fluorescence-activated cell sorter analysis of eGFP mean fluorescence intensity was performed on BALB/3T3 cells 24 h after the transfection of plasmids expressing wild-type (WT) and mutated L-eGFP fusion proteins.
Localization of the mRNA pool in the cell was performed by oligo(dT)-probed in situ hybridization as described previously (6, 28).
Polypyrimidine tract binding protein (PTB) was localized in infected cells by indirect immunofluorescence as described by Paul and Michiels (25).
IRF-3 dimerization and Nup98 immunoblotting.
L929 cells were infected with 2 PFU per cell of WT or L mutant KJ6 derivatives or mock infected. IRF-3 dimerization was assessed as described previously (16).
Nup98 immunoblotting was performed as described by Park et al. (24). Briefly, at indicated times, crude cell lysates were harvested in Laemmli buffer and equal amounts of lysates were resolved on an 8% sodium dodecyl sulfate-polyacrylamide gel, transferred to a polyvinylidene fluoride membrane, and subjected to Western blot analysis with a rat anti-Nup98 antibody (2H10; Sigma Aldrich) used at 1/1,000 and an anti-rat antibody coupled to horseradish peroxidase (Dako). As a loading control, β-actin was detected with a mouse anti-β-actin monoclonal antibody (AC-15; Sigma Aldrich) used at 1/2,000 and a secondary anti-mouse antibody coupled to horseradish peroxidase (Dako). The signal was detected by chemiluminescence (Supersignal; Pierce).
Mice and infections.
Female 3-week-old FVB/N mice were purchased from Charles River Laboratories. Handling of mice and experimental procedures were conducted in accordance with national and institutional guidelines for animal care and use. Infections were initiated in mice by intracranial injection of 40 μl of serum-free medium containing 105 PFU of the indicated virus derived from the DA1 strain. Control mice were injected with 40 μl of serum-free culture medium. At 45 days postinfection, total RNA was extracted from the spinal cords of infected mice as previously described (25).
Real-time quantitative reverse transcription (RT)-PCR.
Total RNA preparations, RT, and real-time PCRs were performed as previously described (25). The sequences of the primers used are shown in Table 2.
RESULTS
Selection of randomly generated L mutants lacking cytotoxicity by retrovirus-mediated transduction.
We and others previously observed that L expression was cytotoxic (2, 28). Owing to this toxicity, in spite of many efforts, all of our attempts to generate stably transfected or transduced cells expressing L or L-eGFP constructs were unsuccessful (data not shown). Here, we exploited this toxicity of L-eGFP fusions to select nontoxic L mutants after random mutagenesis of the L moiety of the construct.
To limit the selection of frameshift mutations, mutagenic PCR was performed on an L coding sequence lacking the two potential initiation codons present in the L* reading frame which overlaps the L coding region (29, 32). The PCR product was cloned in frame with eGFP, under the control of a tetracycline-inducible promoter, into a retroviral vector (Fig. 1). A retroviral vector library carrying the mutated L-eGFP fusions was thus generated and used to transduce BALB/3T3-rtTA cells stably expressing the tetracycline-inducible rtTA transcriptional activator. Two days after transduction, L-eGFP gene expression was induced by treatment of the cells with 5 μg/ml doxycycline for 72 h. Cells displaying green fluorescence were sorted by flow cytometry and cloned (Fig. 1). Only cells transduced with the construct were expected to display green fluorescence. On the one hand, cells receiving a WT L-eGFP construct or a construct carrying mutations that failed to affect L toxicity were expected to die and thus to be counterselected. On the other hand, stop codon or frameshift mutations in the L moiety of the fusion were expected to prevent eGFP expression. Thus, growing cells expressing eGFP were expected to carry mutations that eliminated L toxicity. After growth, the L coding region of such clones was PCR amplified from genomic DNA and sequenced.
L coding regions could be amplified from 74 out of 145 growing clones. Twenty-one mutants with different L sequences were obtained (Tables 3 and 4). That the mutagenesis procedure was saturating or close to saturation was suggested by the facts that (i) several identical mutants were selected from two independent mutagenesis experiments and (ii) mutations in a single codon (M60) yielded selected mutants with four different coding capacities: T, L, V, and I. Interestingly, all of the selected mutations clustered in the C-terminal part of the L sequence, between codons M60 and M73 (Fig. 2 and Table 3). Six of the 21 mutants possessed more than one mutation, but at least one of these mutations was located in the M60-to-M73 domain (Table 4). This C-terminal domain of the TMEV L sequence is conserved in viruses from the Theilovirus group but is lacking in the EMCV L sequence. We thus propose to name this domain the Theilo domain (Fig. 2).
TABLE 3.
Selected single mutations
| Position and amino acid change | Nucleotide change | na |
|---|---|---|
| M60 | ||
| M60L | ATG→TTG | 1 |
| M60V | ATG→GTG | 2 + 2 |
| M60T | ATG→ACG | 1 + 1 |
| M60I | ATG→ATT | 5 |
| E61, E61G | GAA→GGA | 2 |
| W62, W62R | TGG→CGG | 2 + 2 |
| T63 | ||
| T63A | ACT→GCT | 1 |
| T63P | ACT→CCT | 4 |
| L65, L65*b | TTA→TAA | 3 + 4 |
| P66 | ||
| P66S | CCG→TCG | 1 |
| P66L | CCG→CTG | 3 + 2 |
| D70 | ||
| D70G | GAT→GGT | 4 + 1 |
| D70E | GAT→GAG | 4 |
| M73, M73T | ATG→ACG | 6 |
n + n means that the mutation was obtained in two independent mutagenesis experiments.
* stands for the stop codon. It is not clear how this mutation yielded eGFP-positive clones. We hypothesize that insertion of the stop codon at this position might induce translation reinitiation at AUG 73.
TABLE 4.
Selected multiple mutations
| Amino acid changesa | n |
|---|---|
| K4N, R69H, M73K | 1 |
| Y7N, L38L, P66S | 1 |
| A16G, E32G, E58N, V68E | 1 |
| D18V, P66L | 1 |
| L26*, L67P | 3 |
| W49G, M60T | 4 |
| E61*, T63T | 3 |
* stands for the stop codon. It is not clear how this mutation yielded eGFP-positive clones. We hypothesize that insertion of the stop codon at this position might induce translation reinitiation at AUG 73. Underlined changes occur in the defined Theilo domain.
Selected mutations in the C-terminal Theilo domain of L impair L-mediated inhibition of mRNA export and shutoff of host cell protein expression.
The M60V, W62R, T63A, and P66S mutations were selected for further analysis. The M60V and W62R mutations were selected because cells expressing the corresponding L-eGFP fusions exhibited high fluorescence intensity and fair morphology, suggesting minimal L toxicity. The T63A mutation was selected because it affected a Thr residue that corresponded to a potential phosphorylation site. However, the selected cell clone harboring this mutation grew very poorly, suggesting that this mutation only partly affected L activity. Additional mutations, generated by site-directed mutagenesis, were included in the analysis (outlined in dark gray in the L sequence shown on Fig. 2). These included (i) the zinc finger mutation obtained previously (34); (ii) the 3D-3A mutation obtained by substituting Ala residues for Asp residues D44, D45, and D46; (iii) the 3S-3A mutation obtained by substituting Ala residues for Ser residues S51, S53, and S54; and (iv) the potential phosphomimetic mutation of T63 (T63D) (Fig. 2).
To characterize the impact of the selected mutations on the activities of the L protein, the various mutated L sequences were introduced into the pcDNA3 vector, allowing expression of the L protein alone (pcDNA3 series); into the same vector as an L-eGFP fusion protein (L-eGFP series); or as a bicistronic construct, allowing coexpression of L and eGFP (L-IRES-eGFP series) (Table 1).
As a first clue that L mutants selected after random mutagenesis also lost toxicity when expressed without eGFP, the pcDNA3 derivatives expressing L variants alone were transfected into BALB/3T3 cells and the toxicity of the mutants was tested by measuring LDH activity in the supernatant of transfected cells. As expected, all of the mutants selected after random mutagenesis displayed reduced toxicity. Of the mutants constructed, the 3S-3A mutant showed intermediate toxicity, while the T63D mutant retained WT toxicity (Table 5).
TABLE 5.
LDH activity in the culture medium of BALB/3T3 cells transfected with L-expressing plasmids
| Expressed L variant | LDH activitya (mean ± SD) |
|---|---|
| WT | 3.19 ± 0.19 |
| Zn | 1.16 ± 0.44 |
| Δ6-67 | 1.00 ± 0.17 |
| M60V | 1.02 ± 0.23 |
| W62R | 1.06 ± 0.57 |
| T63A | 1.38 ± 0.53 |
| T63D | 3.25 ± 0.34 |
| P66S | 1.17 ± 0.16 |
| 3D-3A | 1.51 ± 0.12 |
| 3S-3A | 3.19 ± 1.04 |
LDH activity was measured in the culture supernatant of BALB/3T3 cells transfected for 72 h with pcDNA3 derivatives expressing the constructs indicated. Activity was measured in six samples and standardized to that of the Δ6-67 construct.
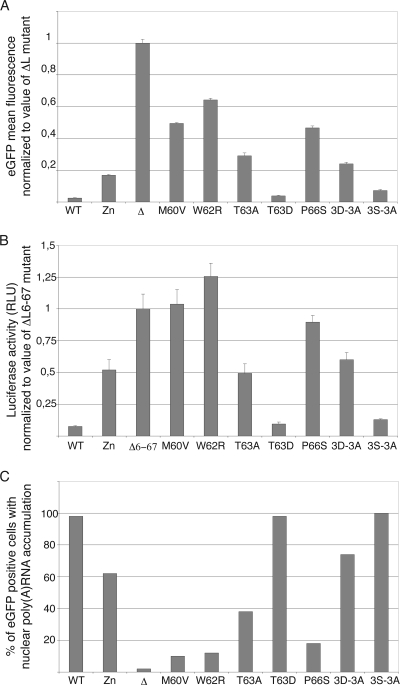
Shutoff of protein expression by the L protein was quantified either by measuring the mean fluorescence intensity of cells transfected with the L-eGFP fusion-expressing constructs or by measuring inhibition of luciferase activity in cells cotransfected with pCS41 and the pcDNA3 derivatives expressing the L variants (Fig. 3A and B).
FIG. 3.
Effects of transiently expressed mutated L proteins on reporter gene expression and on mRNA export from the nucleus. (A) Inhibition of L-eGFP expression by the L moiety of the fusion protein: eGFP mean fluorescence intensity was measured in BALB/3T3 cells 24 h after transfection of the constructs expressing L-eGFP fusions. Histograms show the means and standard deviations of fluorescence intensities measured in eGFP-positive cells, normalized to the average of the control value (ΔL mutant). Shown is a representative experiment done in triplicate. (B) Inhibition of luciferase expression by mutant L proteins: luciferase activity was measured in BALB/3T3 cells 7 h after cotransfection of pCS41 (expressing luciferase) and the L protein expression plasmids indicated (pcDNA3 series). Histograms show the means and standard deviations of the relative luciferase activities normalized to the average of the control value (ΔL6-67 mutant). Since no internal standard for transfection efficiency was available, the experiment was repeated several times with at least two different plasmid DNA batches for each sample. Shown are the data for a representative experiment done in triplicate. (C) The effect of L mutants on mRNA export from the nucleus was examined by in situ hybridization with an oligo(dT) probe in BALB/3T3 cells transfected for 16 h with the L-IRES-eGFP plasmids. Histograms show the percentage of eGFP-positive cells exhibiting mRNA nuclear retention, as assessed by confocal microscopy. Counts for one representative experiment out of three are presented. RLU, relative light units.
L protein inhibition of mRNA export from the nucleus was measured, by in situ hybridization with an oligo(dT) probe, for cells transfected with plasmids coexpressing the L variants and eGFP (Fig. 3C).
Interestingly, the M60V, W62R, and P66S mutations that were selected after random mutagenesis had a profound impact on the L activities tested. These mutations completely prevented L-mediated inhibition of luciferase or eGFP expression and dramatically reduced L-mediated mRNA nuclear export inhibition. Mutations 3D-3A and T63A had an intermediate, yet important, impact on L activity, as did the previously characterized mutation of the Zn finger. Interestingly, mutation of the Thr 63 residue into the phosphomimetic Asp residue (T63D) fully restored L activity, suggesting that phosphorylation of this residue might indeed regulate L protein activity, as was suggested for the T47 residue of the EMCV L protein (36). Surprisingly, mutation of the Ser cluster (3S-3A mutant) had little impact on L activity, although this cluster is well conserved among TMEV strains. Taken together, our data show that mutations selected after random mutagenesis, more noticeably, mutations M60V, W62R, and P66S, targeted a critical domain of the L protein required for inhibition of mRNA export and for shutoff of protein expression.
To check whether the Theilo domain extends to residue M73, as was defined by mutagenesis and sequence alignments (Fig. 2), we tested the influence of the C-terminal D70G, D70E, and M73T mutations by using the inhibition of luciferase expression assay. Luciferase activities in cells transfected with these mutants were 120% ± 8% (D70G), 114% ± 19% (D70E), and 130% ± 12% (M73T) of that measured in cells transfected with the Δ6-67 mutant, which was taken as a reference. In this series of experiments, the luciferase activity was 8% ± 1% for the WT, 131% ± 21% for M60V, and 123% ± 4% for W62R. The impact of the D70G, D70E, and M73T mutations on L protein activity was thus close to that of the M60V and W62R mutations.
Influence of selected L mutations in the context of viral infections.
To test whether the Theilo domain mutations also affected other functions of the L protein such as the inhibition of cytokine and chemokine gene transcription, the perturbation of nucleocytoplasmic protein trafficking and viral persistence in vivo, mutations M60V, W62R, and T63A were introduced into the viral genome. These mutations were chosen because (i) they greatly affected L activity (M60V and W62R), (ii) they could be inserted into the viral genome without affecting the amino acid sequence of the L* protein encoded by an overlapping alternative reading frame, and (iii) they were distant (positions −16, −14, and −13, respectively) from the L/VP4 cleavage site of the viral polyprotein and were thus not expected to affect this cleavage. These mutations were introduced into plasmids carrying the genomes of viruses DA1 (WT) and KJ6, a DA1 derivative adapted to infect L929 cells with high efficiency (17). The corresponding viruses were produced and titrated on BHK-21 cells in parallel with the WT virus and with the previously constructed Zn and Δ6-67 mutants. In BHK-21 cells, which are reportedly deficient in type I IFN production, L mutant viruses formed plaques that were similar in size to those formed by their respective parental strains (DA1 and KJ6). KJ6 derivatives were also tested for plaque formation on IFN-proficient L929 cells. On the latter cells, KJ6 (WT) and FB11 (T63A) formed medium-size plaques; TM659 (Zn), FB09 (M60V), and FB10 (W62R) formed small plaques; and SB3 (LΔ6-67) formed minute plaques. These observation suggest that the L mutations tested, with the exception of the T63A mutation, affected the IFN antagonist activity of the L protein.
Replication of the mutated viruses in single-cycle L929 cell infection, as measured by real-time RT-PCR, was similar to that of the WT virus (Fig. 4A). Western blot analysis performed on infected L929 cell extracts confirmed that L protein expression and processing were WT for the M60V and T63A mutants. In the case of the W62R mutant, the amount of mature L protein detected by Western blot analysis was reduced, suggesting incomplete processing and/or protein instability (Fig. 4E). Alternatively, this mutation might have decreased the affinity of the antibody for the L protein.
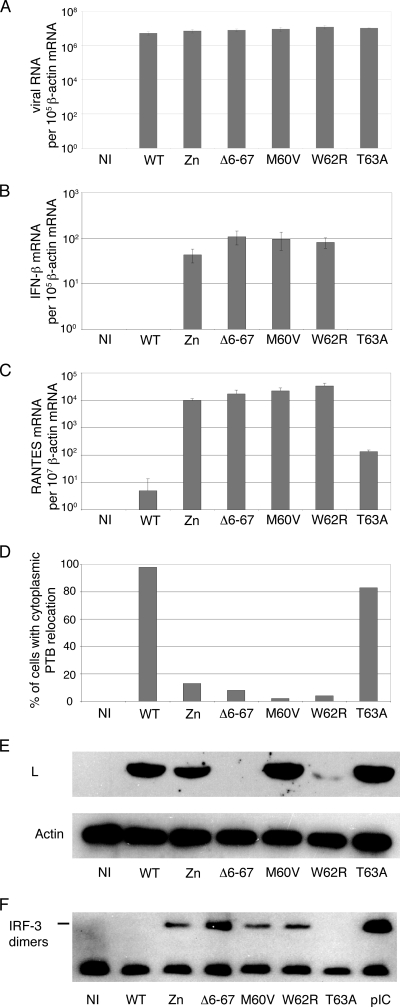
FIG. 4.
Influence of L mutations in the context of viral infections. L929 cells were mock infected (NI) or infected with 2 PFU per cell of KJ6 derivatives expressing the WT L protein or the mutant L proteins indicated. (A to C) Viral RNA, IFN-β mRNA, and RANTES mRNA levels measured at 9 hpi by real-time RT-PCR and normalized to the amount of β-actin mRNA for each sample. Histograms show the means and standard deviations of relative RNA amounts for a triplicate infection experiment. (D) PTB detection by immunofluorescence in L929 cells infected for 5 h. Histograms show counts of cells showing redistribution of PTB to the cytoplasm (images are shown in Fig. 6). (E) Western blot analysis showing L protein and β-actin (control) detection in L929 cells at 16 hpi. (F) Western blot analysis of a nondenaturing gel showing IRF-3 dimer formation in infected L929 cells 9 hpi with the indicated viruses or 7 h after poly(I:C) transfection (pIC).
As shown in Fig. 4B and C, the M60V and W62R point mutations dramatically affected the ability of the virus to block IFN-β or RANTES gene transcription and, accordingly, to inhibit IRF-3 dimerization (Fig. 4F). The effect of these two mutations was almost as important as that of the Δ6-67 deletion and much more prominent than that of the T63A mutation. For most of the phenotypes tested, the Zn mutation appeared to be less effective than the M60V and W62R mutations.
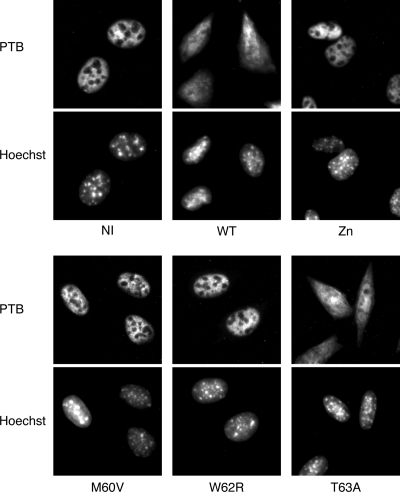
Perturbation of nucleocytoplasmic trafficking, as measured by the cytoplasmic redistribution of the nuclear PTB, was also reduced strongly for the M60V and W62R mutant viruses, to a lesser extent for the Zn mutant, and hardly, if at all, for the T63A mutant (Fig. 5).
FIG. 5.
PTB distribution in infected cells. PTB localization was examined by immunofluorescence in L929 cells infected for 5 h with WT and L mutant KJ6 derivatives. Cell nuclei were stained with Hoechst 33258. NI, mock infected.
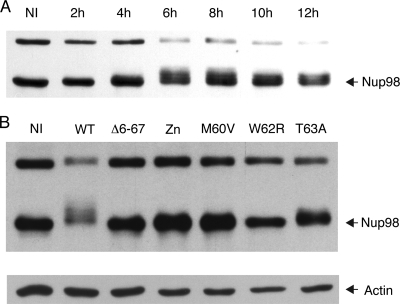
The L proteins of both EMCV and TMEV were recently shown to trigger hyperphosphorylation of nucleoporins (3, 27, 28). Influence of L mutations on Nup98 phosphorylation paralleled the effect observed for the other phenotypes. Infection of L929 cells with KJ6 (LWT) strongly induced Nup98 phosphorylation, which peaked at 6 to 8 hpi (Fig. 6A). Nup98 phosphorylation was detectable at 6 and 8 hpi in cells infected with the T63A mutant (although to a much lesser extent than in the case of the WT virus) and not detectable in cells infected with the M60V and W62R mutants (Fig. 6B and data not shown).
FIG. 6.
Phosphorylation of Nup98 in cells infected with L mutant viruses. Lysates from L929 cells infected with 2 PFU per cell of L mutant KJ6 derivatives were analyzed by Western blot analysis to determine the phosphorylation state of Nup98. (A) Kinetics of Nup98 phosphorylation in L929 cells infected with KJ6 (LWT). (B) Nup98 detection in cell lysates taken at 8 hpi with the indicated mutant KJ6 derivatives. The band detected at a higher molecular weight likely corresponds to the 186-kDa precursor of Nup98 and Nup96 (13). β-Actin was detected as a loading control. NI, mock infected.
Finally, we checked the ability of the mutants to persist in the spinal cords of infected FVB/N mice. Viral loads were measured by real-time RT-PCR in spinal cord RNA samples collected 45 days postinfection. As expected, the M60V and W62R mutations affected viral persistence, as did the Zn mutation. Viruses carrying the T63A mutation were less attenuated and only exhibited a moderate decrease in viral load compared to the WT virus (Fig. 7).
FIG. 7.
Persistence of L mutant viruses in vivo. Viral RNA levels detected by real-time RT-PCR in the spinal cords of FVB/N mice infected for 45 days with 105 PFU of the DA1 virus derivatives indicated. Columns show the results for individual mice. ND, not detected; NI, mock infected.
Important viral load variation from mouse to mouse suggested the emergence and selection of revertant viruses that regained a functional L protein. However, sequencing of viral L coding regions that were RT-PCR amplified from spinal cord RNA samples confirmed the nature of the infecting viruses and failed to reveal the presence of revertants.
DISCUSSION
We used random mutagenesis and a retrovirus transduction strategy to select TMEV L protein mutants that lost their toxicity. Although mutations were selected by using an L-eGFP fusion, selected mutations also impaired L protein function when expressed without eGFP or when introduced back into the viral genome. The screening strategy appeared to be very efficient, as it allowed the identification of point mutations that turned out to have a major impact on the L protein functions tested, including viral persistence.
Interestingly, selected mutations clustered in a 14-amino-acid region that defines a new functional domain of L, close to the C terminus of the protein. This was unexpected because this domain is not conserved in the L proteins of EMCV strains. It was also surprising that the screening procedure failed to yield zinc finger mutations, as mutations in this domain are known to have a major influence on L function (26, 27, 34, 36). However, we observed previously that the Zn mutant had some residual activity (28) and we believe that this residual activity might keep L toxicity too high to allow its stable expression in cells. Among the mutations selected after random mutagenesis, only the T63A mutation had a residual activity that was comparable to or even higher than that of the Zn mutant (Fig. 3). On the one hand, the cell clone expressing this T63A mutant was moribund when collected after the screening procedure; possibly, this cell clone also expressed very low amounts of the protein. On the other hand, this mutation had a greater relative impact on L protein activity when expressed by transfection than when expressed in the viral context (compare Fig. 3 and 4).
Among the mutations tested, M60V, W62R, and P66S showed the most dramatic effect on L protein functions. This effect was more pronounced than that of the zinc finger mutation or that of the 3D-3A mutation, affecting three conserved acidic residues of the central domain of the protein. Thus, our mutants define a new domain of the protein that is critically important for function. This domain, extending from M60 to M73, is highly conserved in all of the TMEV strains sequenced to date (Fig. 2). Interestingly, in spite of some variation upstream of M60, the defined domain is also conserved in the recently described strains of Saffold virus (1, 4, 8, 12, 18, 20, 35), a highly prevalent human Cardiovirus related to TMEV (alignment in Fig. 2). However, this domain is lacking in the EMCV L protein. Thus, the identified cluster forms an extra microdomain conserved in Theilovirus L proteins but lacking in the EMCV L protein. Accordingly, we called this domain the Theilo domain.
A goal of this work was to test whether the various functions defined for the TMEV L protein are coupled. It is striking that mutations in the Theilo domain affected all of the functions of the L protein tested. In view of the strongly diverging physicochemical properties of tryptophan and arginine, the W62R mutation could well affect the overall folding of the protein. In contrast, the M60V mutation is not expected to modify the global folding of the protein. This mutation failed to affect virus infectivity and L protein production or stability (Fig. 4E). Yet, the M60V mutation profoundly impacted all of the L protein activities tested. On the other hand, the mutation that affects the Zn finger of the protein also impairs these functions. This suggests that the activity of the L protein depends on the cooperation of its N- and C-terminal domains. Resolution of the Cardiovirus L protein three-dimensional structure, which is in progress (9), will be valuable in the further interpretation of the data.
The data also suggest that activities as seemingly unrelated as transcriptional inhibition of cytokine genes and hyperphosphorylation of nucleoporins might be connected. Indeed, none of the mutations clearly allowed the uncoupling of L protein phenotypes. Influence of the Zn mutation on mRNA export inhibition was relatively modest compared to its effect on cytokine expression. However, it is not clear whether such small differences reflect functional specificities of the Zn finger and of other domains of the proteins or whether they simply reflect sensitivity differences in the functional tests that were used.
In conclusion, mutations selected after random mutagenesis define a new domain of the Theiler's virus L protein that is critical for activity. This domain likely cooperates with the Zn finger domain of the protein to accomplish the many activities of the protein, which appear to be connected. Since this domain is conserved in the L protein of Saffold virus, it can be anticipated that the critical residues defined in this work would represent excellent targets for Saffold virus attenuation with the prospect of producing a vaccinal strain.
Acknowledgments
We are indebted to André Tonon for outstanding knowledge and expert help in fluorescence-activated cell sorter analysis and cell sorting. We thank Muriel Minet for always excellent technical assistance.
C.R. was a fellow of the Belgian FNRS (National Fund for Scientific Research). F.S. was supported by an FSR fellowship from the Université catholique de Louvain. S.V.H. was supported by a mosaic grant from the Netherlands Organization for Scientific Research (NWO-017.002.025). This work was supported by the National Fund for Medical Scientific Research (FRSM), by Crédits aux chercheurs of the FNRS, by the FSR of the University of Louvain, by the DIANE program of the Belgian Walloon region, and by Actions de Recherche Concertées, Communauté Française de Belgique. Work in the laboratory of F.J.M.V.K. was partly supported by a grant from the Netherlands Organization for Scientific Research (NWO-VIDI-917.46.305).
Footnotes
Published ahead of print on 26 August 2009.
REFERENCES
- 1.Abed, Y., and G. Boivin. 2008. New Saffold cardioviruses in 3 children, Canada. Emerg. Infect. Dis. 14:834-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baida, G., B. Popko, R. L. Wollmann, S. Stavrou, W. Lin, M. Tretiakova, T. N. Krausz, and R. P. Roos. 2008. A subgenomic segment of Theiler's murine encephalomyelitis virus RNA causes demyelination. J. Virol. 82:5879-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardina, M. V., P. V. Lidsky, E. V. Sheval, K. V. Fominykh, F. J. van Kuppeveld, V. Y. Polyakov, and V. I. Agol. 2009. Mengovirus-induced rearrangement of the nuclear pore complex: hijacking cellular phosphorylation machinery. J. Virol. 83:3150-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blinkova, O., A. Kapoor, J. Victoria, M. Jones, N. Wolfe, A. Naeem, S. Shaukat, S. Sharif, M. M. Alam, M. Angez, S. Zaidi, and E. L. Delwart. 2009. Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J. Virol. 83:4631-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahic, M., J. F. Bureau, and T. Michiels. 2005. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu. Rev. Microbiol. 59:279-298. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty, P., N. Satterly, and B. M. Fontoura. 2006. Nuclear export assays for poly(A) RNAs. Methods 39:363-369. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. H., W. P. Kong, and R. P. Roos. 1995. The leader peptide of Theiler's murine encephalomyelitis virus is a zinc-binding protein. J. Virol. 69:8076-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, C. Y., A. L. Greninger, K. Kanada, T. Kwok, K. F. Fischer, C. Runckel, J. K. Louie, C. A. Glaser, S. Yagi, D. P. Schnurr, T. D. Haggerty, J. Parsonnet, D. Ganem, and J. L. DeRisi. 2008. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc. Natl. Acad. Sci. USA 105:14124-14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornilescu, C. C., F. W. Porter, K. Q. Zhao, A. C. Palmenberg, and J. L. Markley. 2008. NMR structure of the mengovirus leader protein zinc-finger domain. FEBS Lett. 582:896-900. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, J. B., A. M. Pappenheimer, and S. Richardson. 1952. Observations on encephalomyelitis of mice (DA strain). J. Exp. Med. 96:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delhaye, S., V. van Pesch, and T. Michiels. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78:4357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drexler, J. F., L. K. Luna, A. Stocker, P. S. Almeida, T. C. Ribeiro, N. Petersen, P. Herzog, C. Pedroso, H. I. Huppertz, C. Ribeiro Hda, Jr., S. Baumgarte, and C. Drosten. 2008. Circulation of 3 lineages of a novel Saffold cardiovirus in humans. Emerg. Infect. Dis. 14:1398-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontoura, B. M., G. Blobel, and M. J. Matunis. 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144:1097-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossen, M., S. Freundlieb, G. Bender, G. Muller, W. Hillen, and H. Bujard. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268:1766-1769. [DOI] [PubMed] [Google Scholar]
- 16.Hato, S. V., C. Ricour, B. M. Schulte, K. H. Lanke, M. de Bruijni, J. Zoll, W. J. Melchers, T. Michiels, and F. J. van Kuppeveld. 2007. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 9:2921-2930. [DOI] [PubMed] [Google Scholar]
- 17.Jnaoui, K., and T. Michiels. 1998. Adaptation of Theiler's virus to L929 cells: mutations in the putative receptor binding site on the capsid map to neutralization sites and modulate viral persistence. Virology 244:397-404. [DOI] [PubMed] [Google Scholar]
- 18.Jones, M. S., V. V. Lukashov, R. D. Ganac, and D. P. Schnurr. 2007. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J. Clin. Microbiol. 45:2144-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, W. P., G. D. Ghadge, and R. P. Roos. 1994. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc. Natl. Acad. Sci. USA 91:1796-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, Z., A. S. Kumar, M. S. Jones, N. J. Knowles, and H. L. Lipton. 2008. Phylogenetic analysis of the species Theilovirus: emerging murine and human pathogens. J. Virol. 82:11545-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lidsky, P. V., S. Hato, M. V. Bardina, A. G. Aminev, A. C. Palmenberg, E. V. Sheval, V. Y. Polyakov, F. J. van Kuppeveld, and V. I. Agol. 2006. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 80:2705-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAllister, A., F. Tangy, C. Aubert, and M. Brahic. 1989. Molecular cloning of the complete genome of Theiler's virus, strain DA, and production of infectious transcripts. Microb. Pathog. 7:381-388. [DOI] [PubMed] [Google Scholar]
- 23.Michiels, T., V. Dejong, R. Rodrigus, and C. Shaw-Jackson. 1997. Protein 2A is not required for Theiler's virus replication. J. Virol. 71:9549-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, N., P. Katikaneni, T. Skern, and K. E. Gustin. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 82:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul, S., and T. Michiels. 2006. Cardiovirus leader proteins are functionally interchangeable and have evolved to adapt to virus replication fitness. J. Gen. Virol. 87:1237-1246. [DOI] [PubMed] [Google Scholar]
- 26.Porter, F. W., Y. A. Bochkov, A. J. Albee, C. Wiese, and A. C. Palmenberg. 2006. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 103:12417-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter, F. W., and A. C. Palmenberg. 2009. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 83:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricour, C., S. Delhaye, S. V. Hato, T. D. Olenyik, B. Michel, F. J. van Kuppeveld, K. E. Gustin, and T. Michiels. 2009. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein. J. Gen. Virol. 90:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roos, R. P., W. P. Kong, and B. L. Semler. 1989. Polyprotein processing of Theiler's murine encephalomyelitis virus. J. Virol. 63:5344-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommereyns, C., S. Paul, P. Staeheli, and T. Michiels. 2008. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Eyll, O., and T. Michiels. 2000. Influence of the Theiler's virus L* protein on macrophage infection, viral persistence, and neurovirulence. J. Virol. 74:9071-9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Eyll, O., and T. Michiels. 2002. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J. Virol. 76:10665-10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Pesch, V., and T. Michiels. 2003. Characterization of interferon-alpha 13, a novel constitutive murine interferon-alpha subtype. J. Biol. Chem. 278:46321-46328. [DOI] [PubMed] [Google Scholar]
- 34.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zoll, J., S. Erkens Hulshof, K. Lanke, F. Verduyn Lunel, W. Melchers, E. Schoondermark-van de Ven, M. Roivanen, J. Galama, and F. J. van Kuppeveld. 2009. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 5:e1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoll, J., W. J. Melchers, J. M. Galama, and F. J. van Kuppeveld. 2002. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-κB. J. Virol. 76:9664-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]