Abstract
The human recombinase hRad51 is a key protein for the maintenance of genome integrity and for cancer development. Polymerization and depolymerization of hRad51 on duplex DNA were studied here using a new generation of magnetic tweezers, measuring DNA twist in real time with a resolution of 5°. Our results combined with earlier structural information suggest that DNA is somewhat less extended by hRad51 than by RecA (4.5 vs. 5.1 Å per base pair) and untwisted by 18.2° per base pair. They also confirm a stoichiometry of 3–4 bp per protein in the hRad51-dsDNA nucleoprotein filament. At odds with earlier claims, we show that after initial deposition of a multimeric nucleus, nucleoprotein filament growth occurs by addition/release of single proteins, involving DNA twisting steps of 65° ± 5°. Simple numeric simulations show that this mechanism is an efficient way to minimize nucleoprotein filament defects. Nucleoprotein filament growth from a preformed nucleus was observed at hRad51 concentrations down to 10 nM, whereas nucleation was never observed below 100 nM in the same buffer. This behavior can be associated with the different stoichiometries of nucleation and growth. It may be instrumental in vivo to permit efficient continuation of strand exchange by hRad51 alone while requiring additional proteins such as Rad52 for its initiation, thus keeping the latter under the strict control of regulatory pathways.
Keywords: homologous recombination, magnetic tweezers, nucleation and growth, single molecule, mechanoenzyme
Homologous recombination (HR) is the main pathway for accurate repair of DNA double-strand breaks and maintenance of genome integrity. It is also essential for overcoming stalled replication forks. Rad51 is the key protein of HR and the recombinational DNA repair process in eukaryotes (1–3). First, Rad51 polymerizes at or near one end of a DNA break, forming a right-handed helical nucleoprotein filament. This nucleoprotein filament then searches the genome for a homologous DNA sequence. Once the nucleoprotein filament and homologous sequence are synapsed, new DNA synthesis can proceed using the homologous sequence as template. Finally, this structure is resolved to repair the DNA break. Despite intense efforts, some divergences remain about the formation of the nucleoprotein filament. Nucleoprotein filaments assembled onto ssDNA or dsDNA in the presence of ATP as a cofactor display very similar helical structures on electron microscopy (EM), with a rise per base pair of 4.7 Å (4). This structure also looks much like that achieved with RecA, the bacterial homologue of Rad51 (5). The stoichiometry of both proteins has been shown to depend on experimental conditions (6). Reported values range from 3 to 7 bp per protein for RecA (5, 7–9) and 2 to 3 and 4 to 5 bp per protein for Rad51 (4, 10–12). The kinetics of RecA and Rad51 polymerization were recently the subject of several single-molecule studies (13–21). There is consensus about a nucleation and growth mechanism for both RecA (14–16, 22) and Rad51 (19–21), and about a highly cooperative nucleation step requiring the simultaneous binding of several proteins. The stoichiometry of this nucleus, however, is not well established for Rad51, and values ranging from 2–3 (21) to 4–6 (19, 20) protein monomers were proposed. The existence of a similar cooperativity during the growth phase remains subject to debate: using single-molecule FRET, Joo et al. reached the conclusion that RecA binds to dsDNA by monomers (17), whereas from magnetic tweezers (MT) experiments and Monte Carlo simulations, van der Heijden et al. concluded that the Rad51 binding unit is a tetramer or a pentamer, as for nucleation (19).
To shed new light on these open questions, we applied here a new generation of single-molecule nanomanipulation devices, free rotation magnetic tweezers (FRMT), to follow the polymerization of human Rad51 (hRad51) onto DNA. In contrast with conventional MT, which lock torsion of DNA and measure its length, FRMT directly measure the torsional state of DNA during polymerization. Because the binding of one hRad51 is expected to stretch the dsDNA molecule on the order of nanometers and unwind the dsDNA by approximately 30°–75° [estimated from the twist of 18.6 bp per turn and a stoichiometry between 2 and 5 bp per protein, measured in yeast Rad51 filament (10)], a measurement of the torsional state should be a more accurate reporter of protein binding than elongation.
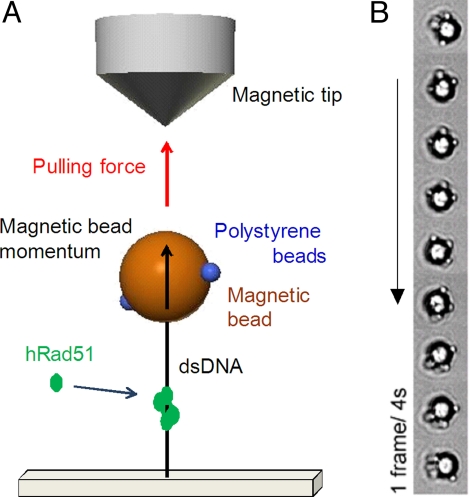
The twisting of DNA during an enzymatic process was already studied by 2 different methods. In a study by Gore et al. (23), a rotor bead “tracer” was used to analyze the DNA gyrase mechanochemics; this technique required a complex DNA construct, and its angular resolution was limited to several tens of degrees. The FRMT system (Fig. 1A) uses a simpler concept, first proposed to study the activity of polymerase (24). As in MT, a DNA molecule is attached at one end to a surface and at the other end to a magnetic bead pulled by a magnetic field gradient. Unlike in MT, the direction of the magnetic field is collinear to the extension of DNA to allow free rotation. The attachment of small nanobeads onto the large magnetic bead allows us to track its angular position. As compared with the first version proposed by Harada et al. (24), we optimized the DNA–bead construct and tracking algorithm (see SI Text and Figs. S1–S6), improving angular and time resolution down to 5° and 40 ms, respectively. As in MT, the pulling force was measured from transverse fluctuations of the bead position (25).
Fig. 1.
Experimental design. (A) A single DNA molecule is held between a coverslip and a magnetic bead, which is pulled by a cylindrical permanent magnet coupled with a conical iron tip. Magnetic field and force are vertical, allowing the bead to rotate around the vertical axis. Small polystyrene beads are grafted to the magnetic bead to allow angle tracking. (B) Snapshots of the clockwise rotation of a DNA–bead, visualized in bright field, during hRad51 polymerization. The nucleoprotein filament was nucleated at 250 nM hRad51 concentration, and then hRad51 concentration was decreased to 75 nM. In the displayed sequence the molecule performs approximately ¾ turn in 32 s (1 frame every 4 s, from top to bottom). The black rim with a white center corresponds to the magnetic bead, and the smaller white spots at the periphery correspond to the polystyrene particles.
Results
Polymerization Initiation and Extension as a Function of Concentration.
Upon addition of hRad51 at a relatively high concentration (250 nM), in a buffer containing 100 μM ATP and 2 mM Mg2+, to a DNA–bead construct involving a single unnicked dsDNA, stretched typically at 0.5 pN, the magnetic bead started rotating clockwise (Fig. 1B and Movie S1). This corresponds to the unwinding of the DNA right-handed double-helix by hRad51 polymerization. Under conditions suitable for polymerization, the bead rotated typically for more than 1 h at a relatively constant speed of up to 2 rpm. The initiation of polymerization becomes less frequent upon decreasing hRad51 concentration, and we never observed any bead rotation when the experiment was started at concentrations lower than 100 nM. However, when the polymerization was initiated at 250 nM hRad51, and the microchannel was then flushed with solutions containing hRad51 at concentrations at which nucleation was never observed (10, 25, 50, and 75 nM), or was rare (100, 125, and 150 nM), rotation continued clockwise, suggesting that growth of an existing nucleus can proceed at a much lower hRad51 concentration than required for nucleation.
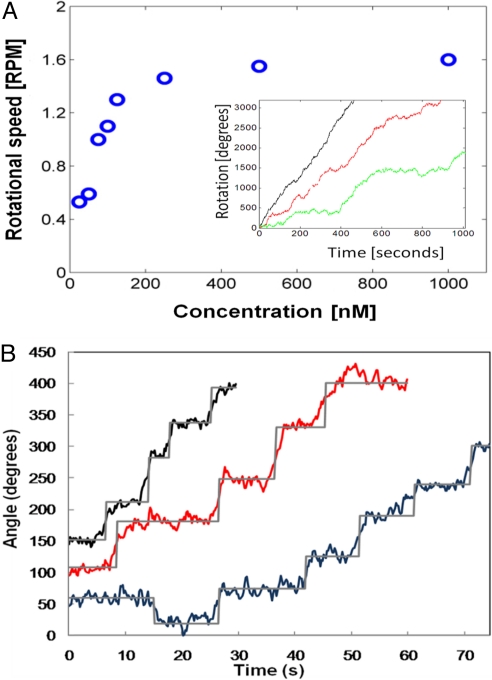
Three typical bead rotation traces, at 100 nM (black), 50 nM (red), and 10 nM (green) hRad51 concentrations are shown in Fig. 2A (Inset), together with the bead rotation rate at different hRad51 concentrations.
Fig. 2.
Typical rotating traces of DNA–beads. (A) Rotation rate of the bead at different hRad51 concentrations. Inset: Traces of 3 DNA–beads in the presence of 100 nM (black), 50 nM (red), and 10 nM (green) hRad51. Because of the viscous drag of the bead, the speed saturates at high hRad51 concentration. (B) Stepping traces of 3 different DNA–beads at 10 nM (black), 75 nM (red), and 50 nM (blue) [hRad51]. Sixty-five-degree steps are emphasized by the gray trace issued from a semiautomated step-finding algorithm (see text). [Warning: the rotation seems faster at 10 nM, but averaging polymerization speed on such a short period is meaningless owing to the strong polymerization speed fluctuations apparent in A (see text).]
We observe that the average rotational speed increases with the concentration of hRad51 in solution, then saturates when the concentration exceeds 200–300 nM. This contrasts with our previous report (20) in which the magnetic bead was blocked in torsion by the MT and the DNA molecule untwisted manually. We attribute this behavior to the viscous drag of the bead, which opposes the formation of the nucleoprotein filament and becomes the limiting factor at high hRad51 concentrations. It considerably slows down polymerization and prevents us from reaching full coverage of the DNA molecule. In practice, most of the experiments only showed a partial coverage of the DNA, because reaching the full coverage in FRMT, even at high hRad51 concentration, would take typically approximately 24 h, with a high risk of losing the molecule during this period.
Also note that although the average speed increases with hRad51 concentration, the instantaneous speed undergoes strong fluctuations. We attribute this to the presence of a residual horizontal magnetic field, which creates a periodic potential for the bead's rotation. This potential may induce temporary arrests of the rotation, especially at low hRad51 concentrations, thus creating strong fluctuations of the polymerization velocity on a short time scale.
Eventually, we did not see any rotation when the ATP was replaced by adenylyl-imidodiphosphate (AMP-PNP) or ATPγS. We previously observed (20) that nonhydrolyzable ATP analogs considerably slowed down the polymerization in conventional MT, suggesting that the association energy is weaker with these cofactors, or the association energy barrier higher. It is thus possible that polymerization of hRad51 in the presence of a nonhydrolyzable ATP analog is too small to overcome the torque induced by the residual horizontal magnetic field of the FRMT (see above). We also attempted polymerization in the presence of Ca2+ but could not observe polymerization over more than 10 attempts. In MT, we had seen that polymerization of hRad51 occurred at a similar rate on negatively twisted duplex DNA. However, the conditions of the present experiments are different, notably because of the existence of a torque opposing polymerization. Generally speaking, FRMT experiments have a low success yield; therefore, we cannot ascertain at this point whether we did not observe polymerization because we did not find the right conditions or because of a physical reason yet to be understood.
Twisting Steps at Low Concentrations.
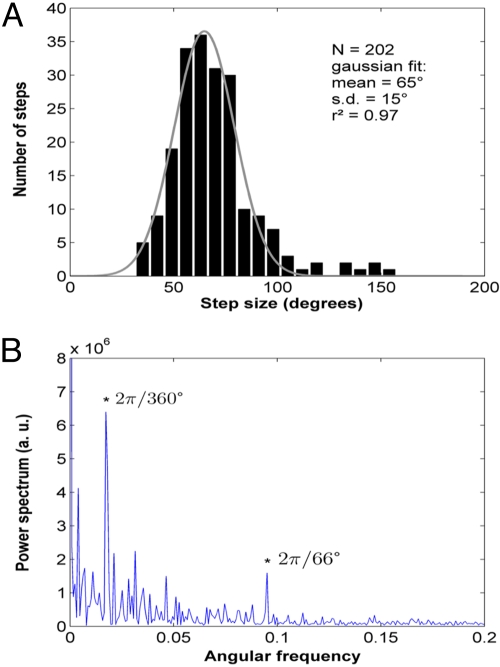
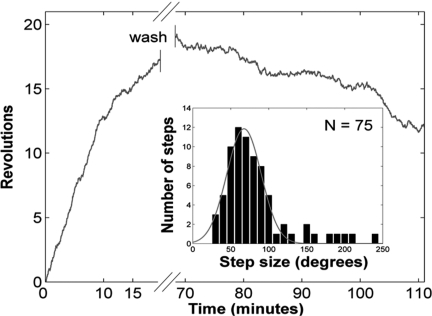
To focus on the mechanism of nucleoprotein filament growth, we “zoomed in” the polymerization profiles, corresponding to experiments in which the polymerization was initiated at 250 nM hRad51 and kept at lower concentrations. Below 100 nM hRad51, the rotation of the bead produced a stepping pattern (Fig. 2B and Movie S1), reminiscent of the behavior of molecular motors (26). We analyzed these steps by a “semiautomated” method using a manual setting of the time boundaries of plateaus and obtained a monomodal step-size distribution, with a mean step size of approximately 65° irrespective of hRad51 concentration. In Fig. 3A we present a histogram gathering all steps observed (this corresponds to 202 steps within a total rotation of 26,200°, from 5 different molecules and hRad51 concentrations from 10 to 50 nM). The histogram was well fitted by a normal distribution (r2 = 0.97) with a mean step size of 65° ± 15° (standard deviation). Eight steps (4% of total) are too large to fit the normal distribution.
Fig. 3.
Step size during the polymerization. (A) Histogram of a total of 202 steps from 5 different DNA–bead constructs rotating at hRad51 concentrations ranging from 10 to 50 nM. The histogram of step sizes was fitted to a normal distribution, yielding a mean of 65° and a standard deviation of 15°. (B) Fourier power spectrum of the pairwise distribution calculated from a rotating trace at 10 nM. Two peaks depart from noise. They correspond to angles 66° and 360°, respectively. This suggests the existence of steps of 66° and 360°. The latter is assigned to an experimental bias created by a residual horizontal magnetic field.
We cross-checked these results with an independent method less prone to operator bias, performing a pairwise rotation correlation followed by Fourier transform. An example of a Fourier power spectrum obtained from rotation data acquired at 10 nM hRad51 is given in Fig. 3B. Although noise increases for small angular frequencies, 2 peaks clearly show up above this noise, at 2π/360 and 2 π/66 (corresponding to favored rotation angles of 360° and 66°, respectively). The correlation at 360° corresponds to a full rotation. An independent analysis performed on naked DNA (see SI) shows that this correlation stems from a macroscopic horizontal component of the magnetic field and does not have a molecular origin. The other peak, however, appeared in the same angular range of 60°–70° for all experiments performed between 10 and 50 nM hRad51 after nucleoprotein filament initiation, and was absent from Fourier spectra of data obtained at hRad51 concentrations above 100 nM. We thus associate this peak with the steps visually observed in Fig. 2B and with hRad51 polymerization.
Finally, we applied to our data a modified version of the fluctuation analysis recently used to analyze kinesin stepping (27) (see SI Text, “Fluctuation Analysis”). For an undamped Poisson stepping process, fluctuations about the average speed reflect the underlying process stochasticity (i.e., the number of independent steps and their size). For simple and independent steps, which involve only 1 rate-limiting process per step and no cooperativity between steps, the angular step size α should be:
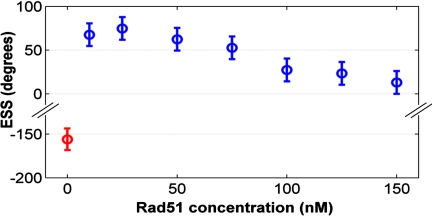
where θ(t) is angular position, and angle brackets represent average over all positions. Physically, α represents the size perfectly stochastic steps should have to reach the same level of fluctuations as observed experimentally. In the following we call it effective stochastic step (ESS) to reflect this property. This fluctuation analysis was performed on all our data from 0 nM (depolymerization experiments; Fig. 4, red) to 150 nM (Fig. 4, blue): at low hRad51 concentrations (≤75 nM), we obtained a stable ESS around 64° ± 13°. The high consistency between the step sizes obtained using 3 different data analysis approaches provides strong evidence in favor of a polymerization by steps around 65° at hRad51 concentrations up to 75 nM.
Fig. 4.
ESS (see text) calculated for polymerization at hRad51 concentrations ranging from 10 nM to 150 nM (blue) and depolymerization at 0 nM (red, negative value).
At higher hRad51 concentrations, the ESS in Fig. 4 decreased to approximately 20° at 150 nM. Notably, this also corresponds to situations in which one can no more identify steps in the motion, either by Fourier analysis or by step finding. We interpret this as follows: when hRad51 concentration increases, the mean interval between 2 hRad51 binding events decreases below the response time of the bead. This smoothes rotation and decreases the randomness associated with steps (for a purely viscous motion, the ESS would be 0). Note that a decrease in ESS could not be accounted for protein binding as multimers, which would increase the ESS.
The distribution of waiting time between steps, and its evolution with hRad51 concentration, would support a growth by addition of single monomers. Unfortunately, in our setup the residual horizontal magnetic field seriously biases waiting times depending on the “uphill” or “downhill” side of the step regarding this external field. This bias is much stronger on the step dwell time than on the step size (for which it tends to widen slightly the distribution, without affecting the average value).
Depolymerization in hRad51-Free Conditions.
After thorough rinsing with hRad51- and nucleotide-free buffer, DNA–beads rotated counterclockwise (Fig. 5). The same buffer, supplemented with 1 mM of ADP, completely inhibits this counterclockwise rotation of the bead. These observations indicate that the ADP release induces a rewinding of the DNA molecule toward its initial torsional state. We associate this behavior to a depolymerization of hRad51 from the DNA after ATP hydrolysis (see SI Text). The average rotation speed, approximately 0.2 rpm, is much smaller than for polymerization and corresponds to a rate of 0.018 hRad51 bp−1 s−1. We applied the same step analysis as for polymerization and detected 75 analyzable steps from 3 different DNA–beads, for a total angular rotation of 8,550°. A histogram of these steps was fitted to a normal distribution (r2 = 0.95), yielding a mean step size of 67° and a standard deviation of 23° (Fig. 5, Inset). Eight steps (11%) were too large to fit the normal distribution. Applying fluctuation analysis to depolymerization data yielded an ESS of −150° ± 15° (Fig. 4).
Fig. 5.
Rotating trace of a full experiment. After approximately 20 min of clockwise rotation (polymerization) at 100 nM hRad51, rotation became irregular and very slow for approximately 50 min until the bead finally stopped (this latter section was cut in the graph to more clearly display the other sections). After intense rinsing, rotation started counterclockwise at a slower rate. We associate this reverse rotation to the depolymerization of hRad51 from the nucleoprotein filament formed during the first phase. Inset: histogram of step sizes for 75 steps obtained during depolymerization on 3 different molecules. The best-fit normal distribution has a mean of 67° and standard deviation of 23°.
Discussion
Polymerization Threshold.
We observed that the polymerization of hRad51 on dsDNA requires a minimal concentration of approximately 100 nM to initiate. This is consistent with previous results independently obtained both with conventional MT (20) and in single-molecule videomicroscopy (21) confirming the existence of a threshold concentration for hRad51 polymerization initiation. The threshold in ref. 21 seems rather to be approximately 50 nM, twice as low as that observed here, but this discrepancy may be attributed to minor differences in experimental conditions: in ref. 21, 2 mM calcium was used in the buffer, whereas we used 2 mM magnesium, which seems a somewhat less efficient co-ion for hRad51 binding. In additions, the flow used in ref. 21 to keep molecules extended involves a stretching, which is nonuniform but can be high enough, especially at some places, to significantly increase hRad51 binding rate. Besides this minor discrepancy, data in refs. 20 and 21 both suggest that filament nucleation rate depends on hRad51 concentration as a power law with an exponent significantly larger than 1. Conversely, the growth of a preexisting filament is linear in concentration. Our results, showing that nucleation becomes rare at low concentrations, whereas growth of a prenucleated filament can proceed, are qualitatively consistent with these observations.
Polymerization Kinetics.
At high hRad51 concentrations, we observed a constant rotation speed, suggesting a polymerization dominated by sequential addition of proteins to one end of a preexisting nucleoprotein filament. It was previously proposed that RecA polymerization on DNA involves a slow nucleation followed by a faster directional growth (14, 22). More recent MT investigations of hRad51 polymerization on dsDNA also reported, at 100 nM hRad51 in the same buffer as used here, a length increase linear in time (20). That work also proved that the hRad51 polymerization happens with a discrete and small number of simultaneously growing fronts. Finally, recent videomicroscopy experiments very directly confirmed that polymerization of hRad51 on dsDNA occurs by a mechanism involving a slow nucleation followed by a polymerization from the preformed nuclei (21). This latter article also reported, in low hRad51 concentration situations involving only a few growing fronts, intermittent growth with rather extensive polymerization pauses. Such pauses could also explain the large fluctuations in the polymerization speed, observed here at low hRad51 concentration.
Thus, our experiments are qualitatively consistent with those performed in MT or with videomicroscopy. The major advantage of the FRMT is their angular resolution, allowing us to observe molecular events that yield variations of the molecule length too small to be detected in MT.
Rotation Stepping.
We observed individual twisting steps of 65° ± 5° for polymerization of hRad51 at low concentrations (from 10 to 75 nM) and for depolymerization in the absence of hRad51 and ATP in the buffer. Earlier works based on EM experiments showed that RecA and Rad51 unwind the DNA double helix by approximately 15° per bp (5, 10). The DNA/Rad51 stoichiometries were rather variable from one report to the other, ranging from 2 to 5 bp per protein. This corresponds to an expected unwinding between 30° and 75° per protein. Our results are thus consistent with a nucleoprotein filament growth occurring by addition of individual hRad51 monomers, as previously reported for RecA (17) and incompatible with the coordinated addition of 4.3 ± 0.5 monomers, as recently proposed in a study of hRad51 polymerization on dsDNA measured by MT (19). The latter would involve steps between 142° and 324°. We do not have a certain explanation for this discrepancy. We note, however, that this conclusion was reached (19) through a rather indirect analysis, involving a comparison of experimental data with simulation results through a 2-step fitting procedure: first, Monte-Carlo simulations of several nucleation and growth models, involving either monomers or pentamers for each of these mechanisms, were fitted to the data at a given hRad51 concentration, and the nucleation and growth constants, knucl and kgrowth, respectively, were determined from this fit. In a second step, nucleation and growth constants obtained at different hRad51 concentrations were fitted to the Hill model (28) to get the number of proteins involved in each of these steps. We doubt that such a procedure, applied to real data, is accurate enough to yield an independent measure of knucl and kgrowth in the relevant concentration range. At concentrations higher than 150 nM hRad51, nucleation and growth are concomitant. Thus, differences in the ratio knucl/kgrowth should affect only the very beginning of the curve. In MT experiments, however, this part of the kinetics is not known very accurately, because at high hRad51 concentrations polymerization starts during the buffer exchange period (this point is clearly visible upon detailed observation of the curves). This buffer exchange requires a flow that makes accurate length measurement impossible, and it typically lasts several tens of seconds because of buffer front dispersion by the flow's Poiseuille profile (29).
As another potential explanation for this discrepancy, the simulations in ref. 19 assume that once a nucleus is formed, it grows indefinitely. However, ref. 21 clearly demonstrated that filaments continuously grow on a finite length only, generally not exceeding 5 kbp. Thus, the Monte Carlo model should have included a filament termination step resulting in filament segments smaller than 5 kbp. This limitation of the model may have induced a bias in the fit, incorrectly yielding an apparent growth by addition of pentamers. Finally, in ref. 20 we observed a very dramatic change in the shape of the polymerization curves with hRad51 concentration, which would not be consistent with a mechanism in which nucleation and growth have the same concentration dependence.
Biological Relevance of a Polymerization by Monomers.
The formation of the nucleoprotein filament involves 2 phases: the nucleation of a seed (20, 21) and, as shown above, an elongation by addition of single hRad51 monomers. This mechanism has 2 consequences for the dynamics of the nucleoprotein filament. First, high hRad51 concentrations favor nucleation, whereas elongation requires much lower concentrations. In particular, we have shown that, once a nucleus is formed, polymerization can continue at hRad51 concentrations as low as 10 nM, typically 10 times smaller than those necessary for spontaneous polymerization initiation. In vivo, the initiation of HR is strongly regulated and involves other proteins associated to the Brca2 and Rad52 pathway (30, 31). These 2 proteins, in particular, seem necessary for HR initiation in mammalian cells (32, 33). The possibility, demonstrated here, of continuing polymerization at hRad51 concentrations much too low to support nucleation, could thus allow a continuation of polymerization (which is necessary for strand exchange on long distances) by hRad51 alone, while keeping the nucleation of nucleoprotein filaments under the strict control of the DNA damage signaling pathway.
Second, growth by monomers produces filaments with defects less numerous and of smaller size than a hypothetical elongation by addition of pentamers (see SI Text, “Nucleation and Growth Monte Carlo Simulations”) (34, 35). For both monomer- and pentamer-based growth, the number of defects is minimal when the (uncatalyzed) nucleoprotein assembly is dominated by elongation, a situation that should prevail in vivo (see above discussion). We notice, however, that the final number of uncovered base pairs is lower when the elongation happens by addition of single monomers, over a large range of growth/nucleation rates. This should thus yield a more efficient nucleoprotein filament.
Structural Parameters of the Nucleoprotein Filament Revisited.
The change in DNA twist associated with hRad51 binding was directly measured here. It can be combined with earlier EM and structural data (4, 10, 36, 37) to sharpen our view of the nucleoprotein filament structure, and in particular to confirm the state of DNA, which is not directly seen on EM. Regarding protein organization, the results obtained by EM are very consistent in different reports, with a filament pitch of 98 ± 2 Å and 6.18 ± 0.04 proteins per turn. These values probably represent sections of the nucleoprotein filament organized into a defect-free “canonical” structure. From our results, the DNA fragment contained in a full turn of the nucleoprotein filament is unwound by 6.18 × 65° = 402° as compared with B-DNA. Assuming that the DNA and the protein helix have the same twist in the nucleoprotein filament (i.e., by definition 360° per turn), we obtain the total twist that the DNA fragment comprised in 1 turn of the nucleoprotein filament had before its incorporation: 402° + 360° = 762°. Using as an input the helicity of B-DNA, 10.4 bp per turn, we thus deduce that this fragment of DNA contained in 1 nucleoprotein filament turn consists of 10.4 × 762/360 = 22 ± 1.3 bp. Consequently, within the nucleoprotein filament, the DNA raises 4.45 ± 0.25 Å per base pair and is unwound by 18.2° ± 1° per base pair. This yields a stoichiometry of 3.6 ± 0.3 bp per hRad51. These values are within the (rather scattered) range of previously published data, but they confirm or suggest significant differences with RecA [for which the corresponding parameters are 5.1 Å (rise per base pair), 15° (twist per base pair), and 3 bp per RecA (stoichiometry), respectively (5, 7)]. Considering the error involved in these calculations, the stoichiometry measured here is consistent with integer values of 3 and 4 bp per hRad51. As an alternative interpretation, we note that recent crystallographic data on a Rad51 mutant, I345T (36), suggest that the nucleoprotein filament does not have a strict helical 6-fold symmetry but is rather a helix of Rad51 dimers. Indeed, our results would better fit an actual stoichiometry of 7 bp per hRad51 dimer than integer stoichiometries of 3 or 4 bp per monomer.
Depolymerization.
Regarding depolymerization, the ESS we measured was much higher in absolute value than that obtained for polymerization, suggesting either a depolymerization by multimers or correlations between depolymerization steps. This is indeed in qualitative agreement with recent conclusions of single-molecule videomicroscopy studies (38), which showed that hRad51 indeed depolymerizes from dsDNA by multimeric “bursts.” These investigators proposed the following interpretation: ATP hydrolysis occurs at random inside a nucleoprotein filament, but it does not induce depolymerization as long as the site at which this hydrolysis occurs is flanked by 2 hRad51 molecules. When hydrolysis occurs on a terminal hRad51, however, the stability of the protein is insufficient and it disassembles. All ADP-bound proteins, directly following this released “cap,” can then depolymerize readily until reaching the next ATP-bound protein. For the early depolymerization steps observed in our experiments, disassembly pauses would correspond to the removal of 2 to 3 hRad51 monomers on average and yield an ESS of 2 to 3 times 65°.
Conclusions
In summary, the new type of FRMT experiments developed and used in this study provided original structural and kinetic data for hRad51/DNA nucleoprotein filaments. Combined with EM data, our results suggest that hRad51 unwinds the DNA by 18.2° per base pair and that the hRad51 nucleoprotein filament is less extended (4.5 Å per base pare) as compared with RecA (15° per base pair and 5.1 Å per base pair). We demonstrated that, after nucleation, hRad51-dsDNA nucleoprotein filaments grow by addition of single hRad51 monomers, corresponding to individual rotation steps of 65° ± 5°. Using Monte Carlo simulations, we quantitatively confirmed that this is an efficient way to minimize the number of defects in the nucleoprotein filament. We also observed that polymerization onto the end of a preexisting nucleus can proceed extensively at hRad51 concentrations far too low to allow spontaneous nucleation. On molecular grounds, this result is a consequence of the difference between nucleation and growth rates. Physiologically, it may be instrumental in vivo to allow efficient recombination on long DNA sequences, while keeping the nucleation of polymerization, and thus the initiation of strand exchange, under the strict control of other facilitating proteins belonging to the Brca2 regulation pathway.
Materials and Methods
Microchannel Preparation.
A polydimethylsiloxane microchannel 2 cm long, 2 mm wide, and approximately 100 μm high was prepared according to our previous reports (15, 20) and placed onto a glass coverslip of 24 mm × 40 mm (Erie Scientific) treated with antidigoxigenin (Roche) for subsequent binding of digoxigenin-labeled DNA molecules. Before first use of the channel, 10 mg/mL of BSA was injected into the channel and incubated overnight at 4 °C to minimize subsequent protein adsorption.
DNA Constructs.
The DNA molecule held in the MT was a 14,400-bp PCR fragment generated from a λ DNA template (15, 20). This molecule was ligated at one end to a multidigoxigenin-labeled DNA fragment of 600 bp and at the other end to a multibiotin-labeled fragment of approximately 820 bp.
Human Rad51 Proteins.
Human Rad51 proteins were purified as described previously (20). The protein concentration was determined using the Bio-Rad protein assay kit using BSA as the standard protein. The DNA strand exchange and DNA-dependent ATPase activities were verified.
Protocol of DNA/hRad51 Twisting Experiment.
The biotinylated dsDNA molecules were bound to streptavidin-coated 2.8-μm magnetic beads (Dynal Biotech) in binding buffer [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, and 50 mM NaCl]. The dsDNA/magnetic bead complexes were incubated in the microchannel at room temperature for 10 min. After incubation, most of the unbound beads were washed out from the channel with TE-BSA buffer [10 mM Tris–HCl, 1 mM EDTA (pH 7.5), and 0.2 mg/mL BSA]. Next, biotin-polystyrene nanobeads (GKX01; G. Kisker GbR, 810 nm in diameter), suspended in binding buffer, were introduced into the channel and incubated until a few nanobeads on average were attached to each magnetic bead. After washing out excess nanobeads with TE-BSA buffer, the permanent magnet with a conical tip at its extremity was lowered toward the top of the microchannel, to exert a vertical force on the magnetic beads. The force was calibrated using lateral fluctuations of beads (25). Finally, hRad51 in solution in Rec2 buffer [2 mM MgCl2, 15 mM Tris–HCl (pH 7.5), 25 mM NaCl, 1 mM DTT, and 0.05% Tween 20], supplemented with 100 μM ATP, was flown into the microchannel under gravity (20-cm height difference between the feed reservoir and the exit reservoir) until full buffer exchange (10 min). Then the flow was stopped and the cell scanned for rotating beads. Beads can be directly attached to the surface, attached by several DNA molecules, or by a nicked DNA, so this scan can take typically from 10 to 60 min. The duplex DNA can be covered up to 15% during this “blind” period.
Data Analysis by “Manual” Step Finding in the Real Space.
First, for the “manual” approach, putative steps in the rotation trace are selected visually, the time boundaries before and after plateaus are defined by the operator, and the mean angular position is calculated for both plateaus. The difference between these values gives the step size.
Error Analysis.
The standard deviation δθ for the experimental step-size distributions, ranging from 10° to 23°, is a reflection of Brownian noise and not a measure of the experimental error. Assuming that this Brownian noise is the only source of deviation from the step size, the error should be δθ/ ≈ 1°. This is certainly too optimistic, because experimental causes of error also exist. Comparing data obtained on independent molecules, obtained on independent days, and with different analysis methods (see below), we estimate the real error to 5°.
Data Analysis by Correlation.
The following correlation analysis was applied to several series of data. For each pair of points, θ(ti) and θ(j), we calculate the distance dij = < θ(ti) - θ(tj) >, with i ≠ j. The pairwise correlation of positions is defined as the distribution P(dij) of dij. This correlation is then Fourier-transformed using the MATLAB fast Fourier transform algorithm, yielding power spectra such as that presented in Fig. 3B. For data obtained at concentrations lower than 100 nM, 2 main peaks appear. We attribute the first one, around 2π/360°, to a residual macroscopic horizontal component of the magnetic field, imposing a preferred orientation for the beads. Because the magnetic coupling energy, usually around 4 KbT (for potential energy confining the magnetic beads angular fluctuations; see SI), is significantly smaller than those involved in hRad51 polymerization, this coupling did not significantly affect our results.
Supplementary Material
Acknowledgments.
We thank Marie Dutreix, Jacques Prost, and Paolo Pierobon for fruitful discussions and experimental help; and the editor and reviewers for providing constructive comments and suggestions to improve this article. This work was funded in part by Agence Nationale pour la Recherche programmes Programme Nanosciences et Nanotechnologies and Physico Chimie du Vivant. M.T. is supported by Grant 3862 from the Association pour la Recherche sur le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902234106/DCSupplemental.
References
- 1.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 2.Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 4.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad5l protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stasiak A, Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 6.Lauder SD, Kowalczykowski SC. Asymmetry in the RecA protein-DNA filament. J Biol Chem. 1991;266:5450–5458. [PubMed] [Google Scholar]
- 7.Di Capua E, Engel A, Stasiak A, Koller TH. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J Mol Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi M, Kubista M, Norden B. Binding stoichiometry and structure of RecA-DNA complexes studied by flow linear dichroism and fluorescence spectroscopy. J Mol Biol. 1989;205:137–147. doi: 10.1016/0022-2836(89)90371-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA–ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 11.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 12.Tombline G, Fishel R. Biochemical characterization of the human RAD51 protein. J Biol Chem. 2002;277:14417–14425. doi: 10.1074/jbc.M109915200. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein IJ, Greene EC. Single molecule studies of homologous recombination. Mol Biosyst. 2008;4:1094–1104. doi: 10.1039/b811681b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivashankar GV, Feingold M, Krichevsky O, Libchaber A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: The role of ATP hydrolysis. Proc Natl Acad Sci USA. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulconis R, et al. Twisting and untwisting a single DNA molecule covered by RecA protein. Biophys J. 2004;87:2552–2563. doi: 10.1529/biophysj.104.043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 17.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 18.Prasad TK, Yeykal CC, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 19.Van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miné J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51–DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilario J, Amitani I, Bashkin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cazenave C, Toulmé JJ, Hélène C. Binding of RecA protein to single-stranded nucleic acids: Spectroscopic studies using fluorescent polynucleotides. EMBO J. 1983;2:2247–2251. doi: 10.1002/j.1460-2075.1983.tb01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gore J, et al. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada Y, et al. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 25.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 26.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 27.Svoboda K, Mitra PP, Block SM. Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc Natl Acad Sci USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill AV. The heat produced in contracture and muscular tone. J Physiol. 1910;40:389–403. doi: 10.1113/jphysiol.1910.sp001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bancaud A, Wagner G, Dorfman KD, Viovy JL. Measurement of the surface concentration for bioassay kinetics in microchannels. Anal Chem. 2005;77:833–839. doi: 10.1021/ac048996+. [DOI] [PubMed] [Google Scholar]
- 30.Sung P, Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat Rev Mol Cel Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 31.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 32.Navadgi VM, Shukla A, Vempati RK, Rao BJ. DNA mediated disassembly of hRad51 and hRad52 proteins and recruitment of hRad51 to ssDNA by hRad52. FEBS J. 2006;273:199–207. doi: 10.1111/j.1742-4658.2005.05058.x. [DOI] [PubMed] [Google Scholar]
- 33.San Filippo J, et al. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epstein IR. Kinetics of large-ligand binding to one-dimensional lattices: Theory of irreversible binding. Biopolymers. 1979;18:765–788. [Google Scholar]
- 35.Epstein IR. Kinetics of nucleic acid-large ligand interactions: Exact Monte Carlo treatment and limiting cases of reversible binding. Biopolymers. 1979;18:2037–2050. doi: 10.1002/bip.1979.360180815. [DOI] [PubMed] [Google Scholar]
- 36.Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Mameren J, et al. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2008;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.