Abstract
Deletion of the acyltransferases responsible for triglyceride and steryl ester synthesis in Saccharomyces cerevisiae serves as a genetic model of diseases where lipid overload is a component. The yeast mutants lack detectable neutral lipids and cytoplasmic lipid droplets and are strikingly sensitive to unsaturated fatty acids. Expression of human diacylglycerol acyltransferase 2 in the yeast mutants was sufficient to reverse these phenotypes. Similar to mammalian cells, fatty acid-mediated death in yeast is apoptotic and presaged by transcriptional induction of stress-response pathways, elevated oxidative stress, and activation of the unfolded protein response. To identify pathways that protect cells from lipid excess, we performed genetic interaction and transcriptional profiling screens with the yeast acyltransferase mutants. We thus identified diacylglycerol kinase-mediated phosphatidic acid biosynthesis and production of phosphatidylcholine via methylation of phosphatidylethanolamine as modifiers of lipotoxicity. Accordingly, the combined ablation of phospholipid and triglyceride biosynthesis increased sensitivity to saturated fatty acids. Similarly, normal sphingolipid biosynthesis and vesicular transport were required for optimal growth upon denudation of triglyceride biosynthesis and also mediated resistance to exogenous fatty acids. In metazoans, many of these processes are implicated in insulin secretion thus linking lipotoxicity with early aspects of pancreatic β-cell dysfunction, diabetes, and the metabolic syndrome.
Introduction
Lipotoxicity, the phenomenon of cell death due to lipid imbalance, is a common consequence of obesity and likely influences many of the morbidities associated with this syndrome, including diabetes and atherosclerosis. The mechanisms by which an imbalance in lipid metabolism culminates in cell death differ for each offending metabolite and surprisingly are poorly understood. For example, saturated and unsaturated fatty acids are pro-apoptotic in most cell types (1) but achieve this through different pathways (for review see Ref. 2). The cytotoxicity of saturated fatty acids such as palmitate likely arises from elevated ceramide levels and/or reactive oxygen species (3, 4). By contrast, the accumulation of oleate, an unsaturated fatty acid, promotes apoptosis through activation of serine/threonine protein phosphatases such as PP2Cα/β (5). Similarly, subcellular cholesterol accumulation induces cellular stress responses such as the unfolded protein response pathway, apoptosis, and necrosis (6).
Any process that limits the accumulation of lipid alcohols (such as diacylglycerol and sterols) or fatty acids is likely to be cytoprotective. One such reaction is their mutual co-esterification to produce neutral lipids (e.g. triglyceride (TG)8 or steryl ester (SE)). Subsequently, these lipids are sequestered in cytoplasmic lipid droplets (CLDs), the preponderance of which correlates with lipotoxic states such as obesity and fatty liver disease (7). To date, three acyltransferase gene families independently direct the terminal and committed steps of neutral lipid synthesis in eukaryotes (8). In mammals, the esterification of sterol is accomplished by members of the acyl-CoA:cholesterol acyltransferase (ACAT) family or in serum by lecithin-cholesterol acyltransferase. ACAT members carry out the reaction in an acyl-CoA-dependent fashion, whereas lecithin-cholesterol acyltransferase uses phospholipids as an acyl donor. Diacylglycerol (DAG) esterification is primarily the activity of DGAT2 (a member of the diacylglycerol acyltransferase (DGAT) 2 gene family) or DGAT1 (a member of the ACAT family).
In Saccharomyces cerevisiae, the ACAT-related enzymes Are1p and Are2p are responsible for the esterification of sterols (9–12). DAG esterification during the logarithmic phase of growth is mediated by Lro1p (a lecithin-cholesterol acyltransferase ortholog that uses DAG and phospholipids as substrates (13, 14)), whereas DGA1, the sole representative of the DGAT2 gene family in this organism, directs synthesis of TG during stationary phase (11, 15, 16). Yeast strains that lack all four acyltransferase genes are viable but completely lack neutral lipids and thus present a genetically malleable system to investigate fatty acid-induced cytotoxicity (17).
We show here that elimination of neutral lipid biosynthesis results in contrasting effects of saturated and unsaturated fatty acids on cellular metabolism and viability and that this primarily reflects differential channeling of fatty acids into phospholipids. We describe the use of genetic interaction screens and transcriptional profiling to identify pathways that interact with the neutral lipid acyltransferases (see also Ref. 18). Defects in several components of endoplasmic reticulum homeostasis impose limitations on growth of ARE1, ARE2, LRO1, or DGA1 mutants, implicating them as evolutionarily conserved components of lipid homeostasis that protect a cell from lipotoxicity.
EXPERIMENTAL PROCEDURES
General
Media preparations (yeast extract, peptone, dextrose; YPD, synthetic; SCD), molecular and yeast genetic procedures, DNA modification, and oligonucleotide purifications (QIAquick gel extraction kit, Qiagen) were according to conventional (19) or the manufacturers' protocols.
Yeast Strains and Growth, Plasmids, and Transformation
The yeast strains used in this study (supplemental Table S1) were derived from W303 (20) or s288C (Open Biosystems). Strains with deletions in OPI3 or CHO and CYB5 or OLE1 were obtained from Susan Henry (21) and Charles Martin (22, 23), respectively. Synthetic genetic interactions with neutral lipid acyltransferase genes were determined from an ER-Golgi epistatic SGA map (24). Fatty acid growth studies were performed using YPD or SCD supplemented with 0.6% ethanol/tyloxapol (5:1, v/v) and the indicated amounts of fatty acids (Sigma or Nu-Check Prep Inc; 10% (w/v) stock in ethanol). For cell viability assays, aliquots from 2-ml YPD precultures were washed twice in distilled H2O and plated as serial dilutions or used to inoculate YPD or YPD oleate such that the starting optical density was 0.4 (A600 nm). Cultures were grown for 18 h, plated onto YPD, incubated for 2 days at 30 °C, and colonies counted. 30° growth curves were obtained using a Microbiology Workstation Bioscreen C (Thermo Electron Corp.) and Research Express Bioscreen C software (Transgalactic Ltd.). Cultures (three isolates per genotype) were normalized to an A600 of 0.1, and 10 μl of each strain was added to 290 μl of media per well. The requirement for the PEMT pathway was determined on media lacking inositol and choline (SMM-I).
Plasmid pIU2555 (from M. Bard and M. Valochovic) was digested with BamHI and SalI, and the 1.8-kb fragment containing GFP in-frame with ERG27 was inserted at the BamHI and SalI sites of pRS242-GPD (ATCC) resulting in pRS424-GPD-GFP-ERG27. Sec63-GFP was expressed using a SEC63-GFP expression construct. A full-length DGAT2 cDNA was expressed from the GAL1/10 promoter in pRS424-GP (25).
Lipid Analysis and Metabolic Labeling
[9,10-3H]Oleic acid and [9,10-3H]palmitic acid (PerkinElmer Life Sciences) pulse labeling in exponential phase was performed with 0.1 μCi/ml[3H]oleate or 0.5 μCi/ml [3H]palmitate in YPD ± fatty acids at 30 °C for 1 h (11). Strains transformed with hDGAT2 were grown to mid-log phase in 2% galactose, 1% raffinose media. Lipids were isolated by hexane/isopropyl alcohol (2:1) organic extraction of dried cell pellets and resolved by TLC in petroleum ether/diethyl ether/acetic acid (84:15:1). Phospholipids were further resolved with a second solvent of chloroform/methane/acetic acid/water (60:50:1:4) (26). Lipids were detected using iodine staining and quantified via scintillation counting. Capillary gas chromatography analysis of fatty acid composition following esterification was performed on a Hewlett-Packard 5890A equipped with a SupelcoTM 10 fused silica capillary column (30 m, 0.25 mm inner diameter), essentially as described (27).
DNA Microarray Analysis
RNA was extracted from yeast cells grown at 30 °C to ∼0.650 (A600 nm) in YPD or in YPD plus 0.01 mm oleate and used to prepare cDNA for hybridization to Affymetrix S98Yeast GeneChip arrays (28). Results are in triplicate, and each set was normalized using Robust Multichip Average (29, 30) followed by comparison analysis using the LIMMA module in conjunction with affylmGUI (31–33) found in the R software package (Linear Models for Microarray Data; version 2.2.0).
Verification of Gene Expression Changes
RNA was prepared from mutant cells as above and used to generate first-strand cDNA (SuperScript First-strand Synthesis System, Invitrogen). Real time PCRs were performed with the MyiQ single-color real time PCR detection system (Bio-Rad), SYBR Green 2× Supermix (Bio-Rad), and the indicated primers. Expression levels were calculated relative to ACT1 using the MyiQ real time detection software (34). Northern blots were performed using 15 μg of RNA (28), hybridized with PCR-generated gene-specific probes, and analyzed after overnight exposure using a phosphorimager (GE Healthcare).
Fluorescence Microscopy
Yeast cells were grown in appropriate media supplemented with 20× adenine to minimize autofluorescence of the vacuole associated with ade2 mutants. For CLD analysis, cells were stained with Nile Red (1 μg/ml) or envisaged for GFP reporters using a long pass GFP filter (excitation 440 nm). Cell viability using the FUN1 stain was determined using Yeast LIVE/DEAD viability kit (Molecular Probes). Apoptosis was assessed by annexin V binding for phosphatidylserine externalization and propidium iodide staining using an ApoAlert annexin V-FITC apoptosis kit (BD Biosciences) (35). Reactive oxygen species were detected using dihydrorhodamine 123 (DHR 123; Sigma) and visualized with a fluorescein filter (36). 4′,6-Diamidino-2-phenylindole (nuclei and mitochondria), MDY-64, LysoSensor Green DND-189, 5-carboxy- and 6-carboxy-2′,7′-dichlorofluorescein-diacetate and 7-amino-4-chloromethylcoumarin-l-arginine amide (vacuolar membranes, pH, and proteolytic function, respectively) and calcofluor white (cell wall) were used to stain yeast cells for the indicated structures. Microscopic analysis was performed on a Zeiss Axiovert 200 M using a 63× oil immersion objective. All images were taken using a Hamamatsu Orca-ER camera.
RESULTS
Phenotypes of CLD-deficient Strains
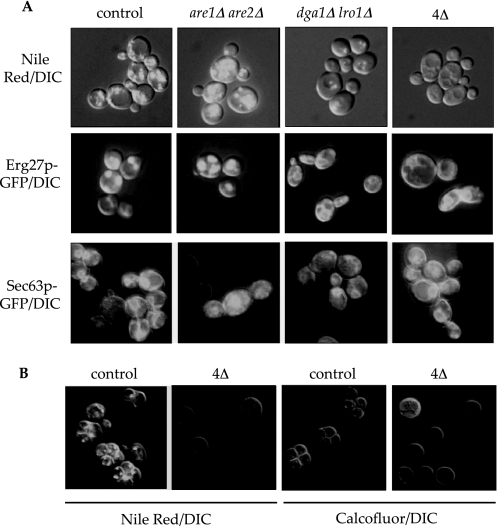
Neutral lipids represent a safe harbor for cytotoxic lipids enabling cells to efficiently store lipid precursors in a biologically inert form. We and others have described viable yeast strains with combined deletions of the ARE1, ARE2, LRO1, and DGA1 genes that lack neutral lipids and CLDs (11, 15, 16) (confirmed in Fig. 1A). Surprisingly, the sterol content of membranes, fluid phase endocytosis, and pH and the proteolytic capability of the vacuole in the mutants were equivalent to control strains as determined by staining with lipophilic dyes (Filipin, MDY-64, LysoSensor Green DND-189, carboxy-DCFDA, and CMAC-Arg, respectively; data not shown). By contrast, we discovered that the protein composition of the CLD organelle is responsive to changes in neutral lipids. 3-Keto sterol reductase (ERG27) fused to GFP (37) was coincident with Nile red staining of lipid droplets in control cells and unaffected by loss of sterol esterification (Fig. 1A). However, loss of TG synthesis in the dga1Δ lro1Δ strains or complete loss of neutral lipids in the quadruple mutant resulted in Erg27-GFP retention at the ER (Fig. 1A).
FIGURE 1.
Neutral lipid deposition, marker protein, and sporulation in acyltransferase mutants. A, saturated cultures of the indicated genotypes were stained with Nile red (neutral lipids/CLDs) or imaged for Erg27-GFP (CLD proteins) or Sec63-GFP (endoplasmic reticulum) as indicated. DIC, differential interference contrast. B, diploid strains homozygous for the indicated mutations were sporulated under conventional conditions and stained with Nile red (CLDs) or calcofluor white (cell wall). All fluorescence images were merged with the differential interference contrast image to outline the cell and are representative of several fields of view during multiple experiments. The quadruple acyltransferase mutant is designated as 4Δ.
Another critical role of neutral lipid biosynthesis is to provide important membrane and energy reserves, most likely in response to nutrient limitation to the cell. Under such circumstances, diploid yeast differentiate (sporulate) to the haploid state and accumulate neutral lipids, both within and between daughter cells, the latter presumably of maternal origin (Fig. 1B) (38–40). We generated homozygous quadruple acyltransferase mutants (2n) that were completely deficient in neutral lipid accumulation and observed a severe block in spore formation (Fig. 1B). After 8 days in nitrogen-deficient media, 4.3% of quadruple mutant cells had sporulated versus 62.5% of control diploid cells (averaged over multiple fields of view). This suggests a requirement for neutral lipids and/or CLDs or alternatively an inhibitory effect of lipid substrate accumulation during early sporulation, perhaps at entry into meiosis.
Compensatory Pathways for Neutral Lipid Deficiency
The viability of mutants in the neutral lipid biosynthetic pathway was surprising given the key role these enzymes play in metabolite storage and detoxification. We reasoned this is likely due to regulatory responses to an overabundance of the precursors to these reactions, i.e. alcohols (sterols or diacylglycerols) and fatty acids, as well as the utilization of alternate systems that bypass the toxic effects of free fatty acids. In mammalian systems this homeostasis initiates at the transcriptional level (41, 42). We therefore profiled the transcriptional status of are1Δ are2Δ (SE-deficient), lro1Δ dga1Δ (TG-deficient), and are1Δ are2Δ lro1Δ dga1Δ (lacking neutral lipids) strains using Affymetrix oligonucleotide microarrays (supplemental Tables S2–S5). The data, generated in triplicate, were analyzed using linear modeling (see “Experimental Procedures”).
Global Gene Expression Changes as a Result of Steryl Ester or Triglyceride Deficiency
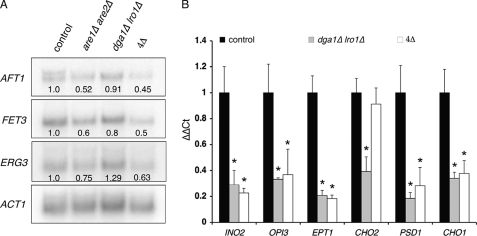
Yeast cells deficient in sterol esterification have reduced ergosterol biosynthetic rates and maintain free ergosterol at levels equivalent to control cells (9, 43). Surprisingly, there was no evidence of statistically significant transcriptional regulation of the sterol biosynthetic pathway in sterol esterification-defective strains. The major pathways influenced by loss of sterol esterification determine or respond to membrane status (e.g. down-regulation of vesicular transporters such as Uso1p, Cog7p, and Dsl1p). We also observed a statistically significant decrease in the expression of several components of iron metabolism such as the iron-responsive transcriptional regulator AFT1 and Aft1p target genes such as FET3. These results (supplemental Tables S2 and S3) were confirmed via Northern blotting (Fig. 2A). Many steps in ergosterol biosynthesis incorporate iron into their structure in various forms as follows: as a mono- or binuclear species, as a part of an Fe-S cluster, or as heme groups (44). These observations indicate an unexpected feedback interaction between iron and sterol homeostasis that may confer an alternate avenue by which sterol biosynthesis is regulated.
FIGURE 2.
Transcriptional responses to loss of neutral lipid biosynthesis. A, Northern blot analysis of selected genes involved in iron or ergosterol metabolism using 20 μg of RNA isolated from cells with indicated lesions in NL biosynthetic pathways. Transcription of actin was used as a loading control, and values under each signal represent gene: ACT1 ratios relative to control cells. B, qRT-PCR measurements of expression of genes involved in phospholipid metabolism as a consequence of losing TG (dga1Δ lro1Δ) or NL (dga1Δ lro1Δ are1Δ are2Δ: 4Δ) biosynthesis. ΔΔCt representing the change in mRNA expression (after ACT1 normalization) relative to wild-type strains was calculated as 2−(Gene CtMutant−ACT1 CtMutant)−(Gene CtControl− ACT1 CtControl), where “Gene” represents the mRNA under study and “Ct” is the number of thermal cycles needed to cross threshold. Asterisks indicate significant difference from control strains (p ≤ 0.05; Student's t test, n = 3).
Triglyceride deficient dga1Δ lro1Δ mutants exhibited a statistically significant reduction in multiple key components of phospholipid homeostasis relative to control strains. The positive transcriptional regulator INO2 and its targets CHO1, CHO2, PSD1, OPI3, and EPT1 were down-regulated in the mutant cells (supplemental Tables S2 and S4 and Fig. 2B). There is an intimate relationship between triglycerides and phospholipids; both biosynthetic pathways converge through common intermediates (diacylglycerol and fatty acids); thus it is not surprising that they are coordinately regulated.
Global Gene Expression Changes Due to Neutral Lipid Deficiency
Total ablation of neutral lipid biosynthesis led to many changes in gene expression (supplemental Tables S2 and S5), many of which were confirmed by Northern hybridizations or quantitative real time PCR (Fig. 2). Consistent with the are1Δ are2Δ or dga1Δ lro1Δ double mutants, the quadruple mutant strain also displayed decreased expression of genes involved in iron and phospholipid metabolism. Additionally, we observed statistically significant decreases in several genes responsible for ergosterol biosynthesis, namely ERG3, ERG4, and ERG5 (supplemental Table S6 and Fig. 2A). Thus it appears that loss of the CLD compartment, as arises when neutral lipids are absent, has a more significant impact on the lipid transcriptome than does loss of steryl ester or TG alone.
Exogenous Fatty Acids Are Lipotoxic
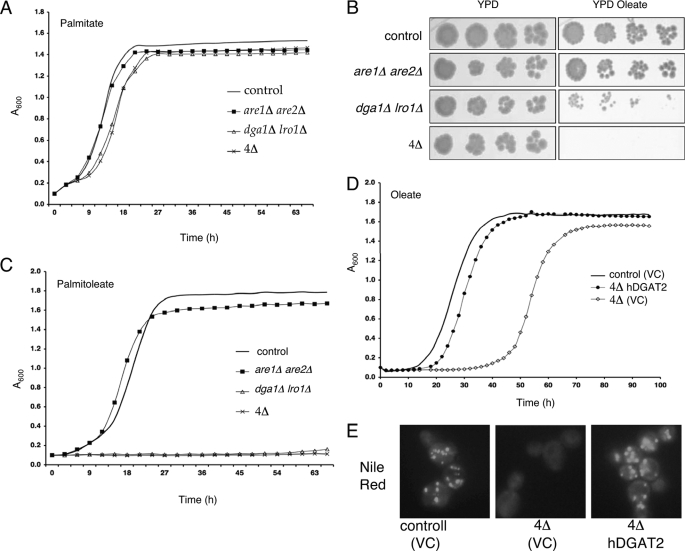
To provoke a viability response to neutral lipid deficiency, we incubated yeast cells with increasing concentrations of saturated or unsaturated free fatty acids, reasoning that these were common substrates to all of the enzymes under study and would reach toxic levels in the deletion strains. All cells, regardless of genotype, absorbed equivalent levels of free fatty acids (∼5 pmol/mg cell dry weight, based on radiolabeled metabolic incorporation studies). The saturated fatty acids palmitate (C16:0) and stearate (C18:0, data not shown) had no detectable impact on the viability of these strains (Fig. 3A). By contrast, unsaturated fatty acids such as oleate (C18:1) were toxic to strains lacking normal levels of triglyceride synthesis (Fig. 3B). This was more striking in the quadruple deletion strains lacking neutral lipid biosynthesis, where severe growth inhibition was observed with concentrations as low as 0.1 mm (data not shown). The shortening of the chain length (e.g. palmitoleate, C16:1, see Fig. 3C) or increasing the degree of unsaturation (e.g. linoleate, 18:2, data not shown) further exacerbated this lipotoxicity. In the case of oleate treatment, a pronounced lag phase was followed by a virtually normal growth rate. Palmitoleate toxicity is more severe; the cells never enter log phase of growth. Interestingly, are1Δ are2Δ strains lacking sterol esterification were unaffected by exogenous fatty acids.
FIGURE 3.
Exogenous free fatty acids are cytotoxic to strains lacking triglycerides. A, growth curve analysis of control and mutant strains in YPD + 0.5 mm palmitate. B, control and mutant strains were plated in serial dilutions on YPD ± 0.5 mm oleate. Results are representative of at least three experiments performed in triplicate. C, growth curve analysis of control and mutant strains in YPD + 0.5 mm palmitoleate. D, growth curve analysis of control and are1Δ are2Δ dga1Δ lro1Δ (4Δ) strains harboring hDGAT2 in media supplemented with 0.5 mm oleate. E, Nile red fluorescence detecting cytoplasmic neutral lipid deposition in control or are1Δ are2Δ dga1Δ lro1Δ (4Δ) strains harboring hDGAT2. VC = vector control.
Expression of Human DGAT2 Prevents Lipotoxicity
The mechanism that underlies the death of UFA-treated neutral lipid-deficient yeast cells is likely multifactorial. To confirm that restoration of TG biosynthesis was sufficient to restore cell viability, we expressed a human DGAT2 cDNA in these mutants (Fig. 3, D and E), an approach used previously to characterize mammalian neutral lipid biosynthesis (25). Expression of human DGAT2 in these yeast strains resulted in a marked elevation of TG synthesis (supplemental Fig. S1), complete rescue of the oleate-induced inviability of the quadruple deletion strain (Fig. 3D), and deposition of NL in cytoplasmic lipid droplets as observed by Nile red staining (Fig. 3E). This is a critical observation that suggests it is the accumulation of TG precursors or loss of the CLD compartment that limits the growth of the TG-deficient strains.
Transcriptional Response to UFA Challenge
To assess the primary cellular response to fatty acids in the context of neutral lipid deficiency, we assessed global gene expression changes between control and are1Δ are2Δ lro1Δ dga1Δ strains grown for 6 h in sub-lethal concentrations of oleate (0.01 mm, see supplemental Table S6). Differential gene expression between control and mutant cells was determined by p value (p ≤ 0.05) or by using the T-profiler algorithm, which correlates microarray expression data with DNA sequences immediately upstream of the ORFs (45) (supplemental Fig. S2). Despite the absence of a growth phenotype at this concentration of UFA, expression of all genes classified under the GO category “stress response” were increased relative to control strains (supplemental Fig. S2A). Four consensus motifs associated with differential gene expression summarize the data (Table 1). This includes an overall higher expression of genes containing the stress response-related RLM1 (366 ORFs) and STRE (960 ORFs) consensus motifs compared with that of all other genes on the microarray (positive T-value). Concomitant with the increase in stress-responsive genes was a general decrease of genes involved in transcription, RNA, ribosome, and protein metabolism (supplemental Fig. S2B), including RNA polymerase III subunits RPC17 and RPC34. The rRPE and PAC consensus motifs found on genes associated with rRNA and ribosome biosynthesis (1297 and 248 ORFs, respectively) displayed a negative T-profiler value indicating an overall decrease in expression of genes within this category (Table 1). Thus, even at sub-lethal concentrations, it is clear that neutral lipid-deficient cells initiate a general stress response to accommodate toxic fatty acids such as oleate.
TABLE 1.
Consensus motifs associated with statistically significant (t test) transcriptional changes in are1Δ are2Δ dga1Δ lro1Δ strains after growth in 0.01 m oleate
The T-value represents the change in the average activity of sets of genes based on Gene Ontology, chromatin immunoprecipitation-chip experiments, and known transcription factor consensus motifs (45).
| Motif | Transcription factor | T-value | ORFs |
|---|---|---|---|
| TAWWWWTAGM | RLM1 | 4.61 | 366 |
| CCCCT | STRE | 3.61 | 960 |
| AAAATTT | rRPE | −5.54 | 1297 |
| CGATGAG | PAC | −6.81 | 248 |
Lipoapoptosis in Budding Yeast
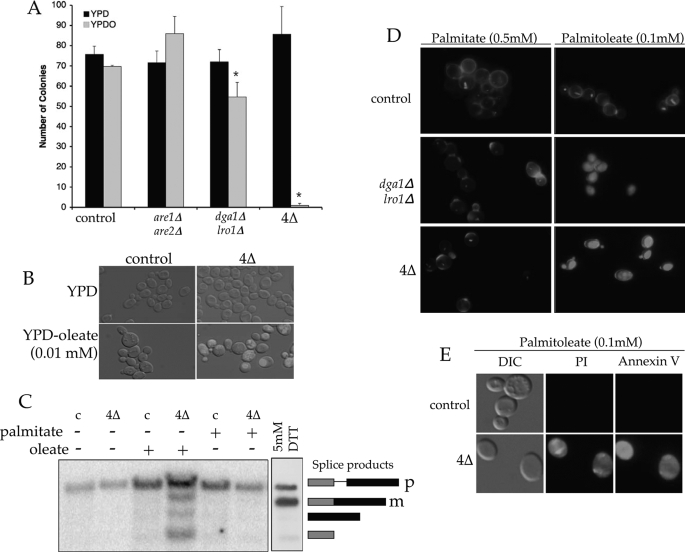
Programmed cell death as a consequence of surplus free fatty acids has been described in several organisms (3, 46) and is likely to play a major role in the metabolic syndrome of obesity, diabetes, and atherosclerosis. We therefore investigated the mechanism of cell death in yeast acyltransferase mutants treated with oleate or palmitoleate (Fig. 4). Senescence as opposed to quiescence (i.e. toxicity versus starvation) is the predominant outcome of UFA treatment; colony formation on fatty acid-free media following oleate preincubation (0.5 mm) was markedly impaired in cells lacking neutral lipids (Fig. 4A).
FIGURE 4.
Mechanisms of lipid-mediated cell death. A, colony-forming assay (means of triplicates ± S.D.) on YPD plates after growth in either liquid YPD or YPD containing 0.5 mm oleate (YPDO). Asterisks indicate significant difference (p ≤ 0.05; Student's t test). B, accumulation of reactive oxygen species in acyltransferase mutants grown in the presence of 0.01 mm oleate, as visualized by DHR 123. C, Northern blot analysis examining the induction of the UPR via IRE1-mediated HAC1 splicing in RNA extracts isolated from control (c) or dga1Δ lro1Δ are1Δ are2Δ (4Δ) cells grown in YPD ± 0.05 mm fatty acid. 5 mm Dithiothreitol (DTT) was used to induce the UPR as indicated by generation of the mature (m) spliced form of HAC1 mRNA from its precursor (p). Lower molecular weight bands represent splicing intermediates. D, cellular metabolism and viability as assessed by FUN-1 (20 μm; internal staining) of normal or mutant cells grown in YPD plus palmitate or palmitoleate for 18 h. The cell wall was visualized using calcofluor white (10 μm; cell perimeter). E, fluoroscopic assays of apoptosis in control or neutral lipid mutant cells grown in YPD ± 0.1 mm palmitoleate using PI (2 μg/ml) and annexin V (1 μg/ml).
The accumulation of reactive oxygen species (ROS) is an early indicator and possible cause of cell death in most organisms. To assess the impact of such molecules in lipotoxicity in yeast, we stained cells with DHR 123 (Fig. 4B). DHR 123-positive staining in the quadruple acyltransferase mutants treated with sub-lethal levels of oleate (0.01 mm) was absent from control strains or any cell incubated in the presence of saturated fatty acids suggesting that ROS accumulation is an early component of UFA-mediated cell death in this yeast mutant.
The integrity of the ER in mammalian cells is a major component of cell homeostasis and often responds to stressors, such as ROS accumulation, by activation of the unfolded protein response (UPR) transcriptional cascade. In turn this can trigger apoptosis (47). In yeast and humans a key initiating event in this response is the Ire1p-mediated mRNA splicing of a transcription factor (Hac1p in yeast and XBP1 in metazoans) that activates numerous metabolic events, including phospholipid biosynthesis and sterol esterification (for example, human ACAT2 and yeast ARE1 are both induced by the UPR). We assessed splicing of HAC1 as a reporter of the UPR in strains lacking neutral lipid biosynthesis, by Northern hybridization (Fig. 4C). The presence of sub-toxic levels of oleate (0.05 mm) clearly induced the splicing of HAC1 and thus the UPR; however, SFA at a 10-fold higher concentration had no effect on this aspect of ER quality control.
To further investigate the mechanism of cell death, we assessed the impact of fatty acids using the yeast cell viability stain FUN1 (Fig. 4D). In control cells, FUN1 is transported to the vacuole and converted into a cylindrical intravacuolar structure. This process has been shown to require mitochondrial ATP as addition of uncoupling agents prevents cylindrical intravacuolar structure formation (48). By contrast, triglyceride- and neutral lipid-deficient strains exhibited diffuse FUN1 staining after growth in 0.05 mm UFA, suggesting metabolic debilitation due to mitochondrial dysfunction. Finally, we assessed the role of programmed cell death in this phenomenon by propidium iodide (PI) staining of cell nuclei and phosphatidylserine exposure at the outer leaflet of the plasma membrane as judged by annexin V binding (Fig. 4E). PI staining alone is indicative of necrosis; however, the combination of PI staining and phosphatidylserine exposure indicates a late stage of apoptosis (35). TG-deficient strains clearly underwent lipoapoptosis after treatment with UFAs (0.1 mm). Treatment of any strain with saturated fatty acids was without phenotype by these assays.
Fate of Fatty Acids in the Absence of Neutral Lipid Synthesis
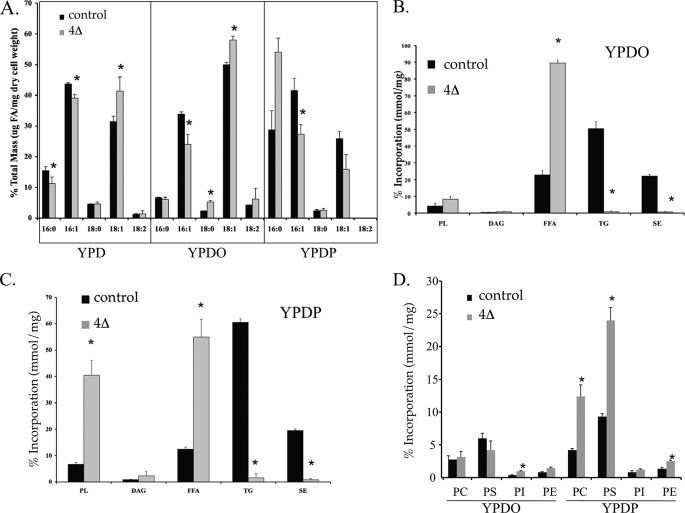
Saturated or unsaturated fatty acid cytotoxicity is cell type-dependent in mammalian systems and arises by different mechanisms (2, 49–51). To investigate the mechanism of lipotoxicity in NL-deficient yeast, we assessed the metabolism of oleate or palmitate in control and mutant cells treated with unlabeled fatty acids for 6 h (Fig. 5). This time course had no detectable impact on growth rates. Using capillary gas chromatography (Fig. 5A), we observed a remodeling of endogenous fatty acids in the NL-deficient strains such that oleate accumulated at the expense of palmitate and palmitoleate, consistent with elevated fatty acid elongation and desaturation. The short term inclusion of exogenous UFA (0.5 mm oleate) markedly and specifically elevated the UFA composition of all cells, with greater impact when TG synthesis was genetically ablated. A similar bolus application of 0.5 mm palmitate to cells specifically and significantly elevated cellular palmitate with little impact on UFA suggesting that elongation and desaturation of exogenous fatty acids did not occur in this time period with this quantity of SFA. In control cells, 70–80% of fatty acids were incorporated into neutral lipids (TG and SE), regardless of saturation (Fig. 5, B and C). However, in cells lacking the ability to synthesize neutral lipids, the metabolism of UFA versus SFA was markedly divergent. In the mutant cells, ∼40% of palmitate was preferentially incorporated into phospholipids (Fig. 5B), mainly phosphatidylcholine and phosphatidylserine (a 2–3-fold elevation relative to control cells, see Fig. 5D). Phosphatidylcholine mass in the NL-deficient strain was also elevated ∼2-fold (supplemental Fig. S3). By contrast, oleate incorporation into phospholipids in the quadruple mutant strain was similar to wild-type cells (∼6%, see Fig. 5C). As a consequence, ∼90% of total cell-associated counts accumulated as free oleate in the quadruple mutant.
FIGURE 5.
Phospholipid synthesis is protective against lipotoxicity. A, fatty acid species as measured by capillary gas chromatography. The percentage of major long chain fatty acid species was measured at mid-log in control and mutant (4Δ) strains after growth in YPD, YPD + 0.5 mm of oleate (YPDO), or palmitate (YPDP) for 6 h. The data are the average of two experiments, in duplicate. Lipid synthesis (expressed as a percentage of total) in control and dga1Δ lro1Δ are1Δ are2Δ strains grown in 0.5 mm palmitate (YPDP; B) or oleate (YPDO; C) until mid-log followed by a 1 h [3H]palmitic acid or [3H]oleic acid pulse, respectively. D, phospholipid synthesis (expressed as a percentage of total lipid) after growth of control and dga1Δ lro1Δ are1Δ are2Δ strains in 0.5 mm oleate or palmitate until mid-log followed by a 1-h [3H]oleic acid or [3H]palmitic acid pulse, respectively. Asterisks indicate significant difference (p ≤ 0.05). PL, phospholipid; FFA, free fatty acid; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PS, phosphatidylserine.
Epistatic Interactions with NL Biosynthetic Genes
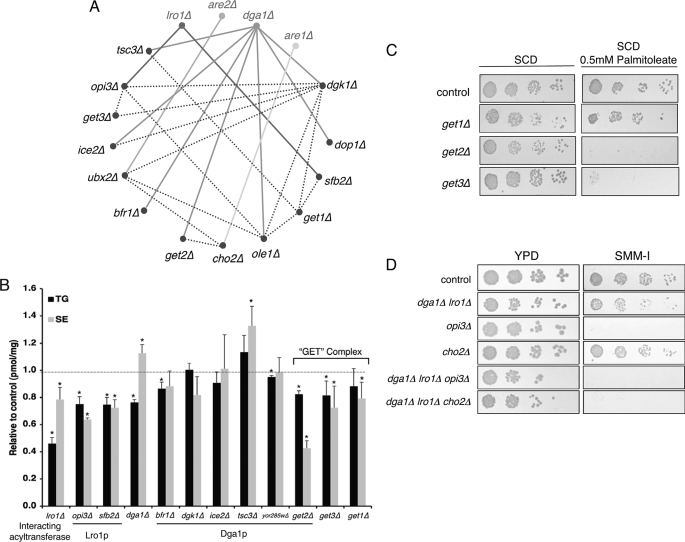
We present here a genetic model of lipotoxicity in which cells lacking the genes for neutral lipid biosynthesis succumb to the toxic effects of substrate accumulation. The successful exploitation of this paradigm rests on the application of genetic screens to identify interacting pathways and thus new facets of lipid homeostasis. Numerous interactions between genes encoding early secretory pathway proteins have been observed in synthetic lethality screens (e.g. Epistatic Mini Array Profiling (eMAP) (24)). We focused on mutations that were growth-impaired when combined with lesions in any one of the neutral lipid biosynthetic pathways (eMAP score less than −3, see Table 2 and Fig. 6A). The majority of interactions were with DGA1, likely because it encodes the predominant pathway for TG synthesis during the late logarithmic/stationary phase. Strains deleted at these loci were then assessed for alterations in lipid metabolism (Fig. 6B) or sensitivity to exogenous UFA (0.5 mm palmitoleate, see Table 2).
TABLE 2.
Epistatic interactions with acyltransferase genes
Viable, negatively interacting single mutants (eMAP score less than − 3) were grown in YPD or YPD + 0.5 mm palmitoleate (YPDPO). Growth is expressed as time taken for each single mutant to reach t½,max (normalized to wild type) in each media condition. Growth <1.0 = decreased growth rates. OLE1 and DOP1 are essential genes and were studied as decreased abundance by mRNA perturbation alleles (24). Strains deleted for ARE1, ARE2, DGA1, or LRO1 grew equivalently to control strains on YPD and YPD + palmitoleate.
| Acyltransferase | Interacting gene (ORF) | Interacting gene function | e-MAP score | Growth rate of deletion strain |
|
|---|---|---|---|---|---|
| YPD | YPDPO | ||||
| ARE1 | CHO2(YGR157W) | Phosphatidylcholine biosynthesis | −4.47 | 1.13 ± 0.15 | 1.40 ± 0.04a |
| ARE2 | UBX2(YML013W) | Protein catabolism (ubiquitin-mediated) | −3.06 | 1.16 ± 0.11 | 0.91 ± 0.01 |
| LRO1 | OPI3(YJR073C) | Phosphatidylcholine biosynthesis | −3.84 | 1.05 ± 0.12 | 1.30 ± 0.03a |
| SFB2(YNL049C) | COPII vesicle-mediated transport (SEC24 family) | −3.09 | 1.10 ± 0.11 | 1.14 ± 0.04a | |
| DGA1 | BFR1(YOR198C) | mRNA metabolism/vesicle-mediated transport | −8.16 | 1.01 ± 0.10 | 1.20 ± 0.07a |
| DOP1(YDR141C) (essential) | Membrane organization | −6.82 | 0.98 ± 0.01 | 0.95 ± 0.01 | |
| GET2(YER083C) | Golgi-ER vesicle-mediated transport | −6.17 | 0.97 ± 0.08 | 0.55 ± 0.01a | |
| DGK1(YOR311C) | Diacylglycerol kinase | −5.76 | 1.15 ± 0.18 | 0.99 ± 0.03 | |
| TSC3(YBR058C-A) | Sphingolipid biosynthesis | −4.04 | 1.16 ± 0.09 | 0.49 ± 0.04a | |
| YOR285W | Unknown | −3.94 | 1.04 ± 0.11 | 1.03 ± 0.08 | |
| OLE1(YGL055W) (essential) | Stearoyl-CoA desaturase | −3.32 | 0.96 ± 0.02 | 0.93 ± 0.05 | |
| ICE2(YIL090W) | ER organization | −3.15 | 1.03 ± 0.02 | 0.92 ± 0.04 | |
a Significant change is indicated (p value <0.05; Student's ttest).
FIGURE 6.
Epistatic interactions of neutral lipid biosynthetic genes. A, network visualization (Osprey spoke diagram) of interactions between ER-Golgi proteins that exhibit synthetic viability phenotypes (eMAP score less than −3 (24)) with acyltransferase mutants. Connecting lines indicate the double mutant was significantly less viable than either parent. Interactions with DGA1, LRO1, ARE1, and ARE2 are exhibited by solid lines. Dotted lines indicate genetic interactions among the other genes in this subset. B, TG and SE levels relative to control strains after 1 h of [3H]oleic acid pulse in mutants shown to interact with DGA1 or LRO1 deletions. Asterisks indicate significant difference from control strains (p ≤ 0.05; Student's t test, n = 3). C, control and GET-complex mutants were plated at serial dilutions on SCD or SCD + palmitoleate (0.5 mm). D, strains with the indicated mutations in phosphatidylethanolamine methylation and neutral lipid biosynthetic pathways were plated at serial dilutions on YPD or media lacking inositol and choline (SMM-I). Results are representative of at least three experiments.
Consistent with the aforementioned studies on the role of phospholipids in evading lipotoxicity, we observed many genetic interactions between the acyltransferases and mediators of phospholipid metabolism. DGK1 encodes diacylglycerol kinase that mediates the formation of phosphatidate from diacylglycerols (52, 53) and thus shares a substrate with its synthetic partner, the diacylglycerol acyltransferase encoded by DGA1 (Fig. 6A). Similarly, strains deleted for TSC3, encoding a regulatory subunit of serine palmitoyltransferase, the first step in sphingolipid synthesis, are strikingly sensitive to exogenous UFA and synthetically compromised by loss of DGA1. The production of PC by sequential methylation of phosphatidylethanolamine is mediated by the CHO2- and OPI3-encoded methyltransferases (54). These reactions are altered transcriptionally (Fig. 2) and display epistatic interactions with ARE1 and LRO1 (Fig. 6A). As anticipated, dga1Δ lro1Δ opi3Δ mutants exhibited decreased growth under basal conditions when compared with control strains or single mutants (Fig. 6D). Moreover, deletion of either OPI3 or CHO2 in the TG-deficient (dga1Δ lro1Δ) background reduced tolerance to 0.5 mm palmitate (Table 3 and supplemental Fig. S4). Finally, dga1Δ lro1Δ strains display a reduction in viability on minimal media lacking inositol and choline, where the methyltransferase pathway becomes limiting for synthesis of PC. This was markedly exacerbated when combined with cho2Δ mutants (Fig. 6D).
TABLE 3.
TG mutants harboring a defective PEMT pathway exhibit sensitivity to palmitate during the logarithmic growth phase
Growth is expressed as time taken for each mutant to reach t½,max (normalized to wild type; growth <1.0 = decreased growth rates). Growth curves for this experiment are described in supplemental Fig. S4.
| Strain | YPD | YPD + 0.5 mm palmitate |
|---|---|---|
| Control | 1.0 ± 0.0 | 1.0 ± 0.0 |
| dga1Δ lro1Δ | 1.0 ± 0.02 | 0.9 ± 0.05 |
| dga1Δ lro1Δ opi3Δ | 0.95 ± 0.10 | 0.66 ± 0.11a |
| dga1Δ lro1Δ cho2Δ | 1.0 ± 0.03 | 0.64 ± 0.15a |
a Significant change is indicated (p value <0.05; Student's t test).
Other gene deletions were epistatic with mutations in the acyltransferases (Table 2 and Fig. 6A) and also impacted lipid metabolism. Strains lacking the COPII-vesicle coat protein encoded by SFB2 or bearing lesions in the UBX2 gene (involved in ER-associated degradation) synthesize less triglyceride and were growth-compromised when combined with LRO1 or ARE2 deletions, respectively (Fig. 6). Strains deleted for mediators of vesicular transport (BFR1 and GET2) displayed altered incorporation of fatty acids into neutral lipids (Fig. 6B) and altered growth in response to exogenous UFA (Table 2). Get2p along with the GET1 and GET3 gene products form the GET complex required for protein transit between ER and Golgi. Given our observations with get2Δ strains, we also assessed GET1 and GET3 deletion mutants. Loss of any component of this complex compromised oleate incorporation into TG or SE (Fig. 6B); however, only GET3 deletions resembled get2Δ strains with respect to palmitoleate sensitivity (Fig. 6C).
DISCUSSION
The model described here facilitates the manipulation of genetic and environmental pressures that lead to cellular dysfunction due to lipid excess. This phenomenon is among a pallet of events associated with syndromes such as obesity, type 2 diabetes, and atherosclerosis. Disease onset reflects the metabolic saturation or genetic deficiency of cellular homeostatic mechanisms that normally protect the body from a lipid surplus. Oleate-induced lipotoxicity arises due to interruption of a protein phosphorylation/dephosphorylation cycle (5), whereas saturated fatty acids, such as palmitate, induce toxicity by elevating ceramide and/or reactive oxygen species (3, 55). In Chinese hamster ovary cells, saturated fatty acid toxicity is overcome in the presence of oleate by channeling into TG biosynthetic pathways (56). Diminution of TG biosynthesis renders saturated and unsaturated fatty acids equally toxic (56).
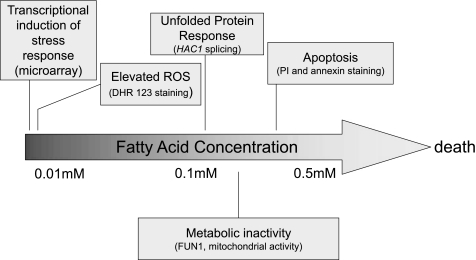
The genetic ablation of the yeast neutral lipid biosynthetic pathway and consequential loss of the CLD compartment combined with the addition of exogenous UFAs mimic the acute lipid overload (lipotoxicity) that often besets nonadipose tissues. By assessing the response of these yeast strains to increasing concentrations of UFA, we have characterized the events that lead to cell death. The cascade initiates with a generalized transcriptional stress response followed by elevated reactive oxygen species, induction of the unfolded protein response, metabolic dysfunction, and apoptosis (Fig. 7). These results suggest that lipotoxicity studies in yeast are relevant to higher eukaryotes and represent a suitable model to study this process in greater detail.
FIGURE 7.
Progression of unsaturated fatty acid-mediated lipotoxicity in the absence of NL biosynthesis. The mechanisms underlying lipid-mediated cell death are depicted as a function of exogenous fatty acid concentrations. In yeast and in mammals, unsaturated fatty acids are toxic in the absence of neutral lipid biosynthesis. Upon further deprivation of lipid synthesis (e.g. phospholipids), saturated fatty acids also become toxic to yeast. The fluorescent stains PI, DHR 123, and FUN1 were used to visualize fragmented DNA, ROS, and metabolic states, respectively (see text).
It is unclear if the transcriptional and biochemical effects associated with neutral lipid deficiency result from the loss of the lipid molecules or are by-products of the loss of CLDs. This cellular compartment, consisting of a neutral lipid (commonly TG or SE) core encased in a polar lipid monolayer embedded with proteins, exists in the cytosol of all eukaryotes (57, 58) and in the ER-lumen of specialized cells such as hepatocytes. In the budding yeast, S. cerevisiae, we have investigated the impact of losing this depot by ablating neutral lipid biosynthesis. We demonstrate that the production of a core neutral lipid is the initiating event in CLD biogenesis. Moreover, the major regulator of CLD formation is the synthesis of TG; SE deprivation does not significantly impact production of CLDs, although it is sufficient for their formation. The finding that the composition of the CLD core (TG versus SE) impacts its protein composition is unprecedented. However, protein translocation between subcellular compartments is a common and effective mechanism for activation or deactivation of reactions. Indeed, complete loss of the CLD compartment modulates sterol levels by retention of ergosterol biosynthetic enzymes at the ER (43).
In contrast to the striking impact of even sub-physiological levels of unsaturated fatty acids (0.05 mm oleate here, as compared with ∼0.5 mm in human serum (59)), exogenous saturated fatty acids (up to its solubility of 0.5 mm) had no effect on the viability of the acyltransferase-deficient yeast strains. We propose that differential channeling of SFA into phospholipids (Fig. 5) is a significant contributor to the evasion of lipotoxicity. This hypothesis is supported by several observations, the first of which is the existence of an epistatic growth interaction between the LRO1-encoded acyltransferase and OPI3, a gene involved in the synthesis of phosphatidylcholine. Strains lacking TG and PEMTs encoded by OPI3 or CHO2 displayed an ∼2-fold growth defect in the presence of 0.5 mm palmitate. Additionally, when exposed to minimal media lacking inositol and choline (thereby obligating a functional PEMT pathway for the synthesis of PC), dga1Δ lro1Δ strains showed a decreased growth capacity relative to control strains. Interestingly, the majority of mammalian tissues, with the notable exception of the liver, lacks the ability to methylate phosphatidylethanolamine (60), perhaps explaining the sensitivity of many mammalian cells to SFA.
The alternate use of exogenous SFA as substrates for phospholipid biosynthesis by dga1Δ lro1Δ strains clearly demonstrates a symbiosis between phospholipids and triglycerides. One of the most striking epistatic relationships is the synthetic interaction of the pathways defined by DGK1 and DGA1. Both enzymes modify diacylglycerol by phosphorylation or esterification, respectively, and thus control the flux of fatty acids into phospholipids or triacylglycerol. The loss of both pathways would thereby expose a cell to distortions in DAG that could lead to inviability. Unexpectedly, DGK1 is an interaction hub for this subset of genes; the great majority of genes that interact with the sterol and DAG acyltransferases also interact with DGK1 (Fig. 6A). Similarly, Tsc3p, a positive regulator of serine-palmitoyltransferase activity (61), the first reaction in sphingolipid biosynthesis, was required for optimal growth in the context of impaired neutral lipid production or exposure to palmitoleate. This phenomenon is further supported by transcriptional changes to many genes involved in phospholipid formation in the TG-deficient strains. We conclude that the synthesis of neutral lipids acts as a buffering mechanism allowing cells to maintain viability in the face of excess lipid exposure and that phospholipid synthesis, particularly for saturated fatty acids, represents an alternate salvage point.
The ability of neutral lipid-deficient yeast to utilize the phospholipid biosynthetic machinery to maintain cellular homeostasis in the presence of toxic fatty acids prompted us to search for other compensatory pathways. Deletion of GET2, encoding a subunit of the GET complex, a multicomponent pathway for Golgi-ER (i.e. retrograde) transport, genetically interacts with a DGA1 deletion to compromise growth. Moreover, loss of GET2 or GET3 confers sensitivity to exogenous UFA and reduced steryl ester or TG biosynthesis. GET mutations are exceptionally pleiotropic, with phenotypes as diverse as metal sensitivity, mitochondrial fragmentation, temperature sensitivity, and defective sporulation. This has been explained based on a role of the GET proteins and their metazoan counterparts in the insertion of “tail-anchored” proteins into ER membranes (62). Our results link this process to lipid metabolism in terms of how a eukaryotic cell deals with toxic levels of fatty acids.
We reasoned that the identification of pathways that respond or compensate for defects in neutral lipid synthesis could lead to a better comprehension of CLD formation in all cells and the pathogenesis and treatment of human illnesses that have lipotoxicity as a component. A high number of the yeast genes identified here have human orthologs, suggesting the pathways are conserved through evolution. This includes diacylglycerol kinase, the phosphatidylethanolamine N-methyltransferases, and GET3, which is orthologous to human ASNA1. Intriguingly, PEMT activity is elevated in diabetic states (63) and the ASNA1 gene product is required for insulin secretion in murine and nematode models (64). Our findings thus provide a mechanistic link between lipotoxicity and insulin secretion that was not previously apparent. We hypothesize that our approach to developing S. cerevisiae as a model of lipid-mediated cell death has identified new components of human diseases such as type II diabetes where lipid overload is implicated as causative.
Supplementary Material
Acknowledgments
We appreciate the assistance of Maya Schuldiner and Harmen Bussemaker and a National Science Foundation Award 0090286 on Development, Evaluation, and Dissemination of Methods for the Analysis of Gene Expression by Microarrays (to David Allison, University of Birmingham, Birmingham, AL).
This work was supported, in whole or in part, by National Institutes of Health Grant DK54320 (to S. L. S.) and Grant 2P30-DK63608-07 from NIDDK (pilot award from Diabetes and Endocrinology Research Center to Columbia University). This work was also supported by the American Diabetes Association and the American Heart Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S6.
- TG
- triacylglycerol
- ACAT
- acyl-CoA:cholesterol acyltransferase
- CLD
- cytoplasmic lipid droplet
- DAG
- diacylglycerols
- DGAT
- Diacylglycerol Acyltransferase
- SE
- steryl ester
- ORF
- open reading frame
- GFP
- green fluorescent protein
- ROS
- reactive oxygen species
- DHR
- dihydrorhodamine
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum
- PI
- propidium iodide
- eMAP
- Epistatic Mini Array Profiling
- UFA
- unsaturated fatty acid
- SFA
- saturated fatty acid
- NL
- neutral lipid.
REFERENCES
- 1.McGarry J. D. (2002) Diabetes 51, 7–18 [DOI] [PubMed] [Google Scholar]
- 2.Garbarino J., Sturley S. L. (2009) Curr. Opin. Clin. Nutr. Metab. Care 12, 110–116 [DOI] [PubMed] [Google Scholar]
- 3.Listenberger L. L., Ory D. S., Schaffer J. E. (2001) J. Biol. Chem. 276, 14890–14895 [DOI] [PubMed] [Google Scholar]
- 4.Holland W. L., Brozinick J. T., Wang L. P., Hawkins E. D., Sargent K. M., Liu Y., Narra K., Hoehn K. L., Knotts T. A., Siesky A., Nelson D. H., Karathanasis S. K., Fontenot G. K., Birnbaum M. J., Summers S. A. (2007) Cell Metab. 5, 167–179 [DOI] [PubMed] [Google Scholar]
- 5.Schwarz S., Hufnagel B., Dworak M., Klumpp S., Krieglstein J. (2006) Apoptosis 11, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 6.Feng B., Yao P. M., Li Y., Devlin C. M., Zhang D., Harding H. P., Sweeney M., Rong J. X., Kuriakose G., Fisher E. A., Marks A. R., Ron D., Tabas I. (2003) Nat. Cell Biol. 5, 781–792 [DOI] [PubMed] [Google Scholar]
- 7.Chang B. H., Chan L. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1465–G1468 [DOI] [PubMed] [Google Scholar]
- 8.Turkish A., Sturley S. L. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G953–G957 [DOI] [PubMed] [Google Scholar]
- 9.Yang H., Bard M., Bruner D. A., Gleeson A., Deckelbaum R. J., Aljinovic G., Pohl T. M., Rothstein R., Sturley S. L. (1996) Science 272, 1353–1356 [DOI] [PubMed] [Google Scholar]
- 10.Oelkers P., Behari A., Cromley D., Billheimer J. T., Sturley S. L. (1998) J. Biol. Chem. 273, 26765–26771 [DOI] [PubMed] [Google Scholar]
- 11.Oelkers P., Cromley D., Padamsee M., Billheimer J. T., Sturley S. L. (2002) J. Biol. Chem. 277, 8877–8881 [DOI] [PubMed] [Google Scholar]
- 12.Sandager L., Dahlqvist A., Banaú A., Ståhl U., Lenman M., Gustavsson M., Stymne S. (2000) Biochem. Soc. Trans. 28, 700–702 [PubMed] [Google Scholar]
- 13.Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., Sturley S. L. (2000) J. Biol. Chem. 275, 15609–15612 [DOI] [PubMed] [Google Scholar]
- 14.Dahlqvist A., Stahl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandager L., Gustavsson M. H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. (2002) J. Biol. Chem. 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- 16.Sorger D., Daum G. (2002) J. Bacteriol. 184, 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garbarino J., Sturley S. L. (2005) Biochem. Soc. Trans. 33, 1182–1185 [DOI] [PubMed] [Google Scholar]
- 18.Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., Natter K., Kohlwein S. D. (2009) J. Biol. Chem. 284, 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1998) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- 20.Orr-Weaver T. L., Szostak J. W., Rothstein R. J. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaynor P. M., Gill T., Toutenhoofd S., Summers E. F., McGraw P., Homann M. J., Henry S. A., Carman G. M. (1991) Biochim. Biophys. Acta 1090, 326–332 [DOI] [PubMed] [Google Scholar]
- 22.Mitchell A. G., Martin C. E. (1995) J. Biol. Chem. 270, 29766–29772 [DOI] [PubMed] [Google Scholar]
- 23.Stukey J. E., McDonough V. M., Martin C. E. (1989) J. Biol. Chem. 264, 16537–16544 [PubMed] [Google Scholar]
- 24.Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., Krogan N. J. (2005) Cell 123, 507–519 [DOI] [PubMed] [Google Scholar]
- 25.Turkish A. R., Henneberry A. L., Cromley D., Padamsee M., Oelkers P., Bazzi H., Christiano A. M., Billheimer J. T., Sturley S. L. (2005) J. Biol. Chem. 280, 14755–14764 [DOI] [PubMed] [Google Scholar]
- 26.Cartwright I. J. (1993) Methods in Molecular Biology, Vol. 19, pp. 153–167, Humana Press, Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 27.Hultin M., Müllertz A., Zundel M. A., Olivecrona G., Hansen T. T., Deckelbaum R. J., Carpentier Y. A., Olivecrona T. (1994) J. Lipid Res. 35, 1850–1860 [PubMed] [Google Scholar]
- 28.Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., Sturley S. L. (2002) J. Biol. Chem. 277, 32466–32472 [DOI] [PubMed] [Google Scholar]
- 29.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry R. A., Ooi S. L., Wu Z., Boeke J. D. (2003) Stat. Appl. Genet. Mol. Biol. 2, Article 1 [DOI] [PubMed] [Google Scholar]
- 31.Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article 3 [DOI] [PubMed] [Google Scholar]
- 32.Wettenhall J. M., Smyth G. K. (2004) Bioinformatics 20, 3705–3706 [DOI] [PubMed] [Google Scholar]
- 33.Wettenhall J. M., Simpson K. M., Satterley K., Smyth G. K. (2006) Bioinformatics 22, 897–899 [DOI] [PubMed] [Google Scholar]
- 34.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 35.Madeo F., Fröhlich E., Fröhlich K. U. (1997) J. Cell Biol. 139, 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madeo F., Fröhlich E., Ligr M., Grey M., Sigrist S. J., Wolf D. H., Fröhlich K. U. (1999) J. Cell Biol. 145, 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mo C., Milla P., Athenstaedt K., Ott R., Balliano G., Daum G., Bard M. (2003) Biochim. Biophys. Acta 1633, 68–74 [DOI] [PubMed] [Google Scholar]
- 38.Esposito M. S., Esposito R. E., Arnaud M., Halvorson H. O. (1969) J. Bacteriology 100, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law S. W., Burton D. N. (1976) Can. J. Microbiol. 22, 1710–1715 [DOI] [PubMed] [Google Scholar]
- 40.Henry S. A., Halvorson H. O. (1973) J. Bacteriol. 114, 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ou J., Tu H., Shan B., Luk A., DeBose-Boyd R. A., Bashmakov Y., Goldstein J. L., Brown M. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. (2001) J. Biol. Chem. 276, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 43.Sorger D., Athenstaedt K., Hrastnik C., Daum G. (2004) J. Biol. Chem. 279, 31190–31196 [DOI] [PubMed] [Google Scholar]
- 44.Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003) FEMS Microbiol. Rev. 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 45.Boorsma A., Foat B. C., Vis D., Klis F., Bussemaker H. J. (2005) Nucleic Acids Res. 33, W592–W5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q., Chieu H. K., Low C. P., Zhang S., Heng C. K., Yang H. (2003) J. Biol. Chem. 278, 47145–47155 [DOI] [PubMed] [Google Scholar]
- 47.Patil C., Walter P. (2001) Curr. Opin. Cell Biol. 13, 349–355 [DOI] [PubMed] [Google Scholar]
- 48.Millard P. J., Roth B. L., Thi H. P., Yue S. T., Haugland R. P. (1997) Appl. Environ. Microbiol. 63, 2897–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kudo T., Wu J., Ogawa Y., Suga S., Hasegawa N., Suda T., Mizukami H., Yagihashi S., Wakui M. (2006) J. Pharmacol. Exp. Ther. 318, 1203–1210 [DOI] [PubMed] [Google Scholar]
- 50.Cury-Boaventura M. F., Curi R. (2005) Clin. Sci. 108, 245–253 [DOI] [PubMed] [Google Scholar]
- 51.Cury-Boaventura M. F., Pompéia C., Curi R. (2004) Clin. Nutr. 23, 721–732 [DOI] [PubMed] [Google Scholar]
- 52.Han G. S., O'Hara L., Carman G. M., Siniossoglou S. (2008) J. Biol. Chem. 283, 20433–20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han G. S., O'Hara L., Siniossoglou S., Carman G. M. (2008) J. Biol. Chem. 283, 20443–20453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carman G. M., Zeimetz G. M. (1996) J. Biol. Chem. 271, 13293–13296 [DOI] [PubMed] [Google Scholar]
- 55.Shimabukuro M., Zhou Y. T., Levi M., Unger R. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2498–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S., Schaffer J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Packter N. M., Olukoshi E. R. (1995) Arch. Microbiol. 164, 420–427 [DOI] [PubMed] [Google Scholar]
- 58.Zweytick D., Athenstaedt K., Daum G. (2000) Biochim. Biophys. Acta 1469, 101–120 [DOI] [PubMed] [Google Scholar]
- 59.Roden M., Price T. B., Perseghin G., Petersen K. F., Rothman D. L., Cline G. W., Shulman G. I. (1996) J. Clin. Invest. 97, 2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walkey C. J., Cui Z., Agellon L. B., Vance D. E. (1996) J. Lipid Res. 37, 2341–2350 [PubMed] [Google Scholar]
- 61.Gable K., Slife H., Bacikova D., Monaghan E., Dunn T. M. (2000) J. Biol. Chem. 275, 7597–7603 [DOI] [PubMed] [Google Scholar]
- 62.Stefanovic S., Hegde R. S. (2007) Cell 128, 1147–1159 [DOI] [PubMed] [Google Scholar]
- 63.Hartz C. S., Nieman K. M., Jacobs R. L., Vance D. E., Schalinske K. L. (2006) J. Nutr. 136, 3005–3009 [DOI] [PubMed] [Google Scholar]
- 64.Kao G., Nordenson C., Still M., Rönnlund A., Tuck S., Naredi P. (2007) Cell 128, 577–587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.