Abstract
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is well characterized for its role in antagonizing the phosphoinositide 3-kinase pathway. Previous studies using size-exclusion chromatography demonstrated PTEN recruitment into high molecular mass complexes and hypothesized that PTEN phosphorylation status and PDZ binding domain may be required for such complex formation. In this study, we set out to test the structural requirements for PTEN complex assembly and identify the component(s) of the PTEN complex(es). Our results demonstrated that the PTEN catalytic function and PDZ binding domain are not absolutely required for its complex formation. On the other hand, PTEN phosphorylation status has a significant impact on its complex assembly. Our results further demonstrate enrichment of the PTEN complex in nuclear lysates, suggesting a mechanism through which PTEN phosphorylation may regulate its complex assembly. These results prompted further characterization of other protein components within the PTEN complex(es). Using size-exclusion chromatography and two-dimensional difference gel electrophoresis followed by mass spectrometry analysis, we identified heterogeneous nuclear ribonucleoprotein C (hnRNP C) as a novel protein recruited to higher molecular mass fractions in the presence of PTEN. Further analysis indicates that endogenous hnRNP C and PTEN interact and co-localize within the nucleus, suggesting a potential role for PTEN, alongside hnRNP C, in RNA regulation.
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN)4 was cloned in 1997 (1–3) and has been well characterized for its tumor-suppressive role by dephosphorylating phosphatidylinositol 3,4,5-trisphosphate to phosphatidylinositol 4,5-bisphosphate and antagonizing the phosphoinositide 3-kinase pathway (4–7). PTEN also regulates cell migration, cell cycle progression, DNA damage response, and chromosome stability independently of its lipid phosphatase activity through its potential protein phosphatase activity and/or protein-protein interaction (8–11) (for recent reviews, see 12–14).
PTEN is composed of an N-terminal catalytic domain and a C-terminal regulatory domain. The catalytic domain contains a conserved signature motif (HCXXGXXR) found in dual-specific protein phosphatases, and mutations within this catalytic domain, including the C124S mutation, are known to abrogate PTEN catalytic activity (4). The C terminus of PTEN contains a PDZ (PDS-95/Disc-large/Zo-1) binding domain, which interacts with PDZ-containing proteins such as MAGI-1b, MAGI-2, MAGI-3, hDLG, hMAST and NHERF (15–19). In addition to the PDZ binding domain, several key serine and threonine phosphorylation sites (Ser380, Thr382, Thr383, and Ser385) at the PTEN C terminus are reported to play an important role in regulating its stability, localization, and activity (20–26).
Recent studies suggest that PTEN may function within higher molecular mass complexes. Through size-exclusion chromatography, Vazquez et al. (27) demonstrated that PTEN can be separated into two populations: a monomeric hyperphosphorylated subpopulation and a higher molecular mass hypophosphorylated subpopulation. It was hypothesized that PTEN in its dephosphorylated form can interact with PDZ-containing proteins such as hDLG and be recruited into a higher molecular mass complex. Although the components within PTEN complex(es) have not been systematically studied and purified, MAGI-2, hDLG (27), NHERF2, PDGFR (19), NEP (28), and MVP (29) have been identified as potential components of the PTEN complex using the same size-exclusion chromatography methodology.
In this paper, we aim to (i) investigate the essential elements of PTEN required for its complex formation and (ii) dissect the components of the PTEN-associated complex(es). Our results indicate that PTEN catalytic activity or its PDZ binding domain is not absolutely required for complex assembly. PTEN phosphorylation status on amino acids Ser380, Thr382, Thr383, and Ser385, on the other hand, has a significant role in complex formation. In addition, we demonstrate that the PTEN complex is enriched in nuclear lysates, which suggests a mechanism through which phosphorylation can regulate complex assembly. Using two-dimensional difference gel electrophoresis (DIGE) analysis and comparing proteins present in higher molecular mass fractions in the presence and absence of PTEN followed by mass spectrometry analysis, we have identified heterogeneous nuclear ribonucleoprotein C (hnRNP C) as a potential component within the PTEN complex. PTEN and hnRNP C are shown here to interact and co-localize in the nucleus. We hypothesize that the PTEN and hnRNP C complex may play a role in RNA regulation.
EXPERIMENTAL PROCEDURES
Constructs
PTEN expression plasmids pSG5L A4-PTEN (S380A, T382A, T383A, and S385A), pSG5L E4-PTEN (S380E, T382E, T383E, and S385E), and NLS-PTEN were generous gifts from Dr. W. R. Sellers. The pCMV FLAG WT-PTEN and pCMV FLAG ΔPDZ-PTEN (1–400) constructs were made by cloning WT-PTEN and ΔPDZ-PTEN, lacking the 3 amino acids of the PDZ binding domain at the C terminus, into the pCMV-Tag2B vector (Stratagene) using BamHI and EcoRI sites. Constructs used to make the PC3 PTEN-inducible lines were generated using the pLenti4/V5-DEST Gateway Vector system (Invitrogen). WT and C124S PTEN cDNAs were recombined to entry vectors followed by recombination to destination, lenti vectors.
Cell Lines and Treatment Conditions
For generation of PC3 WT-PTEN- and C124S-PTEN-inducible cell lines, PC3 parental cells were transiently transfected with either WT or C124S-PTEN pLenti4/V5-DEST clones in conjunction with pLenti/TR (tet repressor). Generation of stable lines was then achieved using co-selection with blasticidin and zeocin. The PC3-inducible and PC3 parental cells were maintained in RPMI 1640 medium and HeLa cells in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 100 units/ml penicillin-streptomycin (Invitrogen). For H2O2 treatments, cells were incubated with 1 mm H2O2 and harvested after 1 h. For PTEN induction, 2 μg/ml doxycycline (Dox) was added to the PC3 PTEN-inducible cells, and proteins were harvested 36–48 h after treatment.
Nuclear/Cytosol Fractionation
HeLa cells were harvested, and the nuclear and cytoplasmic extracts were prepared using the NE-PER extraction reagents (Thermo Scientific). For NLS-PTEN-transfected PC3 cells, fractionation was performed using the Nuclear Extract kit (Active Motif). Following that, 2 mg of nuclear lysate and 7 mg of cytosolic lysate were loaded on the gel filtration column.
Transfection and Western Blot Analysis
Constructs were transfected into PC3 cells using the Lipofectamine Plus reagents (Invitrogen), and 48 h after transfection cells were lysed in radioimmune precipitation assay buffer (50 mm Tris, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, and 0.5% sodium deoxycholate) containing phosphatase (Sigma) and protease inhibitors (Roche Applied Science). Proteins were quantitated, and after preparation in Laemmli buffer containing β-mercaptoethanol, equal amounts were run on SDS-polyacrylamide gels with 4% stacking and 10% resolving layers. Proteins were transferred overnight onto nitrocellulose and after blocking with 2% milk incubated with antibodies made in 5% bovine serum albumin overnight. Following washing and incubation for 1 h with secondary antibodies conjugated to horseradish peroxidase (GE Healthcare), the blots were further washed and developed using ECL reagents (Pierce). The following antibodies were used for immunoblotting: Pan Actin (A5060; Sigma), AKT (9272; Cell Signaling), P-AKT (9271; Cell Signaling), GRP78 (3183; Cell Signaling), HDAC (sc-7872; Santa Cruz Biotechnology), hDLG (610874; BD Biosciences), hnRNP C (ab10294; Abcam), PTEN (9552, 9556; Cell Signaling), PTEN P380 (9551; Cell Signaling), and V5 (46-0705; Invitrogen).
Co-immunoprecipitation
For whole lysate co-immunoprecipitation, cells were lysed in immunoprecipitation buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40), and 1 mg of protein in immunoprecipitation buffer was precleared using A/G Plus agarose beads (Upstate) and incubated overnight with IgG control (sc-2028; Santa Cruz Biotechnologies) or PTEN (sc-6818; Santa Cruz Biotechnologies) antibodies. Samples were washed three times with the immunoprecipitation buffer, and proteins were eluted by boiling in Laemmli buffer containing β-mercaptoethanol followed by Western blot analysis. For co-immunoprecipitation done on nuclear and cytoplasmic fractions, cells were fractionated (as described above), and 200 μg of nuclear and 1 mg of cytoplasmic lysates were used.
Immunofluorescence Staining
Cells were grown on coverslips in 6-well plates, and PC3 WT-PTEN-inducible cells were induced for 36 h. After fixing with methanol, samples were blocked with 0.5% normal goat serum in 1% phosphate-buffered saline/Tween 20 and incubated with primary antibodies: hnRNP C (ab10294; Abcam) and PTEN (9559; Cell Signaling) for 1–2 h followed by incubation with secondary antibodies conjugated with Alexa Fluor 594 or 488 (Molecular Probes). The coverslips were mounted using mounting medium (Vector Laboratories).
Gel Filtration
Cells were lysed in TNN buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 5 mm EDTA) containing protease inhibitors (Roche Applied Science) and phosphatase inhibitor mixtures (Sigma). After lysis and centrifugation, samples were applied to a Sephacryl S-300 column (Amersham Biosciences) and eluted with phosphate-buffered saline in 2-ml fraction volumes with a 0.5 ml/min flow rate. For immunoblot analysis, proteins in each fraction were precipitated using cold acetone and resuspended in Laemmli buffer containing β-mercaptoethanol followed by Western blot analysis as described previously. For quantitative immunoblotting, fractions were concentrated using Amicon Ultra Centrifugal filter devices (UFC8 003 24; Millipore). Bradford assay was performed on these fractions, and an equal amount of protein was loaded on the SDS-polyacrylamide gels.
DIGE Analysis
PC3 WT-PTEN-induced and uninduced samples were lysed in lysis buffer (50 mm Tris, pH 7.0, 150 mm NaCl, 0.5% Nonidet P-40, phosphatase inhibitors, protease inhibitors, 50 μg/ml DNase I, 20 μg/ml RNase A) and applied to a Sephacryl S-300 column. Samples were eluted with EB (20 mm Tris, pH 7, 150 mm NaCl, 20 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride). Protein fractions 19 and 20 from PTEN-induced and uninduced samples were selected and concentrated by using Microcon ultracentrifugal filtration devices (3-kDa molecular mass cutoff). Samples were precipitated with a two-dimensional Clean-up kit (GE Healthcare) and suspended in labeling buffer (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 30 mm Tris, pH 8.5). Protein concentration in the induced and uninduced fractions (19 and 20, respectively) was determined with the two-dimensional Quant protein quantitation kit (GE Healthcare), and fractions 19 and 20 from induced and uninduced samples were combined, respectively. Protein samples were labeled according to the manufacturer's minimum labeling protocol (GE Healthcare) with the N-hydroxysuccinimidyl ester derivatives of the cyanide dyes Cy2, Cy3, and Cy5. Briefly, each sample was divided into three equal volumes containing a 30 μg of proteins each and individually labeled with 1 μl (240 pmol/μl) of Cy3 and Cy5, while the remaining 30 μg of each induced and uninduced protein samples were pooled and labeled with Cy2. Equal amounts of Cy3-labeled and Cy5-labeled, induced and uninduced protein samples were combined with the Cy2-labeled pool sample. The labeled samples were applied to immobilized pH 3–11 nonlinear gradient isoelectric focusing gel strips, and isoelectric focusing of passively rehydrated immobilized pH gradient strips was performed on an Ettan IPGphor II apparatus. After isoelectric focusing, immobilized pH gradient strips were equilibrated in equilibration buffer containing 50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, 1% dithiothreitol for 15 min at room temperature with gentle shaking and then reequilibrated in the equilibration buffer containing 4% iodoacetamide instead of dithiothreitol. Immobilized pH gradient strips were placed on top of the SDS gradient gels (12–16%) for the second dimension separation. Gel images of the Cy2, Cy3, Cy5, and Sypro Ruby were scanned by using a Typhoon TRIO Variable Mode Imager (GE Healthcare).
RESULTS
PTEN Catalytic Activity and PDZ Binding Domain Are Not Absolutely Required for PTEN Complex Assembly
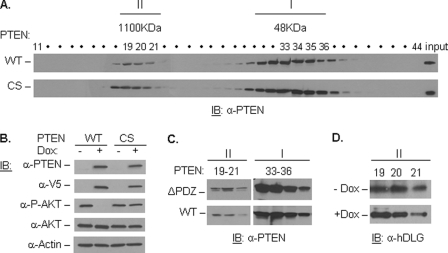
Many point mutations associated with human cancer are found in the catalytic domain of PTEN (30). To test whether PTEN catalytic activity is required for its complex formation, we performed size-exclusion chromatography analysis using PC3-inducible cell lines carrying either WT or the catalytic inactive C124S (CS) PTEN construct. Consistent with previous studies (19, 27, 28), PTEN can be detected in two main fractions: a lower molecular mass monomer (depicted as I) and a higher molecular mass complex (depicted as II) (Fig. 1A). Importantly, both WT and CS PTEN are able to form higher molecular mass complexes at comparable efficiency, indicating that the catalytic activity of PTEN is not essential for its complex assembly. As validation, the V5-tagged WT and CS mutant PTEN could be detected in the presence of the tetracycline analog, Dox, and WT but not CS mutant PTEN could reduce endogenous AKT phosphorylation upon Dox treatment (Fig. 1B).
FIGURE 1.
PTEN catalytic activity and PDZ binding domain are not absolutely required for complex assembly. A, PC3 WT- and CS (inactive)-PTEN-inducible cells were induced with Dox and run through a gel filtration column. Fractions were collected and immunoblotted with anti-PTEN antibody. Higher molecular mass and monomeric fractions are depicted as II and I, respectively. B, PC3 WT- and CS-PTEN-inducible cells were treated with 2 μg/ml Dox for 36 h, and cell lysates were immunoblotted (IB) with antibodies against PTEN, V5 tag and the PTEN downstream target, P-AKT, and total AKT, using anti-actin as a loading control. C, PC3 cells were transfected with WT- or ΔPDZ-PTEN (PTEN lacking the PDZ binding domain) and run on a gel filtration column. Complex (19–21) and monomer (33–36) fractions were collected and immunoblotted with PTEN antibody. D, complex fractions (19–21) from PC3 WT-PTEN-induced (+Dox) and uninduced (−Dox) were immunoblotted with an antibody against hDLG.
The PDZ binding domain has been implicated in facilitating the interaction of PTEN with several PDZ domain-containing proteins, such as hDLG and NHERF2 (18, 19). In addition, these proteins have been shown to co-fractionate with PTEN in high molecular mass fractions (19, 27). To test the importance of the PTEN PDZ binding domain in facilitating complex assembly, we transfected WT-PTEN (pCMV FLAG WT-PTEN) and PTEN with a truncated PDZ binding domain (pCMV FLAG ΔPDZ-PTEN) into the PTEN-null PC3 parental cells. Interestingly, we observed no significant change in complex assembly efficiency of ΔPDZ-PTEN (Fig. 1C). Because hDLG has been suggested as a potential component of the PTEN complex through interaction with its PDZ binding domain, we tested the recruitment of hDLG into high molecular mass fractions under PTEN-induced and uninduced conditions. As shown, in our PC3-inducible cells, hDLG recruitment to high molecular mass fractions takes place independently of PTEN status (Fig. 1D). Although we cannot rule out hDLG as a component of the PTEN complex, our result indicates that PTEN is not required for hDLG to be recruited to the high molecular mass complex. Moreover, our results suggest that neither the catalytic activity nor the PDZ binding domain of PTEN is essential for its complex formation.
Phospho-mimicking of PTEN on Ser380, Thr382, Thr383, and Ser385 Significantly Diminishes Complex Assembly
It has been proposed that PTEN activity and localization are regulated through phosphorylation and dephosphorylation of the Ser/Thr residues 380, 382, 383, and 385; we therefore tested whether PTEN phosphorylation status on these four residues affects complex formation. Size-exclusion chromatography and Western blot analysis were performed using PC3 WT-PTEN-induced cells, and phosphorylated PTEN was detected with P380PTEN antibody (rabbit) followed by stripping and reprobing with total PTEN (mouse) antibody. The results demonstrated that phosphorylated PTEN was present in both the monomer (I) and complex (II) fractions (Fig. 2A). However, similar to the observation made by Vazquez et al. (27), quantitative analysis demonstrated that phosphorylated PTEN is preferentially enriched in the monomeric fractions (Fig. 2B). To determine whether complex assembly depends on phosphorylation of Ser/Thr residues 380, 382, 383, and 385, we transfected PTEN-null PC3 parental cells with either A4- PTEN (S380A, T382A, T383A, and S385A), mimicking dephosphorylated PTEN, or E4-PTEN (S380E, T382E, T383E, and S385E), mimicking phosphorylated PTEN and applied the proteins to a size-exclusion column. As shown in Fig. 2C, although both mutants can participate in complex assembly, there is significantly less E4-PTEN mutant being recruited into high molecular mass fractions. Using monomeric PTEN levels as controls for protein stability, we found 6- and 2.7-fold less E4-PTEN proteins being recruited to fractions 19 and 20, respectively, suggesting that Ser380, Thr382, Thr383, and Ser385 are important for PTEN complex assembly.
FIGURE 2.
Dephosphorylation of PTEN on Ser380, Thr382, Thr383, and Ser385 is critical for complex assembly. A, PC3 WT-PTEN-induced cells were run through a gel filtration column, and fractions were collected and immunoblotted (IB) with anti-phospho-PTEN (P380) and total PTEN antibodies. B, relative proportion of P380PTEN to unphosphorylated PTEN (calculated by subtracting P380PTEN from total PTEN) was quantified from A for each fraction by densitometry analysis. C, PC3 cells were transfected with E4 (4 Ser/Thr amino acids being mutated to glutamic acid) and A4 (4 Ser/Thr amino acids being mutated to alanine) PTEN mutant constructs. After gel filtration, complex (19–22) and monomer (34–37) fractions were collected and immunoblotted with PTEN antibody.
PTEN Complex Is Enriched in the Nucleus
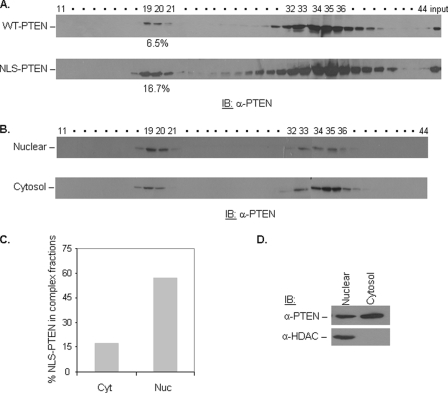
Our previous work suggests that the phosphorylation status of PTEN may regulate its nuclear localization. Specifically, dephosphorylation of PTEN at Ser380 leads to increased nuclear retention (26). To address whether the enrichment of unphosphorylated PTEN in the higher molecular mass fractions is due to the preferential enrichment of the complex in the nucleus, the complex-forming potentials of WT versus NLS-PTEN, in which PTEN is fused to a nuclear localization sequence (26), were directly compared. In this experiment, NLS-PTEN or WT-PTEN was transfected into the PTEN-null PC3 cell line, and lysates were applied to a size-exclusion column. NLS-PTEN has significantly increased complex-forming capability compared with that of WT-PTEN (Fig. 3A). Because PTEN contains several nuclear exclusion signals, (31) and therefore, a significant portion of NLS-PTEN can also be found outside of the nucleus, we addressed this issue by performing nuclear and cytoplasmic fractionation followed by size-exclusion column. Although PTEN is present in high and low molecular mass fractions in both extracts, quantitative analysis, by normalizing the amount of PTEN protein in the complex fractions to total PTEN, demonstrated that PTEN complex is significantly enriched in the nuclear fraction (Fig. 3B). These results suggest that a larger proportion of PTEN high molecular mass complex is in the nucleus. PTEN complexes found in the cytosolic compartment either belong to different complexes or are due to a contamination of nuclear/cytosol pools, which can happen during the fractionation process.
FIGURE 3.
PTEN complex is enriched in the nucleus. A, WT-PTEN and NLS-PTEN were transfected into PC3 cells. Cell lysates were applied to a gel filtration column. Fractions were collected and subjected to immunoblotting (IB) with anti-PTEN antibody. The percentage of PTEN in the complex fractions was calculated by dividing the amount of PTEN in the high molecular mass fractions by total PTEN (complex + monomer). The amount of PTEN in these fractions was determined through densitometry analysis. B, NLS-PTEN was transfected into PC3 cells, the nuclear and cytoplasmic fractions were prepared and run through a gel filtration column, and fractions were collected and immunoblotted with anti-PTEN antibody. C, percentage of NLS-PTEN in the complex fractions was calculated for nuclear (Nuc) and cytosolic (Cyt) extracts as described in A and depicted in a bar graph. D, nuclear and cytosolic fractions (column input) were immunoblotted with anti-PTEN and anti-HDAC (nuclear marker) antibodies.
Identification of Components within PTEN-associated Complex via DIGE Analysis
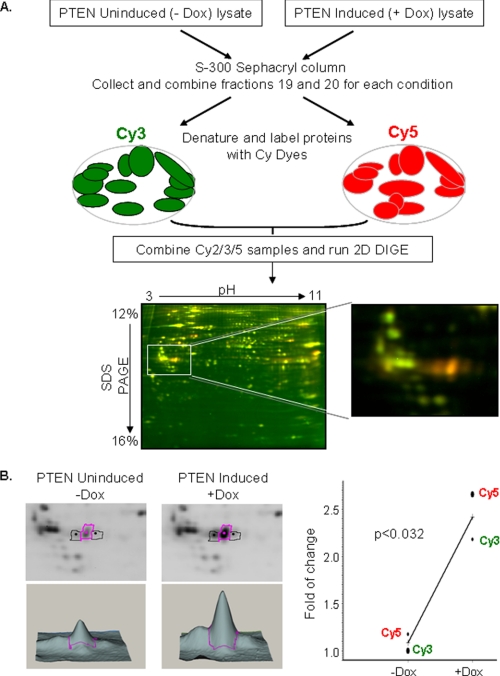
To characterize the PTEN complex further and identify major components associated with it, we employed a two-dimensional DIGE approach. In this method, proteins from two or more samples are labeled with either Cy3 or Cy5 fluorescent dyes, resolved via two-dimensional gel electrophoresis, and differentially expressed proteins are identified using mass spectrometry analysis (32, 33). As shown in Fig. 4A, complex fractions 19 and 20 from PC3 WT-PTEN-inducible cells in the presence (+Dox) and absence (−Dox) of Dox were collected and labeled with Cy5 and Cy3, respectively. The pool of these two samples was labeled with Cy2 and served as a standard control for normalization and quantitation of Cy5- and Cy3-labeled samples. The samples were combined and resolved on two-dimensional DIGE gels. Fig. 4A shows one of the representative two-dimensional DIGE images, where false-colored Cy5 (red) or Cy3 (green) spots indicate proteins that are potentially recruited to or excluded from the high molecular mass complex in the presence of PTEN, respectively, whereas Cy3/Cy5 (yellow) spots identify those proteins in the higher molecular mass fraction whose presence is independent of PTEN. Following this methodology, several proteins with altered abundance were discovered and further identified using mass spectrometric analysis. HnRNP C was one of the proteins identified through this methodology (supplemental Fig. S1) and was enriched by 2.2-fold in the presence of PTEN (Fig. 4B). To eliminate any CyDye labeling bias, a CyDye-swapping approach was used, and the PTEN-induced and uninduced samples were reversely labeled with Cy3 and Cy5, respectively. The results further validate that hnRNP C is enriched in PTEN+ fractions (Fig. 4B).
FIGURE 4.
Schematic outline of isolation and two-dimensional DIGE analysis of the PTEN-associated protein complex. A, proteins from PC3 WT-PTEN-inducible cells under induced (+Dox) and uninduced (−Dox) conditions were extracted and separated using the same parameters through a gel filtration Sephacryl S-300 column. PTEN-associated complex fractions 19 and 20 in the presence of Dox (PTEN-induced) and in the absence of Dox (PTEN uninduced) were combined and labeled with Cy5 and Cy3, respectively, whereas the pool of these two samples was labeled with Cy2. Protein samples were combined, and isoelectric focusing was performed on pH 3–11 nonlinear strips and then separated on 12–16% SDS gradient gels. False-colored Cy5 (red)-labeled PTEN-induced and Cy3 (green)-labeled PTEN uninduced representative gel images are shown. B, three-dimensional profiles and the graphical view of fold changes are shown for hnRNP C protein under PTEN-induced and uninduced conditions.
HnRNP C Is a Candidate Component of the PTEN-associated Complex
Following identification of hnRNP C, PC3 WT-PTEN-induced cells fractionated through size-exclusion chromatography were immunoblotted with hnRNP C and PTEN. The hnRNP C antibody recognized two bands corresponding to hnRNP C1 and C2 (41 and 43 kDa, respectively), which represent alternatively spliced variants of the same gene (34). Interestingly, PTEN and hnRNP C co-fractionated only in the high molecular mass fractions (Fig. 5A). By immunoblotting of higher molecular mass fractions (19 and 20) in the presence or absence of PTEN, we demonstrated increased recruitment of hnRNP C into these fractions (Fig. 5B). In addition, comparing hnRNP C levels in fractions 17–26 under PTEN-induced and uninduced conditions showed a shift of hnRNP C toward higher molecular mass fractions in the presence of PTEN (Fig. 5C). To rule out the possibility that PTEN regulates overall hnRNP C protein levels, whole lysates from PC3 WT-PTEN-induced and uninduced cells were subjected to immunoblot analysis. No change in the levels of hnRNP C was detected under PTEN-induced and uninduced conditions (Fig. 5D), suggesting that PTEN does not alter hnRNP C expression.
FIGURE 5.
HnRNP C is a candidate component of the PTEN-associated complex. A, PC3 WT-PTEN-inducible cells were treated with Dox and run through a gel filtration column. Fractions were collected and immunoblotted (IB) with hnRNP C and PTEN antibodies. B, fractions 19 and 20 from the PC3 WT-PTEN-inducible cells in the presence and absence of Dox were immunoblotted with PTEN and hnRNP C. The histogram on the right represents the quantitative densitometry analysis of bands shown on the left. C, fractions 17–26 from PC3 WT-PTEN-inducible cells under induced and uninduced conditions were run on SDS-polyacrylamide gels and immunoblotted for hnRNP C and GRP78 control. D, whole lysates from PC3 WT-PTEN-inducible cells in the presence and absence of Dox were analyzed with the indicated antibodies.
PTEN Interacts and Co-localizes with HnRNP C in the Nucleus
To investigate the interaction of PTEN and hnRNP C, co-immunoprecipitation was performed using PC3 PTEN-induced cells with an antibody specific to the N terminus of PTEN and IgG control. The results demonstrated an interaction between PTEN and hnRNP C in these cells (Fig. 6A). The interaction between PTEN and hnRNP C was also present when PC3 cells were transiently transfected with WT-PTEN but absent with the C terminus of PTEN (supplemental Fig. S2). This suggests that the C terminus of PTEN on its own is not sufficient for complex formation with hnRNP C. Interaction of PTEN and hnRNP C was further validated in HeLa cells expressing endogenous levels of both proteins (Fig. 6B). We hypothesized that the interaction between PTEN and hnRNP C is taking place in the nucleus because of the higher relative levels of hnRNP C found in the nucleus and enrichment of PTEN-associated complex in nuclear lysates (Fig. 3). To test this, we used hydrogen peroxide to trigger nuclear retention of PTEN (26). Hydrogen peroxide treatment resulted in higher nuclear PTEN (Fig. 6C) and increased the interaction between PTEN and hnRNP C (Fig. 6B). To test further the interaction of PTEN and hnRNP C in the nucleus, nuclear and cytoplasmic fractions from HeLa cells were immunoprecipitated with PTEN and IgG control. Immunoblotting for hnRNP C showed specific interaction between endogenous hnRNP C and PTEN in the nuclear compartment (Fig. 6D). Further analysis using immunofluorescent-labeled antibodies showed the two proteins co-localized within the nucleus (Fig. 6E). As a negative control, PC3 WT-PTEN uninduced cells (−Dox) were used and showed undetectable PTEN levels and no co-localization between PTEN and hnRNP C. Taken together, our data show a physical interaction between PTEN and hnRNP C in the nucleus, which can be further enhanced with stimulation known to promote PTEN nuclear retention, such as H2O2 treatment.
FIGURE 6.
Physical interaction of PTEN and HnRNP C. A, PC3 WT-PTEN-inducible cells treated with Dox were immunoprecipitated (IP) using PTEN and IgG control antibodies and immunoblotted (IB) with PTEN and hnRNP C. B, HeLa cells under control conditions or treated with 1 mm H2O2 for 1 h were immunoprecipitated with PTEN and IgG antibodies and immunoblotted with anti-PTEN and -hnRNP C antibodies. Bottom, quantitation of hnRNP C immunoprecipitated with PTEN antibody in the presence and absence of H2O2, (normalized to IgG controls). C, cytoplasmic (Cyt) and nuclear (Nuc) fractions from HeLa cells untreated and treated with 1 mm H2O2 for 1 h were immunoblotted with the indicated antibodies. D, cytoplasmic and nuclear fractions from HeLa cells were immunoprecipitated using PTEN and IgG control antibodies and immunoblotted with PTEN and hnRNP C antibodies. The nuclear and cytoplasmic inputs were immunoblotted with anti-PTEN, anti-hnRNP C, and anti-HDAC antibodies. E, PTEN and hnRNP C co-localize in the nucleus. PC3 WT-PTEN-inducible cells with and without Dox were co-stained with anti-PTEN and -hnRNP C antibodies and counterstained with DAPI.
To address the functional significance of the interaction between PTEN and hnRNP C, we have investigated a possible role for hnRNP C in interacting with PTEN and regulating its lipid phosphatase activity by knockdown of hnRNP C expression in PC3 WT-PTEN-inducible cells. Compared with the uninduced cells, hnRNP C knockdown did not lead to detectable changes in either total PTEN or P-AKT levels (supplemental Fig. S3A). In addition, hnRNP C small interfering RNA did not decrease nuclear PTEN levels (supplemental Fig. S3B), suggesting that hnRNP C is not affecting PTEN localization and lipid phosphatase activity. The functional significance of this interaction and the regulation of hnRNP C function by PTEN remain to be studied further.
DISCUSSION
In this study, we set to understand the key elements involved in PTEN complex assembly. Using inducible PTEN expression in PTEN-null cells, we demonstrated that the catalytic activity of PTEN is not absolutely required for its higher molecular mass assembly because the catalytically dead mutant, C124S, has similar complex assembly capability compared with the WT-PTEN protein. Previous reports have suggested that the PTEN complex forms through interaction with PDZ-containing proteins, such as NHERF2 and hDLG (19, 27). By deleting the key amino acids of the PTEN PDZ binding domain, we showed that the absence of a functional PDZ domain does not prevent PTEN from being recruited to higher molecular mass fractions. These results open up the possibility that PTEN can exist in multiple high molecular mass complexes, and the PTEN PDZ binding domain may be required for the formation of certain complexes but dispensable for others. Interestingly, the presence of hDLG in the higher molecular mass complex is independent of PTEN.
PTEN phosphorylation at its C-terminal tail has been shown to play a critical role in its regulation (20–26, 35), and previously it has been suggested that phosphorylation of PTEN in this region, measured by P380 antibody, negatively regulates its complex assembly (27). Our results agree with these findings and demonstrate that phosphorylated PTEN is enriched in monomeric fractions and that a phospho-mimicking mutation of PTEN on Ser380, Thr382, Thr383, and Ser385 (E4-PTEN mutant) leads to decreased complex assembly. To identify further the mechanism through which phosphorylation of PTEN may regulate its complex assembly, we investigated the cellular localization of the PTEN complex. A previous study demonstrated that PTEN nuclear retention is positively regulated through dephosphorylation of its P380 residue. Our results showed that NLS-PTEN has increased complex-forming potential and such an increase is due, at least in part, to the enrichment of the PTEN-associated complex in the nuclear fraction.
Characterization of novel PTEN-interacting partners is critical in understanding PTEN tumor-suppressive functions. To find new components to the PTEN-associated complex(es), DIGE was used to identify those proteins specifically being recruited to the complex in the presence of PTEN. Our approach led to the identification of hnRNP C. Interestingly, in a pulldown experiment performed by Crockett et al. (36), a novel protein similar to hnRNP C was identified as an interacting partner of PTEN, but the interaction was not validated. Our results demonstrated that hnRNP C and PTEN co-fractionate only in high molecular mass complexes and that hnRNP C is significantly recruited into these fractions in the presence of PTEN. Further analysis using co-immunoprecipitation and co-localization experiments showed complex assembly between hnRNP C and PTEN in the nucleus.
Our results have identified hnRNP C as a component of the PTEN nuclear complex and show an interaction between these two proteins. To take an initial step in understanding the functional significance of this interaction, we have investigated the effect of hnRNP C on PTEN function. Our results demonstrate that under hnRNP C knockdown conditions, no significant changes in PTEN, P-AKT, and nuclear PTEN levels can be detected, suggesting that the interaction between these two proteins might be more important in regulating hnRNP C functions.
HnRNP C is a very abundant nuclear protein and binds to RNA through its RNA binding domains (34, 37). It is a component of the spliceosome complex (38), and a recent study using a hnRNP C knockdown approach identified BCL2L12 and PCBP4 as genes whose splicing is regulated by hnRNP C (39). Additional functions, such as regulating telomerase activity (40), XIAP translation through the IRES promoter (41), and response to hydrogen peroxide stress (42, 43) have been attributed to hnRNP C. The interaction of PTEN with hnRNP C and the complex assembly may be important in regulating these known hnRNP C functions.
Supplementary Material
Acknowledgments
We thank Drs. Reginald Hill and Jing Jiao for helpful comments, Drs. Samson Chow and Thomas Wilkinson for allowing us to use their HPLC instrument, and Dr. Michael Haykinson for an initial consultation on two-dimensional DIGE experiments.
This work is supported, in whole or in part, by National Institutes of Health Grants P50 CA092131 and R01 CA107166 (to H. W.), National Cancer Institute/National Institutes of Health Grant U01 CA84128-06, the Prostate Cancer Foundation, and Department of Defense Grant PC031130 (to H. W.). The UCLA Mass Spectrometry and Proteomics Technology Center was established with a grant from the W. M. Keck Foundation. The functional proteomics laboratory (DIGE Shared Resource) was supported by the Jonsson Comprehensive Cancer Center, the dean's office at the David Geffen School of Medicine, and a Chancellor's Bioscience Facility grant, UCLA.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” additional references, and Figs. S1–S3.
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CS
- C124S
- DIGE
- difference gel electrophoresis
- Dox
- doxycycline
- hnRNP C
- heterogeneous nuclear ribonucleoprotein C
- NLS
- nuclear localization sequence
- WT
- wild-type.
REFERENCES
- 1.Li D. M., Sun H. (1997) Cancer Res. 57, 2124–2129 [PubMed] [Google Scholar]
- 2.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R., Bigner S. H., Giovanella B. C., Ittmann M., Tycko B., Hibshoosh H., Wigler M. H., Parsons R. (1997) Science 275, 1943–1947 [DOI] [PubMed] [Google Scholar]
- 3.Steck P. A., Pershouse M. A., Jasser S. A., Yung W. K., Lin H., Ligon A. H., Langford L. A., Baumgard M. L., Hattier T., Davis T., Frye C., Hu R., Swedlund B., Teng D. H., Tavtigian S. V. (1997) Nat. Genet. 15, 356–362 [DOI] [PubMed] [Google Scholar]
- 4.Maehama T., Dixon J. E. (1998) J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 5.Myers M. P., Pass I., Batty I. H., Van der Kaay J., Stolarov J. P., Hemmings B. A., Wigler M. H., Downes C. P., Tonks N. K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stambolic V., Suzuki A., de la Pompa J. L., Brothers G. M., Mirtsos C., Sasaki T., Ruland J., Penninger J. M., Siderovski D. P., Mak T. W. (1998) Cell 95, 29–39 [DOI] [PubMed] [Google Scholar]
- 7.Sun H., Lesche R., Li D. M., Liliental J., Zhang H., Gao J., Gavrilova N., Mueller B., Liu X., Wu H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura M., Gu J., Matsumoto K., Aota S., Parsons R., Yamada K. M. (1998) Science 280, 1614–1617 [DOI] [PubMed] [Google Scholar]
- 9.Weng L. P., Brown J. L., Eng C. (2001) Hum. Mol. Genet. 10, 599–604 [DOI] [PubMed] [Google Scholar]
- 10.Radu A., Neubauer V., Akagi T., Hanafusa H., Georgescu M. M. (2003) Mol. Cell. Biol. 23, 6139–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen W. H., Balajee A. S., Wang J., Wu H., Eng C., Pandolfi P. P., Yin Y. (2007) Cell 128, 157–170 [DOI] [PubMed] [Google Scholar]
- 12.Tamguney T., Stokoe D. (2007) J. Cell Sci. 120, 4071–4079 [DOI] [PubMed] [Google Scholar]
- 13.Carracedo A., Salmena L., Pandolfi P. P. (2008) Cell 133, 550–551 [DOI] [PubMed] [Google Scholar]
- 14.Salmena L., Carracedo A., Pandolfi P. P. (2008) Cell 133, 403–414 [DOI] [PubMed] [Google Scholar]
- 15.Kotelevets L., van Hengel J., Bruyneel E., Mareel M., van Roy F., Chastre E. (2005) FASEB J. 19, 115–117 [DOI] [PubMed] [Google Scholar]
- 16.Wu X., Hepner K., Castelino-Prabhu S., Do D., Kaye M. B., Yuan X. J., Wood J., Ross C., Sawyers C. L., Whang Y. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y., Dowbenko D., Spencer S., Laura R., Lee J., Gu Q., Lasky L. A. (2000) J. Biol. Chem. 275, 21477–21485 [DOI] [PubMed] [Google Scholar]
- 18.Adey N. B., Huang L., Ormonde P. A., Baumgard M. L., Pero R., Byreddy D. V., Tavtigian S. V., Bartel P. L. (2000) Cancer Res. 60, 35–37 [PubMed] [Google Scholar]
- 19.Takahashi Y., Morales F. C., Kreimann E. L., Georgescu M. M. (2006) EMBO J. 25, 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres J., Pulido R. (2001) J. Biol. Chem. 276, 993–998 [DOI] [PubMed] [Google Scholar]
- 21.Georgescu M. M., Kirsch K. H., Akagi T., Shishido T., Hanafusa H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10182–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres J., Rodriguez J., Myers M. P., Valiente M., Graves J. D., Tonks N. K., Pulido R. (2003) J. Biol. Chem. 278, 30652–30660 [DOI] [PubMed] [Google Scholar]
- 23.Vazquez F., Ramaswamy S., Nakamura N., Sellers W. R. (2000) Mol. Cell. Biol. 20, 5010–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. (2004) Science 303, 1179–1181 [DOI] [PubMed] [Google Scholar]
- 25.Odriozola L., Singh G., Hoang T., Chan A. M. (2007) J. Biol. Chem. 282, 23306–23315 [DOI] [PubMed] [Google Scholar]
- 26.Chang C. J., Mulholland D. J., Valamehr B., Mosessian S., Sellers W. R., Wu H. (2008) Mol. Cell. Biol. 28, 3281–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez F., Grossman S. R., Takahashi Y., Rokas M. V., Nakamura N., Sellers W. R. (2001) J. Biol. Chem. 276, 48627–48630 [DOI] [PubMed] [Google Scholar]
- 28.Sumitomo M., Iwase A., Zheng R., Navarro D., Kaminetzky D., Shen R., Georgescu M. M., Nanus D. M. (2004) Cancer Cell 5, 67–78 [DOI] [PubMed] [Google Scholar]
- 29.Yu Z., Fotouhi-Ardakani N., Wu L., Maoui M., Wang S., Banville D., Shen S. H. (2002) J. Biol. Chem. 277, 40247–40252 [DOI] [PubMed] [Google Scholar]
- 30.Ali I. U., Schriml L. M., Dean M. (1999) J. Natl. Cancer Inst. 91, 1922–1932 [DOI] [PubMed] [Google Scholar]
- 31.Gil A., Andrés-Pons A., Fernández E., Valiente M., Torres J., Cervera J., Pulido R. (2006) Mol. Biol. Cell 17, 4002–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unlü M., Morgan M. E., Minden J. S. (1997) Electrophoresis 18, 2071–2077 [DOI] [PubMed] [Google Scholar]
- 33.Tonge R., Shaw J., Middleton B., Rowlinson R., Rayner S., Young J., Pognan F., Hawkins E., Currie I., Davison M. (2001) Proteomics 1, 377–396 [DOI] [PubMed] [Google Scholar]
- 34.Swanson M. S., Nakagawa T. Y., LeVan K., Dreyfuss G. (1987) Mol. Cell. Biol. 7, 1731–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller S. J., Lou D. Y., Seldin D. C., Lane W. S., Neel B. G. (2002) FEBS Lett. 528, 145–153 [DOI] [PubMed] [Google Scholar]
- 36.Crockett D. K., Fillmore G. C., Elenitoba-Johnson K. S., Lim M. S. (2005) Proteomics 5, 1250–1262 [DOI] [PubMed] [Google Scholar]
- 37.Görlach M., Wittekind M., Beckman R. A., Mueller L., Dreyfuss G. (1992) EMBO J. 11, 3289–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. (1986) Science 231, 1534–1539 [DOI] [PubMed] [Google Scholar]
- 39.Venables J. P., Koh C. S., Froehlich U., Lapointe E., Couture S., Inkel L., Bramard A., Paquet E. R., Watier V., Durand M., Lucier J. F., Gervais-Bird J., Tremblay K., Prinos P., Klinck R., Elela S. A., Chabot B. (2008) Mol. Cell. Biol. 28, 6033–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ford L. P., Suh J. M., Wright W. E., Shay J. W. (2000) Mol. Cell. Biol. 20, 9084–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holcík M., Gordon B. W., Korneluk R. G. (2003) Mol. Cell. Biol. 23, 280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone J. R., Collins T. (2002) J. Biol. Chem. 277, 15621–15628 [DOI] [PubMed] [Google Scholar]
- 43.Hossain M. N., Fuji M., Miki K., Endoh M., Ayusawa D. (2007) Mol. Cell. Biochem. 296, 151–157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.