Abstract
Mouse T cell clone 2C recognizes two different major histocompatibility (MHC) ligands, the self MHC Kb and the allogeneic MHC Ld. Two distinct peptides, SIY (SIYRYYGL) and QL9 (QLSPFPFDL), act as strong and specific agonists when bound to Kb and Ld, respectively. To explore further the mechanisms involved in peptide potency and specificity, here we examined a collection of single amino acid peptide variants of SIY and QL9 for 1) T cell activity, 2) binding to their respective MHC, and 3) binding to the 2C T cell receptor (TCR) and high affinity TCR mutants. Characterization of SIY binding to MHC Kb revealed significant effects of three SIY residues that were clearly embedded within the Kb molecule. In contrast, QL9 binding to MHC Ld was influenced by the majority of peptide side chains, distributed across the entire length of the peptide. Binding of the SIY-Kb complex to the TCR involved three SIY residues that were pointed toward the TCR, whereas again the majority of QL9 residues influenced binding of TCRs, and thus the QL9 residues had impacts on both Ld and TCR binding. In general, the magnitude of T cell activity mediated by a peptide variant was influenced more by peptide binding to MHC than by binding the TCR, especially for higher affinity TCRs. Findings with both systems, but QL9-Ld in particular, suggest that many single-residue substitutions, introduced into peptides to improve their binding to MHC and thus their vaccine potential, could impair T cell reactivity due to their dual impact on TCR binding.
Introduction
Elimination of virus-infected cells or cancer cells by cytotoxic T lymphocytes is governed by interactions between an αβ heterodimeric T cell receptor (TCR)2 and a short, processed, peptide that is bound to a product of the major histocompatibility complex (peptide-MHC) (1). It is well established that a single TCR is capable of recognizing multiple distinct peptide-MHC ligands while maintaining exquisite specificity for each (2, 3).
Ultimately, both the specificity and potency of a peptide derives from its interactions with the MHC product and with the TCR. The importance of understanding the role of each peptide residue in eliciting T cell activity can be seen in the considerable effort toward the development of peptide vaccines for the treatment of infection diseases or cancer (4). These efforts have often attempted to enhance the activity of a peptide by generating peptide variants with improved binding for the MHC product (5–7). This approach is absolutely dependent on the ability of T cells to react well not only with the peptide variant but with the native peptide that is presented by infected cells or by the cancer cells. In this regard, it is important to understand whether changes to a peptide impact not only MHC product binding but also binding by the TCR.
Multiple genes and extensive polymorphism in the MHC enable different MHC products to bind and present a distinct set of peptides (8). For example, the mouse MHC products Kb and Ld differ by 30 amino acid residues in the peptide binding grooves, and each protein binds to a diverse array of peptides that differ in their anchor motifs (8, 9). Structural studies showed that Kb binding peptides lie flat in the peptide binding groove, whereas the main chain of Ld binding peptides is bulged (10–14). This bulge has been attributed to bulky aromatic residues (Trp-73 and Tyr-99) that line the floor of the peptide binding groove (13–15). These differences between Kb and Ld are thought to force Ld binding peptides to use N- and C-terminal residues as anchors, whereas Kb binding peptides use a central residue and a C-terminal residue.
It is unclear if these distinctly different peptide binding modes also yield differences in the mechanisms by which TCR binding and specificity are achieved. In some cases, studies have measured the effects on TCR binding of peptides that are substituted at TCR contact sites; these peptide variants have been called altered peptide ligands (16). For example, surface plasmon resonance was used to measure binding of the MHC I-Ek-specific TCR 2B4 to peptide MCC variants (17). However, in many of the cases, TCR 2B4 was unable to detect these peptide variants due to the low affinity of the wild type interaction (KD = 5 μm). In another example, binding of the MHC I-Ab-specific TCR B3K508 to peptide 3K variants could not be detected due to a 10-fold or greater reduction in binding affinity (18). Thus, although it is possible to occasionally measure quantitatively the effect of peptide substitution on TCR binding, often it is not possible due to inherently low (KD values of 1–100 μm) binding affinities.
In order to examine in more detail the quantitative impacts of peptide substitutions on both MHC and TCR binding, here we used the mouse 2C T cell system, including the wild type 2C TCR, and two higher affinity mutants of the 2C TCR called m67 and m6 (19, 20). The system is advantageous because 2C recognizes as strong agonist two completely different peptide-MHC ligands, SIY (SIYRYYGL) and QL9 (QLSPFPFDL), when bound to Kb and Ld, respectively (21, 22). In addition, there are high resolution crystal structures of both the 2C TCR-SIY-Kb complex (10) and the 2C TCR-QL9-Ld complex (14) in order to interpret the binding results with single alanine substitutions of the peptides.
We show that the activity of T cell clone 2C was impacted by alanine substitution of almost every residue of its two strong agonist peptides, SIY and QL9. We then assessed whether these reductions in T cell function were due to 1) reduced binding of the peptide variant to MHC, 2) reduced binding of the TCR to the peptide variant Kb or Ld complexes, or 3) effects on both MHC and TCR binding. Characterization of SIY binding to Kb revealed significant effects of alanine substitution on three SIY residues that were clearly buried within the Kb molecule. In contrast, QL9 binding to MHC Ld was influenced by alanine substitution of the majority of the peptide side chains, distributed across the entire length of the peptide.
To measure the quantitative impact that each alanine substitution had on TCR binding, the two TCRs, m67 and m6, that recognize SIY-Kb or QL9-Ld with high (nanomolar) affinity were used (19, 20). From these experiments, we determined the changes in binding free energies (ΔΔG) for each alanine substitution in order to identify peptide SIY and QL9 “hot spot” residues (i.e. alanine-substituted residues that reduce TCR binding by ≥10-fold) (23). Binding of the SIY-Kb complex to the TCR involved only three SIY residues that were pointed toward the TCR. In contrast, the majority of QL9 residues influenced binding of the TCR m6. Thus, in the Ld system, QL9 peptide residues influenced binding to both Ld and the TCR.
We also observed that, in general, the magnitude of reduction in T cell activity mediated by a peptide variant was influenced more by peptide binding to MHC than by binding the TCR, especially if the TCR was of higher affinity, above a particular threshold. We conclude that T cell specificity for these peptides was achieved by different strategies, dictated largely by the ways in which Kb and Ld bound to and presented the peptides. Furthermore, the dual impact of many of the QL9 residues on both Ld binding and TCR binding suggests that one must be cautious in assuming that changes in peptide residues to improve MHC binding will not have an impact on T cell recognition. In other words, current vaccine formulations that include modified peptides should consider whether the responding T cells will be capable of optimal activation by the native peptide antigens.
EXPERIMENTAL PROCEDURES
Peptides
Peptides (QL9, QLSPFPFDL; SIY, SIYRYYGL; MCMV, YPHFMPTNL; OVA, SIINFEKL; QL9 alanine variants; and SIY alanine variants) were synthesized by the Penn State Macromolecular Core Facility (Hershey, PA) using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. Peptides were purified using C18 reverse phase column chromatography with a linear elution gradient of 0–60% acetonitrile containing 0.1% trifluoroacetic acid. Peptide mass was verified by electrospray ionization mass spectrometry (University of Illinois, Urbana-Champaign). Concentrations were determined by quantitative amino acid analysis (University of California, Davis) or by spectrophotometric analysis.
Cloning, Expression, and Refolding of Soluble Single-chain TCRs
Two high affinity 2C T cell receptors m67 and m6 (19, 20) were cloned and expressed in Escherichia coli as single-chain (Vβ8.2-linker-Vα3.1) constructs (scTCR), each with a 15-amino acid BirA substrate cloned at the C terminus as described previously (24). These two high affinity TCRs differ from the wild type 2C TCR by 5 amino acids in the CDR3α loop (2C, SGFASAL; m67, SLERPYL; m6, SHQGRYL). Genes for the single-chain TCRs m67 and m6 were amplified from the yeast display plasmid called pCT302 using a forward primer (NcoI site and a His6 tag) and a reverse primer (EcoRI site and 15-amino acid BirA substrate GLNDIFEAQKIEWHE). This single-chain construct was then cloned into the E. coli expression plasmid pET-28a for the expression of single-chain TCR in the form of inclusion bodies.
Both the scTCR-pET-28a and pBIRAcm plasmids were co-transformed with E. coli BL21 (DE3) (Stratagene). The pBIRAcm plasmid contains genes for the expression of BirA, an E. coli enzyme that site-specifically biotinylates a lysine within the 15-amino acid substrate (generously provided by John Cronan, University of Illinois, Urbana, IL). Briefly, E. coli cells containing 100 μg/ml kanamycin (pET-28a) and 10 μg/ml chloramphenicol (pBIRAcm) were cultured to an A600 of 0.4–0.6. At this time, the pBIRAcm plasmid was induced with a 1:1000 dilution of a 20% l-arabinose solution, and a final concentration of 50 μm d-biotin was added to the cells. Cells were further cultured to an A600 of 1.0. scTCR-pET-28a was induced with 0.7 mm isopropyl 1-thio-β-d-galactopyranoside, and cells were cultured for an additional 2 h at 37 °C. Cells were suspended in lysis buffer (50 mm Tris-HCl, 1% Triton-X-100, 100 mm NaCl, 0.1% sodium azide, 10 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, pH 8.0) and subjected to microfluidization, and the inclusion body pellets were washed three times with osmotic shock buffer (20 mm Tris-HCl, 2.5 mm EDTA, 0.5% Triton X-100, pH 8.0) and three times without Triton X-100. Inclusion bodies (600–700 mg) that contained the single-chain TCR were solubilized in 6 m guanidine hydrochloride and then added dropwise to 400 ml of refold buffer (3 m urea, 50 mm Tris, 2 mm reduced glutathione, 0.2 mm oxidized glutathione, pH 8.0). Dilution buffer (200 mm NaCl and 50 mm Tris, pH 8.0) was added dropwise to the sample over 24 h until a volume of 2.4 liters was reached. After incubation for another 24 h at 4 °C, Ni2+-nitrilotriacetic acid beads (Qiagen) were added, and the solution was incubated for 24 h. The single-chain TCR was eluted from Ni2+-nitrilotriacetic acid-agarose (Qiagen) with 500 mm imidizole. TCR was purified further using a Superdex-200 size exclusion column (GE Healthcare).
T Cell Activation Assays
Wild type 2C and high affinity 2C T cell transfectants m67 and m6 were incubated with Kb- or Ld-positive cells and various concentrations of peptide SIY and QL9 alanine variants. T cell activation was measured by assaying for levels of IL-2 release, as described (25). Briefly, T cell transfectants (7.5 × 104) were incubated with T2-Kb (7.5 × 104) or T2-Ld (7.5 × 104) along with various concentrations of peptide for 20–24 h at 37 °C and 5% CO2. Supernatant was harvested, and levels of IL-2 were measured in an enzyme-linked immunosorbent assay type format. Results were plotted as percentage of maximal IL-2 release = ((A450 sample − A450(no peptide))/(Max A450(sample) − A450(no peptide))) × 100; signal obtained from no peptide was similar to that obtained for the null peptides MCMV or OVA. Binding curves were generated in GraphPad Prism by plotting the percentage of maximal IL-2 release against peptide concentration. The concentrations of peptide yielding 50% maximal IL-2 release (SD50) were calculated using non-linear regression (sigmoidal fitting; GraphPad Prism) of the activation curves.
MHC Stabilization Assays
Detection of up-regulated MHC Ld on the cell surface by peptide QL9 (QLSPFPFDL) or QL9 alanine variants was measured as described (26). Briefly, 3 × 105 T2-Ld were incubated with various concentrations of peptide QL9 alanine variants for 3 h at 37 °C. Cells were stained with 20 μg/ml 30-5-7 from ascites fluid, an mAb specific for the α1 and α2 domains of MHC Ld (27). Cells were washed two times with PBS, 0.5% BSA and then stained with secondary reagent (1 μg/ml) phycoerythrin-labeled goat anti-mouse IgG (Southern Biotech). After one wash with PBS, 0.5% BSA, cells were analyzed with a Coulter Epics XL Flow Cytometer (Beckman Coulter). Curves were generated by plotting mean fluorescent units against peptide concentration, and binding doses yielding 50% maximum cell surface levels (BD50) were determined by non-linear regression (sigmoidal fitting; GraphPad Prism). BD50 values represent the concentration of peptide required for half-maximal up-regulation of MHC on the cell surface.
For detection of up-regulated MHC Kb on the cell surface by peptide SIY (SIYRYYGL) or SIY alanine variants, the mouse H-2Kb-positive cell line RMA-S was incubated overnight at room temperature to induce the up-regulation of empty MHC on the cell surface. RMA-S cells (105) were incubated with various concentrations of peptide SIY alanine variants for 2 h at 37 °C. Cells were stained with 50 μl of 10 μg/ml ascites-purified B.8.24.3, an mAb specific for the α1 and α2 domains of MHC Kb. Cells were washed two times with PBS, 0.5% BSA and then stained with the secondary reagent (20 μg/ml) Alexa 488-labeled goat anti-mouse IgG (Invitrogen). After one wash with PBS, 0.5% BSA, cells were analyzed with a Coulter Epics XL Flow Cytometer (Beckman Coulter). BD50 values were calculated as described above.
Peptide-Kb Cell Surface Lifetimes
Levels of peptide SIY (SIYRYYGL) and SIY alanine variants remaining on the cell surface of T2-Kb were monitored over time as described previously (28). Briefly, 2 × 105 T2-Kb were incubated with saturating levels (100 μm) of peptide SIY or alanine variants for 2 h at 37 °C. Cells were washed two times with PBS, 0.5% BSA and suspended with 1 μm null peptide OVA (SIINFEKL) at 4 °C. Cells were placed at 37 °C (time 0), and at various times later, cells were removed, washed, and stored at 4 °C until staining with specific soluble tetrameric 2C-m67. Soluble single-chain high affinity 2C TCR m67 was coupled to streptavidin-phycoerthryin through a C-terminal BirA substrate as described (24). Cells were stained with 50 μl of 200 nm tetrameric TCR m67 for 45 min at 4 °C. Cells were washed with PBS, 0.5% BSA and then analyzed with a Coulter Epics XL Flow Cytometer (Beckman Coulter). Cell surface lifetime curves were generated by plotting mean fluorescent unit (MFU) values against time (h) in GraphPad Prism. Linear regression of the lifetime curves (one-phase exponential decay; GraphPad Prism) was used to determine the cell surface half-life (t½ = ln 2/koff). As a control, signal obtained from the null peptide OVA was used to subtract background from SIY measurements; signal obtained from no peptide was similar to OVA.
TCR Binding Analysis
Soluble high affinity 2C TCRs m67 and m6 were used to detect peptide SIY or QL9 alanine variants loaded onto cells expressing either MHC Kb or Ld, respectively. T2-Ld cells (105) were incubated with saturating levels (100 μm) of wild type peptide QL9, alanine variants, or null peptide MCMV for 3 h at 37 °C. Cells were stained with various concentrations of biotinylated 2C TCR m6 for 40 min at 4 °C. Cells were washed two times with PBS, 0.5% BSA and then stained with secondary reagent (2 μg/ml) streptavidin-phycoerythrin (BD Pharmingen) for 15 min at 4 °C. Cells were analyzed with a Coulter Epics XL Flow Cytometer (Beckman Coulter). Total Ld levels were also measured by detection with the anti-Ld mAb (30-5-7) (27). Cells were stained with (15 μg/ml) 30-5-7, purified from ascites, for 40 min at 4 °C. Cells were washed twice with PBS, 0.5% BSA and then stained with (1 μg/ml) phycoerythrin-labeled goat anti-mouse IgG (Southern Biotech) for 30 min at 4 °C. Cells were analyzed with a Coulter Epics XL flow cytometer (Beckman). Binding curves were generated by plotting MFU against TCR concentration in GraphPad Prism. MFU values were adjusted by dividing with the following correction factor (CF), CF = (MFUvariant/MFUQL9), where MFU values represent values obtained by measuring for levels of Ld with 30-5-7 as described earlier. Binding constants (KD) were determined by non-linear regression of the binding curves, which were used to determine ΔΔG (kcal/mol). ΔΔG = RTln(KD(mut)/KD(WT)), where RT = (1.987 × 10−3 kcal/mol × K) × (277.15K), where R is the gas constant at 1.987 × 10−3 kcal/mol, T is the temperature in Kelvin, and KD(mut) and KD(WT) are the equilibrium dissociation constants for the peptide variant and wild type peptide, respectively.
T2-Kb cells (105) were incubated with saturating levels (100 μm) of wild type peptide SIY and SIY alanine variants for 2 h at 37 °C. Cells were stained with (50 μl) various concentrations of biotinylated TCR m67 for 45 min at 4 °C. Cells were washed two times with PBS, 0.5% BSA and then stained with 50 μl of secondary reagent (2 μg/ml) phycoerythrin-labeled streptavidin (BD Pharmingen) for 20 min at 4 °C. Cells were analyzed with a Coulter Epics XL flow cytometer (Beckman Coulter). Data were analyzed as described above.
RESULTS
Structural Analysis of Peptides SIY and QL9
The 2C T cell receptor was originally isolated from the mouse cytotoxic T lymphocyte clone 2C (29) and recognizes two MHC ligands, the self MHC Kb and the allogeneic MHC Ld. Two distinct peptides, SIY (SIYRYYGL) and QL9 (QLSPFPFDL), act as strong and specific agonists when bound to Kb and Ld, respectively (21, 22). Peptide QL9 was derived from the ubiquitous enzyme α-ketoglutarate dehydrogenase (30, 31), and peptide SIY was identified from a combinatorial peptide library (22). Furthermore, the crystal structures have been solved for 2C TCR complexed with both SIY-Kb and QL9-Ld (10, 14).
The structures showed that peptide SIY lies flat in the Kb peptide-binding groove, clearly revealing residues whose side chains point either toward the 2C TCR (Arg-4, Tyr-6, and Gly-7) or MHC Kb (Tyr-3, Tyr-5, and Leu-8) (Fig. 1A). In contrast, the main chain of peptide QL9 is bulged due to bulky amino acid residues that line the floor of the Ld binding groove (Trp-73 and Tyr-99) (Fig. 1B). In addition, the side chains of several peptide QL9 residues (Pro-4, Phe-5, Phe-7, and Asp-8) are located at the interface of the Ld helices and contact both the 2C TCR and MHC Ld (Fig. 1B). Having the structures of both complexes, we were in a position to interpret the functional impact on TCR/MHC binding of each side chain of the peptides by examining single-site alanine variants of SIY and QL9.
FIGURE 1.
Structural features of the 2C/SIY-Kb and 2C/QL9-Ld complexes, focusing on peptides SIY and QL9. A, side view of the 2C-SIY-Kb complex (Protein Data Bank code 1G6R). Peptide SIY (SIYRYYGL) is represented in orange. The Vα and Vβ domains of 2C TCR are represented in red and blue, respectively. Mouse class I MHC Kb is represented in green. B, side view of the 2C-QL9-Ld complex (Protein Data Bank code 2Oi9). Peptide QL9 (QLSPFPFDL) is represented in orange. The Vα and Vβ domains of 2C TCR are represented in red and blue, respectively. Mouse class I MHC Ld is represented in green.
T Cell Activity Mediated by Peptide SIY and QL9 Variants
T cell activation assays were performed to measure effects of peptide SIY and QL9 residues on T cell function. This involved the stimulation of stable T cell lines transfected with either wild type 2C TCR or high affinity TCRs m67 and m6. 2C, m67, and m6 T cell activation was measured by the amount of IL-2 released upon stimulation with alanine variants of peptides SIY or QL9. For these experiments, 2C, m67, and m6 TCR transfectants of the mouse 58−/− T cell hybridoma line were used. The lines express similar levels of TCR and the co-receptor CD8αβ on the cell surface (data not shown). The co-receptor CD8 enhances binding between TCR and pMHC interactions and is required for interactions much lower than 1 μm, such as the 2C TCR-SIY-Kb interaction (32–35). For this reason, the CD8+ T cells were used to enhance the sensitivity of T cell activation involving the peptide alanine variants. These T cell hybridoma transfectants express only H-2k; thus, neither Kb nor Ld are expressed on the surface to complicate presentation of peptides by the antigen-presenting cells.
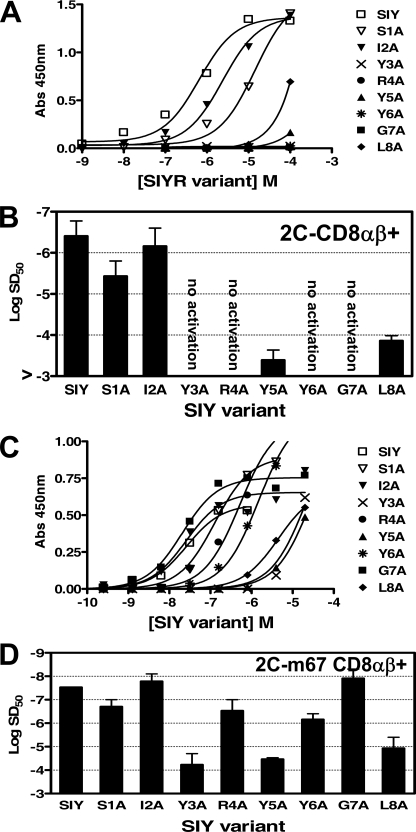
T cells with the WT 2C TCR were incubated with the antigen-presenting cell T2-Kb and various concentrations of peptide SIY alanine variants (Fig. 2A). Alanine substitution at every position from 3 to 8 of peptide SIY reduced the activity by over 100-fold (Fig. 2A). There was no detectable activity even at 100 μm peptide for the Y3A, R4A, Y6A, and G7A variants; hence, SD50 values were estimated to be >10−3 m for these variants (Fig. 2B). Most of these non-stimulatory SIY variants (Y3A, R4A, and G7A) were capable of stimulating 2C CTL in a cytolytic response assay (36), supporting the notion that other T cell responses have a lower threshold of activity than IL-2 release (37).
FIGURE 2.
Activation of wild type 2C or high affinity mutant T cell transfectants with peptide SIY alanine variants. A, the 2C T cell transfectant (CD8αβ+) was stimulated in the presence of T2-Kb and various concentrations of peptide SIY (SIYRYYGL) alanine variants. Activation was measured by assaying for levels of cytokine IL-2 release in an enzyme-linked immunosorbent assay. B, sensitization doses, SD50, determined from non-linear regression of the activation curves in A. Error bars, S.D. values averaged from four independent experiments. C, the m67 T cell transfectant (CD8αβ+) was stimulated in the presence of T2-Kb and various concentrations of peptide SIY alanine variants. D, SD50 values determined from non-linear regression of the activation curves in C. Error bars, S.D. values averaged from four independent experiments.
The high affinity 2C T cell transfectant m67 was stimulated with each peptide SIY alanine variant (Fig. 2C). The stimulation by all variants is presumably directly due to the fact that TCR m67 recognizes SIY-Kb with about 1,000-fold higher affinity (KD = 7 nm) than the WT 2C TCR (KD = 22 μm) (38) and that the m67 affinity for the variants is above the affinity threshold for T cell activity. Nevertheless, the levels of m67 T cell activity were reduced 100–1,000-fold for alanine substitutions at the SIY residues involved in Kb binding (Tyr3, Tyr5, and Leu8; see below) (Fig. 2D). Variants S1A, R4A, and Y6A showed about 10-fold reduced activity, whereas the I2A and G7A variants showed minimal effects on activity.
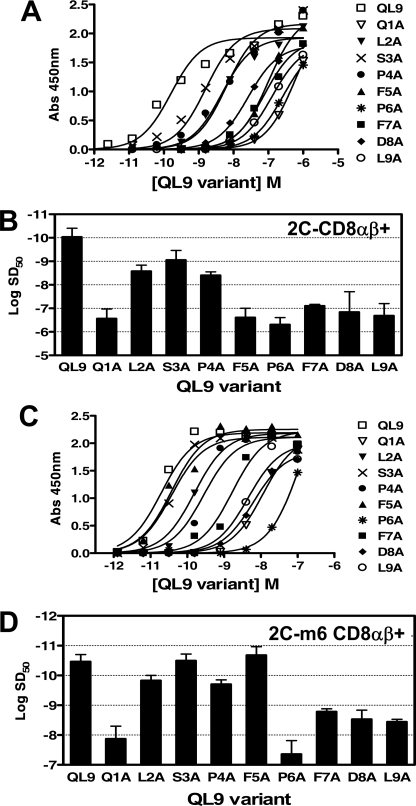
Similar functional analyses were performed with T cell hybridomas expressing the wild type 2C and high affinity TCR m6, using QL9 alanine variants and the T2-Ld as antigen-presenting cells. T cells with the WT 2C TCR were incubated with the antigen-presenting cell T2-Ld and various concentrations of peptide QL9 alanine variants (Fig. 3A). In the case of QL9, all of the variants were capable of stimulating the 2C CD8+ transfectant, unlike the case of SIY. This is probably due to the higher affinity of the wild type 2C TCR for QL9-Ld (KD = 1.2 μm) compared with the affinity for SIY-Kb (KD = 22 μm) (38). Nevertheless, 2C T cell activity was reduced, compared with wild type QL9, for each of the variants. Activity was reduced 1,000–10,000-fold for the Q1A, F5A, P6A, F7A, D8A, and L9A variants (Fig. 3B).
FIGURE 3.
Activation of wild type 2C or high affinity mutant T cell transfectants with peptide QL9 alanine variants. A, the 2C T cell transfectant (CD8αβ+) was stimulated in the presence of T2-Ld and various concentrations of peptide QL9 (QLSPFPFDL) alanine variants. B, sensitization doses, SD50, determined from non-linear regression of the activation curves in A. Error bars, S.D. values averaged from two independent experiments. C, the m6 T cell transfectant (CD8αβ+) was stimulated in the presence of T2-Ld and various concentrations of peptide QL9 alanine variants. D, SD50 values determined from non-linear regression of the activation curves in C. Error bars, S.D. values averaged from two independent experiments.
The T cell transfectant expressing the high affinity 2C TCR m6 was also stimulated by each of the QL9 alanine variants loaded onto T2-Ld (Fig. 3C). Overall, the impact of the substitutions was less than that of the WT 2C TCR transfectant; e.g. activation of m6 T cells was reduced 100–1,000-fold for the Q1A and P6A variants, and 10–100-fold for the F7A, D8A, and L9A variants (Fig. 3D). Substitutions at positions L2, S3, P4, and F5 had only modest or no detectable impact on activity.
Binding of SIY and QL9 Variants to MHC Kb and Ld
In order to determine whether the reduced T cell activity of the alanine substitutions was due to binding to the MHC product, to the TCR, or to both, we first examined the ability of the variants to bind to Kb or Ld. The levels of Kb or Ld stabilized on the cell surface of RMA-S or T2-Ld cells, respectively, were measured at various concentrations of the alanine variants (26). The stabilization assay is a well characterized method for comparing the relative binding affinities of peptides because the more stable peptide-MHC interactions lead to enhanced surface levels of the MHC at lower peptide concentrations.
Levels of Kb, stabilized on RMA-S with SIY alanine variants, were detected with an mAb specific for Kb (Fig. 4A). The BD50 values were determined for each peptide. Compared with the wild type SIY peptide, Kb binding of variants Y3A, Y5A, and L8A was reduced greater than 20-fold (Fig. 4B). Alanine substitution of all of the other SIY residues showed little impact on Kb binding.
FIGURE 4.
Analysis of SIY and alanine variants binding to MHC Kb. A, detection of MHC Kb up-regulation on the surface of RMA-S with the addition of peptide SIY (SIYRYYGL) alanine variants. Levels of MHC Kb were detected with anti-Kb antibody B.8.24.3 and flow cytometry. MFU or the fluorescence signal above a no peptide background were plotted against peptide concentration. B, binding doses, BD50, determined from non-linear regression of the stabilization curves in A. BD50 values represent the concentration of peptide required to up-regulate half-maximal MHC Kb complexes. Error bars, S.D. values averaged from two independent experiments. C, levels of peptide SIY and alanine variants in complex with MHC Kb on the surface of T2-Kb were monitored at the indicated time points by detection with soluble tetrameric 2C-m67 coupled to streptavidin-phycoerythrin, and binding was detected with flow cytometry. Data are plotted as percentage of maximal peptide-MHC, which represents the percentage of peptide-Kb remaining on the cell surface at a specific time. Percentage of maximal peptide Kb = ((MFUsample − MFUnull OVA)/(Max MFUsample − MFUnull OVA)) × 100.
Because we have a soluble high affinity TCR that binds to SIY-Kb (and the alanine variants of SIY; see below), we were also in a position to specifically determine the cell surface lifetimes of the SIY-Kb complexes, providing an additional quantitative assessment of their Kb binding potential. Cell surface lifetimes were measured by loading a saturating level of the SIY variant and then examining the remaining SIY-Kb on the surface at various times later, as described previously (28). The pep-Kb complexes were detected with soluble high affinity 2C TCR m67 tetramers that bind specifically to SIY-Kb with high affinity (24). In this assay, a t½ of 10 h was measured for the WT SIY, a value similar to that measured previously (28). The same three peptide variants that exhibited reduced ability to stabilize Kb also showed reduced cell surface lifetimes, with a half-life (t½) of ∼30 min for SIY variants Y3A, Y5A, and L8A (Fig. 4C). Thus, both the Kb stabilization assay and cell surface lifetime experiments show that SIY residues Tyr-3, Tyr-5, and Leu-8 contribute considerable energy to Kb binding.
The stabilization assay was also performed with QL9 variants and Ld. In this case, levels of Ld stabilized on the T2-Ld cell line in the presence of various concentrations of the alanine variants of QL9, were detected with a mAb specific for Ld (Fig. 5A). Substitution of the N- and C-terminal residues, Q1A and L9A, showed the most significant effects on Ld stabilization (30–100-fold) (Fig. 5, A and B). Alanine residues substituted at all other positions of QL9, except F5, showed moderate but significant effects on Ld binding (3–10-fold reductions in BD50). Because the cell surface lifetime of the WT QL9-Ld complex is already very short (t½ = 13 min) (28), the lifetimes of the alanine variant complexes were not examined.
FIGURE 5.
Analysis of QL9 and alanine variants binding to MHC Ld. A, detection of MHC Ld up-regulation on the surface of T2-Ld with the addition of peptide QL9 (QLSPFPFDL) and alanine variants. Levels of MHC Ld were detected with anti-Ld antibody 30-5-7 and flow cytometry. MFU values, or the fluorescence signal above a no peptide background, were plotted against peptide concentration. B, binding doses, BD50, determined from non-linear regression of the stabilization curves in A. BD50 values represent the concentration of peptide required to up-regulate half-maximal MHC Ld complexes. Error bars, S.D. values averaged from two independent experiments.
Binding of the TCRs to SIY and QL9 Variant Complexes
In order to complete the analysis of whether the reduced T cell activity of the alanine substitutions was due to binding to the MHC product, to the TCR, or to both, we examined the effect of the peptide substitutions on TCR binding. Because the wild type 2C TCR recognizes SIY-Kb and QL9-Ld with relatively low affinities (KD values of 22 and 1.6 μm, respectively) (38), we used the high affinity TCRs m6 and m67 as probes for determining the exact quantitative impact of each alanine substitution. It is possible that the proximity of the high affinity CDR3α mutations with peptide could skew binding energies, compared with the WT 2C TCR and that the strengths of specific interactions between CDR3α and a peptide residue(s) differ compared with the WT 2C CDR3α. Nevertheless, we believe that the high affinity measurements are reasonable surrogates for the WT 2C TCR for several reasons. First, the docking of the high affinity TCRs on their respective peptide-MHC ligands is virtually identical to the WT 2C TCR (14, 38). Second, there was a strong linear relationship for activity between WT 2C and high affinity m6 T cells when comparing the different peptide variants. Hence, peptide fine specificity was not compromised significantly by introducing mutations into the CDR3α loop of the high affinity m6 TCR. Third, high affinity TCR m33 that binds SIY-Kb and is very similar to m67 (CDR3α sequences: LHRPA for m33 and LERPY for m67) was recently subjected to alanine scanning in which various CDR residues were substituted with alanine (35). The results showed that m33 and 2C had very similar binding energies, implying that their binding landscape on SIY-Kb are nearly identical. Although we believe that the high affinity variants of 2C TCR are valid surrogates of the WT 2C TCR, this does not imply that other TCRs will show identical interactions with each of the SIY or QL9 peptide residues.
A quantitative binding analysis was used to determine ΔΔG values of interactions between each residue of peptides SIY and QL9 and their respective high affinity 2C TCRs, m67 and m6. To measure TCR binding, various concentrations of soluble biotinylated TCR m67 were used to detect T2-Kb incubated in the presence of saturating levels of each SIY alanine variant (Fig. 6A). This approach avoids having to purify and immobilize each of the pep-Kb complexes for surface plasmon resonance, and it allows the same cell surface complexes to be analyzed for TCR binding as were used to assess T cell activity. Peptide SIY variants Y3A, R4A, Y6A, and G7A exhibited significantly reduced TCR m67 binding compared with WT SIY, as indicated by a change in ΔΔG ≥1.3 kcal/mol (Fig. 6B). Hence, SIY Tyr-3, Arg-4, Tyr-6, and Gly-7 were designated “hot spot” residues for binding to TCR m67.
FIGURE 6.
Analysis of peptide SIY and QL9 alanine variants binding to high affinity 2C TCRs m67 and m6. A, titrations of T2-Kb pulsed with 10 μm peptide SIY (SIYRYYGL) alanine variants and various concentrations of soluble biotinylated 2C TCR m67. Binding was detected with streptavidin-phycoerythrin and flow cytometry. MFU values, or fluorescence intensity above a no peptide background, were converted to percentage of maximal TCR binding as follows, % maximal TCR binding = (MFUsample/MFUmax) × 100. B, ΔΔG, determined from the binding constants (KD) in A. Error bars, S.D. values averaged from two independent experiments (no calculated S.D. is shown for SIY R4A because no binding was detected in one of the experiments). C, titrations of T2-Ld pulsed with 10 μm peptide QL9 (QLSPFPFDL) alanine variants along with various concentrations of soluble biotinylated 2C TCR m6. D, ΔΔG, determined from the binding constants (KD) shown in C. Error bars, S.D. values averaged from two independent experiments.
To measure TCR binding to single amino acid variants of QL9, various concentrations of soluble biotinylated TCR m6 were used to detect T2-Ld incubated in the presence of saturating levels of QL9 alanine variants (Fig. 6C). Peptide QL9 variants F5A, F7A, and D8A exhibited significantly reduced TCR m6 binding compared with wild type QL9, as indicated by a change in ΔΔG ≥1.3 kcal/mol (Fig. 6D). However, three other QL9 peptide variants (Q1A, P4A, and P6A) also showed significant effects on binding by the m6 TCR, with free energy effects in the range of 0.5–1.0 kcal/mol (Fig. 6D). Accordingly, the binding effects in the case of the m6/QL9-Ld system seemed to be distributed more across the entire peptide, compared with the SIY-Kb system.
DISCUSSION
The specificity and ultimately the potency of a peptide antigen for a T cell are due to its interactions with both the MHC product and the TCR. These interactions have often been depicted by showing peptide side-chain residues that point toward the MHC product and thereby serve as “anchor” residues; other peptide side-chain residues have been depicted as pointing toward the TCR, thereby providing a minimal binding energy and conferring antigen specificity to the T cell. Is this a reasonable view of an antigenic peptide, or is it overly simplistic? Here we were able to examine in detail this question, by comparing two quite different peptide-MHC ligands (SIY-Kb and QL9-Ld) that bind to the same TCR, 2C. In addition to comparing the Kb and Ld systems, we were able to assess directly the quantitative impact of MHC binding versus TCR binding on T cell activity.
Our salient findings are that the Kb system follows more closely the often depicted representation in which a peptide has residues that impact either MHC or TCR. In contrast, the Ld system generates its specificity and potency by having many residues that are involved in both MHC and TCR binding. It could be argued that the differences observed relate more to recognition of an allogeneic ligand (Ld in the case of 2C) versus a syngeneic ligand (Kb in the case of 2C) than they do to an inherent difference between Ld and Kb. For example, it has often been suggested that there is a reduction in peptide specificity for recognition of allogeneic MHC compared with syngeneic ligands (3, 39). Alternatively, it is possible that these differences have to do with an affinity threshold for T cell activation (e.g. allogeneic TCRs have higher affinity and thus appear less peptide dependent) rather than with a difference between TCR binding to peptides associated with syngeneic versus allogeneic ligands. Although additional TCR-pep-Kb and TCR-pep-Ld systems will need to be examined to clarify these issues, the evidence here suggests that distinct differences in the way that Kb and Ld bind and present peptides influence the way in which TCR engages the entire complex.
From a potential clinical perspective, the findings that QL9 variants often impacted both Ld binding and TCR binding indicate that for some MHC alleles, one must consider whether a peptide variant optimized for MHC product binding will also have a negative impact on T cell recognition (4). Thus, design of peptide variants for the purpose of improving binding to a specific MHC allele should assess whether TCR recognition and thus T cell activity might be impacted in a negative manner.
The correlates of MHC and TCR binding can be considered in light of the structures of the corresponding complexes by examining the peptide residues that have contacts with either the TCR or the MHC (Fig. 7). In this representation, the number of atomic interactions between a peptide residue and either the TCR or the MHC is shown by lines (for this purpose, an interaction was defined as two atoms involved in hydrogen bonds or van der Waals with atomic distances of ≤4.5 Å). Both the 2C TCR-QL9-Ld (Fig. 7B) and m6 TCR-QL9-Ld (Fig. 7C) complexes are shown in order to illustrate that the contacts were similar for the low and high affinity complexes, within the limits of the resolution of the respective structures.
FIGURE 7.
Peptide SIY and QL9 contact maps representing the number of atomic interactions with TCR and MHC. A, contact map represents number of atomic interactions between each residue of peptide SIY (SIYRYYGL) and either the 2C TCR or MHC Kb using the structure of 2C-SIY-Kb (Protein Data Bank code 1G6R). Interactions were determined using the program “Contacts” of the Collaborative Computational Project Number 4 (CCP4 Suite). An interaction was defined as two atoms involved in hydrogen bonds or van der Waals with atomic distances of ≤4.5 Å. B, contact map represents the number of atomic interactions between each residue of peptide QL9 (QLSPFPFDL) and the 2C TCR or the MHC Ld using the structure of 2C/QL9-Ld (Protein Data Bank code 2Oi9). C, contact map represents the number of atomic interactions between each residue of peptide QL9 and the high affinity 2C-m6 TCR or MHC Ld using the structure of m6/QL9-Ld (Protein Data Bank code 2E7L).
Overall, this analysis shows that there is not a strict correlation between the observed atomic distances and the energetic consequences of the interactions (as has been noted in many “alanine scans” (e.g. see Ref. 23)). Furthermore, it is impossible to predict, from the structural analysis, the indirect effects of peptide substitutions that occur through changes in conformations distal to the single-site substitution. For example, based on the structure of 2C TCR-SIY-Kb (Fig. 7A), substitution of Tyr-3 of SIY might be predicted to have an effect on Kb binding (as it did; Fig. 4), but one would not have predicted that it would have an effect of almost 2 kcal/mol on TCR binding (Fig. 6B). Similarly, several residues in QL9, including Gln-1, Pro-4, and Pro-6, might also not have been predicted to impact TCR binding, but substitution of each reduced the binding affinity substantially (∼1 kcal/mol).
We believe that these results will extrapolate to other peptides because there are conserved features. Peptides that bind to Kb contain the predominant octamer motif: Y3, Y/F5, and L/M8 (8, 11, 12, 40–42). In addition, peptides that bind to Ld often have a proline at position 2, and a hydrophobic residue at the C terminus (9, 13, 43, 44), yet this MHC Ld binding motif is not conserved for all peptides, including peptides QL9 (QLSPFPFDL) and tum− (TQNHRALDL) (14, 44).
Previous studies have described the impact of changing residues that point toward the MHC but have typically not examined binding by the TCR. An exception is an analogous study using a high affinity TCR against a class II ligand. Substitution of several Hb peptide residues involved in binding to I-Ek resulted in reduced binding by the high affinity TCR (45).
Aside from understanding the impact of peptide residues involved in the Kb system versus the Ld system, what is the magnitude of effects on activity of residues that influence only MHC binding compared with those that influence only TCR binding? To address this question, it is useful to examine SIY residues Tyr-5 and Leu-8, both well characterized anchor residues for Kb, and both of the alanine substitutions at these positions had no effect on TCR binding by the m67 TCR. These substitutions also reduced the cell surface lifetime of the Kb complexes from 10 to 0.5 h. In the case of both 2C and m67 T cells, the activity of these two peptides was reduced about 1,000-fold. In the case of the QL9-Ld system, the only residue where alanine substitution had an impact only on Ld binding (and not TCR binding) was the anchor position L9. The L9A variant also showed a reduction in activity for both the 2C and m6 T cells by about 1,000-fold. Together, these findings suggest that the C-terminal anchor residue of a potential peptide vaccine may be the optimal position to enhance MHC binding and immunogenicity (46) without influencing TCR binding appreciably.
Conversely, to address the magnitude of effects when only TCR binding is impacted by a substitution, it is useful to examine SIY variants R4A and Y6A that affected only binding by the TCR and not binding to Kb. Based on the m67 TCR binding analyses, the Arg-4 and Tyr-6 side chains each contributed about 2.7 kcal/mol to the interaction with TCR. In the case of the 2C WT interaction, this would reduce the affinity from a KD value of 22 μm to about 3 mm. Clearly, this is below the threshold required for T cell activity, which explains why the 2C T cells are not active with R4A or Y6A. In contrast, this reduces the affinity of the m67 TCR from a KD value of 7 nm to about 1 μm. This affinity is above the threshold required for CD8-mediated activity; thus, there is only about a 10–20-fold reduction in the activity of the R4A and Y6A variants for the m67 T cells.
A similar result was observed with the QL9-Ld system, where F5A was the only variant that influenced TCR m6 binding but not Ld binding. The Phe-5 side chain had an impact of about 2.3 kcal/mol on m6 TCR binding. In the case of the 2C WT interaction, this would reduce the affinity from a KD value of 1.6 μm to about 100 μm. TCRs with KD values in this range have been shown previously to allow some T cell activity (47, 48), and in fact 2C T cells exhibited activity with F5A, albeit 3,000-fold reduced compared with QL9. In contrast, this reduces the affinity of the m6 TCR from a KD value of 20 nm to about 1 μm, well above the affinity threshold for maximal activity in the presence of CD8. Hence, there was no effect of the F5A substitution on the activity of m6 T cells.
Finally, we were in a position to examine structurally how peptide residues might have impacted binding to both the MHC product and the TCR. In the case of the SIY-Kb system, the single most significant variant in this regard was Y3A. We suggest that the Y3A substitution may affect TCR binding indirectly, because Tyr-3 is embedded in pocket D of Kb, through numerous interactions. Arg-155 of Kb, one of the residues that contacts Tyr-3, also interacts with Tyr-31 of the 2C TCR (Fig. 8A), a residue that contributes significant binding free energy to the interaction with SIY-Kb (49). Thus, we propose that this network of interactions is disrupted with the Y3A substitution.
FIGURE 8.
Structural features of SIY and QL9 peptide residues that appear to act indirectly on binding by the TCR. A, position of peptide SIY residue Tyr3 (orange) in the 2C TCR-SIY-Kb complex (Protein Data Bank code 1G6R). A portion of the Vα domain of the 2C TCR is represented in red, showing CDR1α residue Tyr-31. Class I MHC Kb is represented in green, showing residues Glu-152 and Arg-155. B, position of peptide QL9 residue Gln-1 (orange) in the m6 TCR-QL9 Gln-1-Ld complex (Protein Data Bank code 2E7L). A portion of the Vα domain of the 2C TCR is represented in red, showing CDR1α residues Thr-29 and Tyr-31. The class I MHC Ld is represented in green, showing residues Tyr-159 and Glu-163.
Past studies have emphasized the significance of the C terminus of peptide QL9 (PFDL) in binding to Ld and recognition by the 2C CTL (50, 51). Here, we show that the N terminus of peptide QL9, in particular Gln-1, also influences binding to both the MHC product and the TCR. The N-terminal glutamine of QL9 makes multiple contacts with Ld in the 2C-QL9-Ld and m6-QL9-Ld complexes (14). This suggests that the effects on TCR binding arise indirectly either from perturbations of the peptide conformation associated with Ld binding or from perturbations of the Ld molecule in regions that contact the TCR. For example, Gln-1 of QL9 contacts Tyr-159 of Ld, which in turn contacts both Thr-29 and Tyr-31 in CDR1α of m6 (Fig. 8B). These two TCR residues were shown to contribute significant binding energy to the interaction of the 2C TCR and QL9-Ld (52), and substitution of Gln-1 has been shown previously to impact Ld binding and 2C activity (53). Thus again, we propose that this network of interactions is disrupted with the Q1A substitution. Indirect binding effects have been observed in the past for CTL recognition of peptide-induced conformational changes in the MHC product (e.g. Refs. 54 and 55), including studies that showed that peptides p2Ca and tum− influenced binding of the Ld-α1-specific monoclonal antibody B22/249 (56).
In conclusion, almost every residue of the SIY and QL9 peptides contributed to the specificity of 2C T cells. Even for positions in which the alanine substitution had minimal effect, it is likely that substitution of larger bulky side chains would also show a significant negative impact (18, 26). The interactions of each side chain with the TCR, the MHC, or both serve as the origin of this specificity. Thus, the TCR-MHC system has evolved to take full advantage of the structural and mechanistic aspects of the interactions to optimize specificity of the foreign peptide.
Acknowledgments
We thank the University of Illinois Flow Cytometry Facility for assistance and Phil Holler for providing the transfected T cell lines.
This work was supported, in whole or in part, by National Institutes of Health Grant GM55767 (to D. M. K.) and AI048540 (to K. C. G.). This work was also supported by a National Science Foundation predoctoral fellowship (to L. A. C.).
- TCR
- T cell receptor
- MHC
- major histocompatibility complex
- IL
- interleukin
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- MFU
- mean fluorescent unit(s)
- WT
- wild type
- SD50
- concentration(s) of peptide yielding 50% maximal IL-2 release
- BD50
- binding dose(s) yielding 50% maximum cell surface levels
- ΔΔG
- binding free energy
- mAb
- monoclonal antibody.
REFERENCES
- 1.Davis M. M., Bjorkman P. J. (1988) Nature 334, 395–402 [DOI] [PubMed] [Google Scholar]
- 2.Wucherpfennig K. W., Allen P. M., Celada F., Cohen I. R., De Boer R., Garcia K. C., Goldstein B., Greenspan R., Hafler D., Hodgkin P., Huseby E. S., Krakauer D. C., Nemazee D., Perelson A. S., Pinilla C., Strong R. K., Sercarz E. E. (2007) Semin. Immunol. 19, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felix N. J., Donermeyer D. L., Horvath S., Walters J. J., Gross M. L., Suri A., Allen P. M. (2007) Nat. Immunol. 8, 388–397 [DOI] [PubMed] [Google Scholar]
- 4.Iero M., Filipazzi P., Castelli C., Belli F., Valdagni R., Parmiani G., Patuzzo R., Santinami M., Rivoltini L. (2009) Cancer Immunol. Immunother. 58, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borbulevych O. Y., Baxter T. K., Yu Z., Restifo N. P., Baker B. M. (2005) J. Immunol. 174, 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valmori D., Fonteneau J. F., Lizana C. M., Gervois N., Liénard D., Rimoldi D., Jongeneel V., Jotereau F., Cerottini J. C., Romero P. (1998) J. Immunol. 160, 1750–1758 [PubMed] [Google Scholar]
- 7.Parkhurst M. R., Salgaller M. L., Southwood S., Robbins P. F., Sette A., Rosenberg S. A., Kawakami Y. (1996) J. Immunol. 157, 2539–2548 [PubMed] [Google Scholar]
- 8.Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. (1991) Nature 351, 290–296 [DOI] [PubMed] [Google Scholar]
- 9.Corr M., Boyd L. F., Frankel S. R., Kozlowski S., Padlan E. A., Margulies D. H. (1992) J. Exp. Med. 176, 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degano M., Garcia K. C., Apostolopoulos V., Rudolph M. G., Teyton L., Wilson I. A. (2000) Immunity 12, 251–261 [DOI] [PubMed] [Google Scholar]
- 11.Fremont D. H., Stura E. A., Matsumura M., Peterson P. A., Wilson I. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2479–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W., Young A. C., Imarai M., Nathenson S. G., Sacchettini J. C. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8403–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balendiran G. K., Solheim J. C., Young A. C., Hansen T. H., Nathenson S. G., Sacchettini J. C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6880–6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colf L. A., Bankovich A. J., Hanick N. A., Bowerman N. A., Jones L. L., Kranz D. M., Garcia K. C. (2007) Cell 129, 135–146 [DOI] [PubMed] [Google Scholar]
- 15.Speir J. A., Garcia K. C., Brunmark A., Degano M., Peterson P. A., Teyton L., Wilson I. A. (1998) Immunity 8, 553–562 [DOI] [PubMed] [Google Scholar]
- 16.Sloan-Lancaster J., Allen P. M. (1996) Annu. Rev. Immunol. 14, 1–27 [DOI] [PubMed] [Google Scholar]
- 17.Wu L. C., Tuot D. S., Lyons D. S., Garcia K. C., Davis M. M. (2002) Nature 418, 552–556 [DOI] [PubMed] [Google Scholar]
- 18.Huseby E. S., Crawford F., White J., Marrack P., Kappler J. W. (2006) Nat. Immunol. 7, 1191–1199 [DOI] [PubMed] [Google Scholar]
- 19.Holler P. D., Chlewicki L. K., Kranz D. M. (2003) Nat. Immunol. 4, 55–62 [DOI] [PubMed] [Google Scholar]
- 20.Holler P. D., Holman P. O., Shusta E. V., O'Herrin S., Wittrup K. D., Kranz D. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5387–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sykulev Y., Brunmark A., Tsomides T. J., Kageyama S., Jackson M., Peterson P. A., Eisen H. N. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 11487–11491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udaka K., Wiesmüller K. H., Kienle S., Jung G., Walden P. (1996) J. Immunol. 157, 670–678 [PubMed] [Google Scholar]
- 23.DeLano W. L. (2002) Curr. Opin. Struct. Biol. 12, 14–20 [DOI] [PubMed] [Google Scholar]
- 24.Zhang B., Bowerman N. A., Salama J. K., Schmidt H., Spiotto M. T., Schietinger A., Yu P., Fu Y. X., Weichselbaum R. R., Rowley D. A., Kranz D. M., Schreiber H. (2007) J. Exp. Med. 204, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chervin A. S., Aggen D. H., Raseman J. M., Kranz D. M. (2008) J. Immunol. Methods 339, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlueter C. J., Manning T. C., Schodin B. A., Kranz D. M. (1996) J. Immunol. 157, 4478–4485 [PubMed] [Google Scholar]
- 27.Lie W. R., Myers N. B., Gorka J., Rubocki R. J., Connolly J. M., Hansen T. H. (1990) Nature 344, 439–441 [DOI] [PubMed] [Google Scholar]
- 28.Brophy S. E., Jones L. L., Holler P. D., Kranz D. M. (2007) Mol. Immunol. 44, 2184–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranz D. M., Sherman D. H., Sitkovsky M. V., Pasternack M. S., Eisen H. N. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udaka K., Tsomides T. J., Eisen H. N. (1992) Cell 69, 989–998 [DOI] [PubMed] [Google Scholar]
- 31.Udaka K., Tsomides T. J., Walden P., Fukusen N., Eisen H. N. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11272–11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniels M. A., Jameson S. C. (2000) J. Exp. Med. 191, 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho B. K., Lian K. C., Lee P., Brunmark A., McKinley C., Chen J., Kranz D. M., Eisen H. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1723–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holler P. D., Kranz D. M. (2003) Immunity 18, 255–264 [DOI] [PubMed] [Google Scholar]
- 35.Chervin A. S., Stone J. D., Holler P. D., Bai A., Chen J., Eisen H. N., Kranz D. M. (2009) J. Immunol. 183, 1166–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brock R., Wiesmüller K. H., Jung G., Walden P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13108–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone J. D., Chervin A. S., Kranz D. M. (2009) Immunology 126, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones L. L., Colf L. A., Stone J. D., Garcia K. C., Kranz D. M. (2008) J. Immunol. 181, 6255–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bevan M. J. (1984) Immunol. Today 5, 128–130 [DOI] [PubMed] [Google Scholar]
- 40.Saito Y., Peterson P. A., Matsumura M. (1993) J. Biol. Chem. 268, 21309–21317 [PubMed] [Google Scholar]
- 41.Johansen T. E., McCullough K., Catipovic B., Su X. M., Amzel M., Schneck J. P. (1997) Scand. J. Immunol. 46, 137–146 [DOI] [PubMed] [Google Scholar]
- 42.van Bleek G. M., Nathenson S. G. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11032–11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergmann C. C., Stohlman S. A. (1996) J. Virol. 70, 3252–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson R. A., Lee D. R. (1996) J. Immunol. 156, 4266–4273 [PubMed] [Google Scholar]
- 45.Donermeyer D. L., Weber K. S., Kranz D. M., Allen P. M. (2006) J. Immunol. 177, 6911–6919 [DOI] [PubMed] [Google Scholar]
- 46.Allen P. M., Matsueda G. R., Adams S., Freeman J., Roof R. W., Lambert L., Unanue E. R. (1989) Int. Immunol. 1, 141–150 [DOI] [PubMed] [Google Scholar]
- 47.Sykulev Y., Brunmark A., Jackson M., Cohen R. J., Peterson P. A., Eisen H. N. (1994) Immunity 1, 15–22 [DOI] [PubMed] [Google Scholar]
- 48.Bowerman N. A., Crofts T. S., Chlewicki L., Do P., M. B. B., Garcia K. C., Kranz D. M. (2009) Mol. Immunol. 46, 3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee P. U., Churchill H. R., Daniels M., Jameson S. C., Kranz D. M. (2000) J. Exp. Med. 191, 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sykulev Y., Vugmeyster Y., Brunmark A., Ploegh H. L., Eisen H. N. (1998) Immunity 9, 475–483 [DOI] [PubMed] [Google Scholar]
- 51.Gillanders W. E., Hanson H. L., Rubocki R. J., Hansen T. H., Connolly J. M. (1997) Int. Immunol. 9, 81–89 [DOI] [PubMed] [Google Scholar]
- 52.Manning T. C., Schlueter C. J., Brodnicki T. C., Parke E. A., Speir J. A., Garcia K. C., Teyton L., Wilson I. A., Kranz D. M. (1998) Immunity 8, 413–425 [DOI] [PubMed] [Google Scholar]
- 53.Hornell T. M., Martin S. M., Myers N. B., Connolly J. M. (2001) J. Immunol. 167, 4207–4214 [DOI] [PubMed] [Google Scholar]
- 54.Rohren E. M., McCormick D. J., Pease L. R. (1994) J. Immunol. 152, 5337–5343 [PubMed] [Google Scholar]
- 55.Bluestone J. A., Kaliyaperumal A., Jameson S., Miller S., Dick R., 2nd (1993) J. Immunol. 151, 3943–3953 [PubMed] [Google Scholar]
- 56.Solheim J. C., Carreno B. M., Smith J. D., Gorka J., Myers N. B., Wen Z., Martinko J. M., Lee D. R., Hansen T. H. (1993) J. Immunol. 151, 5387–5397 [PubMed] [Google Scholar]