Abstract
Asparaginase depletes circulating asparagine and glutamine, activating amino acid deprivation responses (AADR) such as phosphorylation of eukaryotic initiation factor 2 (p-eIF2) leading to increased mRNA levels of asparagine synthetase and CCAAT/enhancer-binding protein β homologous protein (CHOP) and decreased mammalian target of rapamycin complex 1 (mTORC1) signaling. The objectives of this study were to assess the role of the eIF2 kinases and protein kinase R-like endoplasmic reticulum resident kinase (PERK) in controlling AADR to asparaginase and to compare the effects of asparaginase on mTORC1 to that of rapamycin. In experiment 1, asparaginase increased hepatic p-eIF2 in wild-type mice and mice with a liver-specific PERK deletion but not in GCN2 null mice nor in GCN2-PERK double null livers. In experiment 2, wild-type and GCN2 null mice were treated with asparaginase (3 IU per g of body weight), rapamycin (2 mg per kg of body weight), or both. In wild-type mice, asparaginase but not rapamycin increased p-eIF2, p-ERK1/2, p-Akt, and mRNA levels of asparagine synthetase and CHOP in liver. Asparaginase and rapamycin each inhibited mTORC1 signaling in liver and pancreas but maximally together. In GCN2 null livers, all responses to asparaginase were precluded except CHOP mRNA expression, which remained partially elevated. Interestingly, rapamycin blocked CHOP induction by asparaginase in both wild-type and GCN2 null livers. These results indicate that GCN2 is required for activation of AADR to asparaginase in liver. Rapamycin modifies the hepatic AADR to asparaginase by preventing CHOP induction while maximizing inhibition of mTORC1.
Introduction
The enzyme l-asparaginase has been used to treat both pediatric and adult forms of acute lymphoblastic leukemia for over 40 years (1). Asparaginase cleaves the nonessential amino acid asparagine into aspartic acid plus ammonia. The tumor-killing properties of asparaginase are ascribed to dependence on circulating asparagine for growth of the leukemic lymphoblast (2). The form of asparaginase most commonly administered to patients is purified from Escherichia coli. Treatment with asparaginase is oftentimes accompanied by secondary complications, including hepatic dysfunction, neurologic seizures, pancreatitis, and immunosuppression (3). Work by this group and others (4–6) suggest that these effects are largely due to the inherent glutaminase activity of the enzyme, causing depletion of the conditionally essential amino acid glutamine.
Asparaginase reduces protein synthesis by increasing phosphorylation of eukaryotic initiation factor 2 (eIF2)2 (4). Phosphorylation of eIF2 (p-eIF2) is catalyzed by a family of four protein kinases that are differentially sensitive to various cell stressors (7). During amino acid deprivation, the eIF2 kinase called general control nonderepressible 2 (GCN2) is activated by intracellular accumulation of deacylated tRNA (8). The accumulation of misfolded proteins within the endoplasmic reticulum (ER) activates the eIF2 kinase called protein kinase R-like endoplasmic reticulum kinase (PERK) (9, 10), whereas viral infection stimulates protein kinase R, and heme deprivation triggers heme-controlled repressor. In addition to reducing global protein synthesis, p-eIF2 stimulates gene-specific translation of activating transcription factor 4 (ATF4), which induces the expression of genes such as asparagine synthetase (ASNS) to relieve the particular cell stress. If the stress cannot be relieved, gene expression then switches to promoting cell death via transcription of the pro-apoptotic transcription factor, CCAAT/enhancer-binding protein homologous protein (CHOP, also known as GADD153) (11). Simultaneous control of global and gene-specific translation directed by p-eIF2 is collectively referred to as the integrated stress response (ISR) (7, 11). In this study, we examined the role of GCN2 versus PERK in mediating activation of the ISR by asparaginase. To address this question, we generated mice with PERK disrupted in liver using a conditional gene deletion strategy. These mice were then crossed with GCN2 null mice to produce mice with both GCN2 and PERK deleted in liver that were subsequently characterized for changes in the ISR in response to asparaginase.
Amino acid depletion is also sensed by the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway, which regulates mRNA translation via phosphorylation of translational repressor eIF4E-binding protein 1 (4E-BP1) and the p70 ribosomal S6 kinase 1 (S6K1) (12). Phosphorylation of 4E-BP1 facilitates cap-dependent mRNA translation, whereas phosphorylation of S6K1 plays important roles in ribosome biogenesis, control of cell size, and assembly of the translation preinitiation complex (13). Amino acid starvation inhibits phosphorylation of 4E-BP1 and S6K1 similar to treatment with the mTOR inhibitor rapamycin, yet microarray analysis shows that these two conditions are distinct from each other (14, 15). On the basis of these earlier observations, we wished to address whether the combination of rapamycin and asparaginase is a more potent mTORC1 inhibitor than either agent alone.
The existence of cross-talk between the ISR and mTORC1 pathway in mammals is also important to consider as our laboratory reported that mTORC1 signaling is derepressed in GCN2 null mice fed a leucine-devoid diet (16). Furthermore, rapamycin is reported to block induction of CHOP by amino acid deprivation in mouse fibroblasts (17), highlighting a role for the mTORC1 pathway in modulating a commonly used biomarker of the ISR. In this work, show that although both asparaginase and rapamycin down-regulate mTORC1 signaling, only asparaginase activates the ISR. Furthermore, nearly all amino acid deprivation responses (AADR) to asparaginase required GCN2, with the exception of CHOP, which asparaginase increased in a rapamycin-sensitive fashion.
EXPERIMENTAL PROCEDURES
Materials
p-eIF2α antibody was purchased from Cell Signaling Technology (Beverly, MA). Results were normalized for total eIF2α (Santa Cruz Biotechnology). Primary antibodies for phosphorylation of eIF4E-binding protein-1 (4E-BP1), ribosomal protein S6 kinase (S6K1), and actin were purchased from Bethyl Laboratories. Antibodies against REDD1, p-Akt (Ser473), Akt, p-eEF2, eEF2, p-p44/42 MAPK (p-ERK1/2), and p-PDK1 were obtained from Cell Signaling Technology. ERK1/2 antibody was purchased from Santa Cruz Biotechnology. Secondary antibody was purchased from The Jackson Laboratory. Experimental animal diet used in this study was a standard commercial rodent food based on AIN-93 standards (18) consisting of 18% protein and 4% fat (7017 NIH-31 Open Formula Mouse/Rat Sterilizable diet, Harlan Teklad).
Measurement of l-Asparaginase Activity
The activity of experimental l-asparaginase derived from E. coli (Elspar® product from Merck) was determined by the Nesslerization technique, as described previously (4, 19). Briefly, the production of ammonia by l-asparaginase over time was expressed relative to the slope of known ammonia standards. The resulting value represented the activity of the enzyme in international units, in which 1 IU equaled the amount of enzyme that catalyzed the formation of 1 μmol of ammonia per min. Enzyme activity of l-asparaginase was determined prior to administration.
Animals
The following study protocol was approved by the Institutional Care and Use Committee at the Indiana University School of Medicine, Evansville. In these studies, mice were maintained on a 12-h light:dark cycle and given unrestricted access to food and water over the course of the experiment. In experiment 1, mice homozygous for the LoxP allele of PERK were mated with albumin-driven Cre recombinase transgenic mice (AlbCre+) (creation of mice with floxed PERK are described in Ref. 20). The AlbCre+ transgene-bearing offspring were bred to homozygocity for the LoxP allele of PERK to create a liver-specific deletion of PERK (AlbCre+PERKf/f) and produce Cre-negative wild-type littermates (AlbCre-PERKf/f). GCN2−/− mice (described below) were then bred to these lines, and progeny were brother-sister-mated to homozygocity so as to create a GCN2 null mouse possessing PERK knockdown in liver only. Male and female young adult mice (six mice per treatment group and equal sex distribution) were injected intraperitoneally once daily with either phosphate-buffered saline (PBS) or PBS containing an enzyme activity of 3 IU of Elspar® l-asparaginase per g of body weight, and tissues were collected 6 h after injection. In experiment 2, 6–8-week-old male and female C57BL/6J (GCN2+/+) mice and mice deleted for GCN2 (GCN2−/−; backcrossed onto C57BL/6J 10 generations) were randomly sorted (six animals per treatment group and equal sex distribution) into one of four treatment groups as follows: saline only, asparaginase only, rapamycin only, or rapamycin plus asparaginase. At the start of the experiment, mice were first administered by intraperitoneal injection either rapamycin at a dose of 2 mg per kg of body weight or an equal volume of PBS. Thirty minutes later, mice were then injected with either PBS or PBS containing an enzyme activity of 3 IU of Elspar® l-asparaginase per g of body weight. Body weight was measured prior to the first injection and at the end of study. All mice lost a slight amount of body weight between injection and tissue collection (∼5% or less), consistent with the study being conducted during the light cycle. Food intake was measured throughout the experiment and was similar between strains and among all treatment groups. Mice were killed 8 h after administration of l-asparaginase. Liver and pancreas were quickly dissected, rinsed in ice-cold PBS, weighed, and frozen immediately in liquid nitrogen.
Genotyping
Tissue samples (ear punch or tail snippet, liver, pancreas, and spleen) were digested overnight with proteinase K, and total DNA was extracted using a commercial kit (DNeasy tissue kit, Qiagen). Mice were genotyped for GCN2 expression by PCR as described previously (21). To examine expression of both Cre recombinase and floxed PERK, multiplex PCR was carried out using the following primer sets: Cre forward, 5′-CCTGGAAAATGCTTCTGTCCGTTT-3′, and Cre reverse, 5′-GAGTTGATAGCTGGCTGGTGGCAGATG-3′; flPERK forward, 5′-CACTCTGGCTTTCACTCCTCACAG-3′, and flPERK reverse, 5′-GTCTTACAAAAAGGAGGAAGGTGGAA-3′. Each primer (1 μl of 10 μm stock) was aliquoted into a PCR tube and combined with TaqPCR Master Mix (Qiagen), water, and 150 ng of template DNA. PCR cycle conditions were as follows: 94 °C for 5 min and then 39 cycles of 94 °C for 45 s, 58 °C for 45 s, and 70 °C for 1 min, finishing with 70 °C for 10 min. Samples were resolved on a 1.2% agarose gel, and bands were detected using a Kodak 4000MM Multimodal Imager with Carestream Health wide angle excitation (535) and emission (600) filters to visualize ethidium bromide.
Tissue Preparation for Immunoblot Analysis
Tissues were homogenized, as described previously (4), using a glass-on-glass homogenizer in 7 volumes of buffer A consisting of (in mm) 20 HEPES (pH 7.4), 100 KCl, 0.2 EDTA, 2 EGTA, 1 dithiothreitol, 50 sodium fluoride, 50 β-glycerophosphate, 0.1 phenylmethylsulfonyl fluoride, 1 benzamidine, and 0.5 sodium orthovanadate. The homogenates were immediately centrifuged at 10,000 × g for 10 min at 4 °C for analysis of protein expression and phosphorylation state as described below. Samples were size-fractionated on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (Immobilon-P, Millipore, MA). Blots were developed with enhanced chemiluminescence (Amersham Biosciences). Protein expression was analyzed using a Kodak 4000MM Multimodal Imager, and band density was quantitated using Kodak Molecular Imaging software (version 4.0.4).
Immunoblot Analysis
Phosphorylation of eIF2α was assessed using an antibody that recognizes the protein only when it is phosphorylated at Ser51. Results were normalized for total eIF2α. Phosphorylation of 4E-BP1 and S6K1 were measured as a decrease in mobility during SDS-PAGE, detected by immunoblot as described previously (22). REDD1 protein expression was normalized to actin. Phosphorylation of Akt at Ser473 was normalized for total Akt. Phosphorylation of ERK1/2 was assessed using an antibody that recognizes ERK1/2 when phosphorylated at ERK1 (Thr202 and Tyr204) and/or ERK2 (Thr185 and Tyr187). Results were normalized for total ERK1/2.
Reverse Transcription and Real Time PCR of ASNS and CHOP
Total RNA was extracted from frozen tissue using TriReagentTM (Molecular Research Center, Inc., Cincinnati, OH) followed by DNase treatment (VersaGene DNase kit, Gentra Systems). To inactivate the reaction, the samples were heated to 70 °C for 5 min. The A260:280 ratio was between 1.8 and 2.0 following RNA clean up (RNeasy mini kit) (Qiagen). mRNA expression was determined by quantitative PCR using TaqMan reagents. 1 μg of RNA solutions was used for reverse transcription using high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. TaqMan gene expression master mix and TaqMan gene expression assays (Applied Biosystems) were used for the PCR step. Amplification and detection were performed using the StepOnePlus real time PCR system (Applied Biosystems) with the following profile: 1 cycle at 50 °C for 2 min, 95 °C for 10 min, and 40 cycles each at 95 °C for 15 s and 60 °C for 1 min. 10 ng of cDNA was used per reaction in a 10-μl reaction volume. All samples were run in triplicates. 18 S ribosomal RNA was chosen as a suitable normalization control gene. Results were obtained by the comparative Ct method, using ΔCt (the value obtained by subtracting the Ct value of ASNS or CHOP from the Ct value of 18 S ribosome mRNA of individual sample). Specifically, the quantity of target mRNA relative to 18 S ribosome mRNA was expressed as 2−(ΔΔCt). Results are expressed as fold change with respect to the experimental control.
Statistics
All data were analyzed by the STATISTICA statistical software (StatSoft, Inc.). Data were analyzed using one-way, two-way, or three-way analysis of variance (ANOVA) to assess main and interaction effects, with “Group” as the independent variable for one-way ANOVA (Fig. 1B); “Drug” and “Genotype” as the independent variables for two-way ANOVA (Fig. 1, C and D; supplemental Fig. S1, B and C); and “Rapamycin,” “Asparaginase,” and “Genotype” as the independent variables for three-way ANOVA (Figs. 2–4, supplemental Fig. S1, A and D, and supplemental Fig. S2). When a significant main or interaction effect was detected, differences among treatment groups were assessed with Duncan's multiple range post hoc test. The level of significance was set at p < 0.05 for all statistical tests.
FIGURE 1.
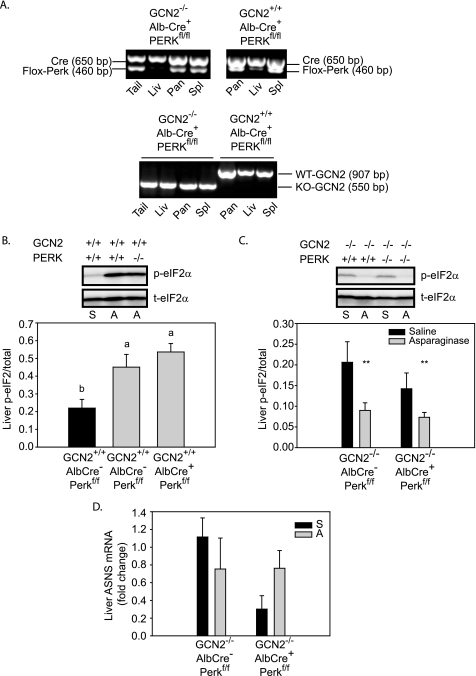
A, AlbCre-mediated knockdown of floxed PERK (AlbCre+, PERKfl/fl) in liver (Liv) but not pancreas (Pan), spleen (Spl), or tail of GCN2+/+ and GCN2−/− mice (upper panel). GCN2 was deleted in the liver, pancreas, and spleen of GCN2−/−-AlbCre+-PERKfl/fl mice but not GCN2+/+-AlbCre+-PERKfl/fl mice (lower panel). WT, wild type; KO, knock-out. B, asparaginase increased phosphorylation of eIF2 in the liver of both wild-type (GCN2+/+-AlbCre−-PERKfl/fl) and PERK knockdown (GCN2+/+-AlbCre+-PERKfl/fl) mice. Means not sharing same lowercase letter are different from each other (by one-way ANOVA), p < 0.05. C, phosphorylation of eIF2 is reduced in the liver of GCN2 null mice both with and without AlbCre-mediated PERK knockdown following asparaginase injection. **, main effect of asparaginase to increase p-eIF2 (by two-way ANOVA), p < 0.05. D, hepatic ASNS mRNA expression was not significantly increased following asparaginase treatment in GCN2 null mice with or without AlbCre-mediated deletion of floxed PERK in liver. Abbreviations: S, saline; A, asparaginase.
FIGURE 2.
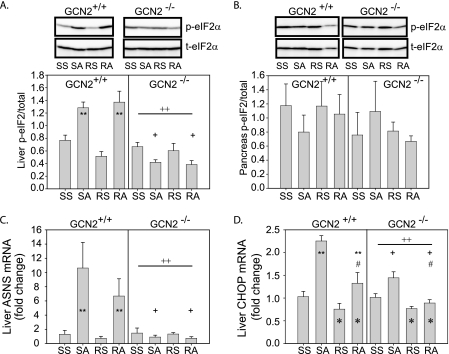
GCN2 is required to activate the integrated stress response in the liver but not pancreas of mice treated with asparaginase. A, phosphorylation of eIF2 in the liver of GCN2+/+ and GCN2−/− mice. **, asparaginase increased p-eIF2 in GCN2+/+ mice, p < 0.05; ++, loss of GCN2 reduced p-eIF2 independent of drug treatment, p < 0.05; +, effect of asparaginase to increase p-eIF2 required GCN2, p < 0.05. B, phosphorylation of eIF2 in the pancreas of GCN2+/+ and GCN2−/− mice was not altered by either drug or genotype. C, hepatic ASNS mRNA expression in GCN2+/+ and GCN2−/− mice. **, asparaginase increased ASNS mRNA in GCN2+/+ mice, p < 0.05; ++, loss of GCN2 reduced ASNS mRNA independent of drug treatment, p < 0.05; +, effect of asparaginase to increase ASNS mRNA depended on GCN2, p < 0.05. D, hepatic CHOP/GADD153 mRNA expression in GCN2+/+ and GCN2−/− mice. **, asparaginase increased CHOP mRNA in GCN2+/+ mice, p < 0.05; ++, loss of GCN2 reduced CHOP mRNA independent of drug treatment, p < 0.05; +, effect of asparaginase to increase CHOP mRNA required GCN2, p < 0.05; *, rapamycin reduced CHOP mRNA independent of genotype, p < 0.05; #, effect of asparaginase was blunted by rapamycin, p < 0.05. Abbreviations: SS, saline then saline; SA, saline then asparaginase; RS, rapamycin then saline; RA, rapamycin then asparaginase.
FIGURE 3.
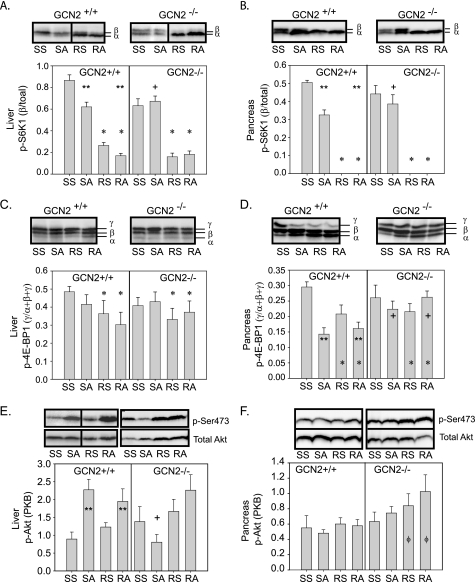
Asparaginase and rapamycin impart tissue-specific effects on the phosphorylation of S6K1, 4E-BP1, and Akt. A and B, reduced p-S6K1 in liver and pancreas by asparaginase required GCN2. **, asparaginase reduced p-S6K1 in GCN2+/+ mice, p < 0.05; *, rapamycin reduced p-S6K1 independent of genotype, p < 0.05; +, effect of asparaginase to decrease p-S6K1 depended on GCN2, p < 0.05. C, hepatic 4E-BP1 phosphorylation is reduced by rapamycin independent of genotype. *, p < 0.05. D, 4E-BP1 phosphorylation in pancreas was reduced by asparaginase and rapamycin. **, asparaginase reduced p-4E-BP1 in GCN2+/+ mice, p < 0.05; *, rapamycin reduced p-4E-BP1 independent of genotype, p < 0.05; +, effect of asparaginase to decrease p-4E-BP1 depended on GCN2, p < 0.05, E, deletion of GCN2 blocked induction of hepatic p-Akt/PKB by asparaginase. **, asparaginase increased p-Akt in GCN2+/+ mice, p < 0.05; +, effect of asparaginase to increase p-Akt depended on GCN2, p < 0.05, F, phosphorylation of Akt/PKB in pancreas. ϕ, rapamycin increased p-Akt only when in combination with GCN2 deletion, p < 0.05. Abbreviations: SS, saline then saline; SA, saline then asparaginase; RS, rapamycin then saline; RA, rapamycin then asparaginase.
FIGURE 4.
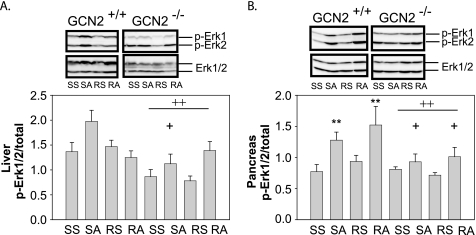
GCN2 is required to increase phosphorylation of ERK1/2 in the liver (A) and pancreas (B) of mice treated with asparaginase. A, ++, loss of GCN2 reduced overall hepatic p-ERK1/2 independent of drug treatments, p < 0.05; **, asparaginase increased p-ERK1/2 in GCN2+/+ mice, p < 0.05; +, effect of asparaginase to increase p-ERK1/2 depended on GCN2, p < 0.05. Abbreviations: SS, saline then saline; SA, saline then asparaginase; RS, rapamycin then saline; RA, rapamycin then asparaginase.
RESULTS
Phosphorylation of eIF2 by Asparaginase Requires GCN2 and Not PERK
Previously, we showed that treatment with asparaginase activates p-eIF2 in liver and spleen but not pancreas (4). In that study, a slight increase in p-eIF2 was detectable in the liver of GCN2−/− mice 1–6 h after asparaginase injection. Considering a recent paper reporting that asparaginase temporarily induces protein aggregation in hepatocytes (23), we further examined the contribution of PERK to p-eIF2 6 h after asparaginase treatment in liver. Mice with a liver-specific knockdown of PERK (produced by AlbCre-mediated deletion of floxed PERK (20); see Fig. 1A) showed robust hepatic p-eIF2 to asparaginase that was similar to AlbCre-negative control mice treated with asparaginase (Fig. 1B). GCN2 null mice were then bred to AlbCre-positive and -negative mice expressing floxed PERK to produce GCN2 null mice with PERK additionally knocked down in liver (Fig. 1A). Mice possessing GCN2-PERK double null livers demonstrated reduced p-eIF2 following asparaginase treatment as compared with saline-injected mice (Fig. 1C). Furthermore, induction of ASNS mRNA expression to asparaginase was similarly precluded in both GCN2 null livers and GCN2-PERK double null livers (compare values in Fig. 1D with SS control group in Fig. 2C). Thus, p-eIF2 and the ISR in response to asparaginase are mediated by GCN2 and not PERK.
Asparaginase but Not Rapamycin Activates the Integrated Stress Response in Liver via GCN2
Previously, we demonstrated asparaginase to activate the ISR in liver, indicated by reduced hepatic protein synthesis alongside induction of p-eIF2, ASNS mRNA, and CHOP protein expression (4). In this study, asparaginase increased p-eIF2 and ASNS mRNA expression in the liver of GCN2+/+ but not GCN2−/− mice (Fig. 2, A and C). Hepatic CHOP mRNA levels were significantly increased in GCN2+/+ mice and slightly less so in GCN2−/− mice following asparaginase (Fig. 2D). Rapamycin prevented hepatic CHOP induction in both strains (Fig. 2D) but had no effect on hepatic p-eIF2 and ASNS mRNA expression. In the pancreas of both GCN2+/+ and GCN2−/− mice, neither asparaginase nor rapamycin altered p-eIF2 and mRNA expression of ASNS (Fig. 2B and supplemental Fig. S1). Asparaginase did not increase expression of ATF3 nor the molecular chaperone BiP/Grp78 and did not induce caspase-3 cleavage in the livers of GCN2+/+ and GCN2−/− mice (supplemental Fig. S1).
Signaling Downstream of mTORC1 Is Derepressed in GCN2−/− Mice Treated with Asparaginase but Not Rapamycin
In the liver and pancreas of GCN2+/+ mice, both rapamycin and asparaginase reduced phosphorylation of S6K1 (p-S6K1) (Fig. 3, A and B), with rapamycin being the more potent of the two agents. On the other hand, whereas rapamycin reduced p-S6K1 in both strains, asparaginase was effective in GCN2+/+ mice only. A similar pattern in 4E-BP1 phosphorylation was seen with respect to asparaginase and rapamycin in GCN2+/+ and GCN2−/− mice, although the effects were much more modest (Fig. 3, C and D). To better understand why GCN2−/− mice demonstrate derepression of mTORC1 signaling following asparaginase treatment, we examined expression of the mTOR inhibitor, REDD1 (regulated in development and DNA damage responses 1). In response to a variety of cell stress conditions, REDD1 is transcriptionally up-regulated, reducing mTORC1 activity. On the other hand, loss of REDD1 protein by its rapid degradation and/or reduced expression leads to increased mTORC1 activity (24). However, neither asparaginase nor rapamycin altered REDD1 protein expression in the livers of GCN2+/+ and GCN2−/− mice (supplemental Fig. S2). In addition, no specific treatment or strain effects were observed in the phosphorylation of PDK1 (Ser241) or eEF-2 (Thr56) (supplemental Fig. S2).
Phosphorylation of Akt/PKB by Asparaginase and Rapamycin Is Modulated by GCN2
In certain cell lines, inactivation of S6K1 by rapamycin can activate the insulin/insulin-like growth factor-I/phosphatidylinositol 3-kinase (PI3K) axis through stabilization of insulin receptor substrate 1, leading to an increase in p-Akt/PKB(Ser473) (25). In the liver of GCN2+/+ mice, asparaginase increased p-Akt (Fig. 3E), but the effect of asparaginase alone was blocked in GCN2 null mice. Considering that phosphorylation of Akt is associated with protection of cells from apoptosis (26), the loss of p-Akt in the liver of GCN2 null mice treated with asparaginase suggests that these mice may be less capable of adapting to asparaginase upon continued asparaginase treatment. Rapamycin increased p-Akt(Ser473) in pancreas, but this effect depended on loss of GCN2 (Fig. 3, E and F).
Increased Phosphorylation of ERK1/2 by Asparaginase Requires GCN2
Amino acid starvation activates the ERK1/2 pathway to stimulate autophagy. A relationship between ERK1/2 signaling and GCN2 is suggested by a previous report showing that inhibition of MEK activation blocks p-eIF2 and ATF4 synthesis triggered by amino acid limitation in hepatocytes (27). In this study, asparaginase increased p-ERK1/2 in liver and pancreas but only in GCN2+/+ mice (Fig. 4, A and B). Rapamycin did not significantly alter asparaginase-induced p-ERK1/2 in either tissue.
DISCUSSION
This study produced several novel and clinically important findings. First, the data show that GCN2 and not PERK is required for activation of p-eIF2 by asparaginase in the liver of mice. Second, GCN2 is necessary not only for induction of hepatic p-eIF2 by asparaginase but also for a variety of AADR, such as down-regulation of mTORC1 and activation of p-Akt and p-ERK1/2. Third, GCN2 is not necessary for increased CHOP mRNA expression in liver by asparaginase, and finally, induction of CHOP by asparaginase is rapamycin-sensitive. These findings advance our understanding of the basic biological mechanism of asparaginase action in tissues in vivo and further reveal the critical role of GCN2 in managing the stress response to this chemotherapeutic agent.
GCN2 Is Required for Activation of Hepatic ISR by Asparaginase
Previously, we reported that asparaginase does not induce phosphorylation of PERK between 15 min and 6 h after a single injection (4). The decision to further explore the role of PERK in mediating p-eIF2 by asparaginase was influenced by a recent paper proposing that asparaginase induces a temporary conformational disease state (23). In that report (23), the authors show that asparaginase impairs secretion of antithrombin and α1-antitrypsin by HepG2 cell and causes significant intracellular retention and accumulation of glycoproteins in mouse secretory tissues. Furthermore, microscopic analysis shows the formation of intracellular aggregates within dilated endoplasmic reticulum (ER) cisterns in liver, pancreas, and brain. These findings imply that asparaginase may be causing ER stress and thus activating PERK as part of the unfolded protein response (UPR), as is reported in the case of α1-antitrypsin deficiency (28). Although it is clear that a single injection of asparaginase induces the ISR, we are unable to observe full activation of the tripartite UPR. Evaluation of caspase-3 cleavage, BiP/Grp78 expression, and XBP-1 splicing revealed no differences between treatment groups following a single injection of asparaginase (supplemental Fig. S1 and data not shown). Nevertheless, in our laboratory, multiple injections of asparaginase over several days results in histological features consistent with the findings of Corral and co-workers (23) suggesting induction of the UPR or an ER overload response.3 Considering that this study is focused on an early point in time, it is possible that intracellular build-up of protein aggregates may not have occurred within the time boundaries of this acute study. A recent study comparing the distinct sets of genes regulated in mouse liver by GCN2 versus PERK proposes that a gradient of p-eIF2 based on the subcellular localization of the eIF2 kinase and the particular stress being sensed results in a different subset of regulated genes being activated (29). Following this line of thinking, we argue that in the case of asparaginase, the primary stress is clearly that of amino acid deprivation, sensed by GCN2. Continued absence of necessary substrate leading to faulty synthesis of nascent peptides may then lead to misfolding of protein and ER stress or overload at some later point in time. As such, further study evaluating a secondary or auxiliary role for PERK or other arms of the UPR during longer term asparaginase treatment is warranted.
Combination of Asparaginase and Rapamycin Maximally Inhibits mTORC1 Signaling
The most well known mTORC1 inhibitor is the immunosuppressant rapamycin (30). Recent studies testing anticancer properties of mTOR inhibitors report that rapamycin and its structural analogs can sensitize cancer cells to other chemotherapy agents (31, 32). Understanding this and knowing that both asparaginase and rapamycin inhibit downstream signaling to S6K1 and 4E-BP1, it was of interest to understand how the combination would affect mTORC1 signaling. Based on studies reporting that the stress response to amino acid starvation only partially overlaps with rapamycin (14, 15), we hypothesized that the combination of asparaginase and rapamycin would maximally inhibit p-S6K1 and p-4E-BP1. Overall, the current results are consistent with this hypothesis and suggest that additional testing of rapamycin in combination with asparaginase on cancer cell lines to improve tumor killing is worthy of further consideration, particularly because the combination of asparaginase and rapamycin was not acutely toxic in the whole animal.
GCN2 Is Necessary for Down-regulation of mTORC1 by Asparaginase but Not Rapamycin
In this study, loss of GCN2 precluded dephosphorylation of S6K1 by asparaginase but not rapamycin. This agrees with our previous findings in which derepression of mTORC1 signaling is reported in GCN2 null livers during dietary leucine starvation (16) and indicates that the relationship between GCN2 and mTORC1 is specific to conditions that are sensed by GCN2 (e.g. amino acid limitation). Although a full explanation for this effect remains elusive, the data suggest that the REDD1 repressor is not the primary regulating factor because REDD1 protein expression was similar across treatment groups in both strains. This finding conflicts with recent data demonstrating that REDD1 expression is up-regulated in several cell lines during ER stress through a mechanism involving activation of the PERK/p-eIF2/ATF4 pathway (33). In this study, activation of the ISR by asparaginase did not induce REDD1 in normal mouse liver nor did the loss of GCN2 prevent an increase in REDD1. There are no data demonstrating REDD1 to be specifically up-regulated by the GCN2/p-eIF2 pathway, and so perhaps REDD1 expression is not responsive to amino acid starvation. Alternatively, intrinsic differences between cell lines and tissues from intact animals are plausible to consider. Finally, considering that the half-life of the REDD1 mRNA is quite short (≤5 min) (24), future time course analysis may help clarify the contribution of REDD1 to mTORC1 signaling following asparaginase.
Induction of CHOP by Asparaginase Is GCN2-independent and Rapamycin-sensitive
Over the past several years, an increasing number of investigations have described CHOP as a specific marker of the ISR and even the UPR. This pattern continues despite the fact that previous data suggest that multiple signaling pathways lead to the expression of CHOP mRNA under conditions of nutrient deprivation and DNA damage (34). In the case of amino acid deprivation, induction of CHOP is inhibited by rapamycin and by the PI3K inhibitor LY294002 in cells in culture (17). In accordance with this, these findings suggest that asparaginase incorporates the PI3K/mTOR pathway to induce CHOP. Furthermore, we demonstrate for the first time that CHOP requires both eIF2 phosphorylation and mTORC1 signaling for maximal induction by asparaginase. The current data highlight the role of the PI3K/mTOR pathway in regulating CHOP and point out potential risk in using CHOP as a unique marker of the UPR because increased expression of CHOP can occur in the absence of increased p-eIF2 as reported previously (4).
Activation of ERK1/2 Pathway by Asparaginase Requires GCN2
The MAPK are a group of serine/threonine kinases activated in response to a variety of extracellular stimuli. The three major MAPK pathways in mammalian cells are the p38 MAPK, the JNK, and the MEK/ERK. Amino acid depletion has been shown to activate the MEK/ERK pathway, which subsequently serves to stimulate autophagy. Thiaville et al. (27) reports interdependence between activation of the MEK/ERK pathway and the ISR during nutrient stress. Following amino acid deprivation in HepG2 cells, ERK1/2 phosphorylation increases. Treatment with the MEK inhibitor PD98059 completely blocks induction of p-eIF2 and production of ASNS mRNA by histidine deprivation, whereas inhibitors to the p38 MAPK and JNK are without effect. In addition, ERK1/2 phosphorylation did not occur in GCN2 null mouse embryonic fibroblasts deprived of histidine. These data support and extend these findings to the in vivo condition by demonstrating that activation of ERK1/2 by asparaginase is dependent on GCN2 in liver and pancreas. It is worth noting that activation of the MEK/ERK pathway and down-regulation of mTORC1 both promote the induction of autophagy (35). Very little information about autophagy and asparaginase is available, and very little is known about the role of mammalian GCN2 in regulating autophagy, although in yeast the role of GCN2 in regulating autophagy is well documented (36). As such, it is of future interest to understand if asparaginase activates autophagy and, if so, how the absence of GCN2 would modulate this process.
In summary, GCN2 is required for induction of p-eIF2 and the ISR alongside repression of mTOR signaling in response to asparaginase. The mechanism by which asparaginase inhibits mTORC1 differs from that of rapamycin, resulting in maximal inhibition by the combination. Rapamycin precludes induction of CHOP and p-ERK1/2 by asparaginase, suggesting important cross-talk exists between the GCN2/p-eIF2, MEK/ERK, and PI3K/mTOR pathways. Further study comparing AADR to asparaginase in GCN2−/− tissues versus GCN2−/− tumors will lead to novel and improved chemotherapeutic agents and approaches.
Supplementary Material
Acknowledgment
We acknowledge the technical assistance of James Elliott (Applied Biosystems).
This work was supported, in whole or in part, by National Institutes of Health Grant GM49164 (to R. C. W.), the American Institute for Cancer Research (to T. G. A.), a research enhancement grant from the Indiana University School of Medicine (to T. G. A.), and an American Cancer Society institutional grant (to T. G. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
P. Bunpo, J. Cundiff, and T. Anthony, unpublished observations.
- eIF2
- eukaryotic initiation factor 2
- p-eIF2
- phosphorylation of eukaryotic initiation factor 2
- mTORC1
- mammalian target of rapamycin complex 1
- GCN2
- general control nonderepressible 2
- PERK
- protein kinase R-like endoplasmic reticulum resident kinase
- S6K1
- 70-kDa ribosomal protein S6 kinase
- 4E-BP1
- eukaryotic initiation factor 4E-binding protein 1
- CHOP/GADD153
- CAAT/enhancer-binding protein homologous protein/growth arrest DNA damage-inducible gene 153
- ISR
- integrated stress response
- UPR
- unfolded protein response
- ATF
- activating transcription factor
- ASNS
- asparagine synthetase
- MAPK
- mitogen-activated protein kinases
- MEK/ERK
- MAPK kinase/extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinases
- PI3K
- phosphatidylinositol 3-kinase
- Akt/PKB
- protein kinase B
- REDD1
- regulated in development and DNA damage responses 1
- ANOVA
- analysis of variance
- ISR
- integrated stress response
- PBS
- phosphate-buffered saline
- mTOR
- mammalian target of rapamycin
- ER
- endoplasmic reticulum
- AADR
- amino acid deprivation responses.
REFERENCES
- 1.Hill J. M., Roberts J., Loeb E., Khan A., MacLellan A., Hill R. W. (1967) JAMA 202, 882–888 [PubMed] [Google Scholar]
- 2.Su N., Pan Y. X., Zhou M., Harvey R. C., Hunger S. P., Kilberg M. S. (2008) Pediatr. Blood Cancer 50, 274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holcenberg J. (2004) J. Pediatr. Hematol. Oncol. 26, 273–274 [DOI] [PubMed] [Google Scholar]
- 4.Reinert R. B., Oberle L. M., Wek S. A., Bunpo P., Wang X. P., Mileva I., Goodwin L. O., Aldrich C. J., Durden D. L., McNurlan M. A., Wek R. C., Anthony T. G. (2006) J. Biol. Chem. 281, 31222–31233 [DOI] [PubMed] [Google Scholar]
- 5.Bunpo P., Murray B., Cundiff J., Brizius E., Aldrich C. J., Anthony T. G. (2008) J. Nutr. 138, 338–343 [DOI] [PubMed] [Google Scholar]
- 6.Ollenschläger G., Roth E., Linkesch W., Jansen S., Simmel A., Mödder B. (1988) Eur. J. Clin. Invest. 18, 512–516 [DOI] [PubMed] [Google Scholar]
- 7.Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 8.Kimball S., Anthony T., Cavener D., Jefferson L. (2004) in Topics in Current Genetics: Nutrient-induced Responses in Eukaryotic Cells (Winderickx P., Taylor J. eds) pp. 113– 130, Springer-Verlag, Berlin [Google Scholar]
- 9.Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 10.Sood R., Porter A. C., Ma K., Quilliam L. A., Wek R. C. (2000) Biochem. J. 346, 281–293 [PMC free article] [PubMed] [Google Scholar]
- 11.Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 12.Shah O. J., Anthony J. C., Kimball S. R., Jefferson L. S. (2000) Am. J. Physiol. Endocrinol. Metab. 279, E715–E729 [DOI] [PubMed] [Google Scholar]
- 13.Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. (2004) Mol. Cell. Biol. 24, 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng T., Golub T. R., Sabatini D. M. (2002) Mol. Cell. Biol. 22, 5575–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deval C., Chaveroux C., Maurin A. C., Cherasse Y., Parry L., Carraro V., Milenkovic D., Ferrara M., Bruhat A., Jousse C., Fafournoux P. (2009) FEBS J. 276, 707–718 [DOI] [PubMed] [Google Scholar]
- 16.Anthony T. G., McDaniel B. J., Byerley R. L., McGrath B. C., Cavener D. R., McNurlan M. A., Wek R. C. (2004) J. Biol. Chem. 279, 36553–36561 [DOI] [PubMed] [Google Scholar]
- 17.Entingh A. J., Law B. K., Moses H. L. (2001) Endocrinology 142, 221–228 [DOI] [PubMed] [Google Scholar]
- 18.Reeves P. G., Nielsen F. H., Fahey G. C., Jr. (1993) J. Nutr. 123, 1939–1951 [DOI] [PubMed] [Google Scholar]
- 19.Distasio J. A., Niederman R. A., Kafkewitz D., Goodman D. (1976) J. Biol. Chem. 251, 6929–6933 [PubMed] [Google Scholar]
- 20.Bobrovnikova-Marjon E., Hatzivassiliou G., Grigoriadou C., Romero M., Cavener D. R., Thompson C. B., Diehl J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16314–16319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P., McGrath B. C., Reinert J., Olsen D. S., Lei L., Gill S., Wek S. A., Vattem K. M., Wek R. C., Kimball S. R., Jefferson L. S., Cavener D. R. (2002) Mol. Cell. Biol. 22, 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anthony T. G., Reiter A. K., Anthony J. C., Kimball S. R., Jefferson L. S. (2001) Am. J. Physiol. Endocrinol. Metab. 281, E430–E439 [DOI] [PubMed] [Google Scholar]
- 23.Hernández-Espinosa D., Miñano A., Martínez C., Pérez-Ceballos E., Heras I., Fuster J. L., Vicente V., Corral J. (2006) Am. J. Pathol. 169, 142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimball S. R., Do A. N., Kutzler L., Cavener D. R., Jefferson L. S. (2008) J. Biol. Chem. 283, 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han E. K., Leverson J. D., McGonigal T., Shah O. J., Woods K. W., Hunter T., Giranda V. L., Luo Y. (2007) Oncogene 26, 5655–5661 [DOI] [PubMed] [Google Scholar]
- 26.Song G., Ouyang G., Bao S. (2005) J. Cell. Mol. Med. 9, 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiaville M. M., Pan Y. X., Gjymishka A., Zhong C., Kaufman R. J., Kilberg M. S. (2008) J. Biol. Chem. 283, 10848–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawless M. W., Greene C. M., Mulgrew A., Taggart C. C., O'Neill S. J., McElvaney N. G. (2004) J. Immunol. 172, 5722–5726 [DOI] [PubMed] [Google Scholar]
- 29.Dang Do A. N., Kimball S. R., Cavener D. R., Jefferson L. S. (2009) Physiol. Genomics 38, 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., Avruch J. (1998) J. Biol. Chem. 273, 14484–14494 [DOI] [PubMed] [Google Scholar]
- 31.Smolewski P. (2006) Expert Opin. Investig. Drugs 15, 1201–1227 [DOI] [PubMed] [Google Scholar]
- 32.Janus A., Linke A., Cebula B., Robak T., Smolewski P. (2009) Anticancer Drugs 20, 693–701 [DOI] [PubMed] [Google Scholar]
- 33.Whitney M. L., Jefferson L. S., Kimball S. R. (2009) Biochem. Biophys. Res. Commun. 379, 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q., Lau S. S., Monks T. J. (1999) Biochem. J. 341, 225–231 [PMC free article] [PubMed] [Google Scholar]
- 35.Aoki H., Takada Y., Kondo S., Sawaya R., Aggarwal B. B., Kondo Y. (2007) Mol. Pharmacol. 72, 29–39 [DOI] [PubMed] [Google Scholar]
- 36.Tallóczy Z., Jiang W., Virgin H. W., 4th, Leib D. A., Scheuner D., Kaufman R. J., Eskelinen E. L., Levine B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.