Abstract
Antibody affinity limits sensitivity of detection in many areas of biology and medicine. High affinity usually depends on achieving the optimal combination of the natural 20 amino acids in the antibody binding site. Here, we investigate the effect on recognition of protein targets of placing an unnatural electrophile adjacent to the target binding site. We positioned a weak electrophile, acrylamide, near the binding site between an affibody, a non-immunoglobulin binding scaffold, and its protein target. The proximity between cysteine, lysine, or histidine on the target protein drove covalent bond formation to the electrophile on the affibody. Covalent bonds did not form to a non-interacting point mutant of the target, and there was minimal cross-reactivity with serum, cell lysate, or when imaging at the cell surface. Electrophilic affibodies showed more stable protein imaging at the surface of mammalian cells, and the sensitivity of protein detection in an immunoassay improved by two orders of magnitude. Thus electrophilic affibodies combined good specificity with improved detection of protein targets.
Introduction
Human blood contains a panoply of proteins below the detection limit of standard immunoassays, including early markers of cancer or neurodegeneration (1, 2). Immunoassays have advanced enormously over the last 60 years, opening up whole new fields of research from the ability to detect lower and lower abundance species (3). Nearly all enzyme-linked immunosorbent assays (ELISAs)2 stop working at low picomolar concentration of target (4), but recently, methods such as nanowires (5) or the bio-barcode assay (6) have made leaps in detection sensitivity. These advances mean that amplification is not the limitation on immunoassay sensitivity any longer; it is the antibody itself that is the limiting factor (4). Antibody binding strength correlates with protection from disease agents (7, 8) and can be a limiting factor in live cell imaging, since antibody off-rates are on the order of 30 min (9–11) but lifetimes of cell surface proteins are often ∼1 day.
Affinity-matured antibodies generally have micromolar to nanomolar monovalent affinity for their target (9). It has proved very difficult to generate antibodies of subnanomolar monovalent affinity. This may be because of a limit to the affinity of antibodies that can be selected in an immunized animal, as a result of the kinetics of B cell stimulation (11). In vitro selection methods based on phage display (12), mRNA/ribosome display (13), or yeast cell surface display (14) have sometimes managed to obtain femtomolar affinity antibodies. There are a few examples of antibodies that form covalent bonds to artificial small molecule antigens (15–17), but these strategies, such as the antibody made by Meares' group to an electrophilic metal chelator, do not enable recognition of protein antigens composed of the regular 20 amino acids. Immunizing mice with electrophilic antigens generated an antibody that formed SDS-stable complexes with its target, but the chemistry of the interaction is unknown, and this antibody has not yet shown its applicability to imaging or to improving detection sensitivity (18). Recently, an antibody was engineered to contain a metal binding site and formed an extremely stable complex with an endogenous protein target. However, complex formation only proceeded to 10% completion, while the specificity of the reaction in complex biological systems has not been demonstrated (19).
Here, instead of an antibody, we employ an affibody, a robust low molecular weight non-immunoglobulin scaffold. Affibodies are based on a three-helix bundle of 58 amino acids, where 13 surface residues can be mutated and selected for binding, which can easily be expressed in the cytosol of bacteria (20). Proximity has been shown to increase reaction rate more than a millionfold, such that small molecules that bear weak electrophiles, which do not react with the many other nucleophiles at millimolar concentrations, can react rapidly with apposed nucleophiles on interacting biomolecules (17, 21–25). The strategy we explore here is to introduce a weak electrophile onto the affibody, adjacent to the target binding site, such that proximity to nucleophilic amino acid side chains (e.g. Cys, Lys, and His) present on the target could drive specific covalent bond formation (see Fig. 1A).
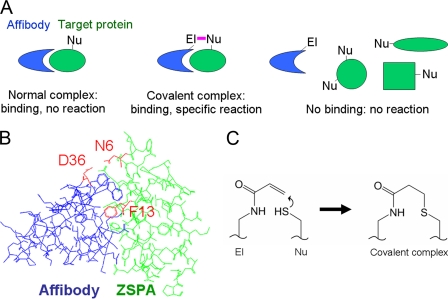
FIGURE 1.
Electrophilic affibody strategy. A, principle of electrophilic affibody reactivity. We start with a pre-existing affibody (blue) that binds a protein target (green). We then introduce a weak electrophile (El) at the side of the target binding site, so that the electrophile is aligned with a nucleophilic residue (Nu) on the target protein (e.g. Lys, His, or Cys). This alignment drives covalent bond formation. If the electrophile is in the wrong position in the complex or if the protein does not bind the affibody, reaction should not occur. B, model affibody-target interaction. The crystal structure of the complex between affibody (blue) and ZSPA (green) is shown (from Protein Data Bank code 1LP1). We positioned the electrophile at Asp-36 on the affibody by reacting with EBA and tested alternative nucleophiles at Asn-6 on ZSPA. Phe-13 of ZSPA was mutated to Glu to disrupt the binding interface. C, reaction between acrylamide electrophile on affibody and a nucleophile, in this case cysteine, on the target protein.
EXPERIMENTAL PROCEDURES
Cloning
The Z domain of Staphylococcus aureus Protein A (ZSPA) was obtained by PCR from pBS1479, a kind gift of K. Nasmyth (Oxford University), using the primers 5′-CATTGGATCCGGGCGTGGACAACAAATTCAAC and 5′-GATCCTCGAGGCCTTTCGGCGCCTGAGCATC and was inserted at the BamHI and XhoI sites of pET21b (Novagen), to give pET21b-ZSPA. To generate MBP-ZSPA, we amplified maltose binding protein (MBP) from pMAL (New England Biolabs) using the primers 5′-CAAGCATATGAAAATCGAAGAAG and 5′-CGAACCCGAGCTCGAATTAGTCTG and then amplified ZSPA from pET21b-ZSPA using the primers 5′-CTAATTCGAGCTCGGGTTCGATGGTGGACAACAAATTC and 5′-CAAGCTCGAGGCCTTTCGGCGC. The two fragments were joined by overlap extension PCR, and MBP-ZSPA was cloned into the NdeI and XhoI sites of pET21b.
Affibody D36C was generated from pET21b-ZSPA by inverse PCR (26), using the primers 5′-AGGTTCGGCAGGGTAACGATTTCACGACCAGCAACAGACAGTTCTTTGTTGAATTTGTTG and 5′-GAACGACCCGCAGAAAAAAGCTTTCATCTTCTCTCTGTGGGATGACCCAAGCCAAAGCGC to give affibody ZSPA-1 (27), followed by addition of the acceptor peptide, a short target for BirA biotinylation (28), to the N terminus by inverse PCR using the primers 5′-TATCGTTCAGGCCACCCATTTGCTGTCCACC and 5′-TCTTCGAGGCCCAGAAGATCGAGTGGCACGAGGGCCGGGATCCGGGCGTGGAC, and introduction of the D36C mutation using the primer 5′-CATCTTCTCTCTGTGGTGTGACCCAAGCCAAAGCG and its reverse complement.
Site-directed mutants were made using the QuikChangeTM (Stratagene) protocol but with KOD Hot Start DNA polymerase (Merck). We verified all constructs and mutations by sequencing.
We constructed ZSPA-TM, for cell surface expression of ZSPA, by PCR from pET21b-ZSPA using the primers 5′-CAGTGGATCCGGAATGGTGGACAACAAATTC and 5′-CTGAGTCGACTCCCTCGAGGCCTTTCGGCGC, digesting the PCR product with BamHI and SalI and ligating into the BglII and SalI sites of pDisplay (Invitrogen). Point mutations in ZSPA, MBP-ZSPA, and ZSPA-TM were made using the following primers and their reverse complements: ZSPA N6C, 5′- GGCGTGGACAACAAATTCTGCAAAGAACAACAAAACGCG; ZSPA-TM N6C and MBP-ZSPA N6C, 5′-GATGGTGGACAACAAATTCTGCAAAGAACAACAAAACGC; MBP-ZSPA N6D, 5′-GATGGTGGACAACAAATTCGACAAAGAACAACAAAACGC; MBP-ZSPA N6E, 5′-GATGGTGGACAACAAATTCGAGAAAGAACAACAAAACGCGTTC; MBP-ZSPA N6H, 5′-GATGGTGGACAACAAATTCCACAAAGAACAACAAAACGC; MBP-ZSPA N6K, 5′-GATGGTGGACAACAAATTCAAGAAAGAACAACAAAACGCG; ZSPA E24C,E25Q (referred to as CQ), 5′-CCTAACTTAAACTGCCAACAACGAAACGCCTTCATC; F13E into MBP-ZSPA variants, ZSPA CQ, and ZSPA-TM N6C, 5′-CAACAAAGAACAACAAAACGCGGAGTATGAGATCTTACATTTACCTAACTTAAACG. The nuclear co-transfection marker pECFP-H2B (human histone H2B fused to enhanced cyan fluorescent protein) has been described previously (29).
Protein Expression
All proteins were expressed using E. coli BL21 DE3 RIPL cells (Stratagene), grown in LB with 0.8% glucose and 0.1 mg/ml ampicillin. We diluted overnight cultures 100-fold, grew at 37 °C to A600 0.5, and induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 30 °C. Proteins were purified by nickel affinity chromatography, using standard methods, and dialyzed into phosphate-buffered saline (PBS). Protein concentration was determined from A280, using the extinction coefficient predicted by ExPASy ProtParam.
Protein Derivatization
Affibody D36C, ZSPA CQ, or ZSPA CQ F13E was reduced with 5 mm Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, dissolved in PBS and adjusted to pH 8.0, from Pierce) for 2 h at 37 °C, before conjugating with either 60 mm N,N′-ethylene bisacrylamide (EBA, 0.6 m stock dissolved in H2O and 5% DMF, Aldrich) for a further 2 h at 37 °C, or 10 mm iodoacetamide (Alfa Aesar) for 30 min at 37 °C. We removed free EBA, iodoacetamide, and TCEP by dialysis in PBS, before storing at 4 °C and using the protein conjugates within 2 weeks. We biotinylated affibody D36C (typically at 200 μm) in PBS with 5 mm MgCl2, 10 μm BirA, 1 mm ATP, and 300 μm biotin for 1 h at 25 °C and then dialyzed in PBS.
Tetravalent His6-tagged core streptavidin was purified as described (30) and fluorescence-labeled with Alexa Fluor 555 succinimidyl ester (Invitrogen) according to manufacturer's instructions, using a 10-fold molar excess of dye. Unreacted dye was removed by gel filtration and dialysis.
We assessed the fraction of free cysteine by mixing affibody D36C unmodified, modified with iodoacetamide, or modified with EBA in PBS, pH 8.0, with an equal volume of 32 mm CuCl2 and incubating for 2 h at 37 °C, so that free cysteines formed disulfide bonds (31). Samples were heated at 95 °C for 7 min in non-reducing SDS loading buffer on a Bio-Rad C1000 thermal cycler before SDS-PAGE.
Protein Cross-linking
75 μm affibody-EBA was incubated in PBS at the indicated pH with 19 μm each MBP-ZSPA mutant at 37 °C. Time courses were stopped by placing at −80 °C. We adjusted buffer pH to 8.3 or 8.8 using 0.6 m bicine, pH 8.5 or pH 9.0, respectively (Fig. 2C). Cysteine-containing proteins were reduced with TCEP prior to cross-linking. Reduced l-glutathione (Alfa Aesar) was dissolved in PBS, pH 8.0, and added at the indicated concentrations to 37.5 μm affibody-EBA with 19 μm MBP-ZSPA N6C in PBS, pH 8.0, for 5 h at 37 °C. 75 μm ZSPA CQ, ZSPA CQ-EBA, or ZSPA CQ F13E-EBA was incubated with 0.5 mg/ml mouse monoclonal anti-green fluorescent protein IgG2a antibody JL-8 (Clontech) in PBS, pH 8.7, for 5 h at 37 °C, before SDS-PAGE.
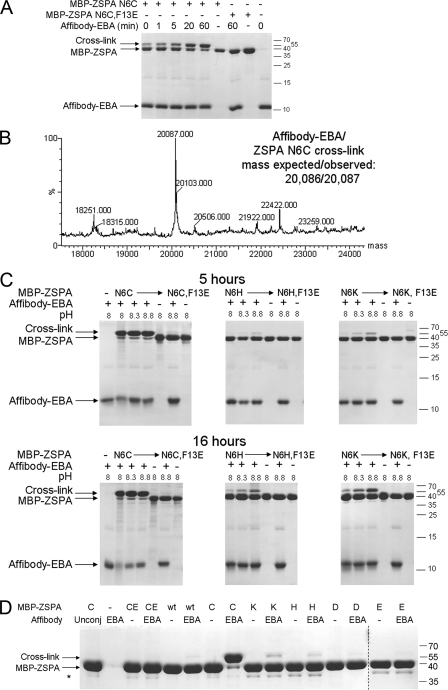
FIGURE 2.
Electrophilic affibody covalent bond formation. A, speed of covalent bond formation to N6C. We incubated affibody-EBA with MBP-ZSPA N6C for the indicated times at 37 °C at pH 8.0 and analyzed cross-linking by SDS-PAGE. A point mutant of MBP-ZSPA that disrupted the initial non-covalent interaction (N6C F13E) was a negative control. Bands corresponding to affibody-EBA, MBP-ZSPA, and cross-links are indicated. B, mass spectrometry to verify cross-link between affibody-EBA and ZSPA N6C. C, pH-dependence of reactivity. We incubated affibody-EBA with MBP-ZSPA N6C, N6H, or N6K for 5 h (top row) or 16 h (bottom row) at pH 8.0, 8.3, or 8.8 and determined cross-linking by SDS-PAGE. Negative controls are shown without affibody-EBA, without any MBP-ZSPA, or with non-interacting F13E variants. D, reactivity of all potential nucleophilic side chains with affibody-EBA. MBP-ZSPA wild-type (wt) or with various mutations at position Asn-6 was incubated with or without affibody-EBA at pH 8.3 for 4 h at 37 °C and analyzed by SDS-PAGE and Coomassie staining. Lane 1 is a control where affibody D36C is unmodified with EBA (Unconj). CE is the N6C F13E non-interacting negative control. An asterisk relates to small amounts of a proteolytic fragment from cleavage of the linker between MBP and ZSPA. The dotted line indicates samples run on a different gel at the same time.
SDS-PAGE
SDS-PAGE was performed on 8, 10, or 18% polyacrylamide gels, using an XCell SureLock (Invitrogen) at 200 V on ice with precooled running buffer. Samples were heated at 95 °C for 7 min in SDS loading buffer with 10 mm dithiothreitol before loading. Band intensities were quantified using a ChemiDoc XRS imager and QuantityOne (version 4.6) software (Bio-Rad).
Immunoblotting
10% fetal calf serum (PAA Laboratories) was incubated with 5 μm affibody-EBA and 1 μm MBP-ZSPA N6K in PBS, pH 8.0, for 5 h at 37 °C. To test affibody-EBA reactivity with cell lysate, HeLa cells were trypsinized and washed once in growth medium and once in PBS, and the pellet was lysed in PBS, pH 8.0, with 5 mm EDTA, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (complete, mini, EDTA-free; Merck Biosciences). Post-nuclear supernatant was diluted 4-fold (60,000 cell equivalents per lane) and incubated with 1 μm MBP-ZSPA N6K and 1 μm affibody-EBA at pH 8.0 for 5 h at 37 °C. We blotted with antibody to the T7 tag (Novagen) at 1 μg/ml in PBS/2.5% skimmed milk for 1 h. The membrane was incubated with 1:1,000 anti-mouse IgG-horseradish peroxidase (Sigma) in PBST (PBS with 0.05% Tween 20) for 40 min, washed four times for 5 min in PBST, and developed using SuperSignal West Pico (Pierce).
Mass Spectrometry
We performed mass spectrometry with ZSPA variants, rather than MBP-ZSPA variants, so that the lower molecular weight would allow a more precise measurement. After reduction with TCEP, in PBS, pH 8.0, we incubated 234 μm affibody-EBA or 200 μm affibody-iodoacetamide with 195 μm ZSPA N6C for 1.5 h at 37 °C. Proteins were dialyzed into MilliQ water using drop dialysis on V series membranes (mixed cellulose esters membrane, pore size 0.025 μm, Millipore) and then dithiothreitol was added to 5 mm. Liquid chromatography-electrospray ionization-mass spectrometry, was performed by the Oxford Protein Production Facility using an Ultima HPLC (Dionex) with a reverse-phase C4 precolumn (Anachem) connected to a quadrupole time-of-flight micromass spectrometer (Waters). The m/z spectrum was converted to a molecular mass profile using maximum entropy processing. Predicted masses were calculated by ExPASy ProtParam.
ELISA
Proteins were coated at the indicated concentrations in 100 μl PBS with 10 mm dithiothreitol on Costar 9018 high capacity binding EIA/RIA polystyrene plates overnight at 4 °C. Plates were washed 3× with PBST containing 0.1% 2-mercaptoethanol (PBSTM) and blocked with 300 μl PBS with 2.5% bovine serum albumin (BSA) for 48–72 h at 4 °C. Plates were further washed twice with PBSTM and twice with PBST and then 50 μl 2 μm affibody D36C, or affibody-EBA in PBS with 1% BSA and 10 mm TCEP at pH 8.0 was added. After 1 h at 25 °C on a rocker, plates were washed with PBSTM for 4 × 8 min and once quickly with PBST, and 200 μl 1 μg/ml ImmunoPure streptavidin horseradish peroxidase (Pierce) diluted in PBS with 1% BSA was added for 1.5 h at 25 °C on a rocker. We washed 3× with PBST for 5 min each time and then developed with 200 μl 0.4 mg/ml o-phenylene diamine in phosphate-citrate buffer (103 mm dibasic sodium phosphate with 48.6 mm citric acid, pH 5.0), measuring A450 using a Spectramax M5 plate reader (Molecular Devices, Ltd.).
For Fig. 3D, after plating and blocking as above, samples were washed with PBSTM 2× and then with PBST 2× for 5 min at 25 °C. 1:100 goat anti-mouse IgG-horseradish peroxidase (Sigma) in PBS adjusted with bicine to pH 8.8 was added for 5 h at 25 °C. The plate was then washed 4× with PBST for 5 min each time and developed as above.
FIGURE 3.
Reaction of electrophilic ZSPA with immunoglobulin. A, design of electrophilic ZSPA. Guided by the Protein Data Bank structure 1FC2, ZSPA E24 (marked in red) was mutated to Cys, so that EBA attached here should be in proximity with the conserved lysine 317 (marked in red) on the antibody heavy chain. To block the non-covalent interaction, as a negative control, Phe-13 (marked in orange) was mutated to Glu. B, conservation of the target lysine (marked in red) in different antibody classes from various species. C, electrophilic ZSPA reaction with antibody. ZSPA CQ-EBA was incubated with antibody (Ab) and analyzed by SDS-PAGE and Coomassie staining. Negative controls were with unmodified ZSPA CQ (Unconj) or ZSPA CQ F13E. D, F13E mutation in ZSPA blocks binding to antibody. Plates were coated with ZSPA CQ or ZSPA CQ F13E and probed by ELISA with goat anti-mouse IgG-horseradish peroxidase. Means of triplicate measurements are shown ± 1 S.D.
Cell Culture, Labeling, and Microscopy
COS7 and HeLa cells were grown in RPMI with 10% fetal calf serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cells were transfected using 1 μl TurboFect (Fermentas), 0.5 μg of ZSPA- TM N6C or ZSPA-TM N6C F13E and 50 ng co-transfection marker H2B-ECFP per well of a 48-well plate, according to manufacturer's instructions. For imaging, 24 h after transfection, we washed cells twice in PBS with 5 mm MgCl2 (PBS/Mg), incubated with 3 μm biotinylated affibody D36C with or without EBA in PBS/Mg with 1% BSA and 10 mm TCEP for 15 min at 37 °C, washed twice in PBS/Mg and incubated for 30 min at 37 °C in PBS 10 mm TCEP. TCEP prevented the possibility of disulfide bond formation between affibody D36C and N6C. After an additional wash in PBS/Mg, we stained cells with 0.1 μm streptavidin-Alexa Fluor 555 in PBS/Mg with 1% BSA for 10 min at 4 °C, washed twice in ice-cold PBS/Mg, and imaged live. Cells were imaged using a wide field DeltaVision Core fluorescent microscope (Applied Precision) with a 40× oil immersion lens. ECFP (436DF20 excitation, 480DF40 emission, Chroma 86002 version 1 dichroic), Alexa Fluor 555 (540D420 excitation, 600DF50 emission, Chroma 84100bs polychroic), and bright field images were collected and analyzed using softWoRx version 3.6.2 software. Typical exposure times were 0.1–1.0 s, and fluorescence images were background corrected. Different samples in the same experiment were prepared, imaged, and analyzed under identical conditions.
RESULTS
Electrophilic Affibodies Form Covalent Bonds to Protein Targets
The affibody used in this study forms a non-covalent complex with its target, ZSPA (32). Guided by the structures of this interaction (27, 33), a cysteine was introduced near the target binding site (affibody D36C, Fig. 1B), allowing attachment of an acrylamide electrophile at a unique site on the affibody by labeling the cysteine with EBA. We verified derivatization of D36C, since cysteine modification prevented copper-induced disulfide formation (supplemental Fig. 1), and EBA modification of the affibody was confirmed by mass spectrometry (supplemental Fig. 2A). We hypothesized that a nucleophilic amino acid present at position 6 on ZSPA would be well placed to undergo Michael addition with the acrylamide on affibody D36C in the protein complex (7.5 Å between β-carbons of Asp-36 and Asn-6, while EBA spans 11 Å) (Fig. 1C). ZSPA and its mutants were fused to MBP to clearly distinguish affibody-ZSPA heterodimers from any homodimers. Cysteine is the most nucleophilic side chain, and so N6C was tested first. We incubated MBP-ZSPA N6C with affibody-EBA and saw rapid and high yielding covalent bond formation by SDS-PAGE (Fig. 2A). Even for the 0 min sample, where the proteins were mixed and immediately placed at −80 °C, there was some cross-linking. The identity of this cross-link was confirmed by mass spectrometry (Fig. 2B). To show that covalent bond formation depended on specific interaction between affibody-EBA and the ZSPA mutant, we introduced the F13E mutation into MBP-ZSPA N6C (Fig. 1B). By ELISA, F13E abolished the interaction of MBP-ZSPA N6C with affibody D36C (see Fig. 5). MBP-ZSPA N6C F13E did not form a covalent bond to affibody-EBA (Fig. 2A). Also, a negative control for the mass spectrometry, where affibody D36C was labeled with iodoacetamide instead of EBA, did not form a covalent bond to ZSPA N6C (supplemental Fig. 2B).
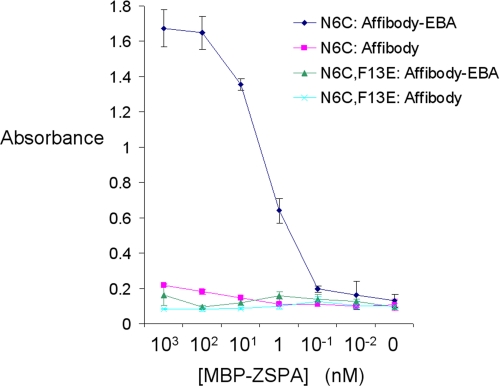
FIGURE 5.
Electrophilic affibody improves target detection sensitivity. We coated plates with varying concentrations of MBP-ZSPA N6C or N6C F13E and probed by ELISA using affibody D36C with or without EBA. Results of triplicate measurements are shown ± 1 S.D. Some error bars are too small to be visible.
Because cysteine is a rare surface amino acid, we were pleased to find that lysine or histidine also allowed covalent bond formation (Fig. 2C). As anticipated from known patterns of acrylamide reactivity (34), reaction with lysine and histidine was less efficient than with cysteine, requiring 5 h for appreciable bond formation, although the extent of reaction was increased at slightly higher pH (Fig. 2C). Aspartate or glutamate at N-6 showed little reactivity (Fig. 2D), consistent with their low nucleophilicity (34). A trace of reactivity was seen for wild-type MBP-ZSPA (Fig. 2D): this most likely relates to attack by Lys-4 or Lys-7, which are more distant from EBA but protein flexibility may allow slow reaction (27, 33). Unconjugated affibody D36C did not form a covalent bond to MBP-ZSPA N6C (Fig. 2D, lane 1). Thus, covalent bond formation depended on initial non-covalent binding, as well as the electrophile EBA.
Covalent Complex Formation with an Endogenous Protein
We established the generality of our proximity-directed electrophile ligation strategy by designing an alternative covalent protein-protein interaction, to an endogenous unmutated protein. Protein A is a central tool in antibody analysis and purification (35). Protein A can bind to antibodies at both Fc and Fab domains, but ZSPA only binds to the Fc domain (35). We generated a variant of ZSPA that could form a covalent bond to an unmodified antibody. We expressed an E24C E25Q (“CQ”) mutant of ZSPA, so that EBA attached at position 24 would be in proximity with a surface lysine on the antibody heavy chain (36) (Fig. 3A). This lysine is conserved in multiple immunoglobulin classes (Fig. 3B). Since Glu-25 on ZSPA could potentially also react with the EBA, Glu-25 was changed to Gln, to give ZSPA CQ (E24C and E25Q). Electrophilic ZSPA formed a covalent bond to the IgG2a antibody heavy chain, as determined by SDS-PAGE after boiling with SDS (Fig. 3C). Without EBA no cross-link formed. We verified the specificity of the ZSPA-antibody cross-link by introducing an F13E mutation (Fig. 3A) in ZSPA CQ. F13E disrupted the non-covalent interaction between ZSPA and antibody, based on ELISA (Fig. 3D) and also abolished the covalent interaction with the antibody (Fig. 3C).
Electrophilic Affibodies Label Their Target Specifically
A key issue for electrophilic affibodies is whether covalent bond formation is accompanied by a loss of specificity. First, we assessed covalent complex formation in the presence of high concentrations of competing nucleophile, namely reduced glutathione, which is present at ∼0.5 mm in serum (37) and contains a free thiol that could potentially react with EBA. In the absence of glutathione, cross-linking between affibody-EBA and MBP-ZSPA N6C occurred with 89% yield (Fig. 4A). Glutathione up to 2.5 mm did not prevent covalent bond formation (Fig. 4A), corroborating that the electrophile only reacted efficiently with nucleophiles in the presence of an initial non-covalent interaction.
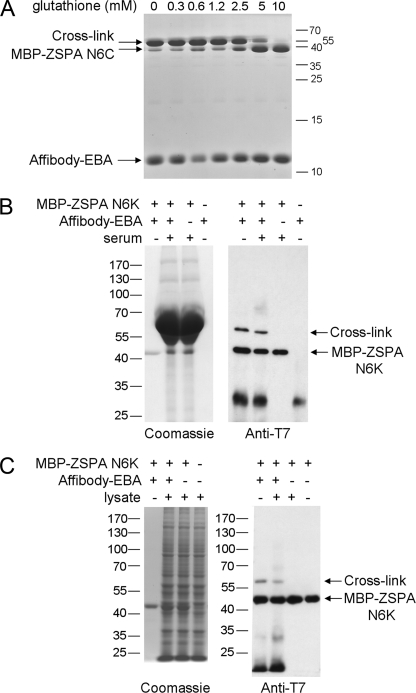
FIGURE 4.
Electrophilic affibody binds its target specifically. A, nucleophiles free in solution inhibit covalent cross-linking weakly. We incubated affibody-EBA with MBP-ZSPA N6C in the presence of varying concentrations of reduced glutathione at pH 8.0 for 5 h at 37 °C and analyzed complex formation by SDS-PAGE and Coomassie staining. B, electrophilic affibody binds its target specifically in the presence of serum. Affibody-EBA was incubated with MBP-ZSPA N6K alone or in the presence of fetal calf serum. We imaged total proteins present by SDS-PAGE and Coomassie staining (left panel), whereas we detected complexes containing MBP-ZSPA N6K or that had reacted with affibody-EBA by immunoblotting with anti-T7 tag antibody (right panel). The band at ∼30 kDa is a minor impurity of the affibody-EBA sample not visible by Coomassie staining. C, electrophilic affibody binds its target specifically in the presence of cell lysate. Affibody-EBA was incubated with MBP-ZSPA N6K alone or in the presence of HeLa cell lysate and analyzed as in B.
Secondly, we tested specificity in a diverse mixture of proteins. Affibody-EBA was incubated with MBP-ZSPA N6K in the presence of serum (Fig. 4B) or mammalian cell lysate (Fig. 4C), and covalent reaction was analyzed by immunoblotting. The specific affibody-ZSPA cross-link still formed in each case, but affibody-EBA showed little cross-reactivity with the diverse range of other proteins present seen by Coomassie staining. Thus, covalent bond formation by electrophilic affibodies was still compatible with high specificity.
Electrophilic Affibodies Improve Target Detection Sensitivity
A principal motivation for increasing the stability of affibody-target interactions is to increase the sensitivity of immunoassays. We tested the effect of electrophile modification on the sensitivity of MBP-ZSPA N6C detection by ELISA (Fig. 5). Unmodified affibody D36C only detected MBP-ZSPA N6C weakly (lowest concentration of N6C significantly different from N6C F13E was 10 nm, two-tailed t test, p < 0.001), but EBA modification improved detection 100-fold (lowest concentration of N6C significantly different from N6C F13E was 0.1 nm, two-tailed t test, p < 0.05). Even at the highest concentration of the non-interacting variant MBP-ZSPA N6C F13E, binding of affibody-EBA was low (Fig. 5), further supporting the specificity of covalent bond formation.
Electrophilic Affibodies Allow More Stable Cellular Imaging
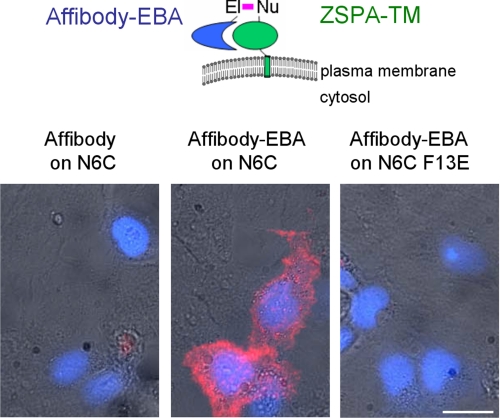
Antibody dissociation is often an obstacle in live cell imaging. Thus we tested the use of electrophilic affibodies for stable protein labeling at the surface of mammalian cells. ZSPA was targeted to the cell surface by fusion to a signal sequence and the transmembrane helix of platelet-derived growth factor receptor (to give ZSPA-TM). We detected binding of biotinylated affibody to ZSPA-TM N6C using dye-labeled streptavidin. Affibody-EBA bound to COS7 cells expressing ZSPA-TM N6C but not to neighboring untransfected cells, despite the other diverse proteins present on the cell surface, providing a stringent test of electrophilic affibody specificity (Fig. 6). The F13E mutation abolished cell surface binding of affibody-EBA, showing that labeling depended on the initial non-covalent interaction. 30 min after labeling, there was little signal from the unmodified affibody, whereas the signal from affibody-EBA was still strong (Fig. 6). Thus, electrophilic affibodies improved stability for cellular imaging, while showing good specificity.
FIGURE 6.
Electrophilic affibody improves stability and has high specificity at the mammalian cell surface. COS7 cells expressing ZSPA-TM N6C or N6C F13E were labeled with biotinylated affibody D36C with or without EBA. Affibody binding was detected with streptavidin-Alexa Fluor 555 (red), and this staining is overlaid with cyan fluorescent protein co-transfection marker (blue) and bright field images. Scale bar, 20 μm.
DISCUSSION
Here, we showed covalent bonding of an affibody to its target protein by site-specific incorporation of a weak electrophile on the affibody. We investigated the general applicability of electrophilic affibodies by testing the reactivity with different target residues, specificity, and impact for in vitro detection and cellular imaging.
Electrophilic affibodies rapidly formed covalent bonds with ∼90% coupling yield if a cysteine was apposed on the target protein. Cysteine is infrequently present on the surface of proteins (<1% of surface residues) (38) and most cysteines form disulfides in proteins that are secreted or at the cell surface. However, in many cases, these disulfides can break to form free thiols with important roles, such as on CD4 for human immunodeficiency virus entry, on integrins for platelet aggregation, and on plasmin for angiogenesis (39). Nevertheless, it was important that we found that the electrophilic affibody could also form covalent bonds to surface histidine and lysine, which are abundant on protein surfaces (together ∼13% of surface residues) (38). Given the range of potential nucleophilic amino acids on target proteins, our strategy to form covalent protein-protein interactions should be applicable to the majority of protein targets and not just those with surface cysteines. Reaction with lysine and histidine occurred at pH 8.0 (Figs. 2 and 4), which is still compatible with cell viability, but diagnostic assays could be run at arbitrary pH to further increase reactivity.
Immunoglobulins do not have surface cysteines (40), and we showed the generality of our approach by making an electrophilic Protein A variant that could covalently react with an unmodified antibody. Antibodies themselves are an important target for detection, either via secondary antibodies for laboratory assays, or to diagnose viral infection (41) or cancer (42). A version of Protein G with a photoreactive group for covalent conjugation to antibodies has recently been described (43), although the requirement for UV irradiation will be limiting in some applications.
Numerous examples in biological research and medicine, from aspirin to penicillin, show that covalent bond formation is compatible with high specificity (22, 24, 25, 44–46). Here, electrophilic affibodies showed good specificity in five different contexts: to a point mutant of their cognate target, at the mammalian cell surface, and in the presence of glutathione, serum, or cell lysate. This specificity was accompanied by improved functionality for ELISA and for imaging of a transfected surface protein in mammalian cells.
We chose affibody-ZSPA for our initial demonstration of covalent protein complex formation because of the structural information about the interaction and the ease of expression of both partners. The protein-protein interaction studied here is in fact weak (Kd 1.8 μm) (47), and so covalent bond formation should be easier to achieve for antibody complexes of nanomolar Kd. In the ideal situation (infinite affinity), the affibody would react faster than the half-time for dissociation (17).
To extend the applicability of electrophilic affibodies or antibodies, it will be valuable to explore linkers of different lengths and flexibilities as well as alternative electrophiles (44), to increase the speed and yield of covalent bond formation, particularly to lysine and histidine. Even without structural information, testing a few different linkers and positions for electrophile attachment may allow the generation of antibodies that form specific covalent interactions, with the goal of stable cell imaging of endogenous proteins and lowering the detection threshold of serum markers of disease (1–3).
Supplementary Material
Acknowledgment
We thank Anthony Walsh for assistance.
This work was supported by the Biotechnology and Biological Sciences Research Council, Engineering and Physical Sciences Research Council, Medical Research Council, and Ministry of Defence through the Bionanotechnology Interdisciplinary Research Centre, by the Oxford University Department of Biochemistry, and by Worcester College, Oxford.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- ELISA
- enzyme-linked immunosorbent assay
- BSA
- bovine serum albumin
- EBA
- ethylene bisacrylamide
- MBP
- maltose binding protein
- TCEP
- Tris(2-carboxyethyl)phosphine hydrochloride
- ZSPA
- Z domain of S. aureus Protein A.
REFERENCES
- 1.Parekh D. J., Ankerst D. P., Troyer D., Srivastava S., Thompson I. M. (2007) J. Urol. 178, 2252–2259 [DOI] [PubMed] [Google Scholar]
- 2.Georganopoulou D. G., Chang L., Nam J. M., Thaxton C. S., Mufson E. J., Klein W. L., Mirkin C. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2273–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng M. M., Cuda G., Bunimovich Y. L., Gaspari M., Heath J. R., Hill H. D., Mirkin C. A., Nijdam A. J., Terracciano R., Thundat T., Ferrari M. (2006) Curr. Opin. Chem. Biol. 10, 11–19 [DOI] [PubMed] [Google Scholar]
- 4.Jackson T. M., Ekins R. P. (1986) J. Immunol. Methods 87, 13–20 [DOI] [PubMed] [Google Scholar]
- 5.Zheng G., Patolsky F., Cui Y., Wang W. U., Lieber C. M. (2005) Nat. Biotechnol. 23, 1294–1301 [DOI] [PubMed] [Google Scholar]
- 6.Nam J. M., Stoeva S. I., Mirkin C. A. (2004) J. Am. Chem. Soc. 126, 5932–5933 [DOI] [PubMed] [Google Scholar]
- 7.Maynard J. A., Maassen C. B., Leppla S. H., Brasky K., Patterson J. L., Iverson B. L., Georgiou G. (2002) Nat. Biotechnol. 20, 597–601 [DOI] [PubMed] [Google Scholar]
- 8.Weisman L. E. (2009) Curr. Opin. Mol. Ther. 11, 208–218 [PubMed] [Google Scholar]
- 9.Houk K. N., Leach A. G., Kim S. P., Zhang X. (2003) Angew. Chem. Int. Ed. Engl. 42, 4872–4897 [DOI] [PubMed] [Google Scholar]
- 10.Schwesinger F., Ros R., Strunz T., Anselmetti D., Güntherodt H. J., Honegger A., Jermutus L., Tiefenauer L., Pluckthun A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foote J., Eisen H. N. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1254–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths A. D., Duncan A. R. (1998) Curr. Opin. Biotechnol. 9, 102–108 [DOI] [PubMed] [Google Scholar]
- 13.Lipovsek D., Plückthun A. (2004) J. Immunol. Methods 290, 51–67 [DOI] [PubMed] [Google Scholar]
- 14.Boder E. T., Midelfort K. S., Wittrup K. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10701–10705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleet G. W., Knowles J. R., Porter R. R. (1972) Biochem. J. 128, 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbas C. F., 3rd, Heine A., Zhong G., Hoffmann T., Gramatikova S., Björnestedt R., List B., Anderson J., Stura E. A., Wilson I. A., Lerner R. A. (1997) Science 278, 2085–2092 [DOI] [PubMed] [Google Scholar]
- 17.Chmura A. J., Orton M. S., Meares C. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8480–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama Y., Mitsuda Y., Taguchi H., Planque S., Salas M., Hanson C. V., Paul S. (2007) J. Biol. Chem. 282, 31250–31256 [DOI] [PubMed] [Google Scholar]
- 19.Trisler K., Looger L. L., Sharma V., Baker M., Benson D. E., Trauger S., Schultz P. G., Smider V. V. (2007) J. Biol. Chem. 282, 26344–26353 [DOI] [PubMed] [Google Scholar]
- 20.Orlova A., Feldwisch J., Abrahmsén L., Tolmachev V. (2007) Cancer Biother. Radiopharm. 22, 573–584 [DOI] [PubMed] [Google Scholar]
- 21.Li X., Liu D. R. (2004) Angew. Chem. Int. Ed. Engl. 43, 4848–4870 [DOI] [PubMed] [Google Scholar]
- 22.Levitsky K., Boersma M. D., Ciolli C. J., Belshaw P. J. (2005) Chembiochem. 6, 890–899 [DOI] [PubMed] [Google Scholar]
- 23.Fry D. W., Bridges A. J., Denny W. A., Doherty A., Greis K. D., Hicks J. L., Hook K. E., Keller P. R., Leopold W. R., Loo J. A., McNamara D. J., Nelson J. M., Sherwood V., Smaill J. B., Trumpp-Kallmeyer S., Dobrusin E. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen M. S., Zhang C., Shokat K. M., Taunton J. (2005) Science 308, 1318–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukiji S., Miyagawa M., Takaoka Y., Tamura T., Hamachi I. (2009) Nat. Chem. Biol. 5, 341–343 [DOI] [PubMed] [Google Scholar]
- 26.Gama L., Breitwieser G. E. (2002) Methods Mol. Biol. 182, 77–83 [DOI] [PubMed] [Google Scholar]
- 27.Högbom M., Eklund M., Nygren P. A., Nordlund P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3191–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckett D., Kovaleva E., Schatz P. J. (1999) Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platani M., Goldberg I., Lamond A. I., Swedlow J. R. (2002) Nat. Cell Biol. 4, 502–508 [DOI] [PubMed] [Google Scholar]
- 30.Howarth M., Ting A. Y. (2008) Nat. Protoc. 3, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed A. K., Schaffer S. W., Wetlaufer D. B. (1975) J. Biol. Chem. 250, 8477–8482 [PubMed] [Google Scholar]
- 32.Eklund M., Axelsson L., Uhlén M., Nygren P. A. (2002) Proteins 48, 454–462 [DOI] [PubMed] [Google Scholar]
- 33.Wahlberg E., Lendel C., Helgstrand M., Allard P., Dincbas-Renqvist V., Hedqvist A., Berglund H., Nygren P. A., Härd T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LoPachin R. M., Barber D. S., Gavin T. (2008) Toxicol. Sci. 104, 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hober S., Nord K., Linhult M. (2007) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 848, 40–47 [DOI] [PubMed] [Google Scholar]
- 36.Deisenhofer J. (1981) Biochemistry 20, 2361–2370 [PubMed] [Google Scholar]
- 37.Ji L. L., Katz A., Fu R., Griffiths M., Spencer M. (1993) J. Appl. Physiol. 74, 788–792 [DOI] [PubMed] [Google Scholar]
- 38.Fukuchi S., Nishikawa K. (2001) J. Mol. Biol. 309, 835–843 [DOI] [PubMed] [Google Scholar]
- 39.Schmidt B., Ho L., Hogg P. J. (2006) Biochemistry 45, 7429–7433 [DOI] [PubMed] [Google Scholar]
- 40.Alberini C. M., Bet P., Milstein C., Sitia R. (1990) Nature 347, 485–487 [DOI] [PubMed] [Google Scholar]
- 41.Allain J. P. (2003) Transfus. Clin. Biol. 10, 1–5 [DOI] [PubMed] [Google Scholar]
- 42.Qiu J., Hanash S. (2009) Clin. Lab. Med. 29, 31–46 [DOI] [PubMed] [Google Scholar]
- 43.Jung Y., Lee J. M., Kim J. W., Yoon J., Cho H., Chung B. H. (2009) Anal. Chem. 81, 936–942 [DOI] [PubMed] [Google Scholar]
- 44.Evans M. J., Saghatelian A., Sorensen E. J., Cravatt B. F. (2005) Nat. Biotechnol. 23, 1303–1307 [DOI] [PubMed] [Google Scholar]
- 45.Way J. C. (2000) Curr. Opin. Chem. Biol. 4, 40–46 [DOI] [PubMed] [Google Scholar]
- 46.Erlanson D. A., Braisted A. C., Raphael D. R., Randal M., Stroud R. M., Gordon E. M., Wells J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9367–9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lendel C., Dincbas-Renqvist V., Flores A., Wahlberg E., Dogan J., Nygren P. A., Härd T. (2004) Protein Sci. 13, 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.