Abstract
The Arabidopsis thaliana histone H2A-1 is important for Agrobacterium tumefaciens–mediated plant transformation. Mutation of HTA1, the gene encoding histone H2A-1, results in decreased T-DNA integration into the genome of Arabidopsis roots, whereas overexpression of HTA1 increases transformation frequency. To understand the mechanism by which HTA1 enhances transformation, we investigated the effects of overexpression of numerous Arabidopsis histones on transformation and transgene expression. Transgenic Arabidopsis containing cDNAs encoding histone H2A (HTA), histone H4 (HFO), and histone H3-11 (HTR11) displayed increased transformation susceptibility, whereas histone H2B (HTB) and most histone H3 (HTR) cDNAs did not increase transformation. A parallel increase in transient gene expression was observed when histone HTA, HFO, or HTR11 overexpression constructs were cotransfected with double- or single-stranded forms of a gusA gene into tobacco (Nicotiana tabacum) protoplasts. However, these cDNAs did not increase expression of a previously integrated transgene. We identified the N-terminal 39 amino acids of H2A-1 as sufficient to increase transient transgene expression in plants. After transfection, transgene DNA accumulates more rapidly in the presence of HTA1 than with a control construction. Our results suggest that certain histones enhance transgene expression, protect incoming transgene DNA during the initial stages of transformation, and subsequently increase the efficiency of Agrobacterium-mediated transformation.
INTRODUCTION
Agrobacterium tumefaciens–mediated plant genetic transformation is an important experimental tool for investigation of various aspects of plant biology and for agricultural biotechnology. Development of this transformation technology represents the culmination of many decades of effort to improve tissue culture and plant genetic engineering techniques. Agrobacterium-mediated transformation is based on a conjugative transfer-like process, which eventually takes the T-DNA to the host cell nucleus (Gelvin, 2000, 2003a, 2009; Tzfira and Citovsky, 2003; Citovsky et al., 2007). After attachment of the bacterium to plant cells and induction of Agrobacterium virulence (vir) genes, a single-stranded DNA (the T-strand) is processed from the resident tumor-inducing or root-inducing plasmid and transported from the bacterium to the plant cell. In addition, a number of Virulence effector proteins, including VirD2 (attached to the T-strand), the single-strand DNA binding protein VirE2, plus VirE3, VirD5, and VirF are also transferred to the plant (Otten et al., 1984; Stahl et al., 1998; Vergunst et al., 2000, 2003, 2005; Schrammeijer et al., 2003). These proteins are likely involved in protecting T-DNA from nuclease digestion, directing T-DNA to the nucleus, stripping proteins from the T-strand prior to integration, and integrating T-DNA into the plant genome. T-DNA integration occurs randomly into genomic DNA by the process of illegitimate recombination (Matsumoto et al., 1990; Gheysen et al., 1991; Mayerhofer et al., 1991; Tinland, 1996; Tzfira and Citovsky, 2000, 2003; Gelvin, 2003b; Kim et al., 2007). The level of transgene expression may depend upon the site of integration into the host genome and the associated chromatin structure of this region (Day et al., 2000; Tzfira and Citovsky, 2002; Kohli et al., 2003; Lacroix et al., 2006; Gelvin and Kim, 2007).
During the past several years, T-DNA integration research has identified plant and bacterial proteins that are involved in the integration process. Using yeast and Arabidopsis thaliana as models, several host genes involved in T-DNA integration have been identified, although many others likely await discovery (Li et al., 2005b; Endo et al., 2006). Research in this area will lead not only to the identification of proteins involved in the integration process, but will also result in elucidation of the precise molecular mechanism of T-DNA integration. In addition, understanding the molecular pathway(s) of T-DNA integration will likely lead to the development of new tools and approaches for controlling this process.
An increasing number of reports have emphasized the roles of chromatin proteins, including histones, in the regulation of gene expression (Kornberg, 1977; Luger et al., 1997; Verbsky and Richards, 2001; Kouzarides, 2007; Tremethick, 2007). In particular, histone modifications, such as acetylation and methylation, may regulate gene expression (Shilatifard, 2006; Zhang et al., 2006; Chan et al., 2007; Shahbazian and Grunstein, 2007; Zilberman et al., 2007). Minor histone variants may contribute to diverse nuclear functions by directing distinct or unique chromatin architectures (Talbert et al., 2002). These more recent findings have once again brought histones into the limelight as important mediators of cellular events.
A forward genetic screen identified an Arabidopsis histone H2A variant (H2A-1, encoded by HTA1) as important for T-DNA integration (Mysore et al., 2000a). A T-DNA insertion into the 3′-untranslated region of the histone HTA1 gene shows a large reduction in stable but not transient Agrobacterium-mediated transformation efficiency (Mysore et al., 2000a). Other Arabidopsis rat (resistant to Agrobacterium transformation) mutants, identified following T-DNA mutagenesis or RNA interference expression, disrupt expression of genes involved in chromatin structure and remodeling (Zhu et al., 2003b; Crane and Gelvin, 2007). Some of these mutants show high transient but low stable transformation efficiency, suggesting that, in these mutants, the rat phenotype most likely results from inhibition of T-DNA integration. In addition, several studies have suggested a role for histone H2A in directing T-DNA to the site of integration (Li et al., 2005a; Loyter et al., 2005), perhaps by altering host chromatin structure (Mysore et al., 2000a). Although characterization of the roles of these proteins in the transformation process will require intensive investigation, the diversity of the genes identified so far indicates a significant plant contribution to the Agrobacterium-mediated transformation process.
Recently, genetic and biochemical investigations have revealed chromatin proteins that may influence T-DNA integration (Endo et al., 2006; Crane and Gelvin, 2007; Gelvin and Kim, 2007). It has been suggested that VIP1 (VirE2 interacting protein 1) protein can target T-DNA to host plant chromatin by interaction with histones (Li et al., 2005a; Loyter et al., 2005). Using viral-induced gene silencing, Anand et al. (2007) showed that decreased expression of some Nicotiana benthamiana chromatin genes could inhibit subsequent Agrobacterium-mediated transformation. In particular, blocking expression of genes encoding histone H2A and H3 decreased transformation. Whereas silencing HTA genes resulted in a decrease in cell division, inhibition of HTR expression had relatively little effect on plant growth but specifically inhibited T-DNA integration (Anand et al., 2007).
Taken together, these studies indicate that intracellular T-DNA transport and integration into the plant genome depends on numerous host factors, but how chromatin proteins influence this process has not yet been elucidated.
Here, we show that introduction of overexpression constructs for particular Arabidopsis histones into plants increases subsequent retransformation frequency. A parallel increase in transient gene expression was observed when histone HTA, HFO, or HTR11 overexpression constructs were cotransfected with a plant-active gusA gene into tobacco (Nicotiana tabacum) BY-2 protoplasts. We demonstrate that nuclear localization of these histone proteins is not necessarily required to enhance transgene expression and that overexpression of histone H2A-1 does not increase expression of a previously integrated transgene. Rather, several histones apparently protect incoming transgenes from nuclease digestion within the cell. Based on our observations, we propose a mechanism by which several histones increase the frequency of transformation by enhancing transgene stability during the initial stages of transformation.
RESULTS
Overexpression of Several Arabidopsis Histone cDNAs Increases Susceptibility to Agrobacterium-Mediated Transformation
In previous studies, we showed an important role for the Arabidopsis histone H2A-1, encoded by HTA1, in Agrobacterium-mediated transformation. Disruption of this gene results in decreased transformation due to reduced frequency of T-DNA integration into the plant genome (Nam et al., 1999; Mysore et al., 2000a). Expression of HTA1 in plants coincides with the tissues and cell types that are most susceptible to transformation (Yi et al., 2002). Overexpression of HTA1 genomic or cDNAs results in increased transformation of Arabidopsis root segments (Mysore et al., 2000a; Yi et al., 2006).
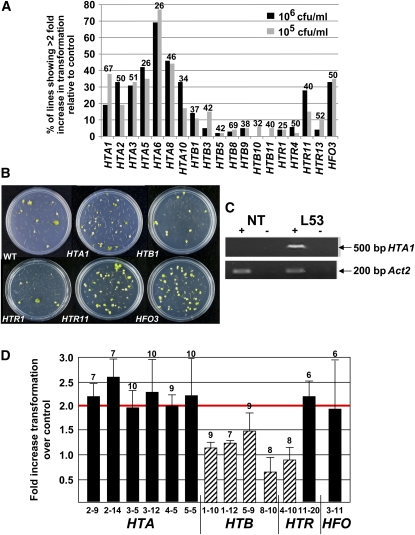
The Arabidopsis genome comprises 47 genes that encode 33 different core histone proteins (13 HTA genes that encode 13 different H2A proteins, 11 HTB genes that encode 11 different H2B proteins, 15 HTR genes that encode eight different H3 proteins, and eight HFO genes that encode 1 H4 protein; www.chromdb.org; see Supplemental Figure 1 online). To test whether overexpression of histones other than HTA1 may influence Agrobacterium-mediated transformation, we generated transgenic Arabidopsis lines designed to overexpress various representative core histone cDNAs and assayed roots of these T1 generation transgenic lines for transformation susceptibility. We chose to overexpress these histone cDNAs because they represent various clades of histone proteins from each core histone group. For most histone cDNAs, we assayed >50 independent transgenic events (see Supplemental Figure 2 online for detailed transformation data; note that Supplemental Figure 2 contains data from only one representative set out of several transformation sets). We subsequently quantified the number of lines displaying enhanced transformation compared with that of wild-type control plants. For these experiments, we inoculated >100 root segments per plant each at two different Agrobacterium concentrations (105 and 106 colony-forming units (cfu)/mL). Particular histone constructs were considered to enhance transformation if >25% of the lines showed a minimum of twofold increased transformation for at least one bacterial inoculum concentration. In no instance did we observe any obvious growth or developmental differences between the various transgenic lines containing histone cDNA expression cassettes and wild-type plants.
Figure 1C shows, as an example, that the HTA1 transgene is expressed in roots of transgenic plants containing this cDNA expression cassette. Figure 1A summarizes the results of the transformation assays. Expression of all seven tested HTA histone cDNAs and the HFO histone cDNA resulted in increased transformation of the derived transgenic lines. By contrast, none of the seven representative HTB cDNAs nor three of the four HTR cDNAs affected transformation frequency. We did, however, observe an increase in transformation when the histone H3 cDNA HTR11 was expressed in the plants. Figure 1B shows representative plates of tumors growing on inoculated transgenic Arabidopsis root segments.
Figure 1.
Overexpression of Specific Arabidopsis Core Histone cDNAs Increases Agrobacterium-Mediated Transformation.
(A) Roots segments from T1 generation plants containing the indicated histone cDNAs, as well as root segments from Wassilewskija (Ws) wild-type plants, were inoculated with either 105 or 106 cfu/mL of the tumorigenic strain Agrobacterium A208. After 2 d, roots segments were transferred to Murashige and Skoog (MS) medium containing 100 μg/mL Timentin to kill bacteria. The percentage of root segments developing tumors was scored after 1 month. A histone gene was considered to increase transformation efficiency if at least 25% of the lines containing that particular cDNA showed a minimum twofold increase in transformation efficiency relative to that of wild-type plants. Numbers above each bar represent the number of independent T1 generation transgenic plants assayed for each histone construction, as depicted in Supplemental Figure 2 online. Additional sets of transformation data were also collected, with similar results.
(B) After 1 month, representative plates of tumors growing on inoculated transgenic Arabidopsis root segments were photographed.
(C) RNA was extracted from a nontransformed line (NT) and transgenic line L53 (which has increased susceptibility to Agrobacterium-mediated transformation and contains a HTA1 cDNA expression cassette) and assayed for HTA1 and Act2 mRNA by RT-PCR. The HTA1 primers were designed to amplify only HTA1 transgene mRNA and not endogenous HTA1 mRNA. +, reverse transcriptase present in the reaction mix; −, reverse transcriptase absent in the reaction mix.
(D) Roots of T2 generation plants from selected lines showing ≥2-fold increased transformation in T1 generation plants (HTA lines 2-9, 2-14, 3-5, 3-12, 4-5, and 5-5, HTR line 11-20, and HFO line 3-11; solid bars), lines not showing increased transformation in T1 generation plants (HTB lines 1-10, 1-12, 5-9, and 8-10, and HTR line 4-10; diagonal striped bars), and wild-type plants were assayed for transformation susceptibility as described above. Numbers above each bar indicate the number of individual plants assayed from each line. Line numbers are designated as follows: the first number identifies the histone gene, and the second number indicates the particular transgenic line. Error bars indicate se.
To confirm these results, we investigated the transformation susceptibility, relative to wild-type control plants, of numerous T2 generation transgenic lines containing various histone cDNAs. Figure 1D shows that T2 generation lines derived from transgenic plants that were hypersusceptible to Agrobacterium-mediated transformation remained hypersusceptible, whereas lines derived from plants that did not display increased transformation susceptibility in the T1 generation likewise did not display hypersusceptibility in the T2 generation.
The results of these experiments indicate that some histone cDNAs, in addition to the previously described HTA1, could enhance root transformation when overexpressed in transgenic Arabidopsis plants.
Overexpression of Particular Histone cDNAs Can Enhance Transgene Expression
Because mutation the histone H2A gene HTA1 decreased transformation of Arabidopsis by reducing T-DNA integration (Mysore et al., 2000a), we initially hypothesized that overexpression of histone genes increased transformation by increasing T-DNA integration. However, we observed that additional copies of HTA1 could increase transient as well as stable transformation (L.-Y. Lee, S.J. Johnson, X. Sui, and S.B. Gelvin, unpublished data). Transient transformation does not require T-DNA integration. We therefore speculated that overexpression of particular histones increased transformation by increasing reporter and selectable marker transgene expression.
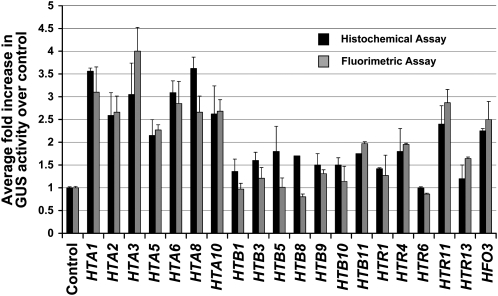
In order to unlink the effects of histone genes on Agrobacterium-mediated transformation from the effects on transgene expression, we conducted transgene expression studies in tobacco BY-2 protoplasts. To this end, we assayed both β-glucuronidase (GUS)-specific activity and the percentage of cells showing GUS activity 24 h after coelectroporation of a gusA gene and either a histone cDNA expression cassette or an empty vector control plasmid (Jefferson et al., 1987; Mysore et al., 1998).
Figure 2 shows the results of these studies. When coelectroporated into BY-2 protoplasts with a gusA gene, all tested HTA cDNAs and the HFO cDNA caused a more than twofold increase in GUS activity relative to that of the empty vector control plasmid. None of the tested HTB nor most HTR cDNAs caused an increase in GUS activity more than twofold over that of the control. Only one of the histone H3 cDNAs, HTR11, caused a more than twofold increase in GUS activity.
Figure 2.
Overexpression of Specific Histone cDNAs Increases gusA Transgene Expression in Transiently Transfected Tobacco BY-2 Protoplasts.
Tobacco BY-2 protoplasts were cotransfected with a gusA reporter gene and either a histone cDNA expression cassette or an empty vector control plasmid. Histochemical and fluorimetric assays were performed 24 h after transfection. For histochemical analysis, >1000 cells were examined for each treatment to determine the percentage of cells staining blue with X-gluc. The number reported is the average percent of blue cells for each line divided by the percent of blue cells for the empty vector control. Fluorimetric assays measured GUS-specific activity as average fluorescence intensity of each line divided by the fluorescence intensity of the empty vector control. Numbers represent GUS activity relative to that of the control. Error bars indicate se of at least three biological replicates.
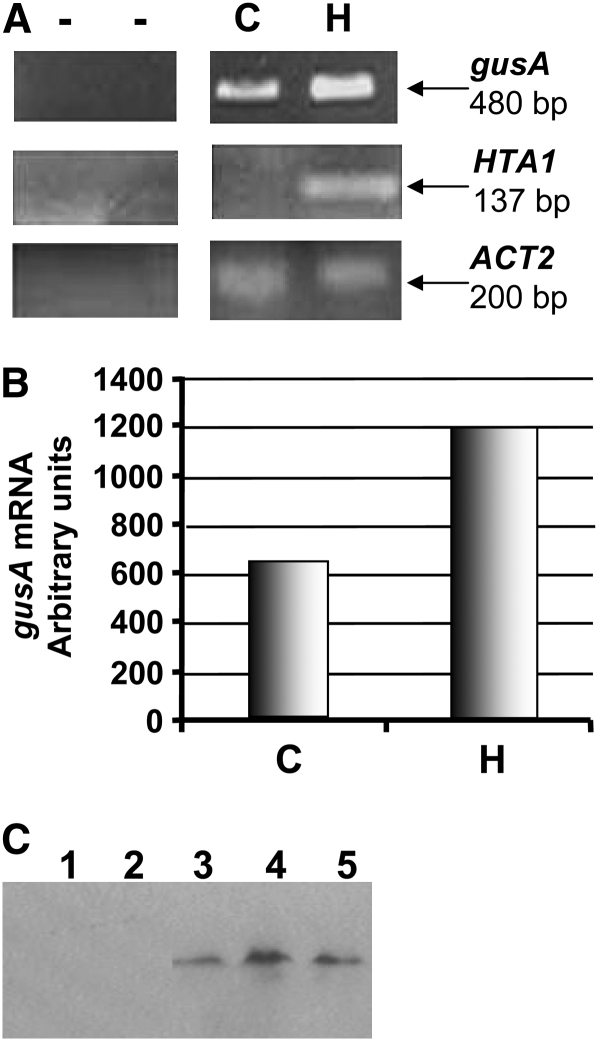
We used RT-PCR and gene-specific primers to investigate expression of HTA1 and gusA in transfected protoplasts. Figure 3A shows expression of both of these genes. Figure 3B shows that, relative to coelectroporation with an empty vector control and normalized to endogenous actin mRNA, the HTA1 cDNA caused an approximately twofold increase in gusA mRNA in tobacco BY-2 protoplasts. These data support the approximately threefold increase in GUS-specific activity effected by the HTA1 cDNA (Figure 2).
Figure 3.
gusA mRNA Steady State Levels Increase When Tobacco BY-2 Protoplasts Are Cotransfected with a HTA1 cDNA.
(A) Tobacco BY-2 protoplasts were cotransfected with a gusA expression cassette and either a HTA1 cDNA expression cassette or an empty vector control plasmid. After 24 h, RNA was extracted and subjected to RT-PCR. gusA mRNA levels were determined in cells cotransfected with an empty vector control plasmid (C) or a HTA1 cDNA expression cassette (H). gusA mRNA levels were normalized relative to actin mRNA levels.
(B) Graphical representation of the increase in gusA mRNA after coelectroporation of the gusA expression cassette and either an empty vector or a HTA1 cDNA expression cassette.
(C) Proteins were extracted from tobacco BY-2 protoplasts 24 h after electroporation of an untagged HTA1 gene (lanes 1 and 2, representing two independent experiments) or a gene encoding a HTA1-T7 tag fusion protein (lanes 3 to 5, representing three independent experiments). Total cellular proteins were subjected to protein gel blot analysis using antibodies directed against the T7 tag. Inclusion of an HTA1-YFP gene in each electroporation experiment, followed by fluorescent imaging of the cells, indicated that electroporation was successful for each experiment.
To show that introduction of the histone gene into tobacco BY-2 protoplasts results in increased histone protein production, we tagged the H2A-1 protein with a T7 epitope. Cotransfection of HTA1-T7 with HTA1-YFP resulted both in yellow fluorescent protein (YFP) fluorescence (data not shown) and expression of H2A-1-T7 protein, as detected by protein blot analysis using anti-T7 epitope antibodies (Figure 3C, lanes 3 to 5). As a control for antibody specificity, we cotransfected BY-2 protoplasts with HTA1 and HTA1-YFP. Although the cells showed YFP fluorescence, indicating successful transfection, we could not detect untagged H2A-1 using this antibody (Figure 3C, lanes 1 and 2). The T7 tag did not affect the ability of histone H2A-1 to restore transformation proficiency to the rat5 histone hta1 mutant nor its ability to increase transgene expression in transiently transfected tobacco BY-2 protoplasts (see Supplemental Figure 3 online).
Our data indicate that there was a perfect correlation among histone cDNAs that caused a more than twofold increase in transient transgene expression and those that increased stable transformation of Arabidopsis roots.
The Histone H2A-1 N-Terminal Domain Can Increase Transgene Expression
All core histones contain a highly structured C-terminal histonefold domain and a highly charged but less structured N-terminal domain that emerges from the histone core. This N-terminal region contains large DNA interaction surfaces involved in processes including transcriptional activation, silencing, chromatin assembly, and DNA replication, and it may play critical roles in both the organization and posttranslational modification of chromatin (Peterson and Laniel, 2004; Mariño-Ramírez et al., 2005). Histone N-terminal domains may also function to control the translational positioning of nucleosomes in vitro (Parra and Wyrick, 2007) and can mediate internucleosome interactions that are required for the formation of higher-order chromatin structures.
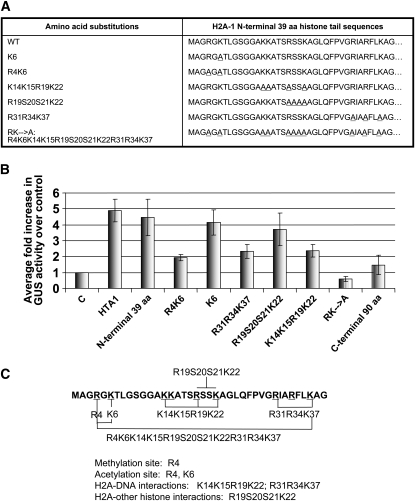
Our initial experiments indicated that expression of full-length histone H2A-1 increased expression of a gusA reporter transgene in BY-2 tobacco protoplasts. We subsequently investigated the importance of the N-terminal and C-terminal domains of H2A-1 for increasing transgene expression. We transfected BY-2 tobacco protoplasts with a gusA reporter gene plus either an empty vector control plasmid, a plasmid encoding the N-terminal 39 amino acids, or a plasmid encoding the C-terminal amino acids (40 to 130) of H2A-1. We measured GUS activity 24 h after transfection. Figure 4B shows that the N-terminal 39 amino acids of H2A-1 increased GUS activity ∼4.5-fold relative to that of the control, whereas the C-terminal domain of H2A-1 showed no such increase in GUS activity.
Figure 4.
The N-Terminal 39 Amino Acids of Histone H2A-1 Are Sufficient to Increase gusA Transgene Expression in Tobacco BY-2 Protoplasts.
(A) Amino acid sequence substitutions in the N-terminal H2A-1 region. Underlined amino acids in the right panel were substituted by Ala.
(B) Relative GUS activity in protoplasts cotransfected with a gusA expression cassette and various H2A-1 wild-type and mutant peptides. Numbers represent GUS activity relative to the empty vector control. Transfected cells were stained histochemically with X-gluc and examined microscopically 24 h later. More than 1000 cells were examined for each treatment. Error bars indicate se of at least three biological replicates.
(C) The first 39 amino acids of the histone H2A-1 protein. Specific amino acid mutations within this region are marked. Amino acids involved in acetylation, histone–DNA interactions, and histone–histone interactions are indicated.
The N-terminal domain of H2A-1 is characterized by a high content of positively charged amino acids. Lys residue 6 (Lys-6) can be acetylated; such acetylation often correlates with transcriptional activity of genes associated with this modified histone. Arg residue 4 (Arg-4) can be acetylated or methylated. To identify amino acid sequences in the N-terminal domain of H2A-1 that effect increased transgene expression, we constructed a series of vectors in which particular amino acids were substituted with Ala. Figure 4A lists these substitutions. Plasmids encoding these mutant H2A-1 N-terminal peptides were cotransfected with a gusA gene into tobacco BY-2 protoplasts and assayed 24 h later for GUS activity. Figure 4B shows that mutation of K6 and R19S20S21K22 (involved in histone acetylation and histone–histone interactions, respectively; Luger et al., 1997; Luger and Richmond, 1998) had relatively little effect on the ability of the N-terminal H2A-1 protein to increase transgene expression. Mutation of R4K6, K14K15R19K22, and R31R34K37 (involved in histone methylation and histone-DNA interactions) had a more severe effect. Although these mutant peptides could increase GUS activity relative to that of the empty vector control, they were only approximately half as effective as was the wild-type N-terminal H2A-1 peptide. By contrast, when all basic amino acids were substituted with Ala (RK→A), the resulting mutant H2A-1 N-terminal peptide slightly decreased GUS activity relative to the empty vector control.
The ability of N-terminal H2A-1 peptides to enhance transgene expression is not related to the overall positive charge of the peptide. For example, the R4K6, R31R34K37, and K14K15R19K22 peptides were approximately equally effective in increasing transgene expression, despite the fact that they bear dissimilar charges. Rather, specific amino acids are important for this effect. Figure 4C shows the amino acid sequence of the H2A-1 protein, indicating various domains important for methylation, acetylation, histone-DNA binding, and histone–histone interactions. We note that amino acids involved in histone–DNA interactions are important for increasing transgene expression. In addition, Arg-4, a potential site for acetylation and methylation, also appears important.
Subcellular Localization of Histone H2A-1 Variants in BY-2 Tobacco Cells and Protoplasts
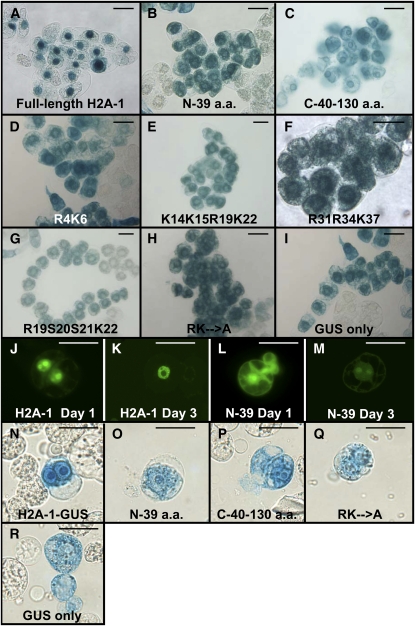
Because histones are normally associated with chromatin in the nucleus, whereas transgene DNA must first traverse the cytoplasm to reach its final nuclear destination, we sought to determine where the effect of histones on transgene expression and transformation may occur. To identify the subcellular site of localization of full-length and mutant H2A-1 peptides in plant cells, we fused the various H2A-1 coding regions to a gusA reporter gene. Agrobacterium strains containing these constructions within the T-DNA were used to generate stable BY-2 tobacco cell lines expressing the various fusion proteins. The cells were stained with X-gluc and examined microscopically for GUS activity. Figure 5A shows that full-length H2A-1 localized predominantly to the nucleus, whereas peptides containing the H2A-1 N- and C-terminal domains are localized additionally to the cytoplasm. There was a tendency, however, for these latter two peptides to accumulate in the region surrounding the nuclei. This perinuclear localization was also noted for various N-terminal mutant peptides.
Figure 5.
Subcellular Localization of Full-Length Histone H2A-1 and Various Mutant Peptides Fused to Either GUS or YFP Reporter Proteins.
(A) to (I) X-gluc staining of stably transformed tobacco BY-2 cell lines expressing the indicated GUS fusion proteins. Note that expression of an unfused gusA gene results in cytoplasmic staining of the cells (I), whereas expression of a full-length H2A-1-GUS fusion protein results in predominantly nuclear staining (A). Expression of the various H2A-1 mutant peptides results in predominantly cytoplasmic, especially perinuclear, staining ([B] to [H]).
(J) to (M) Epifluorescence images of tobacco BY-2 protoplasts 1 d ([J] and [L]) or 3 d ([K] or [M]) after transfection with the indicated constructions. Note that the full-length H2A-1-YFP fusion protein localizes predominantly to the nucleus ([J] and [K]) but that the N-terminal H2A-1-YFP fusion protein localizes throughout the cell ([L] and [M]). YFP fluorescence is shown in yellow-green.
(N) to (R) X-gluc staining of transfected tobacco BY-2 protoplasts expressing the indicated GUS fusion proteins 1 d after transfection.
Bars = 50 μM.
The intracellular localization of the H2A-1-GUS fusion proteins depicted in Figures 5A to 5I was determined several weeks after the initial transformation events that established these BY-2 lines. However, we had conducted our transgene expression analyses 24 h after transfection. We therefore examined the intracellular localization of full-length H2A-1 and N-terminal H2A-1 (N-39) YFP fusion proteins in electroporated BY-2 protoplasts within the first few days after transfection. Figures 5J and 5K show that, for the 3 d examined, the full-length H2A-1 YFP fusion protein localized predominantly to the nucleus, although some cytoplasmic localization occurred. However, the localization of the N-terminal H2A-2 YFP fusion protein differed during this initial transfection period. On days 1 and 3 following transfection, the protein localized throughout the cell, including in the cytoplasm, nucleus, and nucleolus. This latter observation confirmed the expression pattern of the N-terminal H2A-1 GUS fusion protein in the BY-2 cell line. Thus, on day 1 when the effect of the various histone proteins on transgene expression was assayed, full-length H2A-1 was predominantly but not exclusively in the nucleus, whereas the N-terminal H2A-1 peptide localized throughout the cell.
The molecular masses of the H2A-1 and N-terminal H2A-1 YFP fusion proteins are ∼41.3 and 31.3 kD, respectively. They are therefore below the nuclear pore exclusion size and may thus diffuse into the nucleus (Stewart et al., 2007). To test whether the various histone derivatives passively diffused into the nucleus, we fused H2A-1 and several derivatives to the GUS protein, thus generating GUS fusion proteins that exceed the nuclear pore exclusion size limitation. Figures 5N to 5R show the localization of these various GUS fusion constructions 1 d following transfection. As observed for the experiments described above, full-length H2A-1 fused to GUS localized predominantly, but not exclusively, to the nucleus, whereas the N- and C-terminal regions of H2A-1 fused to GUS localized to both the nuclear and cytoplasmic subcellular compartments. The RK→A H2A-1 mutant and GUS protein only localized to the cytoplasm (the light-blue staining over the nuclear areas following transfection of these latter two constructions likely results from cytoplasmic GUS activity surrounding the nucleus). We obtained similar localization results on days 2 and 3 following transfection. Thus, full-length H2A-1 and the N-terminal 39–amino acid H2A-1 fragment are present both in the cytoplasm and in the nucleus during the first day after transfection.
Histone H2A-1 Does Not Increase Expression of Previously Integrated Transgenes
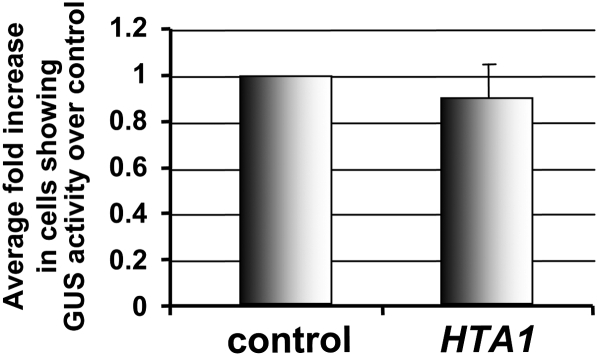
We investigated whether HTA1 could increase transgene expression of an integrated gusA reporter gene. We generated a BY-2 tobacco line that stably expressed a gusA gene. Protoplasts isolated from this line were electroporated with either an empty vector or a HTA1 cDNA expression construction and assayed 24 h later for GUS activity. Figure 6 shows that GUS activity in these protoplasts was approximately equal in cells transfected with the empty vector or with the HTA1 cDNA. Thus, HTA1 could not increase activity of a previously integrated and expressing gusA transgene.
Figure 6.
Overexpression of HTA1 in Stably Transformed Tobacco BY-2 Cells Does Not Increase Activity of a Previously Integrated gusA Gene.
Protoplasts isolated from a BY-2 line stably expressing a gusA gene were transfected with either a control empty vector or HTA1 expression constructions. After 24 h, the cells were stained with X-gluc, and the percentage of cells showing GUS activity was determined. Numbers represent GUS activity relative to the control. More than 1000 cells were examined for each treatment. Error bars indicate se of three biological replicates.
We additionally asked whether HTA1 could reverse silencing of an integrated transgene. We had previously identified two Arabidopsis mutants, rat4 and rat14, that are resistant to Agrobacterium transformation (Zhu et al., 2003a, 2003b). During the course of characterizing these mutants, we noted that the nptII selectable marker transgene had silenced in these lines, which became kanamycin sensitive. We introduced into these mutants a HTA1 cDNA expression cassette using a flower dip transformation protocol (Clough and Bent, 1998). We had previously shown that most rat mutants, although resistant to somatic cell transformation, are highly susceptible to flower dip transformation (Mysore et al., 2000b). When plated on medium containing kanamycin, the derived transgenic lines remained sensitive to this antibiotic. Thus, a HTA1 cDNA expression cassette could not increase expression either of a previously integrated and expressing gusA transgene or of a previously integrated but silenced nptII transgene.
Histone H2A-1 Protects Incoming DNA from Nuclease Degradation
Our observations indicated that several histones could increase expression of incoming but not previously integrated transgene DNA. We therefore hypothesized that overexpression of various histones could protect incoming transgene DNA and that increased transgene stability was the cause of increased transgene expression.
To test this hypothesis, we conducted electroporation experiments similar to those described above. We coelectroporated tobacco BY-2 protoplasts with a plasmid encoding a gusA reporter transgene and either a control empty vector or a vector containing various histone cDNAs. At various times after transfection, we harvested cells and treated them with DNase I to degrade any DNA bound to the outside of the cells. We subsequently isolated DNA from these cells and conducted PCR analyses with primers directed against the gusA transgene. To normalize our results for the amount of isolated DNA, we also conducted PCR reactions using primers directed against an endogenous tobacco actin gene. Supplemental Figure 4 online shows that our quantifications were linear with the number of PCR cycles over the cycle times investigated.
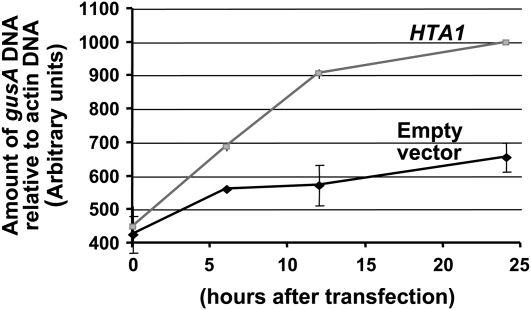
Figure 7 shows that the amount of gusA DNA within BY-2 protoplasts increased with time after electroporation. After 24 h, the rate of accumulation of gusA DNA was approximately twofold greater when a HTA1 cDNA was coelectroporated compared with that following coelectroporation with a control empty vector.
Figure 7.
Rate of Accumulation of gusA Double-Stranded DNA in Tobacco BY-2 Protoplasts Cotransfected with Either a HTA1 cDNA or an Empty Vector Control Plasmid.
Protoplasts were electroporated with a gusA transgene, plus either an empty vector or a HTA1 expression construction. After various periods of time, samples were treated with DNase I, following which DNA was extracted. PCR was conducted to determine the amount of gusA DNA within the cell and normalized to the amount of actin DNA. Error bars indicate se of three biological replicates.
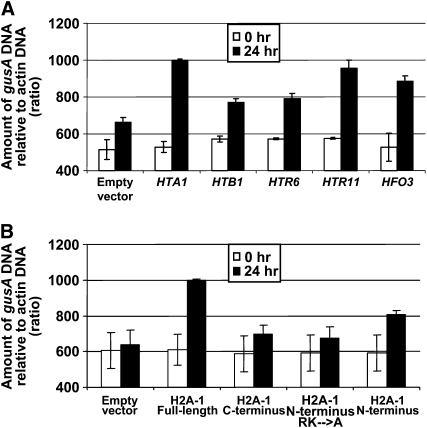
We next examined the effect of histones other than H2A-1 on an introduced gusA gene. As representative histones, we selected two that had a relatively minimal effect on increasing Agrobacterium-mediated transformation and transgene activity (histones H2B-1 and H3-6) and two that had a greater effect on these processes (histones H3-11 and H4-3). Figure 8A shows that introduction of histone HTA1, HTR11, and HFO3 cDNAs resulted in greater levels of a cointroduced gusA gene than did the HTB1 and HTR6 cDNAs. In addition, Figure 8B shows that introduction of cDNAs encoding full-length and the N-terminal 39–amino acid fragment of histone H2A-1 could also result in levels of an introduced gusA DNA greater than could the C-terminal 90–amino acid region or a mutant N-terminal fragment of H2A-1 that had all basic amino acids converted to Ala.
Figure 8.
Accumulation of Double-Stranded gusA DNA in Tobacco BY-2 Protoplasts following Cotransfection with Constructions Expressing Various Histone cDNAs or Mutations of HTA1.
Tobacco protoplasts were cotransfected with a gusA expression construction and either an empty vector control plasmid or constructions expressing various histone cDNAs or histone mutations. At 0 and 24 h after transfection, samples were treated with DNase I and DNA was extracted. PCR was conducted to determine the amount of gusA DNA within the cell, relative to actin DNA. The relative intensities of the ethidium-stained gel bands were determined by densitometric scanning, using Lab Works 4.1 software. Intensity is in arbitrary units, starting at 400. Error bars indicate se of two or three biological replicates.
(A) Empty vector, empty vector + gusA gene; HTA1, HTA1 + gusA gene; HTB1, HTB1 + gusA gene; HTR6, HTR 6 + gusA gene; HTR11, HTR11 + gusA gene; HFO, HFO3 + gusA gene.
(B) Empty vector, empty vector + gusA gene; H2A-1 full-length, full-length H2A-1 + gusA gene; H2A-1 C terminus, C-terminal region of H2A-1 + gusA gene; H2A-1 N terminus, N-terminal region of H2A-1 + gusA gene; H2A-1 N terminus RK→A, first 39 amino acids of H2A-1 in which all basic amino acids were converted to Ala + gusA gene.
Taken together, our data indicate that the same histones (or histone H2A-1 mutants) that increased both Agrobacterium-mediated transformation and transgene expression could increase the steady state level of introduced gusA DNA to a greater extent than could those histones (or histone H2A-1 mutants) that had a lesser effect on these two processes.
Introduction of a HTA1 cDNA Can Increase Expression of a Single-Stranded Coelectroporated gusA Gene
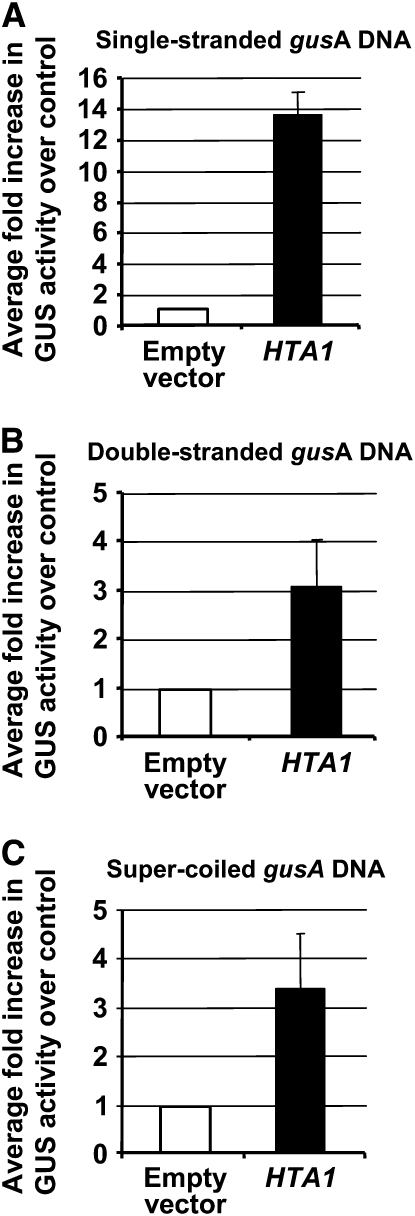
During Agrobacterium-mediated plant transformation, a single-stranded form of T-DNA, termed the T-strand, is transferred from the bacterium to the plant, where it is converted to a double-stranded form in the nucleus (Tinland et al., 1994; Yusibov et al., 1994; Narasimhulu et al., 1996; Mysore et al., 1998; Vergunst and Hooykaas, 1998). Thus, T-DNA remains single stranded in the plant cytoplasm. In order to mimic the process better of Agrobacterium-mediated transformation, we introduced single-stranded gusA DNA (in the form of denatured linear DNA) into tobacco BY-2 protoplasts, along with double-stranded forms of either an empty vector control or a HTA1 cDNA. We subsequently measured GUS activity 24 h after transfection. As a control, we used either linear or super-coiled forms of double-stranded gusA DNA. Figure 9 shows that cotransfection of a HTA1 cDNA expression cassette seemingly had an even greater effect upon GUS activity directed by a single-stranded template than it did on double-stranded gusA templates. However, it should be recognized that in these experiments the absolute percentage of cells showing GUS activity was relatively small when single-strand DNA was introduced (7.2 to 7.8%) compared with when double-strand DNA was introduced (18 to 20%). For all forms of introduced DNA tested, cotransfection with the HTA1 gene resulted in 70 to 78% of the cells showing GUS activity. Thus, the greater relative increase in GUS expression when cotransfecting the HTA1 gene with single-strand DNA compared with double-stranded DNA likely reflects the lower stability of single-stranded DNA within the plant cell when not bound to histones.
Figure 9.
Overexpression of a HTA1 cDNA Can Increase Expression of Various Forms of a Coelectroporated gusA DNA.
Tobacco BY-2 protoplasts were cotransfected with various conformations of a gusA reporter gene and either a histone cDNA expression cassette or an empty vector control plasmid. After 24 h, the percentage of cells expressing GUS activity (X-gluc staining) was determined. The data are presented as the percentage of stained cells relative to that of the control.
(A) Single-stranded gusA transgene.
(B) Double-stranded gusA transgene.
(C) Super-coiled gusA transgene. More than 1000 cells were examined for each treatment. Error bars indicate se.
Introduction of a HTA1 cDNA Protects Incoming Single-Stranded DNA from Nuclease Degradation
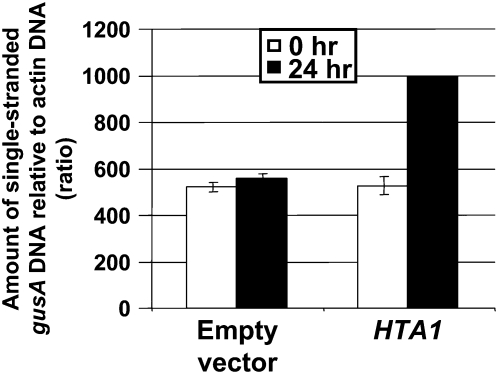
Our transient expression analyses indicated that overexpression of a HTA1 cDNA increased expression of incoming single-stranded or double-stranded gusA DNA when coelectroporated into BY-2 tobacco protoplasts. To test whether histones can protect and stabilize incoming single-stranded DNA, we conducted electroporation experiments similar to those described above. We coelectroporated tobacco BY-2 protoplasts with single-stranded gusA DNA and either a control empty vector or a vector containing a HTA1 cDNA. We treated the protoplasts with DNase I and processed the samples as described above. Figure 10 shows that the accumulation of gusA DNA was twofold greater when a HTA1 cDNA was coelectroporated compared with that following coelectroporation with a control empty vector. Taken together with the results reported above, these data indicate that cotransfection of cells with a HTA1 cDNA results in increased levels of reporter gene DNA introduced either in single- or double-stranded DNA form.
Figure 10.
Accumulation of Single-Stranded gusA DNA in Tobacco BY-2 Protoplasts following Cotransfection with HTA1.
Tobacco BY-2 protoplasts were cotransfected with single-stranded gusA DNA and either a histone cDNA expression cassette or an empty vector control plasmid. At 0 and 24 h after transfection, samples were treated with DNase I and DNA was extracted. PCR was conducted to determine the amount of gusA DNA within the cell, relative to actin DNA. Error bars indicate se of two or three biological replicates.
Histone H2A-1 Cannot Substitute for VirE2 during Agrobacterium-Mediated Plant Transformation
Agrobacterium VirE2 protein binds to single-stranded DNA (Gietl et al., 1987; Christie et al., 1988; Citovsky et al., 1988, 1989; Das, 1988) and protects transferred T-strands from nucleolytic degradation in the plant (Yusibov et al., 1994; Rossi et al., 1996). Because histone H2A-1 may also protect single-stranded T-DNA from degradation, we tested whether a HTA1 gene could compensate for loss of VirE2 during Agrobacterium-mediated plant transformation. We infected root segments of a transgenic Arabidopsis plant containing a HTA1 expression cassette with 108 cfu/mL of an Agrobacterium virE2 mutant and scored the efficiency of tumor formation 4 weeks later. Table 1 shows that the presence of this HTA1 expression cassette could not compensate for loss of VirE2 production by Agrobacterium.
Table 1.
Transgenic Arabidopsis Containing an HTA1 Expression Cassette Cannot Be Transformed by a virE2 Mutant Agrobacterium Strain
| Agrobacterium Straina | Wild-Type Arabidopsis (n)bc | HTA1 Arabidopsis (n)bc |
|---|---|---|
| Agrobacterium A348 | 41.8% (1298) | NDd |
|
Agrobacterium A348∷mx358 |
0.1% (1280) |
0% (1010) |
A348, wild-type A. tumefaciens; mx358, virE2 mutant of A. tumefaciens A348.
Percentage of root segments showing tumors.
Number of root segments analyzed.
ND, not determined.
DISCUSSION
Several Arabidopsis Histone Genes Enhance Agrobacterium-Mediated Transformation and Transgene Expression
The core histones H2A, H2B, H3, and H4 are among the most abundant and highly conserved proteins in eukaryotic organisms (Kornberg, 1977; Luger et al., 1997; Jenuwein and Allis, 2001; Loidl, 2004; Mariño-Ramírez et al., 2005; March-Diaz et al., 2008). A total of 47 core histone genes have been identified in the Arabidopsis genome (www.chromdb.org). Some genes within a family encode evolutionarily conserved variants that display significant differences in primary sequence. Increasing evidence indicates that these histone variants have specialized functions; therefore, their incorporation into chromatin at specific regions of the genome or loci may confer specific regulatory or structural features (Kamakaka and Beggins, 2005; March-Diaz et al., 2008; Jin et al., 2009). Histone H2A has the largest number of variants, which include H2A-X (Arabidopsis HTA3 and HTA5) and H2A-Z (Arabidopsis HTA8, HTA9, and HTA11; Raisner and Madhani, 2006; Yi et al., 2006). In plants, as in other eukaryotes, histone H3 variants include H3.2 and H3.3 (Arabidopsis HTR12), which is found in the centromeric region. The N-terminal tails of these histone H3 variants are extremely divergent and share no sequence similarity with those of the other canonical histones. Histone H4 is the most conserved histone and does not have sequence variants (Zhang et al., 2005).
The results described in this study indicate that particular histone cDNAs enhance both Agrobacterium-mediated transformation and transient transgene expression in plant cells. Tested histone genes that effect these increases include seven HTA, one of five tested HTR (HTR11), and one HFO cDNAs. However, none of the seven tested HTB, nor four of the five tested HTR cDNAs, substantially (more than twofold) increase either transformation or transgene expression. Importantly, there was a perfect correlation between those histones that increased transformation and those that increased transgene expression (Figures 1 and 2; see Supplemental Figure 2 online). In addition, the increase in transient GUS activity caused by HTA1 was associated with a parallel increase in gusA mRNA in tobacco BY-2 protoplasts (Figure 3).
Mutation of HTA1 in Arabidopsis severely reduced integration of T-DNA into the plant genome, thus causing a rat phenotype (Mysore et al., 2000a). Mutant Arabidopsis lines containing T-DNA insertions in or near several other core histone genes, including HTA2, HTA3, HTA11, HTB5, HTB6, HTR5/4, HFO3, and HFO4, also reduced transformation frequency to various extents (Zhu et al., 2003b), although we have not yet determined the specific stage of transformation that was disrupted. Our observations that overexpression of HTA1 increased transformation frequency (Mysore et al., 2000a; Yi et al., 2006) suggested to us that increased T-DNA integration may be the cause of this hypersusceptibility to Agrobacterium-mediated transformation. However, the results of this study and other work (L.-Y. Lee, S.J. Johnson, X. Sui, and S.B. Gelvin, unpublished data) indicate that although overexpression of several core histone genes may affect T-DNA integration, a major effect is to increase transgene expression.
Histone-Mediated Enhancement of Transgene Expression Results from an Increase in Incoming Transgene DNA
Our data suggest that the enhancement of transgene expression and Agrobacterium-mediated transformation effected by overexpression of particular histones results from increased stability of DNA in the plant cell, possibly by protecting incoming DNA from nuclease degradation (Figure 7). This explanation is consistent with our observations that expression of previously integrated transgenes cannot be increased (or activated when silenced) by overexpression of histone genes. Our observation that gusA DNA actually increases over time in transfected BY-2 protoplasts suggests that this DNA is replicating within the plant cells and that particular histones either protect the template from nuclease degradation or serve a more active role in recruiting replication enzymes to the incoming DNA. Because T-DNA is introduced from Agrobacterium into plant cells as a single-stranded DNA form and must integrate into and replicate along with plant chromosomal DNA, conversion of single-stranded T-strand DNA to a double-stranded DNA form is necessary. Conversion of T-strands into a double-stranded form prior to T-DNA integration has been well documented (Narasimhulu et al., 1996; Vergunst and Hooykaas, 1998).
Histones are basic proteins that are known for their DNA binding capacity. They may interact with DNA via electrostatic interactions between the negatively charged phosphate DNA backbone and numerous positively charged Lys and Arg residues. We conducted our protoplast electroporation analyses using two different gusA transgene substrates: single- or double-stranded transgene DNA. In the nucleus, histones are usually associated with the formation of nucleosomes on double-stranded DNA (Luger et al., 1997; Luger and Richmond, 1998; Carruthers and Hansen, 2000; Mariño-Ramírez et al., 2005). However, the ability of histones to interact with single-stranded DNA has been well documented (Palter et al., 1979; Schlaeger and Knippers, 1979).
We have shown that our HTA1 cDNA expression cassette generates HTA1 transcripts both in transgenic Arabidopsis roots and in tobacco BY-2 protoplasts (Figures 1C and 3A). We have additionally demonstrated that we have increased histone H2A-1 protein in transfected BY-2 cells. Antibodies directed against histone H2A-1 would likely cross-react with endogenous H2A proteins. We therefore generated a construction with a T7 peptide tag on the C terminus of H2A-1. When transformed into the rat5 (HTA1) mutant, this construction complements the mutant HTA1 gene and restores transformation competence to rat5 roots. It also increases gusA transgene expression in transfected protoplasts to the same extent as does untagged H2A-1 (see Supplemental Figure 3 online). Thus, the H2A-1-T7 fusion protein is biologically active for both processes examined in this study, crown gall tumorigenesis and increased transgene expression. Additionally, our ability to visualize GUS and YFP fusions to mutant histone H2A proteins strongly suggests that these mutant proteins are stable in plant cells.
Our studies indicated that particular histones, including H2A-1, H3-11, and H4-3, have the ability to stabilize incoming gusA DNA to a greater extent than do histones H2B-1 or H3-4 (Figure 8A). However, there does not appear to be any consistent properties of these histones that would explain why certain ones increase transformation and transgene expression and others do not. For example, particular histones can form interacting dimers (H2A-H2B and H3-H4). In addition, histones H3 and H4 are more basic, and their amino acid sequences are more highly conserved than are histones H2A and H2B. Thus, the least and most basic histones (H2A and H4) are those that increase Agrobacterium-mediated transformation and transgene expression to the greatest extent. We note, however, that histone H3-11, the only histone H3 variant that increased transformation and transgene expression, is also the only tested H3 variant that lacks Lys residues at positions 9 and 27. Methylation of these Lys residues is often associated with repressive chromatin (Gendrel et al., 2002; Li et al., 2008). We also note that residue R4 in both histones H2A and in histone H4 can potentially be methylated or acetylated (Branscombe et al., 2001; Strahl et al., 2001; Wang et al., 2001; Hyllus et al., 2007; Pei et al., 2007). Modification of these important residues may influence the ability of these histones to affect the level of transformation or transgene expression. We are currently conducting experiments to test this hypothesis.
Increased transgene expression and stabilization of incoming transgene DNA may not necessarily depend upon nuclear localization of histones. Translational fusions of full-length H2A-1 to GUS or YFP reporter proteins localized predominantly to the nucleus, whereas peptides containing only the H2A-1 N-terminal domain localized to both the nucleus and to the cytoplasm (Figure 5). Both full-length and the N-terminal region of H2A-1 enhance transgene expression and stabilize incoming transgene DNA. Because full-length H2A-1 localized predominantly to nuclei at early transfection times, our data suggest that the protective effect of H2A-1 (and other histone proteins) may additionally extend to the nuclear compartment.
Structural analyses indicate that the H2A N-terminal domain contains motifs involved in both histone–histone interactions and histone–DNA interactions (Luger et al., 1997). The crystal structure of the core histone octamer indicates that the N-terminal region of H2A constitutes one of the larger DNA interaction surfaces. This region forms two short α-helices (residues 17 to 21 and 27 to 37) interrupted by a short loop that allows the two helices to anchor three adjacent phosphates along one strand of the DNA (Luger et al., 1997). The importance of this N-terminal domain in H2A–DNA interactions may explain the enhanced rate of accumulation of gusA DNA when coelectroporated with cDNAs encoding the full-length or the N-terminal 39–amino acid fragment of histone H2A-1. When domains of basic amino acids (K14K15R19K22 and R31R34K37) important for H2A–DNA interaction were converted to Ala, the resulting N-terminal H2A peptide lost the ability to protect incoming transgene DNA and to increase transgene expression. These functions were not altered by conversion of the amino acids R19S20S21K22 (which play a role in histone–histone interactions) to Ala. We speculate that the N-terminal region of H2A-1 in particular protects incoming gusA transgene DNA, thus providing increased template for transgene expression.
Histones Can Increase Transgene Expression in Several Transfection Systems
Plant histone H2A proteins form a distinct group that contains conserved N-terminal and C-terminal regions. The N-terminal region 39 amino acids are important for DNA–histone and histone–histone interactions within and/or between nucleosomes and for interaction with nonhistone proteins (Ren and Govorsky, 2003).
Previous studies in animal systems have shown the importance of the first 37 amino acids of histone H2A (corresponding to the first 39 amino acids of Arabidopsis H2A-1) for increasing the efficiency of DNA delivery and enhancing expression of a transgene when different histone peptides were transfected along with a reporter gene in cell cultures (Balicki et al., 2002). Two mechanisms were suggested as being responsible for this effect: (1) electrostatically driven DNA binding and condensation by histones and (2) nuclear import of histone H2A-DNA polyplexes via nuclear localization signals in the protein. Transfection of DNA in a complex with histone H1 also increases the efficiency of delivery of DNA, RNA, and small interfering RNA into immortalized and primary cells; the transfection efficiency using histone H1 was comparable to or even greater than that of liposome-based systems (Puebla et al., 2003). These authors assessed the ability of various histone H1 fragments to condense DNA and to mediate transgene delivery into primary and established neuroblastoma cells. H1.4F, a fragment containing the N-terminal and middle portions of the protein corresponding to amino acids 1 to 129, mediated the highest transfection efficiency. Other studies demonstrated that histone H2A, but not H2B, H3, or H4, could increase transgene transfection frequency in a number of animal cell lines (Balicki et al., 2002; Kaouass et al., 2006).
The N-terminal domain of human histone H2A amino acids 1 to 26 or 1 to 37 (fused to a β-galactosidase reporter protein) localized to the nucleus of animal cells (Baake et al., 2001). By contrast, GUS and YFP fusions to the N-terminal domain (amino acids 1 to 39) of Arabidopsis histone H2A-1 fractionate to both the nucleus and cytoplasm. Human histone H2A contains nuclear localization signal sequences that interact with importin/transportin proteins that mediate nuclear import (Baake et al., 2001; Mulhausser et al., 2001). These nuclear localization signals map to both the N-terminal tail and central core domains.
In addition to functions involving nuclear localization, the N-terminal domain of histone H2A plays critical roles in regulating chromatin structure and gene transcription. Genome-wide expression profiling was used to identify yeast genes whose expression was altered by mutation or deletion of the N-terminal domain of histone H2A. These experimental results indicated that a small subdomain, 16 to 20 amino acid residues from the N terminus, is required for transcriptional repression of a BNA2 reporter gene (Parra and Wyrick, 2007). Our results indicate that specific regions within the N-terminal region of H2A-1 are important for increasing both transgene expression and stability. For example, mutation of basic amino acids, such as the K14K15R19K22 and R31R34K37 motifs, which correspond to the DNA binding domain region, affect transgene expression more than does mutation of Lys-6 (a potential acetylation site) or R19S20S21K22 (a motif important for histone–histone interactions). It is interesting to note that Arg-4, an amino acid important for the ability of the N-terminal peptide of H2A-1 to enhance transgene expression, can be both methylated and acetylated (Branscombe et al., 2001; Strahl et al., 2001; Hyllus et al., 2007; Pei et al., 2007). These results suggest that the ability of the N-terminal domain of H2A-1 to increase transgene expression is more related to functions important for DNA binding than to the overall net positive charge of the peptide.
Overexpression of a HTA1 cDNA Can Increase Expression of a Single-Stranded Coelectroporated gusA Transgene
Several studies indicate that T-DNA is transferred to plants as a single-stranded molecule that is then made double-stranded in the nucleus prior to or during the process of integration into the plant genome (Tinland, 1996; Tzfira and Citovsky, 2000; Brunaud et al., 2002; Gelvin 2003a). Following transfer of T-DNA to the plant cytoplasm, it likely forms complexes with various transferred Agrobacterium virulence effector proteins, including VirE2, a single-stranded DNA binding protein (Gietl et al., 1987; Christie et al., 1988; Citovsky et al., 1988, 1992; Das, 1988) and VirF, which may mediate proteolysis of VirE2 in the nucleus (Schrammeijer et al., 2001; Tzfira et al., 2004). In addition, plant-encoded proteins, such as VIP1 and importin α, may form supercomplexes with the T-strand and help target T-DNA to the nucleus (Ballas and Citovsky, 1997; Tzfira et al., 2001; Bhattacharjee et al., 2008; Lee et al., 2008). The timing and subcellular location of VirE2 interaction with T-strands remains unknown. Although some evidence suggests that VirE2 may interact with T-strands in the cytoplasm and help direct nuclear localization, other data suggest that VirE2 may remain in the plasma membrane and pick up T-strand DNA as it enters the plant cell (Dumas et al., 2001; Duckely and Hohn, 2003; Duckely et al., 2005). It is possible that overexpressed histones can also interact with T-strands and help in both nuclear targeting and protection against nucleolytic degradation. This model is consistent with our protoplast electroporation data. Although certain histones can stabilize T-DNA within the plant cell, they cannot fulfill all the roles of VirE2 because a histone H2A-1 overexpressing plant could not compensate for loss of VirE2 from during Agrobacterium-mediated transformation (Table 1).
Our past results indicated that histone H2A-1 is important for stable but not transient transformation (Mysore et al., 2000a). The results described in this article, which indicate that overexpression of particular histones increases Agrobacterium-mediated transformation and transient transgene expression by protecting incoming transgene DNA, therefore pose a conundrum. If wild-type levels of histones other than H2A-1 are sufficient for transient Agrobacterium-mediated transformation (as they are in the rat5 mutant), why are they not sufficient for stable transformation? Although our data do not specifically address this paradox, they suggest that histone H2A-1, in addition to its protective effect, may also play a role in T-DNA integration. Thus, histones may play one role in the context of the plant cytoplasm and nucleoplasm and another role in the context of plant chromatin.
Genetic transformation can be defined as a process by which DNA is introduced into a cell. Transformation can be transient (i.e., the DNA does not integrate into the host genome and transgene expression is eventually lost) or stable (i.e., the introduced DNA integrates into the host chromosomes and may, unless silenced, express from this integrated position). Increased transgene expression (especially of reporter and selection marker genes) can give the appearance of increased transformation if such expression allows more facile recovery of transgenic events. We propose that hyperexpression of certain histone genes increases expression of incoming transgenes by protecting the DNA, permitting the DNA to express to a greater extent and thus facilitating the selection of transformed cells. Additionally, increased expression of certain histone genes may protect incoming T-DNA transferred from Agrobacterium to plant cells, thus providing more substrate DNA for integration into the host genome and, consequently, resulting in a higher likelihood of stable transformation.
METHODS
Cloning Histone cDNAs
cDNAs for the histone genes HTA1 (E1701), HTA6 (E2272), and HTA8 (E2267) were excised from their respective pBluescript vectors (Yi et al., 2006) using EcoRI and XbaI and cloned into the same sites of the T-DNA binary vector pE2786 (see below). cDNAs for the histone genes HTB1 (C00142), HTB3 (U14625), HTR4 (U12616), and HFO3 (U1196) were obtained from the Arabidopsis Stock Center (The Ohio State University), excised from their respective vectors using EcoRI and XbaI, and cloned into these same sites of pE2786. Other histone cDNAs were PCR amplified and cloned into pBlueScript KS− as described by Yi et al. (2006). cDNAs were amplified using either previously published primers (Yi et al., 2006) or primers listed in Supplemental Table 1 online.
Plasmid Constructions
Arabidopsis thaliana histone cDNAs were cloned into the T-DNA binary expression vector pE2786 as described in Supplemental Table 1 online. pE2786 was constructed by cloning a PstI fragment from pRTL2-GUS (E1295; Restrepo et al., 1990), containing a cauliflower mosaic virus double 35S promoter-gusA-CaMV 35S terminator expression cassette, into the PstI site of pCB302 (Xiang et al., 1999). Histone cDNA clones were cloned into the EcoRI-XbaI sites of this plasmid, replacing the gusA gene with the histone cDNAs. These binary vectors were separately introduced into Agrobacterium tumefaciens GV3101 (Koncz and Schell, 1986) by electroporation.
Mutagenesis of the HTA1 cDNA and Generation of GUS and YFP Fusion Proteins
All histone H2A-1 variants were generated by mutagenesis of particular amino acids to Ala using a Promega GeneEditor in vitro site-directed mutagenesis system (kit Q9280). Mutagenic oligonucleotides and primers used to amplify various portions of HTA1 are listed in Supplemental Table 2 online. Each histone mutation was confirmed by sequencing. The amplified fragments containing the various mutations were fused with a gusA gene that had been amplified using the primers 5′-AAACTAGTATGGTCCGTCCTGTAGAAACC-3′ and 5′-AATCTAGATCATTGTTTGCCTCCCTGCTG-3′ and subsequently cloned into the SmaI site of pBluescript KS− (from which the SpeI site had been removed). This was accomplished by digestion of the mutated amplicons with SalI and SpeI and cloning into the SalI and SpeI sites of the gusA plasmid described above. Supplemental Table 1 online describes the plasmids used for transfection experiments. The resulting HTA1-gusA fusion genes were released from these plasmids by digestion with SalI and XbaI and cloned into the corresponding sites of the T-DNA binary vector pE1774 under the control of a superpromoter (Lee et al., 2007). These plasmids were introduced into Agrobacterium EHA105 (Hood et al., 1993) by electroporation and used to generate stable tobacco (Nicotiana tabacum) BY-2 transgenic lines. YFP fusions to full-length and the N-terminal H2A-1 proteins were made as follows: HTA1 full-length and HTA1 N-terminal cDNAs were excised from pE2924 and pE2932, respectively, using SalI and SpeI. The 5′ overhanging nucleotides were made blunt using the Klenow fragment of DNA polymerase and deoxynucleotide triphosphate. The plasmid pSAT6-EYFP-N1 (pE3225; Citovsky et al., 2006) was digested with XhoI and BamHI, and the 5′ overhanging nucleotides filled in similarly. The blunted HTA1 full-length and N-terminal coding regions were ligated into the blunted pSAT6-EYFP-N1 plasmid, generating pSAT6-H2A-EYFP (pE3467) and pSAT6-39aaH2A-EYFP (pE3468).
Generation of Arabidopsis Plants Overexpressing Histone cDNAs
Agrobacterium strains containing the histone cDNA expression cassettes in the binary vector pE2786 were used to generate transgenic Arabidopsis plants (ecotype Ws) using a floral dip method (Clough and Bent, 1998). T0 generation seeds were germinated on B5 medium (GIBCO) containing 100 μg/mL timentin (GlaxoSmithKline) and 20 μg/mL phosphinothricin (ppt; Duchefa) to select for transgenic plants.
Root Tumorigenesis Assay
We conducted root tumorigenesis assays as described previously (Nam et al., 1999; Mysore et al., 2000b; Yi et al., 2002), with the exception that the infecting bacterial concentration was either 105 or 106 cfu/mL. In brief, Arabidopsis plants, initially selected on B5 medium containing timentin and ppt, were grown in baby food jars containing B5 medium for 2 to 3 weeks at 25°C. Root segments (2 to 5 mm) were infected with the tumorigenic strain Agrobacterium A208. After 2 d cocultivation, root segments were separated onto solidified MS medium (Murashige and Skoog, 1962) lacking phytohormones but containing 100 μg/mL timentin. Tumors were scored 3 to 4 weeks later. To investigate whether overexpression of HTA1 could compensate for loss of Agrobacterium VirE2, root segments of wild-type (ecotype Ws) or HTA1-overexpressing plants were inoculated with 108 cfu wild-type Agrobacterium A348 or the virE2 mutant Agrobacterium A348∷mx358 (Stachel and Nester, 1986). Tumorigenesis was determined as described above.
Analysis of Transient GUS Expression in Tobacco BY-2 Protoplasts
Protoplasts were isolated from 5-d-old N. tabacum BY-2 suspension cells. Suspension cultures were grown at 23°C with shaking (130 rpm) in a medium containing MS salts supplemented with 1 mg/L thiamine-HCl, 370 mg/L KH2PO4, 30 g/L sucrose, and 2 mg/L 2,4-D, pH 5.7. Cells were subcultured once per week by adding 2.5 to 3 mL of inoculum to 50 mL of fresh medium in 250-mL Erlenmeyer flasks. The 50 mL of suspension culture was centrifuged at 500 rpm at room temperature for 5 min in a Sorvall GLC-2 centrifuge, and the pellet (15 mL packed cell volume) was resuspended in 50 mL of protoplast isolation solution containing 7.4 g/L CaCl2·2H2A, 1 g/L NaOAc, and 45 g/L mannitol supplemented with 1.2% cellulose R10 (Onazuka) and 0.6% Macerozyme (Duchefa), pH 5.7. Approximately 15 mL of suspension culture was transferred into three 20 × 100-mm sterile Petri dishes and incubated in the dark with a gentle shaking (40 rpm) at room temperature for 4 h. The protoplasts were washed twice with protoplast isolation solution, and the pellet was resuspended in 50 mL of floating solution (99 mg/L myo-inositol, 2.88 g/L l-proline, 100 mg/L enzymatic casein hydrolysate, 102.6 g/L sucrose, 97.6 mg/L MES buffer, 4.3 g/L MS salts, 1 mg/L thiamine-HCl, and 370 mg/L KH2PO4, pH 5.7). Protoplasts (floating on the top of the solution) were transferred to a new tube and resuspended in 50 mL of electroporation solution (10 mM NaCl, 4 mM CaCl2·2H2O, 120 mM KCl, 10 mM HEPES, and 0.6 M mannitol, pH 7.2). Aliquots of protoplasts containing ∼3 × 106 cells/mL were used for electroporation. Thirty to fifty micrograms of each plasmid DNA were added to 300 μL of protoplasts in a tube and placed on ice. The electroporation was conducted using a Bio-Rad Gene Pulser apparatus at 0.16 kV, with the pulse controller set to infinity and the capacitance extender set to 960 μFD. After 10 min incubation on ice, the protoplasts were transferred into 10 mL of BY-2 culture medium supplemented with 0.4 M mannitol and incubated overnight. The cells were collected in tubes and assayed for GUS activity 12 to 24 h after transfection.
Histochemical Assay for GUS Activity
Histochemical staining was performed according to Jefferson et al. (1987) with modifications. Protoplasts were incubated in GUS staining solution composed of 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucoronide (X-gluc), 100 mM sodium phosphate, pH 7.0, 0.1 mM K3[Fe(CN6)], 0.1 mM K4[Fe(CN6)], 0.3 M mannitol, and 4.3 g/L MS salts and incubated at 37°C overnight. Stained cells were visualized using a Nikon Optiphot phase contrast microscope and scored for GUS expression by counting blue staining cells. For each transfection experiment, a minimum of 1000 cells was scored; each experiment was repeated three times. For subcellular localization of various HTA1-gusA constructs, tobacco BY-2 protoplasts were transfected with 50 μg DNA and the cells were stained with X-gluc after 1, 2, or 3 d as described by Mysore et al. (1998).
Quantitative GUS Fluorimetric Assay
GUS activity was measured by monitoring cleavage of 4-methylumbelliferyl β-d-glucuronide (4-MUG; Jefferson et al., 1987; Gallagher, 1992). Total protein was isolated by grinding the protoplasts in extraction buffer (50 mM phosphate buffer, pH 7.0, 10 mM 2-mercaptoethanol, 10 mM Na2EDTA, and 0.1% Triton X-100). The extract was centrifuged at 15,000g at 4°C for 10 min. The supernatant solution was collected and the protein concentration was measured according to Bradford (1976). All protein extracts were stored at −80°C prior to performing fluorimetric analysis. Ten micrograms of total protein was added to each well of a 96-well microtiter plate containing GUS assay buffer (4 mM 4-MUG in extraction buffer) in a 200-μL total reaction volume. The reaction was monitored using a Gemini XSP spectrophotometer plate reader (Molecular Devices) at 37°C with fluorescence measurements each 15 min for 2 h. The excitation and emission wavelengths were 365 and 455 nm, respectively. Each assay was performed in duplicate. Relative GUS activity was calculated as the ratio between the average fluorescence intensity (slope of the line denoting the rate of cleavage of 4-MUG) of each line divided by the fluorescence intensity (slope) of the empty vector control.
Determination of gusA and HTA1 mRNA Levels in Tobacco BY-2 Protoplasts and Arabidopsis Roots
Total RNA was isolated from BY-2 protoplasts after transfection and from roots of Arabidopsis plants expressing HTA1 using TRIzol reagent (Life Technologies). To remove contaminating genomic DNA, total RNA (6 μg) was incubated with 2 units TURBO DNaseI (Ambion) for 30 min at 37°C. First-strand cDNA was synthesized using a Promega reverse transcription system according to the manufacturer's instructions. The PCR reaction was performed in a 50-μL total volume with primers specific for the gusA or HTA1 transgenes. RT-PCR using primers directed against actin mRNA was used as internal standard. Primer sequences were as follows: for gusA, 5′-AGCGTATCGTGCGTTTCG-3′ and 5′-AGAGGTGCGGATTCACCACTTGC-3′; for HTA1, 5′-GCTTCTTGGAGATGTGACGATTGC-3′ and 5′-GAGAGACGTGTTCTATATCATTGTGT-3′ for detection of HTA1 transcripts in BY-2 tobacco protoplasts; or 5-CAATGGCTGGTCGTGGAAAAAC-3′ and 5′-CGGCAACAGGATTCAATCTTAAGAAA-3′ for detection of HTA1 transcripts in Arabidopsis roots overexpressing HTA1; for actin, 5′-CTAAGCTCTCAAGATCAAAGGCTTA-3′ and 5′-ACTAAAACGCAAAAGGAAAGCGGTT-3′. Each gene was amplified using the following conditions: 94°C for 5 min, followed by 25 to 40 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 4 min. PCR products were separated by electrophoresis through 1.0% agarose gels and photographs of the ethidium bromide–stained gel scanned using a UVP Bio Imaging System (GDS-8000 System). Each assay was conducted with a minimum of three biological replicates. To assure that reactions were within the linear range of amplification, samples were analyzed after 20, 25, and 30 cycles. Results were similar at each of these amplification cycles.
Determination of HTA1-T7 Protein Levels in Tobacco BY-2 Protoplasts
Protoplasts were coelectroporated with a HTA1-YFP construct and plasmids designed to express either an untagged or a T7-tagged HTA1 gene. Twenty-four hours after transfection, protoplasts were collected and lysed in 2× protein loading buffer (100 mM Tris-HCl, pH 6.8, 200 mM DTT, 4% SDS, and 20% glycerol). The samples were boiled for 10 min, and the supernatant solution was used for determination of protein concentration. Total protein (2.5 μg) was fractionated by electrophoresis through a 15% SDS-polyacrylamide gel and blotted onto a nitrocellulose membrane (PALL; Gelman Laboratory). Detection of the T7-tagged H2A-1 protein was performed using T7-Tag antibody HRP conjugate (Novagen) and visualized using Western Blotting Luminol Reagent (Santa Cruz Biotechnology).
Determination of gusA DNA Levels in Tobacco BY-2 Protoplasts
Protoplasts were electroporated with a gusA expression cassette and either a histone cDNA expression cassette or an empty vector control plasmid as described above. Cells (1 to 2.5 × 105) were collected at various times (0, 6, 12, and 24 h) after transfection and were treated with 10 μg/mL DNaseI in BY-2 cell suspension media supplemented with 20 mM MgCl2 in order to digest DNA outside of the cells. The reaction was performed for 30 min at room temperature, following which the cells were washed twice in 1 mL BY-2 media containing 20 mM EDTA. DNA was isolated by a CTAB method as described by Doyle and Doyle (1990). PCR was used to measure the amount of gusA DNA in each sample. To normalize the reaction for total DNA, we conducted PCR using primers directed against a tobacco actin gene. The primers (forward and reverse) used were as follows: for gusA, 5′-AGCGTATCGTGCGTTTCG-3′ and 5′-AGAGGTGCGGATTCACCACTTGC-3′; for actin, 5′-AGTGGCGGTTCGACTATGTTTCC-3′ and 5-TCTGCCTTTGCAATCCACATCTGC-3′. Each gene was amplified using the following conditions: 94°C for 5 min, followed by 25 to 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 4 min. The PCR products were fractioned by electrophoresis through a 1% agarose gel, stained with ethidium bromide, and the DNA was visualized on a UV light box. The amount of DNA in each sample was measured after scanning a photograph of the gel with a UVP Bio Imaging System (GDS-8000) and using Lab Works 4.1 software. Each assay was conducted with a minimum of three biological replicates. To assure that reactions were within the linear range of amplification, samples were analyzed after 20, 25, and 30 cycles. Results were similar at each of these amplification cycles. The amount of gusA DNA from each sample was normalized to the amount of actin DNA. To investigate whether overexpression of HTA1 would increase the amount of gusA DNA introduced as single-stranded DNA, we performed the same analysis as described above. However, the gusA plasmid was first digested with HindIII to excise the gusA expression cassette, followed by DNA purification, denaturation for 5 min at 95°C, and incubation for 5 min on ice prior to electroporation.
Nuclear Localization of Wild-Type and Mutant Histone H2A-1 Peptides
We generated translational fusions between wild-type or mutant histone HTA1 genes and either a gusA-intron gene or an enhanced YFP gene. The construction of these plasmids is described in Supplemental Table 1 online. The gusA-intron and enhanced YFP genes are described by Narasimhulu et al. (1996) and Citovsky et al. (2006), respectively. Agrobacterium EHA105 containing the various histone HTA1-gusA cDNAs, described in Supplemental Table 2 online, were used to transform tobacco BY-2 cells. After transformation, transgenic cells were selected on media containing 20 μg/mL hygromicin and assayed for localization of GUS activity by staining with X-gluc as described above. Cells were visualized using an Olympus VANOX-T AH-2 light microscope and photographed using a SPOT RT CCD camera (Diagnostic Instruments). Plasmids encoding histone-enhanced YFP fusion proteins were individually electroporated into tobacco BY-2 protoplasts and visualized using a Nikon Eclipse E600 fluorescence microscope or a Zeiss LSM510 Meta confocal microscope 24 to 72 h later.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HTA1, Entrez GeneID 835553, Locus Link At5g54640; HTA2, Entrez GeneID 828831, Locus Link At4g27230; HTA3, Entrez GeneID 841910, Locus Link At1g54690; HTA5, Entrez GeneID 837409, Locus Link At1g08880; HTA6, Entrez GeneID 836109, Locus Link At5g59870; HTA8, Entrez GeneID 818463, Locus Link At2g38810; HTA10, Entrez GeneID 841528, Locus Link At1g51060; HTB1, Entrez GeneID 837293, Locus Link At1g07790; HTB3, Entrez GeneID 817421, Locus Link At2g28720; HTB5, Entrez GeneID 818324, Locus Link At2g37470; HTB8, Entrez GeneID 837338, Locus Link At1g08170; HTB9, Entrez GeneID 823741, Locus Link At3g45980; HTB10, Entrez GeneID 831878, Locus Link At5g02570; HTB11, Entrez GeneID 823746, Locus Link At3g46030; HTR1, Locus Link At5g65360; HTR4, Entrez GeneID 830164, Locus Link At4g40030; HTR6, Entrez GeneID 837897, Locus Link At1g13370; HTR11, Entrez GeneID 836661, Locus Link At5g65350; HTR13, Entrez GeneID 830903, Locus Link At5g10390; HFO3, Entrez GeneID 817423, Locus Link At2g28740.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Arabidopsis Core Histone Protein Sequence Alignment.
Supplemental Figure 2. Agrobacterium-Mediated Transformation of Transgenic Arabidopsis Plants Containing cDNAs Encoding Various Core Histone Genes.
Supplemental Figure 3. A HTA1-T7 Transgene Can Restore Transformation Susceptibility to the rat5 hta1 Mutant, and Both HTA1 and HTA1-T7 Genes Increase gusA Transgene Expression to Approximately the Same Extents.
Supplemental Figure 4. PCR Analysis of gusA Transgene DNA in Transfected Tobacco BY-2 Protoplasts.
Supplemental Table 1. T-DNA Binary Vector Histone Strains and Primers to Amplify Histone cDNAs.
Supplemental Table 2. Mutagenic Oligonucleotides and Primers Used to Amplify HTA1 Gene Fragments.
Supplemental Table 3. Histone H2A-1 Mutation Strains.
Supplementary Material
Acknowledgments
We thank Simran Bhullar for generating and assaying some of the initial histone cDNA transgenic plants, Nagesh Sardesai for helping with the microscopy, and Walt Ream for critical reading of the manuscript. This work was supported by grants from the National Science Foundation (2010-0418709-MCB), the Biotechnology Research and Development Corporation, the Consortium for Plant Biotechnology Research, and a P30 grant to the Purdue Cancer Center. G.N.T. was supported in part by a Fulbright Foundation Postdoctoral Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stanton B. Gelvin (gelvin@bilbo.bio.purdue.edu).
Online version contains Web-only data.
References
- Anand, A., Zarir, V., Ryu, C.M., Kang, L., del-Pozo, O., Martin, G.B., and Mysore, K.S. (2007). Identification of plant genes involved in Agrobacterium-mediated transformation by using virus induced gene silencing as a functional genomics tool. Mol. Plant Microbe Interact. 20 41–52. [DOI] [PubMed] [Google Scholar]
- Baake, M., Doenecke, D., and Albig, W. (2001). Characterisation of nuclear localisation signals of the four human core histones. J. Cell. Biochem. 81 333–346. [PubMed] [Google Scholar]
- Balicki, D., Putnam, C.D., Scaria, P.V., and Beutler, E. (2002). Structure and function correlation in histone H2A peptide-mediated gene transfer. Proc. Natl. Acad. Sci. USA 99 7467–7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas, N., and Citovsky, V. (1997). Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. USA 94 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Lee LY, Oltmanns H, Cao H, Veena, Cuperus J, Gelvin SB. (2008). AtImpa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 20 2661–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Branscombe, T.L., Frankel, A., Lee, J.H., Cook, J.R., Yang, Z., Pestka, S., and Clarke, S. (2001). PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276 32971–32976. [DOI] [PubMed] [Google Scholar]
- Brunaud, V., et al. (2002). T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 3 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers, L., and Hansen, J.C. (2000). The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275 37285–37290. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Henderson, I.R., and Jacobsen, S.E. (2007). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360. [DOI] [PubMed] [Google Scholar]
- Christie, P.J., Ward, J.E., Winans, S.C., and Nester, E.W. (1988). The Agrobacterium tumefaciens virE2 gene product is a single-stranded DNA-binding protein that associates with T-DNA. J. Bacteriol. 170 2659–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V., De Vos, G., and Zambryski, P. (1988). Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science 240 501–504. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Kozlovsky, S.V., Lacroix, B., Zaltsman, A., Dafny-Yelin, M., Vyas, S., Tovkach, A., and Tzfira, T. (2007). Biological systems of the host cell involved in Agrobacterium infection. Cell. Microbiol. 9 9–20. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Lee, L.-Y., Vyas, S., Glick, E., Chen, M.-H., Vainstein, A., Gafni, Y., Gelvin, S.B., and Tzfira, T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362 1120–1131. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Wong, M.L., and Zambryski, P. (1989). Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc. Natl. Acad. Sci. USA 86 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V., Zupan, J., Warnick, D., and Zambryski, P. (1992). Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256 1802–1805. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Crane, Y.M., and Gelvin, S.B. (2007). RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. USA 104 15156–15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A. (1988). Agrobacterium tumefaciens virE operon encodes a single-stranded DNA-binding protein. Proc. Natl. Acad. Sci. USA 85 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, C.D., Lee, E., Kobayashi, J., Holappa, L.D., Albert, H., and Ow, D.W. (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, J.L. (1990). Isolation of plant DNA from fresh tissue. Focus 12 13–15. [Google Scholar]
- Duckely, M., Oomen, C., Axthelm, F., Van Gelder, P., Waksman, G., and Engel, A. (2005). The VirE1VirE2 complex of Agrobacterium tumefaciens interacts with single-stranded DNA and forms channels. Mol. Microbiol. 58 1130–1142. [DOI] [PubMed] [Google Scholar]
- Duckely, M., and Hohn, B. (2003). The VirE2 protein of Agrobacterium tumefaciens: The Yin and Yang of T-DNA transfer. FEMS Microbiol. Lett. 223 1–6. [DOI] [PubMed] [Google Scholar]
- Dumas, F., Duckely, M., Pelczar, P., Van Gelder, P., and Hohn, B. (2001). An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc. Natl. Acad. Sci. USA 98 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M., Ishikawa, Y., Osakabe, K., Nakayama, S., Kaya, H., Araki, T., Shibahara, K., Abe, K., Ichikawa, H., Valentine, L., Hohn, B., and Toki, S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25 5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, S.R. (1992). Quantitation of GUS activity by fluorometry. In GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, S.R. Gallagher, ed (San Diego: Academic Press), pp. 47–59.
- Gelvin, S.B. (2000). Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 223–256. [DOI] [PubMed] [Google Scholar]
- Gelvin, S.B. (2003. a). Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin, S.B. (2003. b). Improving plant genetic engineering by manipulating the host. Trends Biotechnol. 21 95–98. [DOI] [PubMed] [Google Scholar]
- Gelvin, S.B. (2009). Agrobacterium in the genomics age. Plant Physiol. 150 1665–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin, S.B., and Kim, S.-I. (2007). Effect of chromatin upon Agrobacterium T-DNA integration and transgene expression. Biochim. Biophys. Acta 1769 410–421. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.-V., Lippman, Z., Yordan, C., Colot, V., and Marteinssen, R. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gheysen, G., Villarroel, R., and Van Montagu, M. (1991). Illegitimate recombination in plants: A model for T-DNA integration. Genes Dev. 5 287–297. [DOI] [PubMed] [Google Scholar]
- Gietl, C., Koukolikova-Nicola, Z., and Hohn, B. (1987). Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc. Natl. Acad. Sci. USA 84 9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, E.E., Gelvin, S.B., Melchers, L.S., and Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2 208–218. [Google Scholar]
- Hyllus, D., Stein, C., Schnabel, K., Schiltz, E., Imhof, A., Dou, Y., Hsieh, J., and Bauer, U.M. (2007). PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21 3369–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1064–1080. [DOI] [PubMed] [Google Scholar]
- Jin, C., Zang, C., Wei, G., Cui, K., Peng, W., Zhao, K., and Felsenfeld, G. (2009). H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet. 10.1038/ng.409. [DOI] [PMC free article] [PubMed]
- Kamakaka, R.T., and Beggins, S. (2005). Histone variants: Deviants? Genes Dev. 19 295–316. [DOI] [PubMed] [Google Scholar]
- Kaouass, M., Beaulieu, R., and Balicki, D. (2006). Histonefection: Novel and potent non-viral gene delivery. J. Control. Release 113 245–254. [DOI] [PubMed] [Google Scholar]
- Kim SI, Veena, Gelvin SB. (2007). Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 51 779–791. [DOI] [PubMed] [Google Scholar]
- Kohli, A., Twyman, R.M., Abranches, R., Wegel, E., Stoger, E., and Christou, P. (2003). Transgene integration, organization and interaction in plants. Plant Mol. Biol. 52 247–258. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kornberg, R.D. (1977). Structure of chromatin. Annu. Rev. Biochem. 46 931–954. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128 693–705. [DOI] [PubMed] [Google Scholar]
- Lacroix, B., Li, J., Tzfira, T., and Citovsky, V. (2006). Will you let me use your nucleus? How Agrobacterium gets its T-DNA expressed in the host plant cell. Can. J. Physiol. Pharmacol. 84 333–345. [DOI] [PubMed] [Google Scholar]
- Lee, L.-Y., Fang, M.-J., Kuang, L.-Y., and Gelvin, S.B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L.-Y., Kononov, M.E., Bassuner, B., Frame, B.R., Wang, K., and Gelvin, S.B. (2007). Novel plant transformation vectors containing the superpromoter. Plant Physiol. 145 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Krychevsky, A., Vaidya, M., Tzfira, T., and Citovsky, V. (2005. a). Uncoupling of the function of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. USA 102 5733–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Vaidya, M., White, C., Vainstein, A., Citovsky, V., and Tzfira, T. (2005. b). Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. USA 102 19231–19236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., et al. (2008). High-resolution mapping of epigenetic modification of the rice genome uncovers interplay between DNA methylaton, histone methylation, and gene expression. Plant Cell 20 259–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, P. (2004). A plant dialect of the histone language. Trends Plant Sci. 9 84–90. [DOI] [PubMed] [Google Scholar]
- Loyter, A., Rosenbluh, J., Zakai, N., Li, J., Kozlovsky, S.V., Tzfira, T., and Citovsky, V. (2005). The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol. 138 1318–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K., Armin, W., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A° resolution. Nature 389 251–260. [DOI] [PubMed] [Google Scholar]
- Luger, K., and Richmond, T. (1998). DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8 33–40. [DOI] [PubMed] [Google Scholar]
- March-Diaz, R., Garcia-Dominguez, M., Lozano-Juste, J., Leon, J., Florencio, F.J., and Reyes, J.C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53 475–487. [DOI] [PubMed] [Google Scholar]
- Mariño-Ramírez, L., Kann, M.G., Shoemaker, B.A., and Landsman, D. (2005). Histone structure and nucleosome stability. Expert Rev. Proteomics 2 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, S., Ito, Y., Hosoi, T., Takahashi, Y., and Machida, Y. (1990). Integration of chromosome: Possible involvement of DNA homology between T-DNA and plant DNA. Mol. Gen. Genet. 224 289–302. [DOI] [PubMed] [Google Scholar]
- Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Bakkeren, G., Crameri, A., Angelis, K., Redei, G.P., Schell, J., Hohn, B., and Koncz, C. (1991). T-DNA integration: A mode of illegitimate recombination in plants. EMBO J. 10 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhausser, P., Muller, E.C., Otto, A., and Kutay, U. (2001). Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–475. [Google Scholar]
- Mysore, K.S., Bassuner, B., Deng, X.-B., Darbinian, N.S., Motchoulski, A., Ream, W., and Gelvin, S.B. (1998). Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 11 668–683. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Kumar, C.T., and Gelvin, S.B. (2000. b). Arabidopsis ecotypes and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 21 9–16. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Nam, J., and Gelvin, S.B. (2000. a). An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 97 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., Zheng, C., Knue, M., Matthysse, A.G., and Gelvin, S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to Agrobacterium transformation. Mol. Gen. Genet. 261 429–438. [DOI] [PubMed] [Google Scholar]
- Narasimhulu, S.B., Deng, X.-B., Sarria, R., and Gelvin, S.B. (1996). Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten, L., DeGreve, H., Leemans, J., Hain, R., Hooykaas, P., and Schell, J. (1984). Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195 159–163. [Google Scholar]
- Palter, K.B., Foe, V.E., and Alberts, B.M. (1979). Evidence for the formation of nucleosome-like histone complexes on single-stranded DNA. Cell 18 451–467. [DOI] [PubMed] [Google Scholar]
- Parra, M.A., and Wyrick, J.J. (2007). Regulation of gene transcription by the histone H2A N-terminal domain. Mol. Cell. Biol. 27 7641–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Y., Niu, L., Lu, F., Liu, C., Zhai, J., Kong, X., and Cao, X. (2007). Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C., and Laniel, M.A. (2004). Histone and histone modification. Curr. Biol. 14 546–551. [DOI] [PubMed] [Google Scholar]
- Puebla, I., Esseghir, S., Mortlock, A., Brown, A., Crisanti, A., and Low, W. (2003). A recombinant H1 histone-based system for efficient delivery of nucleic acids. J. Biotechnol. 105 215–226. [DOI] [PubMed] [Google Scholar]
- Raisner, R.M., and Madhani, H.D. (2006). Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 16 119–124. [DOI] [PubMed] [Google Scholar]
- Ren, Q., and Govorsky, M.A. (2003). The nonessential H2A N-terminal tail can function as an essential charge patch on the H2A.Z variant N-terminal tail. Mol. Cell. Biol. 23 2778–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 10 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, L., Hohn, B., and Tinland, B. (1996). Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 93 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeger, E.J., and Knippers, R. (1979). DNA-histone interaction in the vicinity of replication points. Nucleic Acids Res. 6 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer, B., den Dulk-Ras, A., Vergunst, A.C., Jacome, E.J., and Hooykaas, P.J.J. (2003). Analysis of Vir protein translocation from Agrobacterum tumefaciens using Saccharomyces cerevisiae as a model: Evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 31 860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer, B., Risseeuw, E., Pansegrau, Q., Regensburg-Tuink, T.J.-G., Crosby, W.L., and Hooykaas, P.J.J. (2001). Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homolog of the yeast Skp1 protein. Curr. Biol. 11 258–262. [DOI] [PubMed] [Google Scholar]
- Shahbazian, M.D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76 75–100. [DOI] [PubMed] [Google Scholar]
- Shilatifard, A. (2006). Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 75 243–269. [DOI] [PubMed] [Google Scholar]
- Stachel, S.E., and Nester, E.W. (1986). The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, L.E., Jacobs, A., and Binns, A.N. (1998). The conjugal intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent export of VirE2. J. Bacteriol. 180 3933–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, C.L., Roux, K.J., and Burke, B. (2007). Blurring the boundary: The nuclear envelope extends its reach. Science 318 1408–1412. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., Briggs, S.D., Brame, C.J., Caldwell, J.A., Koh, S.S., Ma, H., Cook, R.G., Shabanowitz, J., Hunt, D.F., Stallcup, M.R., and Allis, C.D. (2001). Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11 996–1000. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland, B. (1996). The integration of T-DNA into plant genomes. Trends Plant Sci. 1 178–184. [Google Scholar]
- Tinland, B., Hohn, B., and Puchta, H. (1994). Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl. Acad. Sci. USA 91 8000–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremethick, D.J. (2007). Higher-order structures of chromatin: The elusive 30 nm fiber. Cell 128 651–654. [DOI] [PubMed] [Google Scholar]
- Tzfira, T., and Citovsky, V. (2000). From host recognition to T-DNA integration: The function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol. Plant Pathol. 1 201–212. [DOI] [PubMed] [Google Scholar]
- Tzfira, T., and Citovsky, V. (2002). Partners-in-infection: Host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12 121–129. [DOI] [PubMed] [Google Scholar]
- Tzfira, T., and Citovsky, V. (2003). The Agrobacterium plant cell interaction. Taking biology lessons from a bug. Plant Physiol. 133 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira, T., Vaidya, M., and Citovsky, V. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20 3596–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira, T., Vaidya, M., and Citovsky, V. (2004). Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431 87–92. [DOI] [PubMed] [Google Scholar]
- Verbsky, M.L., and Richards, E.J. (2001). Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4 494–500. [DOI] [PubMed] [Google Scholar]
- Vergunst, A.C., and Hooykaas, P.J.J. (1998). Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol. Biol. 38 393–406. [DOI] [PubMed] [Google Scholar]
- Vergunst, A.C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C.M.T., Regensburg-Tuink, T.J.G., and Hooykaas, P.J.J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290 979–982. [DOI] [PubMed] [Google Scholar]
- Vergunst, A.C., van Lier, M.C.M., den Dulk-Ras, A., and Hooykaas, P.J.J. (2003). Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 133 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst, A.C., van Lier, M.C.M., den Dulk-Ras, A., Stuve, T.A.G., Ouwehand, A., and Hooykaas, P.J.J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA 102 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Huang, Z.Q., Xia, L., Feng, Q., Erdjument-Bromage, H., Strahl, B.D., Briggs, S.D., Allis, C.D., Wong, J., Tempst, P., and Zhang, Y. (2001). Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293 853–857. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40 711–717. [DOI] [PubMed] [Google Scholar]
- Yi, H., Mysore, K.S., and Gelvin, S.B. (2002). Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 32 285–298. [DOI] [PubMed] [Google Scholar]
- Yi, H., Sardesai N, Fujinuma T, Chan CW, Veena, Gelvin SB. (2006). Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 18 1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov, V.M., Steck, T.R., Gupta, V., and Gelvin, S.B. (1994). Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. USA 91 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K., Sridhar, V.V., Zhu, J., Kapoor, A., and Zhu, J-K. (2005). Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2 e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W.-L., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Nam, J., Carpita, N.C., Matthysse, A.G., and Gelvin, S.B. (2003. a). Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol. 133 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., et al. (2003. b). Identification of Arabidopsis rat mutants. Plant Physiol. 132 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.