Abstract
Eutrophication leading to hypoxic water conditions has become a major problem in aquatic systems worldwide. Monitoring the levels and biological effects of lowered oxygen levels in aquatic systems may provide data useful in management of natural aquatic environments. Fishes represent an economically important resource that is subject to hypoxia exposure effects. Due to the extreme diversity of fish species and their habitats, fishes in general have evolved unique capabilities to modulate gene expression patterns in response to hypoxic stress. Recent studies have attempted to document quantitative changes in gene expression patterns induced in various fish species in response to reduced dissolved oxygen levels. From a management perspective, the goal of these studies is to provide a more complete characterization of hypoxia responsive genes in fish, as molecular indicators (biomarkers) of ecosystem hypoxic stress.
The molecular genetic response to hypoxia is highly complex and overlaps with other stress responses making it difficult to identify hypoxia specific responses using traditional single gene or low throughput approaches. Therefore, recent approaches have been aimed at developing functional genomic (e.g. high density microarray and real-time PCR) and proteomic (two-dimensional fluorescence difference in gel electrophoresis coupled with mass spectrometry based peptide identification) technologies that employ fish species. Many of the fish species utilized in these studies do not have the advantages of underlying genome resources (i.e., genome or transcriptome sequences). Efforts have attempted to establish correlations between discreet molecular responses elicited by fish in response to hypoxia and changes in the genetic profiles of stressed organs or tissues. Notable progress in these areas has been made using several different versions of either cDNA or oligonucleotide based microarrays to profile changes in gene expression patterns in response to hypoxic stress.
Due to these efforts, hundreds of hypoxia responsive genes have been identified both from laboratory reared aquaria fish and from feral fish derived from both fresh and saltwater habitats. Herein, we review these reports and the emergence of hypoxia biomarker development in aquatic species. We also include some of our own recent results using the medaka (Oryzias latipes) as a model to define genetic profiles of hypoxia exposure.
Keywords: Aquatic, Biomarker, Fish, Genomics, Hypoxia, Microarray
Introduction
Hypoxia and its biological importance
Most organisms require molecular oxygen (O2) to support their essential metabolic processes. The major usage of cellular O2 is by mitochondria, where the oxygen serves as the final electron acceptor in oxidative phosphorylation for ATP formation (Voet and Voet, 1995). In addition, some 10 to 15% of total intracellular O2 is consumed in a myriad of cellular reactions, including reactions catalyzed by mono- and dioxygenases, oxidases, and peroxidases (Zhang et al., 1999; Wenger, 2000). Therefore, hypoxia (or reduced oxygen tension) does not only inhibit oxidative phosphorylation, but also may affect many other oxygen-requiring reactions. Exposure to hypoxic conditions can thus adversely affect a broad range of biochemical, physiological, developmental and behavioral processes, including respiration, growth, metabolism, reproduction and locomotion (Clayton, 1993; Heath, 1995). Furthermore, it has been demonstrated that pathological symptoms, including development of certain types of cancer (Rofstad and Maseide, 1999; Rofstad, 2000), heart disease (Petkova et al., 2000) and stroke are associated with hypoxia (Khedr et al., 2000).
Recent studies have shown that, similar to endocrine disruptors (Kavlock, 1999), hypoxia can also affect endocrine function in both mammals (De Angelis et al., 1996) and fish (Wu et al., 2003) by affecting the brain-hypothalamic-pituitary-gonad axis. Hypoxic effects on the endocrine system can occur at all stages of hormone synthesis, storage, delivery, and function. For example, studies using mammalian models have shown that hypoxia depresses secretion of testosterone (Kouchiyama et al., 1989; Martin and Costa, 1992), a principal sex steroid that regulates gametogenesis and is controlled by the hypothalamic-pituitary-testicular axis (Redding and Patino, 1993). Hormonal disruption in turn, alters sexual behavior (Hermans et al., 1993). In female rats, the percentage of estrous rats was significantly higher, while fertility was lower when they were subjected to hypoxia (Martin and Costa, 1992). In addition, it has been shown that pulmonary fibrosis can be the result of exposure to chronic hypoxia, thereby suppressing sex hormones and lending further support to hypoxia suppression of the hypothalamo-pituitary-testicular axis (Semple et al., 1984).
In addition to its effects on hormones, hypoxia also influences the biochemical composition of reproductive organs in male rats (testis, epididymis and vas deferens; Riar et al., 1979). This results in reduced gonad weight, epididymis secretory activity, and sperm quality in adult rats (Riar et al., 1979). Female fishes living in a hypoxic environment, defined as having a dissolved oxygen (DO) concentration of < 2.0 mg l−1, have fewer mature ovaries and produce eggs with smaller yolks (Breitburg, 2002).
Overall, hypoxia has profound effects on various reproductive processes including puberty, fertility, ovarian cycle, and fetal development in different species of animals (Ducsay, 1999; Kavlock, 1999). It is therefore not surprising that recent research efforts have been focused on physiological, developmental and reproductive impairment resulting from hypoxia (Wu et al., 2003; Shang and Wu, 2004; Shang et al., 2006).
Euthrophication and hypoxia
Rapid urban and development over the last several decades has resulted in increased nutrient input into freshwater and marine systems and a phenomenon commonly known as eutrophication. Nutrient (e.g., nitrogen) run-off into streams and rivers may eventually manifest itself as stimulating plant growth and leading to algal blooms. The algae in the bloom eventually die and are subjected to oxidative decay by microorganisms in the water at depths. This decay results in depletion of dissolved oxygen and can lead to hypoxia or “dead zones” in aquatic environments. From a global perspective over the past 20 years the size, severity, and persistence of hypoxic dead zones due to eutrophication resulting from river outflows has been observed to be increasing rapidly (Rosenberg and Loo, 1988; Rosenberg et al., 1992; Justić et al., 1993; Diaz and Rosenberg, 1995; Mason, 1998; Diaz, 2001). Today, hypoxic events affecting thousands of square kilometers of water have been reported worldwide, and some coastal areas (e.g., Black Sea) have become permanently hypoxic (Justić et al., 1993; Diaz and Rosenberg, 1995; Diaz, 2001). Indeed, hypoxia caused by eutrophication is now regarded as one of the most serious threats to coastal marine ecosystems (Goldberg, 1995; McIntyre, 1995; Malakoff, 1998; Wu, 1999; Diaz, 2001). Mass mortality of invertebrates and fish, changes in ecosystem structure, altered migration and spawning patterns, reduction in habitat area, increased susceptibility to predation, susceptibility to infection, and changes in food resources have been reported in hypoxic waters worldwide (Wu, 1999; Naqvi et al., 2000).
Hypoxia biomarkers
Monitoring of oxygen levels in aquatic systems may be required as an early warning signal to protect the health of natural aquatic environments (McNamara and Dildy, 1999). However, since oxygen concentrations vary tremendously with space and time, frequent or continual monitoring of oxygen levels over large areas and over long periods of time would be required. Often, this is not cost-effective and is impractical in open water areas because of the unpredictability of hypoxic events and rapid changes in climatic conditions.
Biomarkers are molecular, biochemical, cellular, or physiological responses that are used as indicators of environmental change. Responses at the molecular level tend to be more sensitive and usually occur earlier than those at higher levels of biological organization (e.g. growth inhibition, changes in rate of development, and reduced reproductive potential). Using changes in the levels and types of stress-proteins or the response to hypoxia at the level of product synthesis (transcription or translation), as biomarkers has been suggested by many investigators (Farr and Dunn, 1999; Bierkens, 2000). Conceivably, hypoxic stress may alter the expression patterns of select genes, which in turn may alter the expression patterns of many other genes as part of a regulatory cascade aimed at ameliorating pathological effects. Thus, accurate measure of quantitative changes in gene expression patterns of hypoxia-responsive gene sets may be employed to identify molecular biomarkers for early detection and characterization of hypoxic stress. Surprisingly, although attempts have been made to identify a consistent set of hypoxia biomarkers in aquatic systems for over a decade (Forlin et al., 1996; Wu and Lam, 1997), there has been little progress in applying hypoxia biomarkers to monitoring and management regimens.
Physiological and biochemical responses of fish to hypoxia
There are many inherent difficulties in developing useful and informative biomarkers for hypoxia, most notably an overall paucity in our understanding of hypoxia-specific physiological and biochemical responses of aquatic organisms. Physiological responses of fishes to hypoxia are beyond the scope of this review and have been extensively reviewed elsewhere (Randall and Perry, 1992; Jensen et al., 1993; Hochachka et al., 1996; Nikinmaa and Salama, 1998). General responses of fish to hypoxia indicate that molecular genetic modulation of gene cascades must certainly occur. For example, it has been shown that different fish species may have alternative energetic strategies to cope with hypoxia (Lutz and Nilsson 1997; Krumschnabel et al., 2000). A common adaptive strategy (adopted by virtually all anoxia-tolerant vertebrates and invertebrates) is a drastic decrease in the rate of ATP consumption, usually referred to as metabolic depression (Nilsson, 1995). However, metabolic depression is not the first response to hypoxic conditions for most fish and occurs only after prolonged and severe hypoxia exposure. For example, oxygen consumption of the rainbow trout (Salmo gairdneri) remains unchanged over a wide range of oxygen levels in the water, but declines by over 30% of normoxic values when the DO drops to 0.87 mg l−1 (i.e., below 80 Torr); however, metabolic depression and increased anaerobic metabolism do not occur until the DO levels are further reduced to 0.32 mg l−1 (below 30 Torr; Boutilier et al., 1988).
At the protein level, when subjected to extended periods of extreme hypoxia, changes in enzyme activities, especially that of lactate dehydrogenase (LDH), occur in the myocardium of hagfish to support anaerobic glycolysis (Sidell, 1983). Increased activities of pyruvate kinase (both free and bound forms) were reported in the brain of striped mullet (Mullus barbatus), while the bound form disappeared after exposure to hypoxia for 1.5 h (Lushchak, 1993). The ratio of pyruvate kinase to citrate synthase (PK CS−1) correlates well with the hypoxia tolerance of heart tissue in different fish species. For example, PK CS−1 was significantly higher in Sebastolobus alascanus, a species inhabiting open, well-oxygenated water, as compared with Scorpaena guttat, a species living in shallow water where oxygen depletion is more likely to occur (Yang and Somero, 1993). A study on 12 species of Serrasalmidae showed that species having LDH B-like subunits are able to maintain oxidative metabolism even in hypoxic waters. Furthermore, tissue distribution patterns of LDH in Cichlasoma amazonarum correlated well with oxygen tension prevailing at the sampling location (Almeida-Val et al., 1995). The ability to regulate expression of LDH in response to oxygen availability enables this fish to survive in hypoxic environments (Almeida-Val and Farias, 1995). The level of two key ‘markers’ of glycolytic and oxidative flux capacity, LDH (lactate dehydrogenase, EC 1.1.1.27) and MDH (malate dehydrogenase, EC 1.1.1.37), was measured in four organs (white muscle, heart, liver, and brain) from different-sized Astronotus ocellatus, one of the most hypoxia tolerant fish of the Amazon. Enzyme levels correlated with body size, thus increasing the anaerobic potential positively with growth and demonstrating that there is a scaling effect on hypoxia tolerance in fishes (Almeida-Val et al., 2000).
Many teleosts release catecholamine hormones, adrenaline and noradrenaline into their circulation in response to hypoxia (Randall and Perry, 1992). Thus, activities of enzymes and proteins related to biosynthesis and metabolic degradation of catecholamines, as well as catecholamine receptors, might be expected to change under hypoxic conditions. Hypoxia also enhances the proliferation and differentiation of juvenile red blood cells (RBCs) in pronephros of flounder (Pleuronectes flesus), with an increase of up to 37% in the number of RBCs being reported (Soldatov et al., 1994).
Although efforts have been made to elucidate hypoxia response mechanisms in fish and other organisms and a unifying theory of hypoxia tolerance proposed (Hochachka et al., 1996), we still have much to learn if we are to achieve the goal of finding hypoxia specific response factors that can be used to develop assays that elucidate various levels and durations of hypoxic events. The complex physiological processes leading to the fish response to hypoxia are likely to be regulated by the combined activities of many genes (Nikinmaa and Rees, 2005). Investigation of how many divergent genes coordinate an organism’s response to hypoxia will offer insights into identification of specific hypoxia responsive factors that may be used to profile environmental hypoxia.
Genomic approaches
Genomic approaches include use of cDNA libraries, differential display methodologies, serial analysis of gene expression (SAGE) techniques, and cDNA or oligonucleotide microarrays. Of these technologies, microarrays are most capable of performing large-scale gene expression screens to investigate the molecular and physiological mechanisms underlying hypoxia and to identify candidate hypoxia biomarkers.
Gracey et al. (2001) performed pioneering studies using a cDNA microarray to profile gene expression responses in wild-caught mudskippers (Gillichthys mirabilis) from saltwater habitats that experience hypoxia. This study is the first to demonstrate that cDNA microarray technology can be used to understand physiological mechanisms of a stress response in a non-model organism for which genomic sequence data are unavailable. Gracey et al.’s array was fabricated from PCR-amplified cDNA inserts of several different cDNA libraries that were constructed using hypoxia stressed mudskippers (Gracey et al., 2001). These libraries included four distinct suppression subtractive hybridization cDNA (SSH-cDNA) libraries, one normalized cDNA library, and one full-length captured cDNA library. Hypoxia responsive genes identified in this study included those related to: (1) ATP metabolism; (2) locomotion and contraction; (3) protein translation; (4) iron metabolism; (5) antigrowth and proliferation; and (6) amino acid metabolism. It is evident from Gracey et al.’s (2001) results that molecular genetic responses to hypoxia are both tissue specific and temporally up- or down-regulated. For example, some genes involved in major energy-consuming processes such as protein synthesis and locomotion were down-regulated in muscle tissue at the onset of hypoxia. While under hypoxia stress, other sets of genes associated with anaerobic ATP production (gluconeogenesis) were up-regulated in liver tissue compared with skeletal muscle. The mudskipper microarray-based study represented an important first step toward understanding the mechanisms of the hypoxia response at a genome-wide scale in fish. However, surprisingly, many genes that are known to be responsive to hypoxia in mammals, such as c-fos, jun B, vascular endothelial growth factor (vegf) and its receptor (vegfr), glucose transporters (gluts) (Zhang et al., 1999; Zhang et al., 2003), CREB-binding protein (CBP) and p300-interacting transactivator (Arany et al., 1996), were not identified as modulated in the mudskipper studies. However, this may have been due to the lack of appropriate representation on this type of array.
As a vertebrate experimental model, laboratory reared zebrafish (Danio rerio) offer several advantages over mammalian models such as small genome size, short generation time, external development allowing embryo manipulation, and optical transparency of embryos. The established conserved genomic synteny between zebrafish and human genomes and conservation of gene structure and function, make zebrafish an excellent system to develop models of human disease and for high throughput screening of genes involved in response to environmental perturbation (Lieschke and Currie, 2007).
Zebrafish microarrays have been applied to document the response to hypoxia exposure (Ton et al., 2003). The zebrafish array employed by Ton et al. (2002) was composed of 4,512 unique zebrafish cDNAs. Unlike most vertebrates that require continuous exposure to oxygen, zebrafish embryos may survive 24 h of anoxia (Padilla and Roth, 2001). Ton et al. (2002, 2003) used their zebrafish cDNA microarray to examine the molecular genetic response of zebrafish embryos to hypoxic stress at discreet developmental stages. They observed that hypoxia exposure resulted in a substantial reduction in embryo motility (i.e., whole body movement) and heartbeat rate. Accordingly, there was also a strong coordinated down-regulation of genes that encode both skeletal and cardiac contractile proteins. Also, cell cycle regulatory genes, such as cyclin G1 and proliferating cell nuclear antigen (PCNA) appeared repressed in hypoxic embryos relative to normoxic ones. This study characterized molecular mechanisms of the zebrafish embryo response to anoxia and showed that embryos became developmentally arrested in the S and G2 phases of the cell cycle. Ton et al.’s (2002 (2003) results represent an early application of zebrafish cDNA microarrays to provide a global view of changes in gene expression patterns during embryonic development and hypoxia exposure.
Although the studies carried out by Gracey et al. (2001) and Ton et al. (2002, 2003) served to illuminate gene expression responses to hypoxia at the genome level, both of these studies employed hypoxia exposures to measure an acute fish response at a single end-point. In order to understand mechanisms of the long-term adaptive response to hypoxia in fish, Van der Meer et al. (2005) employed cDNA microarrays to investigate hypoxia-induced changes in the expression of 15,532 genes in the gills of zebrafish exposed to hypoxia. Starting from air saturated water and 28°C (DO concentration ≈ 8 mg l−1), they designed a specific hypoxia exposure system to step down the DO levels for 4 d of exposure from 80–90% (≈ 6.4–7.2 mg l−1) to 60% (≈ 4.8 mg l−1), 40% (≈ 3.2 mg l−1), 20% (≈ 1.6 mg l−1), and finally, 10% air saturation (≈ 0.8 mg l−1). Results from this study showed 367 differentially expressed genes, of which 117 appeared to be induced by hypoxia and 250 showed a down-regulated expression pattern. Van der Meer et al. (2005) also identified several novel hypoxia-dependent changes in gene expression that were related to physiological adaptation to low environmental oxygen. Examples include genes coding for proteins such as monocarboxylate transporter (mct4), responsible for transport of metabolites like pyruvate and lactate; myoglobin, which increases the oxygen diffusion rate through tissues; and two genes previously associated with human metabolic disorders that affect cholesterol trafficking and degradation, the Niemann-Pick disease gene C (NPC) and lysosomal acid lipase (LAL; cholesterol esterase), which were both up-regulated by hypoxia.
All of the above early examples used microarrays composed of PCR amplified cDNAs having different lengths to characterize fish responses to hypoxia. We now know that cDNA arrays such as these may lead to substantial errors in the output microarray data. This is in part because it is difficult to generate reliable mismatch controls that will assess the specificity of hybridization for each feature on the array (Relógio et al., 2002). Also, array spotting of variable length cDNAs leads to many random contacts between cDNA molecules and the array matrix, resulting in non-uniform hybridization due to steric interference between the probe and target. Further, the cDNA product of PCR amplification needs to be denatured for spotting and both the degrees of denaturation and resultant foldback structures that may form at renaturation are difficult to control (Kunz et al., 2004). All these effects and others make cDNA arrays more difficult to work with than oligonucleotide microarrays.
Like zebrafish, the Japanese medaka (Oryzias latipes) is a valuable laboratory reared small aquaria fish model system. The genome size of medaka is about one-half that of zebrafish (Wakamatsu et al., 2001) and like zebrafish, the eggs and embryos are transparent so all developmental stages may be observed. Medaka has a long tradition of use in environmental toxicity testing. Some medaka species (Oryzias javanicus) exhibit a wide tolerance range of salinity and may live in saltwater or freshwater (Imai et al., 2007). These characteristics make establishment of medaka-based biomarkers attractive because they may be applied to many diverse environments.
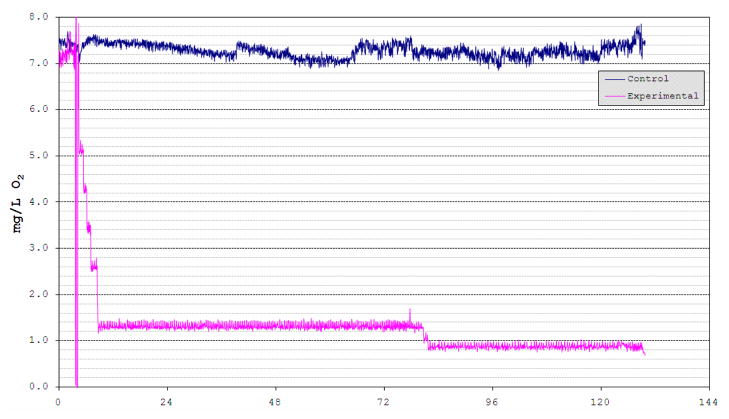
A high-density oligonucleotide (60 mer) microarray containing features representing 8,046 medaka unigenes was designed and constructed in our laboratory (Ju et al., 2007). Using this microarray, gene expression patterns in response to hypoxia were detected in several tissues. At the onset of our experiment, DO in the hypoxic tanks was slowly (over 5 h) brought to 1.3–1.4 mg l−1, then held for three days, followed by two days at 0.8–0.9 mg l−1 until fish were sacrificed and dissected for experimental analyses. The normoxic tanks (immediately adjacent to the hypoxic aquaria) were maintained at 7.3–8.0 mg l−1 throughout the experiment by passing compressed air through air stones into the tank water (Fig. 1). In Table 1 we present results from our hypoxia studies using medaka and this array. Overall for three tissues analyzed there were 372 genes in the brain, 444 in the gill and 515 in the liver that exhibited modulated and differential expression in medaka exposed to hypoxia. These genes were identified using Significant Analyses of Microarray (SAM, Tusher et. al, 2001) and setting a false detection cutoff value of zero (i.e., q = 0%, for full details, see Ju et. al., 2007). Among up-regulated genes, 9, 21, and 20 genes showed the same expression patterns between brain and gill, brain and liver, and gill and liver, respectively. Similarly, among the down-regulated genes, 65, 24, and 26 genes were found to show the same response between brain and gill, brain and liver, and gill and liver, respectively. Overall, greater overlap in hypoxia responsive genes was observed for the down-regulated genes than for the up-regulated genes among these three tissues (Table 1).
Figure 1.
Dissolved oxygen changes in hypoxia tank and control tank during medaka hypoxia experiments. Two 20 gal aquaria were programmed to become hypoxic and one 20 gal control aquarium was immediately adjacent to the others. Each tank was monitored and maintained under pre-set oxygen level profiles using an OxyCycler oxygen control system (Model F84DO, BioSpherix, NY, USA) specifically designed for aquaria. The control tank (blue line) was maintained at 7.3–8.0 mg l−1 DO throughout the experiment by bubbling compressed air through air stones into the tank water. The oxygen level in the hypoxic tanks (pink line) was slowly (over 5 h) brought to 2.5 mg l−1 by bubbling compressed air or nitrogen through air stones, then dropped to 2.0, 1.5 and finally 0.8 mg l−1 over each day and held at 0.8 mg l−1 for 5 d when the fish were sacrificed for experimental analyses.
Table 1.
Medaka tissue specificity of gene responses to hypoxia as assessed by the 8K microarray. Detailed information and a searchable index of the array features that exhibited response to hypoxia exposure is available at the following link: http://xiphophorus.org/medaka_array/medaka_up_down_request.asp.
| Brain | Gill | Liver | |||||
|---|---|---|---|---|---|---|---|
| Tissue | Up | Down | Up | Down | Up | Down | |
| A | Brain | 223 | 149 | 242 | 202 | 242 | 273 |
| B | Brain + | 9 | 65 | 21 | 24 | ||
| C | Gill + | 20 | 26 | ||||
| D | Brain + Gill + | 2 | 7 | ||||
| E | Total | 372 | 313 | 515 | |||
A: Number of genes responsive to hypoxia in brain, gill and liver.
B, C, D: Number of overlapping genes responsive to hypoxia simultaneously in both brain and gill (B, middle), brain and liver (B, right) and gill and liver (C), respectively.
D: Overlapping genes responsive to hypoxia simultaneously in all three tissues.
E: Total number of genes responsive (up or down) to hypoxia in each tissue.
Two ESTs were commonly up-regulated in all three tissues analyzed (brain, gill, and liver) upon medaka exposure to hypoxia, and seven genes, including Fas apoptotic inhibitory molecule 2 (FAIM2), were down-regulated in these three tissues in response to hypoxic conditions. This suggested an increase in the sensitivity of cells to the onset of apoptosis in response to hypoxia. Several well-known hif-1 (hypoxia inducible factor 1) regulated genes (Semenza, 2000) such as glut, ldh-a, and pyruvate kinase were up-regulated in brain. Aldolase was up-regulated in medaka liver upon hypoxia exposure consistent with the activation of HIF-1-mediated response system (Maxwell, 2005).
Full access to gene targets and array features responsive to hypoxia in the above medaka studies are available at http://147.26.215.207/medaka_array/medaka_up_down_request.asp or simply by going the www.xiphphorus.org web site and looking under “database” section for “JEMBE special Issue”. Here the user will find all 1,386 unique medaka genes found to be up- or down-regulated in response to hypoxic conditions in the three tissues examined (liver, gill, and brain). One may use this site to search for regulated genes within one or multiple tissues after selecting the desired criteria from input boxes supplied. Criteria connected by “AND” will display results that appear in both of the specified criteria, while criteria connected by “OR” will display results that appear in one or both of the specified criteria. Use “NOT” to exclude a specific criterion from the search. The results provided consist of the Gene ID, BLAST X ID, and fold change observed in hypoxic compared to normoxic conditions. Negative values represent fold changes in hypoxia responsive genes that exhibited down-regulation when compared to normoxic gene expression levels. Positive values represent fold changes that exhibited up-regulation in gene expression compared with normoxic controls. For additional information such as the oligonucleotide target sequence, GenBank link, and the feature location on the Medaka array, click on the Gene ID.
In these studies, we categorized the medaka hypoxia differentially-expressed genes into known Gene Ontology (GO) groups using the Swiss-Prot database (Bairoch and Boeckmann, 1994; Boeckmann et al., 2003), the Database for Annotation and Visualization and Integrated Discovery (DAVID) (Dennis et al., 2003), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000; Wixon and Kell, 2000) databases. The number of GO groups identified (see Table 2) for down-regulated genes was higher than for up-regulated GO group genes in brain and gill. As may have been expected, metabolism was the largest GO group, containing 32 and 39 down-regulated genes in brain and liver, respectively. There were also 57 metabolism GO group genes up-regulated in liver. Most of the down-regulated genes in brain and gill were grouped into metabolism, catabolism, RNA (RNA metabolism, processing, and/or splicing), and protein (protein catabolism, metabolism, and/or transport), indicating an overall slow-down in general metabolic processing in these tissues. These results are consistent with the early results of Gracey et al (2001).
Table 2.
Number of ontology groups of medaka hypoxia responsive genes. B: Brain, G: Gill, L: Liver.
| GO group | Up 1 | Down2 |
|---|---|---|
| Cell maintenance | 28(L) | |
| Cytoskeleton | 8(L) | |
| Fatty acid metabolism | 3(L) | |
| Intracellular signaling cascade | 6(B) | |
| Metabolism | 57(L) | 32(B)/39(L) |
| mRNA metabolism | 4(L) | 4(B)/3(G)/5(L) |
| mRNA processing | 4(L) | 4(B)/4(L) |
| Neurogenesis | 4(B) | |
| Neurotransmitter transport | 2(B) | |
| Oxygen and reactive oxygen species metabolism | 4(L) | |
| Protein biosynthesis | 5(G)/7(L) | |
| Protein catabolism | 5(B) | |
| Protein metabolism | 27(L) | 14(B)/20(L) |
| Protein modification | 10(L) | |
| Response to endogenous stimulus | 3(B) | |
| Response to hormone stimulus | 2(B) | |
| RNA metabolism | 8(L) | 5(B)/5(L) |
| RNA processing | 6(L) | 4(B)/5(L) |
| RNA splicing | 3(L) | 4(B)/3(L) |
| rRNA metabolism | 2(G) | |
| rRNA processing | 2(G) | |
| Superoxide metabolism | 2(L) | |
| Translation | 2(G) | 3(B) |
| Transport | 38(L) | 8(B)/11(L) |
| Ubiquitin cycle | 6(L) | |
| Ubiquitin-dependent protein catabolism | 3(L) | |
| Total | 15(B)/2(G)/181(L) | 85(B)/12(G)/129(L) |
Number of up-regulated genes.
Number of down-regulated genes.
Hypoxia resulted in more up-regulated genes (181) than down-regulated ones (129) in the medaka liver; but fewer up-regulated genes (15 and 2) than down-regulated genes (85 and 12) in the medaka brain and gill, respectively. Two biological pathways were found significantly dysregulated in medaka upon hypoxic exposure. The ubiquitin-proteasome pathway was down-regulated, while the phosphatidylinositol signaling pathway was up-regulated.
The down-regulated ubiquitin-proteasome pathway is an energy consuming pathway (Kimura et al., 2003). Several components of this pathway, such as the 26S proteasome regulatory subunit, proteasome 26S subunit ATPase, and other proteasome related genes such as proteasome subunit β type 9 and proteasome activator 28-β subunit, were all found to be down-regulated in both the brain and gill tissue upon hypoxia exposure. Down-regulation of this pathway is related to the suppression of major energy-consuming processes.
The up-regulated phosphatidylinositol signaling pathway is represented by a key gene, phospholipase C1, which was found to be induced in medaka liver upon exposure to hypoxia. This up-regulation may promote inositol production resulting in increased detoxification rates for liver tissues.
Several fish homologues of human disease associated genes were also demonstrated to be responsive to hypoxia in medaka, such as: BGH3 (related to visual handicap, Stewart et al., 1999), ZIC2 (related to deformation or damage in brain tissue, Yang et al., 2000), EI2BD (related to brain disease, Van der Knaap et al., 2002), ITM2B (related to Alzheimer’s diseases, Vidal et al., 1999), ATNG (related to dominant renal hypomagnesemia, Meij et al., 2000), MOG (related to dysfunction of the myelin sheath and cell–cell communication, Pham-Dinh et al., 1994), and p21-Rac1 (related to chronic granulomatous disease, Leusen et al., 1996). Our results showed that hypoxia affected a wide range of physiological processes in medaka, down-regulating general organismal metabolic pathways and activating energy-saving mechanisms.
Many varied physiological studies have shown that different species of fish have alternative energetic strategies to cope with hypoxia (Lutz and Nilsson 1997; Krumschnabel et al., 2000). Comparative profiling of gene expression patterns in response to hypoxia among different and varied fish species will elucidate the different physiological pathways and gene networks involved in the hypoxia stress response.
Proteomic approaches
The application of genomic approaches (mostly cDNA microarray) has provided extremely valuable information regarding the transcriptional control of a wide range of genes and pathways discussed above. However, proteins, rather than genes and mRNAs, are responsible for carrying out cellular functions. Extrapolation of changes in the levels of certain transcripts that result in corresponding changes in protein abundance (e.g. post-translational modifications) cannot necessarily be made by using only genomic approaches (Lilley and Griffiths, 2003). Therefore, proteomic approaches to determine changes in protein levels expressed by a genome in a specified tissue or organ and comparison of potential alterations in the abundance of these proteins in response to environmental perturbations are important additions to the genomic approaches.
Bosworth et al. (2005) used zebrafish skeletal muscle as a model to characterize protein expression patterns in response to hypoxia (0.17 mg l−1 DO for 24 h). They employed silver staining to resolve the protein spots after 2D-PAGE protein separation. They concluded that hypoxia did not change the general pattern of protein expression, with the exception of six low abundance proteins. 2D-PAGE coupled with silver staining has been the major separation technique used in proteomics, allowing the resolution of several thousand proteins in a single sample; however, the limitations of this technique are low sensitivity, reduced dynamic range and gel-to-gel variability (White et al., 2004).
Two-dimensional fluorescence-based difference gel electrophoresis (2D-DIGE), which allows co-separation of control and experimental samples pre-labeled with distinct fluorescent dyes, circumvents many of the issues associated with traditional 2D-PAGE and produces more sensitive and quantitative proteomic analyses (Marouga et al., 2005). We applied 2D-DIGE methods in combination with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF-MS) for resolution of protein spectra for downstream identification using post-source decay fragmentation to successfully identify six abundant proteins and related isozymes that were up-regulated (> 1.49, p < 0.005) in hypoxia exposed medaka brain tissues (Oehlers et al., 2007). These proteins included two hemoglobin β subunits, four carbonic anhydrase forms, calbindin, aldolase, succinate dehydrogenase, and glutathione-S-transferase. These proteins have all been associated in previous studies (Hochachka et al., 1996; Christako et al., 1989; Hochachka and Lutz, 2001; Yenari et al., 2001; Wu et al., 2002) with hypoxia/ischemia in other animals, and thus represent a good source of hypoxia markers in brain tissue.
Bioinformatics and data management
The goal of applications of genomic and proteomic approaches in fish hypoxia studies is to provide a genome-level understanding of the complex functional changes associated with the hypoxia response in fish. This understanding will help to identify overlapping and unique pathways that may be good candidates of hypoxia biomarkers. Never before have scientists conceptually or technically been able to perform such genome-wide gene expression screening and analyses to understand molecular mechanisms of the hypoxia response in fishes. Even with the limitations of current genomic and proteomic approaches, there is still a tremendous ability to envision novel relationships between the fish genome and proteome upon hypoxia exposure. Currently, hundreds of hypoxia responsive genes and proteins (many of them are novel and/or unknown genes and proteins) have been identified using laboratory reared small aquaria fish models or feral fish from both fresh and saltwater habitats. The rapid pace of characterization of genomic and proteomic approaches and the resulting extreme diversity of fish species coupled with the data complexity pose special challenges. In particular, new methods for structuring and searching fish hypoxia related genomic and proteomic databases to retrieve orthologus groups of fish genes based upon well-known pathways, functional classifications, and specific gene networks should be further developed. As an example, we have placed information on the features of our 8K medaka oligonucleotide microarray on a web page: (http://xiphophorus.org/Medaka_Array/Medaka_Slides.asp).
Conclusions and perspective
Current genome scale gene expression profiling results complement the existing physiological and biochemical information on hypoxia responsiveness of fish.
In order to further integrate, codify, and link the function of hypoxia responsive genes identified by genomic and proteomic approaches to higher-level biological effects, many questions may need to be addressed: (1) To what extent are these genes involved in regulating cell proliferation under hypoxic stress? (2) Are these genes regulated and influenced by other cellular proteins under hypoxic stress? (3) What are the rate-limiting factors that control the activity of these genes? (4) What roles do these genes play under acute or prolonged hypoxia in different fish organs? (5) Do these novel genes play some unknown role in hypoxia responses apart from metabolic depression, glycolysis, and apoptosis? (6) Are the genes identified functionally related to those in the mammalian counterparts?
Most hypoxia studies have been performed using fish models exposed to a single stressor condition. However, in the natural environment, aquatic hypoxia is frequently associated with changes in temperature, food availability, or xenobiotic exposure, each of which may interact with hypoxia responses in fish. The combination of effects from these stressors with hypoxia may be very complex due to antagonism, synergism, or additive responses. Some stressors, such as starvation and oxidative chemical exposure may lead to very similar gene responses in fish. The future challenge will be to use genomic and proteomic approaches to further distinguish specific responses to hypoxia from changes in gene expression caused by other stressors. Molecular response(s) to hypoxia is (are) very complex and presently, no single gene is known to respond solely to hypoxia in a linear fashion. The c-fos gene was suggested as a potential biomarker for hypoxia (Erickson and Millhorn 1991); however, its expression is affected by many different factors (Hill and Treisman, 2000) and its response to hypoxia is nonlinear. Therefore, the use of patterns of hypoxia-responsive genes, rather than individual gene biomarkers, may be used to produce reliable assays. These are subjects for current and future study.
Acknowledgments
We thank Leon P. Oehlers for technical support on hypoxic proteomic experiments as well as the staff at the Xiphophorus Genetic Stock Center (Markita Savage and Leona Hazlewood) for fish culture and dissection assistance. We also wish to thank Roxie Smeal, Al Martinez, Mikki Boswell, and Rachell Booth for their assistance in preparation of this manuscript. This work was supported by NOAA National Ocean Service (grants NA04-NOS4261162 and NA06-NOS4260118); the NIH - National Center for Research Resources (P40-RR17072), and the Roy F. and Joanne Cole Mitte Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida-Val VMF, Farias IP. Respiration in fish of the Amazon: Metabolic adjustments to chronic hypoxia. In: Almeida-Val VMF, Randall DJ, editors. Physiology and biochemistry of the fishes of the Amazon. Instituto Nacional de Pesquisas da Amazônia; Manaus: 1995. pp. 257–270. [Google Scholar]
- Almeida-Val VMF, Farias IP, Silva MN, Duncan WP, Val AL. Biochemical adjustments to hypoxia by Amazon cichlids. Braz J Med Biol Res. 1995;28:1257–1263. [PubMed] [Google Scholar]
- Almeida-Val VMF, Val AL, Duncan WP, Souza FCA, Paula-Silva MN, Land S. Scaling effects on hypoxia tolerance in the Amazon fish Astronotus ocellatus (Perciformes: Cichlidae): contribution of tissue enzyme levels. Comp Biochem Physiol B. 2000;125:219–226. doi: 10.1016/s0305-0491(99)00172-8. [DOI] [PubMed] [Google Scholar]
- Arany Z, Huang LE, Eckner R, Bhattachary S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank: current status. Nucleic Acids Res. 1994;22:3578–3580. [PMC free article] [PubMed] [Google Scholar]
- Bierkens JG. Applications and pitfalls of stress-proteins in biomonitoring. Toxicology. 2000;153:61–72. doi: 10.1016/s0300-483x(00)00304-8. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosworth CA, 4th, Chou CW, Cole RB, Rees BB. Protein expression patterns in zebrafish skeletal muscle: initial characterization and the effects of hypoxic exposure. Proteomics. 2005;5:1362–1371. doi: 10.1002/pmic.200401002. [DOI] [PubMed] [Google Scholar]
- Boutilier RG, Dobson G, Hoeger U, Randall DJ. Acute exposure to graded levels of hypoxia in rainbow trout (Salmo gairdneri): metabolic and respiratory adaptations. Respir Physiol. 1988;71:69–82. doi: 10.1016/0034-5687(88)90116-8. [DOI] [PubMed] [Google Scholar]
- Breitburg D. Effects of hypoxia, and the balance between enrichment, on coastal fishes and fisheries. Estuaries. 2002;25:767–781. [Google Scholar]
- Christako S, Garielides C, Rhoten WB. Vitamin D-dependent calcium binding proteins: chemistry, distribution, functional considerations, and molecular biology. Endocr Rev. 1989;10:3–26. doi: 10.1210/edrv-10-1-3. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Mudskippers. In: Ansell AD, Gibson RN, Barnes M, editors. Oceanography and marine biology: An annual review. University College London Press; London: 1993. pp. 507–577. [Google Scholar]
- De Angelis C, Ferri C, Urbani L, Farrace S. Effect of acute exposure to hypoxia on electrolytes and water metabolism regulatory hormones. Aviat Space Environ Med. 1996;67:746–750. [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Diaz RJ. Overview of hypoxia around the world. J Environ Qual. 2001;30:275–281. doi: 10.2134/jeq2001.302275x. [DOI] [PubMed] [Google Scholar]
- Diaz RJ, Rosenberg R. Marine benthic hypoxia: a review of its ecological effects and the behavioural response of benthic marcofauna. Oceanogr Mar Biol, Annu Rev. 1995;33:245–303. [Google Scholar]
- Ducsay CA. Hypoxia effect on reproduction. In: Knobil E, Neil DJ, editors. Encyclopedia of reproduction. 2. Academic Press; London: 1999. pp. 769–775. [Google Scholar]
- Erickson JT, Millhorn DE. Fos-like protein is induced in neurons of the medulla oblongata after stimulation of the carotid sinus nerve in awake and anesthetized rats. Brain Res. 1991;567:11–24. doi: 10.1016/0006-8993(91)91430-9. [DOI] [PubMed] [Google Scholar]
- Farr S, Dunn RT., 2nd Concise review: gene expression applied to toxicology. Toxicol Sci. 1999;50:1–9. doi: 10.1093/toxsci/50.1.1. [DOI] [PubMed] [Google Scholar]
- Forlin L, Baden SP, Eriksson S, Granmo A, Lindesjoo E, Magnusson K, Ekelund R, Esselin A, Sturve J. Effects of contaminants in roundnose grenadier (Coryphaenoides rupestris) and Norway lobster (Nephrops norvegicus) and contaminant levels in mussel (Mytilus edulis) in the Skagerrak and Kattegat compared to the Faroe Islands. J Sea Res. 1996;35:209–222. [Google Scholar]
- Goldberg ED. Emerging problems in the coastal zone for the twenty-first century. Mar Pollut Bull. 1995;31:152–158. [Google Scholar]
- Gracey AY, Troll JV, Somero G. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc Natl Acad Sci USA. 2001;98:1993–1998. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AG. Water pollution and fish physiology. 2. CRC Press; Florida: 1995. p. 359. [Google Scholar]
- Hermans RH, McGivern RF, Chen W, Longo LD. Altered adult sexual behavior in the male rat following chronic prenatal hypoxia. Neurotoxicol Teratol. 1993;15:353–363. doi: 10.1016/0892-0362(93)90051-o. [DOI] [PubMed] [Google Scholar]
- Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 2000;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Lutz PL. Mechanism, origin, and evolution of anoxia tolerance in animals. Comp Biochem Physiol B. 2001;130:435–459. doi: 10.1016/s1096-4959(01)00408-0. [DOI] [PubMed] [Google Scholar]
- Imai S, Koyama J, Fujii K. Effects of estrone on full life cycle of Java medaka (Oryzias javanicus), a new marine test fish. Environ Toxicol Chem. 2007;26:726–731. doi: 10.1897/05-539r2.1. [DOI] [PubMed] [Google Scholar]
- Jensen BF, Nikinmaa M, Weber RE. Environmental perturbations of oxygen transport in teleost fishes: causes, consequences and compensations. In: Rankin JC, Jensen FB, editors. Fish ecophysiology. Hapman and Hall; London: 1993. pp. 161–179. [Google Scholar]
- Ju Z, Wells MC, Heater SJ, Walter RB. Multiple tissue gene expression analyses in Japanese medaka (Oryzias latipes) exposed to hypoxia. Comp Biochem Physiol C. 2007;145:134–144. doi: 10.1016/j.cbpc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Justić D, Rabalais NN, Turner RE, Wiseman WJ., Jr Seasonal coupling between river born nutrients, net productivity and hypoxia. Mar Pollut Bull. 1993;26:184–189. [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock RJ. Overview of endocrine disruptor research activity in the United States. Chemosphere. 1999;39:1227–1236. doi: 10.1016/s0045-6535(99)00190-3. [DOI] [PubMed] [Google Scholar]
- Khedr EM, Shinawy EO, Khedr TY, Ali AA, Awad EM. Assessment of corticodiaphragmatic pathway and pulmonary function in acute ischemic stroke patients. Eur J Neurol. 2000;7:323–330. doi: 10.1046/j.1468-1331.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- Kimura T, Ishizuka H, Yoshida A, Morii M, Takeguchi N, Asano S. Quantity and quality control of gastric proton pump in the endoplasmic reticulum by ubiquitin/proteasome system. Biochemistry. 2003;42:4771–4779. doi: 10.1021/bi020513d. [DOI] [PubMed] [Google Scholar]
- Kouchiyama S, Shinozaki T, Masuyama S, Tatsumi K, Kimura H, Kuriyama T. Depression of testosterone secretion in male patients with respiratory failure. Nihon Kyobu Shikkan Gakkai Zasshi. 1989;27:345–351. [PubMed] [Google Scholar]
- Krumschnabel G, Schwarzbaum PJ, Lisch J, Biasi C, Wieser W. Oxygen-dependent energetics of anoxia-tolerant and anoxia-intolerant hepatocytes. J Exp Biol. 2000;203:951–959. doi: 10.1242/jeb.203.5.951. [DOI] [PubMed] [Google Scholar]
- Kunz M, Ibrahim SM, Koczan D, Scheid S, Thiesen HJ, Gross G. DNA microarray technology and its applications in dermatology. Exp Dermatol. 2004;13:593–606. doi: 10.1111/j.0906-6705.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- Leusen JH, de Klein A, Hilarius PM, Ahlin A, Palmblad J, Smith CI, Diekmann D, Hall A, Verhoeven AJ, Roos D. Disturbed interaction of p21-rac with mutated p67-phox causes chronic granulomatous disease. J Exp Med. 1996;184:1243–1249. doi: 10.1084/jem.184.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007:353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lilley KS, Griffiths DR. Proteomics in Drosophila melanogaster. Briefings in Functional Genomics and Proteomics. 2003;2:106–113. doi: 10.1093/bfgp/2.2.106. [DOI] [PubMed] [Google Scholar]
- Lushchak VI. Free and bound pyruvate kinase from fish brain: properties and redistribution after hypoxia. Biochem Mol Biol Int. 1993;29:1103–1109. [PubMed] [Google Scholar]
- Lutz PL, Nilsson GE. Contrasting strategies for anoxic brain survival--glycolysis up or down. J Exp Biol. 1997;200:411–419. doi: 10.1242/jeb.200.2.411. [DOI] [PubMed] [Google Scholar]
- Malakoff D. Death by suffocation in the Gulf of Mexico. Science. 1998;281:190–193. [Google Scholar]
- Marouga R, David S, Hawkins E. The development of the DIGE system, 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- Martin IH, Costa LE. Reproductive function in female rats submitted to chronic hypobaric hypoxia. Arch Int Physiol Biochim Biophys. 1992;100:327–330. doi: 10.3109/13813459209000720. [DOI] [PubMed] [Google Scholar]
- Mason WJ., Jr Macrobenthic monitoring in the Lower St. Johns River, Florida. Environ Monit Assess. 1998;50:101–130. [Google Scholar]
- Maxwell PH. Hypoxia-inducible factor as a physiological regulator. Exp Physiol. 2005;90:791–797. doi: 10.1113/expphysiol.2005.030924. [DOI] [PubMed] [Google Scholar]
- McIntyre AD. Human impact on the oceans: The 1990s and beyond. Mar Pollut Bull. 1995;31:147–151. [Google Scholar]
- McNamara HM, Dildy GA., 3rd Continuous intrapartum pH, pO2, pCO2, and SpO2 monitoring. Obstet Gynecol Clin North Am. 1999;26:671–693. doi: 10.1016/s0889-8545(05)70106-6. [DOI] [PubMed] [Google Scholar]
- Meij IC, Koenderink JB, van Bokhoven H, Assink KF, Groenestege WT, de Pont JJ, Bindels RJ, Monnens LA, van den Heuvel LP, Knoers NV. Dominant isolated renal magnesium loss is caused by misrouting of the Na(+), K(+)-ATPase gamma-subunit. Nat Genet. 2000;26:265–266. doi: 10.1038/81543. [DOI] [PubMed] [Google Scholar]
- Naqvi SW, Jayakumar DA, Narvekar PV, Naik H, Sarma VV, D’Souza W, Joseph S, George MD. Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature. 2000;408:346–349. doi: 10.1038/35042551. [DOI] [PubMed] [Google Scholar]
- Nikinmaa M, Rees BB. Oxygen-dependent gene expression in fishes. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1079–R1090. doi: 10.1152/ajpregu.00626.2004. [DOI] [PubMed] [Google Scholar]
- Nikinmaa M, Salama A. Oxygen transport in fish. In: Perry SF, Tufts B, editors. Fish physiology, vol.17. Fish respiration. Academic Press; New York: 1998. pp. 141–185. [Google Scholar]
- Nilsson GE. O2 availability: brain defense mechanisms. In: Hochachka PW, Mommsen TP, editors. Biochemistry and molecular biology of fishes, vol. 5 Environmental and ecological biochemistry. Elsevier Science; Amsterdam: 1995. pp. 19–43. [Google Scholar]
- Oehlers LP, Perez AN, Walter RB. Detection of hypoxia-related proteins in medaka (Oryzias latipes) brain tissue by difference gel electrophoresis and de novo sequencing of 4-sulfophenyl isothiocyanate-derivatized peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Comp Biochem Physiol C. 2007;145:120–133. doi: 10.1016/j.cbpc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Padilla PA, Roth MB. Oxygen deprivation causes suspended animation in the zebrafish embryo. Proc Natl Acad Sci USA. 2001;98:7331–7335. doi: 10.1073/pnas.131213198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova SB, Ashton A, Bouzah B, Huang H, Pestell RG, Tanowitz HB. Cell cycle molecules and diseases of the cardiovascular system. Front Biosci. 2000;5:452–460. doi: 10.2741/petkova. [DOI] [PubMed] [Google Scholar]
- Pham-Dinh D, Allinquant B, Ruberg M, Della Gaspera B, Nussbaum JL, Dautigny A. Characterization and expression of the cDNA coding for the human myelin/oligodendrocyte glycoprotein. J Neurochem. 1994;63:2353–2356. doi: 10.1046/j.1471-4159.1994.63062353.x. [DOI] [PubMed] [Google Scholar]
- Randall DJ, Perry SF. Catecholamine. In: Hoar WS, Randall DJ, Farrell AP, editors. Fish physiology. 12B. Academic Press; New York: 1992. pp. 255–299. [Google Scholar]
- Redding JM, Patino R. Reproductive physiology. In: Evans DH, editor. The physiology of fishes. CRC Press; Florida: 1993. pp. 503–534. [Google Scholar]
- Relógio A, Schwager C, Richter A, Ansorge W, Valcárcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riar SS, Malhotra MS, Bhat KS, Parshad R. Changes in reproductive organs of male rats on exposure to hypoxia (6,060 m) and cold (-5 degrees C) Int J Biometeorol. 1979;23:137–149. doi: 10.1007/BF01560094. [DOI] [PubMed] [Google Scholar]
- Rofstad EK. Microenvironment-induced cancer metastasis. Int J Radiat Biol. 2000;76:589–605. doi: 10.1080/095530000138259. [DOI] [PubMed] [Google Scholar]
- Rofstad EK, Måseide K. Radiobiological and immunohistochemical assessment of hypoxia in human melanoma xenografts: acute and chronic hypoxia in individual tumours. Int J Radiat Biol. 1999;75:1377–1393. doi: 10.1080/095530099139250. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Loo LO. Marine eutrophication induced oxygen deficiency: Effects on soft bottom fauna, western Sweden. Ophelia. 1988;29:213–225. [Google Scholar]
- Rosenberg R, Loo LO, Möller P. Hypoxia, salinity and temperature as structuring factors for marine benthic communities in a eutrophic area. Neth J Sea Res. 1992;30:121–129. [Google Scholar]
- Semenza GL. Surviving ischemia: adaptive responses mediated by hypoxia-inducible factor 1. J Clin Invest. 2000a;106:809–812. doi: 10.1172/JCI11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000b;88:1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: using two hands to flip the angiogenic switch. Cancer Metastasis Rev. 2000c;19:59–65. doi: 10.1023/a:1026544214667. [DOI] [PubMed] [Google Scholar]
- Semple PD, Beastall GH, Brown TM, Stirling KW, Mills RJ, Watson WS. Sex hormone suppression and sexual impotence in hypoxic pulmonary fibrosis. Thorax. 1984;39:46–51. doi: 10.1136/thx.39.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang EH, Wu RS. Aquatic hypoxia is a teratogen and affects fish embryonic development. Environ Sci Technol. 2004;38:4763–4767. doi: 10.1021/es0496423. [DOI] [PubMed] [Google Scholar]
- Shang EH, Yu RM, Wu RS. Hypoxia affects sex differentiation and development, leading to a male-dominated population in zebrafish (Danio rerio) Environ Sci Technol. 2006;40:3118–3122. doi: 10.1021/es0522579. [DOI] [PubMed] [Google Scholar]
- Sidell BD. Cardiac metabolism in the Myxinidae: physiological and phylogenetic considerations. Comp Biochem Physiol A. 1983;76:495–505. doi: 10.1016/0300-9629(83)90452-8. [DOI] [PubMed] [Google Scholar]
- Soldatov AA, Rusinova OS, Trusevich VV, Zvesdina TF. Effect of hypoxia on biochemical parameters of Scorpaena erythrocytes. Ukr Biokhim Zh. 1994;66:115–118. [PubMed] [Google Scholar]
- Stewart HS, Ridgway AE, Dixon MJ, Bonshek R, Parveen R, Black G. Heterogeneity in granular corneal dystrophy: identification of three causative mutations in the TGFBI (BIGH3) gene-lessons for corneal amyloidogenesis. Human Mutat. 1999;14:126–132. doi: 10.1002/(SICI)1098-1004(1999)14:2<126::AID-HUMU4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Ton C, Stamatiou D, Dzau VJ, Liew CC. Construction of a zebrafish cDNA microarray: gene expression profiling of the zebrafish during development. Biochem Biophys Res Commun. 2002;296:1134–1142. doi: 10.1016/s0006-291x(02)02010-7. [DOI] [PubMed] [Google Scholar]
- Ton C, Stamatiou D, Liew CC. Gene expression profile of zebrafish exposed to hypoxia during development. Physiol Genomics. 2003;13:97–106. doi: 10.1152/physiolgenomics.00128.2002. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Knaap MS, Leegwater PA, Konst AA, Visser A, Naidu S, Oudejans CB, Schutgens RB, Pronk JC. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol. 2002;51:264–270. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- Van der Meer DL, van den Thillart GE, Witte F, de Bakker MA, Besser J, Richardson MK, Spaink HP, Leito JT, Bagowski CP. Gene expression profiling of the long-term adaptive response to hypoxia in the gills of adult zebrafish. Am J Physiol Regul Integr Comp Physiol. 2005;289:1512–1519. doi: 10.1152/ajpregu.00089.2005. [DOI] [PubMed] [Google Scholar]
- Vidal R, Frangione B, Rostagno A, Mead S, Revesz T, Plant G, Ghiso J. A stop-codon mutation in the BRI gene associated with familial British dementia. Nature. 1999;399:776–781. doi: 10.1038/21637. [DOI] [PubMed] [Google Scholar]
- Voet D, Voet JG. Biochemistry. 2. Wiley and Sons; New York: 1995. [Google Scholar]
- Wakamatsu Y, Pristyaznhyuk S, Kinoshita M, Tanaka M, Ozato K. The see-through medaka: A fish model that is transparent throughout life. Proc Natl Acad Sci USA. 2001;98:10046–10050. doi: 10.1073/pnas.181204298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH. Mammalian oxygen sensing, signaling and gene regulation. J Exp Biol. 2000;203:1253–1263. doi: 10.1242/jeb.203.8.1253. [DOI] [PubMed] [Google Scholar]
- White IR, Pickford R, Wood J, Skehel JM, Gangadharan B, Cutler P. A statistical comparison of silver and SYPRO Ruby staining for proteomic analysis. Electrophoresis. 2004;25:3048–54. doi: 10.1002/elps.200405947. [DOI] [PubMed] [Google Scholar]
- Wixon J, Kell D. The Kyoto encyclopedia of genes and genomes-KEGG. Yeast. 2000;17:48–55. doi: 10.1002/(SICI)1097-0061(200004)17:1<48::AID-YEA2>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MJ, Lai LW, Lien YH. Cytoprotective effects of calbindin-D (28k) against antimycin-A induced hypoxic injury in proximal tubular cells. Life Sci. 2002;21:559–569. doi: 10.1016/s0024-3205(02)01710-1. [DOI] [PubMed] [Google Scholar]
- Wu RSS. Eutrophication, trace organics and water-borne pathogens: Pressing problems and challenge. Mar Pollut Bull. 1999;39:11–22. [Google Scholar]
- Wu RSS, Lam PKS. Glucose-6-phosphate dehydrogenase and lactate dehydrogenase in the green-lipped muscle (Perna virids): Possible biomarkers for hypoxia in the marine environment. Water Res. 1997;11:2797–2801. [Google Scholar]
- Wu RSS, Zhou BS, Randall DJ, Woo NY, Lam PK. Aquatic hypoxia is a disrupter and impairs fish reproduction. Environ Sci Technol. 2003;37:1137–1141. doi: 10.1021/es0258327. [DOI] [PubMed] [Google Scholar]
- Yang TH, Somero GN. Effects of feeding and food deprivation on oxygen consumption, muscle protein concentration and activities of energy metabolism enzymes in muscle and brain of shallow-living (Scorpaena guttata) and deep-living (Sebastolobus alascanus) scorpaenid fishes. J Exp Biol. 1993;181:213–232. [Google Scholar]
- Yang Y, Hwang CK, Junn E, Lee G, Mouradian MM. ZIC2 and Sp3 repress Sp1-induced activation of the human D1A dopamine receptor gene. J Biol Chem. 2000;275:38863–38869. doi: 10.1074/jbc.M007906200. [DOI] [PubMed] [Google Scholar]
- Yenari MA, Minami M, Sun GH, Meier TJ, Kunis DM, McLaughlin JR, Sapolsky DY, Steinberg RM. Calbindin d28k overexpression protects striatal neurons from transient focal cerebral ischemia. Stroke. 2001;32:1028–1035. doi: 10.1161/01.str.32.4.1028. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Behrooz A, Ismail-Beigi F. Regulation of glucose transport by hypoxia. Am J Kidney Dis. 1999;34:189–202. doi: 10.1016/s0272-6386(99)70131-9. [DOI] [PubMed] [Google Scholar]
- Zhang ZP, Wu RSS, Mok HO, Wang YL, Poon WW, Cheng SH, Kong RY. Isolation, characterization and expression analysis of a hypoxia-responsive glucose transporter gene from the grass carp, Ctenopharyngodon idellus. Eur J Biochem. 2003;270:3010–3017. doi: 10.1046/j.1432-1033.2003.03678.x. [DOI] [PubMed] [Google Scholar]