Summary
Epidermal stem cells are of major importance for tissue homeostasis, wound repair, tumor initiation, and gene therapy. Here we describe an in vivo regeneration assay to test for the ability of keratinocyte progenitors to maintain an epidermis over the long term in vivo. Limiting dilution analysis of epidermal repopulating units in this in vivo regeneration assay at sequential time points allows the frequency of short term (transit amplifying cell) and long term (stem cell) repopulating cells to be quantified.
Keywords: Stem cell, transit amplifying cell, epidermis, limiting dilution, skin, regeneration
1. Introduction
In vivo assessment of epidermal stem cell function and frequency has been well recognized as an important goal [1–3]. We have developed and used a long term repopulating assay to test for sustained tissue regeneration and maintenance in vivo. This may be considered the most rigorous definition of an epidermal stem cell. For this assay dissociated keratinocytes regenerate a differentiated epidermis on top of dermal fibroblasts seeded on the subcutaneous fascia of immunodeficient mice. GFP negative keratinocytes serve to ensure the production of an intact differentiated epidermis despite variations in the numbers of GFP positive cells in the test population. For the test population, a range of dilutions of GFP positive keratinocytes is used. Mixtures of test keratinocytes (GFP positive) are seeded into chambers along with a constant number of GFP negative keratinocytes, and the presence of GFP positive epidermal repopulating units is assessed 2 to 30 weeks after epidermal regeneration (Fig. 1). At each assessment the epidermis is scored as positive or negative, for the presence or absence of a cluster of GFP positive cells. By seeding a range of doses of GFP positive keratinocytes in this repopulating assay and waiting until all transit amplifying cells and their progeny have differentiated and been lost from the epidermis, limiting dilution analysis allows the frequency of cells with long-term repopulating ability in a given population to be quantified [3].
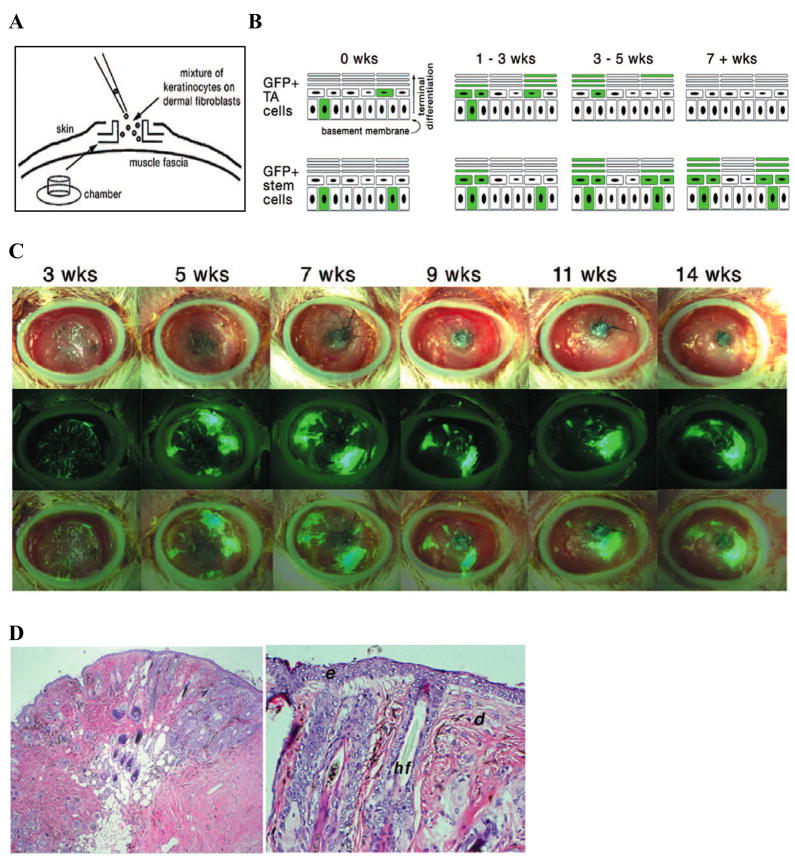
Figure 1.
Limiting dilution analysis for the determination of short term (TA) and long term (stem) repopulating cell frequency. (A) Diagram of the 6 mm diameter silicone chamber implanted onto the dorsal fascia of a NODSCID mouse into which the keratinocyte and fibroblast populations are seeded. (B) Schematic of epidermal layers where GFP positive units derived from short term repopulating cells (TA cells) disappear after 3 to 5 weeks (top row) or GFP positive cells derived from long term repopulating cells (stem cells) persist for 9 to 30 weeks. (C) Maintenance of GFP positive repopulating units up to 14 weeks. Top panels show bright-field images of regenerated skin, middle panels show epifluorescence images taken at the same time, and bottom panels show an overlay of the GFP positive repopulating units on the regenerated skin. (D) Hematoxylin and eosin staining of regenerated skin in cross section (10x, 20x). d: dermis, e: epidermis, hf: hair follicle.
Table 1 is presented as an example of how this type of assay can be used to compare the frequency of stem cells in various populations of keratinocytes. As seen in Table 1, keratinocytes that rapidly adhere to collagen were implanted at a range of doses. At the highest dose of 240,000 keratinocytes, all chambers were positive for GFP positive repopulating units at all time points. At the lowest doses, of 1,900 and 7,500 keratinocytes, no chambers had GFP positive repopulating units at any time point. At intermediate doses, (e.g. 60,000 keratinocytes), it can be seen that, while at early time points (3 weeks) 9 of 10 chambers contained GFP positive repopulating units, at later time points (e.g., 9 weeks) only 5 of the 10 chambers remained positive. At sequential time points the repopulating frequency decreases until only long-term repopulating cells remain and the repopulating unit frequency remains constant thereafter. Using Poisson distribution statistics the calculated frequency of repopulating units corresponds to the derived cell dose at which 37% of the tests yield a negative response (L-Calc software v1.1, www.stemcell.com). For the study in Table 1, we relied on previous work that found that the stem cell frequency of total unsorted cells at 9 weeks was 1 in 30,000 keratinocytes [3]. As can be seen for the dose of 30,000 total unsorted keratinocytes at 9 weeks (Table 1), 4 out of 10 chambers are negative, consistent with Poisson statistic predictions. For each population of cells to be tested (for example, rapidly adherent cells, not-rapidly-adherent cells, unsorted cells) the doses to be used experimentally will depend on the expected long term repopulating unit frequency.
Table 1.
Frequency of repopulating units in rapidly adherent, not rapidly adherent, and unsorted murine keratinocytes over time.1
| Number of GFP+ cells2 | 3 weeks | 5 weeks | 7 weeks | 9 weeks | |
|---|---|---|---|---|---|
| Rapidly Adherent Cells | 1,900 | 0/1 | 0/1 | 0/1 | 0/1 |
| 7,500 | 0/7 | 0/7 | 0/7 | 0/7 | |
| 30,000 | 5/9 | 4/9 | 4/9 | 4/9 | |
| 60,000 | 9/10 | 8/10 | 5/10 | 5/10 | |
| 120,000 | 8/10 | 7/10 | 7/10 | 7/10 | |
| 240,000 | 5/5 | 5/5 | 5/5 | 5/5 | |
| Repopulating Unit Frequency | 1 in 47,501 | 1 in 63,657 | 1 in 81,635 | 1 in 81,635 | |
| (± Standard Error) | (37,476–60,209) | (50,236–80,665) | (64,151–103,884) | (64,151–103,884) | |
| (95% Confidence Interval) | (29,848–75,596) | (40,021–101,253) | (50,900–130,929) | (50,900–130,929) | |
| Not rapidly Adherent Cells | 7,500 | 0/3 | 0/3 | 0/3 | 0/3 |
| 30,000 | 8/9 | 6/9 | 5/9 | 5/9 | |
| 60,000 | 9/10 | 9/10 | 8/10 | 8/10 | |
| 120,000 | 7/7 | 7/7 | 7/7 | 7/7 | |
| 240,000 | 5/5 | 5/5 | 5/5 | 5/5 | |
| Repopulating Unit Frequency | 1 in 21,094 | 1 in 27,630 | 1 in 36,196 | 1 in 36,196 | |
| (± Standard Error) | (15,888–28,006) | (21,147–36,100) | (27,962–46,856) | (27,962–46,856) | |
| (95% Confidence Interval) | (12,103–36,765) | (16,360–46,664) | (21,825–60,031) | (21,825–60,031) | |
| Unsorted Cells | 30,000 | 8/10 | 7/10 | 6/10 | 6/10 |
| Repopulating Unit Frequency | 1 in 18,640 | 1 in 24,918 | 1 in 32,741 | 1 in 32,741 | |
| (± Standard Error) | (12,583–27,613) | (16,683–37,218) | (21,455–49,964) | (21,455–49,964) | |
| (95% Confidence Interval) | (8,629–40,627) | (11,350–54,704) | (14,299–74,969) | (14,299–74,969) | |
Adapted from [17]
Added to chamber with 2 million GFP-negative cells.
Phenotypic analysis of hematopoietic stem cells in in vivo transplantation assays has allowed separation of long-term repopulating cells from cells detected in colony forming assays [4–11]. These types of studies have defined a hierarchy of hematopoietic stem cell phenotypes (see [12] Figure 1). Primitive progenitors that represent the closest stem cell descendents which can be prospectively isolated from the true stem cell by flow cytometry, are still multipotent, yet already have a decline in self-renewal capacity, underscoring long-term repopulating ability as the sine qua non of a stem cell [13, 14]. These progenitors produce distinct highly proliferative colonies, which can differentiate into specific lineages, but unlike a true stem cell are unable to repopulate all hematopoietic lineages for the life of the animal. Thus in vivo transplantation assays have long been the gold standard for the study of hematopoietic stem cells, and after almost 20 years remain so [15].
One important question regarding functional assays for stem cells is what duration of repopulation distinguishes the true epidermal stem cell from a short-term repopulating cell. In the epidermis, the short-term repopulating cells are termed transit amplifying cells. Cell cycle duration has been estimated to be 4 to 5 days and transit amplifying cells go through approximately 3 divisions [16], before terminally differentiating. Our initial studies showed that there is no further decline in repopulating cell frequency after 7 weeks, indicating that at this point transit amplifying cells and their progeny have differentiated and been lost from the epidermis, and we are assaying the true long term repopulating epidermal stem cell [3, 17]. Thus in all subsequent epidermal stem cell studies we have selected an endpoint of 9 weeks or later to ensure the study of stem cells rather than short term repopulating transit amplifying cells.
As noted in similar hematopoietic transplantation assays [18], estimates of stem cell frequency are most certainly underestimates since the detection efficiency of the assay procedure is not known, but is almost certain to be less than 1. In recognition of this we term the GFP positive clusters of cells repopulation units rather than epidermal stem cell units. This does not undermine the value of comparing the relative frequency of progenitor cells in different cell populations. While this assay estimates that 1 in 10,000 basal cells is a truly primitive epidermal stem cell, similar to stem cell frequencies in other tissues [11, 18, 19], previous work on the epidermis showed that 1 in 10 basal cells was a colony forming stem cell (for review, see [20]). However, more recently it has been shown that colony forming cells do not all represent stem cells [1, 17, 20]. These findings lead us to believe that stem cell frequency is significantly less than previously thought [20–23]. Using the in vivo transplantation assay described here multiple studies have reported that the frequency of epidermal stem cells in young and in neonatal murine epidermis is approximately 1 in 10,000 basal cells [3, 17, 24] strengthening the argument that the frequency of epidermal stem cells is similar to that of other somatic stem cell populations.
In this chapter we describe a method to determine the frequency of short and long term repopulating epidermal progenitors in vivo using limiting dilution analysis. First, primary keratinocytes are isolated from GFP positive and GFP negative neonatal murine epidermis. Then fibroblasts are isolated from the GFP negative skin. Next, a range of doses of GFP positive test keratinocytes are prepared and left on ice, while silicone chambers are implanted onto the fascia of NODSCID mice. Fibroblasts and then keratinocytes are seeded into the chambers. The regenerated epidermis is imaged over time and analyzed for presence or absence of GFP positive repopulating units. Analysis of the positive and negative results is performed using Poisson statistics for limiting dilution analysis, and allows a quantitative analysis of the repopulating unit frequency. The strengths of this assay are the long term functional nature of the repopulation carried out in vivo, which allows for the distinction between true long term repopulating stem cells and short term repopulating (transit amplifying) cells, and the ability to quantify the number of long term repopulating epidermal stem cells.
2. Materials
2.1 Tissue and Reagents
C57BL/6-TgN(ACTbEGFP)1Osb mice (Jackson Laboratories)
NODSCID mice (Jackson Laboratories)
Ketamine (100 mg/mL)
Aceprozamine Maleate (10 mg/mL; Henry Schein)
Sulfatrim (Actavis)
Forceps, fine-tip scissors, #15 disposable scalpels
Tegaderm and Coban (3M)
Forane (Baxter)
CNT-07 complete medium (Cell-N-Tech)
Hibiclens (Regent Medical)
HBSS-CMF (Invitrogen)
HBSS-CMF with 5x PSA (Penicillin, Streptomycin & Amphotericin; Invitrogen)
0.05% trypsin-EDTA (Invitrogen)
TNS (Trypsin Neutralizing Solution: HBSS-CMF + 5% chelexed FBS) (see Subheading 2.2)
Dispase (25 U/ml in HBSS-CMF; BD Biosciences)
1% Collagenase Type 1A (Sigma; C9891) (see Subheading 2.2)
Hemacytometer (Fisher)
Trypan blue (Sigma)
UV lamp (Long wave UV (365 nm) filter; Spectroline)
Silicone chambers (6-mm internal diameter; Renner GmbH, Germany; http://www.renner-gmbh.de)
Epifluorescence stereomicroscope (Stemi SV; Carl Zeiss, Inc.) with UV 488 nm filter
L-calc software (Stemsoft; www.stemsoft.com)
2.2 Reagent Preparation
Prepare chelexed FBS by mixing 100 g of Chelex 100 resin (200–400 mesh; Bio-Rad) with 500 mL FBS and stir at room temperature for 1 hour. Let settle and filter (0.2 uM).
Prepare a 1% (1 mg/mL) solution of Collagenase Type 1A (Sigma #C9891) in HBSS-CMF. Alilquot and store at −20° C. Use within 6 months.
3. Methods
3.1 Primary Isolation of Neonatal Murine Keratinocytes
Using the long-wave UV lamp to determine whether pups are GFP positive, collect 3 to 4 day-old GFP positive and 3 to 4 day-old GFP negative neonates. (see Note 1)
Humanely euthanize neonates as per an approved IACUC protocol.
Using forceps, pinch skin and make a small incision with fine-tip scissors.
Remove truncal skin from neonates and place in CNT-07 medium at 4° C.
Scrape and remove the subcutaneous fat and wash once (1–2 minutes) in 10% Hibiclens and then twice in HBSS with 5x PSA (1–2 minutes).
Place skin, epidermis up, in a 35 mm dish with 3 mL dispase, and incubate 24 hours at 4° C, or 2.5 hours at 37° C.
Remove skin carefully from dispase and gently peel epidermis from dermis. To collect the dermal fibroblasts proceed to Subheading 3.2.
Place the epidermis in 2 mL of pre-warmed 0.05% trypsin-EDTA and incubate for 20 minutes at 37° C followed by gentle tapping and shaking to encourage keratinocyte separation from the stratum corneum.
Neutralize the trypsin with 6 mL TNS, then separate the keratinocytes from the stratum corneum by centrifugation at 500 rpm for 10 minutes. Remove the supernatant and resuspend the cell pellet in CNT-07 medium.
Using the hemacytometer and trypan-blue, count the cells and determine the percentage of dead cells; approximately 3 to 4 million keratinocytes are usually recovered from each neonatal murine epidermis. (see Note 2)
Centrifuge at 500 rpm for 5 minutes and resuspend the GFP positive and GFP negative keratinocyte populations in CNT-07 medium in the appropriate volume. (see Subheading 3.4) Keep cells on ice while chambers are implanted.
3.2 Primary Isolation of Neonatal Murine Dermal Fibroblasts
Following dispase treatment and removal of the epidermis (Subheading 3.1), place the dermis in 2 mL of pre-warmed 0.05% trypsin and incubate for 25 minutes at 37° followed by 2 seconds of vortexing.
Neutralize trypsin with 6 mL TNS and place dermis in empty 100 mm dish.
Using 2 #15 scalpels, cut dermis into 2 mm3 pieces; transfer pieces to 0.25% – 0.5% collagenase (dilute from 1% stock in HBSS-CMF; add 0.3 mM CaCl2 ) Incubate for 1 hour at 37° C; after slight vortexing a homogeneous cell slurry should be obtained.
Add 6 mL of HBSS-CMF and centrifuge at 1000 rpm for 20 minutes. Remove the supernatant and resuspend the cell pellet in CNT-07 medium.
After resuspension, strain cells through 100 uM cell strainer to remove debris and any non-dissociated tissue still remaining.
Using the hemacytometer and trypan-blue, count the cells and determine the percentage of dead cells; approximately 5 to 7 million fibroblasts should be recovered from each pup dermis. (see Note 3)
Centrifuge at 1000 rpm for 10 minutes and resuspend in the appropriate volume of CNT-07 medium. (see Subheading 3.4) Keep cells on ice while chambers are implanted.
3.3 Chamber Implantation
Once the cells have been prepared at the appropriate dilutions and are stored on ice, the chambers can be implanted in preparation for cell transplantation. (see Note 5)
Anesthetize host NODSCID mice by intramuscular injection of ketamine / aceprozamine (100mg/kg ketamine / 10mg/kg acepromazine, usually 0.03–0.05ml of 1mg/ml ketamine / 0.1 mg/ml aceprozamine). The mouse should be fully sedated by 5 to 10 minutes as verified by testing the foot pad reflex. Sedation lasts 1 to 2 hours, providing ample time to implant the chamber and allow the cells to settle and adhere.
Using forceps pinch the dorsal skin in the midline just posterior to the shoulder blades and cut just below the forceps to excise a small ellipse of tissue approximately 5 mm in length.
Fold a sterilized chamber in half and clasp with forceps. Pinch up the skin on the anterior side of the excised site and insert folded chamber with forceps. Hold skin and inserted side of the chamber while using the forceps to bring the posterior side of the excised site around the rest of the chamber. (see Note 6)
To prevent the chamber from moving around on the fascia, wrap the upper body of the mouse with Coban. Cut a small hole in the Coban first to allow the top of the chamber to be exposed.
Cells should be pipetted directly onto the muscle fascia in a total volume of 50 to 70 uL. Fibroblasts are added first, and then 5 to 10 minutes later the keratinocytes are added. (see Subheading 3.4) Place Tegaderm over the chamber and adhere to the Coban.
Deliver Sulfatrim (200 mg sulfamethoxazole and 40 mg trimethoprim/200 mL water; protect from light) and carefully monitor the Coban and Tegaderm daily until the implantation has stabilized.
The Tegaderm should be replaced when necessary for 1 to 2 weeks after chamber implantation, after which time the chamber can be left open to the air.
3.4 Cell Transplantation
Resuspend the GFP negative fibroblasts and keratinocytes at concentrations of 108 cells/mL. In order to ensure formation of a regenerated skin a minimum of 2 x 106 fibroblasts and 2 x 106 keratinocytes should be used, and these doses should be kept constant for each dilution of test keratinocytes.
The doses of test keratinocytes will depend on the expected stem cell frequency. For initial limiting dilution studies choose dilutions that range many orders of magnitude such as 106, 104, 103, and 102. For 106 test keratinocytes, a dilution which should yield all positive responses, resuspend the GFP positive keratinocytes at a concentration of 50 x 106 cells/mL so that 20 uL equals 106 cells. Continue serially diluting the cells at concentrations that yield the desired test keratinocyte number in a 20 uL volume. Keep cells on ice while the chambers are being implanted.
Seed 20 uL of GFP negative fibroblasts into the chamber first, wait 5 · 10 minutes for the cells to settle and adhere, then add 20 uL of GFP negative keratinocytes and 20 uL of GFP positive keratinocytes at the appropriate dilution.
3.5 Epifluorescence Imaging and Limiting Dilution Analysis
Regenerated epidermis can be imaged 1 to 2 weeks after chamber implantation and as desired after that. (see Note 7)
For imaging, anesthetize mice (see Subheading 3.3) and place 100 – 200 uL of sterile PBS on top of the regenerated skin and scab.
After 20 minutes, gently dab away the remaining PBS and soaked scab with sterile kimwipes being careful not to disrupt the regenerated epidermis.
Place the mouse under the microscope and capture both bright-field and fluorescence images. It is important to place each mouse in the same orientation and to use the same exposure times for sequential observations. (see Figure 1C)
For limiting dilution analysis the regenerated epidermis is scored as positive if at least one GFP-positive epidermal cell cluster is detected.
Use the L-Calc software to perform statistical analysis per the manufacturer’s instructions, and determine the repopulating unit frequency. (see Table 1)
The regenerated skin can be excised for immunohistochemical analysis at the end of the imaging period. (see Figure 1D)
Footnotes
Pups that are 3 to 4 days old are optimal for the isolation of both follicular and interfollicular neonatal keratinocytes because separation of the epidermis from the dermis is easier and more complete than at later time points.
3.5 to 4 million keratinocytes can routinely be recovered from one day 4 neonatal skin, and less than 10% of the cells should be dead. Less recovery could be due to the following factors: subcutaneous fat incompletely removed resulting in only a partial epidermal-dermal separation; trypsin not adequately pre-warmed; keratinocytes not adequately separated from stratum corneum; keratinocytes not completely resuspended (still clumped) and lost during filtration
5 to 7 million fibroblasts can routinely be recovered from one day 4 neonatal skin, and less than 10% of the cells should be dead. Less recovery could be due to the following factors: dermis not adequately digested by collagenase (see Note 4); fibroblasts not completely resuspended (still clumped) and lost during filtration.
The collagenase treatment of the dermis is complete when no more tissue pieces are visible and a homogeneous cell slurry is obtained . Collagenase that is not properly prepared or has expired may not work as well.
The prepared cells are viable on ice for several hours, and the number of conditions per experiment will be determined by the number of chambers that can be implanted during that time.
The optimal fit of the chambers depends on the initial size of the ellipse of skin that is excised. If the excision is too large the chamber will have to be sutured in place (use 5.0 absorbable sutures). However, if the initial excision site is too small and/or chamber insertion takes many attempts, the host skin will be irritated and the mice tend to disrupt the chambers. The Coban should be tight but not tight enough to restrict breathing.
Scab removal should not be attempted prior to 2 weeks post chamber implantation. During the initial 2 week regeneration period the epidermis is very fragile and should not be disturbed.
References
- 1.Kolodka TM, Garlick JA, Taichman LB. Evidence for keratinocyte stem cells in vitro: long term engraftment and persistence of transgene expression from retrovirus-transduced keratinocytes. Proc Natl Acad Sci U S A. 1998;95(8):4356–61. doi: 10.1073/pnas.95.8.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li A, et al. Extensive tissue-regenerative capacity of neonatal human keratinocyte stem cells and their progeny. J Clin Invest. 2004;113(3):390–400. doi: 10.1172/JCI19140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider TE, et al. Measuring stem cell frequency in epidermis: a quantitative in vivo functional assay for long-term repopulating cells. Proc Natl Acad Sci U S A. 2003;100(20):11412–7. doi: 10.1073/pnas.2034935100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodgson GS, Bradley TR. Properties of haematopoietic stem cells surviving 5-fluorouracil treatment: evidence for a pre-CFU-S cell? Nature. 1979;281(5730):381–2. doi: 10.1038/281381a0. [DOI] [PubMed] [Google Scholar]
- 5.Lerner C, Harrison DE. 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp Hematol. 1990;18(2):114–8. [PubMed] [Google Scholar]
- 6.Morrison SJ, I, Weissman L. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1(8):661–73. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 7.Randall TD, et al. Expression of murine CD38 defines a population of long-term reconstituting hematopoietic stem cells. Blood. 1996;87(10):4057–67. [PubMed] [Google Scholar]
- 8.Spangrude GJ, Johnson GR. Resting and activated subsets of mouse multipotent hematopoietic stem cells. Proc Natl Acad Sci U S A. 1990;87(19):7433–7. doi: 10.1073/pnas.87.19.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trevisan M, Iscove NN. Phenotypic analysis of murine long-term hemopoietic reconstituting cells quantitated competitively in vivo and comparison with more advanced colony-forming progeny. J Exp Med. 1995;181(1):93–103. doi: 10.1084/jem.181.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Zant G. Studies of hematopoietic stem cells spared by 5-fluorouracil. J Exp Med. 1984;159(3):679–90. doi: 10.1084/jem.159.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissman IL. The road ended up at stem cells. Immunol Rev. 2002;185:159–74. doi: 10.1034/j.1600-065x.2002.18514.x. [DOI] [PubMed] [Google Scholar]
- 12.Terskikh AV, et al. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;102(1):94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- 13.Akashi K, et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–7. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 14.Kondo M, I, Weissman L, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–72. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 15.Purton LE, Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1(3):263–70. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Watt FM. Epidermal stem cells: markers, patterning and the control of stem cell fate. Philos Trans R Soc Lond B Biol Sci. 1998;353(1370):831–7. doi: 10.1098/rstb.1998.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strachan LR, et al. Rapid adhesion to collagen isolates murine keratinocytes with limited long-term repopulating ability in vivo despite high clonogenicity in vitro. Stem Cells. 2008;26(1):235–43. doi: 10.1634/stemcells.2007-0534. [DOI] [PubMed] [Google Scholar]
- 18.Szilvassy SJ, et al. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci U S A. 1990;87(22):8736–40. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 20.Strachan LR, Ghadially R. Tiers of Clonal Organization in the Epidermis: The Epidermal Proliferation Unit Revisited. Stem Cell Rev. 2008 doi: 10.1007/s12015-008-9020-6. [DOI] [PubMed] [Google Scholar]
- 21.Bickenbach JR, Stern MM. Plasticity of epidermal stem cells: survival in various environments. Stem Cell Rev. 2005;1(1):71–7. doi: 10.1385/SCR:1:1:071. [DOI] [PubMed] [Google Scholar]
- 22.Terunuma A, et al. Stem cell activity of human side population and alpha6 integrin-bright keratinocytes defined by a quantitative in vivo assay. Stem Cells. 2007;25(3):664–9. doi: 10.1634/stemcells.2006-0434. [DOI] [PubMed] [Google Scholar]
- 23.Triel C, et al. Side population cells in human and mouse epidermis lack stem cell characteristics. Exp Cell Res. 2004;295(1):79–90. doi: 10.1016/j.yexcr.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 24.Yue L, et al. Epidermal regeneration: an increase in short term repopulating cells allows aged epidermis to keep up with the young. Journal of Investigative Dermatology. 2008;128(S1):S107. [Google Scholar]