Abstract
In growing plant cells, the combined activities of the cytoskeleton, endomembrane, and cell wall biosynthetic systems organize the cytoplasm and define the architecture and growth properties of the cell. These biosynthetic machineries efficiently synthesize, deliver, and recycle the raw materials that support cell expansion. The precise roles of the actin cytoskeleton in these processes are unclear. Certainly, bundles of actin filaments position organelles and are a substrate for long-distance intracellular transport, but the functional linkages between dynamic actin filament arrays and the cell growth machinery are poorly understood. The Arabidopsis (Arabidopsis thaliana) “distorted group” mutants have defined protein complexes that appear to generate and convert small GTPase signals into an Actin-Related Protein2/3 (ARP2/3)-dependent actin filament nucleation response. However, direct biochemical knowledge about Arabidopsis ARP2/3 and its cellular distribution is lacking. In this paper, we provide biochemical evidence for a plant ARP2/3. The plant complex utilizes a conserved assembly mechanism. ARPC4 is the most critical core subunit that controls the assembly and steady-state levels of the complex. ARP2/3 in other systems is believed to be mostly a soluble complex that is locally recruited and activated. Unexpectedly, we find that Arabidopsis ARP2/3 interacts strongly with cell membranes. Membrane binding is linked to complex assembly status and not to the extent to which it is activated. Mutant analyses implicate ARP2 as an important subunit for membrane association.
In plant cells, the actin cytoskeleton forms an intricate network of polymers that organizes the cytoplasm and defines the long-distance intracellular trafficking patterns of the cell. The actin network is critical not only for tip-growing cells (for review, see Cole and Fowler, 2006; Lovy-Wheeler et al., 2007) but also during the coordinated cell expansion that occurs in cells that utilize a diffuse growth mechanism (for review, see Wasteneys and Galway, 2003; Smith and Oppenheimer, 2005). For example, the polarized diffuse growth of leaf trichomes is highly sensitized to actin cytoskeleton disruption (Mathur et al., 1999; Szymanski et al., 1999), and a recent analysis of Arabidopsis (Arabidopsis thaliana) ACTIN mutants revealed widespread cell swelling and isotropic expansion in numerous cell types in the root and shoot (Kandasamy et al., 2009). The actin network is dynamic. The array reorganizes during cell morphogenesis (Braun et al., 1999; Szymanski et al., 1999) and in response to endogenous (Lemichez et al., 2001) and external (Hardham et al., 2007) cues. A major research goal is to better understand not only how plant cells convert G-actin subunits to particular actin filament arrays but also how the actin network interacts with the cellular growth machinery during cell expansion.
This is a difficult problem to solve, because in expanding vacuolated cells the actin array adopts numerous configurations and consists of dense meshworks of cortical actin filaments and bundles (Baluska et al., 2000), thick actin bundles that penetrate the central vacuole (Higaki et al., 2006), and meshworks of filaments and bundles that surround the nucleus and chloroplasts (Kandasamy and Meagher, 1999; Collings et al., 2000). The spatial relationships between these actin networks and localized cell expansion are not obvious. Certainly, the plasma membrane-cell wall interface is a critical location for the regulated delivery and fusion of vesicles containing cell wall polysaccharides. Frequent reports of localized domains of enriched cortical actin signal at regions of presumed localized cell expansion have led to the widely held view that the cortical actin array creates local tracks for vesicle-mediated secretion (for review, see Smith and Oppenheimer, 2005; Hussey et al., 2006). In one study, the dynamics of actin filaments were analyzed in living hypocotyl epidermal cells that utilize a diffuse growth mechanism (Staiger et al., 2009). In this case, individual actin filaments are very unstable and randomly oriented; therefore, the precise relationships between cortical F-actin, vesicle delivery, and cell shape change remain obscure. The best known function for the actin cytoskeleton is that of a track for myosin-dependent vesicle and organelle trafficking (Shimmen, 2007). The actin bundle network mediates the transport of cargo between endomembrane compartments (Geldner et al., 2001; Kim et al., 2005) and the long-distance actomyosin transport of a variety of organelles, including the Golgi (Nebenfuhr et al., 1999; Peremyslov et al., 2008; Prokhnevsky et al., 2008). Generation of distributed (Gutierrez et al., 2009; Timmers et al., 2009) and localized (Wightman and Turner, 2008) actin bundle networks appears to define early steps in the trafficking of Golgi-localized cellulose synthase complexes to the sites of primary and secondary wall synthesis, respectively.
Plant cells employ diverse collections of G-actin-binding proteins, actin filament nucleators, and actin-bundling and cross-linking proteins to generate and remodel the F-actin network (for review, see Staiger and Blanchoin, 2006). One actin filament nucleator, termed the Actin-Related Protein2/3 (ARP2/3) complex, controls numerous aspects of plant morphogenesis and development. The vertebrate complex consists of the actin-related proteins ARP2 and ARP3 and five other unrelated proteins termed ARPC1 to ARPC5, in order of decreasing mass. ARP2/3 in isolation is inactive, but in the presence of proteins termed nucleation-promoting factors such as WAVE/SCAR (for WASP family Verprolin homologous/Suppressor of cAMP Repressor), ARP2/3 is converted into an efficient actin filament-nucleating machine (for review, see Higgs and Pollard, 2001; Welch and Mullins, 2002). In mammalian cells, ARP2/3 activities are linked to membrane dynamics. Keratocytes that crawl persistently on a solid substrate appear to use ARP2/3-generated dendritic actin filament networks at the leading edge to either drive or consolidate plasma membrane protrusion (Pollard and Borisy, 2003; Ji et al., 2008). In many vertebrate cell types, ARP2/3 has a strong punctate intracellular localization (Welch et al., 1997; Strasser et al., 2004), which could reflect hypothesized activities at the Golgi (Stamnes, 2002) or late endosomal (Fucini et al., 2002; Holtta-Vuori et al., 2005) compartment.
Genetic studies in plants reveal nonessential but widespread functions for ARP2/3. In the moss Physcomitrella patens, the ARPC4 and ARPC1 subunit genes are critical during tip growth of protonemal filaments (Harries et al., 2005; Perroud and Quatrano, 2006). In Arabidopsis, loss of either ARP2/3 subunit gene or mutations in WAVE complex genes that positively regulate ARP2/3 cause complicated syndromes, including the loss of polarized diffuse growth throughout the shoot epidermis, defective cell-cell adhesion, and decreased hypocotyl elongation (for review, see Szymanski, 2005). Altered responses to exogenous Suc (Li et al., 2004; Zhang et al., 2008) and reduced root elongation (Dyachok et al., 2008) are also reported for wave and arp2/3 strains. In higher plants, the involvement of ARP2/3 in tip growth and root hair development is more subtle. In Lotus japonicus, mutation of NAP1 and PIR1, known positive regulators of ARP2/3 (Basu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004a), causes incompletely penetrant root hair phenotypes, but in the presence of symbiotic bacteria, the mutants have defective infection threads and reduced root nodule formation. Arabidopsis arp2/3 mutants do not have obvious tip growth defects in pollen tubes or root hairs, but in the presence of GFP:TALIN (Mathur et al., 2003b) and in double mutant combinations with the actin-binding protein CAP1 (Deeks et al., 2007), the effects of ARP2/3 on root hair growth are unmasked.
In Arabidopsis, the genetics of the positive regulation of ARP2/3 are well characterized and appear to occur solely through another heteromeric complex termed WAVE (Eden et al., 2002; for review, see Szymanski, 2005). The putative WAVE/SCAR complex contains five subunits, one of which is the ARP2/3 activator SCAR. Plant SCARs contain conserved N-terminal and C-terminal domains that mediate interactions with other WAVE complex proteins and ARP2/3 activation, respectively (Frank et al., 2004; Basu et al., 2005). In nonplant systems, the regulatory relationships between WAVE and ARP2/3 appear to vary between cell types and species (for review, see Bompard and Caron, 2004; Stradal and Scita, 2006). However, in Arabidopsis, double mutant analyses indicate that WAVE is the sole pathway for ARP2/3 activation and that all subunits positively regulate ARP2/3 (Deeks et al., 2004; Basu et al., 2005; Djakovic et al., 2006). SCAR quadruple mutants are indistinguishable from arp2/3 null plants (Zhang et al., 2008). In moss, BRICK1 and ARP2/3 mutants have similar phenotypes, suggesting conserved regulatory relationships between WAVE and ARP2/3 in the plant kingdom (Harries et al., 2005; Perroud and Quatrano, 2006, 2008).
Despite extensive molecular genetic knowledge about the ARP2/3 pathway and the strong actin cytoskeleton and growth phenotypes of arp2/3 plants, there are few direct data on the existence of the plant complex and its cellular function. There are reports of ARP2/3 localization based on the behavior of individual subunits (Le et al., 2003). In some cases, the results are weakened by the unknown specificity of heterologous ARP2/3 antibodies (Van Gestel et al., 2003; Fiserova et al., 2006). A specific antibody was raised against Silvetia ARP2 (Hable and Kropf, 2005). In developing zygotes, rhizhoid emergence is an early and actin-dependent developmental event, and at this stage a broad subcortical cone of ARP2 signal extends from the nuclear envelope toward the rhizhoid apex (Hable and Kropf, 2005). Double labeling experiments detected considerable overlap between ARP2 and actin, but surprisingly, there was a broad cortical domain of putative organelle-associated distal ARP2 that did not overlap with actin. In tip-growing P. patens chloronema cells, ARPC4 also appears to be membrane associated and localizes to a broad subcortical apical zone (Perroud and Quatrano, 2006). For these localization and genetic studies that rely on individual ARP2/3 subunits, it is important to prove that a plant ARP2/3 complex exists to test for an association of the complex with endomembrane compartments.
In this paper, we provide several lines of evidence for an evolutionarily conserved pathway for ARP2/3 complex assembly in plant cells. These studies are based in part on genetic and biochemical analyses of the putative ARP2/3 subunit gene ARPC4. We found that disruption of the ARPC4 gene caused catastrophic disassembly of the complex and an array of phenotypes that were indistinguishable from known arp2/3 mutants. Chromatography experiments clearly revealed that functional hemagglutinin (HA)-tagged ARPC4 and endogenous ARP3 subunits assemble fully into ARP2/3 complexes. Surprisingly, much of the cellular pool of the plant ARP2/3 complex is membrane associated. An analysis of an extensive collection of wave and arp2/3 mutants allowed us to conclude that the normal association with membranes depended on the presence of ARP2 and the assembly status of the complex but not on the existence of an active pool of ARP2/3 in the cell.
RESULTS
Molecular Characterization of ARPC4
In all known ARP2/3 complexes, ARPC4 is a core subunit that seeds complex assembly (Winter et al., 1999; Gournier et al., 2001). However, in our screens and in a comprehensive mapping study of distorted mutants, there was no locus that mapped to the region of ARPC4 (Schwab et al., 2003). To test the involvement of Arabidopsis ARPC4 in the WAVE-ARP2/3 pathway, we initiated a reverse genetic analysis of the gene using the Salk collection of Arabidopsis T-DNA knockout lines (Alonso et al., 2003). ARPC4 was originally described as a protein fusion to a kinesin-related protein (Mathur et al., 2003a). Subsequently, an ARPC4 cDNA sequence very similar to those of the yeast and human homologs was deduced based on the designed oligonucleotide primer sequences and reverse transcription (RT)-PCR amplification products (Li et al., 2003). However, a 5′ end sequence from a full-length ARPC4 DNA clone (GenBank accession no. CB258042) predicted a different sequence at the N terminus based on the presence of an intron separating the start codon and the second amino acid. We confirmed the accuracy of this alternative gene model, shown in Figure 1A, by testing an array of oligonucleotides for their ability to amplify the 5′ end of ARPC4 mRNA in RT-PCR experiments. Primers C4RT-P4 and C4RT-P5 (Supplemental Table S1) targeting exon 2 amplified a cDNA molecule, while C4RT-P6 (Supplemental Table S1) targeting intron 2 failed to generate an amplification product. DNA sequencing of a cloned ARPC4 cDNA identified a single open reading frame that encodes a polypeptide of 20 kD that is approximately 65% identical to yeast and human ARPC4.
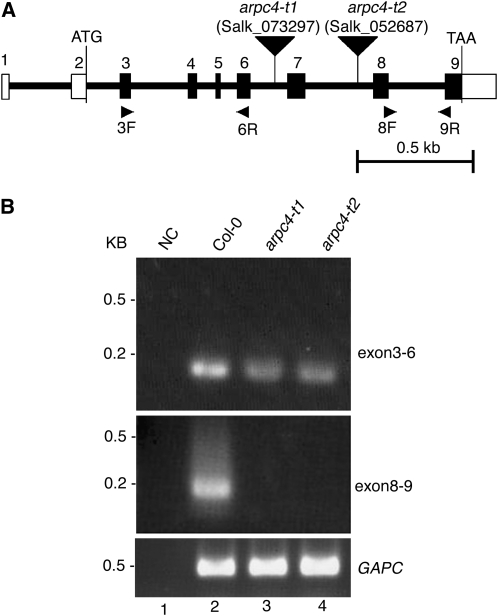
Figure 1.
Physical map of the ARPC4 gene and characterization of the arpc4 T-DNA insertion alleles. A, The positions of exons (numbered vertical rectangles) and introns (thick lines) are represented. The start and stop codons are indicated, and the 5′ and 3′ untranslated regions are labeled with open rectangles. The locations of the arpc4-t1 and arpc4-t2 T-DNA insertions are shown using inverted black triangles. The names and locations of primers used for RT-PCR experiments are also indicated. Bar = 0. 5 kb. B, The T-DNA insertions cause premature transcriptional termination. Transcription of the ARPC4 exons upstream (top panel; using primers 3F and 6R) and downstream (middle panel; using primers 8F and 9R) from the T-DNA was monitored. The quality of the RNA was assayed using primers to detect glyceraldehyde 3-phosphate dehydrogenase subunit C (GAPC; bottom panel). NC is a “no-template” control for the potential contamination of reagents with ARPC4 cDNA. DNA size standards are shown to the left.
If ARPC4 encodes an important ARP2/3 subunit, then mutation of the gene should cause the syndrome of phenotypes displayed by other known distorted mutants. The insertion line SALK_073297 (arpc4-t1) harbors a T-DNA insertion in the sixth intron of ARPC4. The location of the T-DNA was confirmed using the T-DNA-specific oligonucleotide primer LB1 and the ARPC4-specific primer SALK_073297-R (Supplemental Table S1). Lines that were homozygous for the arpc4-t1 allele displayed a typical distorted trichome phenotype (Fig. 2B). Like all other known distorted mutants, arpc4-t1 was recessive, and heterozygous plants gave rise to progeny that segregated very close to the expected Mendelian ratio of 3:1 wild type:mutant. To confirm that the distorted trichome phenotype was caused by a mutation of ARPC4, we used a second independent T-DNA allele (SALK_052687) named arpc4-t2. The location of the arpc4-t2 T-DNA in the seventh intron of the gene was confirmed by PCR using the primers LB1 and SALK_052687-F (Supplemental Table S1). The arpc4-t2 mutation was also recessive. Complementation tests demonstrated that arpc4-t1 and arpc4-t2 were alleles of the same gene, because when the two mutants were crossed, 100% (n = 16) of the F1 progeny had the distorted phenotype.
Figure 2.
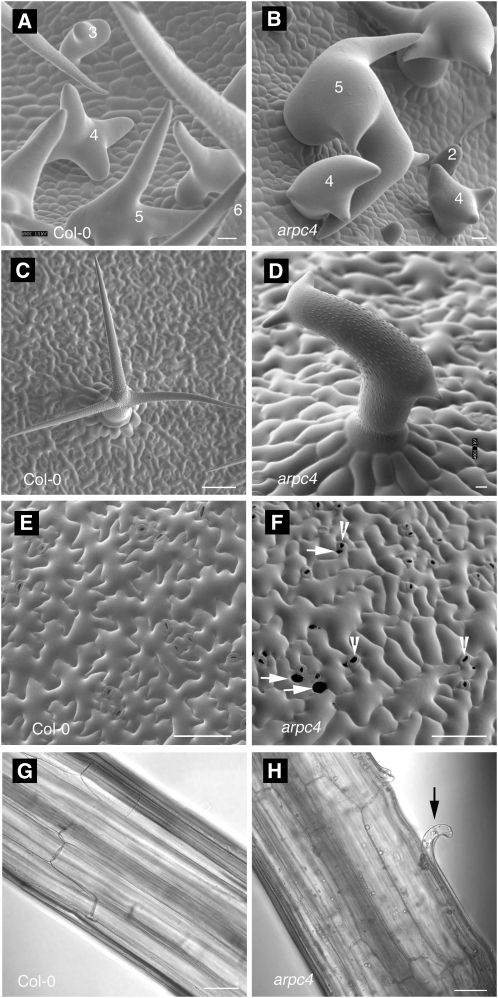
Epidermal cell shape and cell-cell adhesion defects of arpc4 plants. A to D, Upper surface of developing leaves, showing young trichomes at the onset of the mutant phenotype during the transition to stage 4. A, Wild type. B, arpc4. C and D, Mature trichomes on wild-type (C) and arpc4 (D) leaves. E and F, Upper surface of cotyledon pavement cells at 12 d after germination. E, Wild type. F, arpc4. G and H, Cell-cell adhesion defects in etiolated hypocotyl. G, Wild type. H, arpc4. Bars = 50 μm (A, B, E, and F), 10 μm (C and D), and 100 μm (G and H). Numbers in A and B indicate the trichome developmental stages (Szymanski et al., 1998). Arrowheads indicate stomata. Arrows indicate gaps between adjacent pavement cells. The black arrow in H indicates cell adhesion defects on the arpc4 hypocotyl.
The RNA from each genotype was intact, because glyceraldehyde 3-phosphate dehydrogenase subunit C transcripts were easily detected (Fig. 1B, lanes 2–4). The PCR reagents were free from contamination, because in the absence of cDNA template there was no amplification signal (Fig. 1B, lane 1), and there was no detectable contamination of the RNA with genomic DNA, because higher molecular mass PCR products containing introns were not detected (Fig. 1B, lanes 2–4, top panel). Both arpc4 alleles caused premature transcriptional termination, because mRNA containing exons 3 to 6 was detected but the exons downstream from the T-DNA were not (Fig. 1B). Both arpc4 alleles are predicted to be null, because they cause equal phenotypes and these truncated transcripts do not encode the C-terminal amino acids that are required for critical interaction with the other core subunit, ARPC2 (Robinson et al., 2001; El-Assal et al., 2004b). Therefore, the genetic analysis of two independent arpc4 alleles proves that disruption of ARPC4 causes the distorted trichome phenotype.
We examined the phenotypes of the arpc4 alleles in more detail and compared them with the strong ARP2/3 subunit mutants arp3/dis1 (Le et al., 2003) and arpc2/dis2 (El-Assal et al., 2004b). In segregating populations, arpc4-t1 and arpc4-t2 plants displayed completely penetrant and equally severe effects on trichome morphogenesis. Following branch initiation (stage 3), wild-type trichomes transition to a growth phase in which the initiated trichome branches elongate with a blunt tip morphology (stage 4; Fig. 2A; see Fig. 1 in Szymanski et al., 1998). During stage 5, the branch tip acquires a pointed morphology (Fig. 2A), and most cell expansion occurs during this phase (Szymanski et al., 1999). All of the leaf trichomes on arpc4 plants, like other distorted mutants, displayed a loss of polarized growth at the transition to stage 4 and the cells twisted and swelled in an unpredictable manner (Fig. 2, B and D). Compared with the wild type (Fig. 2C), mature arpc4 trichomes (Fig. 2D) also have a greatly decreased trichome branch length (Table I), but arpc4 did not differ from arpc2/dis2 and arp3/dis1.
Table I.
Comparisons of the growth and morphogenesis phenotypes of arpc4 with the wild type and known arp2/3 mutants
Values shown are means ± sd. Asterisks indicate significant differences from the wild type using Student's t test (P < 0.05).
| Sample | Wild Type | arpc4-t1 | arpc2/dis2-1 | arp3/dis1-1 |
|---|---|---|---|---|
| Trichome branch length (μm) | ||||
| Branch 1 | 229.0 ± 79.8 (n = 9) | 73.7 ± 42.4* (n = 17) | 58.3 ± 50.6* (n = 13) | 118.3 ± 68.8* (n = 11) |
| Branch 2 | 197.8 ± 44.3 (n = 9) | 38.2 ± 21.2* (n = 17) | 22.9 ± 12.2* (n = 13) | 40.9 ± 33.6* (n = 11) |
| Branch 3 | 137.1 ± 43.2 (n = 9) | 24.3 ± 11.9* (n = 10) | 17.6 ± 6.7* (n = 13) | 20.1 ± 9.5* (n = 11) |
| Etiolated hypocotyl length (mm) | 18.0 ± 1.5 (n = 10) | 13.6 ± 1.7* (n = 10) | 9.4 ± 1.4* (n = 10) | 12.5 ± 1.8* (n = 10) |
| Gap number per mm2 cotyledon | 0 (n = 9) | 10 (n = 9) | 9 (n = 9) | 7.5 (n = 10) |
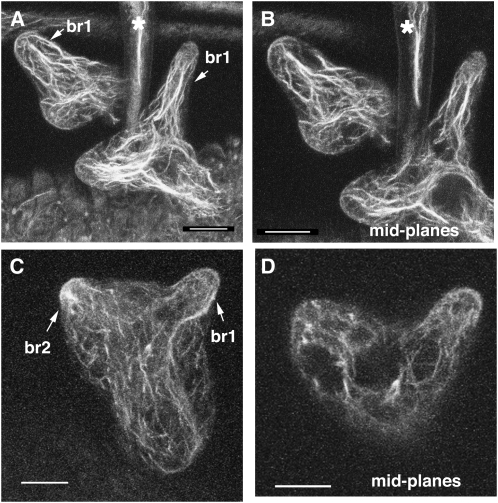
The distorted group genes are expressed throughout the plant (Basu et al., 2005; Zhang et al., 2008), the corresponding mutants have a complex array of growth and developmental phenotypes (Le et al., 2003; Mathur et al., 2003a, 2003b), and we found that arpc4 plants share them all. For example, dark-grown arpc4 plants have a reduced hypocotyl length (Table I). Like other arp2/3 and wave mutants (Li et al., 2003; Brembu et al., 2004), arpc4 pavement cells had a slightly more simple shape compared with the wild type, and cell-to-cell adhesion is affected in both cotyledons (Fig. 2F; Table I) and dark-grown hypocotyls (Fig. 2H). Analyses of the actin phenotypes of distorted trichomes, using either live cell probes or whole mounts of fixed samples, identify a defect in the generation or maintenance of aligned actin bundles in the core cytoplasm of developing branches (Szymanski et al., 1999; Le et al., 2003; Deeks et al., 2004; Basu et al., 2005; Djakovic et al., 2006). The core bundle organization defects in stage 4 trichomes correlate with the severity of cell morphology defects and are likely to be the most relevant. The frequently observed reduction in the quantity of core actin filaments is observed only in stage 5 cells (Le et al., 2003, 2004; Zhang et al., 2005), which is well after the onset of the mutant phenotype. These later stage cytoskeletal defects may reflect an indirect effect of aberrant vacuole positioning or cell morphology. Similar to previous reports (Zhang et al., 2008), we detected aligned core actin filament bundles in nearly all stage 4 wild-type trichomes (19 of 22 branches; Fig. 3, A and B). However, in arpc4 stage 4 trichomes, only nine of 28 stage 4 branches had core actin bundles that were aligned with the cell axis and terminated near the branch apex (Fig. 3, C and D). Based on the identical phenotypes of arpc4 and known wave and arp2/3 mutants, we conclude that ARPC4 functions within the WAVE-ARP2/3 pathway of actin-based morphogenesis.
Figure 3.
The arpc4-t1 mutant causes trichome actin cytoskeleton defects that are indistinguishable from other arp2/3 mutants. A, Maximum projection of confocal image stacks of fixed wild-type trichomes probed with phalloidin. Arrows show trichome branch 1 in two different stage 4 cells. B, Mid planes of wild-type trichome branch 1 from the cells in A. C, Maximum projection of confocal image stacks of arpc4-t1 trichomes, with arrows showing trichome branches 1 and 2. D, Mid planes of arpc4-t1 trichome branches 1 and 2. Stars in A and B indicate a subsection of a mature (stage 6) branch from an adjacent trichome. Bars = 10 μm.
Biochemical Evidence for an Arabidopsis ARP2/3 Complex
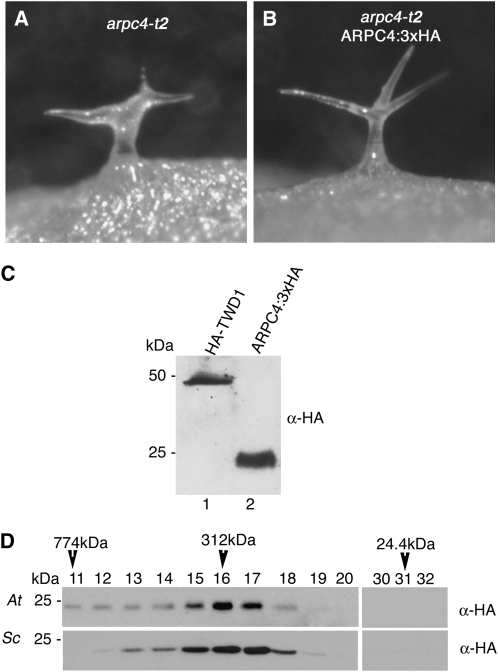
In plants, biochemical evidence for the existence of a plant ARP2/3 complex and its distribution in cells is lacking. Therefore, it was important to develop tools that would allow us to test for the presence of a complex. We tagged the C terminus of ARPC4 with a triple HA, because this region of the protein is unordered and extends outward from the other ARP2/3 subunits (Robinson et al., 2001). We drove ARPC4 expression using its endogenous promoter by cloning ARPC4 and the intergenic sequences between the start codon- and stop codon-flanking genes AT4G14145 and AT4G14150, respectively. To assess the functionality of ARPC4:HA, we used Agrobacterium tumefaciens-mediated transformation to introduce the transgene into arpc4 plants (Clough and Bent, 1998). ARPC4:HA effectively rescued the mutant phenotype, providing further confirmation of the correct identification of ARPC4. An example of a representative ARPC4:HA arpc4-transformed plant is shown in Figure 4B. Transformed line ARPC4:HA-2 had a single T-DNA insert based on a 3:1 segregation of the dominant selectable marker gene and an approximately 3:1 segregation of wild-type (31 plants) to distorted (11 plants) phenotypes. From this segregating population, we generated a stock that was homozygous for the ARPC4:HA transgene and displayed stable rescue activity over multiple generations. We next confirmed that ARPC4:HA retained the epitope tag by probing western blots with a monoclonal anti-HA antibody. The antibody specifically detected an HA-tagged version of TWISTED DWARF1 (HA-TWD1; Geisler et al., 2003; Fig. 4C, lane 1). Western blotting of ARPC4:HA arpc4 extracts detected a single band with a molecular mass of approximately 24 kD, which is very close to what was expected for ARPC4:HA (Fig. 4C, lane 2).
Figure 4.
The epitope-tagged version of ARPC4 expressed with its native promoter rescues the arpc4 phenotype and assembles into an ARP2/3 complex. A, Distorted trichome phenotype of the arpc4-t2 mutant. B, The distorted trichome phenotype in the arpc4-t2 mutant is rescued by ARPC4:HA gene expression driven by its native promoter. C, The HA tag is retained and the ARPC4 protein is detected at the expected size (lane 2). Total, unfractionated HA-TWD1 sample was used as the positive control (lane 1). D, ARPC4-HA protein is a subunit of the ARP2/3 protein complex, the size of which is indistinguishable from the budding yeast ARP2/3. Plant protein microsomes containing ARP2/3 were solubilized with 4% cholate and analyzed by SEC. Column fractions from arpc4-t2 ARPC4:HA rescue line (top panel) and yeast (bottom panel) extracts were probed with anti-HA antibody. The estimated mass of the ARP2/3 complex is labeled above the western-blot panels. At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae.
Although the ARPC4:HA rescue experiments demonstrated clearly the activity of the fusion protein, we wanted to test for the presence of high molecular mass ARP2/3 complexes containing ARPC4:HA. To address this question, we used analytical size-exclusion chromatography (SEC) to approximate the mass of ARPC4:HA-containing complexes. Surprisingly, in our initial cell fractionation assays, we found that Arabidopsis ARP2/3 associated strongly with the membrane fraction (see below). Therefore, the complexes analyzed by SEC were solubilized from a crude microsome fraction (see “Materials and Methods”). Based on its elution profile, the ARPC4:HA complex was estimated to be approximately 310 kD (Fig. 4D, top panel). To compare Arabidopsis ARP2/3 with a known ARP2/3 complex, we analyzed crude extracts from a Saccharomyces cerevisiae strain expressing an epitope-tagged version of ARPC3 (ARC18-HA; a gift from David Drubin). Yeast ARP2/3 complex had a mobility that was nearly identical to that observed for the plant complex (Fig. 4D, bottom panel). Importantly, we failed to detect ARPC4:HA in chromatography fractions that would contain the monomeric (24-kD) form (Fig. 4D). These biochemical and transgenic rescue data indicated that ARPC4:HA assembled into a functional ARP2/3 complex and indicate that the properties of ARPC4:HA accurately reflected the properties of ARP2/3 complexes. Thus far, we have not been able to detect SCAR-dependent ARP2/3 actin filament nucleation activity in the cholate-solubilized extracts.
To further test for the assembly of ARPC4:HA into an ARP2/3 complex, we analyzed its ability to associate with the putative ARP2/3 complex protein ARP3 in a coimmunoprecipitation (coIP) assay. During the process of screening more than a dozen ARP2/3 antibodies that were either heterologous or that we generated against Arabidopsis subunit proteins, we identified a specific antibody that recognized endogenous ARP3. In western-blot experiments, the antibody most strongly recognized a single protein that was preferentially localized in the crude microsome fraction (Fig. 5A, lane 3). The detected protein corresponds to ARP3, because there was no similar signal detected from extracts from arp3/dis1 null plants (Fig. 5A, lanes 2 and 4). In clarified extracts prepared from ARPC4:HA arpc4 plants, we found that endogenous ARP3 and ARPC4-HA associated with each other (Supplemental Fig. S1, top panel, lane 3). The association was specific, because no ARPC4:HA was detected when preimmune serum was used in the coIP assay (Supplemental Fig. S1, lane 2). The coIP and SEC data combined with the identical phenotypes of arpc4 mutants and all other ARP2/3 subunit mutants strongly suggest that ARPC4 and other ARP2/3 subunits assemble into a conserved ARP2/3 complex.
Figure 5.
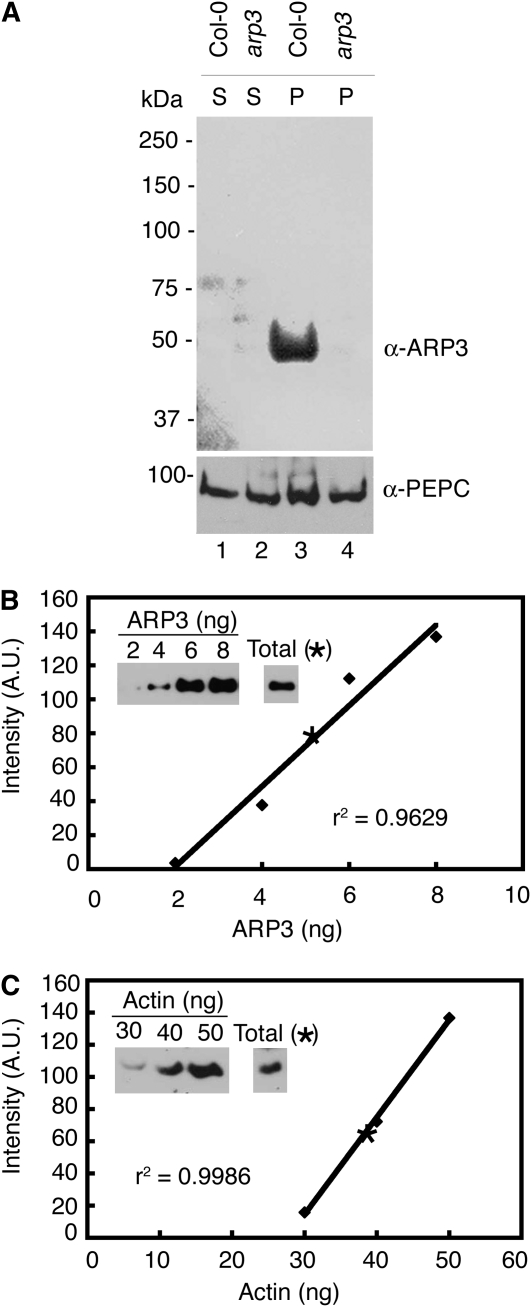
The endogenous ARP2/3 complex is moderately abundant compared with actin. A, The anti-ARP3 antibody is specific and most prominently detects a single protein at the expected mass for ARP3 (Col-0; lane 3). No corresponding band is present when protein extracts from arp3 (dis1) are probed under identical conditions. An antibody against PEPC was used to probe the same filters and demonstrate similar loading between genotypes. P, Pellet; S, soluble. B, The abundance of endogenous ARP3 (see band intensity indicated as a star in the standard curve) as a percentage of total protein was quantified using purified ARP3 inclusion bodies, which was the source of the original antigen, as a standard (see “Materials and Methods”). Western-blot signals were quantified by densitometry using ImageQuant 5.2 software analyzer. C, ACTIN content was also measured as described above. Vertebrate actin was used as a standard. r2 is the correlation coefficient showing how well the standard curve data points fit the line. A.U., Arbitrary unit.
Endogenous ARP2/3 Is Moderately Abundant and Membrane Associated
We used the ARPC4:HA arpc4 line to examine the distribution of ARPC4 in crude plant cell extracts that were subjected to differential centrifugation. We expected most of the protein to reside in the soluble fraction, because established ARP2/3 purification protocols from a variety of organisms typically use this fraction as the source of material (Machesky et al., 1994; Winter et al., 1999; Robinson et al., 2001). However, the common occurrence of punctate, putatively membrane-associated ARP2/3 signal in a variety of motile cell types suggests that vertebrate ARP2/3 has the potential to associate with organelles (Strasser et al., 2004; Shao et al., 2006).
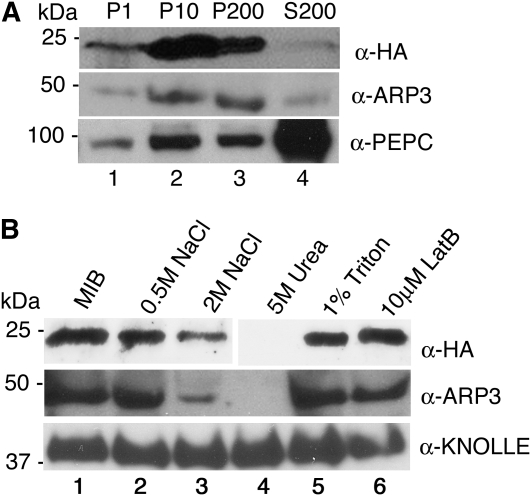
Surprisingly, after homogenization of shoots at 16 to 20 d after germination and filtration of cell debris, we consistently found that most of the ARPC4:HA signal arose from membrane fractions (Fig. 6). The 1,000g pellet fraction containing chloroplasts, nuclei, residual cell debris, and starch grains contained only a small fraction of the total pool of ARP2/3. The vast majority of ARP2/3 was in the higher speed pellet fractions containing crude microsomal membranes (Fig. 6A, lanes 2 and 3). Based on densitometry of immunoblot signal from two independent experiments, only approximately 10% of the protein was present in the soluble fraction after high-speed centrifugation (Fig. 6A, lane 4). The subcellular distribution of endogenous ARP3 was indistinguishable from ARPC4:HA and was also highly enriched in the membrane fractions (Fig. 6A, middle panel, lanes 2 and 3). Phosphoenolpyruvate carboxylase (PEPC) was used as a control to monitor the status of a soluble protein, and as expected, PEPC accumulated in the soluble fraction (Fig. 6A, bottom panel, lane 4).
Figure 6.
The ARP2/3 protein complex is peripherally associated with membrane fractions. A, Plant protein extracts were centrifuged sequentially at different speeds: 1,000g (P1; lane 1), 10,000g (P10; lane 2), and 200,000g to generate final pellet (P200; lane 3) and soluble cytosolic (S200; lane 4) fractions. In each case, the cell fractions were loaded by equal proportion. The fractions from arpc4-t2 ARPC4:HA and wild-type plants were probed with anti-HA (top panel) and anti-ARP3 (middle panel) antibodies, respectively. PEPC, an abundantly soluble protein (bottom panel), was used as an internal control to monitor the soluble protein fractions. B, ARP2/3 is a peripheral membrane complex. Microsomal pellets were resuspended in MIB alone (control; lane 1) or with MIB supplemented with increasing NaCl concentrations (lanes 2 and 3), 5 m urea (lane 4), 1% Triton (lane 5), and the actin filament-depolymerizing drug latrunculin B (LatB; lane 6). After incubation for 30 min under the different conditions, the soluble and membrane-associated fractions were separated by ultracentrifugation and pellets were probed with anti-HA (top panel), anti-ARP3 (middle panel), and anti-KNOLLE to detect the integral membrane syntaxin (bottom panel).
We were also able to measure the abundance of the endogenous ARP3. As expected, we found that ARP3 signal arose exclusively from a molecular mass complex that was indistinguishable in size compared with the complex detected by probing for ARPC4:HA (Fig. 4D). At present, there is no information on the abundance of the plant ARP2/3. We used recombinant His-tagged ARP3 as a standard and conducted quantitative western-blot analysis of total cell extracts prepared from shoots at 16 to 20 d after germination. From these experiments, we estimated that ARP3 was 0.05% of the total protein (Fig. 5B). As a standard for comparison, we probed total Arabidopsis extracts with a maize (Zea mays) actin antibody (Gibbon et al., 1999). The maize antibody efficiently recognizes Arabidopsis, maize, and vertebrate actin (Chaudry et al., 2007). Actin was approximately five times more abundant and constituted 0.23% of the total protein (Fig. 5C). The measured concentration for actin in total shoot extracts was similar to a previously reported value of 0.36% for Arabidopsis rosette leaves (Chaudry et al., 2007). Our data indicate that in cell extracts, ARP2/3 is relatively abundant and that the approximately 5:1 molar ratio of actin to ARP2/3 is lower than the actin-ARP2/3 ratios that have been reported in other species (Pollard et al., 2000). The question of ARP2/3 concentration in the cytosol and in different cell types remains unanswered. Taken together, these data indicate that the Arabidopsis ARP2/3 is a moderately abundant heteromeric complex and that a majority of the protein pool is membrane associated.
To our knowledge, the partitioning of ARP2/3 into soluble and microsomal fractions has not been rigorously tested in other systems. In one example, an accounting of ARP2/3 during its purification from Acanthomoeba indicates that the complex is mostly soluble (Kelleher et al., 1995; Wu and Pollard, 2005). Therefore, we examined in more detail the nature of plant ARP2/3 association with microsomes. ARP2/3 subunits were not partitioned into the microsomal fraction due to nonspecific trapping, because in arpc4, ARP3 is completely solubilized (Fig. 7A, lanes 7 and 8) compared with the marker protein, PEPC (Fig. 7A, bottom panel). We also wanted to rule out the possibility that ARP2/3 was sedimented as part of an aggregated F-actin network. The inability of 1% Triton to solubilize HA-tagged and endogenous ARP2/3 (Fig. 6B, lane 5) is consistent with the idea that the complex is part of a cytoskeleton-associated fraction (Abe and Davies, 1995), but many membrane-associated proteins, including the syntaxin KNOLLE (Fig. 6B), are not solubilized by 1% Triton. Using fluorescence microscopy, we failed to observe any remnants of F-actin after incubation of the microsome fraction with the F-actin-binding probes Alexa 488-phalloidin and rhodamine-phalloidin. Under the same imaging conditions, individual actin filaments polymerized in vitro were easily detected (Basu et al., 2005). Furthermore, treatment of crude microsomes with high concentrations of the actin-depolymerizing drug latrunculin B had no effect on the association of ARP2/3 with membranes (Fig. 6B, lane 6). Instead, microsomal ARP2/3 had properties of a peripheral membrane-associated protein. For example, it could be partially removed from the membrane fraction after incubation with 2 m NaCl and was completely dissociated from membranes by 5 m urea (Fig. 6B, lanes 3 and 4). The integral membrane syntaxin KNOLLE contains a single membrane-spanning segment (Lauber et al., 1997), and neither 2 m salt treatment nor 5 m urea released it from membranes (Fig. 6B, bottom panel). These results suggest that most of the cellular pool of Arabidopsis ARP2/3 exists as a peripheral membrane complex and that the localization to the membrane fraction does not require F-actin.
Figure 7.
The association of plant ARP2/3 with membranes is linked to its assembly and not to the presence of an active pool. A, Cell extracts from the wild type (Col-0) and the indicated genetic mutant backgrounds were separated into crude microsomal membranes (pellet; P) and soluble fractions (soluble; S) by ultracentrifugation. For these experiments, the proportion of soluble to membrane protein that was loaded on SDS-PAGE gels was approximately 1:20. Western blots were probed with anti-ARP3 (top panel) and anti-PEPC (bottom panel) antibodies. B, Microsomal protein from the wild type (Col-0) and the indicated mutant backgrounds was solubilized in 4% cholate and subjected to SEC. Column fractions were TCA precipitated, separated by SDS-PAGE, and blotted with anti-ARP3 antibody.
Assembly-Dependent Association of ARP2/3 with Membranes
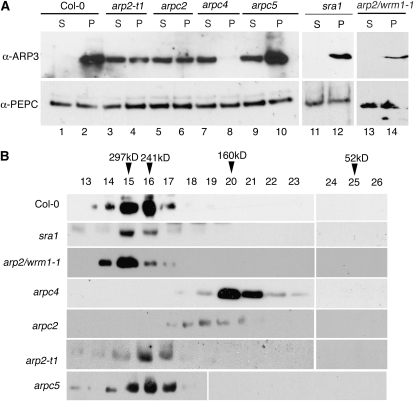
If ARP2/3 is membrane associated, then it is possible that either the assembly of the complex or its activity controls membrane binding. We used a collection of arp2/3 and wave subunit mutants to distinguish these possibilities. Cell extracts from wild-type and mutant cells were prepared, and soluble and crude microsome fractions were isolated by a single high-speed centrifugation step. The solubility of ARP2/3 was monitored by probing soluble and microsome protein fractions with the anti-ARP3 antibody. This experiment was repeated at least two times for each genotype, and the results were very consistent. A representative data set is shown in Figure 7. As expected for wild-type extracts, ARP2/3 was clearly partitioned into the membrane fraction (Fig. 7A, lanes 1 and 2). Interestingly, each of the arp2/3 subunit null lines that were examined had a reduced membrane association of ARP3 (Fig. 7A, lanes 3–10). The effect was specific, because PEPC, an irrelevant marker protein with no known linkage to the actin cytoskeleton, was not affected in the arp2/3 lines (Fig. 7A, bottom panel).
To understand the mechanism by which ARP2/3 associates with the cell membranes, we hypothesized that the membrane association of ARP2/3 was linked to its activation by the WAVE complex. To test this hypothesis, we analyzed the membrane distribution of ARP2/3 in the sra1 background. Loss of the putative WAVE complex protein SRA1 causes a strongly distorted phenotype (Basu et al., 2004; Saedler et al., 2004). SRA1 is an evolutionarily conserved target for Rho family small GTPase signals that may mediate SCAR-dependent activation of ARP2/3 (Eden et al., 2002; Basu et al., 2004, 2008; Steffen et al., 2004). Given that sra1 does not affect the stability of other WAVE/SCAR complex proteins such as NAP1 or SCAR2 (Le et al., 2006), it was not surprising that ARP2/3 was present at normal levels and fully assembled in this background (Fig. 7). Importantly, in sra1 plants, we failed to detect any decrease in the membrane association of ARP2/3 (Fig. 7A, lanes 11 and 12). As an additional test for the membrane association of fully assembled inactive complexes, we analyzed both the microsome association and assembly status of the complex in arp2-1/wrm1-1 (G151D). The arp2-1 complex was fully membrane associated (Fig. 7A, lanes 13 and 14) and displayed a mobility on the SEC column that was indistinguishable from that of the wild-type complex (Fig. 7B). Collectively, the sra1 and arp2-1 data indicated clearly that membrane association was not linked to the ability of the complex to be activated.
We next hypothesized that the assembly status of the complex may be the key to membrane association. Therefore, we analyzed the microsomal association of the complex in different mutant backgrounds lacking individual subunits. ARPC4 is the most critical subunit for the assembly of vertebrate and S. cerevisiae ARP2/3 (Mullins et al., 1997; Winter et al., 1999; Gournier et al., 2001). Consistent with its structural importance, loss of Arabidopsis ARPC4 from cells had the most severe defects in the assembly of ARP3-containing complexes (Fig. 7B). Interestingly, in arpc4, we failed to detect membrane-associated ARP2/3, even though the membrane fraction was overloaded 20-fold compared with the soluble fraction. In the arpc4 background, the overall level of ARP3 was reduced compared with other genotypes (Fig. 7A, lanes 7 and 8). From this, it was apparent that ARP3 was not sufficient for membrane association; however, the reason for a reduced level of ARP3 in arpc4 is not obvious. In Acanthamoeba and Schizosaccharomyces pombe, ARP2/3 subunits are present at nearly equal stoichiometries (Kelleher et al., 1995; Wu and Pollard, 2005). It is possible that the instability of free ARP2/3 subunits and/or subsets of partially assembled complexes maintain equal subunit stoichiometries in the cell, and the reduced level of ARP3 in arpc4 reflects the extreme defects in complex assembly. ARPC2 is a known core subunit of the complex that forms a dimer with ARPC4 (Winter et al., 1999; Gournier et al., 2001). Genetic analyses in Arabidopsis suggest that the ARPC2-ARPC4 interaction is also important for ARP2/3 function (El-Assal et al., 2004b). Consistent with the behavior of yeast and human complex assembly, we found that removal of ARPC2 caused a severe shift of ARP3 toward lower molecular mass that was matched only by removal of ARPC4 (Fig. 7B). Combined with previous yeast two-hybrid data (El-Assal et al., 2004b), our data support a conserved model for ARP2/3 assembly in plants in which the ARPC2-ARPC4 dimer forms a seed around which the other subunits assemble. The greatly increased solubility of ARP3 complexes in the arpc2 background is also consistent with the importance of complex assembly for membrane association (Fig. 7A, lanes 5 and 6).
Upon examination of additional ARP2/3 mutants, it quickly became clear that membrane association did not simply correlate with the sizes of ARP3-containing complexes. Removal of ARP2 had a subtle effect on the size of the complex (Fig. 7B), yet ARP3-containing complexes were mislocalized to the soluble fraction to a similar extent compared with arpc2 (Fig. 7A, lanes 3 and 4). Given that the resolution of our SEC column is approximately 70 kD, the barely detectable shift was consistent with the selective loss of the 44-kD ARP2 subunit. Again, the effects of removal of ARP2 on complex assembly mirrors what has been observed in other species. For example, for both the yeast (Winter et al., 1999; Nolen and Pollard, 2008) and human (Gournier et al., 2001) ARP2/3, loss of the ARP2 subunit does not affect the assembly of any of the other subunits. Recently, mutant ARP2/3 from S. pombe lacking only ARP2 has been purified and crystallized, and removal of ARP2 has very subtle effects on the structure of the complex. However, the partial complex does not respond to nucleation-promoting factors and lacks significant actin filament nucleation activity (Nolen and Pollard, 2008). Given the subtle effect of arp2-t1 null on complex assembly and its relatively strong effect on membrane association, coupled with the fully membrane-associated status of the arp2-1-containing complex, our data suggest that the ARP2 subunit promotes membrane association. In the case of arpc5, we consistently detected a more mild shift of ARP2/3 to the soluble fraction compared with the other mutants (Fig. 7A, lanes 9 and 10). Despite a molecular mass of only 15 kD for ARPC5, SEC analysis of the complex in the arpc5 background showed a broader distribution toward a lower molecular mass (Fig. 7B). This suggests that removal of ARPC5 affects another subunit of the complex in Arabidopsis.
DISCUSSION
Understanding the pathways leading to actin filament nucleation and the function of dynamic actin arrays are long-standing goals in plant cell biology. WAVE-ARP2/3 is implicated as one nucleation pathway that has been adopted throughout the plant kingdom to regulate polarized growth (Frank and Smith, 2002; Le et al., 2003; Li et al., 2003, 2004; Mathur et al., 2003a, 2003b; Harries et al., 2005), Suc signaling (Li et al., 2004; Zhang et al., 2008), and root nodulation (Yokota et al., 2009). In Arabidopsis, progress has been made in characterizing the regulation of ARP2/3: heteromeric complexes function in series to generate (SPIKE1) and translate (WAVE) activating Rho of plants small GTPase signals into an actin-based (ARP2/3) growth response (Basu et al., 2004, 2008). A key missing element of this pathway, and in the field of plant ARP2/3 in general, is biochemical proof of the complex and an understanding of its subcellular distribution. In this paper, we provide genetic, biochemical, and cell biological evidence for the existence of plant ARP2/3. Interestingly, we find that a large fraction of the ARP2/3 pool associates strongly with cell membranes. Membrane binding is linked to the assembly and subunit composition of the complex and not its presumed actin filament nucleation activity in the cell. These unexpected results suggest that the substrate for WAVE complex signaling is an organelle-associated pool of inactive ARP2/3.
These discoveries grew out of molecular genetic analyses of Arabidopsis ARPC4. In yeast and vertebrates, ARPC4 is a core subunit of the complex, and gene disruption experiments in Physcomitrella point to its importance during tip growth (Perroud and Quatrano, 2006). Our genetic analyses demonstrate that Arabidopsis ARPC4 is a bona fide distorted group gene and that its in vivo function is to regulate cell morphogenesis within the well-characterized WAVE-ARP2/3 pathway. This conclusion is based on the identical phenotypes of two independent arpc4 alleles and the ability of ARPC4:HA to rescue arpc4 phenotypes (Fig. 4B). The identical phenotypes of arpc4 plants and the four other known arp2/3 mutants (Le et al., 2003; Li et al., 2003; Mathur et al., 2003a, 2003b; El-Assal et al., 2004b) strongly suggest that ARPC4 is part of an ARP2/3 complex. ARPC4 shares a high degree of amino acid identity with its vertebrate orthologs and also retains a conserved interaction with ARPC2 (El-Assal et al., 2004b). Our ability to coimmunoprecipitate ARPC4:HA and ARP3 (Supplemental Fig. S1) and the strong effects of arpc4 on ARP2/3 assembly and stability (Fig. 7) also prove that ARPC4 is an ARP2/3 subunit.
The availability of an epitope-tagged ARPC4 subunit and a specific ARP3 antibody allowed us to test directly for an ARP2/3 complex and to determine how complex assembly is linked to its subcellular distribution. The results obtained with ARPC4:HA and endogenous ARP3 are equally valid, because both probes monitor the behavior of an ARP2/3 complex and not free subunits. First, SEC experiments showed clearly that ARPC4:HA and endogenous ARP3 are present in high molecular mass complexes that have a mobility that is indistinguishable from yeast ARP2/3 (Fig. 4D). Importantly, we failed to detect a significant pool of monomeric ARPC4:HA or ARP3. In addition, we found that ARPC4:HA and endogenous ARP3 had indistinguishable distributions in cell fractionation experiments (Fig. 6A). The dependence of ARP3 membrane association (Fig. 7A) and complex assembly (Fig. 7B) on several other ARP2/3 subunits provided strong proof that we are following the behavior of a bona fide plant ARP2/3 complex.
Given the highly conserved and functionally interchangeable properties of Arabidopsis, yeast, and human ARP2/3 subunits (Le et al., 2003; Mathur et al., 2003b), it is not surprising that the complex is detected (Fig. 4D) and that it shares a common mechanism for assembly with the yeast and human complexes (Fig. 7B). However, we did not expect to find that a large pool of ARP2/3 associated with cell membranes (Fig. 6). To our knowledge, a large microsomal pool of ARP2/3 has not been previously reported. However, either direct or indirect associations of ARP2/3 with membranes are expected given the wide variety of organelle-based functions for the complex in eukaryotic cells (Eitzen et al., 2002; Fucini et al., 2002; Fehrenbacher et al., 2005; Kaksonen et al., 2005), and the often-observed punctate ARP2/3 localization in mammalian cells reflects a cellular pool of membrane-associated ARP2/3 (Welch et al., 1997; Strasser et al., 2004; Shao et al., 2006). Microsomal ARP2/3 has been reported in plants using a variety of heterologous antibodies (Van Gestel et al., 2003; Fiserova et al., 2006), but the data are conflicting and the extent of membrane association depended on which ARP2/3 subunit antibody was used.
We determined conclusively that both the ARPC4-HA (Fig. 4C) and ARP3 detection was specific (Fig. 5A), but we wanted to rule out the trivial explanation that the association of Arabidopsis ARP2/3 with microsomes was due to denaturation of the complex or nonspecific trapping of the complex within F-actin networks. Nonspecific trapping cannot explain its strong association with membranes, because only a small fraction of the soluble protein PEPC was membrane associated. The complete solubilization of ARP3 in the arpc4 background also argues against a trapping artifact (Fig. 7A, lane 7). The membrane-associated complex is not a denatured aggregate, because the solubilized ARP2/3 is cleanly resolved using SEC (Figs. 4D and 7B) and its migration on the column is sensitive to the loss of individual ARP2/3 subunits (Fig. 7B). Furthermore, the microsomal localization is not caused by an association with actin filament networks, because the crude microsomes have no detectable phalloidin binding and high concentrations of the F-actin-depolymerizing drug latrunculin B had no effect on the association of ARP2/3 with microsomes (Fig. 6B, lane 6). In addition, association of the complex with membranes does not involve ARP2/3-generated actin filaments, because ARP2/3 has a normal membrane association in arp2-1 and sra1 extracts that contain inactive, fully assembled ARP2/3 (Fig. 7).
Instead, we propose that the membrane association of plant ARP2/3 reflects the activity of individual subunits within the complex and that membrane binding may have an in vivo relevance. For example, we find that, compared with the other arp2/3 mutants, the membrane association of ARP2/3 is least affected in the arpc5 background (Fig. 7A, lanes 9 and 10). The arpc5 line has a weaker phenotype compared with all other ARP2/3 subunit mutants (Djakovic et al., 2006). In humans, ARPC5 is a peripheral ARP2/3 subunit, and its removal yields a subcomplex with significant residual nucleation activity (Gournier et al., 2001). Our SEC data are consistent with results obtained for human ARP2/3 assembly, in which ARPC5 recruits ARPC1 into the complex (Gournier et al., 2001). If the reduced mobility of mutant ARP2/3 in arpc5 predominantly reflects the loss of ARPC1 as well, our data suggest that neither ARPC5 nor ARPC1 is essential for efficient membrane association. Because thresholds of ARP2/3 activity define the phenotypic severity in Arabidopsis distorted mutants (Zhang et al., 2008), arpc5 cells may contain partial complexes that retain significant residual membrane-binding and actin filament nucleation activity. The other arp2/3 subunits likely fall too far below the activity threshold to generate a leaky phenotype.
The availability of a collection of WAVE and ARP2/3 subunit mutants allowed us to dissect the mechanisms of membrane association. ARP2/3 partitioning into a microsomal fraction is not linked to its ability to be activated. For example, in the sra1 background, the putative WAVE complex protein NAP1 and the activator SCAR are present (Le et al., 2006), but WAVE does not positively regulate ARP2/3 (Basu et al., 2004). We confirm here that in sra1, it is the activity of ARP2/3 that is affected, rather than its assembly (Fig. 7). Despite its inactive status in sra1, the association of ARP2/3 with microsomes in this mutant background is indistinguishable from the wild type (Fig. 7A, lanes 11 and 12). Likewise, in the arp2-1 (G151D) background, it is the activity and not the assembly of the complex that is clearly affected (Fig. 7), and the mutant complex localizes normally to the microsomal fraction.
Instead, we find that the association of ARP2/3 with membranes correlates with the assembly status and subunit composition of the complex. Interestingly, Arabidopsis ARPC4, like its yeast and human orthologs (Winter et al., 1999; Gournier et al., 2001), is the most critical subunit for complex assembly. In arpc4, partial complexes are found solely in the soluble fraction (Fig. 7). We propose that the complete shift of ARP3 to the soluble fraction in arpc4 (Fig. 7A, lanes 7 and 8) is caused by catastrophic disassembly of the complex and the failure of ARP3 to associate with other subunits that recruit it to a membrane surface. The increased solubility of ARP3 and the severe decrease in the mass of the complex in arpc2 can also be understood in the context of defective complex assembly (Fig. 7, A, lanes 5 and 6, and B). Assembly defects correlate with ARPC2's known function as an ARPC4-binding partner (El-Assal et al., 2004b) and a core subunit for the assembly of yeast and vertebrate ARP2/3 complexes (Winter et al., 1999; Gournier et al., 2001; Robinson et al., 2001).
It is possible that individual ARP2/3 subunits mediate interactions with membranes. Our data point to the involvement of ARP2. In arp2 null strains, partial complexes have a small but detectable decrease in size that is consistent with the selective loss of the ARP2 subunit (Fig. 7B). This result is consistent with a comparative x-ray crystallography study in S. pombe, in which loss of ARP2 does not affect the recruitment of the other subunits into the complex and the lack of ARP2 has subtle effects on the overall structure of the ARP2-lacking complex (Nolen and Pollard, 2008). In the Arabidopsis arp2-t1 background, the complexes are shifted to the soluble fraction to a similar extent compared with arpc2 (Fig. 7A, lanes 3 and 4). The strong membrane association of the arp2 (G151D) point mutant compared with the weak binding of the arp2-t1 null allele indicates that ARP2 participates in membrane association, either by direct lipid-binding activity or by interaction with some other membrane-associated protein that recruits the complex to an organelle surface. Alternatively, loss of ARP2 may cause general conformational changes in the mutant complex that greatly reduce membrane association.
A Cellular Context for the Membrane Association of ARP2/3
The results described above begin to provide some useful clues about the regulation of ARP2/3 in plant cells. In tip-growing cells in moss, highly polarized cell types concentrate ARP2/3 in broad regions of the apex that support tip growth, and this localization likely requires WAVE complex activity (Perroud and Quatrano, 2008). If our Arabidopsis data reflect general features of ARP2/3 in plants, the results suggest that localized accumulation of ARP2/3 may not be due to the local capture and activation of ARP2/3 from a large soluble pool. Instead, combinations of actomyosin-based transport of ARP2/3-containing organelles and the lipid-binding specificity of ARP2/3-containing complexes may regulate localization and could explain the reported punctate localization of ARP2/3 on actin bundles (Fiserova et al., 2006; Maisch et al., 2009). The membrane association of ARP2/3 may have advantages. In large cells, especially highly vacuolated plant cells, diffusion of soluble ARP2/3 may not be an effective solution to distribute and locally activate the complex.
Positive regulation of ARP2/3 by SCAR and the SPIKE1-WAVE pathway presents an additional layer of complexity (Basu et al., 2005, 2008). It will be important to clearly identify the organelles and subcellular locations that contain active pools of ARP2/3 in plant cells. Probes for WAVE complex proteins may be useful in this regard (Dyachok et al., 2008). Although the microsome association of ARP2/3 is somehow linked to its assembly status, it does not equate to an active pool. Our crude estimates indicate that actin and ARP2/3 are at an approximately 5:1 molar ratio in plant extracts (Fig. 5, B and C). Yet, arp2/3 null mutants have very subtle F-actin phenotypes and no obvious effect on the total amount of F-actin in the cell (Mathur et al., 1999; Szymanski et al., 1999; Le et al., 2003). A simple explanation may be that only a small fraction of the total ARP2/3 pool is active at a given moment and particular subsets of ARP2/3-dependent cortical and/or organelle-associated actin filaments are critical during morphogenesis. Perhaps a latent, distributed pool of ARP2/3 is important to the cell, and the function of SPIKE1 and the WAVE complex is to regulate the location, timing, and extent of activation. It remains to be determined if ARP2/3 cycles between membrane compartments and how such an activity might impact so many aspects of plant morphogenesis.
MATERIALS AND METHODS
Plant Strains, Growth Conditions, and Mutant Characterization
The SALK T-DNA insertion lines for Arabidopsis (Arabidopsis thaliana) ARPC4, AT4G14147 (arpc4-t1, SALK_073297; arpc4-t2, SALK_52687), were obtained from the Arabidopsis Biological Resource Center. Both were backcrossed to the wild type two times prior to their use. The arpc2/dis2-1 (El-Assal et al., 2004b), arp3/dis1-1 (Le et al., 2003), arp2/wrm1-t1 (Le et al., 2003), arp2/wrm1-1 (Mathur et al., 2003a), arpc5/crk-1 (Mathur et al., 2003b), and sra1/pir-3 (Basu et al., 2004) alleles have been previously described. For all experiments, the Columbia (Col-0) background was used as the wild type. The soil-grown plants were planted in SunGro Redi-earth plug and seedling mix series growth medium (SunGro Horticulture) that was overlaid on an equal volume of vermiculite under continuous illumination (110 mmol photons m−2 s−1) at 24°C. The trichome morphometry data were collected using soil-grown plants. Aseptically grown plants were seeded on half-strength Murashige and Skoog plates with 1% Suc under continuous illumination (110 mmol photons m−2 s−1) at 22°C. Dark-grown plants were grown aseptically and wrapped with three layers of aluminum foil. T-DNA insertions in the ARPC4 gene were confirmed by PCR using primer pair sequences as described in Supplemental Table S1. To analyze the expression of the ARPC4 gene in mutant backgrounds, total RNA was extracted from the homozygous T-DNA insertion mutants by TRIzol reagent (Molecular Research Center) and then reversed transcribed as described previously (Zhang et al., 2008). Thereafter, the cDNA was used as the template for PCR using ARPC4 cDNA-specific primers (Supplemental Table S1).
Generation of the ARPC4:HA Rescue Construct
To generate the arpc4-t1 rescue line, an ARPC4 genomic fragment was first engineered to contain a unique NotI site prior to the stop codon. The upstream gene fragment was amplified using Pfu Turbo using the primers C4F1 and C4R1 (Supplemental Table S1) and cloned into the vector pCRTOPOII (Invitrogen) to yield pARPC4A. The downstream fragment was amplified using the primers C4F2 and C4R2 (Supplemental Table S1) and cloned into the vector pCRTOPOII to yield pARPC4B. The sequences of both fragments were confirmed by sequencing. The NotI-EcoRI fragment of pARC4B was cloned into pARPC4A, and the modified ARPC4 locus with the introduced NotI site was cloned into pCB302m vector as a SacII/EcoRI fragment. The resulting ARPC4 genomic clone was then engineered to contain a triple HA epitope tag at the extreme C terminus, yielding the plasmid pCB302m-ARPC4:HA, by cloning a triple HA-encoding EagI fragment into the NotI site of this plasmid to yield pCB202-ARPC4-HA. The plasmid was used to transform arpc4-t1 plants using the floral dip method (Clough and Bent, 1998).
Microsome Preparation and Analysis of Membrane Association
The microsomal protein fraction was isolated by Polytron homogenization of 2 g of seedlings (20 d after germination) in 10 mL of microsome isolation buffer [MIB; 20 mm HEPES/KOH, pH 7.2, 50 mm KOAc, 2 mm Mg(OAc)2, 250 mm sorbitol, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1% (v/v) protease inhibitors; Kang et al., 2001]. The homogenate was filtered through a prewetted double layer of Miracloth and centrifuged at 4°C for 30 min as described in the figure legends. Measurements of the fraction of endogenous and HA-tagged ARP2/3 that was soluble versus membrane associated were done by western-blot analysis of equal proportions of the cell fractions that were generated by differential centrifugation. In the experiments that tested for peripheral membrane association and the effects of arp2/3 mutants on the solubility of the complex, the filtered cell lysate was ultracentrifuged (200,000g, 4°C, 1 h) to generate supernatant and pellet fractions. The resulting pellet was suspended in 10 mm Tris-HCl/KOH, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, 1 mm phenylmethylsulfonyl fluoride, and 1% (v/v) protease inhibitors. For the experiments that assayed the effects of loss of ARP2/3 subunits on membrane association, the membrane fraction was overloaded approximately 20-fold compared with the soluble fraction.
SDS-PAGE, Antibodies, and Immunoblotting
For immunoblotting, the proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Schleicher & Schuell) in 38.6 mm Gly, 48 mm Tris, 1.3 mm SDS, and 20% (v/v) ethanol. Immunoblots were incubated in 3% (w/v) milk in TBS-T (50 mm Tris, pH 7.5, 150 mm NaCl, and 0.01% [v/v] Tween 20). For anti-HA blots, Tween 20 was not included. Primary antibodies, anti-ARP3, anti-HA (monoclonal HA.11), anti-ACTIN (a kind gift from Chris Staiger), and anti-PEPC (Rockland Immunochemicals; 1:1,000 dilution) were added and incubated overnight at 4°C. The primary antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit antibodies (1:50,000 dilution; Pierce). Anti-ARP3 antibody was raised in rabbits using full-length His-tagged Arabidopsis ARP3 expressed in Escherichia coli as the antigen (Harlan Bioproducts for Science).
CoIP
For coIP, the 20,000g pellet fraction was resuspended at a final protein concentration of 1 mg mL−1 in 20 mm HEPES/KOH, pH 7.2, 1 mm EDTA, and 1 mm dithiothreitol and solubilized with 4% cholate (Sigma-Aldrich) for 30 min at room temperature. The extract was clarified by ultracentrifugation (200,000g, 4°C, 30 min). The solubilized and clarified extract (300 μg) was added to 200 μL of coIP buffer (20 mm HEPES/KOH, pH 7.2, 150 mm NaCl, 1% Nonidet P-40, and 0.1% SDS) containing 15 μg of anti-ARP3 or preimmune serum bound to protein A beads (Pierce). The reactions were rocked at 4°C for 16 h, and the protein A beads were washed using immunoprecipitation wash buffer (20 mm HEPES/KOH, pH 7.2, 150 mm NaCl, 1% Nonidet P-40, and 0.1% SDS) six times and probed with anti-HA antibody.
Quantification of Endogenous Actin and ARP3 Protein Content
To estimate the amount of actin and ARP3 protein in Arabidopsis cell extracts, known amounts of ARP3 inclusion body and vertebrate G-actin were run via SDS-PAGE along with plant protein extract, blotted with anti-ARP3 and anti-ACTIN antibody, respectively, and quantified by densitometry using ImageQuant 5.2. The purified vertebrate actin standard was quantified by UV light absorbance. The ARP3 inclusion body standard was quantified by densitometry readings of Coomassie Brilliant Blue-stained SDS-PAGE gels containing ARP3 inclusion body bands compared with dilutions of known amounts of vertebrate actin and bovine serum albumin standards run on the same gel. Nearly identical results for ARP3 inclusion body concentration were obtained in both cases. The linear regression analysis from western-blot data of the actin and ARP3 standards was used to estimate the protein content of actin and ARP3 in plant extracts.
F-Actin Localization
F-actin localization and quantification were done according to Le et al. (2003). Images were collected using an MRC Bio-Rad Radiance 2100 confocal microscope with a Nikon 60x PlanApo (numerical aperture 1.2) water-immersion objective. The images were processed and analyzed using Metamorph (Universal Imaging) or ImageJ (http://rsb.info.nih.gov/ij) software. Criteria to score branches with aligned bundles have been described previously (Basu et al., 2005; Zhang et al., 2008); such branches were defined as those branches that contained more than three bundles that terminated within 3 μm of the branch apex and that were aligned clearly with the long axis of the branch.
SEC
For SEC, microsomal fractions from the wild type and different distorted mutant backgrounds were solubilized in 4% cholate and separated on a Sephadex 200HR 10/300 column (GE Healthcare) as described previously (Basu et al., 2008). Briefly, 200-μL column fractions were collected, TCA precipitated in the presence of 0.01% deoxycholate, and analyzed by western blotting using anti-ARP3 or anti-HA antibodies. For the size distribution of ARPC4:HA-containing complexes, SEC employed extracts that were isolated from arpc4-t1 ARPC4:HA-rescued plants. Endogenous ARP3-containing complexes were analyzed using extracts from wild-type and mutant plants as described above. Yeast ARP2/3 was isolated from the strain DDY2920, a kind gift from David Drubin. In this strain, the ARPC3/ARC18 subunit is tagged with an HA epitope. Yeast cells were lysed using glass beads in MIB, and the cytosolic fraction was analyzed using SEC as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ARPC4-HA and ARP3 proteins are associated with each other in vivo.
Supplemental Table S1. Sequences of oligonucleotide primers used in this work.
Supplementary Material
Acknowledgments
We are grateful to David Drubin for the kind gift of an ARPC3:HA-tagged yeast strain and to Chris Staiger for his gift of the actin antibody. Thanks also to the Purdue Cytoskeleton Group for helpful input during the execution of this project.
This work was supported by the National Science Foundation (grant nos. IPB 0110817IBN and MCB 0640872IBN to D.B.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniel B. Szymanski (dszyman@purdue.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe S, Davies E (1995) Methods for isolation and analysis of the cytoskeleton. Methods Cell Biol 50 223–236 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Baluska F, Salaj J, Mathur J, Braun M, Jasper F, Samaj J, Chua NH, Barlow PW, Volkmann D (2000) Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev Biol 227 618–632 [DOI] [PubMed] [Google Scholar]
- Basu D, El-Assal SE, Le J, Mallery EL, Szymanski DB (2004) Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131 4345–4355 [DOI] [PubMed] [Google Scholar]
- Basu D, Le J, El-Assal SE, Huang S, Zhang C, Mallery EL, Koliantz G, Staiger CJ, Szymanski DB (2005) DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17 502–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB (2008) A SPIKE1 signaling complex controls actin-dependent morphogenesis through the WAVE and ARP2/3 complexes. Proc Natl Acad Sci USA 105 4044–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G, Caron E (2004) Regulation of WASP/WAVE proteins: making a long story short. J Cell Biol 166 957–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Baluska F, von Witsch M, Menzel D (1999) Redistribution of actin, profilin and phosphatidylinositol-4,5- bisphosphate in growing and maturing root hairs. Planta 209 435–443 [DOI] [PubMed] [Google Scholar]
- Brembu T, Winge P, Seem M, Bones AM (2004) NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16 2335–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudry F, Guerin C, vonWitsch M, Blanchoin L, Staiger CJ (2007) Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol Biol Cell 18 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cole RA, Fowler JE (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9 579–588 [DOI] [PubMed] [Google Scholar]
- Collings DA, Carter CN, Rink JC, Scott AC, Wyatt SE, Allen NS (2000) Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12 2425–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Kaloriti D, Davies B, Malho R, Hussey PJ (2004) Arabidopsis NAP1 is essential for ARP2/3-dependent trichome morphogenesis. Curr Biol 14 1410–1414 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Rodrigues C, Dimmock S, Ketelaar T, Maciver SK, Malho R, Hussey PJ (2007) Arabidopsis CAP1: a key regulator of actin organisation and development. J Cell Sci 120 2609–2618 [DOI] [PubMed] [Google Scholar]
- Djakovic S, Dyachok J, Burke M, Frank MJ, Smith LG (2006) BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133 1091–1100 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Shao MR, Vaughn K, Bowling A, Facette M, Djakovic S, Clark L, Smith L (2008) Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol Plant 1 990–1006 [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW (2002) Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418 790–793 [DOI] [PubMed] [Google Scholar]
- Eitzen G, Wang L, Thorngren N, Wickner W (2002) Remodeling of organelle-bound actin is required for yeast vacuole fusion. J Cell Biol 158 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal SE, Le J, Basu D, Mallery EL, Szymanski DB (2004. a) Arabidopsis GNARLED encodes a NAP125 homologue that positively regulates ARP2/3. Curr Biol 14 1405–1409 [DOI] [PubMed] [Google Scholar]
- El-Assal SE, Le J, Basu D, Mallery EL, Szymanski DB (2004. b) DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J 38 526–538 [DOI] [PubMed] [Google Scholar]
- Fehrenbacher KL, Boldogh IR, Pon LA (2005) A role for Jsn1p in recruiting the Arp2/3 complex to mitochondria in budding yeast. Mol Biol Cell 16 5094–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J, Schwarzerova K, Petrasek J, Opatrny Z (2006) ARP2 and ARP3 are localized to sites of actin filament nucleation in tobacco BY-2 cells. Protoplasma 227 119–128 [DOI] [PubMed] [Google Scholar]
- Frank M, Egile C, Dyachok J, Djakovic S, Nolasco M, Li R, Smith LG (2004) Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc Natl Acad Sci USA 16 16379–16384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Smith LG (2002) A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12 849–853 [DOI] [PubMed] [Google Scholar]
- Fucini R, Chen J, Sharma C, Kessels M, Stamnes M (2002) Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol Biol Cell 13 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, Dudler R, et al (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell 14 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428 [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Kovar DR, Staiger CJ (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD (2001) Reconstitution of human Arp 2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell 8 1041–1052 [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed]
- Hable WE, Kropf DL (2005) The Arp2/3 complex nucleates actin arrays during zygote polarity establishment and growth. Cell Motil Cytoskeleton 61 9–20 [DOI] [PubMed] [Google Scholar]
- Hardham AR, Jones DA, Takemoto D (2007) Cytoskeleton and cell wall function in penetration resistance. Curr Opin Plant Biol 10 342–348 [DOI] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS (2005) Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Okubo E, Sano T, Hasezawa S (2006) Actin microfilaments regulate vacuolar structures and dynamics: dual observation of actin microfilaments and vacuolar membrane in living tobacco BY-2 cells. Plant Cell Physiol 47 839–852 [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD (2001) Regulation of actin filament network formation through Arp2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70 649–676 [DOI] [PubMed] [Google Scholar]
- Holtta-Vuori M, Alpy F, Tanhuanpaa K, Jokitalo E, Mutka AL, Ikonen E (2005) MLN64 is involved in actin-mediated dynamics of late endocytic organelles. Mol Biol Cell 16 3873–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57 109–125 [DOI] [PubMed] [Google Scholar]
- Ji L, Lim J, Danuser G (2008) Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol 10 1393–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG (2005) A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123 305–320 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2009) A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21 701–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, Meagher RB (1999) Actin-organelle interaction: association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton 44 110–118 [DOI] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY (2001) The Arabidopsis cell-plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol 126 47–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard TD (1995) Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol 131 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I (2005) Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, El-Assal SE, Basu D, Saad ME, Szymanski DB (2003) Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr Biol 13 1341–1347 [DOI] [PubMed] [Google Scholar]
- Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB (2006) Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol 16 895–901 [DOI] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH (2001) Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev 15 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM (2003) The putative Arabidopsis Arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sorefan K, Hemmann G, Bevan MW (2004) Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol 136 3616–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A, Cardenas L, Kunkel JG, Hepler PK (2007) Differential organelle movement on the actin cytoskeleton in lily pollen tubes. Cell Motil Cytoskeleton 64 217–232 [DOI] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD (1994) Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol 127 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch J, Fiserova J, Fischer L, Nick P (2009) Tobacco Arp3 is localized to actin-nucleating sites in vivo. J Exp Bot 60: 603–614 [DOI] [PMC free article] [PubMed]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M (2003. a) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hulskamp M (2003. b) Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130 3137–3146 [DOI] [PubMed] [Google Scholar]
- Mathur J, Spielhofer P, Kost B, Chua N (1999) The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126 5559–5568 [DOI] [PubMed] [Google Scholar]
- Mullins D, Stafford W, Pollard T (1997) Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol 136 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol 121 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Pollard TD (2008) Structure and biochemical properties of fission yeast ARP2/3 complex lacking the ARP2 subunit. J Biol Chem 283 26940–26948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhevsky AI, Avisar D, Dolja VV (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol 146 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud PF, Quatrano RS (2006) The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil Cytoskeleton 63 162–171 [DOI] [PubMed] [Google Scholar]
- Perroud PF, Quatrano RS (2008) BRICK1 is required for apical cell growth in filaments of the moss Physcomitrella patens but not for gametophore morphology. Plant Cell 20 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29 545–576 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112 453–465 [DOI] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand J-B, Higgs HN, Choe S, Pollard TD (2001) Crystal structure of Arp 2/3 complex. Science 294 1679–1684 [DOI] [PubMed] [Google Scholar]
- Saedler R, Zimmermann I, Mutondo M, Hulskamp M (2004) The Arabidopsis KLUNKER gene controls cell shape changes and encodes the AtSRA1 homolog. Plant Mol Biol 56 775–782 [DOI] [PubMed] [Google Scholar]
- Schwab B, Mathur J, Saedler R, Schwarz H, Frey B, Scheidegger C, Hulskamp M (2003) Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: actin controls the spatial organization of microtubules. Mol Genet Genomics 269 350–360 [DOI] [PubMed] [Google Scholar]
- Shao D, Forge A, Munro PMG, Bailly M (2006) Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil Cytoskeleton 63 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T (2007) The sliding theory of cytoplasmic streaming: fifty years of progress. J Plant Res 120 31–43 [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21 271–295 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Blanchoin L (2006) Actin dynamics: old friends with new stories. Curr Opin Plant Biol 9 554–562 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L (2009) Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J Cell Biol 184 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes M (2002) Regulating the actin cytoskeleton during vesicular transport. Curr Opin Cell Biol 14 428–433 [DOI] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TEB (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J 23 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal TE, Scita G (2006) Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol 18 4–10 [DOI] [PubMed] [Google Scholar]
- Strasser GA, Rahim NA, VanderWaal KE, Gertler FB, Lanier LM (2004) Arp2/3 is a negative regulator of growth cone translocation. Neuron 43 81–94 [DOI] [PubMed] [Google Scholar]
- Szymanski DB (2005) Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr Opin Plant Biol 8 103–112 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125 1161–1171 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers J, Vernhettes S, Desprez T, Vincken JP, Visser RG, Trindade LM (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett 583 978–982 [DOI] [PubMed] [Google Scholar]
- Van Gestel K, Slegers H, von Witsch M, Samaj J, Baluska F, Verbelen JP (2003) Immunological evidence for the presence of plant homologues of the actin-related protein Arp3 in tobacco and maize: subcellular localization to actin-enriched pit fields and emerging root hairs. Protoplasma 222 45–52 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME (2003) Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54 691–722 [DOI] [PubMed] [Google Scholar]
- Welch M, DePace A, Verma S, Iwamatsu A, Mitchison T (1997) The human Arp 2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD (2002) Cellular control of actin nucleation. Annu Rev Cell Dev Biol 18 247–288 [DOI] [PubMed] [Google Scholar]
- Wightman R, Turner SR (2008) The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J 54: 794–805 [DOI] [PubMed]
- Winter DC, Choe EY, Li R (1999) Genetic dissection of the budding yeast Arp 2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA 96 7288–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD (2005) Counting cytokinesis proteins globally and locally in fission yeast. Science 310 310–314 [DOI] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, Jurkiewicz A, Rueda P, Radutoiu S, Held M, Hossain MS, Szczyglowski K, Morieri G, et al (2009) Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284 [DOI] [PMC free article] [PubMed]
- Zhang C, Mallery EL, Schlueter J, Huang S, Fan Y, Brankle S, Staiger CJ, Szymanski DB (2008) Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG (2005) IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.