Abstract
Cardiac complications are a common cause of death in individuals with the inherited multisystemic disease myotonic dystrophy type 1 (DM1). A characteristic molecular feature of DM1 is misregulated alternative splicing due to disrupted functioning of the splicing regulators muscleblind-like 1 (MBNL1) and CUG-binding protein 1 (CUGBP1). CUGBP1 is upregulated in DM1 due to PKC pathway activation and subsequent CUGBP1 protein hyperphosphorylation and stabilization. Here, we blocked PKC activity in a heart-specific DM1 mouse model to determine its pathogenic role in DM1. Animals given PKC inhibitors exhibited substantially increased survival that correlated with reduced phosphorylation and decreased steady-state levels of CUGBP1. Functional studies demonstrated that PKC inhibition ameliorated the cardiac conduction defects and contraction abnormalities found in this mouse model. The inhibitor also reduced misregulation of splicing events regulated by CUGBP1 but not those regulated by MBNL1, suggesting distinct roles for these proteins in DM1 cardiac pathogenesis. The PKC inhibitor did not reduce mortality in transgenic mice with heart-specific CUGBP1 upregulation, indicating that PKC inhibition did not have a general protective effect on PKC-independent CUGBP1 increase. Our results suggest that pharmacological blockade of PKC activity mitigates the DM1 cardiac phenotype and provide strong evidence for a role for the PKC pathway in DM1 pathogenesis.
Introduction
Myotonic dystrophy (DM) is the most common form of adult onset muscular dystrophy and the second most common form of muscular dystrophy overall (1). DM is dominantly inherited and affects multiple organs, including skeletal muscle, heart, brain, and the endocrine system (2). In the more common form of DM, DM type 1 (DM1), cardiac involvement occurs in 80% of the patients (3, 4). The cardiac manifestations of DM1 are heterogeneous and include conduction defects, arrhythmia, and dilated cardiomyopathy (5). Due to the complexity of the cardiac disease, treatment strategies are limited. In addition, the molecular events involved in DM1 heart pathogenesis are unknown.
The genetic basis of DM1 is the expansion of CTG repeats in the 3′ untranslated region of the dystrophica myotonia protein kinase (DMPK) gene (2). Nuclear accumulation of the DMPK RNA with expanded CUG repeats triggers events that lead to disruption of developmentally regulated alternative splicing (6), which result in some of the disease symptoms such as myotonia and insulin resistance (7–9). At least 2 families of RNA–binding proteins are implicated in DM1 pathogenesis: CUGBP and ETR3-like proteins (CELF) and muscleblind like (MBNL). Loss of MBNL function and increased levels of the CELF protein, CUG-binding protein 1 (CUGBP1), correlate with at least some of the splicing changes and disease symptoms observed in DM1 patients (9–11).
Expanded CUG repeats bind and sequester MBNL proteins, resulting in their loss of function (12–15). In support of a role for MBNL1 in DM1 pathogenesis, deletion of MBNL1 isoforms that bind to expanded CUG repeats in mice leads to cataracts, myotonia, development-specific splicing changes, and histological changes in skeletal muscle (10). Furthermore, restoration of MBNL1 expression by adeno-associated viral gene delivery in skeletal muscle of mice expressing RNA containing 250 CUG repeats reverses splicing abnormalities and myotonia (16). While the role of MBNL1 in DM1 skeletal muscle pathology is clear, the involvement in DM1 heart pathogenesis remains to be characterized.
In addition to MBNL1 sequestration, expanded CUG repeats activate the PKC signaling pathway, leading to CUGBP1 protein hyperphosphorylation and stabilization (17), consistent with elevated steady-state levels of CUGBP1 in DM1 heart and skeletal muscle tissues (9, 18). Overexpression of CUGBP1 in mouse heart and skeletal muscle leads to DM1 splicing changes and results in embryonic lethality (19, 20), strongly suggesting pathogenic effects in striated muscle. However, the role of CUGBP1 in DM1 cardiac pathogenesis has not yet been investigated.
We previously established an inducible DM1 mouse model, in which a transgene containing the last exon of DMPK with 960 CTG repeats (EpA960) is induced to express CUG repeat–containing RNA [EpA960(R)], after recombination by Cre-mediated removal of concatamerized polyadenylation sites (21). Tamoxifen-inducible and heart-specific EpA960(R) RNA expression was obtained from bitransgenic progeny of EpA960 animals mated to MerCreMer (MCM) animals, which express a tamoxifen-inducible form of Cre in a heart-specific manner (22). Within 3 weeks after induction of EpA960(R) RNA, these mice exhibited high mortality, conduction abnormalities, and systolic and diastolic dysfunction as well as molecular changes seen in DM1 patients, such as colocalization of MBNL1 with RNA foci and reversion of splicing to embryonic patterns (21). Importantly, activated PKCα/βII and increased CUGBP1 levels were evident within 6 hours after induction of expanded CUG RNA expression (17, 21), strongly suggesting that these are primary responses to expression of the toxic CUG repeat–containing RNA that contribute to DM1 pathogenesis.
To determine whether PKC activation is required to elicit the pathogenicity of EpA960(R) RNA, we used the specific PKC inhibitor, Ro-31-8220, to prevent PKCα/βII activation following induction of EpA960(R) RNA. This inhibitor was previously well characterized in mouse heart tissue by its ability to inhibit PKC isozymes and improve cardiac contractility (23). Here, we show that administration of Ro-31-8220 to the heart-specific DM1 mouse model prevents activation of PKCα/βII as well as hyperphosphorylation and upregulation of CUGBP1. Importantly, Ro-31-8220 administration significantly improved mortality and improved cardiac conduction/contractile dysfunction. Consistent with the hypothesis that EpA960(R) RNA–induced pathogenesis is mediated at least in part via upregulation of CUGBP1, aberrant splicing of CUGBP1-regulated pre-mRNA targets was prevented by Ro-31-8220 administration, while MBNL-regulated splicing events were not. These results suggest that MBNL1 and CUGBP1 have distinct roles in DM1 heart pathogenesis. Importantly, administration of Ro-31-8220 did not improve the mortality in a second mouse model, in which CUGBP1 is inducibly overexpressed in the heart, using tetracycline-inducible transgene. These results indicate that PKC inhibition has no effect on cardiopathology caused by PKC-independent elevation of CUGBP1. Our results suggest that activation of the PKC pathway is a primary pathogenic event and that early abrogation of PKC activity ameliorates pathogenic cardiac features of DM1.
Results
The PKC inhibitor Ro-31-8220 significantly improved the mortality rate of animals expressing EpA960(R) RNA.
We first tested whether PKC inhibition would improve mortality of bitransgenic EpA960/MCM animals induced to express EpA960(R) RNA in the heart. PKC activation is detectable by 6 hours following RNA induction (17). To prevent early activation of PKC, Ro-31-8220 was given 2 hours after the initial tamoxifen injection and continued daily for 24 days. As a mock control, tamoxifen-induced EpA960/MCM mice received saline solution daily for 24 days. Eighty percent of the mock-treated mice died within 3 weeks of RNA induction as reported previously, due to arrhythmias and/or contractile dysfunction (Figure 1A, ref. 21, and see below). Strikingly, administration of Ro-31-8220 reduced the mortality of bitransgenic mice to less than 20%, and it had no effect on the survival of control mice (MCM) treated with tamoxifen (Figure 1A).
Figure 1. PKC inhibition improves the mortality rate of mice expressing EpA960(R) RNA.
(A) Kaplan-Meier survival curve on animals receiving the PKC inhibitor, Ro-31-8220. Percentage survival of tamoxifen-induced EpA960/MCM mice treated with Ro-31-8220 (EpA960/MCM, TAM-Ro-31-8220) or mock injected using saline (EpA960/MCM, TAM), compared with control mice, expressing only Cre treated with tamoxifen and Ro-31-8220 (MCM, TAM-Ro-31-8220). (B) Expression of EpA960(R) RNA after Cre-mediated recombination in TAM-induced mice given Ro-31-8220 (TAM-Ro-31-8220) or saline (TAM) was analyzed by quantitative RT-PCR using TaqMan (mean ± SEM). (C) HW/BW ratio of mice from each group (mean ± SEM). *P < 0.05 versus MCM control.
To determine whether Ro-31-8220 affected the expression of EpA960(R) RNA in the heart, we used TaqMan real-time RT-PCR to determine the level of recombined allele. We found that animals given Ro-31-8220 expressed comparable levels of EpA960(R) RNA to the mock-treated animals (Figure 1B). Consistent with this result, the number and distribution of nuclear RNA foci was comparable between animals that received Ro-31-8220 or saline (data not shown). These results indicate that Ro-31-8220 did not reduce the expression of EpA960(R) RNA but rather blocked its pathogenic effects.
Mice expressing EpA960(R) RNA exhibited higher heart weight to body weight (HW/BW) ratios compared with the control MCM mice, consistent with cardiomyopathy observed in these mice (21). We found that the mice that received Ro-31-8220 retained the increased HW/BW ratio (Figure 1C), suggesting that PKC pathway might not be involved in this component of the phenotype. Overall, these results indicate that Ro-31-8220 significantly reduced mortality caused by expanded CUG RNA, without affecting its level of expression.
Ro-31-8220 prevents PKC activation and reduces CUGBP1 hyperphosphorylation and upregulation.
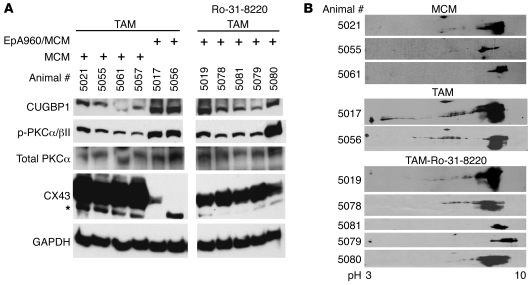
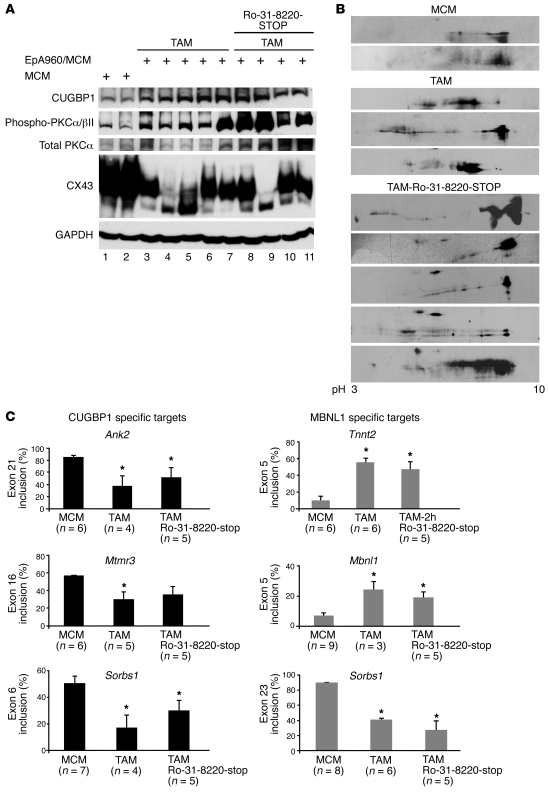
To determine whether Ro-31-8220 inhibited PKCα/βII activation, we used a phospho-specific antibody (Thr638/641) to determine the level of activated PKCα/βII isoforms compared with total PKCα. In the absence of Ro-31-8220, PKCα/βII was activated in heart tissues from mice expressing EpA960(R) RNA compared with MCM control mice (Figure 2A, lanes 5 and 6), as described previously (17). Administration of Ro-31-8220 effectively prevented activation of PKCα/βII and also prevented upregulation of CUGBP1 in heart tissues from 3 of the 5 animals tested (Figure 2A, lanes 8–10). CUGBP1 phosphorylation is analyzed on 2-dimensional (2D) gels, followed by Western blotting of CUGBP1 as described previously (17), because the modifications do not change the mobility of the protein on 1D gels. Analysis of the CUGBP1 phosphorylation by 2D/Western blot demonstrated that hyperphosphorylation was reduced in the majority of the mice that were induced by tamoxifen and treated with the PKC inhibitor (TAM-Ro-31-8220; Figure 2B).
Figure 2. Effect of Ro-31-8220 administration on PKC activation and CUGBP1 and connexin 43 expression.
(A) Western blots of CUGBP1 (50 kDa), phospho-PKCα/βII (75 kDa), total PKC (75 kDa), and connexin 43 (CX43; 43 kDa) in MCM mice and mice induced to express EpA960(R) RNA and treated with saline (TAM) or with Ro-31-8220 (TAM-Ro-31-8220). Ro-31-8220 was administered 2 hours after the first tamoxifen injection. CUGBP1, PKCα/βII, and connexin 43 were detected at the expected molecular weights. The asterisk indicates a cross-reactive band. The samples were run on the same gel. (B) 2D/Western blot analysis of CUGBP1 in heart tissue from MCM, TAM, and TAM-Ro-31-8220 groups. CUGBP1 shifts from basic to acidic pI upon phosphorylation (17). Individual animal numbers are indicated, representing the ear-tag number of each mouse.
Connexin 43 is the major gap junction protein in heart that regulates electric coupling (24). We found that expression of connexin 43 was significantly reduced in heart tissue from animals expressing expanded CUG RNA (Figure 2A, lanes 5 and 6), consistent with results from an independently generated mouse model for DM1 (25). While it is unclear whether loss of connexin 43 is directly responsible for the arrhythmias observed in this DM1 model, decreased connexin 43 expression correlates with arrhythmias in connexin 43 loss-of-function mouse models (26) as well as other genetic and nongenetic models of cardiomyopathy (27). Interestingly, connexin 43 protein levels were partially restored in mice given Ro-31-8220 (Figure 2, lanes 7–11) in comparison with EpA960(R) RNA–expressing mice (Figure 2A, lanes 5 and 6). Collectively, the results suggest that mice expressing EpA960(R) RNA treated with Ro-31-8220 exhibited low PKCα/βII activation, reduced hyperphosphorylation of CUGBP1, reduced CUGBP1 protein levels, and partially preserved connexin 43 protein levels. These results support a central role for PKC in DM1 pathogenesis.
Ro-31-8220 administration prevents cardiac electrophysiology and contractile dysfunction.
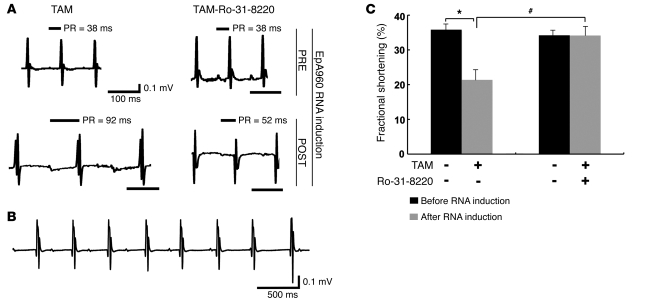
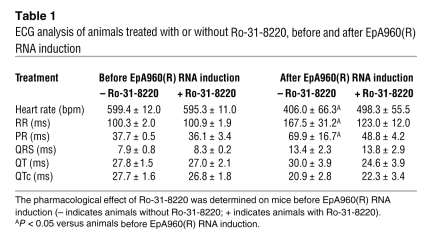
Mice expressing EpA960(R) RNA in the heart displayed dilated cardiomyopathy, systolic and diastolic dysfunction, and conduction abnormalities, including prolonged PR intervals and widening of the QRS complex (21). To determine which aspect(s) of the cardiac phenotype was improved by Ro-31-8220, we first checked its effects on cardiac conduction using ECG telemetry. Ro-31-8220 administration had no effect on heart rate, PR interval, QTc interval, or QRS duration in mice before induction of EpA960(R) RNA by tamoxifen administration (Figure 3A and Table 1), indicating that Ro-31-8220 alone did not affect heart electrophysiology. After induction of EpA960(R) RNA expression, all mice that received mock saline injections displayed prolonged PR intervals (Figure 3A and Table 1), and 1 animal developed a third-degree heart block (Figure 3B). However, mice treated with Ro-31-8220 showed a significantly reduced PR interval prolongation (Figure 3A and Table 1). The mock-treated mice also exhibited widening of the QRS complex as described previously (21), but the differences did not reach the statistical significance (Table 1). The results indicated that Ro-31-8220 administration ameliorated PR prolongation in mice expressing EpA960(R) RNA.
Figure 3. Ro-31-8220 ameliorates cardiac conduction and contractile abnormalities.
(A) Representative ECGs recorded before and after EpA960(R) RNA induction in mice treated with or without Ro-31-8220. Ro-31-8220 given daily for 2 days prior to EpA960(R) RNA induction (PRE) did not affect baseline ECG of mice before EpA960(R) RNA induction. Representative ECGs after EpA960(R) RNA induction (POST) in mice given mock saline injections (TAM) or Ro-31-8220 (TAM-Ro-31-8220). PR intervals are indicated above the tracings. Scale bars below the tracings indicate 100 ms. (B) ECG tracing showing third-degree heart block, with a junctional escape rhythm, from 1 mouse in the TAM group. Scale bar: 500 ms. (C) Percentage fractional shortening in TAM (n = 9) and TAM-Ro-31-8220 (n = 11) groups of EpA960/MCM animals before and after EpA960(R) RNA induction injection (mean ± SEM). *P < 0.05 before and after EpA960(R) RNA induction; #P < 0.05 with and without Ro-31-8220 administration after EpA960(R) RNA induction.
Table 1 .
ECG analysis of animals treated with or without Ro-31-8220, before and after EpA960(R) RNA induction
We also examined whether Ro-31-8220 administration improved contractility of EpA960(R) RNA–expressing mice, using echocardiography to measure fractional shortening. Ro-31-8220 administration did not have an effect on left ventricular function before RNA expression was induced (Figure 3C). Twenty-five days after the induction of EpA960(R) RNA, mice given saline injections exhibited a statistically significant reduction in fractional shortening, while mice treated with Ro-31-8220 exhibited normal fractional shortening (Figure 3C), indicating that Ro-31-8220 prevented the cardiac contraction abnormalities.
Ro-31-8220 prevented misregulation of splicing events regulated by CUGBP1 but not MBNL1.
A major molecular feature of DM1 is misregulation of a subset of alternative splicing events, resulting in the inappropriate expression of embryonic splicing patterns in adult tissues (6). The basis for these splicing transitions is a gain of function for CUGBP1 and a loss of MBNL1 function (1). The first few splicing events characterized in cultured cells were found to be regulated antagonistically by CUGBP1 and MBNL1 (28); however, we have recently shown that in addition to regulating the same splicing events, CUGBP1 and MBNL1 each regulate alternative splicing of separate pre-mRNA targets (29, 30). We identified alternative splicing events that are regulated during mouse heart development that responded only to transgenic overexpression of human CUGBP1 or depletion of endogenous MBNL1 (30). For all of these splicing events, CUGBP1 overexpression or MBNL1 depletion caused reversion to the embryonic splicing pattern. Mice expressing EpA960(R) RNA displayed misregulation of multiple alternative splicing events, including those regulated specifically by CUGBP1 or by MBNL1 (ref. 21 and data not shown).
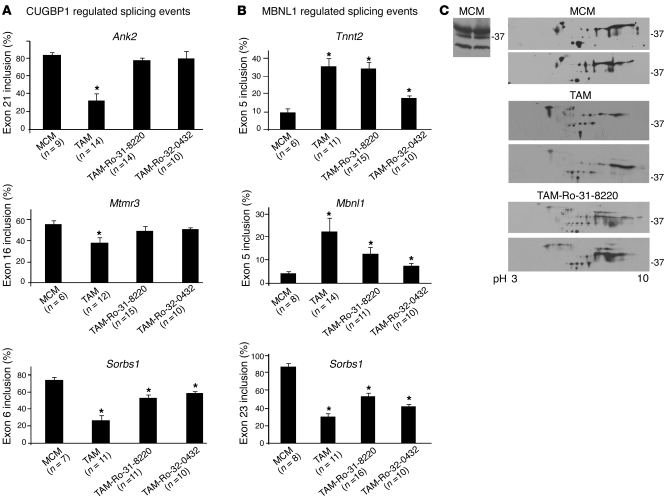
To determine whether PKC inhibition affected misregulated splicing of CUGBP1- and/or MBNL1-regulated pre-mRNA targets, we examined 3 alternative splicing events regulated by CUGBP1 (Ank2 exon 21, Mtmr3 exon 16, and Sorbs1 exon 6) and 3 MBNL1-regulated splicing events (Tnnt2 exon 5, Mbnl1 exon 5, and Sorbs1 exon 23). Mice expressing EpA960(R) RNA treated with saline exhibited the embryonic pattern splicing pattern compared with that of MCM control mice for all 6 splicing events (Figure 4, A and B), which is as expected of a DM1-like phenotype. In mice induced to express EpA960(R) RNA and treated with Ro-31-8220, 2 of the 3 CUGBP1-regulated splicing events (Ank2 exon 21 and Mtmr3 exon 16) were reversed back to normal (Figure 4A), while a third event did not reach statistical significance (Sorbs1 exon 6; Figure 4A). Note that this event was mildly affected by MBNL1 depletion (30). In contrast to CUGBP1-regulated targets, all 3 MBNL1-regulated splicing events remained misregulated in mice treated with Ro-31-8220 (P < 0.05 versus MCM; Figure 4B). It is worth noting that CUGBP1 upregulation has a small but consistent effect on Sorbs1 exon 23 splicing (30). A similar result was obtained using a second PKC inhibitor, Ro-32-0432 (see below). We concluded that Ro-31-8220 does not prevent the alteration of MBNL1-regulated splicing events but significantly ameliorates CUGBP1-mediated splicing abnormalities by preventing CUGBP1 hyperphosphorylation and protein stabilization. The misregulation of MBNL-regulated splicing events was consistent with results indicating that Ro-31-8220 does not affect EpA960(R) RNA expression (Figure 1B).
Figure 4. PKC inhibition affects CUGBP1-mediated misregulated alternative splicing events more than MBNL1-mediated events in induced EpA960/MCM mice.
(A) Ro-31-8220 and Ro-32-0432 prevent misregulation of 2 of 3 tested CUGBP1-regulated splicing events. RT-PCR analysis of CUGBP1-regulated splicing events in MCM control mice given tamoxifen (MCM), mice induced to express EpA960(R) RNA without (TAM) or with (TAM-Ro-31-8220 or TAM-Ro-32-0432) administration of PKC inhibitors. The number of mice used in each splicing event is indicated under the graph bars. Ank2 exon 21, Mtmr3 exon 16, and Sorbs 1 exon 6 are shown with MCM, TAM, Ro-31-8220, and Ro-32-0432 groups (mean ± SEM). *P < 0.05 versus MCM control. (B) Neither Ro-31-8220 nor Ro-32-0432 significantly affects misregulated splicing of MBNL1-specific targets quantified by RT-PCR analysis (mean ± SEM). *P < 0.05 versus MCM control. (C) 2D/Western analysis of MBNL1 in mouse heart reveals extensive posttranslational modification that is unaffected by expression of EpA960(R) RNA or Ro-31-8220 administration. 1D/Western blot analysis of MBNL1 in MCM heart tissues shows 3 MBNL1 isoforms (approximate sizes, 41, 37, 31 kDa) (left panel). 2D/Western blot analysis of MBNL1 in heart tissues from 2 different mice in each group (MCM, TAM, and TAM-Ro-31-8220) (right panel).
To investigate whether EpA960(R) RNA induced MBNL1 posttranslational changes, we performed 2D/Western blot analysis. On both 1D/Western analyses and 2D/Western analyses, our rabbit polyclonal anti-MBNL1 antibody recognized 3 isoforms in adult mouse heart (Figure 4C). When analyzed by 2D/Western blots, each MBNL1 isoform displayed multiple isoforms of different pI values, indicative of posttranslational modifications. We found that MBNL1 patterns were similar in untreated MCM mice and mice induced to express EpA960(R) RNA with or without Ro-31-8220 administration (Figure 4C). The results indicated that MBNL1 is extensively modified in adult mouse heart tissue and that neither expanded CUG RNA nor Ro-31-8220 administration strongly affects these modifications.
A second PKC inhibitor prevents mortality in mice expressing EpA960(R) RNA.
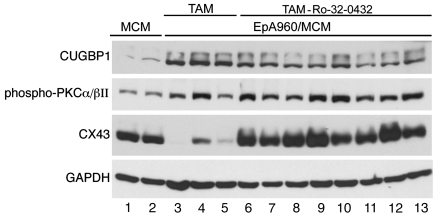
To confirm that amelioration of EpA960(R) RNA–induced cardiac phenotype and misregulation of alternative splicing by Ro-31-8220 was specifically due to PKC inhibition rather than off target effects, we used a second PKC inhibitor, Ro-32-0432. We found that Ro-32-0432 reduced mortality and improved contractile abnormalities but was not as effective as Ro-31-8220 in preventing PR prolongation (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI37976). As for Ro-31-8220, Ro-32-0432 administration also reversed 2 of 3 CUGBP1-splicing events to normal, whereas all 3 MBNL1-splicing events were significantly different from the wild-type pattern observed in MCM mice (Figure 4). Nine out of ten mice survived for 30 days when given Ro-32-0432, with normal left ventricular function, while 7 mice exhibited PR prolongation (Supplemental Table 1). We noticed that steady-state levels and hyperphosphorylation of CUGBP1 were not completely reversed in these mice, correlating with conduction system abnormalities (Figure 5 and Supplemental Figures 1 and 2), but expression of connexin 43 was partially restored (Figure 5). Overall, inhibition of PKC by Ro-32-0432 or Ro-31-8220 correlated with increased survival and improved cardiac function; however, Ro-31-8220 was better at inhibiting PKC and protecting cardiac function. This could be due to the differences in the profiles of PKC isoforms inhibited (31) and/or efficiencies of PKC inhibition by these drugs in vivo.
Figure 5. A second PKC inhibitor modestly inhibits PKC activation and CUGBP1 upregulation.
Western blot analysis of CUGBP1, phospho-PKCα/βII, and connexin 43 in each mouse group is shown (MCM, lanes 1 and 2; TAM, lanes 3–5; and TAM-Ro-32-0432, lanes 6–13). GAPDH was used as a loading control.
PKC inhibition does not ameliorate mortality due to PKC-independent CUGBP1 overexpression.
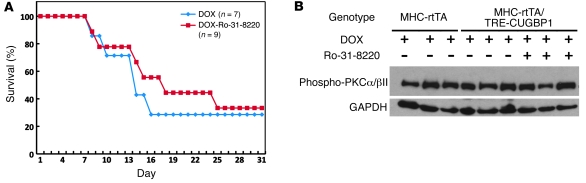
Both of the PKC inhibitors used here (Ro-32-0432 and Ro-31-8220) have been shown to improve cardiac contractility when administered to mice with different cardiac contraction abnormalities (23). To determine whether PKC inhibition could have a general restorative effect on animals expressing EpA960(R) RNA independent of the reduced expression of CUGBP1, we tested the effect of Ro-31-8220 in a different DM1 mouse model, in which human CUGBP1 is expressed in adult heart, using a tetracycline-inducible transgene. These animals exhibit approximately 80% mortality within 3 weeks of induction as well as electrophysiological and contractile abnormalities similar to mice expressing EpA960(R) RNA (our unpublished observations). Ro-31-8220 was administered concomitantly with CUGBP1 induction by doxycycline but did not prevent mortality of CUGBP1-overexpressing mice (Figure 6A). Western blots using phospho-specific antibodies demonstrate that PKCα/βII was not activated by CUGBP1 overexpression (Figure 6B). These results demonstrate that Ro-31-8220 does not ameliorate the effects of CUGBP1 upregulation mediated by a PKC-independent mechanism and strongly suggest that pleiotropic effects of this drug do not explain its effects on mice expressing EpA960(R) RNA.
Figure 6. Ro-31-8220 does not prevent mortality in CUGBP1-overexpressing mice.
(A) Kaplan-Meier survival curve of transgenic animals (MHC-rtTA/TRECUGBP) induced to express CUGBP1, with administration of doxycycline in combination with Ro-31-8220 (DOX+Ro-31-8220) or saline with doxycycline (DOX). Human CUGBP1 was inducibly expressed specifically in hearts of adult mice, using food containing 6 g/kg doxycycline (DOX). (B) PKCα/βII activation in induced MHC-rtTA/TRECUGBP mice, with or without Ro-31-8220 administration. PKC activation was determined by Western blot using phospho-PKCα/βII (75 kDa) antibody. GAPDH was used as a loading control.
Molecular features of DM1 appear after withdrawal of Ro-31-8220.
To determine whether withdrawal of Ro-31-8220 from mice expressing EpA960(R) RNA has long-term protection, a group of 6 animals were given daily injections of Ro-31-8220 for 43 days and then maintained for 22 days without Ro-31-8220 injections (TAM-Ro-31-8220-stop mice; Figure 7). All 6 mice were alive at the end of the study. Molecular analysis demonstrated that PKCα/βII activity was comparable or even higher in heart tissue from mice in which Ro-31-8220 was withdrawn, compared with that in tamoxifen-induced saline-treated mice (Figure 7A, lanes 8–11). Consistent with the appearance of activated PKCα/βII, these animals also expressed high levels of CUGBP1 (Figure 7A, lanes 8–11), which was hyperphosphorylated (Figure 7B). Connexin 43 levels were variable between mice, but overall it appears that withdrawal of Ro-31-8220 did not affect connexin 43 expression in 3 out of 4 mice, suggesting that not all effects of Ro-31-8220 were reversible upon withdrawal.
Figure 7. Molecular features of DM1 reappear after withdrawal of Ro-31-8220.
(A) Western blot analysis of CUGBP1, phospho-PKCα/βII, total PKCα/βII, and connexin 43 in heart tissue from control mice (MCM) (lanes 1 and 2), mice treated with tamoxifen (TAM) (all 5 mice died within 3 weeks) (lanes 3–7), and mice treated with Ro-31-8220 for 43 days then taken off the drug for 22 days (Ro-31-820-STOP) (lanes 8–11). GAPDH was used as a loading control. (B) 2D/Western blot analysis of CUGBP1 in MCM, TAM, and Ro-31-8220-STOP groups. (C) Both CUGBP1- and MBNL1-specific splicing events were misregulated in heart tissues from Ro-31-8220-STOP animals. The number of animals per group for RT-PCR analysis is noted (mean ± SEM). *P < 0.05 versus MCM control.
We next determined whether CUGBP1-regulated splicing events were misregulated in TAM-Ro-31-8220-stop mice. Splicing of Ank2 exon 21 and Sorbs1 exon 6 was misregulated in TAM-Ro-31-8220-stop mice (Figure 7C), and while Mtmr3 exon 16 splicing was misregulated similar to saline-treated tamoxifen mice, it was not significantly different from splicing in the MCM animals that do not express EpA960(R) RNA. As expected, the splicing patterns of all MBNL1 targets were misregulated among animals treated with or without Ro-31-8220 compared with the MCM control group (Figure 7C). Taken together, these results indicate that withdrawal of Ro-31-8220 leads to expression of activated PKC, hyperphosphorylated and upregulated CUGBP1, and misregulation of CUGBP1-specific splicing events. While these mice survived for 22 days without Ro-31-8220, it is unknown whether the mice cardiac physiology is disturbed. In addition, it is possible that EpA960(R) RNA–expressing mice might experience a delayed phenotype after longer periods following Ro-31-8220 withdrawal (see Discussion).
Discussion
Loss of MBNL1 function and elevated CUGBP1 protein levels are 2 major molecular events involved in DM1 pathogenesis (6). However, the individual roles of CUGBP1 and/or MBNL1 in the DM1 cardiac phenotype remain undefined. Loss of specific MBNL1 isoforms in mice recapitulates several features of DM1, such as cataracts, myotonia, and alternative splicing defects in skeletal muscle (10), whereas its role in DM1 heart pathogenesis is unknown. Constitutive overexpression of CUGBP1 in mouse heart and skeletal muscle results in neonatal lethality (19, 20) and inducible expression in adult mouse heart results in arrhythmias, contractile dysfunction, and lethality (our unpublished observations). We have previously demonstrated that the PKC signaling pathway is activated by expression of CUG repeat–containing RNA in cell culture, the DM1 mouse model, and in DM1 heart tissues (17). We also showed that PKC activation is an early event, detectable 6 hours after induction of EpA960(R) RNA in the heart-specific DM1 mouse model (17). In this study, we provide the first evidence to our knowledge that PKC activation plays a pathogenic role in DM1 cardiac pathogenesis and that early blockade of PKC activity ameliorates mortality, arrhythmias, and reduced cardiac output. We found that PKC inhibition suppressed hyperphosphorylation and upregulation of CUGBP1 as well as misregulated splicing of CUGBP1-specific targets. In contrast, neither MBNL1-regulated splicing events nor its posttranslational modifications were affected by the PKC inhibitor administration. Thus, blocking PKC activity may provide a tool to distinguish the different contributions of CUGBP1 and MBNL1 to the DM1 cardiac phenotype.
A role for PKC activation is implicated in the pathogenesis of cardiac hypertrophy, heart failure, and ischemic heart disease (32–35). Genetic or pharmacologic inhibition of PKC activation improves cardiac contractility in several heart failure models (23, 36, 37). The beneficial effects of PKC inhibition in the DM1 mouse model do not appear to be due to a general enhancement of contractility but rather strongly correlate with reduced PKC activation, hyperphosphorylation, and steady-state levels of CUGBP1. The mechanism by which Ro-31-8220 prevents cardiac abnormalities is likely to be mediated by effects on multiple downstream targets of PKC. While a direct cause-effect relationship between decreased CUGBP1 expression and improved cardiac function remains to be established, we demonstrated that PKC inhibition did not reduce mortality of mice overexpressing CUGBP1 by a PKC-independent mechanism (Figure 6), supporting the contention that the protective response to PKC inhibitors is related to the reduced CUGBP1 expression. Overall there is a strong correlation between inhibition of PKC activation, CUGBP1 hyperphosphorylation and stabilization, and normalized splicing of CUGBP1 targets and improved cardiac function.
Two different PKC inhibitors were shown to have protective effects. Both prevented mortality and reduced CUGBP1 hyperphosphorylation but differed in their ability to prevent conduction abnormalities in mice expressing EpA960(R) RNA. This difference is likely to be related to differences in the profiles of PKC isoforms inhibited by Ro-31-8220 and Ro-32-0432 and the differences in the half-life of these drugs in mice. For example, Ro-31-8220 is a more potent inhibitor of PKCε and βII than Ro-32-0432 (31). It also appeared that Ro-31-8220 was more effective at reducing CUGBP1 hyperphosphorylation and steady-state protein levels.
An unexpected observation was that mice can survive for 22 days following withdrawal of Ro-31-8220 administration despite activated PKCα/βII, elevated CUGBP1, and misregulation of CUGBP1-regulated pre-mRNA targets. One possibility is that PKC inhibition “protects” the mice from the acute effects of induced EpA960(R) RNA. DM1 is a chronic and progressive disease, and it is possible that these animals will develop a chronic DM1 heart phenotype over an extended time course, after Ro-31-8220 administration has been halted.
In summary, we have shown that inhibition of PKC activation ameliorates the DM1 cardiac phenotype induced by expression of EpA960(R) RNA, supporting a direct role of the PKC pathway in DM1 pathogenesis. The pathogenic effects of PKC activation include upregulation of CUGBP1 and are likely to include modification of other downstream targets.
Methods
Animal models and Ro-31-8220 administration.
Use of the tamoxifen-inducible heart-specific DM1 mouse model has been described previously (21). The EpA960 line that expresses the highest level of EpA960(R) RNA (line 1323) was used in this study. All EpA960/MCM bitransgenic animals as well as MCM controls were the F1 progeny of pure EpA960 and MCM lines. MCM animals were obtained from J. Molkentin (University of Cincinnati, Cincinnati, Ohio, USA). EpA960(R) RNA was induced in EpA960/MCM bitransgenic animals with 5 consecutive daily intraperitoneal injections of tamoxifen (20 mg/kg/d) (21). The PKC inhibitor Ro-31-8220 (6 mg per kg body weight) and Ro-32-0432 (6 mg per kg body weight) was delivered intraperitoneally in saline solution as described (23). Mock-treated mice were injected with saline solution alone. Ro-31-8220 was delivered daily beginning 2 hours after the first tamoxifen injection. Animals containing a tetracycline-responsive human CUGBP1 transgene (TRECUGBP) were mated to MHCrtTA mice, in which rtTA is expressed using the heart-specific α-MHC promoter (38). MHCrtTA/TRECUGBP and MHCrtTA mice were F1 progeny of TRECUGBP x MHC-rtTA matings. CUGBP1 was induced using food containing 6 g/kg doxycycline. All experiments were conducted in accordance with the NIH Guide for the care and use of laboratory animals and approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Quantification of EpA960(R) expression by real-time PCR.
Total RNA extraction and first-strand cDNA synthesis were described previously (21). Real-time PCR was performed using the TaqMan probe (TTACTCCCACAGCTCCTG) as well as primers EpArecomb-recoF (GCCCCGGCTCTGACT) and EpArecomb-recoR (CAGCACAATAACCAGCACGTT), using TaqMan Universal PCR Master Mix on an ABI Prism 7500 Sequence Detector (Applied Biosystems). Expression of EpA960(R) RNA in each animal was compared with a standard curve generated from a dilution series of cDNA products pooled from several animals. GAPDH was used as internal control, using previously described primers (29).
Sample preparation and 2D gel analysis.
Mouse heart tissue lysates were prepared as described previously (21). Protein concentration was determined by the BCA assay (Pierce). For 1D analysis, 50 μg protein was loaded on 10% SDS-PAGE followed by Western blot. For 2D gel analysis, mouse heart tissues were immediately frozen in liquid nitrogen and pulverized using a mortal and a pestle. Tissue powder was resuspended in 2D urea buffer as described previously (17). Protein concentrations of the cleared lysates were determined by the Bradford assay (Bio-Rad). The 2D/Western blot analysis was performed using 400 μg protein on an Ettan IPGphor Isoelectric Focusing System (GE Healthcare), followed by 10% SDS-PAGE as described previously (17). The predicted pI of unmodified CUGBP1 is 8.5.
Western blots and antibodies.
CUGBP1 (50 kDa) was detected using a monoclonal antibody (3B1) (1:2,500; Upstate). Incubations were overnight at 4°C in 4% blotto (4% dry milk in PBS solution). Anti-mouse light chain–specific IgG-HRP (1:5,000; Jackson ImmunoReaseach Laboratories Inc.) was used as a secondary antibody to avoid detection of heavy chain of mouse IgG (51 kDa). Membranes were washed in PBS with 0.5% Tween 20 for 5–10 minutes 3 times between incubations at room temperature. Activated PKCα/βII was detected using a polyclonal phospho-specific (Thr638/641) PKCα/βII antibody (1:1,000; Cell Signaling Technology). PVDF membranes were then stripped and reprobed with PKCα-specific monoclonal antibody (1:1,000; BD Biosciences — Transduction Laboratories) to determine total PKCα levels. Connexin 43 levels were detected using a polyclonal antibody (1:1,000; Zymed).
RT-PCR splicing analysis.
Total RNA (3–5 μg) and 100 ng of oligo(dT) primer were used for synthesis of first-strand cDNA (21). PCR analysis was performed as described previously (20). Primer sequences for detecting alternative splicing of exon 4 and 5 of Tnnt2, exon 21 of Ank2, exon 16 of Mtmr3, exons 6 and 23 of Sorbs1, and exon 5 of Mbnl1 were described previously (21, 30).
ECG telemetry.
ECG telemetry in nonanesthetized and unrestrained mice was performed using a radiotelemetry implant (Data Sciences International). The implant was placed in the peritoneal cavity with subcutaneously tunneled electrodes in a lead II electrocardiogram configuration. Telemetry was recorded at least 1 week after surgery and prior to tamoxifen injection to establish a baseline. Eight animals were used and given tamoxifen for 5 consecutive days, and half the mice received daily Ro-31-8220 injections for 2 weeks, beginning 2 hours after the first tamoxifen injection. Telemetry was recorded every 2 days and on the last day of Ro-31-8220 injections. ECG data was collected and analyzed using ECG Auto (EMKA technologies). Periods of sinus rhythm were identified and analyzed to determine the heart rate, PR and QTc interval, and QRS duration.
Cardiac function.
Echocardiography was performed on anesthetized mice (1.5% isoflurane mixed with 95% oxygen). Mice were stabilized on a heated platform and taped to ECG electrodes. Evaluation of cardiac function was done using a Visual Sonics Vevo 770 ultra sound, using a 30 MHz probe. Image analysis was done using Visual Sonics software version 2.3.0 as described previously (39).
Statistics.
ANOVA was used to compare means among 3 or more independent groups. For multiple comparisons, Bonferroni’s multiple comparison test was applied. P values of less than 0.05 were considered to be statistically significant. Statistical significance was displayed as an asterisk indicated a difference from control mice (MCM) in the figures.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R01 AR45653, to T.A. Cooper) and the Muscular Dystrophy Association (to T.A. Cooper), an American Heart Association postdoctoral fellowship (to G.-S. Wang), and an American Heart Association Scientist Development grant (to M.N. Kuyumcu-Martinez). M.N. Kuyumcu-Martinez was the recipient of a postdoctoral National Research Service Award from the NIH and a development grant from Muscular Dystrophy Association. X.H.T. Wehrens is a W.M. Keck Foundation distinguished young scholar in medical research and is supported by grants from the NIH/National Heart, Lung and Blood Institute (HL089598-01, R01HL091947), Muscular Dystrophy Association (69238), and the Hankamer Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3797–3806 (2009). doi:10.1172/JCI37976.
References
- 1.Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 2. Harper, P.S. 2001.Myotonic dystrophy . 3rd edition. W.B. Saunders. London, United Kingdom. 436 pp. [Google Scholar]
- 3.Mathieu J., Allard P., Potvin L., Prevost C., Begin P. A 10-year study of mortality in a cohort of patients with myotonic dystrophy. Neurology. 1999;52:1658–1662. doi: 10.1212/wnl.52.8.1658. [DOI] [PubMed] [Google Scholar]
- 4.Groh W.J., et al. Electrocardiographic abnormalities and sudden death in myotonic dystrophy type 1. N. Engl. J. Med. 2008;358:2688–2697. doi: 10.1056/NEJMoa062800. [DOI] [PubMed] [Google Scholar]
- 5.Sovari A.A., Bodine C.K., Farokhi F. Cardiovascular manifestations of myotonic dystrophy-1. Cardiol. Rev. 2007;15:191–194. doi: 10.1097/CRD.0b013e318070d1a7. [DOI] [PubMed] [Google Scholar]
- 6.Kuyumcu-Martinez N.M., Cooper T.A. Misregulation of alternative splicing causes pathogenesis in myotonic dystrophy. Prog. Mol. Subcell. Biol. 2006;44:133–159. doi: 10.1007/978-3-540-34449-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlet-B. N., et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/S1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 8.Mankodi A., et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/S1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 9.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 10.Kanadia R.N., et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 11.Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 12.Mankodi A., et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 13.Fardaei M., Larkin K., Brook J.D., Hamshere M.G. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fardaei M., et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 15.Miller J.W., et al. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanadia R.N., et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady state levels of CUGBP1 in Myotonic Dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timchenko N.A., et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 19.Ho T.H., Bundman D., Armstrong D.L., Cooper T.A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 20.Timchenko N.A., et al. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 21.Wang G.S., Kearney D.L., De Biasi M., Taffet G., Cooper T.A. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohal D.S., et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 23.Hambleton M., et al. Pharmacological- and gene therapy-based inhibition of protein kinase Calpha/beta enhances cardiac contractility and attenuates heart failure. Circulation. 2006;114:574–582. doi: 10.1161/CIRCULATIONAHA.105.592550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy H.S., Fort A.G., Spray D.C. Cardiac connexins: genes to nexus. Adv. Cardiol. 2006;42:1–17. doi: 10.1159/000092550. [DOI] [PubMed] [Google Scholar]
- 25.Yadava R.S., et al. RNA toxicity in myotonic muscular dystrophy induces NKX2-5 expression. Nat. Genet. 2008;40:61–68. doi: 10.1038/ng.2007.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutstein D.E., Morley G.E., Fishman G.I. Conditional gene targeting of connexin43: exploring the consequences of gap junction remodeling in the heart. Cell. Commun. Adhes. 2001;8:345–348. doi: 10.3109/15419060109080751. [DOI] [PubMed] [Google Scholar]
- 27.Lai-Cheong J.E., Arita K., McGrath J.A. Genetic diseases of junctions. J. Invest. Dermatol. 2007;127:2713–2725. doi: 10.1038/sj.jid.5700727. [DOI] [PubMed] [Google Scholar]
- 28.Ho T.H., et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orengo J.P., et al. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalsotra A., et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkinson S.E., Parker P.J. Isoenzyme specificity of bisindolylmaleimides, selective inhibitors of protein kinase C. Biochem. J. . 1993;294:335–337. doi: 10.1042/bj2940335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnsen D.D., et al. Protein kinase C isozymes in hypertension and hypertrophy: insight from SHHF rat hearts. Mol. Cell. Biochem. 2005;270:63–69. doi: 10.1007/s11010-005-3781-x. [DOI] [PubMed] [Google Scholar]
- 33.Sugden P.H. Signalling pathways in cardiac myocyte hypertrophy. Ann. Med. 2001;33:611–622. [PubMed] [Google Scholar]
- 34.Cain B.S., Meldrum D.R., Harken A.H. Protein kinase C in normal and pathologic myocardial states. J. Surg. Res. 1999;81:249–259. doi: 10.1006/jsre.1998.5508. [DOI] [PubMed] [Google Scholar]
- 35.Tsouka V., Markou T., Lazou A. Differential effect of ischemic and pharmacological preconditioning on PKC isoform translocation in adult rat cardiac myocytes. Cell. Physiol. Biochem. 2002;12:315–324. doi: 10.1159/000067901. [DOI] [PubMed] [Google Scholar]
- 36.Braz J.C., et al. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat. Med. 2004;10:248–254. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 37.Hambleton M., et al. Inducible and myocyte-specific inhibition of PKCalpha enhances cardiac contractility and protects against infarction-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H3768–H3771. doi: 10.1152/ajpheart.00486.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valencik M.L., McDonald J.A. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res. 2001;10:269–275. doi: 10.1023/A:1016601928465. [DOI] [PubMed] [Google Scholar]
- 39.Sood S., et al. Intracellular calcium leak due to FKBP12.6 deficiency in mice facilitates the inducibility of atrial fibrillation. Heart Rhythm. . 2008;5:1047–1054. doi: 10.1016/j.hrthm.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.