Abstract
LMP1 induces the phenotypic transformation of fibroblasts and affects regulators of the cell cycle during this process. LMP1 decreases expression of the cyclin-dependent kinase inhibitor p27 and increases the levels and phosphorylation of cyclin-dependent kinase 2 and the retinoblastoma protein. In the present study, the effects of LMP1 on cell cycle progression and the mechanism of p27 downregulation by LMP1 were determined. Although p27 is frequently regulated at the posttranscriptional level during cell cycle progression and in cancer, LMP1 did not decrease ectopically expressed p27. However, LMP1 did decrease p27 RNA levels and inhibited the activity of p27 promoter reporters. The LMP1-regulated promoter element was mapped to a region containing two E2F sites. Electrophoretic mobility shift assays determined that the regulated cis element bound an inhibitory E2F complex containing E2F4 and p130. These findings indicate that LMP1 decreases p27 transcription through effects on E2F family transcription factors. This property likely contributes to the ability of LMP1 to stimulate cell cycle progression.
Epstein-Barr virus (EBV) is a herpesvirus that causes infectious mononucleosis and is associated with a number of malignancies. EBV infection of B cells results in a latent infection and the transformation of B cells in vitro and in vivo. Several proteins are required for EBV-mediated B-cell transformation, including the EBV nuclear antigens (EBNAs), EBNA1, EBNA2, EBNA3A, and EBNA3C, and latent membrane protein 1 (LMP1). LMP1 is expressed in most EBV-associated cancers and is considered the EBV oncogene as it is able to induce fibroblast transformation (34, 60, 83, 84). Expression of LMP1 is frequently detected in nasopharyngeal carcinoma, and LMP1 induces phenotypic changes in epithelial cells and nasopharyngeal carcinoma cell lines, including increased motility, invasion, and migration (12, 45, 60, 66, 73).
LMP1 is a constitutively active tumor necrosis factor receptor homologue. The transmembrane domain of LMP1 mediates self-association in the absence of ligand, and LMP1 is recruited to cholesterol-rich lipid raft domains in the membrane (3, 14, 20, 28, 58, 81). LMP1 initiates signaling from two carboxyl-terminal activating regions (CTAR1 and CTAR2) by recruiting tumor necrosis factor receptor-associated factors (TRAFs) and other adaptors (33). LMP1 induces constitutive signaling of nuclear factor κB (NF-κB), phosphatidylinositol 3-kinase (PI3K), mitogen-activated protein kinase (MAPK), and c-Jun N-terminal kinase (11, 16, 31, 43, 44, 52, 56, 61, 71).
Signaling from CTAR1 is required for EBV B-cell transformation and for LMP1 fibroblast transformation (5, 30, 32, 53, 78). CTAR1 uniquely activates several pathways and proteins and induces both canonical and noncanonical NF-κB, PI3K, and MAPK signaling. Several growth-stimulating proteins upregulated by CTAR1 have been identified, including the epidermal growth factor receptor, TRAF1, and inhibitors of differentiation 1 (Id1) and Id3 (13, 18, 51). LMP1 induces the transcription of the epidermal growth factor receptor, and this property is dependent upon activation of a specific NF-κB complex containing Bcl-3 and dependent upon STAT3 activation (39, 72). The contribution of distinct signaling pathways to fibroblast transformation has been analyzed using a combination of LMP1 CTAR1 mutants and chemical inhibitors. Transformation of fibroblasts required activation of the MAPK-extracellular signal-regulated kinase (ERK) pathways and PI3K, and inhibition of these pathways blocked transformation (44). Inhibition of ERK blocked LMP1-induced epithelial cell motility and invasion (12). In contrast, pharmacological inhibition of NF-κB had minimal effects while inhibition of the expression of Id1 with small interfering RNA slightly impaired the growth of transformed fibroblasts (17, 43). PI3K activation correlated with the regulation of a number of cell cycle proteins associated with G1/S transition (18, 43, 44). During rodent and human fibroblast transformation, LMP1 decreases the levels of the cyclin-dependent kinase inhibitor (CDKI) p27, which binds and inhibits the activity of CDK/cyclin complexes to arrest cell cycle progression. (17, 18, 43-45). The transformed cells also contain increased activated cyclin-dependent kinase 2 (Cdk2) and the phosphorylated inactivated form of retinoblastoma (Rb). Lymphomas of transgenic mice engineered to express LMP1 also have low levels of p27 protein (65), and p27 upregulation is associated with the growth arrest of EBV-positive lymphocyte cell lines (85, 86). Because of their potent growth-inhibitory activity, many cancers have developed mechanisms to downregulate or inactivate the CDKIs (2, 4, 76).
In the current study the mechanism of regulation of p27 by LMP1 was determined. LMP1 and several LMP1 mutants reduced p27 protein expression in Rat-1 cells. However, this did not reflect posttranscriptional regulation of p27, as ectopically expressed p27 was not reduced by LMP1. LMP1 greatly reduced p27KIP1 RNA levels and decreased the activity of p27KIP1 promoter reporters. The cis element regulated by LMP1 was mapped to −900 bp of the promoter that contained E2F consensus sites. According to the results of electrophoretic mobility shift assays (EMSAs), LMP1 affected the migration patterns of probes containing the E2F site, and antibodies for E2F4 and p130 induced new complexes. These data indicate that p27 transcription is regulated by E2F-dependent transcription and that LMP1 induces formation of inhibitory E2F4 and p130-containing complexes. These effects of LMP1 on E2F/pocket proteins likely contribute to the LMP1-mediated stimulation of cell cycle progression. The findings of this study identify reciprocal regulation between p27 and E2F: decreased p27 can increase CDK activity and E2F-dependent transcription and E2F transcription factors can inhibit p27 expression.
MATERIALS AND METHODS
Plasmids.
The cloning of wild-type and mutant LMP1 into retrovirus packaging vector pBabe was previously described (18, 44). LMP1 mutants used in the current study include LMP1-A5 (CTAR1 mutant, TRAF binding domain [P204xQ206xT208] changed to all alanines [A5]), 1-220 (amino acids 1 to 220, CTAR2 deleted), and 1-220-A5 (CTAR1 mutated/CTAR2 deleted).
The cloning of the rat p27KIP1/cyclin-dependent kinase inhibitor 1B gene for specific RNase protection probes was previously described (17). The myc-tagged p27 expression vector, M3-p27, was generated in the same way as the RNase protection probe plasmid except that it was inserted downstream of the triple myc tag of M3-pcDNA3 (40).
The cloning of the full-length p27 promoter reporter construct p27-1501 containing upstream regulatory sequence of p27KIP1/cyclin-dependent kinase inhibitor 1B of Rattus norvegicus was previously described (17). Truncations at the 5′ end of the promoter were constructed using convenient restriction enzyme sites, p27-1066 (BamHI), p27-939 (SacI), and p27-554 (SacI). Putative transcription factor binding sites were determined within the region from −939 to 554 using the online programs TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) (27) and MotifMogul (http://xerad.systemsbiology.net/MotifMogulServer/) (24). Internal deletions within the region from −939 to 554 were created by full-length plasmid PCR mutagenesis with AccuPrime Pfx DNA polymerase (Invitrogen) according to the manufacturer's directions using 5′-phosphorylated primers. A putative FoxA1/3 site was deleted using Foxp27-744′ (GCTAGCCAGAGCAGGTTTGTTGGCAGTCG) and Foxp27-777′ (CAAACGGCCGGAGAGCTGGGG) to create Δ740-774. A putative MZF1 site was deleted using MZF1p27-788′ (CCGTTTGGCTAGTTTGTTTGTCTTATTTTTAATTTCTCCGGGGCCAG) and MZF1p27-812′ (CCGCGGCCAGGCGGAGACCCGGGAATCTAG) to create Δ788-812. Putative E2F sites were deleted with E2Fp27-900′ (GAGGTCGCAGTCCGGAGCGGTGGC) and E2Fp27-924′ (CAGCTGGGCCAGTGAGCTCGGTACCTATCGATAGAGAAATG) to create Δ900-923. Finally, all three regions were deleted with Foxp27-744′ and E2Fp27-924′. Mutated plasmids were ligated and transformed into Escherichia coli. Plasmids were screened for the presence of a newly created restriction site and sequenced.
Transfection and retrovirus transduction.
Cells were transfected with FuGENE 6 (Boehringer Mannheim) according to the manufacturer's directions. Recombinant retroviruses were generated as previously described (64) by transfection of 293T cells with pBabe, wild-type LMP1 or mutant LMP1 plasmids, and VSV-G (pG1-VSV-G)- and gag-pol (pGPZ9)-expressing plasmids. After 24 h medium was changed and cells were incubated at 33°C overnight. The following day culture supernatants were harvested and centrifuged at 1,000 × g for 10 min to remove cells and cellular debris. Rat-1 cells were transduced with clarified 293T culture supernatants overnight with 8 μg/ml Polybrene (Sigma).
Cell culture and stable cell lines.
Cell lines were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with antibiotic-antimycotic mixture and 10% (vol/vol) heat-inactivated fetal bovine serum. Rat-1 rodent fibroblast stable cell lines were established by transduction with retrovirus and selection with puromycin (1 μg/ml; Sigma). Transient selection for p27 expression was accomplished by selection 24 h posttransfection with G418 (500 μg/ml; Mediatech) for 48 h.
RPA.
RNase protection assays (RPAs) were performed using a Direct Protect Lysate RPA kit (Ambion) according to the manufacturer's directions as previously described (17). Briefly, RPA probes were annealed with RNA overnight in lysis buffer. The following day samples were digested with RNase and protease and precipitated. Protected probes were resolved on a 5% polyacrylamide gel, dried, and imaged using a PhosphorImager (Molecular Dynamics). Bands were quantitated using ImageQuant 5.1 (Molecular Dynamics), and p27 RNA levels were determined relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels.
Cell harvesting and Western blotting.
Cell lines were grown in 100-mm tissue culture plates to confluence and harvested. Cells were washed with ice-cold phosphate-buffered saline (Gibco) and lysed with 100 to 250 μl RIPA buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1% deoxycholic acid, protease and phosphatase inhibitors [Pierce]). Cell lysates were clarified by centrifugation and quantitated by Bio-Rad DC protein assay system (Bio-Rad). Samples were then boiled in SDS sample buffer, and indicated amounts of protein were separated using 12% acrylamide SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Li-Cor) for Western blotting analysis. Primary antibodies included actin and hemagglutinin (HA) tag (Cell Signaling), LMP1-specific antibodies (Cao 7E10, Cao 8G3, LMP1 IG6, and Cao 7G8; Ascenion GmbH), p27 (Santa Cruz), and myc tag (Upstate). LMP1 antibodies were detected with biotinylated anti-rat immunoglobulin G (heavy plus light) (16-16-12; Kirkegaard & Perry Laboratories, Inc.) followed by reaction with IRDye 680-labeled streptavidin (Li-Cor). Other bound proteins were detected with IRDye-labeled secondary antibodies by scanning with a Li-Cor Odyssey imaging system. Bands were quantitated using the Li-Cor imaging software.
p27 promoter luciferase assays.
Promoter reporter assays were performed as previously described (17). 293T cells were plated 1:5 into 12-well plates 1 day prior to transfection. Cells were transfected with 0.2 μg of pRL-SV40 (Renilla luciferase) (Promega), 0.2 μg of pGL3-Basic or p27 promoter plasmids, and 0.2 μg of pBabe or LMP1-expressing plasmids. The following day the medium was changed, and 40 h posttransfection cells were harvested and luciferase activity was assayed using the dual-luciferase reporter assay system (Promega) according to the manufacturer's directions. Relative luciferase activity was determined by dividing the firefly luciferase activity of the p27 promoter constructs by the internal control Renilla luciferase activity. Each condition was used in triplicate and replicated in different experiments.
EMSA.
Approximately 5 × 107 cells were trypsinized, washed with ice-cold phosphate-buffered saline, and pelleted. Cell pellets were resuspended in 350 μl buffer A (20 mM HEPES, pH 7.5, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, and protease and phosphatase inhibitors [Pierce]) and incubated on ice for 15 minutes. Cells were lysed by addition of NP-40 (1% final concentration) followed by vortexing for 1 minute. Nuclei were pelleted by centrifugation at 1,000 × g at 4°C for 10 minutes. The nuclear pellet was washed one time in buffer A and solubilized by addition of 1 pellet volume of NE buffer (20 mM Tris, pH 8.0, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% [vol/vol] glycerol), 1/4 pellet volume of 5 M NaCl, and 1 pellet volume of NE buffer followed by vortexing. Clarified nuclear lysates were quantitated as above and used for EMSA.
EMSAs were performed using the EMSA buffer kit for the Odyssey imaging system (Li-Cor) according to the manufacturer's directions. Shifts were performed using 5 μg of nuclear protein bound to labeled probes and were visualized using a Li-Cor Odyssey imaging system. The IRDye 700 infrared dye-labeled oligonucleotides (Integrated DNA Technologies) contained the putative E2F sites of the rat p27KIP1 promoter (CCCTGGGTTTCGCGGGCAAAGACCTGG, forward strand consensus underlined and reverse strand consensus in bold and italics). Competition reactions were performed with a 200-fold molar excess of unlabeled consensus probe (ATTTAAGTTTCGCGCCCTTTCTCAA, consensus underlined) or mutated probe (ATTTAAGgggatataCCTTTCTCAA, mutations in lowercase and underlined). Supershifts were performed by incubation of nuclear lysates in binding buffer with antibodies for 20 min prior to probe addition. Antibodies purchased from Santa Cruz used for supershifts included p107 (sc-318 X), p130 (sc-317 X), Rb (sc-50 X), E2F1 (sc-251 X), E2F4 (sc-512 X), and DP1 (sc-610 X).
RESULTS
In this study, the mechanism of p27 downregulation by LMP1 was determined using LMP1 mutants that differ in their abilities to activate different signaling pathways and induce transformation. LMP1 mutants used in this study include full-length LMP1 in which the TRAF binding domain is mutated to all alanines (LMP1-A5) (mutated CTAR1/wild-type CTAR2), 1-220 (wild-type CTAR1/truncated at amino acid 220 and deletion of CTAR2), and 1-220-A5 (mutated CTAR1/deletion of CTAR2). We have previously shown that LMP1 and 1-220 can transform Rat-1 cells, activate MEK/ERK signaling, and regulate cell cycle markers p27, Cdk2, and Rb. LMP1-A5 is not transforming and does not activate MEK/ERK signaling but still regulates cell cycle markers. 1-220-A5 is not transforming and does not regulate cell cycle proteins. The mechanism of downregulation of p27 by LMP1 is unknown.
Effects of LMP1 on p27 protein.
To determine the effects of LMP1 and LMP1 mutants on p27 protein levels, stable cell lines were examined. Rat-1 cells were stably transduced with control (Babe)-, LMP1-, and LMP1 mutant-expressing retrovirus. Cells were grown to confluence, harvested, and then examined by Western blotting for p27 protein levels (Fig. 1). p27 levels were quantitated relative to actin loading control, and LMP1 and LMP1 mutant expression was confirmed by detection of the HA tag. Full-length LMP1 and LMP1-A5 at about 66 kDa were detected with a typical degradation product at about 35 kDa. 1-220 and 1-220-A5 LMP1 mutants were detected at about 30 kDa. Expression of CTAR1-containing constructs, LMP1 and 1-220, was less than LMP1-A5 and 1-220-A5 expression. Lower LMP1 and 1-220 expression is consistent with increased ubiquitination and proteasomal degradation mediated by CTAR1-TRAF binding and consistent with previous expression of these mutants (44, 62). In agreement with our previous studies, the levels of p27 protein were decreased to approximately 30% of control p27 levels (17, 18, 43-45). The full-length LMP1 mutant with CTAR1 mutated, LMP1-A5, and 1-220 (CTAR2 deleted) also decreased p27 protein to approximately half that of control cell lines while the truncated CTAR1/CTAR2 mutant, 1-220-A5, had minimal effect on p27 protein levels.
FIG. 1.
Downregulation of p27 protein by LMP1 and LMP1 mutants. Control (Babe) and wild-type and mutant LMP1 stable Rat-1 cells were assayed by Western blotting for p27 compared to actin (loading control). Expression of tagged LMP1 constructs was confirmed with HA antibodies. p27 expression was quantitated relative to actin levels using the Li-Cor Odyssey imaging system and indicated as a value relative to Babe cells set as 100. The positions of the molecular weight markers (in thousands) are indicated by the closed arrowheads.
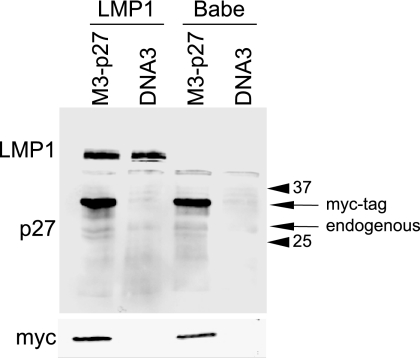
We had previously determined that LMP1 decreased p27 RNA and p27 promoter reporter activity (17). However, p27 is frequently regulated at the posttranscriptional level during cancer development and in cancer cells. LMP1 has been shown to induce proteasome-mediated protein degradation through effects on kinase activation, ubiquitination, and deubiquitination. To determine if LMP1 also affects p27 protein levels, subconfluent Rat-1 control and LMP1 stable cell lines were transfected with vector control (DNA3) or myc-tagged p27 plasmid. Transfected cells were selected 24 h posttransfection with G418 for 24 h and harvested. Western blotting was performed for LMP1 and p27 (Fig. 2). Expression of LMP1 and myc-tagged p27 was confirmed. The myc-tagged p27 (upper arrow) was distinguished from endogenous p27 (lower arrow) by the increase in molecular mass of about 5 kDa due to the tandem triple myc tag and confirmed by Western blotting with myc tag antibody. In agreement with previous unpublished observations, endogenous p27 was barely detectable in subconfluent Rat-1 cells. The levels of the exogenous myc-p27 were equivalent in control and LMP1 stable cell lines. Equal loading was confirmed by Ponceau S staining. These data indicate that LMP1 does not affect p27 protein levels expressed from an ectopic promoter and suggest that LMP1 does not regulate p27 expression by affecting protein stability.
FIG. 2.
Regulation of ectopic p27. The ability of LMP1 to posttranscriptionally regulate ectopically expressed p27 was determined. Control and LMP1 stable cell lines were transfected with myc-tagged p27 and selected for 48 h with G418. Cell lysates from control (Babe) or LMP1 stable cells transfected with vector (DNA3) or p27-expressing plasmids were analyzed by Western blotting for p27 and LMP1 expression. A 7.5-μg amount of protein was loaded per lane. The positions of myc-tagged and endogenous p27 proteins are indicated by the arrows. The positions of the molecular weight markers (in thousands) are indicated by the closed arrowheads.
Regulation of p27 RNA by LMP1 mutants.
To assess the effects of the LMP1 mutants on p27 transcription, RNA RPAs were performed using confluent Rat-1 stable cell lines. RPAs were performed, and values were normalized to that of the housekeeping gene GAPDH. The mean p27/GAPDH expression levels were determined from three independent experiments (Fig. 3). LMP1 reduced p27 RNA to approximately 40% of control p27 levels (17). In LMP1-A5- and 1-220-expressing cells, p27 RNA was reduced to about 40 to 50% of control levels. Surprisingly, p27 RNA was also decreased about 50% with the CTAR1/CTAR2 double mutant 1-220-A5 It is unknown how this doubly deleted LMP1 could affect transcription. However, this nontransforming LMP1 minimally decreased the levels of p27 protein (92% in Fig. 1). These data indicate that LMP1 decreases p27 RNA and that this ability is linked to transformation.
FIG. 3.
Regulation of p27 RNA by LMP1 mutants. RPAs were performed using total RNA from stable cell lines. Fragments protected by rat p27KIP1 RNA probe relative to control (GAPDH) RNA probe were quantitated using a PhosphorImager, and p27 expression is depicted relative to that in control (Babe) stable cells, which was set arbitrarily as 100. Means of three independent experiments are presented with error bars representing the standard deviations.
Regulation of the p27KIP1 promoter by LMP1.
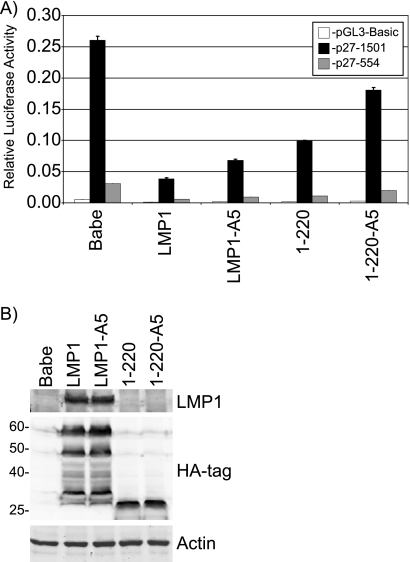
To assess the ability of LMP1 and LMP1 mutants to regulate the p27 promoter, promoter reporter assays were performed. 293T cells were cotransfected with the empty reporter vector (pGL3-Basic) or p27 reporter (p27-1501), transfection control Renilla luciferase, and effector plasmids, encoding LMP1 or LMP1 mutants. Full-length p27 promoter activity was plotted relative to the internal control Renilla luciferase activity (Fig. 4A). In agreement with our previous study (17), LMP1 greatly decreased the activity of the p27 promoter reporter plasmid, indicating that LMP1 regulates the p27KIP1 promoter. LMP1-A5 and 1-220 were slightly impaired compared to wild-type LMP1 in their ability to decrease the promoter activity, and the double mutant, 1-220-A5, reduced the promoter activity by about 30%. Other previously described CTAR1/CTAR2 mutants (44), including 1-208-A5, A5-Y384G, and A5-378stop, also reduced promoter activity by about one-third (data not shown). In parallel, cells were harvested for Western blotting for LMP1 expression (Fig. 4B). Wild-type LMP1 and LMP1 mutants were equally expressed in the reporter assays as determined by blotting with LMP1 and HA tag. Equal loading was confirmed by actin blotting. These data indicate that LMP1 and LMP mutants containing CTAR1 negatively regulate the p27KIP1 promoter similarly to the p27 protein levels while LMP1 mutants lacking CTAR1 and CTAR2 slightly reduce p27 RNA and promoter activity that may not be reflected in the protein levels.
FIG. 4.
Regulation of the p27KIP promoter by LMP1 mutants. Promoter reporter assays were performed by transfection of 293T cells with control pRL-SV40 with pGL3-Basic (empty reporter) or p27 (p27-1501 and p27-554) reporter plasmids and vector control (Babe) or plasmids expressing LMP1 (A). p27 promoter plasmids contain 1,500 and 554 bp of the Rattus norvegicus p27KIP1 promoter relative to the translation start site. Forty hours posttransfection, cells were harvested and dual-luciferase assays were performed. Relative luciferase activity was determined by the firefly luciferase activity of the reporter constructs relative to the control Renilla luciferase activity. Each condition was used in triplicate, and the means of three independent experiments are presented. Error bars represent the standard deviations from the independent experiments. In parallel, cells were harvested for Western blotting (B). Levels of wild-type LMP1 and LMP1 mutants were determined by blotting for LMP1 and HA tag. Actin blotting confirmed equal loading. The positions of the molecular weight markers (in thousands) are indicated.
Previous studies indicated that LMP1 induces two members of the Id family of proteins, Id1 and Id3 (18). The Id proteins are known to inhibit the expression of CDKIs p16 and p21 at the transcriptional level (55, 59, 87). However, we have recently determined that Id1 and/or Id3 overexpression does not regulate p27 RNA or p27 promoter reporter activity (17). This indicates that LMP1 decreases p27 transcription independently from its induction of Id1 and Id3 expression. To map the cis elements required for LMP1 regulation, the promoter was truncated at convenient restriction sites (Fig. 5A). LMP1 and LMP1 mutants were tested for their ability to downregulate p27-554, containing 554 bp of the p27 promoter (Fig. 4A). The reporter p27-554 had little activity in the promoter assay, suggesting that the majority of the activating regions of the p27 promoter were located between bp −555 and −1501. Truncations containing 1,066 (p27-1066) and 939 (p27-939) bp were tested against wild-type LMP1 (Fig. 5B). Since each mutated reporter had slightly different intrinsic activity (Fig. 5B), the activity of each reporter in the presence of LMP1 was normalized to the activity of the reporter with the Babe vector control (Fig. 5C). Both p27-1066 and p27-939 had approximately the same activity as that of the full-length construct and were each inhibited by LMP1 to approximately one-fourth the activity of control reporter (Fig. 5C). This suggests that the region regulated by LMP1 is between −555 and −939 within the p27 promoter.
FIG. 5.
Mutation of cis-acting elements within the p27 promoter. Computer-based algorithms were used to identify putative transcription factor binding sites within the region from −554 to 939 of the p27KIP1 promoter. (A) Putative sites were deleted by PCR to mutate the indicated bases and transcription factor binding sites individually or together. (B) Promoter reporter assays were performed as described for Fig. 4 except that only control (Babe) and LMP1 were used as effector plasmids. (C) Promoter activity of deleted reporters from panel B is depicted relative to activity of the reporter of the control (Babe) effector set to 100.
To determine potential transcription factor binding sites within the region between −555 and −939 of the p27 promoter, online programs, including TFSEARCH and MotifMogul (24, 27), were used to identify putative cis elements. Three potential elements were predicted by more than one program: a forkhead (FOX0) transcription factor family binding site at approximately −750 bp, an MZF1 site at −800 bp, and two E2F sites at approximately −910 bp. PCR primers that flanked the predicted sites were designed, and PCR mutagenesis was performed to delete the putative binding sites (Fig. 5A). Specific sites were deleted in the p27-939 reporter individually: Δ740-774 deleted the FOX0 site, Δ788-812 deleted the MZF1 site, and Δ900-923 deleted the E2F sites. Finally the extreme 5′ and 3′ primers were used to delete all three sites from −740 to −923 (Δ740-923).
Promoter reporter assays were performed with the mutated reporter constructs to identify the cis elements of the p27KIP1 promoter regulated by LMP1. The mutant lacking the FOX0 sites (Δ740-774) had approximately the same activity as that of p27-939 and was downregulated by 75 to 80% by LMP1. However, mutation of the E2F sites (Δ900-923) increased the overall activity of the construct and was only slightly inhibited by LMP1 (Fig. 5B and C). The requirement for this region was confirmed with the mutant having a deletion of the entire region from −740 to −923 (Δ740-923), and the inhibition by LMP1 was impaired (Fig. 5B and C, respectively). Interestingly, this promoter had approximately twice the activity of p27-939 in control cells (Fig. 5B), indicating that the sequences between bp 923 and 939 that contain the E2F sites negatively regulate transcription in the absence of LMP1. Deletion of the predicted MZF1 (Δ788-812) site resulted in complete loss of promoter activity regardless of the presence or absence of LMP1. To ensure that the MZF1 mutant promoter reporter did not contain other mutations that resulted in the loss of activity, the region containing the deletion was subcloned into a new reporter and the site-directed mutagenesis was performed again on a new parental plasmid. Both new constructs also lacked promoter activity (data not shown). The reason for the lack of activity of this construct is unclear and was independent of the presence of LMP1. These data indicate that the p27 promoter is negatively regulated by an element between −900 and −923 bp of the promoter and that this negative regulation is greatly enhanced by LMP1.
Both of the transcription factor binding site algorithms predicted two E2F sites within the −900 to 923 region of the p27 promoter (Fig. 6A). The E2F consensus sequence of TTCGCGC is present in both the forward and reverse orientations in an overlapping element. If this element is important for p27 regulation, then it should be conserved among different species. The p27 promoters of both rat and mouse are entirely conserved at approximately the same position. The E2F-like elements were less conserved in the human p27 promoter but were located at the same relative position as those of the rat and mouse promoters.
FIG. 6.
E2F EMSAs. (A) The positions, directions, and sequences of the putative E2F sites of the rat, mouse, and human p27KIP1 promoters are depicted. The sequences of consensus E2F and E2F4-DP1 sites are indicated. The forward consensus sites are indicated by underlining, and the reverse consensus sites are indicated by bold and italics. Base pair numbers are relative to the translation start site for the p27 protein. (B) EMSAs using control (Babe) and LMP1 nuclear lysates were performed. Complexes were competed with a 200-fold excess of consensus (comp) and mutant oligonucleotides. Supershifts with the indicated antibodies were performed. The arrow indicates the position of nonspecific complexes, and boxes highlight the positions of specific complexes. Arrowheads indicate the positions of the antibody-shifted complexes for E2F4 (closed arrowhead) and p130 (open arrowhead). Lane numbers are indicated below.
To determine if LMP1 regulates E2F DNA binding complexes, EMSAs were performed using nuclear lysates from fractionated control and stable cell lines. Labeled probes corresponding to the E2F sites of the rat p27 promoter were used (Fig. 6A). Shifted probe was detected in both control (Babe) and LMP1 lysates, suggesting that there are multiple shifted complexes (Fig. 6B, lanes 2 and 3). The unshifted probe was barely detectable, suggesting that many forms of E2F can bind the probe and alter its mobility (lane 1). In all experiments, the complexes from LMP1-expressing cells were more intense and migrated in a broader band. This suggests that LMP1 induces more proteins that are able to bind to these sequences. Both the lower and higher complexes were competed with oligonucleotides containing a consensus E2F site (LMP1+comp, lane 4) but not mutated E2F consensus (LMP1+mutant, lane 5). These data confirm that LMP1 increases proteins that bind an E2F site.
To identify specific proteins bound to the probe, the complexes were treated with antibodies to the predicted transcription factors to produce antibody-supershifted complexes. The E2F proteins are a family of transcription factors. E2F1, E2F2, and E2F3a act as activators and are repressed by hypophosphorylated Rb protein. LMP1 induces increased hyperphosphorylated Rb in transformed fibroblasts and tumors from LMP1 transgenic mice (18, 43, 44, 65). Antibody to Rb did not alter the migration of the LMP1-induced complexes (Fig. 6B, lanes 14 and 15), nor did antibody to E2F1 affect the complexes (data not shown).
E2F3b and E2F4-E2F7 act as repressors, and the site present in the rat and mouse promoter closely resembles a consensus defined by hidden Markov modeling (47, 48) for E2F4-DP1 binding, TTCGCGCGAA (Fig. 6A). To determine if E2F4 and DP1 were present in the LMP1-induced complexes, E2F4 and DP1 antibodies were included in the EMSA reaction mixtures. E2F4 antibody induced a shifted complex that was increased in the LMP1 sample (Fig. 6B, lanes 6 and 7). Altered shifted complexes were not detected with the DP1 antibody in LMP1 cells (lanes 8 and 9). To repress transcription, E2F4 binds the pocket protein p107 or p130. Antibody supershifts were performed to determine if p107 or p130 was present in the EMSA complexes. A distinct supershifted band was produced with the p130 antibody, and this complex was considerably more prominent in the LMP1 samples (lanes 10 and 11). Antibody to p107 did not affect the migration of any bands (lanes 12 and 13). The data indicate that LMP1 induces binding of the repressive E2F4 and p130 to the predicted E2F site within the p27 promoter.
DISCUSSION
The data presented in this study define the mechanism of p27 downregulation by LMP1. We have previously shown that several cell cycle markers are affected by LMP1 in fibroblast transformation assays: p27 was decreased, Cdk2 was increased and activated, and Rb was increased and hyperphosphorylated. These same effects have been detected in the tumors of LMP1 transgenic mice and in EBV-transformed cells.
Cell cycle progression is controlled by the activity of cyclins, CDKs, and CDKIs. As a result of this potent growth-inhibitory activity, CDKIs are frequently nonfunctional in many cancers possibly through deletion or mutation (2, 4, 76). Interestingly, during cancer development and in cancer cells p27 is regulated posttranscriptionally and can be degraded or sequestered in the cytoplasm in cancers of the colon, prostate, esophagus, thyroid, ovary, and breast (6, 10, 23, 25, 42, 49, 67, 69, 75). However, in the present study ectopically expressed p27 was not decreased in LMP1-expressing cells. Also, levels of p27 RNA and activity of promoter reporters were decreased to the same extent as p27 protein levels in the presence of LMP1. Together these data indicate that LMP1 primarily affects p27 expression at the transcriptional level. In addition, the data suggest that p27 transcription is decreased in EBV-transformed cells due to effects on E2F and p130 pocket protein function.
Previous LMP1 mutational analyses indicated that activation of PI3K-Akt signaling by LMP1 is required for transformation, and this correlated with the effects of LMP1 on the cell cycle markers p27, Cdk2, and Rb (43, 44). A target of Akt signaling is the forkhead family of transcription factors, and both LMP1 and LMP2 can regulate forkhead transcription factors (68). FOXA1 in conjunction with BRCA1 has been shown to regulate p27 in breast cancer cells (79, 80), and other forkhead family members also regulate p27 transcription (9, 50, 54). Although the putative FOXA1/BRCA1 site is almost entirely conserved between human and rat p27 promoters, the deletion of the FOXA1/BRCA1 site did not affect the decrease in p27 promoter activity by LMP1. In contrast, deletion of the E2F site of the promoter greatly increased baseline promoter function and decreased its inhibition by LMP1 Deletion of the entire region within the promoter had activity similar to that of the E2F deletion alone, suggesting that synergy between E2F factors and forkhead transcription factors is unlikely. Importantly, these data indicate that negative transcriptional regulation of p27 is mediated by members of the E2F family, likely E2F4.
The identified E2F site in the current study is completely conserved between the mouse and rat p27 promoters, and a similar site was observed in the human p27 promoter. The current study examined activity of the rat promoter reporter within human 293T cells. The conservation of sequence and position is suggestive of conserved regulation. Expression of p27 is repressed in other model systems, like LMP1 transgenic mice, and in EBV-transformed cells. Our study would suggest that E2F4 represses the expression of p27 tumors of LMP1 transgenic mice. Future studies will more fully elucidate the regulation of p27 in human cells expressing LMP1 or transformed by EBV.
The p27 promoter deletions are also suggestive of positive regulatory elements. Promoter reporter constructs containing only 554 bp had much less activity than did constructs containing >939 bp. Surprisingly, the deletion of the sequence from bp 788 to 812 abrogated the promoter activity completely while the larger deletion of bp 740 to 923 had increased activity and activity similar to that of the bp 900 to 923 deletion alone. It is unclear why the promoter was inactivated by this smaller deletion of bp 788 to 812. Perhaps the newly juxtaposed sequences created a new site that blocked the recruitment of the normal transcriptional machinery. However, the promoter analyses do indicate that the E2F site negatively regulates the activity of the promoter and that elements between bp 554 and 740 are required for normal or baseline promoter activity.
Deletion of the E2F site resulted in increased promoter activity of the p27 promoter. E2F deletion reporters were reduced by only 25% by LMP1 as opposed to the 75 to 80% reduction for reporters containing the E2F sites. The slight reduction in the reporter in the absence of the E2F site could indicate that multiple factors could be involved in LMP1-induced p27 repression. A recent study has indicated that surrounding sites can influence which E2F factors are recruited to specific promoters (21). These other factors could be responsible for recruitment of repressive E2Fs like E2F4 to the p27 promoter.
Several CTAR1/CTAR2 mutants do not significantly alter p27 protein levels, including 1-220-A5, 1-208-A5, Y384G-A5, 378stop-A5, and 1-231-A5 (current study and reference 44). Each of these mutants reduces p27 RNA levels (1-220-A5, Fig. 3) and p27 reporter activity (1-220-A5, Fig. 4, and others' unpublished observations). Despite the decreased p27 RNA levels and p27 reporter activity, the p27 protein levels remain unchanged. The half-life of p27 protein has been measured to be between 2 and 6 h in actively dividing cells and close to 10 to 12 h in quiescent or arrested cells (38, 46). The discrepancy between p27 protein and RNA levels in cells expressing CTAR1/CTAR2 mutants likely reflects the increased stability of p27 in the contact-inhibited cells.
The EBV proteins LMP1 and EBNA3C regulate similar cell cycle pathways via distinct mechanisms. In the current study LMP1 regulates transcription of p27 RNA while EBNA3C binds Rb in vitro and can cooperate with other oncogenes to induce transformation (57). EBNA3C can also recruit the SCFSkp2 ubiquitin-ligase complex to induce the degradation of both p27 and Rb (36, 37) and block Cdk2/cyclin A-p27 binding to stimulate Cdk2/cyclin A kinase activity (35). Targeting of the cell cycle proteins by different mechanisms suggests the importance of these pathways for EBV-induced transformation and defines mechanisms by which EBV can contribute to malignant transformation.
Typical cell cycle models depict p27 blocking Cdk2 activity, Cdk2 activity targeting Rb for phosphorylation and release of E2F, and E2F transcription enabling cell cycle progression. The findings in the current study suggest that there is a regulatory loop by which p27 expression is negatively regulated by E2F function in LMP1-transformed cells. Several high-throughput screens have found that overexpression of E2F1 leads to increased p27 expression in several different cell lines (26, 29, 77). The most recent of those reports found that addition of serum or overexpression of PI3K blocked E2F1-dependent p27 induction and that the serum effect was blocked by a PI3K inhibitor, LY294002 (26). These data indicate that activation of PI3K signaling negatively regulates p27 transcription. Our previous LMP1 mutational analysis also showed that activation of PI3K signaling correlated with the ability to inhibit p27 expression (43, 44). The previous studies linking E2F1 and increased p27 transcription found E2F1 expressed exogenously to high levels. The data presented here demonstrate binding of an endogenous E2F transcription factor to a specific site in the p27 promoter and suggest that a repressive E2F, E2F4, is recruited to the promoter with the pocket protein p130 to repress p27 transcription. This E2F4/p130 complex may be replaced by an E2F1-containing complex when the promoter is activated, or other factors may also regulate the p27 promoter. Although LMP1 is known to downregulate numerous proteins, the current study is the first to define a repressive transcription complex.
Many studies have demonstrated multiple functions for p27 during the cell cycle (reviewed in reference 1). Although p27 functions as a tumor suppressor by inhibiting Cdk2 complexes, it also stimulates the assembly of Cdk4/and Cdk6/cyclin complexes. Knockout studies confirm an important role for p27 function. In some experimental systems p27+/− mice have a higher tumor incidence than do p27−/− mice. It is unclear whether reduced p27 as a result of LMP1 signaling alters normal cell cycle control that might result in increased genomic instability which could contribute to EBV-associated cancers.
The current study also has implications for the development of EBV-associated cancers. Both the Rb and p53 tumor suppressor pathways are almost always deregulated during malignant transformation. Other DNA tumor viruses encode proteins that regulate these critical pathways (41). Polyomavirus large T antigen, papillomavirus E7, and adenovirus E1A each regulate Rb function and S-phase entry. Polyomavirus large T antigen and papillomavirus E6 regulate p53 function. In these tumors, p53 and Rb are not mutated and their deregulation reflects the activity of the viral oncoproteins. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus also binds both p53 and Rb (74). In EBV-associated tumors Rb and p53 are not mutated, suggesting that EBV deregulates both the Rb and p53 pathways (15, 70). LMP1 specifically inhibits p53-mediated apoptosis through induction of the expression of A20 (22). NF-κB activity induced by LMP1 also blocks apoptosis, and inhibition of NF-κB activity induces apoptosis of EBV-transformed lymphocyte cell lines (7, 8). In a recent study inhibition of MDM2-dependent p53 degradation induced apoptosis of EBV-infected cells that was enhanced by NF-κB inhibition (19). Another EBV protein, EBNA3C, has recently been reported to bind and regulate both p53 and MDM2 (63, 82). The current study suggests that LMP1 regulates the Rb pathway through effects on p27 to stimulate cell cycle progression. Regulation of the Rb and p53 tumor suppressor pathways by LMP1 and/or EBNA3C likely contributes to EBV-induced transformation and to the development of EBV-associated malignancies.
Acknowledgments
This work was supported by NIH grants CA103634, CA32979, and CA19014 to N.R.-T. This work was supported by the H. M. Bligh Cancer Research Laboratories and grant 08-35 of the American Cancer Society, Illinois Division, Inc., to D.N.E.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Abukhdeir, A. M., and B. H. Park. 2008. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev. Mol. Med. 10:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleem, E., and P. Kaldis. 2006. Mouse models of cell cycle regulators: new paradigms. Results Probl. Cell Differ. 42:271-328. [DOI] [PubMed] [Google Scholar]
- 3.Ardila-Osorio, H., C. Pioche-Durieu, F. Puvion-Dutilleul, B. Clausse, J. Wiels, W. Miller, N. Raab-Traub, and P. Busson. 2005. TRAF interactions with raft-like buoyant complexes, better than TRAF rates of degradation, differentiate signaling by CD40 and EBV latent membrane protein 1. Int. J. Cancer 113:267-275. [DOI] [PubMed] [Google Scholar]
- 4.Auerkari, E. I. 2006. Methylation of tumor suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in carcinogenesis. Oral Oncol. 42:5-13. [DOI] [PubMed] [Google Scholar]
- 5.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 6.Baldassarre, G., B. Belletti, P. Bruni, A. Boccia, F. Trapasso, F. Pentimalli, M. V. Barone, G. Chiappetta, M. T. Vento, S. Spiezia, A. Fusco, and G. Viglietto. 1999. Overexpressed cyclin D3 contributes to retaining the growth inhibitor p27 in the cytoplasm of thyroid tumor cells. J. Clin. Investig. 104:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahir-McFarland, E. D., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff. 2000. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 97:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandramohan, V., S. Jeay, S. Pianetti, and G. E. Sonenshein. 2004. Reciprocal control of Forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27Kip1 levels. J. Immunol. 172:5522-5527. [DOI] [PubMed] [Google Scholar]
- 10.Ciaparrone, M., H. Yamamoto, Y. Yao, A. Sgambato, G. Cattoretti, N. Tomita, T. Monden, H. Rotterdam, and I. B. Weinstein. 1998. Localization and expression of p27KIP1 in multistage colorectal carcinogenesis. Cancer Res. 58:114-122. [PubMed] [Google Scholar]
- 11.Dawson, C. W., A. G. Eliopoulos, S. M. Blake, R. Barker, and L. S. Young. 2000. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 272:204-217. [DOI] [PubMed] [Google Scholar]
- 12.Dawson, C. W., L. Laverick, M. A. Morris, G. Tramoutanis, and L. S. Young. 2008. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J. Virol. 82:3654-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devergne, O., E. D. Cahir McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, R. H., and N. Raab-Traub. 1994. Alterations of the p53 gene in Epstein-Barr virus-associated immunodeficiency-related lymphomas. J. Virol. 68:1309-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731-1742. [DOI] [PubMed] [Google Scholar]
- 17.Everly, D. N., Jr., B. A. Mainou, and N. Raab-Traub. 2008. The ID proteins contribute to the growth of rodent fibroblasts during LMP1-mediated transformation. Virology 376:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everly, D. N., Jr., B. A. Mainou, and N. Raab-Traub. 2004. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J. Virol. 78:13470-13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forte, E., and M. A. Luftig. 2009. MDM2-dependent inhibition of p53 is required for Epstein-Barr virus B-cell growth transformation and infected-cell survival. J. Virol. 83:2491-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franken, M., O. Devergne, M. Rosenzweig, B. Annis, E. Kieff, and F. Wang. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 70:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedman, J. A., J. T. Chang, L. Jakoi, and J. R. Nevins. 2009. A combinatorial mechanism for determining the specificity of E2F activation and repression. Oncogene 28:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 70:8653-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita, N., S. Sato, K. Katayama, and T. Tsuruo. 2002. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J. Biol. Chem. 277:28706-28713. [DOI] [PubMed] [Google Scholar]
- 24.Gilchrist, M., V. Thorsson, B. Li, A. G. Rust, M. Korb, J. C. Roach, K. Kennedy, T. Hai, H. Bolouri, and A. Aderem. 2006. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441:173-178. [DOI] [PubMed] [Google Scholar]
- 25.Guo, Y., G. N. Sklar, A. Borkowski, and N. Kyprianou. 1997. Loss of the cyclin-dependent kinase inhibitor p27(Kip1) protein in human prostate cancer correlates with tumor grade. Clin. Cancer Res. 3:2269-2274. [PubMed] [Google Scholar]
- 26.Hallstrom, T. C., S. Mori, and J. R. Nevins. 2008. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 13:11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi, M., K. M. Izumi, and E. Kieff. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc. Natl. Acad. Sci. USA 98:4675-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwanaga, R., H. Komori, S. Ishida, N. Okamura, K. Nakayama, K. I. Nakayama, and K. Ohtani. 2006. Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene 25:1786-1798. [DOI] [PubMed] [Google Scholar]
- 30.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 94:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In B. N. Fields, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, S. E. Straus, and D. M. Knipe (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 34.Kieff, E. D., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. II. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 35.Knight, J. S., and E. S. Robertson. 2004. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J. Virol. 78:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. USA 102:18562-18566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. SCFSkp2 complex targeted by Epstein-Barr virus essential nuclear antigen. Mol. Cell. Biol. 25:1749-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kossatz, U., J. Vervoorts, I. Nickeleit, H. A. Sundberg, J. S. Arthur, M. P. Manns, and N. P. Malek. 2006. C-terminal phosphorylation controls the stability and function of p27kip1. EMBO J. 25:5159-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kung, C. P., and N. Raab-Traub. 2008. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 82:5486-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusano, S., and N. Raab-Traub. 2002. I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Mol. Cell. Biol. 22:6393-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine, A. J. 2009. The common mechanisms of transformation by the small DNA tumor viruses: the inactivation of tumor suppressor gene products: p53. Virology 384:285-293. [DOI] [PubMed] [Google Scholar]
- 42.Liang, J., J. Zubovitz, T. Petrocelli, R. Kotchetkov, M. K. Connor, K. Han, J. H. Lee, S. Ciarallo, C. Catzavelos, R. Beniston, E. Franssen, and J. M. Slingerland. 2002. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med. 8:1153-1160. [DOI] [PubMed] [Google Scholar]
- 43.Mainou, B. A., D. N. Everly, Jr., and N. Raab-Traub. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917-6924. [DOI] [PubMed] [Google Scholar]
- 44.Mainou, B. A., D. N. Everly, Jr., and N. Raab-Traub. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J. Virol. 81:9680-9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mainou, B. A., and N. Raab-Traub. 2006. LMP1 strain variants: biological and molecular properties. J. Virol. 80:6458-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malek, N. P., H. Sundberg, S. McGrew, K. Nakayama, T. R. Kyriakides, and J. M. Roberts. 2001. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature 413:323-327. [DOI] [PubMed] [Google Scholar]
- 47.Marinescu, V. D., I. S. Kohane, and A. Riva. 2005. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 33:D91-D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marinescu, V. D., I. S. Kohane, and A. Riva. 2005. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics 6:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masciullo, V., A. Sgambato, C. Pacilio, B. Pucci, G. Ferrandina, J. Palazzo, A. Carbone, A. Cittadini, S. Mancuso, G. Scambia, and A. Giordano. 1999. Frequent loss of expression of the cyclin-dependent kinase inhibitor p27 in epithelial ovarian cancer. Cancer Res. 59:3790-3794. [PubMed] [Google Scholar]
- 50.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 51.Miller, W. E., H. S. Earp, and N. Raab-Traub. 1995. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J. Virol. 69:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, W. E., G. Mosialos, E. Kieff, and N. Raab-Traub. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J. Virol. 71:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moorthy, R. K., and D. A. Thorley-Lawson. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of Rat-1 fibroblasts. J. Virol. 67:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura, N., S. Ramaswamy, F. Vazquez, S. Signoretti, M. Loda, and W. R. Sellers. 2000. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 20:8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohtani, N., Z. Zebedee, T. J. Huot, J. A. Stinson, M. Sugimoto, Y. Ohashi, A. D. Sharrocks, G. Peters, and E. Hara. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409:1067-1070. [DOI] [PubMed] [Google Scholar]
- 56.Paine, E., R. I. Scheinman, A. S. Baldwin, Jr., and N. Raab-Traub. 1995. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-κ B/Rel family proteins. J. Virol. 69:4572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541-2549. [PubMed] [Google Scholar]
- 58.Pioche-Durieu, C., C. Keryer, S. Souquere, J. Bosq, W. Faigle, D. Loew, M. Hirashima, N. Nishi, J. Middeldorp, and P. Busson. 2005. In nasopharyngeal carcinoma cells, Epstein-Barr virus LMP1 interacts with galectin 9 in membrane raft elements resistant to simvastatin. J. Virol. 79:13326-13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prabhu, S., A. Ignatova, S. T. Park, and X. H. Sun. 1997. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol. Cell. Biol. 17:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raab-Traub, N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431-441. [DOI] [PubMed] [Google Scholar]
- 61.Roberts, M. L., and N. R. Cooper. 1998. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 240:93-99. [DOI] [PubMed] [Google Scholar]
- 62.Rothenberger, S., K. Burns, M. Rousseaux, J. Tschopp, and C. Bron. 2003. Ubiquitination of the Epstein-Barr virus-encoded latent membrane protein 1 depends on the integrity of the TRAF binding site. Oncogene 22:5614-5618. [DOI] [PubMed] [Google Scholar]
- 63.Saha, A., M. Murakami, P. Kumar, B. Bajaj, K. Sims, and E. S. Robertson. 2009. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J. Virol. 83:4652-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shair, K. H., K. M. Bendt, R. H. Edwards, E. C. Bedford, J. N. Nielsen, and N. Raab-Traub. 2007. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog. 3:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shair, K. H., C. I. Schnegg, and N. Raab-Traub. 2008. EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res. 68:6997-7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin, I., F. M. Yakes, F. Rojo, N. Y. Shin, A. V. Bakin, J. Baselga, and C. L. Arteaga. 2002. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat. Med. 8:1145-1152. [DOI] [PubMed] [Google Scholar]
- 68.Shore, A. M., P. C. White, R. C. Hui, A. Essafi, E. W. Lam, M. Rowe, and P. Brennan. 2006. Epstein-Barr virus represses the FoxO1 transcription factor through latent membrane protein 1 and latent membrane protein 2A. J. Virol. 80:11191-11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh, S. P., J. Lipman, H. Goldman, F. H. Ellis, Jr., L. Aizenman, M. G. Cangi, S. Signoretti, D. S. Chiaur, M. Pagano, and M. Loda. 1998. Loss or altered subcellular localization of p27 in Barrett's associated adenocarcinoma. Cancer Res. 58:1730-1735. [PubMed] [Google Scholar]
- 70.Sun, Y., G. Hegamyer, and N. H. Colburn. 1993. Nasopharyngeal carcinoma shows no detectable retinoblastoma susceptibility gene alterations. Oncogene 8:791-795. [PubMed] [Google Scholar]
- 71.Thornburg, N. J., W. Kulwichit, R. H. Edwards, K. H. Shair, K. M. Bendt, and N. Raab-Traub. 2006. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene 25:288-297. [DOI] [PubMed] [Google Scholar]
- 72.Thornburg, N. J., and N. Raab-Traub. 2007. Induction of epidermal growth factor receptor expression by Epstein-Barr virus latent membrane protein 1 C-terminal-activating region 1 is mediated by NF-κB p50 homodimer/Bcl-3 complexes. J. Virol. 81:12954-12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsao, S. W., G. Tramoutanis, C. W. Dawson, A. K. Lo, and D. P. Huang. 2002. The significance of LMP1 expression in nasopharyngeal carcinoma. Semin. Cancer Biol. 12:473-487. [DOI] [PubMed] [Google Scholar]
- 74.Verma, S. C., K. Lan, and E. Robertson. 2007. Structure and function of latency-associated nuclear antigen. Curr. Top. Microbiol. Immunol. 312:101-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viglietto, G., M. L. Motti, P. Bruni, R. M. Melillo, A. D'Alessio, D. Califano, F. Vinci, G. Chiappetta, P. Tsichlis, A. Bellacosa, A. Fusco, and M. Santoro. 2002. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 8:1136-1144. [DOI] [PubMed] [Google Scholar]
- 76.Viglietto, G., M. L. Motti, and A. Fusco. 2002. Understanding p27(kip1) deregulation in cancer: down-regulation or mislocalization. Cell Cycle 1:394-400. [DOI] [PubMed] [Google Scholar]
- 77.Wang, C., X. Hou, S. Mohapatra, Y. Ma, W. D. Cress, W. J. Pledger, and J. Chen. 2005. Activation of p27Kip1 expression by E2F1. A negative feedback mechanism. J. Biol. Chem. 280:12339-12343. [DOI] [PubMed] [Google Scholar]
- 78.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 79.Williamson, E. A., F. Dadmanesh, and H. P. Koeffler. 2002. BRCA1 transactivates the cyclin-dependent kinase inhibitor p27(Kip1). Oncogene 21:3199-3206. [DOI] [PubMed] [Google Scholar]
- 80.Williamson, E. A., I. Wolf, J. O'Kelly, S. Bose, S. Tanosaki, and H. P. Koeffler. 2006. BRCA1 and FOXA1 proteins coregulate the expression of the cell cycle-dependent kinase inhibitor p27(Kip1). Oncogene 25:1391-1399. [DOI] [PubMed] [Google Scholar]
- 81.Yasui, T., M. Luftig, V. Soni, and E. Kieff. 2004. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc. Natl. Acad. Sci. USA 101:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yi, F., A. Saha, M. Murakami, P. Kumar, J. S. Knight, Q. Cai, T. Choudhuri, and E. S. Robertson. 2009. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 388:236-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 84.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 85.Zancai, P., R. Cariati, S. Rizzo, M. Boiocchi, and R. Dolcetti. 1998. Retinoic acid-mediated growth arrest of EBV-immortalized B lymphocytes is associated with multiple changes in G1 regulatory proteins: p27Kip1 up-regulation is a relevant early event. Oncogene 17:1827-1836. [DOI] [PubMed] [Google Scholar]
- 86.Zancai, P., J. Dal Col, S. Piccinin, M. Guidoboni, R. Cariati, S. Rizzo, M. Boiocchi, R. Maestro, and R. Dolcetti. 2005. Retinoic acid stabilizes p27Kip1 in EBV-immortalized lymphoblastoid B cell lines through enhanced proteasome-dependent degradation of the p45Skp2 and Cks1 proteins. Oncogene 24:2483-2494. [DOI] [PubMed] [Google Scholar]
- 87.Zheng, W., H. Wang, L. Xue, Z. Zhang, and T. Tong. 2004. Regulation of cellular senescence and p16(INK4a) expression by Id1 and E47 proteins in human diploid fibroblast. J. Biol. Chem. 279:31524-31532. [DOI] [PubMed] [Google Scholar]