Abstract
Reversible topoisomerase I (Top1)-DNA cleavage complexes are the key DNA lesion induced by anticancer camptothecins (CPTs) (e.g. topotecan and irinotecan) as well as structurally perturbed DNAs (e.g. oxidatively damaged, UV-irradiated, or alkylated DNA). It has been proposed that Top1 cleavage complexes arrest advancing replication forks, triggering the formation of DNA double strand breaks (DSBs) because of replication fork runoff at the Top1 cleavage complex sites on the leading strand. In this study, we show that the formation of replication-dependent DSBs requires the ubiquitin-proteasome pathway in CPT-treated cells. First, the proteasome inhibitor MG-132 specifically inhibited CPT-induced but not ionizing radiation- or hydroxyurea-induced DSBs as revealed by both the neutral comet assay and measurements of the specific DNA damage signals (e.g. γ-H2AX, phosphorylated ataxia telangiectasia mutated (Ser-1981), and phosphorylated Chk2 (Ser-33/35)) that are characteristic for DSBs. Knocking down the 20 S proteasome maturation protein also supported the requirement of the proteasome activity for CPT-induced DSBs. Second, CPT-induced DSB signals were shown to require ubiquitin, ubiquitin-activating enzyme (E1), a CUL-3-based ubiquitin ligase (E3), and the formation of Lys-48-linked polyubiquitin chains on Top1. Third, immunocytochemical studies revealed that the CPT-induced formation of γ-H2AX foci occurred at the replication forks and was attenuated by co-treatment with the proteasome inhibitor MG-132. In the aggregate, these results support a replication fork collision model in which Top1 cleavage complexes at the arrested replication forks are degraded by proteasome prior to replication fork runoff on the leading strand to generate DSBs.
Eukaryotic DNA topoisomerase I (Top1)3 catalyzes the breakage/reunion of DNA by transiently nicking one strand of the DNA duplex through the formation of a reversible Top1-DNA covalent complex (reviewed in Refs. 1–4). Anticancer camptothecins (CPTs) as well as various structurally perturbed DNAs (e.g. UV adducts, abasic sites, base mismatches, uracil incorporation, nicks and gaps, and oxidized DNA lesions) are known to stabilize reversible Top1-DNA covalent complexes, often referred to as Top1 cleavage complexes (reviewed in Refs. 3, 5). Top1 cleavage complexes, in addition to their rapid reversibility, are also characterized by Top1-concealed single strand breaks (SSBs) (6), as their denaturation by SDS or alkali in vitro exposes Top1-linked SSBs in which Top1 molecules are covalently linked to the 3′ phosphoryl ends of SSBs through their active-site tyrosine (6, 7). It is well established that Top1 cleavage complexes are responsible for the antitumor activity of CPTs (3, 5).
The unique nature of Top1 cleavage complexes has led to the proposal that their interactions with cellular machineries (e.g. DNA replication and transcription processes) are needed for the exposure of the Top1-concealed strand breaks in cells (8–11). Consistent with this notion, a robust transcription- and proteasome-dependent mechanism for the intracellular processing of Top1 cleavage complexes into SSBs has been demonstrated (8). It has been suggested that Top1 cleavage complexes arrest elongating RNA polymerases, triggering proteasomal degradation of Top1 (termed Top1 down-regulation) and the concomitant exposure of the Top1-concealed SSBs for repair (8).
The role of active DNA replication in the processing of Top1 cleavage complexes into DNA damage is initially suggested based on the observation that CPTs are exquisitely cytotoxic to S phase cells and simultaneous/transient arrest of DNA synthesis abolishes CPT cytotoxicity (12, 13). Analysis of the CPT-induced aberrant replication intermediates in the SV40 cell-free replication system has led to the suggestion of a replication fork collision model (9, 11). Results from replication runoff studies in HT29 cells and genetic studies in yeast (10, 14, 15) also support such a conclusion. In this model, the reversible Top1 cleavage complexes arrest the advancing replication fork, resulting in the formation of DNA double strand breaks (DSBs) and Top1-DNA cross-links at the collision sites (9, 11, 16). Consistent with this proposed model, many DNA damage signals, such as phosphorylated RPA2, ATM, Chk1, Chk2, p53, NF-κB, and H2AX (γ-H2AX), are detected upon CPT treatment and most of them are replication-dependent (5).
Implicit in the replication fork collision model is that the formation of DSBs at the replication forks occurs as a consequence of replication fork runoff on the leading strand of DNA synthesis (10). Consequently, the processing of Top1 cleavage complexes is not predicted to be necessary for DSB formation as it is for transcription-dependent SSB formation. In this study, we provide evidence showing that the formation of DSBs at the arrested replication forks is dependent on the ubiquitin-proteasome pathway, which involves a CUL-3-based E3 ligase and the formation of Lys-48-linked polyubiquitin chains. These results are most consistent with a modified replication fork collision model in which Top1 cleavage complexes at the arrested replication forks are degraded by proteasome prior to the replication runoff on the leading strand to generate DSBs.
EXPERIMENTAL PROCEDURES
Antibodies, Drugs, and siRNAs
Antibodies against Mrell (Upstate), PRA2 (Oncogene), γ-H2AX (Upstate), Ser(P)-15-p53 (Cell Signaling), Ser(P)-33/35-Chk2 (Cell Signaling), Chk1 (Santa Cruz Biotechnology), and Ser(P)-345-Chk1 (Cell Signaling) were purchased from commercial sources. Anti-Top1 antibodies were obtained from sera of scleroderma 70 patients as described before (17). The hybridoma cell line that produces monoclonal antibody 12G10 (α-tubulin) was obtained from Developmental Studies Hybridoma Bank. The siRNAs targeting POMP (ID SAS1-Hso1-00010430) and CUL-3 (ID SAS1-Hso1-00042272) were purchased from Sigma. Ubiquitin isopeptidase inhibitor I (G5) was purchased from Calbiochem.
Cell Culture
HeLa Tet-On (Clontech) and HT29 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FetalPlex animal serum complex (Gemini Bio-Products, West Sacramento, CA), l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (100 μg/ml) in a 37 °C incubator with 5% CO2. ZR-75-1 stable cell lines that express control, ISG15, or UbcH8 shRNAs (18) were cultured in RPMI medium supplemented with 10% FetalPlex animal serum complex, l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (100 μg/ml) in a 37 °C incubator with 5% CO2. Mouse leukemic cell lines Fm3A and Ts85 (ubiquitin E1 Ts mutant) were cultured in RPMI medium supplemented with 10% FetalPlex animal serum complex, l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (100 μg/ml) in a 30 °C incubator with 5% CO2.
Neutral Comet Assay
The comet assays were performed according to the Trevigen CometAssayTM kit protocol with slight modifications. Cells were pretreated with various inhibitors for 30 min, followed by co-treatment with 25 μm CPT for 1 h. Treated cells were trypsinized with 0.005% trypsin (50× lower than the normal concentration) at 37 °C for 5 min. An equal amount of drug-free medium (with 10% serum) was then added to quench the trypsin activity. The final cell density was about 10,000 cells/ml. 50 μl of the cell suspension was then mixed with 500 μl of 0.5% low melting point agarose (in PBS) (Invitrogen) at 37 °C. 50 μl of the cell/agarose mixture was transferred onto glass slides. Slides were then immersed in pre-chilled lysis buffer (2.5 m NaCl, 100 mm EDTA, 10 mm Tris, pH 10.0, 1% Triton X-100, and 10% DMSO) for 1 h, followed by electrophoresis in 1× TBE buffer at 0.8 V/cm for 8–10 min at room temperature. After electrophoresis, slides were dehydrated in 70% alcohol for 30 min and air-dried overnight. Cells were then rehydrated in H2O and stained with 0.2 μg/ml ethidium bromide for 1 h. Images were visualized under a fluorescence microscope and captured with a CCD camera. The comet tail moment was determined as described (19). The means ± S.E. were obtained from at least 100 cells for each treatment group. Statistical analysis was performed using two-tailed unpaired Student's t test.
Transfection of cDNAs and siRNAs
For overexpression of dominant-negative ubiquitin (K48R-Ub) or HA-tagged ubiquitin (HA-Ub), cells were transfected with the indicated plasmids (obtained from Dr. Cam Patterson, University of North Carolina, Chapel Hill) 24 h prior to CPT treatment using PolyFect transfection reagent (Qiagen). For transfection of siRNAs, 250 pmol of siRNAs were transfected into cells (50% confluent on a 60-mm plate) using Oligofectamine transfection reagent (Invitrogen). After 48 h, transfected cells were split for further experiments.
Top1 Down-regulation Assay
CPT-induced Top1 degradation was monitored by immunoblotting of the alkaline lysates as described previously (8) with slight modification. Briefly, for each 35-mm dish, 100 μl of an alkaline lysis buffer (200 mm NaOH, 2 mm EDTA) was added, and cells were then scraped with a rubber policeman. Alkaline lysates were neutralized by addition of 16 μl of 1 m HCl, 600 mm Tris, pH 8.0, followed by mixing with 13 μl of 10× S7 nuclease buffer (50 mm MgCl2, 50 mm CaCl2, 5 mm dithiothreitol, 1 mm EDTA, and a protease inhibitor mixture) and 60 units of staphylococcal S7 nuclease (20). The digestion was performed on ice for 20 min for releasing Top1 from covalent Top1-DNA complexes. After nuclease digestion, 100 μl of 6× SDS gel sample buffer was added, followed by boiling for 10 min. The samples were then analyzed by SDS-gel electrophoresis and immunoblotted with anti-hTop1 antibody.
Immunoprecipitation
Immunoprecipitation was performed as described previously (21). Briefly, cells were lysed with 400 μl of an alkaline lysis buffer, followed by neutralization with 75 μl of a neutralization buffer (described in Top1 down-regulation assay) with brief sonication. 60 μl of 10× S7 buffer was then added to the cell lysates, followed by S7 nuclease treatment (180 units). Top1 were immunoprecipitated by Top1 antiserum and protein l-conjugated agarose beads.
Immunocytochemistry
Immunostaining of BrdUrd and γ-H2AX was performed according to the previous study (22) with slight modification. Briefly, after 45 min of pulse-labeling with 50 μm BrdUrd in the presence of 1 μm CPT for the last 30 min, cells were fixed in 4% paraformaldehyde for 10 min, followed by a PBS wash, and incubated with methanol for 15 min at −20 °C. Cells were blocked with 5% bovine serum albumin in PBS for 1 h, followed by incubation with mouse anti-γ-H2AX monoclonal antibody in PBS containing 1% bovine serum albumin for 2 h at room temperature. After two PBS washes, cells were incubated with Cy2-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch), followed by three PBS washes and 4% formaldehyde fixation for 10 min. Cells were incubated with 1.5 n HCl for 10 min at 37 °C to denature the DNA. After two PBS washes, cells were blocked with 5% bovine serum albumin in PBS for 30 min, followed by staining with Alexa Fluor 488 mouse anti-BrdUrd antibody (Pharmingen) in PBS for 2 h. After three PBS washes, cells were stained with 4′,6-diamidino-2-phenylindole, mounted, and visualized under the fluorescence microscope, and images were captured with the CCD camera.
RESULTS
CPT-induced DNA DSBs Require Proteasome Activity
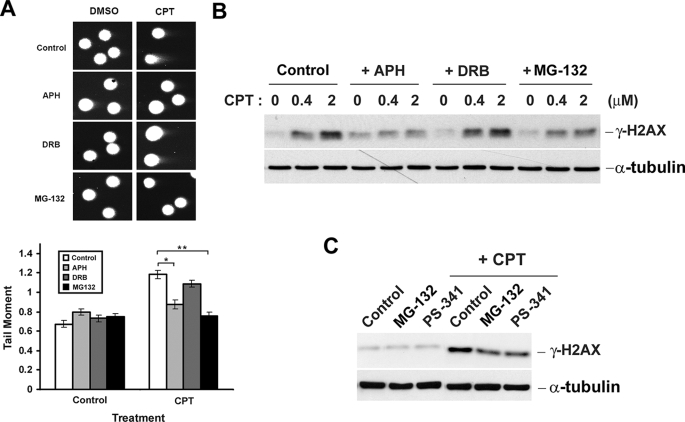
Previous studies have demonstrated that CPT-induced DNA SSBs are transcription-dependent and require proteasome activity (23). To test whether CPT-induced DSBs also require proteasome activity, HeLa cells were treated with CPT in the presence and absence of the proteasome inhibitor, MG-132, and the formation of DSBs was monitored by the neutral comet assay. As shown in Fig. 1A, CPT (1 h of treatment) induced a substantial increase (from 0.7 to 1.2) of the tail moment, indicative of the formation of DNA DSBs. Consistent with previous studies (9, 11, 22), CPT-induced DSBs were largely abolished by co-treatment with the DNA polymerase inhibitor aphidicolin (APH) but not the transcription inhibitor DRB, suggesting that CPT-induced DSBs are exclusively replication-dependent. Interestingly, co-treatment with the proteasome inhibitor MG-132 also largely abolished CPT-induced DSBs in HeLa cells, suggesting a requirement of proteasome activity for CPT-induced DSBs. The involvement of proteasome activity in the formation of DSBs in CPT-treated HeLa cells was also supported by measuring CPT-induced γ-H2AX, a marker for DNA DSBs (24). As in the case of neutral comet assay, CPT-induced (1 h of treatment) γ-H2AX was diminished by co-treatment with the replication inhibitor APH (Fig. 1B) or the proteasome inhibitors MG-132 (Fig. 1B) and PS-341 (bortezomib, a therapeutic proteasome inhibitor) (Fig. 1C), but not the transcription inhibitor DRB (Fig. 1B), suggesting the involvement of both DNA replication and proteasome activity, but not transcription, in the generation of CPT-induced DSBs.
FIGURE 1.
CPT-induced DNA DSBs are diminished by co-treatment with aphidicolin or MG-132. A, HeLa cells were pretreated with APH (5 μm), DRB (100 μm), or MG-132 (6 μm) for 30 min, followed by co-treatment with 5 μm CPT for 1 h. DSBs in samples were analyzed by neutral comet assay as described under “Experimental Procedures.” Upper panel, representative images. Lower panel, histogram of the tail moment plotted against each treatment condition. p values for comparisons (marked * and **) were <0.005 as determined by two-tailed Student's t test. B, HeLa cells were treated with CPT in the presence and absence of metabolic inhibitors as described in A except that cells were treated with increasing concentrations of CPT and cell lysates were immunoblotted with either anti-γ-H2AX (for detecting DSB formation) or anti-α-tubulin antibody (for assessing equal loading). C, HeLa cells were treated with CPT (1 μm) in the presence and absence of MG-132 (6 μm) or PS-341 (1 μm), and cell lysates were immunoblotted as described in B.
CPT-induced DNA Damage Signals Require Proteasome Activity
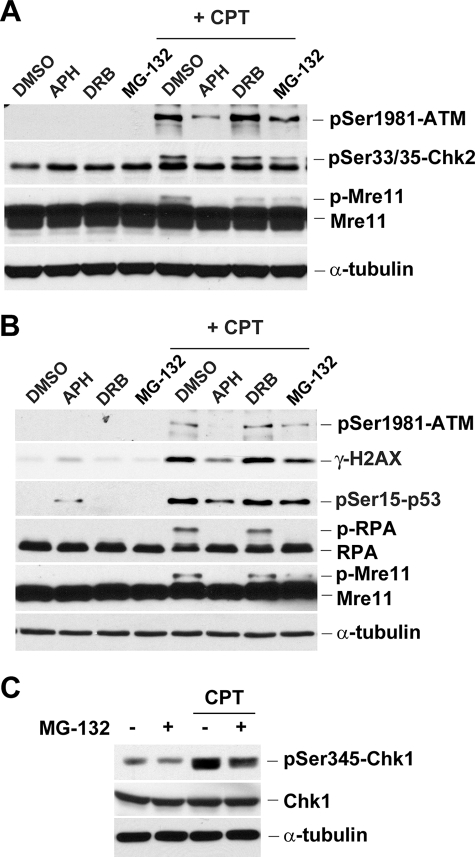
The involvement of proteasome activity in CPT-induced DNA damage was further demonstrated by measuring various DNA damage signals. As shown in Fig. 2A, CPT was shown to induce ATM autophosphorylation (phosphorylation of Ser-1981), Chk2 phosphorylation (phosphorylation of Ser-33/35), and Mre11 phosphorylation in HeLa cells. All these phosphorylations were abolished or reduced by co-treatment with APH and MG-132 but not DRB (Fig. 2A). CPT was also shown to induce Chk1 phosphorylation (Ser-345) in HeLa cells in a proteasome-dependent manner (Fig. 2C). Similar experiments were also performed in HT29 colorectal cancer cells. As shown in Fig. 2B, CPT was shown to induce phosphorylation of ATM (Ser-1981), H2AX (Ser-139), p53 (Ser-15), RPA2 (detected by band shift), and Mre11 (detected by band shift) in HT29 cells. Consistent with previous studies, the phosphorylation of RPA2 and Mre11 appears to be replication-dependent because those band shifts (reflecting the phosphorylated forms of RPA2 and Mre11) induced by CPT were abolished by APH but not DRB (25, 26). Again, these phosphorylations were significantly diminished by co-treatment with MG-132, suggesting the involvement of proteasome in replication-dependent DNA damages induced by CPT.
FIGURE 2.
Multiple DNA damage signals induced by CPT are diminished by aphidicolin and MG-132. HeLa (A) or HT29 (B) cells were pretreated with APH (5 μm), DRB (100 μm), or MG-132 (6 μm) for 30 min, followed by co-treatment with CPT (1 μm) for 1 h. C, HeLa cells were pretreated with MG-132 for 30 min, followed by co-treatment with CPT (1 μm) for 1 h. Cell lysates were immunoblotted with various antibodies as indicated.
Proteasome Activity Is Not Required for DNA Damage Induced by Ionizing Radiation (IR) or Hydroxyurea (HU)
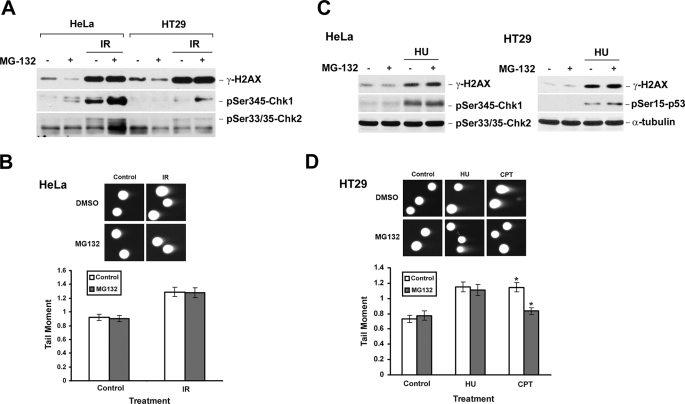
The requirement of proteasome activity appears to be specific for CPT-induced DNA damage. As shown in Fig. 3A, IR-induced DNA damage signals (i.e. γ-H2AX and phosphorylated Chk1 and Chk2), unlike CPT-induced DNA damage signals, were not reduced by MG-132 co-treatment in either HeLa or HT29 cells, suggesting the requirement of proteasome activity is specific for CPT-induced DNA damage. In addition, the amount of IR-induced DNA DSBs, as measured by neutral comet assay, were essentially the same in the presence and absence of MG-132 (Fig. 3B), suggesting that the requirement of proteasome activity for DSB formation is specific for CPT but not IR. Similar experiments were repeated in HeLa and HT29 cells using HU, which is known to arrest the replication forks through depletion of the nucleotide pool. As shown in Fig. 3, C and D, HU-induced DNA damage signals (i.e. γ-H2AX and Ser-15-phosphorylated p53) and DNA DSBs (monitored by neutral comet assay) were unaffected by MG-132. Together, these results suggest that proteasome activity is specifically required for CPT-induced but not IR- or HU-induced DNA damage. It should be noted that Chk1 phosphorylation actually increased, rather than decreased, in IR-irradiated HeLa cells in the presence of MG-132. This phenomenon is consistent with the recent finding that phosphorylated Chk1, which is induced upon DNA damage, is degraded by proteasome (27).
FIGURE 3.
DNA damage signals induced by IR and HU are unaffected by MG-132 co-treatment. A, HeLa or HT29 cells were pretreated with 6 μm MG-132 for 30 min, immediately followed by γ-irradiation at a dose of 10 gray. After 1 h, cells were lysed, and lysates were immunoblotted with antibodies as indicated. B, HeLa cells were pretreated with MG-132 (6 μm) and irradiated at a dose of 20 gray. After 1 h, the amount of DSBs was determined by neutral comet assay as described under “Experimental Procedures.” Upper panel, representative comet images. Lower panel, histogram of the tail moment. C, HeLa or HT29 cells were pretreated with MG-132 (6 μm) for 30 min, followed by co-treatment with HU (2.5 mm) for 1 h. Cell lysates were immunoblotted with specific antibodies as indicated. D, HT29 cells were pretreated with MG-132 (6 μm) for 30 min, followed by co-treatment with HU (10 mm) or CPT (5 μm) for 1 h. The formation of DSBs was monitored by neutral comet assay. The p values for comparison were determined by two-tailed Student's t test. The p value for CPT treatments was less than 0.005 (marked *).
CPT-induced DNA Damage Signals Require the Ubiquitin-Proteasome Pathway; Involvement of Ubiquitin and Ubiquitin E1
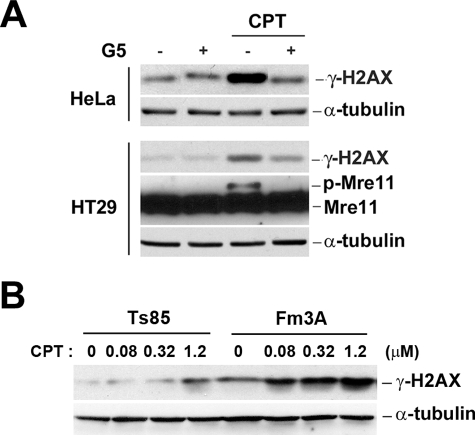
Although the ubiquitin-proteasome pathway is well characterized for the degradation of many proteins, an increasing number of proteins are known to be degraded by proteasome in a ubiquitin-independent manner (reviewed in Ref. 28). To test whether CPT-induced DSBs require all components of the ubiquitin-proteasome pathway, the role of ubiquitin in CPT-induced DNA damage was determined by using a ubiquitin isopeptidase inhibitor I, G5, which is effective in depleting the ubiquitin pool (23, 29). As shown in Fig. 4A (upper panel), CPT-induced DNA γ-H2AX was diminished in HeLa cells pretreated with G5 (1 h pretreatment followed by 1 h co-treatment with CPT, a condition known to deplete the free ubiquitin pool and inhibit CPT-induced Top1 degradation (23)). Similarly, CPT-induced γ-H2AX and phosphorylated Mre11 were diminished in HT29 cells treated with G5 (Fig. 4A, lower panel). These results suggest that CPT-induced DNA damage may involve a ubiquitin-dependent pathway.
FIGURE 4.
CPT-induced DNA damage signals are dependent on ubiquitin and ubiquitin E1. A, HeLa and HT29 cells were pretreated with the ubiquitin isopeptidase inhibitor I, G5 (5 μm), for 1 h to deplete the ubiquitin pool, followed by co-treatment with 1 μm CPT for 1 h. Cell lysates were immunoblotted with specific antibodies as indicated. B, mouse Fm3A (wild type) and Ts85 (E1 ts mutant) cells were incubated at nonpermissive temperature (39 °C) for 30 min, followed by CPT treatment at the same temperature for 1 h. Cell lysates were immunoblotted with specific antibodies as indicated.
To further investigation the involvement of ubiquitin, the role of ubiquitin E1 was also determined using the mouse leukemic Ts85 cell line that harbors a temperature-sensitive ubiquitin E1 and its wild type control cell line, Fm3A (30). As shown in Fig. 4B, CPT-induced γ-H2AX in Fm3A cells was significantly higher than in Ts85 cells at the nonpermissive temperature, 39 °C, suggesting the involvement of ubiquitin E1 in CPT-induced DNA damage.
CPT-induced DNA Damage Signals Require the Ubiquitin-Proteasome Pathway; Involvement of a CUL-3-based Ubiquitin E3 Ligase and the POMP
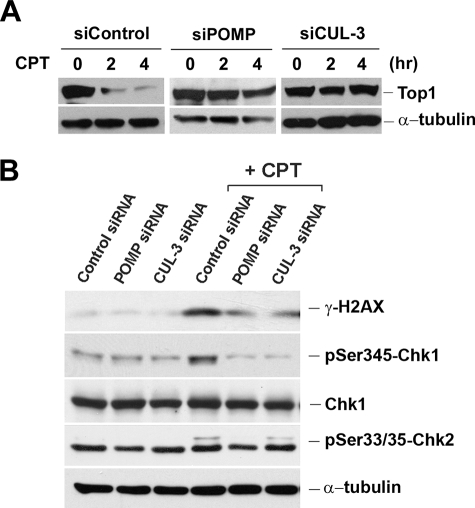
Previous studies have demonstrated that a cullin 3 (CUL-3)-based E3 ligase is involved in degradation of Top1 in CPT-treated cells (31). Although degradation of Top1 in CPT-treated cells is primarily due to a transcription-dependent process, it is possible that a CUL-3-based E3 ligase may also be involved in replication- and proteasome-dependent DNA damage induced by CPT. To test this possibility, CUL-3 was knocked down in HeLa cells by the CUL-3 siRNA. As shown in Fig. 5A, CPT induced rapid degradation of Top1 (Top1 down-regulation) in HeLa cells, a phenomenon that has been well characterized and shown to be transcription- and proteasome-dependent (8, 17, 21). CPT-induced Top1 down-regulation was diminished in HeLa cells treated with CUL-3 siRNA, consistent with the report that Top1 down-regulation requires the CUL-3-based E3 ligase (31). Interestingly, CPT-induced DNA damage signals (e.g. γ-H2AX and phosphorylated Chk2), which are known to be replication-dependent, were also much reduced in CUL-3 siRNA-treated HeLa cells (Fig. 5B), suggesting that replication-dependent DNA damage induced by CPT also requires a CUL-3-based E3 ligase.
FIGURE 5.
CPT-induced DNA damage signals require POMP and a CUL-3-based E3 ligase. A, HeLa cells were transfected with POMP or CUL-3 siRNA for 72 h. Cells were then treated with 25 μm CPT for 2 and 4 h, followed by alkaline hydrolysis and S7 treatment as described under “Experimental Procedures.” Top I levels were determined by immunoblotting. B, siRNA transfection conditions are the same as in A. 72 h post-transfection, HeLa cells were treated with 2 μm CPT for 1 h. Cell lysates were immunoblotted with various antibodies as indicated.
To further evaluate the involvement of proteasome in CPT-induced DNA damage, the proteasome maturation protein (POMP) was also knocked down by POMP siRNA in HeLa cells. POMP is known to facilitate major steps of 20 S proteasome assembly at the endoplasmic reticulum (32). As shown in Fig. 5A, CPT-induced Top1 down-regulation, which is transcription- and proteasome-dependent, was significantly reduced in HeLa cells treated with POMP siRNA, consistent with the proposed role of proteasome in CPT-induced Top1 down-regulation (8). Interestingly, CPT-induced DNA damage signals (e.g. γ-H2AX) (Fig. 5B), which are replication-dependent, were also much reduced in HeLa cells treated with POMP siRNA. This result suggests that the 20 S proteasome assembly is required for CPT-induced, replication-dependent, DNA DSBs, consistent with the notion that the proteasome activity is required for CPT-induced DNA DSBs.
CPT-induced DNA Damage Requires the Formation of Lys-48-linked PolyUb Chains
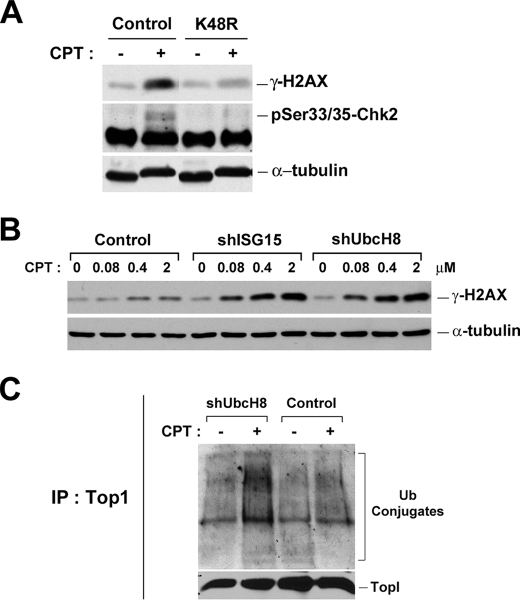
Degradation of proteins by the ubiquitin-proteasome pathway is known to require the formation of polyUb chains through the action of E1/E2/E3 and other factors (reviewed in Ref. 33). A number of lysine residues (e.g. Lys-29, Lys-63, and Lys-48) on ubiquitin are known to form polyUb chains through isopeptide bonds (reviewed in Ref.34), and Lys-48-linked polyUb chains are known to be the major polyUb chains associated with degradation by proteasome (33). In the previous study, we have demonstrated that the formation of Lys-48-, but not Lys-29- and Lys-63-, linked polyUb chains on Top1 is responsible for CPT-induced, transcription-dependent, DNA SSBs (23). To test whether Lys-48-linked polyUb chains are also involved in the CPT-induced, replication-dependent, DSBs, a plasmid expressing the dominant-negative mutant ubiquitin (K48R) was transiently transfected into HeLa cells. As shown in Fig. 6A, overexpression of K48R dominant-negative mutant ubiquitin substantially blocked CPT-induced DNA damage signals (i.e. γ-H2AX and phosphorylated Chk2) in HeLa cells, suggesting the involvement of the formation of Lys-48-linked polyUb chains in CPT-induced, replication-dependent, DNA damage.
FIGURE 6.
CPT-induced DNA damage signals require the formation of Lys-48-linked polyubiquitin chains. A, HeLa cells were transfected with plasmids overexpressing wild type (control) or dominant-negative (K48R) ubiquitin. After 24 h, cells were treated with 2 μm CPT for 1 h. Cell lysates were analyzed by immunoblotting with anti-γ-H2AX or anti-p-Chk2 antibody. B, ZR-75-1 cell lines stably expressing control, ISG15, or UbcH8 shRNA were treated with increasing concentrations of CPT for 1 h. Cell lysates were immunoblotted with specific antibodies as indicated. C, CPT-induced Top1-ubiquitin conjugates in ZR-75-1 cell lines. Cells were transfected with the plasmid encoding HA-Ub for 24 h before being treated with 25 μm CPT for 30 min. Cells were lysed with the alkaline buffer, followed by S7 digestion and immunoprecipitation (IP) with anti-Top1 antisera as described under “Experimental Procedures.”
CPT-induced DNA Damage Is Suppressed by ISG15
Previous studies have demonstrated that ISG15 (interferon-stimulated gene 15, a ubiquitin-like protein), which is elevated in many tumor cells, suppresses the ubiquitin-proteasome pathway by interfering with the formation of polyUb conjugates (35). For example, ZR-75-1 breast cancer cells are known to exhibit a much reduced level of polyUb conjugates due to the elevated ISG15 conjugation pathway (35). Consequently, it is possible to modulate the ubiquitination activity in cells by regulating the expression of components of the ISG15 conjugation pathway (e.g. ISG15 and its E2). To test whether the amounts of CPT-induced DSBs are changed in ZR-75-1 breast cells due to the up-regulation of the ISG15 conjugation pathway (and hence reduced ubiquitination activity), three stable clones of ZR-75-1 cells expressing control shRNA, ISG15 shRNA, and UbcH8 shRNA (the major E2 for ISG15) (18) were employed in this study. As shown in Fig. 6B, ZR-75-1 cells expressing ISG15 and UbcH8 shRNA (labeled shISG15 and shUbcH8, respectively) exhibited much stronger γ-H2AX induction upon CPT treatment compared with control cells expressing the control shRNA, suggesting that CPT-induced DSB signals are increased in cells with the up-regulated ubiquitination activity.
To determine whether the formation of Top1-Ub conjugates was indeed suppressed by the ISG15 conjugation pathway, ZR-75-1 cells expressing UbcH8 shRNA (transiently transfected with the HA-Ub plasmid) were treated with CPT, and the formation of Top1-Ub conjugates examined by Top1 immunoprecipitation was followed by Western blotting with anti-HA antibodies (Fig. 6C). Indeed, the amount of Top1-Ub conjugates was elevated in ZR-75-1 cells expressing UbcH8 shRNA (Fig. 6C, compare 2nd with 4th lanes), suggesting that the formation of Top1-Ub conjugates was suppressed by the ISG15 conjugation pathway. In the aggregate, these results further support the notion that CPT-induced DSBs require the ubiquitination activity. It is interesting to note that ISG15 has recently been demonstrated to be a tumor marker, which is variably expressed in different tumors, and its expression is correlated with disease progression and prognosis (18, 36, 37). Our results thus suggest that the amount of CPT-induced DSBs may vary in different tumor cells depending on the expression status of the ISG15 conjugation pathway.
CPT Induces Proteasome-dependent Formation of DSBs at the Replication Foci
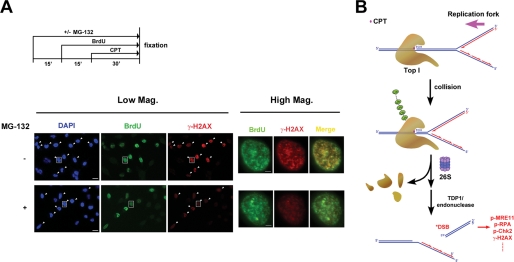
CPT-induced DSBs have been proposed to result from collisions between Top1-DNA covalent adducts and advancing replication forks (10, 11). Indeed, CPT-induced γ-H2AX foci have been shown to co-localize with replication foci in HT29 colorectal cancer cells (22). In this immunocytochemical study, the same population of cells that showed strong staining with BrdUrd (green) (S phase cells) also exhibited strong staining with γ-H2AX (red) (see arrow-pointed cells in Fig. 7A, Low Mag. panels), consistent with the notion that DSBs are generated exclusively in S phase cells upon CPT treatment (22). Interestingly, co-treatment of MG-132 lowered the level of γ-H2AX staining in BrdUrd-positive cells (Fig. 7A, Low Mag. panels), suggesting a requirement of proteasome activity for CPT-induced DSBs in S phase cells. When individual cells (see cells marked by square boxes in Fig. 7A, Low Mag. panels) were viewed at ×5 higher magnification (see Fig. 7A, High Mag. panels), the vast majority of γ-H2AX (DSB) foci (red) were shown to co-localize with BrdUrd (replication) foci (green) as evidenced by the yellowish foci in the merged images, suggesting that the majority of CPT-induced DSBs occurs at the replication sites. Significantly, co-treatment with MG-132 lowered the overall intensity of γ-H2AX staining in replication foci, suggesting that proteasome activity is required for CPT-induced DSBs at the replication sites.
FIGURE 7.
Proteasome-dependent formation of DSBs occurs at the replication foci. A, HeLa cells were pulse-labeled with 50 μm BrdUrd for 45 min in the presence or absence of MG-132 and CPT. MG-132 (6 μm) was added 15 min prior to BrdUrd, whereas CPT was added 15 min after BrdUrd (see the treatment scheme in the upper panel). Cells were then fixed, sequentially immunostained with anti-γ-H2AX and anti-BrdUrd antibodies, and viewed under a fluorescence microscope as described under “Experimental Procedures.” Left panels (Low Mag.): representative images were shown with the same exposure time (viewed with the 20× objective lens). BrdUrd-positive cells were marked by arrows in images of 4′,6-diamidino-2-phenylindole (DAPI)- and γ-H2AX-stained samples (bar, 20 μm). Right panels (High Mag.); magnified images of single cells (marked by squares in the corresponding images in the left panels) (bar, 4 μm). B, replication fork collision model for the involvement of the ubiquitin-proteasome pathway in processing Top1 cleavage complexes into DSBs.
DISCUSSION
The concept that reversible Top1 cleavage complexes can be processed into DNA damage through either a transcription- or replication-dependent event has been proposed and supported by many studies (Refs. 8–10 also reviewed in Refs. 3, 5). Transcription-dependent processing of Top1 cleavage complexes has been shown to generate exclusively SSBs in a ubiquitin/proteasome-dependent manner. Accumulating evidence has suggested a model in which Top1 cleavage complexes arrest the RNA polymerase elongation complexes, triggering proteasomal degradation of Top1 cleavage complexes and concomitant exposure of Top1-concealed SSBs at the arrest sites (8, 23). The ubiquitin-proteasome pathway involved in degradation of Top1 cleavage complexes has been shown to involve the formation of Lys-48-linked polyUb chains on Top1 and the participation of a CUL-3-based E3 ligase (23, 31).
A replication fork collision model has been proposed for the replication-dependent processing event, in which Top1-DNA covalent adducts arrest the replication forks, resulting in the formation of DSBs and Top1-DNA cross-links (9–11). Despite the mechanistic parallels between the two processing events, the ubiquitin-proteasome pathway is not predicted to be involved in the replication-dependent processing event because the formation of DSBs is presumed to be due to replication fork runoff on the leading strand of DNA synthesis (9–11). In this study, we show that the ubiquitin-proteasome pathway is involved in the replication-dependent processing of Top1 cleavage complexes into DSBs. There are two possibilities that could explain the requirement of a proteasome pathway for CPT-induced DSBs in the context of the replication fork collision model. One possibility is that the DSBs formed as the consequence of replication fork runoff are protected by the replication complexes and are not freely available for detection by the DSB-sensing mechanism. Degradation of the replication complexes by proteasome is necessary to reveal the replication complex-protected DSBs. The other possibility is that Top1 cleavage complexes, as a consequence of replication fork arrest, are degraded by the proteasome first to generate peptide-linked SSBs, followed by replication runoff on the leading strand to generate DSBs (see Fig. 7B for such a model). We favor the latter possibility because we have shown in this study that CPT-induced DSB formation is dependent on a CUL-3 based E3 ligase, which is known to be responsible for the degradation of Top1 cleavage complexes (31).
Previous studies have suggested that Top1-DNA covalent adducts could be processed through either TDP1 (tyrosyl-DNA-phosphodiesterase 1) or various endonucleases (e.g. Rad1-Rad9, Mre11, Mus81-Eme1, and Slx4 in yeast, as reviewed in Ref. 38). Our results, which demonstrate the involvement of the ubiquitin-proteasome pathway, could suggest either a third pathway for repairing Top1-DNA covalent adducts or a link between the ubiquitin-proteasome pathway and the TDP1 and/or the endonuclease pathways. For the latter, it is conceivable that the ubiquitin-proteasome pathway acts upstream of the TDP1 and/or the endonuclease pathway. It has been demonstrated that TDP1 can only remove peptide-linked, but not Top1-linked, strand breaks (39–41). Proteasomal degradation of Top1-DNA covalent adducts could generate peptide-linked DNA strand breaks that can serve as the substrates for TDP1. It is less clear whether proteasomal degradation of Top1-DNA covalent adducts is necessary for the endonuclease-mediated removal of Top1 from Top1-DNA covalent adducts because some endonuclease activities are known to be able to remove proteins from covalent protein-DNA adducts (e.g. Mre11 for removing Spo11-DNA covalent adducts (42)).
Recent studies have suggested the involvement of proteasome in DSB repair (43). It has been demonstrated that a component of the “lid” of the 19 S regulatory subunit, human DSS1 (the ortholog of yeast SEM1), is associated with BRAC2 and involved in DSB repair (43). The proteasome inhibitor, epoxomicin, has been demonstrated to be able to shift the balance between two DSB repair pathways, gene conversion and single strand annealing, in human MCF7 cells (44). Proteasome inhibitors have also been shown to sensitize tumor cells to IR and DNA cross-linking agents (45–48). It has been demonstrated that during IR treatment, the inhibition of proteasome suppresses the homologous recombination by impairing the formation of single strand DNA/RPA recombinogenic intermediate, a step involving RPA phosphorylation and BRCA1 foci formation (49). Significantly, proteasome activity has been shown to be required for repair at a late stage (e.g. RPA foci formation), but not for sensing DNA damage at an early stage (e.g. phosphorylation and foci formation of ATM or H2AX), in IR-treated cells (49). Our study has demonstrated that different from the IR treatment case in which the proteasome activity is not required for early DNA damage signals (e.g. γ-H2AX), CPT-induced DSBs and early DNA damage signals are strongly dependent on proteasome activity in the cell lines employed in this study, suggesting an additional and specific role of proteasome in CPT-induced formation of DSBs. It seems quite possible that the ubiquitin-proteasome pathway may play a dual function in CPT-induced DSB repair, being required for its formation at the early stage and also important for its repair at a later stage.
This study shows that MG-132 or POMP siRNA can partially, but not completely, abolish CPT-induced γ-H2AX in HeLa and HT29 cells. This incomplete inhibitory effect could have three possible explanations. One possibility is that the proteasome activity was only partially inactivated by the proteasome inhibitors because of low concentrations of the inhibitors. This does not seem to be the case because increasing the concentrations of the proteasome inhibitors did not result in further inhibition (data not shown). The second possibility is that the orientation of the Top1-DNA covalent complex relative to the replication fork (i.e. leading versus lagging strand of DNA synthesis) may dictate the requirement of the proteasome-dependent processing. It is possible that proteasome-dependent processing is necessary to generate the DSB only when the Top1-DNA covalent complex is nicking the strand that is complementary to the leading strand of DNA synthesis (10, 11). If the Top1-DNA covalent complex is nicking the strand that is complementary to the lagging strand of DNA synthesis, no DSB is formed, but the fork is arrested, which triggers γ-H2AX by a mechanism independent of DSB formation (e.g. see the next possibility of ATR activation at arrested forks). The third possibility, which we favor, is that both proteasome-dependent and proteasome-independent pathways exist for CPT-induced DNA damage signaling. Previous studies have attributed CPT-induced γ-H2AX to both DNA-PK (mostly reflecting DSB formation) and ATR activation (mostly reflecting replication fork arrest) (50). The proteasome-dependent pathway, as demonstrated in this study, could be primarily responsible for the formation of CPT-induced DSBs that activate DNA-PK/ATM as proposed in our model shown in Fig. 7B, although the proteasome-independent pathway could reflect CPT-induced replication fork arrest and hence ATR signaling. Both pathways could contribute to CPT-induced DNA damage signaling (e.g. γ-H2AX formation). Alternatively, the proteasome-independent pathway could also involve an unidentified processing mechanism(s) for Top1-DNA covalent adducts (e.g. endonuclease-mediated processing). Clearly, further studies are necessary to elucidate the molecular components of both proteasome-dependent and -independent pathways that are involved in processing Top1 cleavage complexes into DNA damage at the arrested replication forks.
Acknowledgments
We thank Dr. X. F. Steven Zheng for technical assistance with the microscopy experiments and Dr. Yuan-Chin Tsai for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA39662 (to L. F. L.). This work was also supported by a Department of Defense Idea Award W81XWH-07-1-0407 (to Y. L. L.) and a New Jersey Commission on Cancer Research Grant 08-1079-CCR-EO (to Y. L. L.).
- Top1
- topoisomerase I
- CPT
- camptothecin
- DSB
- double strand break
- POMP
- proteasome maturation protein
- SSB
- single-strand break
- BrdUrd
- bromodeoxyuridine
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- ATM
- ataxia telangiectasia mutated
- HA
- hemagglutinin
- HU
- hydroxyurea
- IR
- ionizing radiation
- PBS
- phosphate-buffered saline
- APH
- aphidicolin
- ATR
- ATM and Rad3-related protein
- DRB
- 5,6-dichloro-1-β-d-ribobenzimidazole
- RPA
- replication protein A
- DNA-PK
- DNA protein kinase.
REFERENCES
- 1.Wang J. C. (1996) Annu. Rev. Biochem. 65, 635–692 [DOI] [PubMed] [Google Scholar]
- 2.Wang J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 3.Li T. K., Liu L. F. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 53–77 [DOI] [PubMed] [Google Scholar]
- 4.Champoux J. J. (2001) Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y. (2006) Nat. Rev. Cancer 6, 789–802 [DOI] [PubMed] [Google Scholar]
- 6.Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. (1985) J. Biol. Chem. 260, 14873–14878 [PubMed] [Google Scholar]
- 7.Hsiang Y. H., Liu L. F. (1988) Cancer Res. 48, 1722–1726 [PubMed] [Google Scholar]
- 8.Desai S. D., Zhang H., Rodriguez-Bauman A., Yang J. M., Wu X., Gounder M. K., Rubin E. H., Liu L. F. (2003) Mol. Cell. Biol. 23, 2341–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiang Y. H., Lihou M. G., Liu L. F. (1989) Cancer Res. 49, 5077–5082 [PubMed] [Google Scholar]
- 10.Strumberg D., Pilon A. A., Smith M., Hickey R., Malkas L., Pommier Y. (2000) Mol. Cell. Biol. 20, 3977–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsao Y. P., Russo A., Nyamuswa G., Silber R., Liu L. F. (1993) Cancer Res. 53, 5908–5914 [PubMed] [Google Scholar]
- 12.D'Arpa P., Beardmore C., Liu L. F. (1990) Cancer Res. 50, 6919–6924 [PubMed] [Google Scholar]
- 13.Holm C., Covey J. M., Kerrigan D., Pommier Y. (1989) Cancer Res. 49, 6365–6368 [PubMed] [Google Scholar]
- 14.Nitiss J. L., Wang J. C. (1996) Mol. Pharmacol. 50, 1095–1102 [PubMed] [Google Scholar]
- 15.Reid R. J., Fiorani P., Sugawara M., Bjornsti M. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., D'Arpa P., Liu L. F. (1990) Cancer Cells 2, 23–27 [PubMed] [Google Scholar]
- 17.Desai S. D., Liu L. F., Vazquez-Abad D., D'Arpa P. (1997) J. Biol. Chem. 272, 24159–24164 [DOI] [PubMed] [Google Scholar]
- 18.Desai S. D., Wood L. M., Tsai Y. C., Hsieh T. S., Marks J. R., Scott G. L., Giovanella B. C., Liu L. F. (2008) Mol. Cancer Ther. 7, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyu Y. L., Kerrigan J. E., Lin C. P., Azarova A. M., Tsai Y. C., Ban Y., Liu L. F. (2007) Cancer Res. 67, 8839–8846 [DOI] [PubMed] [Google Scholar]
- 20.Mao Y., Desai S. D., Ting C. Y., Hwang J., Liu L. F. (2001) J. Biol. Chem. 276, 40652–40658 [DOI] [PubMed] [Google Scholar]
- 21.Desai S. D., Li T. K., Rodriguez-Bauman A., Rubin E. H., Liu L. F. (2001) Cancer Res. 61, 5926–5932 [PubMed] [Google Scholar]
- 22.Seiler J. A., Conti C., Syed A., Aladjem M. I., Pommier Y. (2007) Mol. Cell. Biol. 27, 5806–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C. P., Ban Y., Lyu Y. L., Desai S. D., Liu L. F. (2008) J. Biol. Chem. 283, 21074–21083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiriet C., Hayes J. J. (2005) Mol. Cell 18, 617–622 [DOI] [PubMed] [Google Scholar]
- 25.Shao R. G., Cao C. X., Zhang H., Kohn K. W., Wold M. S., Pommier Y. (1999) EMBO J. 18, 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takemura H., Ashutosh Rao V., Sordet O., Furuta T., Miao Z. H., Meng L., Zhang H., Pommier Y. (2006) J. Biol. Chem. 281, 30814–30823 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. W., Otterness D. M., Chiang G. G., Xie W., Liu Y. C., Mercurio F., Abraham R. T. (2005) Mol. Cell 19, 607–618 [DOI] [PubMed] [Google Scholar]
- 28.Hoyt M. A., Coffino P. (2004) Cell. Mol. Life Sci. 61, 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aleo E., Henderson C. J., Fontanini A., Solazzo B., Brancolini C. (2006) Cancer Res. 66, 9235–9244 [DOI] [PubMed] [Google Scholar]
- 30.Finley D., Ciechanover A., Varshavsky A. (1984) Cell 37, 43–55 [DOI] [PubMed] [Google Scholar]
- 31.Zhang H. F., Tomida A., Koshimizu R., Ogiso Y., Lei S., Tsuruo T. (2004) Cancer Res. 64, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 32.Fricke B., Heink S., Steffen J., Kloetzel P. M., Krüger E. (2007) EMBO Rep. 8, 1170–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 34.Ikeda F., Dikic I. (2008) EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desai S. D., Haas A. L., Wood L. M., Tsai Y. C., Pestka S., Rubin E. H., Saleem A., Nur-E-Kamal A., Liu L. F. (2006) Cancer Res. 66, 921–928 [DOI] [PubMed] [Google Scholar]
- 36.Andersen J. B., Aaboe M., Borden E. C., Goloubeva O. G., Hassel B. A., Orntoft T. F. (2006) Br. J. Cancer 94, 1465–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiessling A., Hogrefe C., Erb S., Bobach C., Fuessel S., Wessjohann L., Seliger B. (2009) Oncogene 28, 2606–2620 [DOI] [PubMed] [Google Scholar]
- 38.Pommier Y., Barcelo J. M., Rao V. A., Sordet O., Jobson A. G., Thibaut L., Miao Z. H., Seiler J. A., Zhang H., Marchand C., Agama K., Nitiss J. L., Redon C. (2006) Prog. Nucleic Acids Res. Mol. Biol. 81, 179–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debéthune L., Kohlhagen G., Grandas A., Pommier Y. (2002) Nucleic Acids Res. 30, 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Interthal H., Chen H. J., Champoux J. J. (2005) J. Biol. Chem. 280, 36518–36528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S. W., Burgin A. B., Jr., Huizenga B. N., Robertson C. A., Yao K. C., Nash H. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11534–11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreau S., Ferguson J. R., Symington L. S. (1999) Mol. Cell. Biol. 19, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogan N. J., Lam M. H., Fillingham J., Keogh M. C., Gebbia M., Li J., Datta N., Cagney G., Buratowski S., Emili A., Greenblatt J. F. (2004) Mol. Cell 16, 1027–1034 [DOI] [PubMed] [Google Scholar]
- 44.Gudmundsdottir K., Lord C. J., Ashworth A. (2007) Oncogene 26, 7601–7606 [DOI] [PubMed] [Google Scholar]
- 45.Mateos M. V., Hernández J. M., Hernández M. T., Gutiérrez N. C., Palomera L., Fuertes M., Díaz-Mediavilla J., Lahuerta J. J., de la Rubia J., Terol M. J., Sureda A., Bargay J., Ribas P., de Arriba F., Alegre A., Oriol A., Carrera D., García-Laraña J., García-Sanz R., Bladé J., Prósper F., Mateo G., Esseltine D. L., van de Velde H., San Miguel J. F. (2006) Blood 108, 2165–2172 [DOI] [PubMed] [Google Scholar]
- 46.Mimnaugh E. G., Yunmbam M. K., Li Q., Bonvini P., Hwang S. G., Trepel J., Reed E., Neckers L. (2000) Biochem. Pharmacol. 60, 1343–1354 [DOI] [PubMed] [Google Scholar]
- 47.Mitsiades N., Mitsiades C. S., Richardson P. G., Poulaki V., Tai Y. T., Chauhan D., Fanourakis G., Gu X., Bailey C., Joseph M., Libermann T. A., Schlossman R., Munshi N. C., Hideshima T., Anderson K. C. (2003) Blood 101, 2377–2380 [DOI] [PubMed] [Google Scholar]
- 48.Pajonk F., van Ophoven A., Weissenberger C., McBride W. H. (2005) BMC Cancer 5, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakawa Y., Sonoda E., Barber L. J., Zeng W., Yokomori K., Kimura H., Niimi A., Lehmann A., Zhao G. Y., Hochegger H., Boulton S. J., Takeda S. (2007) Cancer Res. 67, 8536–8543 [DOI] [PubMed] [Google Scholar]
- 50.Furuta T., Takemura H., Liao Z. Y., Aune G. J., Redon C., Sedelnikova O. A., Pilch D. R., Rogakou E. P., Celeste A., Chen H. T., Nussenzweig A., Aladjem M. I., Bonner W. M., Pommier Y. (2003) J. Biol. Chem. 278, 20303–20312 [DOI] [PubMed] [Google Scholar]