Abstract
Aims/hypothesis
It is widely accepted that production of insulin, glucagon, somatostatin and pancreatic polypeptide in islet cells is specific to beta, alpha, delta and pancreatic polypeptide cells, respectively. We examined whether beta cells express other genes encoding islet hormones.
Methods
Nested RT-PCR was performed on single beta cells of transgenic mice with green fluorescent protein (GFP) driven by mouse insulin I promoter (MIP-GFP).
Results
Only 55% of adult beta cells expressed the insulin gene alone, while others expressed two or more islet hormone genes; 4% expressed all four hormone genes. In embryonic and neonatal cells, 60% to 80% of GFP+ cells co-expressed pancreatic polypeptide and insulin genes in contrast to 29% in adult. To clarify cell fate, we conducted lineage tracing using rat insulin II promoter-cre mice crossed with reporter mice Gt(ROSA)26Sor-loxP-flanked STOP-cassette-GFP. All GFP+ cells expressed insulin I and II genes, and showed similar heterogeneity of co-expression to that seen in MIP-GFP mice. Although we report expression of other hormone genes in a significant proportion of beta cells, our lineage tracing results demonstrate that after inducing InsII (also known as Ins2) expression, beta cell progenitors do not redifferentiate to non-beta cells.
Conclusions/interpretation
This study shows co-expression of multiple hormone genes in beta cells of adult mice as well as in embryos and neonates. This finding could: (1) represent residual expression from beta cell precursors; (2) result from alternative developmental pathways for beta cells; or (3) denote the differentiation potential of these cells. It may be linked to functional heterogeneity. This heterogeneity in gene expression may provide a means to characterise the functional, cellular and developmental heterogeneity seen in beta cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-009-1570-x) contains supplementary material, which is available to authorised users.
Keywords: Heterogeneity, Pancreatic beta cell, Single-cell nested RT-PCR
Introduction
Pancreatic islets have four major types of endocrine cells, those that produce glucagon (alpha cells), insulin (beta cells), somatostatin (delta cells) and pancreatic polypeptide. There has been considerable interest in the heterogeneity of beta cells on a functional level, particularly with insulin secretion [1–7], but heterogeneity of gene expression has received less attention. Previous studies in a variety of species have shown endocrine cells expressing more than one islet hormone gene as islets develop in the fetal state or during neogenesis [8–13]. There are a few examples of beta cells in adult islets with multiple islet hormones by immunostaining. One study found cells double-stained for insulin and somatostatin in an adult mouse model of islet regeneration [14]. With a single-cell gene-profiling method using cDNA amplification and microarray detection on 13 single adult mouse islet cells, Chiang and Melton found one co-expressed insulin, glucagon and pancreatic polypeptide genes and six other cells co-expressed combinations of glucagon, somatostatin and pancreatic polypeptide genes; the other six cells expressed only the insulin gene [13]. To address whether the co-expression of multiple hormone genes in beta cells is a rare event or the norm, and whether it is influenced by developmental stage, we determined the frequency of heterogeneous gene expression in single beta cells from adult, neonatal and fetal mice.
The requirements for such experiments are a good cell separation technique and a sensitive method for measuring gene expression. Flow cytometry has been used with dispersed rat islet cells to obtain beta cell populations that are over 90% pure [15, 16]. Sorting criteria include cell size, cell granularity and autofluorescence. Separation of beta cells using the somewhat specific surface marker neural cell adhesion molecule, did not lead to higher purity [17]. Infection of islets with adenovirus producing green fluorescent protein (GFP) under the control of rat insulin promoter led to more than 95% purity [18], but this approach has the drawback of viral toxicity. This problem can be avoided by using transgenic mice with GFP driven by the mouse insulin I promoter (MIP-GFP) [19]. Noting the demonstration of heterogeneity of gene expression in seemingly homogenous, morphologically defined neural cells [20, 21], we wondered about the possibility of similar findings in beta cells. Using single-cell nested PCR for the amplification of cDNAs, which provides both sensitivity and specificity [22], we found that a large proportion of the beta cells in adult, neonatal and fetal mice expressed multiple islet hormone genes.
Further information about the lineage of these single cells was obtained by the Cre-loxP system [23]. B6.Cg-Tg(Ins2-cre)25Mgn/J (rInsII-cre) mice [24] produce Cre recombinase controlled by a short fragment of the rat insulin II gene promoter. By crossing these rInsII-cre mice with B6;129-Gt(ROSA)26Sortm2Sho/J (ROSA-loxP-stop-loxP-GFP) mice [25], mice were generated in which cells were marked when the insulin promoter is activated. Analysis of these mice demonstrated that cells that were marked upon induction of InsII (also known as Ins2) expression faithfully maintained their differentiated phenotype, even when some of these cells co-expressed other endocrine hormone genes.
Methods
Islets isolation and dispersion
From adult (16-24 weeks old) mice, islets were isolated with collagenase digestion followed by separation with a density gradient, as described previously [26]. After washing, islets were handpicked and immediately dispersed. After washing three times with sterile calcium and magnesium-free PBS, islets were dispersed for 15 min at 37°C in the same buffer containing 1 mg/ml bovine pancreas trypsin (Sigma-Aldrich, St Louis, MO, USA) and 30 μg/ml DNase1 (Roche, Basel Schweiz), being vortexed for 10 s every 5 min. After washing once with RPMI 1640 containing 10% (vol./vol.) fetal bovine serum, cells were centrifuged at 250 g and resuspended in less than 1 ml PBS. Cells were then filtered through a 40 μm filter and stained with propidium iodide (Sigma-Aldrich) prior to cell sorting.
At 15.5 days of gestation (E15.5) or at postnatal day 1 (P1) embryos or neonates respectively were killed by decapitation. Pancreases were removed and minced with a razor blade; the tissue was digested with Liberase for approximately 20 min at 37°C, washed three times with calcium and magnesium-free PBS, and dispersed as above. All procedures on mice were approved by the Institutional Committee on Research Animal Care of the Joslin Diabetes Center.
Cell sorting
Propidium iodide was used for exclusion of dead cells. All samples were analysed on a MoFlo cell sorter with Summit software (Cytomation, Fort Collins, CO, USA). For analysis of islet cells from MIP-GFP mice, the GFP signal was so strong that the neutral density filter was used to reduce brightness.
Analysis of gene expression from total RNA
Double-sorted cells from each population were collected into Trizol (Invitrogen, Carlsbad, CA, USA) and total RNA was extracted following the manufacturer’s protocol. First-strand cDNA was synthesised from 500 ng RNA by using a first-strand synthesis system for RT-PCR (SuperScript 3; Invitrogen) according to the manufacturer’s protocol. All PCR reactions were performed using 35 cycles at 94°C for 60 s, 60°C for 60 s and 72°C for 60 s with gene-specific primers.
Single-cell nested RT-PCR
Multiplex single-cell nested RT-PCR analysis was performed according to the method of Miyamoto et al. [27] with minor modifications. Briefly, double-sorted single cells were deposited into 96-well U-bottom plates (BD, Franklin Lakes, NJ, USA) with 7.5 μl lysis-RT buffer containing five pairs of gene-specific reverse primers (Electronic supplementary material [ESM] Table 1) at final concentration of: 1× first strand buffer (Invitrogen), 10 mmol/l DTT (Invitrogen), 1 mmol/l dNTPs (New England BioLabs, Ipswich, MA, USA), 0.5% (wt/vol.) TritonX-100 (Sigma-Aldrich), 0.1% (wt/vol.) bovine serum albumin, 10 U/μl M-MLV reverse transcriptase (Invitrogen), 0.1 U/μl RNase inhibitor (Invitrogen) and 0.4 μmol/l reverse primers. Cells were lysed by rapid pipetting several times and cell lysates then transferred to 200 μl thin-wall PCR tubes. After incubation at 37°C for 90 min, the samples were incubated at 94°C for 30 s to inactivate the enzyme. The first-round PCR was carried out in the same tube by additing premixed PCR buffer containing the gene-specific forward primers (ESM Table 1) (1× GeneAmp PCR Gold Buffer [Applied Biosystems, Forest City, CA, USA], 2.5 mmol/l MgCl2, AmpliTaq Gold 0.1 U/μl, 0.1 μmol/l forward primers ). The total volume of the first PCR reactions was 30 μl; PCRs were performed using the following variables: one cycle of 5 min at 95°C, then 36 cycles of 30 s at 94°C, 90 s at 60°C and 90 s at 72°C. We replica-plated 0.5 μl of the first-round PCR reactions into new PCR tubes for the second-round PCR, which was carried out separately for each gene with fully nested gene-specific primers (ESM Table 1) (1× GeneAmp PCR Gold Buffer [Applied Biosystems], 2.5 mmol/l MgCl2, AmpliTaq Gold [Applied Biosystems] 0.1 U/μl, 0.25 μmol/l forward and reverse primers). The second-round PCR was performed with the following variables: one cycle of 5 min at 95°C, then 36 cycles of 30 s at 94°C, 90 s at 60°C and 90 s at 72°C. Aliquots of second-round PCR products were then subjected to 2% (wt/vol.) gel electrophoresis. Since the primers are designed to span at least one intron, genomic products can be excluded by their larger size. We used 200 pg of total RNA isolated from mouse islets as the positive control in this study. Double-sorted single B lymphocytes (B220+IgM+) of peripheral blood were used as a negative control.
Results
FCM sorting of GFP+cells from adult MIP-GFP mice
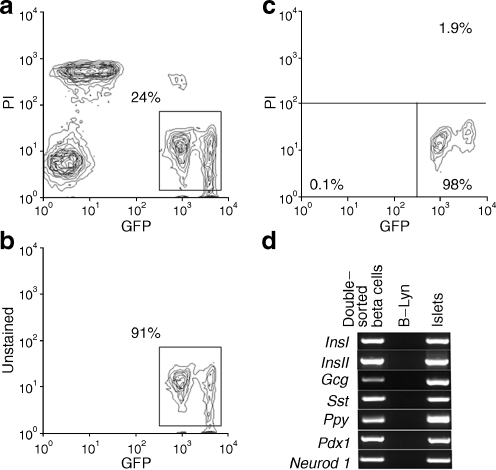
Dispersed islet cells from adult (16–24 week) MIP-GFP mice were sorted using three gates: the first for size and granule-density estimated by forward scatter (FSC) and side scatter, respectively. Beta cells are large and have moderate to high granular density. The second gate employed pulse width and was used to exclude doublets or other cell clusters. The third gate was for GFP and propidium iodide to exclude GFP negative cells and dead cells. By FCM analysis, two populations of GFP+ cells (GFP-medium and GFP-high) could be observed (Fig. 1a–c). We determined the percentage of beta cells in these GFP+ cells by immunostaining with anti-insulin antibody (ESM Methods), finding that 100% of 300 double-sorted GFP+ cells were stained for insulin. These data indicate that the double-sorted GFP+ cells were pure beta cells. To determine the possible presence of cell doublets or clusters in GFP-high population, we estimated cell size and granule-density between these two populations by FSC and side scatter, respectively and found no difference (ESM Fig. 1). The FSC histogram of each population had only one peak, indicating that cells in each of these two populations had similar size and granule-density. Furthermore, there were no differences in the gene expression pattern of islet hormones between these two populations as determined by single-cell nested RT-PCR analysis (ESM Tables 2 and 3). Therefore, propidium iodide-negative and GFP+ populations were combined to obtain living InsI (also known as Ins1)-expressing cells. For further purification of the GFP+ cells, we performed a similar second sorting, again excluding propidium iodide-positive cells (Fig. 1b). After the first sort, the purity of GFP+ cells was 91% (Fig. 1b), increasing to 98% with the second sorting (Fig. 1c).
Fig. 1.
Sorting of GFP+ cells from adult MIP-GFP mice. a First sort with propidium iodide (PI) staining of MIP-GFP islets, (b) second sort without propidium iodide staining of MIP-GFP islets, (c) FCM analysis of double-sorted GFP+ cells and (d) gene expression of sorted beta cells from adult MIP-GFP. The percentages of the cells included in each region vs total FCM-sorted single cells are indicated in the figure. B-Lym, peripheral B lymphocytes
Gene expression analysis of pooled GFP+ cells FCM-sorted from adult MIP-GFP mouse islets
Initially, gene expression of a variety of islet-affiliated genes was determined for this population by conventional RT-PCR. As expected, GFP+ cells sorted from adult MIP-GFP mice expressed both InsI and InsII, as well as Pdx1 and Neurod1 (Fig. 1d). Surprisingly, glucagon, somatostatin and pancreatic polypeptide genes were also expressed in the pooled RNA from these double-sorted GFP+ cells (Fig. 1d).
Establishment of single-cell nested RT-PCR analysis method in pancreatic beta cells
Single-cell nested RT-PCR analysis is a well established technique for studying haemopoietic cells [27], so we applied this to beta cells. With nested PCR [22], mRNA of sorted single cells can be amplified for visualisation after a second PCR. We designed the primers for single-cell nested RT-PCR in different exons to distinguish the PCR products from mRNA from genomic DNA (ESM Table 1). cDNAs from whole islets, used as positive controls, gave correct PCR products for all genes examined. To certify that the FCM-sorted cells were truly single and GFP+, we sorted GFP+ cells from MIP-GFP mice into 96-well plates at a ratio of one cell per well and examined them by bright field and fluorescence microscopy. We did not find any doublets, aggregates or GFP− cells in a total of 228 FCM-sorted cells. Even if some sorted cells were, in fact, aggregates, their frequency in the FCM-sorted cell group must have been very rare (less than 0.44% or 1/228). The contamination of GFP− cells must have been similarly rare (less than 0.44% or 1/228).
We also examined the possibility that the dispersion process might lead to release of RNA that could adhere to the surface of or be endocytosed into beta cells to create an artefact. No islet hormone mRNA contamination was detected using single-cell nested RT-PCR on RNA from sorted GFP+ splenocytes of chicken beta-actin promoter-driven enhanced GFP (C57BL/6-Tg[ACTB-EGFP]1Osb/J) transgenic mice that had been dispersed with islets from wild-type C57/BL/6J mice (ESM, Results). Furthermore, mouse Try1 (also known as Prss1) mRNA, which is highly expressed in acinar cells, was not detected in double-sorted GFP+ cells from adult MIP-GFP mice by single-cell nested RT-PCR (ESM, Results). These data confirm that mRNA contamination from other cells was not a problem.
Next, we estimated the sensitivity of the single-cell nested RT-PCR analysis using template cDNA, the copy number of which was known. The system for InsI used in this study is sensitive enough to detect at least ten copies of cDNA, while those for Gcg, Sst and Ppy can detect one copy (ESM, Results).
Single-cell nested RT-PCR analysis of adult pancreatic beta cell from MIP-GFP
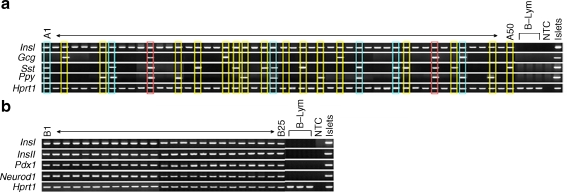
Single-cell nested RT-PCR analysis revealed that all double-sorted GFP+ single cells of adult (16–24 week), P1 neonatal and E15.5 embryonic MIP-GFP mice expressed InsI (Fig. 2a, b, Tables 1, 2 and 3), which confirms the complete (100%) purity of double-sorted single cells from these mice. Mouse beta cells express two non-allelic insulin genes, insulin I and II, both of which make important contributions to insulin synthesis and secretion [28]. Although expression of insulin I gene is restricted to rodent beta cells, InsII mRNA can also be found in brain, thymus, fetal liver and yolk sac [29–31]. A histological study has shown that both proinsulin genes were co-expressed in all beta cells of normal rat pancreas [32]. In our analysis, all insulin I gene-expressing cells from MIP-GFP mice co-expressed InsII (Fig. 2b, Tables 1, 2 and 3). In addition, expression of Pdx1 and Neurod1 was found in every sorted GFP+ cell, further supporting the concept that they are all true beta cells (Fig. 2b, Table 3). Pdx1 is expressed in beta cells and pancreatic ducts [33]; Neurod1 is expressed in beta cells and some alpha cells [34].
Fig. 2.
Multiplex single-cell nested RT-PCR analysis demonstrates the heterogeneity of islet hormone gene expression in single beta cells from adult, P1 neonatal and E15.5 embryonic MIP-GFP mice. a Representative data for InsI, Gcg, Sst and Ppy in single GFP+ beta cells from adult MIP-GFP mice (A1–50). The profiles with two, three and four islet hormone genes expression are indicated by yellow, blue and red outlines, respectively. b Representative data for InsI, InsII, Pdx1 and Neurod1 in single beta cells from adult MIP-GFP mice (B1–25). B-Lym, peripheral B lymphocytes; NTC, no template control
Table 1.
Multiplex single-cell nested RT-PCR analysis demonstrates the heterogeneity of islet hormone genes expression in single beta cells from adult, P1 neonatal and E15.5 embryonic MIP-GFP mice: gene expression at each developmental stage
| Developmental stage | Expression of gene, % (n)a | Total sorted cells (n) | |||||
|---|---|---|---|---|---|---|---|
| InsI | InsII | Gcg | Sst | Ppy | Hprt1 | ||
| E15.5 | 100 (70) | 100 (70) | 13 (9) | 13 (9) | 62 (43) | 100 (70) | (70) |
| P1 | 100 (147) | 100 (147) | 10 (15) | 12 (18) | 80 (117) | 100 (147) | (147) |
| Adult | 100 (476) | 100 (476) | 11 (53) | 29 (138) | 29 (137) | 100 (476) | (476) |
a% is percentage of cells expressing gene indicated, with or without the other genes listed vs total sorted cells at each developmental stage; n is number of cells
Table 2.
Multiplex single-cell nested RT-PCR analysis demonstrates the heterogeneity of islet hormone genes expression in single beta cells from adult, P1 neonatal and E15.5 embryonic MIP-GFP mice: gene expression pattern at each developmental stage
| Expression pattern | Gene | Developmental stage | |||||||
|---|---|---|---|---|---|---|---|---|---|
| InsI | InsII | Gcg | Sst | Ppy | Hprt1 | E15.5, % (n)a | P1, % (n)a | Adult% (n)a | |
| A | + | + | − | − | − | + | 30 (21) | 15 (22) | 55 (262) |
| B | + | + | + | − | − | + | 1 (1) | 3 (5) | 3 (13) |
| C | + | + | − | + | − | + | 7 (5) | 2 (3) | 12 (58) |
| D | + | + | − | − | + | + | 47 (33) | 64 (94) | 10 (48) |
| E | + | + | + | + | − | + | 0 (0) | 0 (0) | 1 (6) |
| F | + | + | + | − | + | + | 9 (6) | 6 (8) | 3 (15) |
| G | + | + | − | + | + | + | 3 (2) | 9 (13) | 12 (55) |
| H | + | + | + | + | + | + | 3 (2) | 1 (2) | 4 (19) |
| Total | 100 (70) | 100 (147) | 100 (476) | ||||||
a% is percentage of cells showing the indicated gene expression pattern vs total sorted cells at each developmental stage; n is number of cells
Table 3.
Multiplex single-cell nested RT-PCR analysis of mRNA expression in single GFP+ cells from adult, P1 neonatal and E15.5 embryonic MIP-GFP mice: gene expression pattern at each developmental stage
| Expression pattern | Gene | Developmental stage | ||||||
|---|---|---|---|---|---|---|---|---|
| InsI | InsII | Pdx1 | Neurod1 | Hprt1 | E15.5, % (n)a | P1, % (n)a | Adult, % (n)a | |
| A | + | + | + | + | + | 100 (70) | 100 (147) | 100 (148) |
| B | − | − | − | − | + | 0 (0) | 0 (0) | 0 (0) |
| Total | 100 (70) | 100 (147) | 100 (148) | |||||
a% is percentage of cells showing the indicated gene expression pattern vs total sorted cells; n is number of cells
By nested RT-PCR, 55% of single GFP+ cells from adult MIP-GFP mice expressed insulin gene without other hormone genes, while 45% co-expressed two or more islet hormone genes. Ins + Gcg + cells accounted for 11%, Ins + Sst + cells 29% and Ins + Ppy + cells 29% of single cells; 4% of cells expressed all four islet hormone genes (Fig. 2a, Tables 1 and 2). It must be emphasised that with immunostaining of pancreatic sections of adult or neonatal pancreas of MIP-GFP mice, islet hormone proteins did not co-localise with insulin (data not shown), as has been found by many others. In aliquots of double-sorted GFP+ cells immunostained with anti-insulin, anti-glucagon, anti-somatostatin or anti-pancreatic polypeptide antibodies separately, all cells stained for insulin were positive for insulin, but none were positive for the other islet hormones. Thus, the presence of other islet hormone protein in beta cells was undetectable by immunostaining.
Furthermore, we determined the expression levels of islet hormone mRNAs in GFP+ cells sorted from adult MIP-GFP islets by quantitative RT-PCR and found that the expression levels of glucagon, somatostatin and pancreatic polypeptide mRNA in the sorted beta cells were 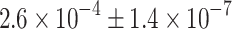 ,
, 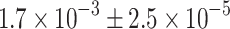 and
and 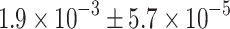 times lower than those in true alpha, delta and pancreatic polypeptide cells, respectively (ESM, Results). Therefore, in beta cells the expression of islet hormone mRNA other than insulin is extremely low and not meaningful for hormone production.
times lower than those in true alpha, delta and pancreatic polypeptide cells, respectively (ESM, Results). Therefore, in beta cells the expression of islet hormone mRNA other than insulin is extremely low and not meaningful for hormone production.
Developmental effect on proportion of cells expressing multiple islet hormone genes
To determine the impact of developmental stage on multiple islet hormone gene expression in beta cells, we performed single-cell nested RT-PCR on GFP+ cells from P1 neonatal and E15.5 embryonic pancreas of MIP-GFP mice (Tables 1 and 2). The percentages of Ins + Gcg + in GFP+ cells in P1 and E15.5 mice were 10% and 13%, respectively, similar to those in adult mice (11%). In contrast, the percentages of Ins + Sst + in GFP+ cells in P1 and E15.5 mice were 12% and 13%, respectively, less than half of those in adult mice (29%). Notably, 80% of GFP+ cells in P1 and 62% in E15.5 expressed pancreatic polypeptide gene in addition to insulin gene (in contrast to only about 29% in adult mice). Of these, 64% in P1 and 47% in E15.5 expressed only pancreatic polypeptide and insulin genes with no other islet hormone (vs 10% in adult animals). Only 1% of GFP+ cells in P1 and 3% in E15.5 expressed all four islet hormone genes.
To estimate the immaturity of individual beta cells, we determined Neurog3 and Pax4 gene expression using single-cell nested RT-PCR (Tables 4 and 5). Previous reports indicate that pancreatic beta cells are differentiated from Neurog3 + cells [35] and Pax4 + cells [36]. Thus, insulin gene-expressing cells that co-express Neurog3 or Pax4 gene could be assumed to be more immature than those do not express these genes. In E15.5 pancreas, 11% of GFP+ cells expressed Neurog3 gene and 53% expressed Pax4 gene as well as InsI gene (Table 4). In P1 pancreas, only 1% of GFP+ cells were InsI + Pax4 +, while in adult pancreas no InsI + Pax4 + cells were observed (Tables 4 and 5). Moreover, we could not find any InsI + Neurog3 + cells in GFP+ cells from P1 or adult mice (Tables 4 and 5). At E15.5 Ppy was expressed in 75% of Ins + Neurog3 + and in 60% of Ins + Neurog3 − cells, and in 65% of Ins + Pax4 + and 58% of Ins + Pax4 − cells (Tables 6 and 7).
Table 4.
Multiplex single-cell nested RT-PCR analysis for genes listed in single GFP+ cells from adult, P1 neonate and E15.5 embryonic MIP-GFP mice: gene expression at each developmental stage
| Developmental stage | Expression of gene, % (n)a | Total sorted cells (n) | |||
|---|---|---|---|---|---|
| InsI | Neurog3 | Pax4 | Hprt1 | ||
| E15.5 | 100 (70) | 11 (8) | 53 (37) | 100 (70) | 70 |
| P1 | 100 (89) | 0 (0) | 1 (1) | 100 (89) | 89 |
| Adult | 100 (77) | 0 (0) | 0 (0) | 100 (77) | 77 |
a% is percentage of cells expressing gene indicated, with or without the other genes listed, vs total sorted cells at each developmental stage; n is number of cells
Table 5.
Multiplex single-cell nested RT-PCR analysis for genes listed in single GFP+ cells from adult, P1 neonate and E15.5 embryonic MIP-GFP mice: gene expression pattern at each developmental stage
| Expression pattern | Gene | Developmental stage | |||||
|---|---|---|---|---|---|---|---|
| InsI | Neurog3 | Pax4 | Hprt1 | E15.5, % (n)a | P1, % (n)a | Adult, % (n)a | |
| A | + | − | − | + | 44 (31) | 99 (88) | 100 (77) |
| B | + | + | − | + | 3 (2) | 0 (0) | 0 (0) |
| C | + | − | + | + | 44 (31) | 1 (1) | 0 (0) |
| D | + | + | + | + | 9 (6) | 0 (0) | 0 (0) |
| Total | 100 (70) | 100 (89) | 100 (77) | ||||
a% is percentage of cells showing the indicated gene expression pattern vs total sorted cells at each developmental stage; n is number of cells
Table 6.
Heterogeneity of islet hormone genes expression in single beta cells from E15.5 embryonic MIP-GFP mice categorised by expression of Neurog3 and Pax4: gene expression in each category
| Category | Expression of gene, % (n) a | Total sorted cells (n) | |||||
|---|---|---|---|---|---|---|---|
| InsI | InsII | Gcg | Sst | Ppy | Hprt1 | ||
| E15.5, % (n) | |||||||
| Neurog3 + | 100 (8) | 100 (8) | 13 (1) | 0 (0) | 75 (6) | 100 (8) | 8 |
| Neurog3 − | 100 (62) | 100 (62) | 13 (8) | 13 (9) | 60 (37) | 100 (62) | 62 |
| Pax4 + | 100 (37) | 100 (37) | 11 (4) | 14 (5) | 65 (24) | 100 (37) | 37 |
| Pax4 − | 100 (33) | 100 (33) | 15 (5) | 12 (4) | 58 (19) | 100 (33) | 33 |
| Total | 100 (70) | 100 (70) | 13 (9) | 13 (9) | 62 (43) | 100 (70) | 70 |
a % is percentage of cells expressing gene indicated, with or without the other genes listed, vs total sorted cells in each category; n is number of cells
Table 7.
Heterogeneity of islet hormone genes expression in single beta cells from E15.5 embryonic MIP-GFP mice categorised by expression of Neurog3 and Pax4: gene expression pattern in each category
| Expression pattern | Gene | Category, % (n)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| InsI | InsII | Gcg | Sst | Ppy | Hprt1 | Neurog3 + | Neurog3 − | Pax4 + | Pax4 − | Total | |
| A | + | + | − | − | − | + | 25 (2) | 31 (19) | 27 (10) | 34 (11) | 30 (21) |
| B | + | + | + | − | − | + | 0 (0) | 2 (1) | 0 (0) | 3 (1) | 1 (1) |
| C | + | + | − | + | − | + | 0 (0) | 8 (5) | 8 (3) | 6 (2) | 7 (5) |
| D | + | + | − | − | + | + | 63 (5) | 45 (28) | 51 (19) | 42 (14) | 47 (33) |
| E | + | + | + | + | − | + | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| F | + | + | + | − | + | + | 12 (1) | 8 (5) | 8 (3) | 9 (3) | 9 (6) |
| G | + | + | − | + | + | + | 0 (0) | 3 (2) | 3 (1) | 3 (1) | 3 (2) |
| H | + | + | + | + | + | + | 0 (0) | 3 (2) | 3 (1) | 3 (1) | 3 (2) |
| Total | 100 (8) | 100 (62) | 100 (37) | 100 (33) | 100 (70) | ||||||
a% is percentage of cells showing the indicated gene expression pattern in each category vs total sorted cells in each category; n is number of cells
Lineage tracing: rInsII-cre: ROSA-loxP-stop-loxP-GFP mice
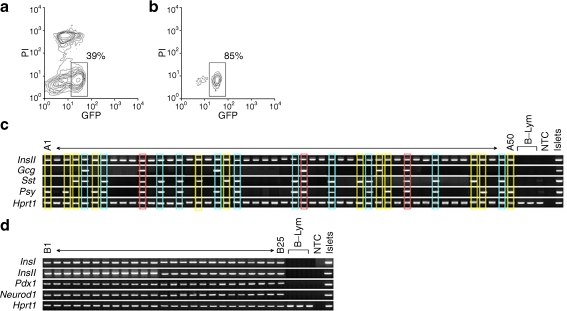
To clarify whether the multiple hormone-expressing beta cells that we detected in MIP-GFP mice differentiate to the other type of pancreatic cells, we performed lineage-tracing analysis using adult rInsII-cre: ROSA-loxP-stop-loxP-GFP mice. We FCM-sorted GFP-producing cells from these adult islets by gating propidium iodide-negative, GFP+ cells (Fig. 3a, b). All GFP+ cells expressed not only InsII but also InsI, Pdx1 and Neurod1 genes (Fig. 3d, Table 8). Therefore, all the cells marked by GFP appeared to be true beta cells. Moreover, 45% of the sorted cells expressed only insulin gene without other islet hormone genes and 55% co-expressed two or more islet hormone genes (Fig. 3c, Tables 9 and 10). InsII + Gcg + cells accounted for 12%, InsII + Sst + cells for 38% and InsII + Ppy + cells for 39% of the single cells; 6% expressed all four islet hormones genes (Fig. 3c, Tables 9 and 10). These proportions of islet hormone gene-expressing cells were quite similar to those of adult MIP-GFP mice.
Fig. 3.
Multiplex single-cell nested RT-PCR analysis demonstrates the heterogeneity of islet hormone gene expression in single GFP+ (beta) cells from adult rInsII-cre: ROSA-loxP-stop-loxP-GFP mice. a First and (b) second sort of rInsII-cre: ROSA-loxP-stop-loxP-GFP mice. c Representative data for InsII, Gcg, Sst, and Ppy in single GFP+ (beta) cells from adult animals as above (a, b) (A1–50). Profiles with two, three and four islet hormone genes expression are indicated by yellow, blue and red outlines, respectively. d Representative data for InsI, InsII, Pdx1 and Neurod1 in single beta cells as above (a, b) (B1–25). The percentages of the cells included in each region vs total FCM-sorted single cells are indicated in the figure. B-Lym, peripheral B lymphocytes; NTC, no template control
Table 8.
Multiplex single-cell nested RT-PCR analysis of mRNA expression as indicated in single GFP+ (beta) cells from adult rInsII-cre: ROSA-loxP-stop-loxP-GFP mice: gene expression pattern
| Expression pattern | Gene | % (n)a | ||||
|---|---|---|---|---|---|---|
| InsI | InsII | Pdx1 | Neurod1 | Hprt1 | ||
| A | + | + | + | + | + | 100 (151) |
| B | − | − | − | − | + | 0 (0) |
| Total | 100 (151) | |||||
a% is percentage of cells showing the indicated gene expression pattern vs total sorted cells; n is number of cells
Table 9.
Multiplex single-cell nested RT-PCR analysis demonstrates heterogeneity of islet hormone gene expression in single GFP+ (beta) cells from adult rInsII-cre:ROSA-loxP-stop-loxP-GFP mice: gene expression
| Gene expression | Gene | Total sorted cells (n) | ||||
|---|---|---|---|---|---|---|
| InsII | Gcg | Sst | Ppy | Hprt1 | ||
| % (n)a | 100 (281) | 12 (34) | 38 (107) | 39 (109) | 100 (281) | 281 |
a% is percentage of cells expressing gene indicated, with or without the other genes listed, vs total sorted cells; n is number of cells
Table 10.
Multiplex single-cell nested RT-PCR analysis demonstrates heterogeneity of islet hormone gene expression in single GFP+ (beta) cells from adult rInsII-cre:ROSA-loxP-stop-loxP-GFP mice: gene expression pattern
| Expression pattern | Gene | % (n)a | ||||
|---|---|---|---|---|---|---|
| InsII | Gcg | Sst | Ppy | Hprt1 | ||
| A | + | − | − | − | + | 45 (127) |
| B | + | + | − | − | + | 3 (7) |
| C | + | − | + | − | + | 12 (34) |
| D | + | − | − | + | + | 12 (34) |
| E | + | + | + | − | + | 1 (4) |
| F | + | + | − | + | + | 2 (6) |
| G | + | − | + | + | + | 19 (52) |
| H | + | + | + | + | + | 6 (17) |
| Total | 100 (281) | |||||
a% is percentage of cells showing the indicated gene expression pattern vs total sorted cells; n is number of cells
Discussion
We performed islet hormone gene profiling to determine the heterogeneity of gene expression of single beta cells and found a significant proportion of adult, neonatal and fetal beta cells co-expressing insulin and one or more other islet hormone genes. We ruled out that this was an artefact due to the presence of cell doublets, aggregates or contamination from GFP− non-beta cells. By FCM analysis, two populations of GFP+ cells (GFP-medium and GFP-high) were observed in islet cells from MIP-GFP mice. All cells of these two populations were single cells and stained with anti-insulin antibody; there were no differences in the gene expression pattern of islet hormones between the two populations as determined by single-cell analysis. Therefore, both GFP+ populations were combined to obtain the pure beta cells used in this study. In these two populations, the InsI mRNA expression levels by quantitative RT-PCR were almost the same. One explanation could be that the insulin promoter may be differentially active in these two populations and that differences in the stability of GFP protein and insulin mRNA (e.g. much longer half-life of insulin mRNA than that of GFP protein) could account for the differences in GFP intensity seen in these populations without any change in insulin message.
Additionally, the use of rInsII-cre: ROSA-loxP-stop-loxP-GFP mice led to the novel demonstration that once cells were marked by insulin II promoter expression, they did not deviate from the path of becoming beta cells, even though some of them expressed multiple other islet hormone genes. All beta cells of MIP-GFP and rInsII-cre: ROSA-loxP-stop-loxP-GFP mice co-expressed insulin I and II genes, which supports the previous immunostaining findings of Blume et al [32]. Interestingly, the pattern of co-expression of multiple hormone genes in adult and embryonic or neonatal beta cells differed, with embryonic and neonatal cells expressing pancreatic polypeptide gene much more frequently.
As described in the introduction, Chiang and Melton found co-expression of more than one islet hormone gene in a small number of single cells [13]. Our findings from 750 single adult beta cells from MIP-GFP and rInsII-cre: ROSA-loxP-stop-loxP-GFP mice demonstrate that the heterogeneity of islet hormone gene expression at the level of single beta cells is the norm and not an artefact of the experimental system, thus extending the preliminary observation of Chiang and Melton. We cannot exclude the possibility that transgene expression in the beta cells of our two models could have influenced these very low levels of gene expression.
Possible explanations for the finding that beta cells expressed multiple islet hormone genes are that the low level of expression of these non-insulin hormone mRNAs: (1) could be a developmental residual of their earlier high expression in beta cell precursors; (2) may reflect the existence of more than one developmental pathway for beta cells, one giving rise to cells expressing only insulin gene and another to those with insulin and other hormone genes; and (3) may be unrelated to development, resulting instead from escape from restrained expression due to some environmental factor. While the low-level expression of these hormone genes in the beta cells could be non-specific noise due to the extreme sensitivity of the technique, the lack of similar expression in control B-lymphocytes indicates that this low-level transcription is a specific, reproducible finding.
The heterogeneity of islet hormone gene expression could be related to the heterogeneity of function found in adult beta cells. Functional heterogeneity of beta cells has been reported with individual beta cells having different thresholds for glucose-stimulated insulin secretion [37, 38]. This cellular heterogeneity may extend to the redox state [4], the threshold for glucose-induced biosynthesis of proinsulin [37] and glucose-induced changes in cytoplasmic free calcium concentrations [39]. A population of dispersed adult rat beta cells that do not respond to glucose has been consistently reported as 10% to 30% of the total [5, 37, 38]. One can speculate that these unresponsive beta cells may be immature, since fetal and neonatal islets lack glucose-stimulated insulin secretion [40].
The concept that beta cell precursors express multiple islet hormone genes is supported by findings of co-staining of insulin and non-insulin islet hormones by immunostaining and gene expression during development [8–13], but what are the implications of these findings? In the haemopoietic system, it has been shown that genes related to multiple lineages are promiscuously expressed preceding commitment [41–44] and that cells with lineage-promiscuous expression contribute to multi-lineage haemopoiesis [27]. Similar promiscuity may occur during islet development [13].
Herrera et al. performed studies of the ontogenetic relationships between the different types of endocrine cells in the islets of Langerhans by generating transgenic mouse embryos, in which cells transcribing glucagon, insulin or pancreatic polypeptide genes were destroyed through the promoter-targeted production of the diphtheria toxin A chain [45]. They concluded that neither glucagon nor insulin gene-expressing cells were essential for the differentiation of the other islet cell types; thus, their results are incompatible with a model of islet development in which cells co-expressing glucagon and insulin gene would be precursors to all islet cells. However, pancreatic polypeptide gene-expressing cells were necessary for the differentiation of beta and delta cells, either because they produced an essential paracrine or endocrine factor, and/or because of a cell-lineage relationship [45, 46]. Furthermore, in order to discriminate between these possibilities, Herrera et al. performed lineage tracing studies and found adult glucagon (alpha) and insulin (beta) cells were derived from cells that had never transcribed the other hormone gene, a finding that confirmed alpha and beta cell lineages to be independent during ontogeny [47, 48]. They also showed that insulin (beta) cell progenitors, but not glucagon (alpha) cell progenitors, transcribed the pancreatic polypeptide gene.
Our data support the hypothesis that expression of pancreatic polypeptide mRNA is a marker of young beta cells since, in the embryo, the proportion of Ppy + cells in Ins + Neurog3 + or Ins + Pax4 + cells was slightly higher than in Ins + Neurog3 − or Ins + Pax4 − cells. However, our data, which showed the co-expression of multiple islet hormone genes, appear to be inconsistent with the model of independent alpha and beta cell lineages suggested by Herrera’s data. Because our beta cells had very low expression of other hormone mRNAs, it is possible that in the glucagon promoter-dependent lineage tracing approach, the production level of either diphtheria toxin A chain or Cre recombinase was insufficient in the beta cells. Also contradictory to the Herrera data is the immunostaining finding that a number of adult islet cells produced both glucagon and pancreatic polypeptide [13].
In conclusion, we found heterogeneity in expression of multiple islet hormone genes in pancreatic beta cells from adult as well as from neonatal and embryonic mice. Co-expression of multiple hormone genes in single cells has been reported at various stages of development as well as under different experimental conditions, raising the possibility of a link between heterogeneity in gene expression and beta cell progenitors expressing multiple islet hormone genes. Similarly, this heterogeneity in gene expression may be linked with observed functional heterogeneity in beta cells. We suggest that analysis of heterogeneity in gene expression may provide the means to characterise the functional, cellular and developmental heterogeneity seen in beta cells.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
(PDF 23 kb)
(PDF 23 kb)
FACS analysis of GFP-high and GFP-medium populations. a FACS analysis of double-sorted GFP+ cells, (b) FSC histogram of GFP-high and (c) GFP-medium populations. d Side scatter histogram of GFP-high and (e) GFP-medium populations. The percentages of the cells included in each region vs. total FACS-sorted single cells are indicated in each region (PDF 720 kb).
Minimum copy number of template cDNA fragment that could be detected using the nested PCR primers for the single-cell nested RT-PCR analysis of InsI, Gcg, Sst and Ppy. NTC, no template control (PDF 278 kb).
(PDF 39 kb).
(PDF 31 kb)
(PDF 36 kb)
(PDF 31 kb)
(PDF 31 kb)
Acknowledgements
The authors thank S. Yatoh for helpful discussions, and J. Lock, J. LaVecchio and G. Buruzula for excellent technical assistance. This study was supported by: the Juvenile Diabetes Research Foundation; the National Institutes of Health (DK66056); the National Institutes of Health (P30 DK36836)—Flow Cytometry Core of Joslin Diabetes and Endocrinology Research Center (DERC); the Diabetes Research and Wellness Foundation.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- FCM
Flow cytometry
- FSC
Forward scatter
- GFP
Green fluorescent protein
- MIP-GFP
Transgenic mice with green fluorescent protein driven by mouse insulin I promoter
- rInsII-cre mice
B6.Cg-Tg(Ins2-cre)25Mgn/J mice
- rInsII-cre: ROSA-loxP-stop-loxP-GFP mice
rInsII-cre mice crossed with ROSA-loxP-stop-loxP-GFP mice
- ROSA-loxP-stop-loxP-GFP mice
B6; 129-Gt(ROSA)26Sortm2Sho/J mice
- E15.5
15.5 days of gestation
- P1
Postnatal day 1
Footnotes
H. Katsuta and T. Akashi contributed equally to this study.
References
- 1.Salomon D, Meda P. Heterogeneity and contact-dependent regulation of hormone secretion by individual B cells. Exp Cell Res. 1986;162:507–520. doi: 10.1016/0014-4827(86)90354-X. [DOI] [PubMed] [Google Scholar]
- 2.Stefan Y, Meda P, Neufeld M, Orci L. Stimulation of insulin secretion reveals heterogeneity of pancreatic B cells in vivo. J Clin Invest. 1987;80:175–183. doi: 10.1172/JCI113045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuit FC, In 't Veld PA, Pipeleers DG. Glucose stimulates proinsulin biosynthesis by a dose-dependent recruitment of pancreatic beta cells. Proc Natl Acad Sci U S A. 1988;85:3865–3869. doi: 10.1073/pnas.85.11.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiekens R, In 't Veld P, Mahler T, Schuit F, van de Winkel M, Pipeleers D. Differences in glucose recognition by individual rat pancreatic β cells are associated with intercellular differences in glucose-induced biosynthetic activity. J Clin Invest. 1992;89:117–125. doi: 10.1172/JCI115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heimberg H, de Vos A, Vandercammen A, Schaftingen E, Pipeleers D, Schuit F. Heterogeneity in glucose sensitivity among pancreatic β-cells is correlated to differences in glucose phosphorylation rather than glucose transport. EMBO J. 1993;12:2873–2879. doi: 10.1002/j.1460-2075.1993.tb05949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung YM, Ahmed I, Sheu L, et al. Electrophysiological characterization of pancreatic islet cells in the mouse insulin promoter-green fluorescent protein mouse. Endocrinology. 2005;146:4766–4775. doi: 10.1210/en.2005-0803. [DOI] [PubMed] [Google Scholar]
- 7.Ling Z, Wang Q, Stange G, In 't Veld P, Pipeleers D. Glibenclamide treatment recruits beta-cell subpopulation into elevated and sustained basal insulin synthetic activity. Diabetes. 2006;55:78–85. doi: 10.2337/diabetes.55.01.06.db05-0820. [DOI] [PubMed] [Google Scholar]
- 8.Alpert S, Hanahan D, Teitelman G. Hybrid insulin genes reveal a developmental lineage for pancreatic endocrine cells and imply a relationship with neurons. Cell. 1988;53:295–308. doi: 10.1016/0092-8674(88)90391-1. [DOI] [PubMed] [Google Scholar]
- 9.de Krijger RR, Aanstoot HJ, Kranenburg G, Reinhard M, Visser WJ, Bruining GJ. The midgestational human fetal pancreas contains cells coexpressing islet hormones. Dev Biol. 1992;153:368–375. doi: 10.1016/0012-1606(92)90121-V. [DOI] [PubMed] [Google Scholar]
- 10.Teitelman G, Alpert S, Polak JM, Martinez A, Hanahan D. Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y but not pancreatic polypeptide. Development. 1993;118:1031–1039. doi: 10.1242/dev.118.4.1031. [DOI] [PubMed] [Google Scholar]
- 11.Yoon KH, Quickel RR, Tatarkiewicz K, et al. Differentiation and expansion of beta cell mass in porcine neonatal pancreatic cell clusters transplanted into nude mice. Cell Transplant. 1999;8:673–689. doi: 10.1177/096368979900800613. [DOI] [PubMed] [Google Scholar]
- 12.Bonner-Weir S, Taneja M, Weir GC, et al. In vitro cultivation of human islets from expanded ductal tissue. Proc Natl Acad Sci U S A. 2000;97:7999–8004. doi: 10.1073/pnas.97.14.7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev Cell. 2003;4:383–393. doi: 10.1016/S1534-5807(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 14.Guz Y, Nasir I, Teitelman G. Regeneration of pancreatic beta cells from intra-islet precursor cells in an experimental model of diabetes. Endocrinology. 2001;142:4956–4968. doi: 10.1210/en.142.11.4956. [DOI] [PubMed] [Google Scholar]
- 15.van de Winkle M, Maes E, Pipeleers D. Islet cell analysis and purification by light scatter and autofluorescence. Biochem Biophys Res Commun. 1982;107:525–532. doi: 10.1016/0006-291X(82)91523-6. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher DJ, Grogan WM, Barras E, Weir GC. Hormone release by islet B cell-enriched and A and D cell-enriched populations prepared by flow cytometry. Endocrinology. 1983;113:1791–1798. doi: 10.1210/endo-113-5-1791. [DOI] [PubMed] [Google Scholar]
- 17.Bernard-Kargar C, Kassis N, Berthault MF, Pralong W, Ktorza A. Sialylated form of the neural cell adhesion molecule (NCAM): a new tool for the identification and sorting of beta-cell subpopulations with different functional activity. Diabetes. 2001;50(Suppl 1):S125–S130. doi: 10.2337/diabetes.50.2007.S125. [DOI] [PubMed] [Google Scholar]
- 18.Meyer C, Grosmann R, Mitrakou A, et al. Effects of autonomic neuropathy on counterregulation and awareness of hypoglycemia in type 1 diabetic patients. Diabetes Care. 1998;21:1960–1966. doi: 10.2337/diacare.21.11.1960. [DOI] [PubMed] [Google Scholar]
- 19.Hara M, Wang X, Kawamura T, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta-cells. Am J Physiol Endocrinol Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 20.Kamme F, Salunga R, Yu J, et al. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/S0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 22.Baier G, Telford D, Gulbins E, Yamada N, Kawakami T, Altman A. Improved specificity of RT-PCR amplifications using nested cDNA primers. Nucleic Acids Res. 1993;21:1329–1330. doi: 10.1093/nar/21.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gannon M, Shiota C, Postic C, Wright CV, Magnuson M. Analysis of the Cre-mediated recombination driven by rat insulin promoter in embryonic and adult mouse pancreas. Genesis. 2000;26:139–142. doi: 10.1002/(SICI)1526-968X(200002)26:2<139::AID-GENE12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.V97.1.324. [DOI] [PubMed] [Google Scholar]
- 26.Gotoh M, Maki T, Kiyoizumi T, Satomi S, Monaco AP. An improved method for isolation of mouse pancreatic islets. Transplantation. 1985;40:437–438. doi: 10.1097/00007890-198510000-00018. [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto T, Iwasaki H, Reizis B, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/S1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 28.Leroux L, Desbois P, Lamotte L, et al. Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes. 2001;50(Suppl 1):S150–S153. doi: 10.2337/diabetes.50.2007.S150. [DOI] [PubMed] [Google Scholar]
- 29.Devaskar SU, Singh BS, Carnaghi LR, Rajakumar PA, Giddings SJ. Insulin II gene expression in rat central nervous system. Regul Pept. 1993;48:55–63. doi: 10.1016/0167-0115(93)90335-6. [DOI] [PubMed] [Google Scholar]
- 30.Alarcon C, Serna J, Perez-Villamil B, de Pablo F. Synthesis and differentially regulated processing of proinsulin in developing chick pancreas, liver and neuroretina. FEBS Lett. 1998;436:361–366. doi: 10.1016/S0014-5793(98)01168-5. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D. Peripheral-antigen-expressing cells in thymic medulla: factors in self-tolerance and autoimmunity. Curr Opin Immunol. 1998;10:656–662. doi: 10.1016/S0952-7915(98)80085-X. [DOI] [PubMed] [Google Scholar]
- 32.Blume N, Petersen JS, Andersen LC, et al. Immature transformed rat islet beta-cells differentially express C-peptides derived from the genes coding for insulin I and II as well as a transfected human insulin gene. Mol Endocrinol. 1992;6:299–307. doi: 10.1210/me.6.2.299. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Stoffers DA, Habener JF, Bonner-Weir S. Exendin-4 stimulates both β-cell replication and neogenesis, resulting in increased β-cell mass and improved glucose tolerance in diabetic rats. Diabetes. 1999;48:2270–2276. doi: 10.2337/diabetes.48.12.2270. [DOI] [PubMed] [Google Scholar]
- 34.Itkin-Ansari P, Marcora E, Geron I, et al. NeuroD1 in the endocrine pancreas: localization and dual function as an activator and repressor. Dev Dyn. 2005;233:946–953. doi: 10.1002/dvdy.20443. [DOI] [PubMed] [Google Scholar]
- 35.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing ß-cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 37.Pipeleers DG. Heterogeneity in pancreatic b-cell population. Diabetes. 1992;41:777–781. doi: 10.2337/diabetes.41.7.777. [DOI] [PubMed] [Google Scholar]
- 38.Bosco D, Meda P, Thorens B, Malaisse WJ. Heterogeneous secretion of individual B cells in response to D-gluclose and to nonglucidic nutrient secretagogues. Am J Physiol. 1995;268:C611–C618. doi: 10.1152/ajpcell.1995.268.3.C611. [DOI] [PubMed] [Google Scholar]
- 39.Jonkers FC, Henquin JC. Measurements of cytoplasmic Ca2+ in islet cell clusters show that glucose rapidly recruits b-cells and gradually increases the individual cell response. Diabetes. 2001;50:540–550. doi: 10.2337/diabetes.50.3.540. [DOI] [PubMed] [Google Scholar]
- 40.Bliss CR, Sharp GW. Glucose-induced insulin release in islets of young rats: time-dependent potentiation and effects of 2-bromostearate. Am J Physiol. 1992;263:E890–E896. doi: 10.1152/ajpendo.1992.263.5.E890. [DOI] [PubMed] [Google Scholar]
- 41.Cheng T, Shen H, Giokas D, Gere J, Tenen DG, Scadden DT. Temporal mapping of gene expression levels during the differentiation of individual primary hematopoietic cells. Proc Natl Acad Sci U S A. 1996;93:13158–13163. doi: 10.1073/pnas.93.23.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu M, Krause D, Greaves M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 43.Delassus S, Titley I, Enver T. Functional and molecular analysis of hematopoietic progenitors derived from the aorta-gonad-mesonephros region of the mouse embryo. Blood. 1999;94:1495–1503. [PubMed] [Google Scholar]
- 44.Akashi K, He X, Chen J, et al. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- 45.Herrera PL, Huarte J, Zufferey R, et al. Ablation of islet endocrine cells by targeted expression of hormone-promoter-driven toxigenes. Proc Natl Acad Sci U S A. 1994;91:12999–13003. doi: 10.1073/pnas.91.26.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrera PL, Huarte J, Sanvito F, Meda P, Orci L, Vassalli JD. Embryogenesis of the murine endocrine pancreas; early expression of pancreatic polypeptide gene. Development. 1991;113:1257–1265. doi: 10.1242/dev.113.4.1257. [DOI] [PubMed] [Google Scholar]
- 47.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 48.Herrera PL, Nepote V, Delacour A. Pancreatic cell lineage analyses in mice. Endocrine. 2002;19:267–278. doi: 10.1385/ENDO:19:3:267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 23 kb)
(PDF 23 kb)
FACS analysis of GFP-high and GFP-medium populations. a FACS analysis of double-sorted GFP+ cells, (b) FSC histogram of GFP-high and (c) GFP-medium populations. d Side scatter histogram of GFP-high and (e) GFP-medium populations. The percentages of the cells included in each region vs. total FACS-sorted single cells are indicated in each region (PDF 720 kb).
Minimum copy number of template cDNA fragment that could be detected using the nested PCR primers for the single-cell nested RT-PCR analysis of InsI, Gcg, Sst and Ppy. NTC, no template control (PDF 278 kb).
(PDF 39 kb).
(PDF 31 kb)
(PDF 36 kb)
(PDF 31 kb)
(PDF 31 kb)