Abstract
Dot1 is a conserved histone methyltransferase that methylates histone H3 on lysine 79. We previously observed that in Saccharomyces cerevisiae, a single DOT1 gene encodes two Dot1 protein species. Here, we show that the relative abundance of the two isoforms changed under nutrient-limiting conditions. A mutagenesis approach showed that the two Dot1 isoforms are produced from two alternative translation start sites as a result of leaky scanning by the ribosome. The leaky scanning was not affected by the 5′- or 3′-untranslated regions of DOT1, indicating that translation initiation is determined by the DOT1 coding sequence. Construction of yeast strains expressing either one of the isoforms showed that both were sufficient for Dot1’s role in global H3K79 methylation and telomeric gene silencing. However, the absence of the long isoform of Dot1 altered the resistance of yeast cells to the chitin-binding drug Calcofluor White, suggesting that the two Dot1 isoforms have a differential function in cell wall biogenesis.
INTRODUCTION
Dot1 is a histone methyltransferase that was originally identified in Saccharomyces cerevisiae. The enzyme, which is conserved from yeast to humans, catalyzes mono-, di- and trimethylation of lysine 79 on histone H3 (H3K79), a residue located on the nucleosome core (1–4). In yeast, Dot1 methylates ∼90% of histone H3 and is involved in gene silencing, activation of the DNA damage response and the pachytene checkpoint in meiosis (1,5–7). In addition, genetic interactions have been reported between DOT1 and different genes involved in cell wall biogenesis, suggesting a function for Dot1 in this process (8,9). In mammals, Dot1 plays a role in Ras-induced gene silencing and aldosterone-induced gene repression as well as in inappropriate gene activation in certain types of leukemia (10–13).
Our antibodies directed against yeast Dot1 detect two protein species on immunoblots, even though the protein is encoded by a single gene (1). Several mechanisms can result in the formation of two different protein species from one gene. First, full-length proteins can be processed by proteases, resulting in the generation of N- or C-terminally truncated forms. Second, several posttranslational modifications such as phosphorylation or ubiquitination can result in an altered mobility of proteins on immunoblots. Finally, different protein isoforms can be generated by alternative translation, which can be caused by variations in the mRNA sequence (for example due to alternative mRNA splicing or alternative transcription start site selection) or by the usage of multiple translation start sites. Translation from multiple start codons can occur when the mRNA contains an internal ribosomal entry site (IRES), or when the ribosome scanning over the mRNA molecule skips the first start codon and subsequently initiates translation from a downstream alternative start codon, a phenomenon called ‘leaky scanning’ (14). Alternatively, the ribosome can bind to the mRNA on the 5′-side of the start codon via the normal mechanism, but then jumps over the first start codon to initiate transcription from a downstream start codon, a process called ribosome shunting (15). Translation start site selection is a highly regulated process that is critical for differential gene regulation in eukaryotes (16). Mutations that affect translation initiation have been associated with a range of genetic diseases in humans (17). However, there are still many open questions regarding the molecular mechanisms of start codon selection (18).
In this study, we determined the mechanism by which the two Dot1 isoforms are generated. We show by protein tagging, protein truncation and mutational analysis that they are the result of leaky scanning by the ribosome. This enabled the construction of yeast strains expressing either one of the isoforms. The two Dot1 isoforms were found to have indistinguishable functions in global methylation and gene silencing. However, they showed distinct functions in resistance to the chitin-binding molecule Calcofluor White (CFW), suggesting that they play different roles in cell wall biogenesis. Our results suggest that leaky scanning of the DOT1 mRNA is affected by sequences downstream of the start codon, whereas the efficiency of translation initiation is usually thought to depend mainly on the 5′-context of the start codon (14). The discovery of unconventional leaky scanning events like the one reported here will be instrumental to unravel the complex mechanisms that regulate translation initiation in yeast and higher eukaryotes.
MATERIALS AND METHODS
Yeast strains, plasmids and media
Yeast strains and plasmids used in this study are listed in Table 1. Yeast media were described previously (1). Silencing assays were performed using media containing 1 g/l 5-fluoroorotic acid (FOA) (Toronto Research Chemicals). CFW sensitivity of cells was tested using YEP media containing 20 µg/ml CFW (Sigma). Cycloheximide (Sigma) was added to the media at a concentration of 50 µg/ml to inhibit protein synthesis. NKI3031 was derived from a cross between BY4740 and BY4742 (19). NKI3049 and NKI3066 were derived from NKI3031 by integration of the GAL1 promoter in front of the DOT1 gene using plasmid pYM-N22 and PCR-mediated gene replacement (20); in NKI3049 the GAL1 promoter was integrated such that 62 bp of the endogenous DOT1 5′-untranslated region was maintained, whereas in NKI3066 the endogenous DOT1 5′-UTR was replaced by GAL1 and vector sequence present in pYM-N22. NKI1060 was constructed from UCC7164 using PCR-mediated gene replacement and plasmid pFA6a-KanMX-PGAL1-3HA (21). The single-copy pLEU2-DOT1 vector (pDOT1) and the multi-copy GAL1-promoter pTRP1-DOT1 plasmid (pFvL18) were described previously (1). Mutations in the DOT1 gene in these plasmids were introduced by Quikchange (Stratagene) or a three-step PCR protocol and verified by sequencing. Details of the mutant DOT1 genes are shown in Figure 2A. pFvL201 was generated by replacing the first methionine of DOT1 in pDOT1 by a single FLAG tag followed by a SGGSGG spacer. pFvL212 was derived from pRS314-DOT1 (single-copy pTRP1-DOT1) by integration of a GSGGSGGS spacer, single HA TAG and TAP tag behind the DOT1 gene; the TAP tag sequence was amplified from pBS1479 (22). pFvL215 was generated by deletion of amino acids 4–20 from pFvL212 by gap-repair. pFvL221 was generated from pFvL18 by integration of a GSGGSGGS linker behind the DOT1 gene. pTW055 was derived from pFvL221 by insertion of the yeast-enhanced citrine tag from plasmid pKT211 (23) behind the linker using an AatII-AscI digest. Mutations in pTW055 were generated by integrating the SalI-EcoRI fragment from pGH03 and pGH04 in SalI-EcoRI digested pTW055 to generate pGH024 and pGH025, respectively.
Table 1.
Yeast strains and plasmids used in this study
| Strain | Genotype | References |
|---|---|---|
| UCC7164 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL | (26) |
| UCC7183 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL dot1Δ::KanMX | (26) |
| NKI1059 | MATa lys2Δ0 trp1Δ63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 ADE2-TEL-VR URA3-TEL-VIIL dot1Δ::KanMX-PGAL1-3HA-DOT1 | (26) |
| NKI1060 | MATa lys2Δ0 trp1Δ63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 ADE2-TEL-VR URA3-TEL-VIIL dot1Δ::KanMX-PGAL1-3HA-dot1(Δ1-17) | This study |
| NKI3031 | MATa leu2Δ0 lys2Δ0 ura3Δ0 | This study |
| NKI3049 | MATa leu2Δ0 lys2Δ0 ura3Δ0 dot1Δ::KanMX-PGAL1-DOT1a | This study |
| NKI3066 | MATa leu2Δ0 lys2Δ0 ura3Δ0 dot1Δ::KanMX-PGAL1-DOT1a | This study |
| YSC-1122 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kre1Δ::KanMX | Open biosystems |
| Plasmid | Description | References |
| pRS315 | LEU2 CEN | (1) |
| pDOT1 | LEU2 CEN DOT1 | (1) |
| pGH01 | LEU2 CEN dot1-M1T | This study |
| pGH02 | LEU2 CEN dot1-M17L | This study |
| pFF018 | LEU2 CEN dot1-I7M | This study |
| pFF025 | LEU2 CEN dot1-initiation mutant 1b | This study |
| pFF026 | LEU2 CEN dot1-initiation mutant 2b | This study |
| pFF027 | LEU2 CEN dot1-initiation mutant 3b | This study |
| pGH18 | LEU2 CEN dot1-frameshift mutant 1c | This study |
| pGH21 | LEU2 CEN dot1-frameshift mutant 2c | This study |
| pFvL201 | LEU2 CEN FLAG-spacer-DOT1d | This study |
| pFF028 | LEU2 CEN DOT1 (A-6C, A04C, G4U, G5U, C6A) | This study |
| pFF029 | LEU2 CEN DOT1 (C60G) | This study |
| pFF030 | LEU2 CEN DOT1 (C6U) | This study |
| pFvL212 | TRP1 CEN DOT1-spacer-HA-TAPe | This study |
| pFvL215 | TRP1 CEN dot1(Δ4-20)-spacer-HA-TAPe | This study |
| pTCG | TRP1 2µ PGAL1 | (1) |
| pFvL18 | TRP1 2µ PGAL1-DOT1 (G8A) | (1) |
| pGH03 | TRP1 2µ PGAL1-dot1-M1T | This study |
| pGH04 | TRP1 2µ PGAL1-dot1-M17L | This study |
| pGH06 | TRP1 2µ PGAL1-DOT1 (G8A, U22A, C23G, A24U) | This study |
| pFvL221 | TRP1 2µ PGAL1-DOT1-linkerf | This study |
| pTW055 | TRP1 2µ PGAL1-DOT1-linker-YECitrinef | This study |
| pGH024 | TRP1 2µ PGAL1-dot1-M1T-linker-YECitrinef | This study |
| pGH025 | TRP1 2µ PGAL1-dot1-M17L-linker-YECitrinef | This study |
aThe DOT1 gene is flanked by its endogenous 5′-context in NKI3049; in NKI3066 the 5′-sequence has been replaced by vector sequence (pYM-N22) and 5′-sequence of the GAL1 gene.
bThe following point mutations were introduced in the three initiation mutants (numbers are relative to A of AUG1): G–3 to A–3 in pFF025; G+5 to C+5 in pFF026 and the double mutation G–3 to A–3 and G+5 to C+5 in pFF027.
cThe following base insertions (bold-faced and underlined) were made in the FSMs:
FSM1 (pGH18): ATG GGC GGT CAA GAA AGT ATA TCA AAA TAA • (TAA, termination codon)
FSM2 (pGH21): ATG GGC GGT CAA GAA AGT ACT ATC AAA TAA • (TAA, termination codon)
dpFvL201 contains a flexible spacer between the FLAG tag and the Dot1 protein, consisting of the amino acids SGGSGG.
epFvL212 and pFvL215 contain a flexible spacer between the Dot1 protein and the HA tag, consisting of the amino acids GSGGSGGS.
fpFvL221, pTW055, pGH024 and pGH025 contain a linker sequence at the very C-terminus of the Dot1 protein, consisting of the amino acids GSGGSGGS.
Figure 2.
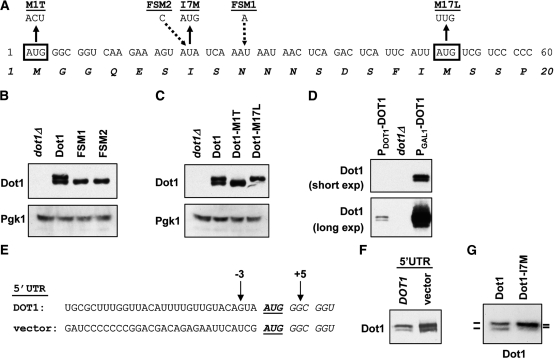
The two Dot1 isoforms are generated by leaky scanning by the ribosome, which is not dependent on the 5′-context of the start codon. (A) Nucleotide and amino acid sequence of the N-terminal part of Dot1. Black boxes show the two potential start codons. The different mutants used in this study are shown underlined. Briefly, in the M1T mutant AUG1 is changed to ACU, in the I7M mutant AUA7 is changed to AUG and in the M17L mutant AUG17 is changed to UUG. FSM1 has an insertion of an A in codon 9 and FSM2 has an insertion of a C in codon 7. Sequence details of the FSMs are shown in Table 1. (B) Immunoblot analysis of FSMs of Dot in a dot1Δ strain (UCC7183) transformed with an empty plasmid (pRS315), a single copy plasmid expressing DOT1 (pDOT1), or a single copy plasmid expressing one of the two FSMs of DOT1 (pGH18 or pGH21). Pgk1 was used as a loading control. (C) Immunoblot analysis of Dot1 expression in dot1Δ cells (UCC7183) containing an empty plasmid, wild-type Dot1, Dot1-M1T or Dot1-M17L (pRS315, pDOT1, pGH01 and pGH02, respectively), showing that mutation of either of the putative start codons leads to expression of a single Dot1 isoform. Pgk1 was used as a loading control. (D) Comparison of Dot1 expression in a wild-type strain (PDOT1-DOT1, UCC7164) and a dot1Δ strain (UCC7183) containing an empty vector (pTCG) or a plasmid overexpressing DOT1 (and 54 bp of its 5′-UTR) from the GAL1 promoter (PGAL1-DOT1, pFvL18). A short and a long exposure of the same blot are shown. (E) Nucleotide sequence around the DOT1 start codon in strains NKI3049 (endogenous DOT1 5′-UTR) and NKI3066 (non-yeast vector 5′-UTR). Thirty nucleotides of 5′-UTR sequence are shown. In strain NKI3049 (DOT1) 62 bp of endogenous DOT1 5′-UTR remains upstream of the first start codon; in strain NKI3066 (vector) the DOT1 5′-UTR has been removed by insertion of the GAL1 promoter and 45 bp of vector sequence of the integration construct pYM-N22. (F) Immunoblot comparison of Dot1 expression in the two strains described in E. (G) Immunoblot analysis of Dot1 expression in a dot1Δ strain (UCC7183) transformed with wild-type Dot1 (pDOT1) or Dot1-I7M (pFF018).
UV irradiation
Exponentially growing cells (OD660 ∼0.5) were arrested in G1/S with 5 µg/ml α-factor for 2 h, spun, resuspended in fresh media containing α-factor and plated on 14 cm Petri dishes. The plates were irradiated with 100 J/m2 UV light of 254 nm using a Stratalinker (Stratagene). Trichloroacetic acid protein extracts (24) were prepared 30 min after irradiation and compared with non-irradiated controls.
Immunoblots
Whole-cell extracts were obtained from ∼5 × 107 cells by the classical glass beads breakage method using 200 μl of glass beads and SUMEB lysis buffer (25) complemented with PMSF (1 mM), benzamidine (5 mM), pepstatin (1 µg/ml), leupeptin (1 µg/ml) and DTT (1 µM). Primary antibody incubations were performed overnight in Tris-buffered saline‐Tween with 2% dry milk. The rabbit polyclonal antibodies against Dot1 (1) and H3K79me1, -me2 and -me3 (26) were described earlier. Commercially available antibodies that were used in this study are H3 (ab1791, Abcam), Pgk1 (A-6457, Invitrogen) and Rad53 (sc-6749, Santa Cruz).
RNA structure prediction
The Vienna RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) was used to predict secondary RNA structures and the effect of mutations in the DOT1 RNA sequence (27). Analysis of different sized fragments from the DOT1 region –10 to + 200 revealed a common predicted structure of the 5′-part of the DOT1 mRNA (–10 to +65).
Confocal microscopy
Log phase cells were fixed for 10 min in 4% formaldehyde in PBS, after which they were incubated with Hoechst 33342 (Invitrogen, 1 μg/ml) for 15 min to stain the nucleus. Cell wall staining was performed by incubation of the cells with ConA-Cy5 for 15 min. Cells were mounted in Vectashield mounting medium (Vector Laboratories). Samples were analyzed with a confocal laser scanning microscope (AOBS, Leica) equipped with a HCX PL APO lbd.bl 63×/NA 1.4 oil corrected objective lens (Leica). The acquisition software used was Leica LCS. Cells were imaged using a 405 nm, a 516 nm and a 633 nm laser to visualize Hoechst, Citrine and Cy5, respectively.
Electron microscopy
Log phase cells were fixed in Karnovsky fixative containing 0.15% Ruthenium Red (BDH chemical) for 5 min at room temperature, for marking the outer membranes of the cells. After washing with 0.1 M cacodylate buffer pH 7.2 the cells were postfixed with 1% osmiumtetroxide, en-block stained with 0.5% uranylacetate (Ultrastain 1, Leica) followed by dehydration series and embedding in EMBed 812 (Aurion). The sections were examined using a CM10 microscope (FEI).
RESULTS
Two isoforms of Dot1 in S. cerevisiae
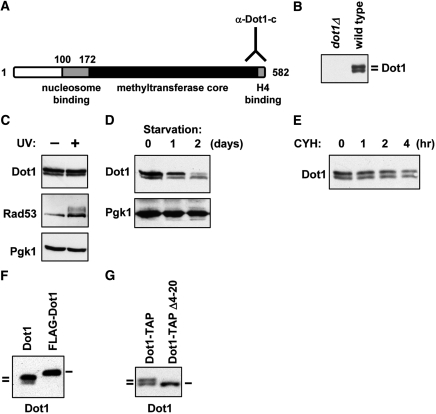
An antibody directed against the C-terminal part of the yeast Dot1 protein (Figure 1A) recognizes two discrete proteins on immunoblots (Figure 1B). The slow-migrating protein is usually more abundant than the fast-migrating protein. The goal of this study was to determine the identity, regulation and the function of the two protein isoforms of Dot1. Since Dot1 is involved in activation of the DNA damage checkpoint upon exposure to UV light (6), we tested whether the relative abundance of the two proteins was affected by DNA damage. Thirty minutes after UV irradiation, there was no detectable difference in relative abundance of the two isoforms, while phosphorylation of the central checkpoint kinase Rad53 confirmed activation of the DNA damage checkpoint (Figure 1C). To investigate whether the production of the two isoforms was regulated by growth conditions, exponentially growing cells were compared with cells arrested by nutrient depletion. After starving the cells for two days, the abundance of the slow-migrating Dot1 protein was reduced while the fast-migrating protein was unaffected (Figure 1D). This change in the ratio of the Dot1 isoforms probably does not reflect a difference in protein stability, since the two isoforms showed approximately the same rate of degradation when protein synthesis was blocked by addition of cycloheximide (Figure 1E). These results show that the relative abundance of the two Dot1 isoforms is dependent on growth conditions.
Figure 1.
Formation of two Dot1 isoforms depends on the N-terminus. (A) Schematic overview of the structure of the yeast Dot1 protein with a lysine-rich putative nucleosome binding domain, a methyltransferase core and a C-terminal domain that binds to histone H4. The Dot1 antibody is directed against the very C-terminus of the protein. Numbering of amino acid residues starts at the first methionine, a residue that is most likely absent from the mature protein due to co-translational cleavage. (B) Immunoblot analysis of Dot1 expression shows that two isoforms are present in log phase cultures. Whole-cell extracts of a dot1Δ strain (UCC7183) transformed with an empty plasmid (pRS315) or a single copy plasmid expressing DOT1 (pDOT1) were analyzed. (C) Immunoblot analysis of Dot1 expression in wild-type cells (NKI3031) following UV irradiation of cells arrested in G1. The mobility shift of Rad53 caused by UV-induced phosphorylation was used to confirm activation of the DNA-damage checkpoint. Pgk1 was used as a loading control. (D) Whole-cell lysates of wild-type strain NKI3031 were prepared from exponentially growing cultures and cultures of starved cells grown overnight or over two nights. Dot1 expression was analyzed by immunoblot and Pgk1 was used as a loading control. (E) To analyze the protein half life of the two Dot1 isoforms, 50 µg/ml cycloheximide was added to a culture of exponentially growing dot1Δ cells transformed with a DOT1 overexpression plasmid (UCC7183 + pFvL18). At the indicated time points aliquots of the culture were taken for immunoblot analysis of Dot1 expression. (F) Immunoblot analysis of Dot1 expression in cells expressing full-length Dot1 without or with an N-terminal FLAG epitope tag (UCC7183 + pDOT1 or pFvL201). (G) Dot1 expression in cells containing full-length Dot1 (UCC7183 + pFvL212) or a deletion mutant of Dot1 (Δ4-20, UCC7183 + pFvL215) was analyzed by immunoblot. Both proteins have a C-terminal TAP tag.
A long and a short Dot1 protein
Our aim was to express the two Dot1 isoforms separately in yeast, to determine whether the existence of the two isoforms and the differential expression is relevant for the function of Dot1. Therefore, first the mechanism of production of the two isoforms was determined. Previous results had shown that the addition of a single N-terminal FLAG tag to the Dot1 protein resulted in a shift in mobility as well as expression of only one Dot1 isoform [Figure 1F, (26)]. In contrast, introduction of a TAP tag at the C-terminus of Dot1 (and the concomitant replacement of the DOT1 3′-UTR with that of the ADH1 gene) did not affect the expression of the two isoforms (Figure 1G). However, an internal deletion of amino acids 4–20 (Dot1Δ4–20) resulted in the expression of only one Dot1 isoform (Figure 1G). These results indicated that the protein sequence or the coding sequence of the first 20 amino acids was involved in the expression of the two isoforms. These results also suggested that the production of two isoforms was not caused by a posttranslational modification of Dot1 because the double band was disrupted by two non-overlapping mutations: N-terminal tagging (which affects the N-terminal residue of Dot1; Figure 1F) as well as an internal deletion (which leaves the first three N-terminal residues of Dot1 intact; Figure 1G).
The Dot1Δ4–20 protein migrated at approximately the same size as the smaller isoform of Dot1 (Figure 1G), suggesting that the slower migrating Dot1 band might represent the full-length protein and the fast-migrating band a Dot1 protein that is missing part of the N-terminus. A smaller protein can be generated by degradation or protein processing of a larger protein or by alternative translation. Inspection of the coding sequence of Dot1 showed that it contains a second in frame AUG codon (AUG17), 16 codons downstream of the first AUG (AUG1) (Figure 2A). Alternative translation initiation at this second start codon would give rise to a Dot1 protein 16 amino acids shorter than the full-length protein. To distinguish between protein processing and alternative translation as the mechanisms responsible for Dot1 isoform production, point mutations were introduced in the DOT1 gene expressed from a single-copy plasmid. The wild-type and mutant plasmids were then introduced in dot1Δ strains. First two frameshift mutants (FSMs) were generated by introduction of an extra base between the first and the second AUG (FSM1 and FSM2, Figure 2A). Both of these insertions result in a stop codon at amino acid position 10, which prevents the generation of full-length Dot1. We reasoned that if the smaller protein is generated by processing of the full-length protein, the frameshift mutations should destroy production of both full length and the smaller Dot1 protein. In contrast, if it is generated by alternative translation, frameshift mutations should not affect production of the smaller Dot1 isoform. As shown in Figure 2B, when the FSMs were expressed in yeast, the short isoform was still present whereas the larger isoform was absent. These experiments rule out the possibility that the smaller Dot1 protein is a processed form of the full-length protein.
Two Dot1 isoforms as a result of alternative translation
To confirm that the two Dot1 isoforms are generated as different translation products from the two start codons present in the 5′-part of the DOT1 coding sequence the first or the second AUG was mutated. Mutation of AUG1 to ACU, changing the initiator Met codon to a non-initiator Thr codon (Dot1-M1T) resulted in the expression of a single protein that had the same size as the small isoform (Figure 2C), confirming that translation can initiate at a downstream start codon. Mutation of AUG17 to UUA, changing the initiator Met codon to a Leu codon (Dot1-M17L) generated a single protein with the same size as the large isoform (Figure 2C), showing that translation starts at AUG1 as well as AUG17. These results suggest that the two isoforms are indeed two translation products starting at M1 or M17, respectively.
Next, the cause of alternative translation was investigated. Two translation products can be generated from two different transcript molecules: one initiated upstream of the first AUG, and one initiated in between the first and second AUG. Alternatively, they can be generated from a single long transcript by alternative start codon usage. To distinguish between these two possibilities, the endogenous DOT1 promoter was replaced by the GAL1 promoter, which is induced when cells are grown in media containing galactose. In this case, 54 bp of the DOT1 5′-UTR was retained to avoid changes in the context of the start codon. When Dot1 was expressed from the GAL1 promoter, the two isoforms were generated in the same ratio as found when the gene was expressed from its endogenous promoter (Figure 2D). Since it is unlikely that two unrelated promoters use identical transcription start sites (i.e. upstream of the first AUG and in between the first and second AUG) and with identical relative frequencies, this result suggests that the two isoforms are not generated by alternative transcription start site selection.
Alternative translation can be caused by ribosome shunting, by leaky scanning of the ribosome or by an IRES. In ribosome shunting, which has mainly been described for viruses, the ribosome binds to the mRNA on the 5′-side of the start codon via the normal mechanism, but then `jumps’ over the first start codon to initiate transcription from a downstream start codon (15). In leaky scanning, the ribosome does scan over the first start codon but does not recognize it in a significant fraction of initiation events (14). An IRES is an mRNA sequence that has special structural properties allowing the recruitment of ribosomes without using the standard mechanism of translation initiation (28).
To address the mechanism of alternative translation, the DOT1 promoter and the complete 5′-UTR were replaced by an exogenous GAL1 promoter and 45 bp of non-yeast vector sequence immediately upstream of the open reading frame of DOT1 (Figure 2E). As a control, the DOT1 promoter was replaced by the GAL1 promoter cassette such that 62 bp of the endogenous DOT1 5′-UTR was retained (Figure 2E). When transcription from the GAL1 promoter was induced, the two isoforms were generated in approximately the same ratio in both strains (Figure 2F), suggesting that the 5′-context of the start codon did not affect the initiation of translation. Therefore, ribosome shunting was not a likely cause of the alternative translation. We note that the overall Dot1 abundance was higher in the presence of the heterologous 5′-UTR (Figure 2F), indicating that the 5′-UTR might affect mRNA stability or transcription.
Most IRES elements are over 100 bp long, even though the minimal IRES modules may be shorter (28). The few bases between the first and second start codon of Dot1 seem insufficient to constitute an IRES. Leaky scanning by the ribosome thus seemed to be the most plausible explanation for the generation of the two isoforms. To verify this, a new start codon was inserted in between AUG1 and AUG17 at position 7 by changing the Ile codon to an initiator Met codon (I7M, Figure 2A). If an IRES element was the cause of the alternative translation, the Dot1-I7M mutation could have two different effects. This mutation might interfere with the activity of the IRES element, resulting in loss of expression of the shorter isoform. Alternatively, the mutation could leave initiation at AUG17 unaffected, which would lead to normal expression of the two isoforms. In contrast, if leaky scanning was the cause of the alternative translation, skipping of AUG1 would lead to translation initiation at the new start codon AUG7 and thereby prevent translation start at AUG17. When the Dot1-I7M mutant was expressed two isoforms were produced, but the smaller one had become slightly larger (Figure 2G). Although the effect of the 10 amino acid size difference was small, we consistently detected a slower migration of the short isoform in the Dot1-I7M mutant in multiple independent experiments (data not shown). This result shows that translation initiated at the new start codon, indicating that the two isoforms of Dot1 result from leaky scanning by the ribosome.
More support for this model comes from the Dot1-M1T and Dot1-M17L mutants in which only one of the isoforms is expressed. In the wild-type DOT1 transcript skipping of the first AUG occurs in a fraction of the translation events. However, if the first AUG is mutated, every ribosome will start translation from AUG17, which is expected to lead to an increase in abundance of the short Dot1 protein. This is indeed what was observed in the Dot1-M1T mutant (Figure 2C). This was not the case in the FSMs, in which AUG1 was still present (Figure 2B). As expected, in the Dot1-M17L mutant the larger isoform was expressed at the same level as in wild-type cells, indicating that mutation of the downstream AUG had no major effects on translation initiation at AUG1 (Figure 2C). We note that no smaller isoforms of Dot1 are detected in this mutant suggesting that no proteins were produced by translation initiation at a downstream start codon such as AUG40.
Mechanism of leaky scanning
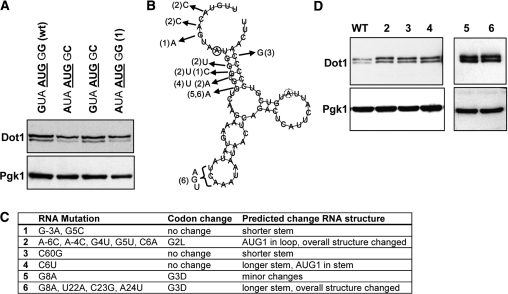
Little is known about factors that affect leaky scanning. The 5′-UTR of the DOT1 gene is unlikely to be involved in determining the efficiency of translation initiation because replacement of the complete DOT1 5′-UTR by an exogenous sequence did not significantly affect the rate of leaky scanning (Figure 2F). The efficiency of translation initiation is thought to be mainly determined by the nucleotide sequence upstream of the start codon, as a consequence of the linear scanning of the ribosome along the mRNA molecule (14). The Kozak consensus sequence defines the optimal context for translation initiation in mammals (29,30), but the requirements in yeast seem less strict (31,32). Recently, a genome-wide computational study showed that there is a strong bias in the yeast genome for an A at position –3 and a C at position +5. Presence of these residues was also correlated with the strength of expression (33). According to these observations AUG1 of the DOT1 gene is not located in an optimal context, with a G at position −3 and a G at position +5 (Figure 2E). In contrast, AUG17 does have an optimal sequence context. To test if the −3 and +5 positions determine the efficiency of translation initiation of AUG1 of DOT1, they were mutated to the optimal A–3 and C + 5, both separately and in combination. However, in these mutants the two Dot1 isoforms were still produced with approximately the same ratio as in wild-type cells (Figure 3A), showing that the sub-optimal proximal context of AUG1 is not the cause of leaky scanning.
Figure 3.
Effect of sequences proximal to the start codon on leaky scanning. (A) Immunoblot analysis of Dot1 expression in strains in which the context of AUG1 has been mutated. The nucleotide sequence is indicated above the lanes. Wild-type cells (UCC7164) are compared with cells harboring the mutation G–3→A–3, G + 5→C + 5, or the double mutation (UCC7183 + pFvL025, pFvL026 and pFvL027, respectively). Numbers are relative to the first nucleotide of AUG1. Pgk1 was used as a loading control. (B) RNA structure of DOT1 mRNA (–10 to +65) predicted by RNAfold (27). Solid circle indicates start of AUG1, dashed circle indicates start of AUG17. Mutant numbers refer to panel C (2–6). (C) Summary of mutations in DOT1 mRNA. (D) Dot1 expression of mutants shown in panel C. Numbers 1–4 are mutants of single copy pDOT1, number 6 is a mutant of PGAL1-DOT1 (number 5) which already contained a G8A mutation.
Another possibility is that the structure of the Dot1 RNA molecule affects the efficiency of translation initiation. Although this structure has not been experimentally determined, the RNA fold algorithm (27) predicts that the region of the mRNA around AUG1 contains a stem loop, which might somehow impair the binding of initiation factors or the polymerase complex to this sequence (Figure 3B). To test this idea, mutations were made that were predicted to disrupt the overall structure of this part of the DOT1 mRNA (Figure 3B and C). Although these mutations slightly increased the expression of Dot1, none of the mutations affected the rate of leaky scanning (Figure 3A and D). This mutagenesis study suggests that inefficient initiation of AUG1 is not caused by sequences or RNA structures proximal to the initiator codon.
Functions of the two Dot1 isoforms
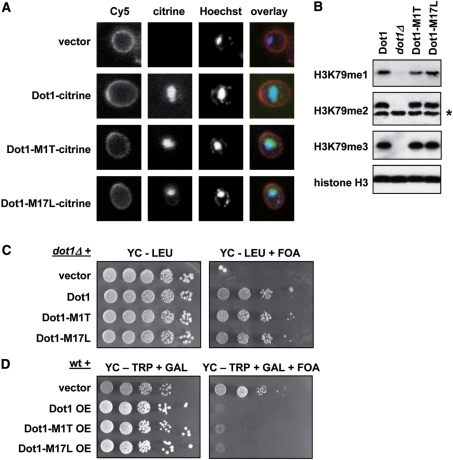
Having addressed the origin of the two Dot1 isoforms, we designed yeast strains that express both or either one of the two isoforms to determine what the function of the Dot1 N-terminus might be. It has been shown that alternative translation in yeast can lead to production of two protein isoforms with or without N-terminal targeting sequences, leading to differences in sub-cellular localization (34–36). To visualize the localization of the two isoforms, plasmids were constructed expressing wild-type Dot1, Dot1-M1T or Dot1-M17L fused to a citrine fluorescent tag (a variant of YFP, 37). Since endogenous Dot1 expression is very low (7,38), the fusion proteins were expressed from a GAL1 promoter. In addition, the yeast cell wall was visualized using ConA-Cy5 and DNA was stained with Hoechst dye. The wild-type Dot1 protein as well as the two mutants showed a diffuse nuclear staining and seemed to be absent from the cytoplasm (Figure 4A), suggesting that the N-terminus of Dot1 does not influence its nuclear localization.
Figure 4.
The N-terminus of Dot1 does not affect localization, H3K79 methylation or telomeric silencing. (A) Analysis of the localization of wild-type Dot1, Dot1-M1T and Dot1-M17L. Log-phase cells overexpressing one of these proteins fused to a fluorescent citrine tag were stained with ConA-Cy5 (cell wall) and Hoechst (DNA) and then imaged using a confocal microscope. For this experiment dot1Δ strain UCC7183 was transformed with pFvL221, pTW055, pGH024 and pGH025. (B) Whole-cell extracts from log-phase cultures were prepared from dot1Δ strain UCC7183 transformed with an empty plasmid or a single copy plasmid expressing Dot1, Dot1-M1T or Dot1-M17L (pRS315, pDOT1, pGH01 and pGH02, respectively). H3K79 methylation and total histone H3 were analyzed by immunoblot. The asterisk indicates a non-specific band recognized by the H3K79me2 antibody. (C) Telomeric silencing of a URA3 reporter gene close to telomere VIIL was measured in the strains described in (B). Cells were plated in 10-fold dilution series on selective media in the presence or absence of 1 g/l 5-FOA. Cells that silence URA3 are resistant to FOA; cells with loss of telomeric silencing express URA3 and are sensitive to FOA. (D) Analysis of telomeric silencing in a wild-type strain (UCC7164) transformed with pTCG, pTCG-DOT1 (pFvL18), pTCG-dot1-M1T (pGH03) or pTCG-dot1-M17L (pGH04). The GAL1 promoter on the pTCG plasmid was induced by growth on selective media containing galactose (GAL) to overexpress (OE) the Dot1 proteins.
Dot1 methylates H3K79 throughout the entire yeast genome and the global levels of this modification determine the strength of telomeric silencing (1,2,26). To investigate whether the N-terminus affects the catalytic activity of Dot1, the levels of methylation of H3K79 were analyzed in cells expressing wild-type Dot1 or only one of the isoforms. Immunoblots using antibodies specific for mono-, di- and trimethylated H3K79 showed no detectable difference in global methylation when one of the Dot1 isoforms was missing (Figure 4B). In line with the fact that no effect on the global levels of H3K79 methylation was detected, either isoform alone was sufficient to complement the telomeric silencing defect in a dot1Δ strain, when expressed from a single copy plasmid (Figure 4C). Both isoforms were also as competent as wild-type Dot1 to disrupt telomeric silencing when overexpressed from the GAL1 promoter on a multi-copy plasmid (Figure 4D). In addition, there was no difference in activation of the DNA damage response after UV irradiation (data not shown).
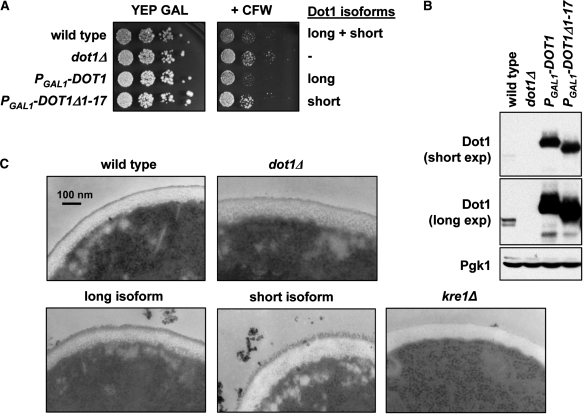
Genetic interactions of DOT1 indicate that Dot1 might have a function in cell wall biology (8,9). To verify this notion, the sensitivity to the antifungal agent CFW was examined of yeast cells with or without Dot1. Altered sensitivity to CFW is an indication of a defect in chitin biosynthesis and/or cell wall biogenesis (39,40). When yeast cells were grown on rich media containing galactose cells lacking Dot1 were more resistant to CFW (Figure 5A), lending support to the idea that Dot1 is required for cell wall function. To examine whether the two Dot1 isoforms might have differential functions in cell wall biogenesis, we tested whether strains expressing either one of the isoforms showed altered sensitivity to CFW. Two strains were constructed in which the GAL1 promoter and a 3xHA tag were integrated either upstream of AUG1 of the endogenous DOT1 coding sequence (PGAL1-3HA-DOT1) or immediately downstream of AUG17, thereby deleting the first 17 amino acids (PGAL1-3HA-DOT1Δ1-17). In both cases, a single protein species was expressed (Figure 5B). These promoter-integration strains were used instead of strains expressing Dot1 from a plasmid since CFW is unstable in synthetic media necessary to ensure propagation of the plasmids. The strain expressing only full-length Dot1 from the GAL1 promoter was as sensitive to CFW as the wild-type strain. In contrast, the strain expressing only the short form of Dot1 was more resistant to CFW, just like a dot1Δ strain (Figure 5A). This result suggests that the N-terminus of Dot1 influences its cell wall-related function.
Figure 5.
Dot1 is involved in cell wall function. (A) Sensitivity to CFW of the following strains: wild type (UCC7164), dot1Δ (UCC7183), PGAL1-3xHA-DOT1 (NKI1059) and PGAL1-3xHA-DOT1Δ1-17 (NKI1060). Cells were plated in 10-fold dilutions on rich media containing 2% galactose in the presence or absence of 20 µg/ml CFW. The image is representative of four independent experiments. (B) Immunoblot analysis of Dot1 expression in the strains described in (A). A long and short exposure is shown to indicate the expression relative to endogenous Dot1. Pgk1 was used as a loading control. (C) The cell wall of the strains described in (A) was imaged using electron microscopy. A kre1Δ strain (YSC-1122) was used as a control.
To further investigate the possible cell wall-related function of Dot1, the sensitivity of wild-type or mutant cells to changes in osmolarity was tested by growing cells on media containing different concentrations of NaCl, sorbitol, glycerol or SDS. However, no differences were observed for any of the conditions (Supplementary Figure 1). To assess a possible function for Dot1 in cell wall biogenesis more directly, electron microscopy was used to visualize the yeast cell wall in wild-type cells, dot1Δ cells and cells expressing only full-length Dot1 or only the short isoform from the GAL1 promoter. As a control, kre1Δ cells were used, which have been reported to lack the outer layer of the cell wall (41), as is also clear from Figure 5C. Whereas the cell wall of wild-type cells and dot1Δ cells has a finely delineated outer layer, this layer was somewhat less structured and more rough in texture in cells overexpressing only full-length Dot1, and especially in cells overexpressing only the short Dot1 isoform (Figure 5C). This result supports the notion that Dot1 affects cell wall metabolism. However, this function of Dot1 is found only when Dot1 is overexpressed and is not differentially affected by the two isoforms.
DISCUSSION
Yeast cells express two isoforms of the H3K79 methyltransferase Dot1. Using mutational analysis, we determined that alternative translation by leaky scanning results in the expression of a full-length protein and a shorter Dot1 protein lacking the first 16 amino acids. Starved cells lose expression of full-length Dot1 and maintain the short Dot1 protein (Figure 1D), suggesting that the leaky scanning of the DOT1 transcript is dependent on growth conditions.
The two isoforms showed indistinguishable localization, bulk H3K79 methylation and silencing properties, indicating that the global activity of Dot1 is not affected by its N-terminus. However, yeast cells expressing either one of the two Dot1 isoforms showed a modest but reproducible difference in sensitivity to CFW. Since CFW has a high affinity for chitin in the cell wall of fungi, cells that are resistant to CFW are thought to have a defect in chitin biosynthesis and/or cell wall biogenesis (39,40). Interestingly, the Dot1 homolog in Drosophila, grappa, has also recently been implicated in stress resistance (42). Our results suggest that cells expressing the wild-type Dot1 products or only the full-length protein are more sensitive to CFW than cells lacking Dot1 or expressing the short Dot1 protein only. Thus, the Dot1Δ1-17 protein does not complement the loss of Dot1, suggesting that the sensitivity to CFW is mediated by the N-terminal part that is missing in the Dot1 protein synthesized after leaky scanning. Inspection of the cell wall by electron microcopy revealed no difference between wild-type and dot1Δ cells, suggesting that the CFW resistance is not caused by large structural re-arrangements of the cell wall. However, the outer layer of the cell wall had a slightly altered structure in cells overexpressing either isoform of Dot1 compared to wild-type or dot1Δ cells. This effect was strongest in cells expressing only the short Dot1 isoform. These results suggest that loss of the Dot1 N-terminus causes subtle changes in cell wall metabolism probably related to chitin deposition, whereas overexpression of Dot1 causes more pronounced changes of the cell wall.
Two previous studies have reported genetic interactions between DOT1 and genes involved in cell wall assembly and chitin synthesis. Deletion of DOT1 is synthetically lethal with deletion of either CHS5, encoding a protein that regulates targeting of the chitin synthase Chs3 to sites of polarized growth, or of GAS1, a gene required for cell wall assembly (8,9). CHS5 also shows genetic interactions with BRE1 and LGE1 (9), which encode subunits of the H2B ubiquitin ligase complex and are required for efficient H3K79 methylation (43,44). However, expression of GAS1 and CHS5 was unaffected in strains lacking Dot1 or expressing either one of the two isoforms in the absence or presence of 20 μg/ml CFW (data not shown). The same was true for four genes that are induced by high CFW concentrations (ARO9, YEA4, FUR4 and TUS1, 45) (data not shown). In addition, in genome-wide expression profiling studies, loss of DOT1 had no or only a very minor effect on genome-wide transcription levels (46,47). We note that in contrast to the functions of Dot1 in telomeric silencing and the DNA damage response (26), our results suggest that the cell wall function of Dot1 is independent of global methylation levels (Figure 4B). Therefore, mechanism by which Dot1 affects the yeast cell wall is still elusive.
The two isoforms of the Dot1 protein in yeast are the result of leaky scanning. Leaky scanning is a common phenomenon in mammals and plants (14) and a few examples have been described in yeast (36,48,49). However, it is unclear how frequent the leaky scanning mechanism is in yeast. In higher eukaryotes, there are strict requirements for the sequence context around the start codon to allow efficient translation initiation. The optimal sequence is defined by the Kozak rule: a purine at position –3 and/or a G at position +4 (14). In yeast, these residues seem to be less critical for the initiation of translation (31,32). However, a recent computational study demonstrated a strong bias in yeast for an A at position –3 and a C at position +5 (33). Our studies show that changes in position −3 and +5 had very little, if any, effect on leaky scanning of AUG1 of DOT1. Moreover, even replacement of the complete DOT1 5′-UTR by an exogenous sequence did not significantly affect the rate of leaky scanning (Figure 2F). Together these results strongly suggest that the 5′-UTR of the DOT1 gene is not a factor in determining the efficiency of translation initiation.
Besides a sub-optimal sequence context, the length of the mRNA leader sequence can also contribute to leaky scanning. Some mRNA transcripts have a 5′-leader sequence that is too short for the ribosome to bind efficiently, leading to skipping of the first start codon (35,50). A leader shorter than 12 nucleotides has been shown to promote leaky scanning, and increasing the leader to more than 20 nucleotides restores proper initiation (51). Recently, the transcription start sites of most yeast genes have been mapped using RNA sequencing (52). This study showed that the mRNA transcript generated from the DOT1 gene has a 5′-leader sequence of ∼80 bps, which is longer than the median of all genes (52) and much longer than the minimal length (51). Thus, it is very unlikely that the first AUG is positioned too close to the 5′-end of the mRNA to be recognized by the ribosome. Translation can also be affected by the 3′-UTR of a transcript. In mammals and plants, binding of a microRNA to its recognition sequence in the 3′-UTR of a target mRNA can inhibit cap-dependent translation initiation by a mechanism that is not yet well-understood (53). In addition, in specific tumor cells, the 3′-UTR of the Her-2 oncogene can override translational inhibition of the Her-2 mRNA, showing that such mechanisms are important in disease (54). In the case of the DOT1 transcript, however, the 3′UTR is not a key factor in translation initiation because introduction of a C-terminal tag and thereby replacing the endogenous DOT1 3′-UTR with the 3′-UTR of the ADH1 gene did not affect the synthesis of the two Dot1 isoforms (Figure 1G). This indicates that leaky scanning of the DOT1 mRNA is affected by features in the open reading frame of the gene. Interestingly, several cases have been reported for a role of downstream coding sequences in translation initiation in other eukaryotes (33,55,56). However, mutagenesis of the sequence proximal to the initiator codon did not affect the rate of leaky scanning (Figure 3), indicating that sequences more downstream are involved.
One possibility is that the structure of the Dot1 RNA molecule affects the efficiency of translation initiation. However, mutation of the sequences immediately downstream of the initiator codon that were predicted to disrupt a putative stem-loop did not affect the rate of leaky scanning (Figure 3). Another possibility might be that the sequence between AUG1 and AUG17 contains several ‘uncommon’ codons for low-abundance tRNAs. If this is the case, production of the long isoform would be energetically more taxing for the cell and leaky scanning might therefore be favorable. However, inspection of the codons between AUG1 and AUG17 suggests that this is unlikely. Only three of the 15 codons have the lowest usage frequency for the amino acid they encode. Furthermore, the average codon usage of the 15 codons (0.024) is higher than the average codon usage for all yeast genes (0.016). Thus, the mechanism by which the open reading frame of the DOT1 gene affects leaky scanning is still unclear.
Recent RNA sequencing of the yeast transcriptome predicts that transcripts of as many as 321 yeast genes (∼6% of the yeast genome) contain small open reading frames (uORFs) upstream of the first in-frame start codon (52). Moreover, uORFs are present in almost half of all human and mouse transcripts (57). A number of uORFs have now been shown to control translation of the downstream gene (16,57) and a key question is how the start codons of the uORFs of a gene are used or skipped to tune the efficiency of protein expression. The frequent occurrence in eukaryotic mRNAs of upstream AUGs or sub-optimal start codons indicates that alternative translation initiation plays an important role in regulation of protein expression and might be more widespread than thought previously (58). The discovery of alternative translation events like the one described here will be instrumental for understanding the mechanisms of translation start site selection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU 6th framework program NOE ‘The Epigenome’ (LSHG-CT-2004-503433); the Leukemia and Lymphoma Society (Special Fellow 3409-04 to F.v.L.); VIDI grant from the Netherlands Organization for Scientific Research (to F.v.L.). Funding for open access charge: Netherlands Organisation for Scientific Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Fornerod, R. Agami and members of the van Leeuwen lab for discussions and/or critical reading of the manuscript, T. van Welsem and N. Ong for technical assistance and help with plasmid constructions.
REFERENCES
- 1.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 2.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 4.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 5.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 7.San Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 9.Lesage G, Shapiro J, Specht CA, Sdicu AM, Menard P, Hussein S, Tong AH, Boone C, Bussey H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J. Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 14.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauro VP, Chappell SA, Dresios J. Analysis of ribosomal shunting during translation initiation in eukaryotic mRNAs. Methods Enzymol. 2007;429:323–354. doi: 10.1016/S0076-6879(07)29015-9. [DOI] [PubMed] [Google Scholar]
- 16.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat. Rev. Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 18.Algire MA, Lorsch JR. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr. Opin. Chem. Biol. 2006;10:480–486. doi: 10.1016/j.cbpa.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 21.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 24.Muzi Falconi M, Piseri A, Ferrari M, Lucchini G, Plevani P, Foiani M. De novo synthesis of budding yeast DNA polymerase alpha and POL1 transcription at the G1/S boundary are not required for entrance into S phase. Proc. Natl Acad. Sci. USA. 1993;90:10519–10523. doi: 10.1073/pnas.90.22.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner R, Cronin S, Leader B, Rine J, Hampton R. Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 1998;9:2611–2626. doi: 10.1091/mbc.9.9.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat. Struct. Mol. Biol. 2008;15:550–557. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 27.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cigan AM, Pabich EK, Donahue TF. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baim SB, Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1988;8:1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porras P, Padilla CA, Krayl M, Voos W, Barcena JA. One single in-frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:16551–16562. doi: 10.1074/jbc.M600790200. [DOI] [PubMed] [Google Scholar]
- 35.Wolfe CL, Lou YC, Hopper AK, Martin NC. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J. Biol. Chem. 1994;269:13361–13366. [PubMed] [Google Scholar]
- 36.Outten CE, Culotta VC. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J. Biol. Chem. 2004;279:7785–7791. doi: 10.1074/jbc.M312421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 38.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 39.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 40.Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 2006;1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 41.Boone C, Sommer SS, Hensel A, Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J. Cell Biol. 1990;110:1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.List O, Togawa T, Tsuda M, Matsuo T, Elard L, Aigaki T. Overexpression of grappa encoding a histone methyltransferase enhances stress resistance in Drosophila. Hereditas. 2009;146:19–28. doi: 10.1111/j.1601-5223.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 43.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 44.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 45.Halbeisen RE, Gerber AP. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol. 2009;7:e105. doi: 10.1371/journal.pbio.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 47.Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 48.Welch EM, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antunez de Mayolo A, Lisby M, Erdeniz N, Thybo T, Mortensen UH, Rothstein R. Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res. 2006;34:2587–2597. doi: 10.1093/nar/gkl280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slusher LB, Gillman EC, Martin NC, Hopper AK. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc. Natl Acad. Sci. USA. 1991;88:9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 52.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 54.Mehta A, Trotta CR, Peltz SW. Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells. Genes Dev. 2006;20:939–953. doi: 10.1101/gad.1388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grunert S, Jackson RJ. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation codons. EMBO J. 1994;13:3618–3630. doi: 10.1002/j.1460-2075.1994.tb06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawant SV, Kiran K, Singh PK, Tuli R. Sequence architecture downstream of the initiator codon enhances gene expression and protein stability in plants. Plant Physiol. 2001;126:1630–1636. doi: 10.1104/pp.126.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.