Abstract
The PAQR family of proteins comprises an intriguing group of newly discovered receptors. Although the agonist is known for 5 of the 11 human PAQRs, most are considered “orphan” receptors. We developed a yeast-based assay system for PAQR receptor activity that can be used to identify agonists for PAQRs of unknown function. Using this system, we found that the proteinaceous hormone adiponectin functions as an agonist of PAQR3, a previously uncharacterized member of this family. This is not surprising given that PAQR3 is most closely related to PAQR1 (AdipoR1) and PAQR2 (AdipoR2), which also sense adiponectin. The identification of adiponectin as an agonist for PAQR3 is of considerable clinical relevance because adiponectin suppresses the proliferation of tumor cells and it has been reported that PAQR3 suppresses tumorigenesis. Thus, the interaction between PAQR3 and adiponectin may help explain the antiproliferative properties of adiponectin.
Keywords: Adiponectin receptor, PAQR3, RKTG, Yeast, AdipoR1
Introduction
The PAQR family of proteins is composed of integral membrane receptors for a diverse array of agonist ligands (1). This superfamily of receptors is unified by homology and can be easily distinguished by the presence of four defining characteristics: 1) at least seven transmembrane domains, 2) an ExxNxxxH motif that precedes TM1, 3) an SxxxHxnD motif that spans the end of TM2 and the beginning of TM3, and 4) an HxxxH motif that precedes TM7(2). There are 11 human PAQRs that can be grouped into three main classes based onsequence comparisons (2). Class I (PAQR1-PAQR4) includes two receptors—AdipoR1/PAQR1 and AdipoR2/PAQR2—for the adipose-derived hormone, adiponectin (3). Class II (PAQR5-PAQR9) includes three receptors—PRα/PAQR7, mPRβ/PAQR8, andmPRγ/PAQR5—for the steroid hormone progesterone (4). Finally, there are two class III receptors—MMD2/PAQR10 and MMD1/PAQR11—for which no agonist is known (5,6). Although it has been reported in the literature that the adiponectin and progesterone receptors are fundamentally different at the structural and mechanistic level, the evidence for this assumption is highly controversial (2,7).
AdipoR1 and AdipoR2 are the subject of intense scientific scrutiny because low circulating levels of their agonist ligand, adiponectin, are believed to play a role in the etiology of type 2 diabetes (3). However, low circulating levels of adiponectin are also a risk factor for other pathological conditions such as cancer (8). As a result, the identification of novel adiponectin receptors is of critical medical significance.
We previously developed a simple assay system that allows for the functional expression of human PAQR receptors in the yeast Saccharomyces cerevisiae (2,9). The basis of this assay is the fact that PAQR receptors from diverse sources, when heterologously expressed in yeast, activate the same downstream signal transduction pathway, presumably by producing the same or similar second messengers. This particular yeast-signaling pathway negatively regulates the expression of a gene called FET3. Using the transcriptional response of FET3 as a reporter for PAQR receptor activity, we demonstrated functional expression of AdipoR1, AdipoR2, mPRα, mPRβ, and mPRγ in yeast (2,9). That is to say, these receptors repress FET3 expression in response to their respective agonist ligands. As proof that our system can be used as a discovery tool to identify novel agonists for orphaned human PAQR receptors, we demonstrated that the two uncharacterized human class II PAQRs—PAQR6 and PAQR9—sense and respond specifically to progesterone. We subsequently renamed PAQR6 andPAQR9 receptors mPRδ and mPRε, respectively (2).
The fact that all five human class II receptors are activated by the same agonist ligand (2) led us to suspect that agonists for orphaned receptors could be predicted on the basis of phylogenetic analysis. Because PAQR1 and PAQR2 respond to adiponectin, we hypothesized that their closest human homologs, PAQR3 and PAQR4, could as well. Herein, we demonstrate that PAQR3, but not PAQR4, can sense and respond to adiponectin, making PAQR3 a likely candidate for a third adiponectin receptor from the PAQR family. This finding is intriguing because PAQR3 has already been identified as a protein that prevents tumorigenesis (10). Thus, these results may help explain the anticancer properties of adiponectin.
Materials and methods
Strains and plasmids
Wild-type BY4742 yeast strain was obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). Yeast were maintained by using standard protocol. Human PAQRs were cloned into pYES260 (N-terminal His tag), pGREG536 (NterminalHA tag) and pGREG575 (N-terminal GFP) vectors as previously described (2,9,11,12). In our system, PAQR functionality is not adversely affected by N-terminal tags. The cloning of yeast Izh2p (osmotin receptor) into pRS316 was also previously described (9,13). These vectors allow for galactose-inducible PAQR expression via the GAL1 promoter. The FET3-lacZ plasmid has been previously described (9,14) and contains the FET3 promoter fused to lacZ.
Receptor activity assay
Iron-deficient Low Iron Medium (LIM-Fe) contains 1 μM Fe3+ and 1 mM EDTA to limit iron-bioavailability (15). When grown in LIM-Fe, FET3-lacZ activity is induced (14). This induction does not occur when PAQRs are activated or overexpressed (2,9); 2% galactose was used as a carbon source to induce full expression of PAQR genes driven by the GAL1 promoter, whereas 0.05% galactose/1.95% raffinose was used for reduced PAQR expression. Thaumatin, recombinant full-length adiponectin (BioVendor Laboratory Medicine, Inc.), or progesterone was added to the growth medium upon inoculation into LIM-Fe. In experiments involving progesterone, untreated cells were actually treated with an equal volume of ethanol used to dissolve the progesterone stock to control for vehicle effects. Briefly, cells were inoculated into LIM-Fe with or without agonist, grown to mid-log phase, and assayed for β-galactosidase activity as previously described (2,9). For individual experiments, each data point has been done in triplicate, and the error bars represent ±1 SD. All experiments were performed at least three times, and a representative experiment is shown. ED50 values were obtained by using the Web-based BioDataFit software using sigmoidal curve fitting (http://www.changbioscience.com/stat/ec50.html).
Protein detection
Western blots were performed as previously described (2) by using a rabbit polyclonal anti-HA or anti-yeast porin primary antibodies and a goat anti-rabbit IgG-HRP conjugate as the secondary antibody. All antibodies are from Santa Cruz Biotechnology.
Adiponectin binding assays
Yeast spheroplasts were prepared as reported by Holcomb et al. (16) and resuspended in 1×PBS at an OD600 of 0.05. Spheroplasts were incubated with (FAM)-labeled adiponectin (Phoenix Pharmauceticals, Inc. #FG-ADI-01-A) at various concentrations for 30 min at 4°C. Bound adiponectin was measured by using a FACS Vantage SE Turbosort (BD Biosciences, San Jose, CA) flow cytometer, which uses Cell Quest software from BD Biosciences to collect and analyze data. First, we determine the number of cells whose fluorescence intensity exceeds the threshold for significance and divide this number by total cells counted, and then we multiply by 100; 50,000 cells were counted for each point. This represents percent of cells with bound adiponectin. For presentation purposes, we subtract percent of control cells with bound adiponectin from percent of cells expressing each PAQR with bound adiponectin.
Sequence analysis
Multiple sequence alignments and phylogenetic trees were produced by using ClustalX with default parameters (17). Trees were visualized by using Tree View (18). PAQR sequences were obtained from either the NCBI or the Joint Genome Institute (http://genome.jgi-psf.org/euk_home.html) websites.
Results
Activation of PAQRs in yeast
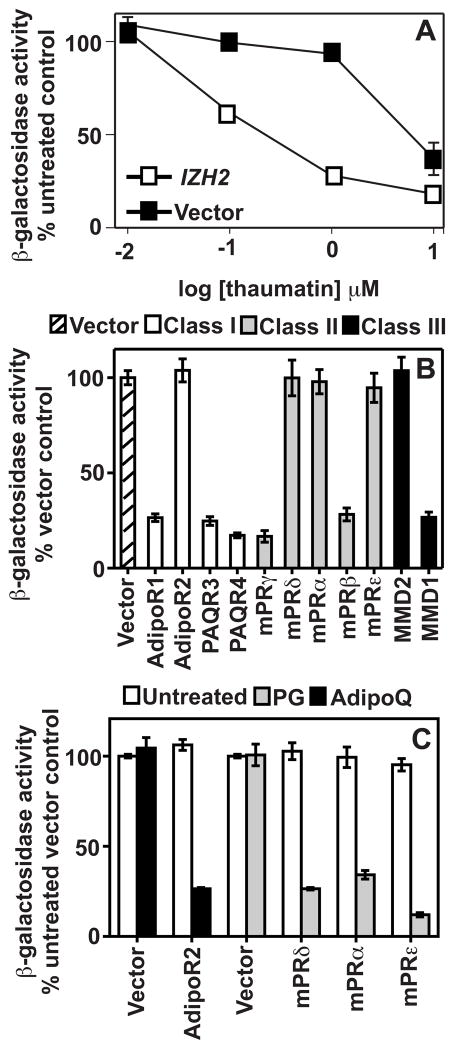
Ordinarily, the FET3 gene is induced by iron deficiency. However, we previously established that overexpression of the yeast PAQR receptor, Izh2p, results in the repression of this gene under conditions where it is supposed to be induced (9). In these experiments, Izh2p was overexpressed by using 2% galactose to fully induce the GAL1 promoter used to drive its expression. It is important to note that this effect was seen in the absence of agonist. It is not uncommon for signaling proteins to transduce signals constitutively when overexpressed due to the presence of basal signaling capability. The fact that Izh2p overexpression affects FET3 suggests that elevated Izh2p expression increases second messenger production above the threshold for activation of downstream pathways independently of the presence of agonist. That said, it is necessary to confirm that the effect of Izh2p on FET3 can be recapitulated at lower receptor expression levels when agonist is added. Izh2p has been shown to function as a receptor for the plant protein osmotin (19). Although osmotin is not commercially available, we obtained thaumatin, a close relative of osmotin from a different plant species (20). Figure 1A shows that in wild-type cells, which express normal levels of Izh2p from the genomic IZH2 locus, thaumatin induced a dose-dependent repression of the FET3-lacZ reporter. Furthermore, we could sensitize FET3-lacZ to the effects of thaumatin by increasing the expression of Izh2p. This was achieved by transforming the wild-type strain with a vector containing a GAL1-driven Izh2p construct and growing the cells in 0.05% galactose. This level of induction was not sufficient to constitutively repress FET3-lacZ, but, as is evident from Figure 1A, the ED50 for thaumatin-dependent FET3-lacZ repression is significantly lower for this strain than for the wild type carrying empty expression vector These results confirm that activation of Izh2p by agonist activates the same pathway as receptor overexpression.
Figure 1.
Functional expression of human PAQRs in yeast. All cells are wild type and are grown in LIM-Fe. (A) Medium contains 0.05% galactose to modestly overexpress GAL1-driven genes. Black squares show FET3-lacZ activity in yeast carrying empty expression vector. This strain carries the genomic copy of IZH2. White squares show activity in yeast carrying a GAL-driven Izh2p overexpression vector in addition to the genomic copy of IZH2. Activity is presented as a percentage of activity measured in a strain that has not been treated with thaumatin. (B) Medium contains 2% galactose to fully induce all human PAQRs. FET3-lacZ activity is presented as a percentage of activity seen in a strain carrying empty expression vector. (C) Medium contains 2% galactose to fully overexpress AdipoR2, mPRα, mPRδ, andmPRε. Cells are exposed to their respective agonists (100 pM adiponectin for AdipoR2 or 10 μM progesterone for mPRδ, mPRα, and mPRε). FET3-lacZ activity is presented as a percentage of activity seen in an untreated strain carrying empty expression vector.
Expression of human PAQRs in yeast
Besides Izh2p, the S. cerevisiae genome encodes three additional class I PAQR proteins called Izh1p, Izh3p, and Izh4p for which no agonist is known (13). We previously discovered that overexpression of any of these other receptors also caused the repression of FET3-lacZ (9). This led us to hypothesize that any PAQR could couple with the yeast-signaling pathway, leading to repression of FET3-lacZ. Indeed, in Figure 1B, we show that several human PAQRs repress FET3-lacZ when overexpressed in yeast by using the GAL1 promoter. Notably, AdipoR1, PAQR3, PAQR4, mPRγ, mPRβ, and MMD1 all repress FET3 when simply overexpressed, whereas AdipoR2, mPRδ, mPRα, mPRε, and MMD2 do not. One interpretation of this is that the latter receptors do not function in yeast. However, when exposed to their respective agonist ligands under these conditions, AdipoR2, mPRδ, mPRα, and mPRε, all elicit FET3-lacZ repression (Fig. 1C). This finding indicates that they are functionally expressed, but their basal signaling is not high enough to activate downstream effectors in the absence of agonist. It should be noted that simple overexpression of PAQR3 in human cells is also sufficient to regulate downstream effectors (10), suggesting that the constitutive activity of human PAQR receptors is not merely an artifact of the yeast expression system.
When the expression of PAQRs is reduced by adding only 0.05% galactose to the medium, none of the receptors, including Izh1p-Izh4p, affected FET3-lacZ expression in the absence of agonist, meaning that they do not produce sufficient second messenger to constitutively activate the downstream pathway (data not shown). Under these conditions, we previously demonstrated that AdipoR1, AdipoR2, mPRα, mPRβ, mPRγ, mPRδ, and mPRε all repress FET3-lacZ in response to their respective agonists (2,9). These findings underscore the benefit of using the yeast system. Yeast do not make or use progesterone or adiponectin, nor do they respond to these hormones when the human PAQRs are not present. Thus, our system can be used to study human PAQRs in isolation from other human receptors that might complicate data analysis when using a homologous expression system like human cell culture. Indeed, with this system, we identified agonists for uncharacterized receptors and determined ED50 values for the activation of seven human PAQRs by their respective agonists (Table 1) (2,9).
Table 1.
Functional Expression of human PAQRs in Yeast
| PAQR | Name | Agonist (ED50) | Functional | Reference |
|---|---|---|---|---|
| PAQR1 | AdipoR1 | Adiponectin (1 ± 1 pM) | Yes | [2] |
| PAQR2 | AdipoR2 | Adiponectin (2 ± 1 pM) | Yes | [2] |
| PAQR3 | RKTG | Adiponectin (26 ± 2 pM) | Yes | This study |
| PAQR4 | - | Unknown | Yes* | This study |
| PAQR5 | mPRγ | Progesterone | Yes | [2] |
| PAQR6 | mPRδ | Progesterone | Yes | [2] |
| PAQR7 | mPRα | Progesterone | Yes | [2] |
| PAQR8 | mPRβ | Progesterone | Yes | [2] |
| PAQR9 | mPRε | Progesterone | Yes | [2] |
| PAQR10 | MMD2 | Unknown | No | This study |
| PAQR11 | MMD1 | Unknown | Yes* | This study |
Functionality assumed because it activates the pathway when overexpressed
Response of class I receptors to adiponectin
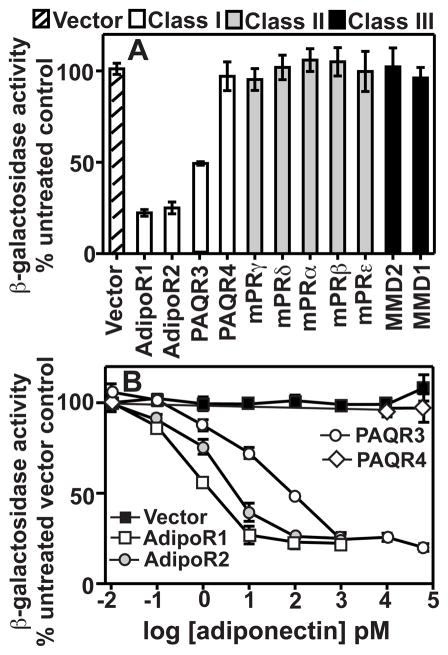
We previously performed a detailed phylogenetic analysis of the PAQR family and demonstrated that PAQR3 and PAQR4 are more closely related to AdipoR1 and AdipoR2 than the other human PAQRs (2). This led us to suspect that PAQR3 and PAQR4 might be adiponectin receptors. Therefore, we analyzed the response of FET3-lacZ to adiponectin in cells expressing all the human PAQRs when only 0.05% galactose was added to the media. Under these conditions, none of the receptors had any basal effect on FET3-lacZ; however, the addition of 100 pM adiponectin caused the repression of FET3-lacZ in cells expressing AdipoR1, AdipoR2, and PAQR3 (Fig. 2A). A dose response curve shows that all three of these receptors respond to adiponectin, but PAQR4 does not (Fig. 2B). Moreover, the ED50 for activation of PAQR3 is higher than for AdipoR1 or AdipoR2 (Table 1). FET3-lacZ did not respond to >60 nM adiponectin in cells carrying empty expression vector. This finding indicates that none of the endogenous yeast PAQRs (Izh1p-Izh4p) can respond to adiponectin. In addition, PAQR4 did not respond to adiponectin at concentrations of >60 nM (Fig. 2B). Because circulating levels of adiponectin are in the low nM range (21), it can be concluded that PAQR4 is unlikely to function as a bona fide adiponectin receptor.
Figure 2.
PAQR3 responds to adiponectin. All cells are wild type and grown in LIM-Fe. (A) Medium contains either 2% galactose for full expression (AdipoR2, mPRδ, mPRα, mPRε, and MMD2) or 0.05% galactose for reduced expression (AdipoR1, PAQR3, PAQR4, mPRγ, mPRβ, and MMD1). The effect of adiponectin on FET3-lacZ in cells carrying each PAQR is presented as a percentage of activity seen in untreated cells. (B) Medium contains 0.05% galactose for reduced PAQR expression. Response of each human class I PAQR to adiponectin is shown. For each expressed PAQR, FET3-lacZ activity is presented as a percentage of activity seen in untreated cells carrying empty expression vector.
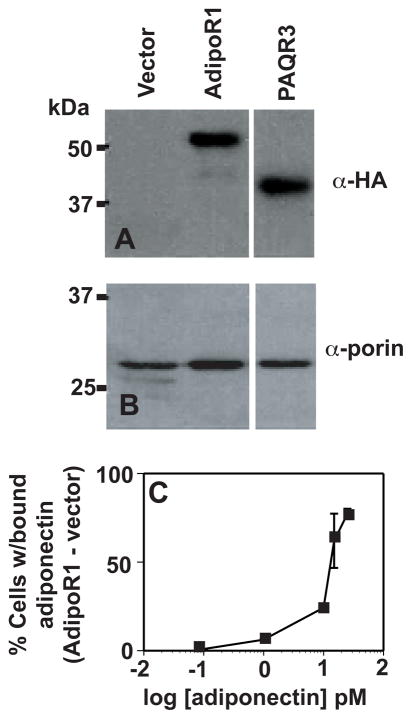
The high ED50 for PAQR3 is an interesting discovery. It is possible that this is an artifact of poor expression in yeast rather than an intrinsic property of the receptor. However, we demonstrate that, despite the large differences in their ED50 values, there is little difference in expression levels for AdipoR1 and PAQR3 (Figs. 3A and 3B). Hence, it is unlikely that the high ED50 for PAQR3 is related to expression levels. To support these ED50 values, we attempted to measure binding of fluorescently labeled adiponectin to spheroplasts of yeast carrying either empty expression vector or the AdipoR1 and PAQR3 expression vectors. Although we could measure increased binding of adiponectin to spheroplasts carrying AdipoR1 overvector control (Fig. 3C), we could not detect increased adiponectin binding to cells carrying PAQR3 (data not shown). A possible explanation for this is that PAQR3 is thought to be primarily Golgi-localized in humans (10), unlike AdipoR1 and AdipoR2, which are cell surface receptors (3). If the same were true in yeast, then it is unlikely that the binding of PAQR3 to adiponectin would be detectable. In fact, expression of GFP-labeled PAQR3 in yeast shows that a significant portion of protein is localized to internal membranes (data not shown), although the exact localization is yet to be determined.
Figure 3.
PAQR3 expression levels. (A) Western blots of total membrane preparations from cells carrying empty expression vector (pGREG536) or HA-tagged AdipoR1 and PAQR3 expression vectors. Blots are probed with an anti-HA antibody. Equal amount of total protein were loaded in each lane. (B) Western blots of the same samples from (A) probed with an antiporin antibody to control for loading of membrane protein extracts. (C) Binding of FAM-labeled adiponectin to spheroplasts of wild-type cells overexpressing AdipoR1. Binding is first determined as a percent of cells with FAM-adiponectin bound for cells carrying either empty expression vector or AdipoR1 expression vector. Data are plotted as percent bound to AdipoR1-expressing cells percent bound to cells—carrying empty vector. Error bars indicate ±1 SD.
Thus, the high ED50 of PAQR3, compared to AdipoR1 and AdipoR2, could be a result of poor localization of PAQR3 to the surface or inefficient delivery of adiponectin to the intracellular compartment where PAQR3 resides. The latter is not beyond the realm of possibility, because, in our assay, cells are exposed to agonist for 16–20 hr—plenty of time for adiponectin to enter the secretory pathway by endocytosis.
Ancient roots of PAQR3
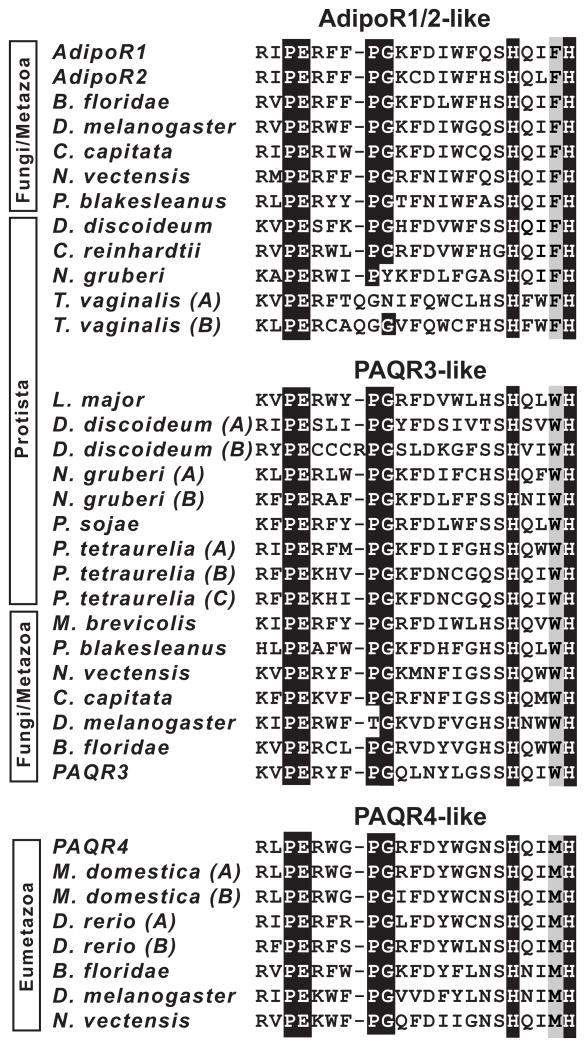
In-depth sequence and phylogenetic analyses of the PAQRs revealed that within class I, AdipoR1 and AdipoR2 are nearly identical (E = 4 × 10128 for pairwise comparisons), whereas PAQR4 is the most divergent (Supplemental Figs. 1, 2, and 3). Class I proteins can be easily identified as being AdipoR1/2-like, PAQR3-like, or PAQR4-like by diagnostic alterations in the motifs that unify the PAQR family. In particular, the conserved PAQR motif that resides between TM6 and TM7 is particularly useful for distinguishing these proteins. This motif is generally PExnPGxnHxxxH (Sup. Fig. 3); however, it is PExnPGxnHxxFH for AdipoR1/2-like proteins, PExnPGxnHxxWH for PAQR3-like proteins, and PExnPGxnHxxMH for PAQR4-likeproteins (Fig. 4). PAQR4-like proteins appear first in eumetazoans. However, both AdipoR1/2-like proteins and PAQR3-like proteins are ubiquitous in the Fungi/Metazoa group. For example, S. cerevisiae has both an AdipoR1/2-like protein (Izh2p) and a PAQR3-like protein of unknown function (Izh3p). Homologs of both these groups can also be found sporadically in protist species.
Figure 4.
Alignment of the third conserved motif in adiponectin receptor homologs. Black shading indicates amino acids that are highly conserved throughout the entire PAQR family. Gray shading indicates amino acids that can be considered diagnostic of AdipoR1/2-like, PAQR3-like, or PAQR4-like proteins. The full names and taxonomic grouping of the organisms from which these sequences are derived are shown in Supplemental Figure 1.
The ancient roots of PAQR3 and its early divergence from AdipoR1/2-like proteins present an interesting dilemma. If AdipoR1, AdipoR2, and PAQR3 all currently function as adiponectin receptors, it suggests that their earliest common ancestor was a receptor for an adiponectin-like protein and that all three proteins coevolved along with adiponectin over time. With this in mind, it is intriguing that the fungal AdipoR1/2-like protein, Izh2p, functions as a receptor for a plant protein with structural similarity to adiponectin (19). Hence, it is possible that the structural components of the agonist/PAQR receptor module have been conserved throughout evolution.
Discussion
The literature regarding the PAQR family is disparate and often confusing. For example, there are reports that the class II progesterone-sensing PAQRs are structurally and mechanistically distinct from the class I adiponectin-sensing PAQRs (22). However, it should be noted that although the evidence for evolutionary relatedness between receptor subtypes is indisputable, the evidence for their mechanistic differences is highly debatable (2). Indeed, we have presented evidence strongly suggesting that PAQR receptors from all three classes, regardless of agonist ligands, are capable of coupling to the same yeast signaling module, suggesting that all PAQRs work by a similar signaling mechanism.
More importantly, we have demonstrated that this yeast-based system is an invaluable tool for identifying potential new agonists for PAQR receptors. As proof of this principle, we demonstrated that heterologous expression of PAQR3 in yeast confers on this organism the ability to sense and respond to adiponectin. Because yeast do not normally respond to adiponectin, this indicates that PAQR3 could function as a third human adiponectin receptor. A possible reason why PAQR3 was not previously identified as an adiponectin receptor is thatPAQR3 is localized to internal membranes (10) and is not detectable by standard binding assays. Although it is clear that adiponectin can function as an agonist for PAQR3 in the yeast system, we should point out that the activation of PAQR3 by adiponectin in the yeast system may not represent the true physiological function of PAQR3. It is possible that PAQR3functions as a receptor for a yet-to-be-identified adiponectin-like protein and that its activation by adiponectin is fortuitous. Considering that there are dozens of uncharacterized adiponectin paralogs encoded by the human genome, this possibility is not outlandish.
Finally, it is important to discuss these findings in light of what is known about the physiology of adiponectin and PAQR3. PAQR3, when overexpressed in human cells, physically interacts with Raf-1 kinase and causes Raf-1 kinase to be sequestered in the Golgi. It has been proposed that this is the mechanism by which PAQR3, also known as RKTG, suppresses carcinogenesis (10). This is extremely important in light of our finding that PAQR3 senses adiponectin, a hormone with anti-tumorigenic properties (8). This means that it is possible that the anticancer effects of adiponectin may be related to its ability to affect Raf kinase through PAQR3. Future studies on mammalian cells will be required to confirm these effects and the true function of PAQR3.
Supplementary Material
Supplemental Figure 1. Phylogenetic tree of class I PAQRs. The tree is rooted with PAQR sequences from chlorphyte plants. On the right is a taxonomic key to the organisms from which these sequences and those in Figure 4 were derived.
Supplemental Figure 2. Multiple-sequence alignment of human and yeast class I PAQRs. Gray shading indicates amino acids that are highly conserved throughout the entire PAQR family. Black shading indicates amino acids that are conserved in AdipoR1 and AdipoR2. Boxes indicate the positions of transmembrane domains (TM). The topological placement of the loop regions (L) is shown.
Supplemental Figure 3. Hydropathy plots for yeast and human class I PAQRs. (A–D) Hydropathy plots for various individual proteins from different groups were generated and aligned (dotted lines). An average hydropathy plot for all members of each group was generated (solid lines). (A) Fungal Izh2p-like proteins. (B) Vertebrate AdipoR1/2-like proteins. (C) Vertebrate PAQR3-like proteins. (D) Vertebrate PAQR4-like proteins. (E) A topological model for all PAQRs with the positions of the three conserved motifs.
Acknowledgments
This work was funded by the University of Florida, Department of Chemistry and the National Institutes of Health (R21DK074812 to TJL).
References
- 1.Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC, Funk WD. PAQR proteins: A novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol. 2005;61(3):372–380. doi: 10.1007/s00239-004-0375-2. [DOI] [PubMed] [Google Scholar]
- 2.Smith JL, Kupchak BR, Garitaonandia I, Hoang LK, Maina AS, Regalla LM, Lyons TJ. Heterologous expression of human mPRalpha, mPRbeta and mPRgamma in yeast confirms their abilityto function as membrane progesterone receptors. Steroids. 2008 doi: 10.1016/j.steroids.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, Murakami K, Ohteki T, Uchida S, Takekawa S, Waki H, Tsuno NH, Shibata Y, Terauchi Y, Froguel P, Tobe K, Koyasu S, Taira K, Kitamura T, Shimizu T, Nagai R, Kadowaki T. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423(6941):762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y, Bond J, Thomas P. Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc Natl Acad Sci U S A. 2003;100(5):2237–2242. doi: 10.1073/pnas.0436133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brauer AU, Nitsch R, Savaskan NE. Identification of macrophage/microglia activationfactor (MAF) associated with late endosomes/lysosomes in microglial cells. FEBS Lett. 2004;563(1–3):41–48. doi: 10.1016/S0014-5793(04)00244-3. [DOI] [PubMed] [Google Scholar]
- 6.Rehli M, Krause SW, Schwarzfischer L, Kreutz M, Andreesen R. Molecular cloning of anovel macrophage maturation-associated transcript encoding a protein with several potentialtransmembrane domains. Biochem Biophys Res Commun. 1995;217(2):661–667. doi: 10.1006/bbrc.1995.2825. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes MS, Brosens JJ, Gellersen B. Honey, we need to talk about the membrane progestin receptors. Steroids. 2008;73(9–10):942–952. doi: 10.1016/j.steroids.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Barb D, Williams CJ, Neuwirth AK, Mantzoros CS. Adiponectin in relation tomalignancies: A review of existing basic research and clinical evidence. Am J Clin Nutr. 2007;86(3):s858–866. doi: 10.1093/ajcn/86.3.858S. [DOI] [PubMed] [Google Scholar]
- 9.Kupchak BR, Garitaonandia I, Villa NY, Mullen MB, Weaver MG, Regalla LM, Kendall EA, Lyons TJ. Probing the mechanism of FET3 repression by Izh2p overexpression. Biochim Biophys Acta. 2007;1773(7):1124–1132. doi: 10.1016/j.bbamcr.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan F, Feng L, He J, Wang X, Jiang X, Zhang Y, Wang Z, Chen Y. RKTG sequestersB-Raf to the Golgi apparatus and inhibits the proliferation and tumorigenicity of human malignantmelanoma cells. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn119. [DOI] [PubMed] [Google Scholar]
- 11.Jansen G, Wu C, Schade B, Thomas DY, Whiteway M. Drag&Drop cloning in yeast. Gene. 2005;344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Melcher K. A modular set of prokaryotic and eukaryotic expression vectors. Anal Biochem. 2000;277(1):109–120. doi: 10.1006/abio.1999.4383. [DOI] [PubMed] [Google Scholar]
- 13.Lyons TJ, Villa NY, Regalla LM, Kupchak BR, Vagstad A, Eide DJ. Metalloregulation of yeast membrane steroid receptor homologs. Proc Natl Acad Sci U S A. 2004;101(15):5506–5511. doi: 10.1073/pnas.0306324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. Embo J. 1996;15(13):3377–3384. [PMC free article] [PubMed] [Google Scholar]
- 15.Eide D, Guarente L. Increased dosage of a transcriptional activator gene enhances iron-limited growth of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138(2):347–354. doi: 10.1099/00221287-138-2-347. [DOI] [PubMed] [Google Scholar]
- 16.Holcomb CL, Etcheverry T, Schekman R. Isolation of secretory vesicles from Saccharomyces cerevisiae. Anal Biochem. 1987;166(2):328–334. doi: 10.1016/0003-2697(87)90581-1. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_Xwindows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 19.Narasimhan ML, Coca MA, Jin J, Yamauchi T, Ito Y, Kadowaki T, Kim KK, Pardo JM, Damsz B, Hasegawa PM, Yun DJ, Bressan RA. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol Cell. 2005;17(2):171–180. doi: 10.1016/j.molcel.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 20.Singh NK, Bracker CA, Hasegawa PM, Handa AK, Buckel S, Hermodson MA, Pfankoch E, Regnier FE, Bressan RA. Characterization of osmotin: A thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiol. 1987;85(2):529–536. doi: 10.1104/pp.85.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lara-Castro C, Luo N, Wallace P, Klein RL, Garvey WT. Adiponectin multimeric complexes and the metabolic syndrome trait cluster. Diabetes. 2006;55(1):249–259. [PubMed] [Google Scholar]
- 22.Thomas P, Pang Y, Dong J, Groenen P, Kelder J, de Vlieg J, Zhu Y, Tubbs C. Steroid and G protein binding characteristics of the sea trout and human progestin membrane receptor alpha subtypes and their evolutionary origins. Endocrinology. 2007;148(2):705–718. doi: 10.1210/en.2006-0974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Phylogenetic tree of class I PAQRs. The tree is rooted with PAQR sequences from chlorphyte plants. On the right is a taxonomic key to the organisms from which these sequences and those in Figure 4 were derived.
Supplemental Figure 2. Multiple-sequence alignment of human and yeast class I PAQRs. Gray shading indicates amino acids that are highly conserved throughout the entire PAQR family. Black shading indicates amino acids that are conserved in AdipoR1 and AdipoR2. Boxes indicate the positions of transmembrane domains (TM). The topological placement of the loop regions (L) is shown.
Supplemental Figure 3. Hydropathy plots for yeast and human class I PAQRs. (A–D) Hydropathy plots for various individual proteins from different groups were generated and aligned (dotted lines). An average hydropathy plot for all members of each group was generated (solid lines). (A) Fungal Izh2p-like proteins. (B) Vertebrate AdipoR1/2-like proteins. (C) Vertebrate PAQR3-like proteins. (D) Vertebrate PAQR4-like proteins. (E) A topological model for all PAQRs with the positions of the three conserved motifs.