Abstract
Brh2, the BRCA2 homolog in Ustilago maydis, plays a crucial role in homologous recombination by controlling Rad51. In turn, Brh2 is governed by Dss1, an intrinsically disordered protein that forms a tight complex with the C-terminal region of Brh2. This region of the protein associating with Dss1 is highly conserved in sequence and by comparison with mammalian BRCA2 corresponds to a part of the DNA-binding domain with characteristic OB folds. The N-terminal region of Brh2 harbors a less-defined, but powerful DNA-binding site, the activity of which is revealed upon deletion of the C-terminal region. Full-length Brh2 complexed with Dss1 binds DNA slowly while the N-terminal fragment binds quickly. The DNA-binding activity of full-length Brh2 appears to correlate with dissociation of Dss1. Addition of Dss1 to the heterotypic Brh2/Dss1 complex attenuates DNA-binding activity, but not by direct competition for the N-terminal DNA binding site. Conversely, the Brh2/Dss1 complex dissociates more quickly when DNA is present. These findings suggest a model in which Brh2 binding to DNA is subject to allosteric regulation by Dss1.
In eukaryotes Rad51 is essential for repair of DNA damage by homologous recombination (1). Rad51 assembles into a complex with single-stranded (ss)1 DNA that has been exposed after some processing of the damaged duplex to form a nucleoprotein filament, which is the catalytically active form of Rad51 for promoting homologous pairing and DNA strand exchange. BRCA2 is an important regulator of Rad51 activity (2, 3) and appears to exert both positive and negative effects on Rad51 filaments (4–7). Biochemical studies with Brh2, the BRCA2 homolog from the fungus Ustilago maydis, have demonstrated that it mediates Rad51 assembly on a protruding 3′-single-strand DNA tail (8) and interacts with Rad51 in other ways to promote post-strand invasion steps in recombination (9). Interaction of Rad51 with Brh2 is through the single BRC element located in the N-terminal region of the protein (10) and a second and less well-defined domain, the CRE, located at the extreme C-terminus (11). These N-terminal and C-terminal domains participate with each other in some as yet unknown way to direct proper Rad51 filament assembly. The process depends on Dss1 (11), a small versatile acidic protein that physically interacts with Brh2 through a region corresponding to the DSS1/DNA-binding domain (DBD) in mammalian BRCA2 (12). The mammalian BRCA2 DBD domain consists of a tandem array of three OB (oligonucleotide/oligosaccharide-binding) folds and a helix-rich domain that is laced to the adjacent OB fold by intertwining with DSS1 (13). Sequence alignment with BRCA2 indicates that a region corresponding to part of the helix-rich domain and the adjacent OB1 and OB2 folds is conserved in Brh2, but that OB3 is absent. In Brh2 36 out of the 40 residues in BRCA2 DBD that contact DSS1 are identical or conserved (12).
The single amino acid change W728A in U. maydis Brh2 abolishes its capacity to bind Dss1 and support DNA repair (11). In the same vein, deletion of the gene encoding Dss1 in U. maydis causes extreme sensitivity to DNA damage and deficiency in recombination, effectively phenocopying the brh2 mutant (12, 14). In view of the physical interaction between Brh2 and Dss1, these genetic findings are consistent with the idea that Dss1 serves as a needed cofactor or as a direct activator of Brh2. Other studies, however, support the view that Dss1 interacts with Brh2 in a more complex way to counteract auto-inhibition. Removal of a tract of 40 amino acids from the C-terminus of Brh2 is enough to cause complete loss of activity in complementing the UV sensitivity of the brh2 mutant. Yet paradoxically, by removing a much longer tract from the C-terminus that encompasses the entire Dss1/DNA-binding domain (the DBD), radiation resistance can be substantially restored not only in the brh2 mutant but also in the dss1 mutant (15). Furthermore, expression of the N-terminal fragment in the brh2 mutant restores Rad51 focus formation and allelic recombination proficiency to an even higher level than in wild type cells (15). These results imply that the DBD deletion removes a negative regulatory function from Brh2 that is countermanded by Dss1.
Functional complementation of the brh2 and dss1 mutants by the Brh2 N-terminal fragment (Brh21–551 or Brh2NT) deleted of the DBD strongly suggests that this truncated protein has an inherent ability to organize Rad51, mediate Rad51 delivery to sites of DNA damage, and support DNA repair independent of Dss1 modulation. To account for all of these functions we supposed that the N-terminal region would contain an intrinsic DNA-binding capacity, and indeed as predicted, we found it has the capacity to bind DNA with an affinity and specificity for structure reflecting the properties of the full-length protein (16). By comparison, the C-terminal region (Brh2505–1075 or Brh2CT) corresponding to the canonical DBD bound DNA less tightly by an order of magnitude, as did the DBD of the mouse BRCA2. These results imply that the primary interaction site in Brh2 for association with DNA resides in the N-terminal fragment.
Taken altogether the model for Brh2 that emerges suggests an architectural arrangement featuring an N-terminal DNA binding domain coupled to a C-terminal regulatory domain that responds to Dss1. What is not clear is how these functional modules communicate. If Dss1 is required for appropriately controlled functional activity in the context of full-length Brh2 protein, then it follows that Dss1 imposes order and/or governs cooperation between the two protein domains. Thus, the hypothesis framed by these observations is that Dss1 exerts control over DNA binding through the N-terminal domain even though the only known site for physical interaction of Dss1 with Brh2 is limited to the C-terminal domain. Here we have investigated the relationship between binding of DNA to Brh2 and association with Dss1.
EXPERIMENTAL PROCEDURES
U. maydis methodology
Manipulations of U. maydis strains, culture methods, gene transfer procedures, survival after UV irradiation have been described previously (17). U. maydis strains included UCM350 (nominal wild type) and UCM591 (Δdss1). Dss1 and the mutant derivative Dss1D37A were expressed from autonomously replicating plasmids with the gap (glyceraldehydes 3-phosphate dehydrogenase) promoter driving expression and with hygromycin resistance as the selectable marker. Dss1D37A was constructed using overlapping oligonucleotides and PCR methodology (18). For spot assays for cell survival cell suspensions of overnight cultures were adjusted to ~2 × 107 per mL, diluted in 10-fold serial dilutions and aliquots (10 μL) of each were spotted on agar medium. Survival is indicated as growth of colonies 3 days after irradiation.
Protein preparations
Brh2 and Brh2CT proteins tagged with MBP in complex with His-tagged Dss1 were purified after overexpression in E. coli as described previously (11, 19). Briefly, purification involved sequential affinity chromatography steps on Ni2+-NTA (nitrilotriacetic acid agarose, Qiagen)) and cross-linked amylose resin (New England Biolabs), in which the protein was eluted specifically with imidazole and maltose, respectively, followed by salt gradient elution off a column of monoQ beads using an AKTA FPLC chromatography system (GE Healthcare). Brh2CT protein devoid of MBP-tag was prepared from a fusion protein which was engineered to contain a tobacco etch virus (TEV) protease recognition site in the linker regions connecting the MBP-tag to Brh2CT as described previously (16). After cleavage with TEV protease, Brh2CT was purified by salt gradient elution from monoQ resin. Brh2NT terminating at residue M551 was purified as a fusion protein with an N-terminal MBP-tag and C-terminal His-tag. Purification followed the same protocol developed for the full-length protein, but included an additional step of salt gradient elution from HiTrap heparin-agarose (GE Healthcare). Brh2/Dss1 complex used in preparation of the Brh2 apoprotein free of Dss1 was purified as above except that it was eluted from amylose resin using 1.0 M α-methylglucoside instead of 10 mM maltose to facilitate rebinding of Brh2 to amylose resin in a subsequent step. After elution of the Brh2/Dss1 complex from the monoQ column, the fraction was brought to 20 mM MgCl2, held at 37° for 1 hr, then loaded on a column of cross-linked amylose at 4°. The column was washed with 50 volumes of buffer A (25 mM Tris-HCl, pH 7.5, 200 mM KCl, 1 mM DTT, 1 mM EDTA, 10% glycerol) containing 20 mM MgCl2 to remove Dss1, then Brh2 was eluted with buffer A containing 10 mM maltose. Proteins were stored in buffer A. His-tagged Dss1 and Dss1D37A were purified after expression in E. coli. The procedure included affinity purification on Ni2+-NTA, gradient elution from monoQ beads, and molecular sieve chromatography on Superose 6 gel filtration column. A form of His-tagged Dss1 (His-PK-Dss1) containing a target sequence that could be phosphorylated and thus labeled with 32P using [γ-32P]ATP and human Protein Kinase A (PKA) catalytic subunit (Sigma-Aldrich) was engineered by inserting an oligonucleotide encoding the PKA amino acid recognition sequence (LRRASV) into the His-Dss1 gene fusion between the hexahistidine coding sequence and Dss1 open reading frame. When MBP-Brh2 and His- PK-Dss1 were co-expressed and purified as heterotypic complex as above, His-PK-Dss1 could be specifically labeled in situ with 32P using PKA catalytic subunit. There was no detectable labeling of the associated MBP-Brh2 protein. Phosphorylation was performed in reactions containing 25 mM Tris-HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 12.5 nM [γ-32P]ATP, 500 nM MBP-Brh2/His-PK-Dss1 and 5 units/ml PKA catalytic subunit for 16 hr at 4°. 32P-labeled MBP-Brh2/ His-PK-Dss1protein was obtained after purification through Ni2+-NTA beads and amylose resin as above, eluted from the latter with α-methylglucoside
DNA binding
DNA binding was assayed by three methods. Electrophoretic mobility shift assays were performed using a single-stranded 60mer oligonucleotide (ss60mer) GCTGCGCAAGGATAGGTCGAATTTTCTCATTTTCCGCCAGCAGTCCACTTCGA TTTAATT (derived from phage φX174 sequence) labeled at the 5′ end with 32P using polynucleotide kinase and [γ-32P]ATP or else the fluorescent dye IRD700 or IRD800 (MWG Biotech AG) as substrate (16). Binding reactions contained 3.3 nM ss60mer and protein as required in reaction buffer (25 mM HEPES, pH 7.5, 60 mM KCl, 2 mM MgCl2). After incubation at 37° glutaraldehyde was added to 0.2%. Fixation was continued for 10 min and quenched with 100 mM Tris-HCl, pH 8.0. Products were monitored after electrophoresis at room temperature in 1% agarose gels cast in 40 mM Tris-acetate, pH 7.6. Gel images were collected with an Odyssey infrared detection platform (LI-COR Biosciences) or with a phosphorimager and quantified with ImageQuaNT software (Molecular Dynamics). Nitrocellulose filter-binding assays were performed using 5′-32P-labeled ss60mer as above but reactions were processed by washing solutions through a double layer of nitrocellulose membrane (BioRad Laboratories) and DEAE-celluose paper (Whatman DE81) in a dot-blot manifold (20). Radiographic images were collected with a phosphorimager and quantified. Fluorescence polarization assays were performed in reactions (100 μL) using 5′-fluorescein labeled ss49mer (MWG Biotech AG) that was derived from plasmid pBluescript II SK+ (Stratagene) and described previously as 49mer(+) (19). Measurements were read at room temperature using a SpectraMax M5 multi-label plate reader (Molecular Devices) 30 minutes after mixing samples in white, flat bottomed 96- well assay plates (Corning). Excitation was set at 492 nm and emission at 525 nm with a 515 nm cutoff filter. G factor was set at 1.0. Anisotropy was determined using internal software of the instrument. All DNA concentrations are expressed in moles of molecules, rather than nucleotide, unless explicitly stated.
Pull-down procedures
Brh2 or Brh2CT complexed with His-Dss1 was added to mixes (60 μL) containing reaction buffer and DNA (ss60mer as above) as required to a final concentration of 550 nM and held at 37°. At specified times the mixes were transferred to a tube containing 30 μL of a settled slurry of affinity beads (Ni2+-NTA or amylose resin as appropriate) and held on ice for 10 min. After brief centrifugation the beads were collected and protein in the supernatant and bound fractions was analyzed by electrophoresis in polyacrylamide gels containing SDS. Protein was visualized by staining with SimplyBlue SafeStain (Invitrogen). When necessary to visualize the DNA, IRD800 ss60mer of identical sequence was added at a molar ratio of 1:50 with respect to unlabeled DNA. DNA was detected with the Odyssey infrared detection platform (LI-COR Biosciences). As an alternative method, Brh2 complexed with 32P-labeled-His-PK-Dss1 was added to reaction mixes containing 3′-biotin-labeled ss60mer (phage φX174 sequence as above, MWG Biotech AG). 3′-biotin-labeled ss60mer annealed with partially complementary 5′-IRD700 labeled 80mer (CGAATTAAATCGAAGTGGACTGCTGGCGGAAAATGAGAAAATTCGACCTAT CCTTGCGCAGCTCGAGAAGCTCTTACTTT) was added as a spike (1:50 molar ratio with respect to ss60mer) to reaction mixes to enable visualization of the DNA. After incubation, mixes (15 μL) were transferred to a tube containing 10 μg streptavidin coated-magnetic particles (Pierce), which were collected with a magnet and the unbound fraction set aside. The beads were washed with two (100 μL) rinses of reaction buffer. Protein (and DNA) in the bound and unbound fractions was analyzed by SDS-gel electrophoresis and visualized by staining, phosphorimaging, and infrared dye detection.
Reconstitution of the Brh2/Dss1 complex
Brh2 apoprotein (30 nM) free of Dss1 was added to mixes (0.5 mL) containing 25 mM Tris-HCl, pH 7.5, 25 mM KCl, 4 mM MgCl2, 1 mM DTT, 10% glycerol with 250 nM His-Dss1. After 1 hr at 4°, 100 μL packed Ni2+-NTA beaded resin was added and the slurry mixed for 10 min. Beads were collected by centrifugation, washed twice (0.5 mL each) with 25 mM Tris-HCl, pH 7.5, 200 mM KCl, 20 mM imidazole, 0.1% NP-40, 10% glycerol, and protein in the supernatant and bound fractions was analyzed by electrophoresis in polyacrylamide gels containing SDS.
RESULTS
Complexes formed by addition of Brh2/Dss1 or Brh2NT to DNA
In the experiments described below we used Brh2NT (Brh21–551), full-length Brh2 (Brh21–1075), and Brh2CT (Brh2505–1075), the latter two necessarily being produced as heterotypic complexes with Dss1 (His-tagged). We have referred to these latter complexes in the past as Brh2 and Brh2CT for simplicity, but to avoid confusion in this present work we will indicate Brh2/Dss1 or Brh2CT/Dss1 when the heterotypic complex was used in reactions. Purification of these proteins as well as Dss1 and their analyses by SDS-gel electrophoresis have been described and documented in previous work (11, 16, 19). We also demonstrated in those previous studies that the affinity tags did not interfere with the biological activity of the proteins as measured by complementation activity.
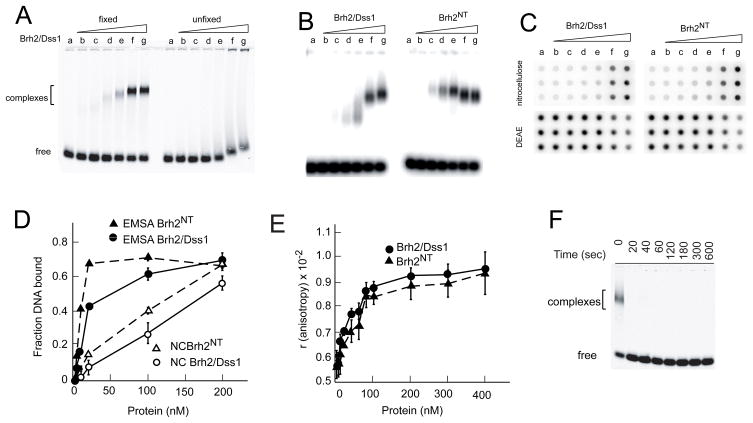
This present work evolved from studies on the DNA structure preference exhibited by Brh2/Dss1 and Brh2NT in binding reactions using the electrophoretic mobility shift assay (16, 19). Experimentation was performed at protein concentrations low enough such that the DNA-binding activity observed was due predominantly to the primary interaction site located in the N-terminal region of Brh2 (16). As substrate we used a single-stranded mixed sequence oligonucleotide of 60 nucleotides 5′-labeled with 32P or IRD fluorophore (16). Mixed sequence oligonucleotide was used as substrate rather than oligo dT, because the latter bound so poorly that complexes were minimally detectable at the protein concentrations used (data not shown). A length of 60 residues was chosen because it was a good substrate in D-loop reactions performed in related studies with Rad51 and it was above an apparent minimum of around 30 residues necessary for efficient complex formation with Brh2 (data not shown). Under our reaction conditions, protein-DNA complexes were unstable. Therefore, we incorporated a glutaraldehyde fixation step so that complexes could be visualized with more clarity (Fig. 1A). Here as in previous studies (16) we found that Brh2/Dss1 and Brh2NT bound natural mixed sequence ss60mer or ss100mer with approximately the same avidity, and formed discrete complexes. At low concentrations of Brh2/Dss1 complexes ran with faster mobility than the Brh2NT-DNA complexes, in spite of the greater mass (Fig. 1B, compare lanes d–g). With increasing concentrations of Brh2/Dss1, DNA complexes with slower mobility formed, while with increasing concentration of Brh2NT the complexes ran more uniformly, and even with slightly faster mobility at higher concentrations. As the mobility shift assay was performed after addition of glutaraldehyde, it is possible that the fixation contributed to anomalies in mobility of the complexes. It is also possible that the protein-DNA complexes changed in conformation as a function of concentration, or that protein-protein interactions contributed to the differences in mobilities, or that DNA secondary structure of the mixed sequence oligonucleotides was contributory.
FIGURE 1.
DNA binding assays. A. Binding reactions containing IRD800 ss60mer and Brh2/Dss1 at the concentrations indicated were incubated for 40 min. Samples were either fixed with glutaraldehyde before electrophoresis or else were run without fixation. Lane a-no protein; b-2 nM; c-4 nM; d-10 nM; e-20 nM; f-100 nM; g-200 nM. B. 32Pss100mer was incubated with Brh2/Dss1 or Brh2NT at the concentrations indicated in A and samples fixed with glutaraldehyde. C. Reaction mixes with 32P-ss60mer incubated with Brh2/Dss1 or Brh2NT at the concentrations indicated in A were passed onto a manifold through a double layer sandwich of nitrocellulose and DEAE-cellulose membrane. D. Quantification of the results in B and C. E. Binding reactions containing 5′-fluorescein-labeled 49mer and Brh2/Dss1 at the indicated concentrations were incubated for 20 min, then anisotropy was determined. F. Sample mixes containing 100 nM Brh2/Dss1 but no DNA were set up. Glutaraldehyde was then added to samples and incubated for the indicated times before initiating the binding reaction by addition of IRD800 ss60mer. After further incubation for 20 min, samples were analyzed by gel electrophoresis.
As an alternative means of determination, we measured DNA binding by the classic nitrocellulose filter retention method (20), which does not rely on a fixation step, but simply traps protein-DNA complexes on a porous membrane of nitrocellulose pretreated to reduce binding of free single-stranded DNA (Fig. 1C). A second layer of DEAE-paper captures the free DNA. An advantage of this assay is that binding reactions can be stopped in seconds simply by passing reaction mixes through the filter to remove uncomplexed components and so can be used to monitor rates of binding. Nevertheless, the method does not give a true measure of equilibrium, can be inefficient at trapping complexes, and does not necessarily yield any additional information about the nature of protein-DNA complexes formed. When performed side by side, the nitrocellulose filter assay was less cumbersome than the mobility shift assay but was less sensitive at detecting complexes formed at lower concentrations of Brh2/Dss1 and Brh2NT (Fig. 1D), and unfortunately gave us no further insight into the basis for the mobility differences noted above.
Finally we used fluorescence polarization as a means to verify binding. An advantage of this method is that observations are made in real time and require no perturbation of the system. A change in anisotropy was evident upon addition of Brh2/Dss1 or Brh2NT to a fluorescein-labeled ss49mer. The magnitudes of change were virtually identical and the binding isotherms were essentially super-imposable. These findings support the notion that the masses of the complexes formed are comparable in the case of Brh2/Dss1 and Brh2NT and also agree with the results from the mobility shift assay that their binding affinities are about the same. The instrument we used was not well-suited for monitoring association rates.
For our purposes the mobility shift assay was excellent for measuring protein- DNA complexes with Brh2/Dss1 and Brh2NT in terms of sensitivity and in discerning differences in the nature of the protein-DNA complexes formed, although the basis of the differences remains unclear. While not ideal for examining association kinetics, the assay works to advantage when the glutaraldehyde fixation step is incorporated as the fixative abolishes Brh2 binding activity within a matter of seconds effectively quenching the reaction (Fig. 1F) as well as stabilizing the complexes (Fig. 1A). Thus, the assay can suffice to detect gross differences in binding rates and can be useful in distinguishing qualitative differences in various types of complexes formed. Knowledge of the basis of the differences will require analysis by a more sophisticated methodology such as surface plasmon resonance, and is beyond the scope of this study.
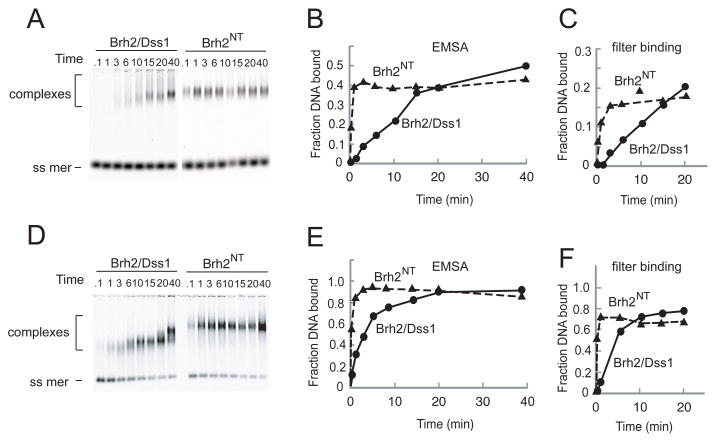
Association of Brh2/Dss1 with DNA is slow compared to Brh2NT
A striking difference that we noted between full length Brh2/Dss1 and Brh2NT in the binding reactions was that full length Brh2/Dss1 bound DNA slower than Brh2NT (Fig. 2). Formation of protein- DNA complexes with two different concentrations of Brh2NT was fast with a half time of seconds and approached a plateau level within 1 min. On the other hand, formation of complexes between full length Brh2/Dss1 and DNA followed apparently hyperbolic association kinetics but at a slower rate that was dependent on the protein concentration. Similar observations were made using both the mobility shift and nitrocellulose filter binding assays (compare Fig. 2B, C with Fig. 2E, F). We had assumed that the association of Brh2/Dss1 and derivatives with DNA was diffusion-controlled and would be rapid as is commonly observed with DNA-binding proteins. The fast association of Brh2NT with DNA was consistent with our expectations for a diffusion-controlled binding reaction, but the slow formation of protein-DNA complexes with the full length Brh2 was surprising and raised the notion that another process was rate-limiting in the binding reaction.
FIGURE 2.
DNA binding rates. Time courses of DNA binding were determined with single-stranded oligonucleotide and Brh2 or Brh2NT. Aliquots were removed from binding reactions at times indicated, mixed with glutaraldehyde, and the DNA analyzed for mobility shift after electrophoresis. The fraction of DNA bound was quantified from the relative intensity of the free and shifted oligomer and is shown graphically below. A. Brh2 or Brh2NT at 20 nM. B. Brh2 or Brh2NT at 100 nM.
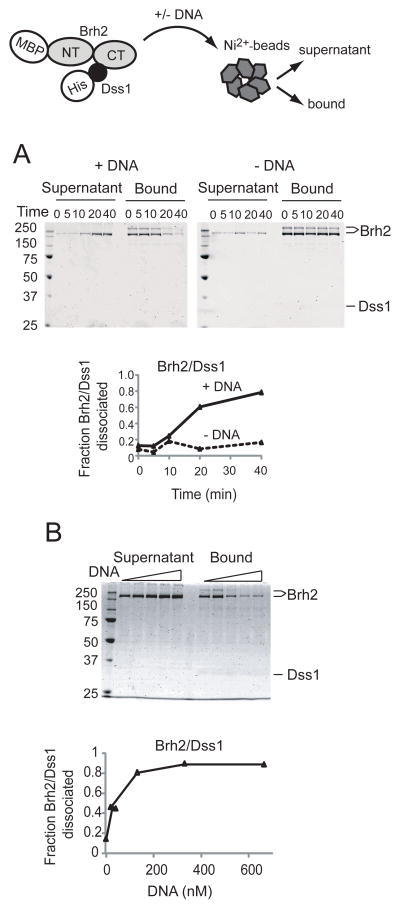
Dissociation of Brh2/Dss1 is enhanced by DNA
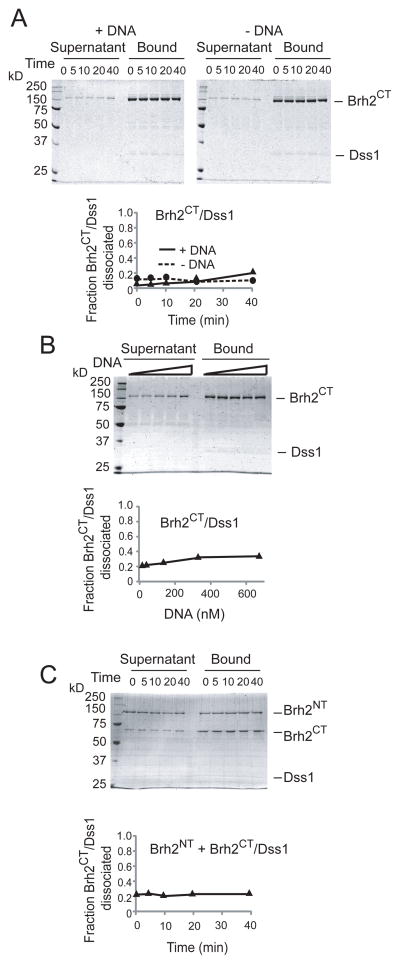
A working model would suggest that a change in state of the protein might be necessary to reveal the strong N-terminal DNA-binding domain. As Brh2 was purified as a heterotypic protein complex with Dss1, we suspected that Dss1 would be most likely responsible for regulating any change in state of Brh2. Therefore, we examined the Brh2/Dss1 complex after addition of DNA. We took advantage of the His-tag on Dss1 as a means for selectively separating Brh2 molecules complexed with Dss1 from those free of Dss1 by a pulldown procedure using Ni2+-nitrilotriacetate agarose (Ni2+-NTA) beads (Fig. 3, schematic). Ni2+-NTA beads were added to a solution containing the Brh2/Dss1 complex, collected by centrifugation, and protein in the supernatant and bound fractions was analyzed by SDS gel electrophoresis and detected by staining the gels with SimplyBlue SafeStain. This stain was advantageous in detecting Dss1, which due to its small size and disordered structure (see below), was problematic in detection and would leach out of gels particularly under the prolonged acidic soaking and washing conditions used in staining with Coomassie brilliant blue. With Brh2/Dss1 samples held on ice there was a small fraction of Brh2 (<10%) that was not captured by the beads. This baseline level represents Brh2/Dss1 complex that simply escaped capture from the Ni2+-NTA beads plus Brh2 molecules in the preparation free of Dss1. After incubation at 37° in buffer containing a modest salt concentration there was only slight dissociation of Brh2/Dss1 complex as evident by the marginal increase in level of Brh2 in the supernatant (Fig. 3A). When DNA was added, however, Brh2 became almost quantitatively dissociated from Dss1 (Fig. 3A). In this particular experiment (Fig. 3A) Dss1 was hardly visible after the staining procedure. With an input of 500 nM Brh2/Dss1 complex, 130 nM ss60mer (~ 1 ss60mer oligonucleotide per 4 Brh2 molecules) was sufficient to promote maximal dissociation (Fig. 3B). Brh2 in the bound fraction was recovered in two forms after elution from the beads as evident in the SDS gels. One form ran with the mass usually observed (~170 kDa), while the second ran with an apparent mass of at least twice that. This is most likely the result of a Ni2+ -mediated oxidative crosslinking reaction that occurred during processing the samples. Such intermolecular crosslinking with His-tagged proteins has been documented (21).
FIGURE 3.
Dissociation of Brh2/Dss1 complexes. MBP-tagged Brh2 (represented as N-terminal and C-terminal lobes) complexes with His-tagged Dss1(black ball) is mixed with Ni2+-NTA beads. A. Brh2/His-Dss1 complex (550 nM) was incubated at 37° in the presence or absence of DNA (130 nM unlabeled ss60mer). At the indicated times (min) Ni2+-NTA beads were added to pull down His-Dss1 either free or complexed with Brh2. Protein composition in the supernatant or bound fractions was determined by SDS gel electrophoresis. Protein was visualized by staining with SimplyBlue Safestain (Invitrogen). The time course of dissociation is shown graphically. B. Reactions mixes contained Brh2/His-Dss1 complex (550 nM) and increasing concentrations of DNA (left to right-20, 40, 130, 330, 660 nM) were incubated for 20 min. The zero point for no DNA added was taken from part A.
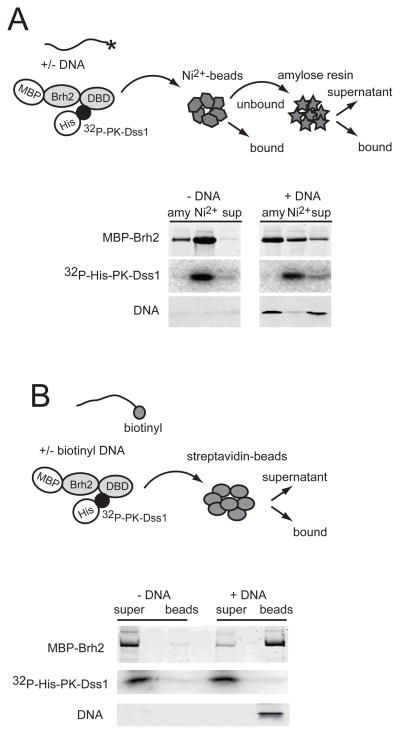
Brh2 bound to DNA is free of Dss1
The above experiment suggested that Dss1 dissociates from Brh2 in the presence of DNA. To investigate this point we asked about the status of Dss1 once DNA became bound to Brh2. Two approaches were taken using a version of Dss1 labeled with 32P so that it could be tracked more easily. For this purpose the protein was engineered to contain a target sequence for protein kinase A that enabled 32P-radiolabeling. With the further addition of fluorescently labeled ss60mer DNA as substrate we were able to track the fate of all three components after mixing. In the first approach, following incubation of the Brh2/Dss1 complex (MBP-Brh2/32P-His-PK-Dss1) with or without DNA, Ni2+- NTA beads were added to trap Dss1 (Fig. 4A). The unbound fraction was collected and was added to amylose resin to trap Brh2. As is evident (Fig. 4A) when no DNA was present, the bulk of the Brh2 was retained on the Ni2+-NTA beads together with the Dss1. Brh2 that was not retained by the Ni2+-NTA beads was pulled down with amylose resin and had little associated Dss1. When DNA was present, the amount of Brh2 retained by the Ni2+-NTA beads was substantially reduced and little DNA was trapped. The bulk of the Brh2 and large fraction of the DNA was trapped by amylose beads. There was little Dss1 in this fraction.
FIGURE 4.
Dss1 is not associated with Brh2 bound to DNA. A. MBP-Brh2 complexed with 32P-labeled His-PK-Dss1 was added to reaction mixes with or without IRD 800 ss60mer DNA. After 20 min each mix was transferred to a tube containing Ni2+-NTA beads (as packed slurry) and held on ice for 10 min. After brief centrifugation the beads were collected and the supernatant containing the unbound protein fraction was then transferred to a tube containing amylose beads. After 10 min, the beads were collected and the bound fractions from both Ni2+-NTA (Ni2+) and amylose (amy) plus the final supernatant (sup) were analyzed for protein and DNA composition after SDS gel electrophoresis. Brh2 was visualized by protein staining, Dss1 by phosphorimaging, and DNA by fluorescence detection. B. MBP-Brh2 complexed with 32P-labeled His-PK-Dss1 was added to reaction mixes with or without 3′-biotin-labeled ss60mer DNA containing IRD700-labeled complementary strand as tracer. After incubation for 40 min mixes were transferred to a tube containing streptavidin coated-magnetic particles. After collecting the beads, protein and DNA in the bound and supernatant fractions were analyzed by SDS-gel electrophoresis.
In the second approach, the DNA was biotin-labeled ss60mer spiked with a small amount hybridized to a partially complementary fluorescently labeled ss80mer (Fig. 4B). After incubation with the Brh2/Dss1 complex, the DNA was trapped on streptavidin-coated beads, and the bound components were analyzed (Fig. 4B). The bulk of the Brh2 was present in the DNA-bound fraction while the bulk of the Dss1 was in the supernatant. Conversely, when no DNA was added, only a background level of Brh2 was associated with the beads. Taken together these results indicate that Dss1 is no longer associated with Brh2 when it is bound to DNA.
Excess Dss1 inhibits binding of Brh2/Dss1 to DNA
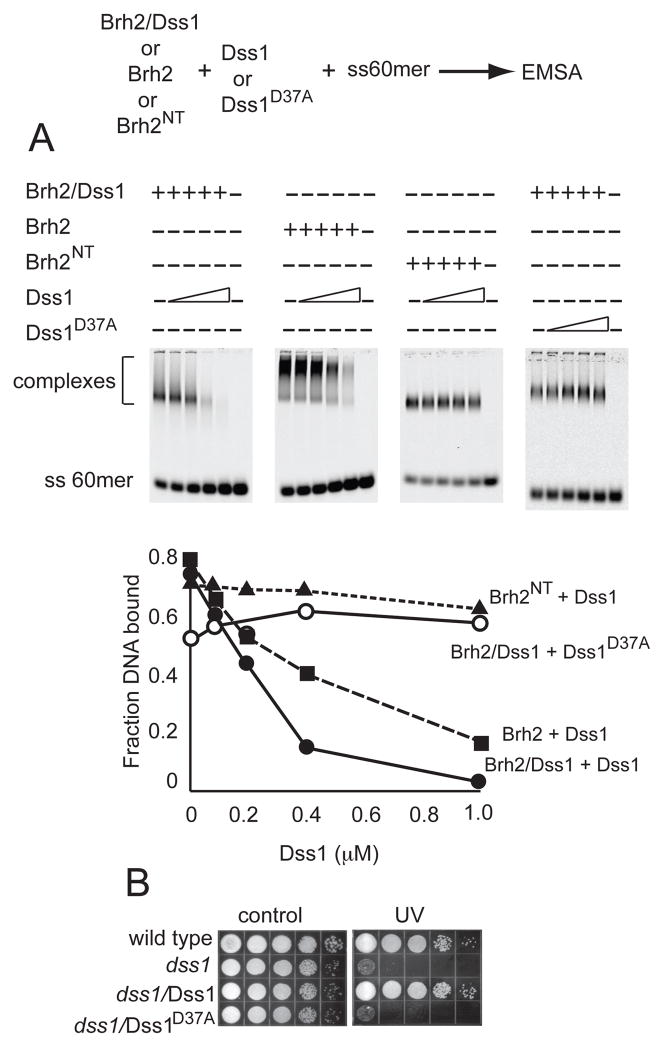
From the crystal structure determination of the the murine BRCA2 DBD, it was found that DSS1 bound to a face of the molecule on the opposite side of the OB fold DNA-binding groove (13). Nevertheless, because Dss1 is a highly acidic polypeptide with interspersed hydrophobic and aromatic residues, it was proposed as a DNA mimic with possible regulatory capability. In view of our localizing the predominant DNA binding domain within Brh2 to the N-terminal region (16), we considered the possibility that Dss1 interacted with this domain in Brh2NT as well as with the canonical C-terminal region corresponding to the face of the DBD opposite the OB-fold DNA binding channel. If Dss1 bound to the DNA-binding domain within the N-terminal region, it would be expected to compete with DNA for binding to Brh2NT. Upon testing this notion we found that increasing the level of Dss1 had no effect on the ability of Brh2NT to bind DNA. Addition of increasing levels of Dss1 to Brh2/Dss1, however, resulted in a corresponding decrease in DNA binding (Fig. 5A). These results suggest that the activity of Dss1 in inhibiting Brh2 from binding DNA is mediated through the C-terminal Dss1-interacting region and is not due simply to competition between Dss1 and DNA for a common binding site.
FIGURE 5.
Dss1 attenuates DNA-binding activity of Brh2/Dss1 complex. A. To DNA binding reaction mixtures containing IRD800 labeled ss60mer (3.3 nM) and Brh2/His- Dss1, Brh2, or Brh2NT (100 nM) was added increasing levels of His-Dss1 or His- Dss1D37A to the lanes as indicated (0.1, 0.2, 0.4, and 1.0 μM, respectively). After incubation for 20 min, DNA binding was determined. B. Survival of dss1 deletion strain expressing Dss1 or Dss1D37A was determined after a UV dose of 120 J/m2.
We asked whether there was specificity in the inhibition of DNA binding by Dss1 or whether the inhibition was due to an electrostatic effect based on the high acidic content of Dss1. To answer this we took advantage of a Dss1 mutant protein altered by change of a single aspartic acid residue to alanine (Dss1D37A). The mutation causes loss of DNA repair activity as determined by failure to complement the radiation sensitivity of the dss1 mutant upon expression of Dss1D37A (Fig. 5B). Residue D37 corresponds to D16 of mammalian DSS1, which contacts K2671 in mouse BRCA2 DBD (or K2750 in human). As the change of D37A reduces the total ionic charge by only 5%, the mutant protein should still retain substantial activity in inhibiting ability of Brh2 to bind DNA if the inhibition is due to a generalized electrostatic repulsion. On the other hand, if regulation of DNA binding by Dss1 is mediated through specific interactions at a functional interface as supported by the intermolecular aspartate-lysine contact in the atomic structure, the mutant should be unable to inhibit DNA binding by Brh2. Experimentally we observed that Dss1D37A did not inhibit Brh2’s DNA-binding activity, indicating that specificity for that particular residue is indeed important. Taken together these observations suggest that the N-terminal DNA-binding domain of Brh2 is modulated by allosteric control through the C-terminal region.
Dissociation of Brh2/Dss1 facilitated by the Brh2 N-terminal region
If binding of DNA through the strong, primary interacting domain in the N-terminal region of Brh2 is negatively regulated by interaction of Dss1 with the C-terminal region, then it seems likely there must be signaling, likely a conformational change, transmitted between the two domains. Consistent with this notion the Brh2CT/Dss1 complex did not dissociate when incubated with DNA (Fig. 6A and B), in contrast to dissociation of Brh2/Dss1 promoted by DNA (Fig. 3). Furthermore, when Brh2NT was added as a separate component to the Brh2CT/Dss1 complex there was no enhanced dissociation of the complex (Fig. 6C). These observations reinforce the notion that DNA binding and Dss1 dissociation are counterbalanced through a mechanism involving structural change that propagates between the N-terminal and C-terminal regions of Brh2.
FIGURE 6.
Brh2 N-terminal domain is required for dissociation of Dss1. A. Brh2CT/His-Dss1 complex (550 nM) was incubated at 37° in the presence or absence of DNA (130 nM unlabeled ss60mer). At the indicated times (min) Ni2+-NTA beads were added to pull down His-Dss1 either free or complexed with Brh2 CT. Protein composition in the supernatant or bound fractions was determined by SDS gel electrophoresis. Protein was visualized by staining with SimplyBlue Safestain (Invitrogen). The time course of dissociation is shown graphically. B. Reactions mixes contained Brh2CT/His-Dss1 complex (550 nM) and increasing concentrations of DNA (left to right-20, 40, 130, 330, 660 nM) were incubated for 20 min. C. Reaction mix contained Brh2CT/His-Dss1 complex (550 nM) and Brh2NT(550 nM). In this case Brh2NT was tagged with MBP and His whereas the MBP-tag was removed from Brh2CT by cleavage with TEV protease. At the times indicated Ni2+-NTA beads were added to pull down His-Dss1 and protein composition in the supernatant and bound fractions was determined.
Brh2 free of Dss1 binds DNA rapidly and re-associates with Dss1
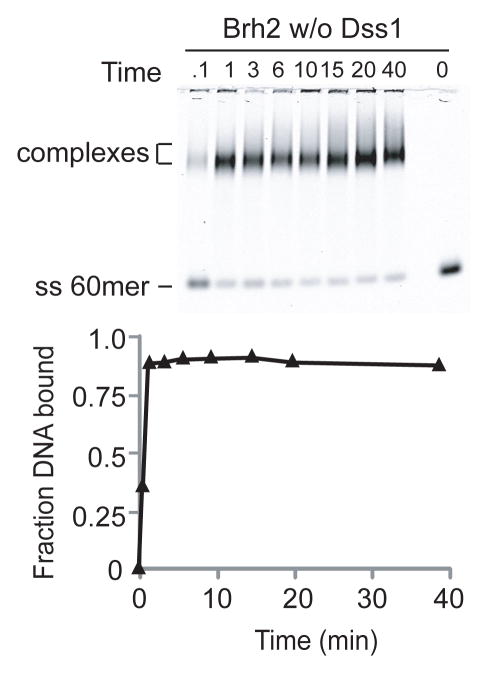
From the initial observations in this study that the strong binding of DNA on addition of Brh2/Dss1 is correlated with dissociation of Dss1 and that excess Dss1 inhibits DNA binding, a two-ligand model is suggested. Namely, DNA and Dss1 negatively regulate each other’s association with Brh2. Corroborating support for this model should be obtainable through simple mass action experiments. Two predictions should be satisfied. First, Brh2 stripped of Dss1 should bind DNA quickly, and second, Dss1 should be capable of associating with Brh2 in the absence of DNA. Unfortunately, obtaining Brh2 free of Dss1 has been problematic because Brh2 expressed in E. coli in the absence of Dss1 is prone to aggregation and is highly susceptible to proteolytic degradation. Brh2 co-expressed with Dss1, however, is not subject to those problems, and exhibits hydrodynamic properties consistent with behavior of a monomeric protein during gel filtration (8, 11). Therefore we used the Brh2/Dss1 complex as starting material and devised a procedure to prepare Brh2 free of Dss1. The approach we took was the empirical one of searching for a condition favoring dissociation of the Brh2/Dss1 complex. Then we took advantage of the affinity tags on Brh2 and Dss1 to isolate Brh2 apoprotein directly or else deplete the preparation of Dss1. Of many conditions tested, the promising one was that divalent cations were effective in promoting dissociation of Brh2/Dss1. We determined that holding the complex in buffer with 20 mM Mg2+ resulted in dissociation of Brh2 from Dss1. By incorporating a Mg2+ incubation step in the purification procedure, coupled with affinity purification, we were able to isolate Brh2 protein largely free of Dss1. Nevertheless, the protein in this form was difficult to work with, as it was highly sensitive to the slightest trace of protease contamination and also appeared prone to aggregation. We have encountered variability in experiments using Brh2 apoprotein and attribute this to its unstable nature.
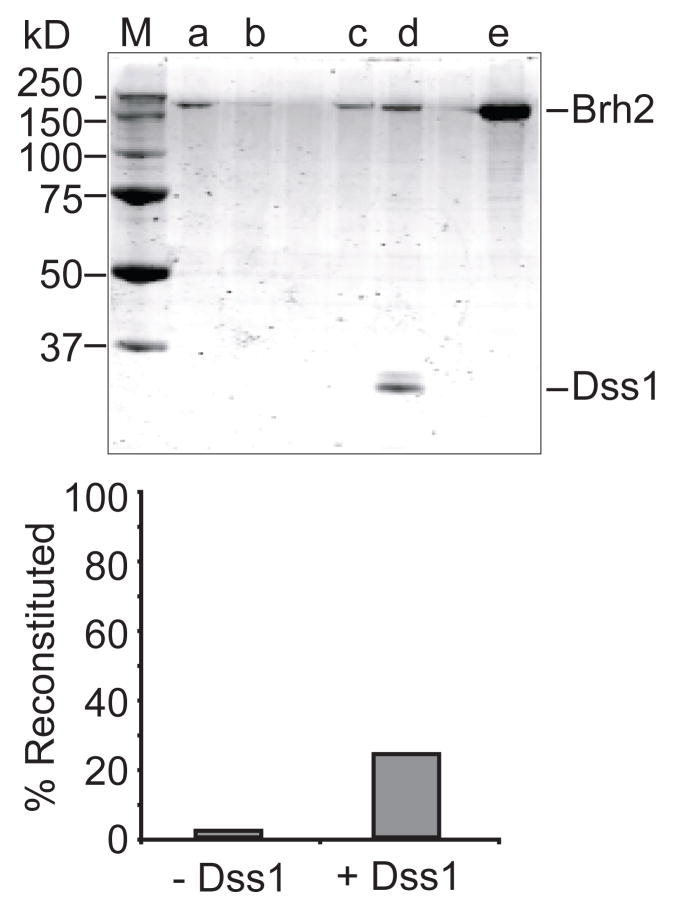
We monitored DNA binding by the Brh2 apoprotein, and indeed observed that the time course exhibited no delay and appeared comparable to that of Brh2NT in the rapid rate of association (Fig. 7). The DNA binding activity of Brh2 apoprotein was also inhibited by addition of Dss1 (Fig. 5). We tested for re-association of His-Dss1 with Brh2 apoprotein using the pulldown procedure with Ni2+-NTA beads to capture His-tagged Dss1. After Brh2 was incubated in a reaction mix containing an 8-fold molar excess of His-Dss1, a substantial fraction of the Brh2 became capable of associating with the Ni2+-NTA resin in a Dss1-dependent manner (Fig. 8). By this procedure we estimated that about 70% of the Brh2 initially added to the reaction tube could be accounted for after recovery in the bound and supernatant fractions. Of this total recovered amount we estimate that 24% had re-associated with His-Dss1 (Fig. 8B). A minor fraction of Brh2 (1.3%) was found associated with the Ni2+-NTA beads in the absence of His-Dss1. Thus, the Brh2 present in the bound fraction is due to specific interaction with His-Dss1. We do not know as yet the factors or conditions that would favor more extensive reassociation of Dss1, but with variation of salt, temperature, divalent cation, etc., it seems likely that the reverse reaction could be optimized empirically. In summary, these observations suggest that an equilibrium between Dss1- bound and Dss1-free forms of Brh2 is important in DNA binding and support the idea that accessibility of Brh2 to DNA is governed by Dss1.
FIGURE 7.
DNA binding rate of Brh2 apoprotein. Time course of DNA binding was determined with IRD800-labeled ss60mer (3.3 nM) and 100 nM Brh2 stripped of Dss1 prepared as described in Material and Methods. Aliquots were removed from binding reactions at times (min) indicated, fixed with glutaraldehyde, and the DNA analyzed for mobility shift after electrophoresis. For the zero time point, DNA was added to a Brh2 reaction mixture already containing glutaraldehyde.
FIGURE 8.
Reconstitution of the Brh2/Dss1 complex. Reaction mixtures containing Brh2 apoprotein were mixed with His-Dss1. After 1 hr at 4°, Ni2+-NTA beads were added, washed, then eluted with SDS sample buffer (80 μL). Aliquots (10 μL) of the supernatant (first wash) and bound fractions were separated by SDS gel electrophoresis. After staining with SimplyBlue Safestain, bands were quantitated using the Odyssey detection platform. Lane a -- supernatant, no Dss1; lane b -- bound, no Dss1; lane c -- supernatant, plus Dss1; lane d -- bound, plus Dss1; lane e -- 12.5% of total Brh2 apoprotein initially added to the reaction. M--size standards.
DISCUSSION
There are two principal conclusions from this study. First, DNA-binding potential of Brh2 is linked to a change in state of the protein that is mediated by Dss1. Second, DNA and Dss1 serve as counterbalancing ligands that attenuate each other’s association with Brh2. These findings suggest that Brh2 function involves dynamic conformational changes in domain configuration that are regulated by Dss1, in essence allostery.
We have established that Dss1 is crucial for proficiency in DNA repair and recombination in U. maydis and made the argument that it serves to regulate Brh2 function (12, 15). That Dss1 serves as a regulator is supported by the observation that variant forms of Brh2 deleted of the Dss1-interacting region, indeed of the entire canonical Dss1/DNA-binding domain, are liberated from an inactive state and exhibit resistance to radiation, enable formation of subnuclear Rad51 foci in cells following DNA damage, and are recombination proficient (15). The N-terminal region of Brh2 has a strong DNA-binding domain with properties that apparently make possible all the DNA-interactive operations attributable to the full-length protein (16). With the added Rad51-interacting BRC element, it would seem that the N-terminal region of Brh2 constitutes a module endowed with all of the abilities to engage Rad51 and to deliver it to DNA (15). The positive and direct role of the N-terminal region of Brh2 in Rad51 presentation appears balanced by forces imposed from the C-terminal region. This is most apparent in studies on recombination in which the frequency of gap repair as well as the rate of spontaneous allelic recombination is higher in cells expressing the Brh2NT as compared to the full-length protein (15). Additional support comes from studies with a mutant defective in the RecQ helicase Blm. Toxic recombination-dependent DNA structures formed during replication stress in the presence of hydroxyurea are not generated in blm brh2 double mutant cells expressing Brh2NT in contrast to the full-length protein (22). In previous work it was found that a second Rad51-interacting element, CRE, unrelated to BRC was present at the extreme C-terminus of Brh2 (11). These elements appear to work together in some way to impose appropriate and controlled recombinational activity, and Dss1 appears to mediate their communication. The present finding showing that Dss1 governs interaction of DNA with the primary binding site within the N-terminus of Brh2 adds another layer of knowledge and some additional insight into mechanism.
We imagine two different models that could account for the observations. In one view Dss1 occludes DNA from binding to the primary N-terminal DNA-binding domain by imposing an unfavorable conformation mediated through the C-terminal region. Here the Dss1-bound form of Brh2 is imagined in equilibrium with the unbound form although the equilibrium lies far to the left (bound form). Upon dissociation of Dss1, the conformation changes and the N-terminal DNA-binding site becomes available for association with DNA. Since Dss1 does not appear to associate with Brh2NT or compete with it for binding DNA, Brh2 is likely in a conformational state when Dss1 is bound that prevents DNA binding to the N-terminal DNA-binding domain. A change resulting from Dss1 dissociation enables DNA binding. An alternative model is that DNA binds to the dominant N-terminal DNA-binding domain and through an induced fit a conformational change ensues that causes ejection of Dss1.
In favor of the latter mechanism (DNA binds first) is the finding that with increasing concentration of DNA added to Brh2/Dss1 complexes, the greater the fraction of Brh2 is dissociated from Dss1. In support of the former mechanism (Dss1 dissociates first) is the finding that binding of DNA to Brh2/Dss1 appears to be slow compared to binding to the Brh2NT, implying that another step is rate-limiting in the DNA binding reaction. We imagine this step is a conformational change concomitant with the dissociation of Dss1. There are no doubt other possibilities, and of course, these views might be over-simplified. Perhaps it is naïve to view DNA and Dss1 as model ligands that are mutually exclusive. Their associations with Brh2 may not be accurately represented in terms of a simple "on" or "off" state when there could be multiple attachment points. In the case of Dss1, in which there could be multidentate association with Brh2, maybe the transient loosening of one site is enough to open contacts for association with DNA without physical separation of Dss1. For instance, it could also be argued that the observed time delay in DNA binding is consistent with the induced fit model in which DNA binds first to force out Dss1. In this case one would have to stipulate that the initial DNA binding is weak so that the protein-DNA encounter complexes formed are not strong enough to be immediately detected by our methods. Then as Dss1 is ejected there would follow the formation of a DNA-bound complex that would become stronger over time as more and more contacts were formed between surfaces. Further experimentation will be required to distinguish between the models.
In summary, it appears that the activation of Brh2 for DNA binding coincides with the dissociation of Dss1. Concomitant with this is conversion of Brh2 to a dimeric or even higher order form (11). The mechanism, however, by which Rad51 delivery to sites of DNA damage is coupled to these structural changes in Brh2 is not understood. It seems evident that interplay between Dss1 and the cognate interaction site within the C-terminal region of Brh2 provides a means for fine-tuning recombination. In the absence of this entire region Rad51 foci persist longer and allelic recombination and gap repair occur at significantly higher frequencies than in wild type cells (15). Given the well-documented relationship between genomic instability and elevated recombination, it can be imagined that such an increase might well be detrimental. We have no information at present about how the DNA-binding domain represented by the C-terminal OB folds contributes to attenuating recombination or how the C-terminal Rad51-interacting element CRE takes part. However, it seems likely that Rad51-filament quality is compromised somehow because cells expressing only the Brh2NT are more sensitive to high doses of UV (15). This is especially apparent in cells also lacking the Rad51 paralog Rec2 (10, 16).
Finally, there is the broader question of why interaction of Brh2 with DNA is regulated by Dss1. Or, put another way, what biological purpose is served by coupling dependence of Brh2 activity on its state of association with Dss1? Bioinformatics analysis with disorder prediction programs such as IUPred (23) or FoldIndex (24) indicates Dss1 to be predominately lacking in secondary structure (data not shown). But as is clear from the crystal structure of the mammalian BRCA2 DBD, regions of Dss1 fold into an ordered state when associated with this interacting partner, and concomitantly impose stability on the partner (13). Thus Dss1 might be considered as a type of chaperone. But based on the above observations it would appear that Dss1 is appropriately and more generally categorized as an intrinsically unstructured or disordered protein, an emerging class of proteins recognized for mediating signaling and regulation of many biological systems (25–27). Besides its role in DNA repair and recombination, Dss1 has been shown to function in mRNA processing (28–30)and to serve as an integral component of the 19S regulatory subunit of the proteasome (31–33). Thus, Dss1 appears to function as a hub linking several diverse cellular networks. Understanding the details of Dss1 interaction with Brh2 will be important for understanding why a crucial DNA repair pathway is dependent on such a regulatory mechanism and could lead to insight into a more general understanding of how DNA repair is coupled to the major cellular systems of RNA and protein processing.
Acknowledgments
We thank Drs. Lorraine Symington, Columbia University, for critical comments on the science and manuscript.
Footnotes
The abbreviations used are: BRC, Rad51-binding element; CRE, C-terminal Rad51-binding element; CT, carboxy-terminal; DBD, DSS1/DNA-binding domain; ds, double-stranded; HEPES, N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid); His-tag, hexahistidine-tag; MBP, maltose binding protein; NT, amino terminal; NTA, nitrilotriacetate agarose; OB, oligonucleotide/oligosaccharide binding; oligo dT, oligothymidylate; ss, single-stranded; UV, ultraviolet
This work was supported in part by grants GM42482 and GM79859 from the National Institutes of Health.
References
- 1.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 2.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 4.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 5.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 7.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc Natl Acad Sci U S A. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 9.Mazloum N, Holloman WK. Second-end capture in DNA double-strand break repair promoted by Brh2 protein of Ustilago maydis. Mol Cell. 2009;33:160–170. doi: 10.1016/j.molcel.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kojic M, Zhou Q, Lisby M, Holloman WK. Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis. Mol Cell Biol. 2006;26:678–688. doi: 10.1128/MCB.26.2.678-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Kojic M, Cao Z, Lisby M, Mazloum NA, Holloman WK. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol Cell Biol. 2007;27:2512–2526. doi: 10.1128/MCB.01907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 14.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 15.Kojic M, Zhou Q, Lisby M, Holloman WK. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol Cell Biol. 2005;25:2547–2557. doi: 10.1128/MCB.25.7.2547-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Kojic M, Holloman WK. DNA-binding domain within Brh2 N-terminus is the primary interaction site for association with DNA. J Biol Chem. 2009;284:8265–8273. doi: 10.1074/jbc.M809226200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojic M, Mao N, Zhou Q, Lisby M, Holloman WK. Compensatory role for Rad52 during recombinational repair in Ustilago maydis. Mol Microbiol. 2008;67:1156–1168. doi: 10.1111/j.1365-2958.2008.06116.x. [DOI] [PubMed] [Google Scholar]
- 18.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci U S A. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fancy DA, Melcher K, Johnston SA, Kodadek T. New chemistry for the study of multiprotein complexes: the six-histidine tag as a receptor for a protein crosslinking reagent. Chem Biol. 1996;3:551–559. doi: 10.1016/s1074-5521(96)90146-5. [DOI] [PubMed] [Google Scholar]
- 22.Mao N, Kojic M, Holloman WK. Role of Blm and collaborating factors in recombination and survival following replication stress in Ustilago maydis. DNA Repair. 2009;8:752–759. doi: 10.1016/j.dnarep.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 24.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 25.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 26.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47:7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, Ideker T, Guthrie C, Krogan NJ. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faza MB, Kemmler S, Jimeno s, Gonzalez-aguilera C, Aguilera A, Hurt E, Panse VG. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- 32.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 33.Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]