Abstract
Myosin is the primary regulator of muscle strength and contractility. Here we show that three myosin genes, Myh6, Myh7, and Myh7b, encode related microRNAs (miRNAs) within their introns, which, in turn, control muscle myosin content, myofiber identity and muscle performance. Within the adult heart, the Myh6 gene, encoding a fast myosin, co-expresses miR-208a, which regulates the expression of two slow myosins and their intronic miRNAs, Myh7/miR-208b and Myh7b/miR-499, respectively. miR-208b and miR-499 are functionally redundant, and play a dominant role in the specification of muscle fiber identity by activating slow and repressing fast myofiber gene programs. The actions of these miRNAs are mediated by a collection of transcriptional repressors of slow myofiber genes. These findings reveal that myosin genes not only encode the major contractile proteins of muscle, but act more broadly to influence muscle function by encoding a network of intronic miRNAs that control muscle gene expression and performance.
Introduction
The speed and force of both myocardial and skeletal muscle contraction are largely dependent on the intrinsic contractile properties of cardiac and skeletal myocytes. Myosin heavy chain (MHC) is the major contractile protein of cardiac and skeletal muscle cells and the primary determinant of the efficiency of muscle contraction. Cardiac and skeletal muscles modulate the expression of myosin genes in response to hormonal signaling and workload to meet physiological demands (Baldwin and Haddad, 2001).
Cardiac contractility depends on the expression of two MHC genes, α- and β-MHC (also known as Myh6 and Myh7, respectively), which are linked in a head-to-tail orientation and are regulated in an antithetical manner (Weiss and Leinwand, 1996). The ratio of α- to β-MHC expression is species-specific and varies in response to developmental, physiological and pathological signaling. In rodents, β-MHC, a slow ATPase, is highly expressed in the embryonic heart, whereas α-MHC, a fast ATPase, is expressed at relatively low levels in the pre-natal heart and is up-regulated during the early postnatal period (Morkin, 2000). Mechanical stress and hypothyroidism induce a shift from α- toward β-MHC composition of the adult heart (Fatkin et al., 2000; Gupta, 2007; Kiriazis and Kranias, 2000; Krenz and Robbins, 2004; Vanderheyden et al., 2008), which correlates with a decline in mechanical performance and contractile efficiency (Lowes et al., 1997; Miyata et al., 2000; Stelzer et al., 2007). Because relatively minor changes in the ratio of these two myosin isoforms can have profound effects on cardiac contractility in human and rodent hearts (Herron and McDonald, 2002; Korte et al., 2005; Rundell et al., 2005; Schiaffino and Reggiani, 1996), and increased expression of β-MHC in the myocardium decreases power output and contributes to depressed systolic function in end-stage heart failure, the mechanisms that regulate MHC gene switching have been the focus of intense interest.
Skeletal myofibers are also characterized by the expression of particular myosin isoforms and other contractile proteins that determine the efficiency of contraction (Bassel-Duby and Olson, 2006). In slow twitch, type I fibers, which display oxidative metabolism and high endurance, β-MHC is the predominant myosin, whereas fast twitch, type II myofibers express fast myosins and are primarily glycolytic and susceptible to fatigue. Thyroid hormone signaling represses β-MHC expression and type I myofiber formation, and promotes the fast myofiber phenotype. However, despite intense study, the exact regulatory mechanism behind myofiber switching in response to thyroid hormone signaling and other cues has remained elusive.
Recently, we reported that an intron of the α-MHC gene encodes a microRNA (miRNA), miR-208, that is required for up-regulation of slow β-MHC in the adult heart in response to stress and hypothyroidism (van Rooij et al., 2007). Given that miR-208 and its host myosin, α-MHC, are only expressed in the heart, these findings raised interesting questions as to whether other miRNAs might control myosin switching and contractile protein gene programs in fast versus slow skeletal muscle.
MiRNAs inhibit mRNA translation or promote mRNA degradation by annealing to complementary sequences in the 3′ untranslated regions of target mRNAs (Bartel, 2004). Individual miRNAs have numerous targets, and individual mRNAs are commonly targeted by multiple miRNAs, providing combinatorial complexity and broad regulatory potential to miRNA:mRNA interactions. miRNAs often target multiple mRNAs with shared functions and in so doing can exert robust control over complex cellular processes through modulation of multiple interrelated targets (Mourelatos et al., 2002; Stefani and Slack, 2008). The thyroid hormone receptor associated protein-1 (Thrap1), which functions as a positive and negative regulator of thyroid hormone signaling, is one target of miR-208 that appears to mediate the functions of this miRNA in the heart (van Rooij et al., 2007; Callis et al., 2009).
In the present study, we show that miR-208 is essential for the expression not only of β-MHC in the heart, but also of a closely related myosin isoform, Myh7b (McGuigan et al., 2004). Remarkably, both of these genes encode slow myosins and contain intronic miRNAs (miR-208b and miR-499, respectively) (Berezikov et al., 2006; Landgraf et al., 2007) that are expressed in cardiac as well as slow skeletal muscle. Through gain- and loss-of-function experiments in mice, we show that these myosin-encoded miRNAs act within a network to control myosin expression and skeletal myofiber phenotypes through the repression of a collection of transcriptional repressors of slow myofiber genes. Thus, myosin genes not only encode the major contractile proteins of muscle, but act more broadly to control muscle gene expression and performance through a network of intronic miRNAs.
Results
A family of miRNAs encoded by myosin genes
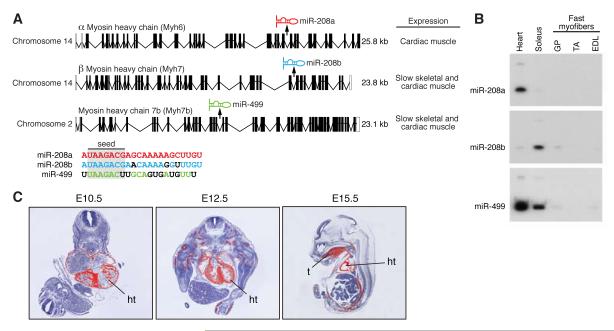
miR-208 is encoded by intron 27 of the mouse α-MHC gene, which is expressed specifically in the heart (Fig. 1A and B). Because of the important role of miR-208 in regulating cardiac gene expression in response to stress (van Rooij et al., 2007; Callis et al., 2009), we scanned other myosin genes for possible intronic miRNAs. Indeed, intron 31 of the mouse β-MHC gene is predicted to encode a closely related miRNA, miR-208b, which has an identical seed sequence to miR-208 and differs at only three nucleotides in the 3′ region (Fig. 1A). miR-208b is co-expressed with β-MHC, showing highest expression in soleus muscle, which is comprised predominantly of slow type I myofibers, and lower expression in adult heart (Fig. 1B). A third member of this miRNA family, miR-499, is encoded by intron 19 of the mouse Myh7b gene (McGuigan et al., 2004), a little studied myosin gene that shares extensive homology with β-MHC (Fig. 1A). Myh7b and miR-499 are highly expressed in the heart and are enriched in the soleus, compared with gastrocnemius/plantaris (G/P), tibialis anterior (TA), or extensor digitorum longus (EDL) muscles, which contain predominantly fast myofibers (Fig. 1B). Similar to β-MHC, Myh7b is expressed in the embryonic heart and skeletal muscle, as revealed by in situ hybridization (Fig. 1C). We refer to this family of miRNAs as MyomiRs, because of their location within and co-expression with their corresponding myosin genes. Henceforth, we refer to miR-208 as miR-208a to distinguish it from miR-208b.
Figure 1. Distinct expression patterns of MyomiRs.
A) Schematic representation of the genomic location of the miRNAs present in the different myosin genes. miR-208a and -208b share a comparable seed region (nt 2-8) but differ at three bases in their 3′ end (indicated in black). The seed region of miR-499 overlaps at 6 nt with miR-208 but differs significantly in the 3′ region.
B) Northern analysis shows that miR-208a expression correlates with the cardiac-specific expression of α-MHC, while miR-208b parallels the predominant slow skeletal expression of β-MHC. miR-499 is co-expressed with Myh7b, a slow myosin that is mainly expressed in the heart and soleus, a slow skeletal muscle, but not in fast myofibers. Label indicates the mature miRNA.
C) In situ hybridization shows that Myh7b is expressed specifically in the heart and somites as early as E12.5 during mouse embryogenesis. Silver grains are pseudocolored red. ht, heart; t, tongue.
Thyroid hormone regulation of Myh7b/miR-499 and β-MHC/miR-208b
Whereas Myh7b/miR-499 and β-MHC/miR-208b expression marks slow, type I myofibers in skeletal muscle, these genes display distinct regulation in the adult mouse heart, with β-MHC/miR-208b being expressed at a low level and induced to a high level in response to stress, and Myh7b/miR-499 being constitutively expressed at a relatively high level. In the adult heart, thyroid hormone (T3) signaling induces α-MHC transcription and represses β-MHC expression (Morkin, 2000; Schuyler and Yarbrough, 1990). Inhibition of T3 biosynthesis with propylthiouracil (PTU) derepresses the expression of β-MHC. To determine whether miR-208b and miR-499 are co-regulated with their corresponding myosin genes, we performed side-by-side comparison of myosin and miRNA expression in response to PTU (Supplemental Fig. 1). Like β-MHC, miR-208b was up-regulated in the hypothyroid state induced PTU, which could be reversed by T3 supplementation. Myh7b and miR-499 were also up-regulated by PTU, but the magnitude of up-regulation was blunted compared with β-MHC/miR-208b because the basal level of Myh7b/miR-499 expression was higher in the heart (Supplemental Fig. 1). Overall, the expression of the MyomiRs paralleled the expression of their host myosin genes during development and in response to T3 signaling, consistent with the conclusion that the MyomiRs are processed out of intronic sequences of their host myosin mRNAs.
miR-208a is required for expression of myh7b / miR-499
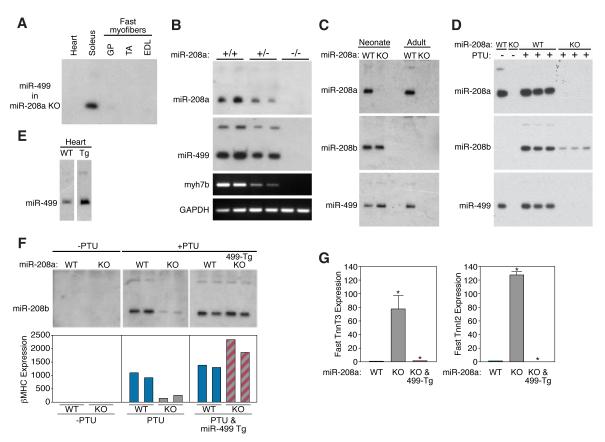
In light of the profound influence of miR-208a on cardiac gene expression (van Rooij et al., 2007), we investigated whether this miRNA might regulate the expression of subordinate miRNAs in the heart, by comparing the miRNA expression profiles in hearts from wild-type and miR-208a−/− mice by microarray analysis (Supplemental Fig. 2). The absence of miR-208a expression in miR-208a−/− hearts served as an internal control for the microarrays. Remarkably, we found that miR-499 expression was extinguished in hearts of miR-208a−/− mice, whereas it was unaffected in the soleus, consistent with the cardiac-specific expression of miR-208a (Fig. 2A). The expression of miR-499 is exquisitely sensitive to the level of miR-208a expression, such that the expression of both miRNAs was reduced by ~50% in hearts from miR-208a+/− mice (Fig. 2B). The expression of miR-499 paralleled that of Myh7b, indicating a regulatory role for miR-208a in the control of myh7b transcription (Fig. 2B).
Figure 2. miR-208a regulates Myh7b / miR-499.
A) Northern blot analysis of heart and skeletal muscle tissue of miR-208a−/− animals shows that miR-499 expression is specifically extinguished in the heart while the expression in soleus remains unaffected. Compare with lower panel of Figure 1B.
B) Northern blot analysis of heart tissue shows that miR-208a regulates the expression of myh7b/miR-499 in a stoichiometric manner. In miR-208a+/− animals, miR-499 and myh7b expression is reduced by 50%, while miR-499 and myh7b expression are eliminated in miR-208a−/− animals. Myh7b and GAPDH were detected by RT-PCR. GAPDH serves as a loading control.
C) Northern blot analysis on cardiac samples of wild-type and miR-208a−/− (KO) neonate and adult mice. miR-208b and miR-499 are expressed normally in neonatal heart of miR-208a KO mice, whereas neither miRNA is expressed in adult miR-208a KO heart.
D) Mice of the indicated miR-208a genotype were treated with PTU, as indicated, and miRNA expression was detected by Northern blot. PTU treatment increases miR-208b in wild-type animals, and this effect is significantly blunted in miR-208a−/− animals.
E) Expression of miR-499 in hearts from wild type and MCK-miR-499 transgenic mice detected by Northern blot.
F) Re-introducing miR-499 with an MCK-miR-499 transgene in the miR-208a−/− background restores expression of miR-208b and β-MHC in response to PTU treatment. Upper panels show Northern blots of hearts from duplicate animals under each condition. Lower panels show β-MHC expression as detected by real time RT-PCR.
G) Fast skeletal muscle troponins (TnnT3 and TnnI2) are up-regulated in hearts of miR-208a−/− animals. Introduction of the MCK-miR-499 transgene into miR-208a−/− mice represses troponin expression as in wild-type. Results represent the average values obtained from two animals as detected by real time PCR.
Although miR-208a is required for up-regulation of β-MHC in the adult heart in response to stress and hypothyroidism (van Rooij et al., 2007; Callis et al., 2009), expression of β-MHC and miR-208b during the neonatal period, is independent of miR-208a (Fig. 2C). Similarly, like the expression of its host gene, β-MHC, miR-208b expression is absent in the adult heart (Fig. 2C). Based on the finding that miR-499 and Myh7b are expressed at comparable levels in wild-type and miR-208a−/− hearts of neonates, while miR-208b is still present, and that miR-499/Myh7b expression is absent in adult miR-208a−/− heart, we conclude that both miR-208a and miR-208b are capable of activating Myh7b/miR-499 expression (Fig. 2C). While PTU up-regulates miR-208b in wild-type hearts, this effect is nearly absent in miR-208a−/− hearts (Fig. 2D); and the slight induction of miR-208b seen in miR-208a−/− animals treated with PTU is apparently insufficient to activate miR-499 expression. Thus, the expression of miR-208b and miR-499 correlates with the expression patterns of their host myosin genes even in the absence of miR-208a. The hearts of adult miR-208a−/− mice are essentially analogous to MyomiR nulls, since neither miR-208b nor miR-499 are expressed at significant levels.
miR-499 can replace the cardiac functions of miR-208a
MiR-208a is required for both the up-regulation of β-MHC in the adult heart and the expression of Myh7b/miR-499. We investigated whether miR-499 could replace the functions of miR-208a by generating transgenic mice that expressed miR-499 under control of MCK regulatory elements, which direct expression in the heart and fast skeletal muscle. Multiple transgenic lines were obtained that expressed miR-499 in the heart at levels approximately 3-fold above the endogenous level (Fig. 2E). As shown in Figure 2F, transgenic expression of miR-499 in the heart of miR-208a−/− mice was sufficient to restore the expression of β-MHC and miR-208b to normal levels in the presence of PTU. Genetic deletion of miR-208a also results in the expression of fast skeletal muscle genes, such as the fast troponins TnnT3 and TnnI2, in the heart (van Rooij et al., 2007); and transgenic expression of miR-499 in the absence of miR-208a repressed cardiac expression of fast skeletal muscle genes (Fig. 2G). The finding miR-499 alone is sufficient to replace the functions of miR-208a in the heart suggests that miR-499 is a downstream mediator of miR-208a actions.
Generation of miR-499 and miR-208b null mice
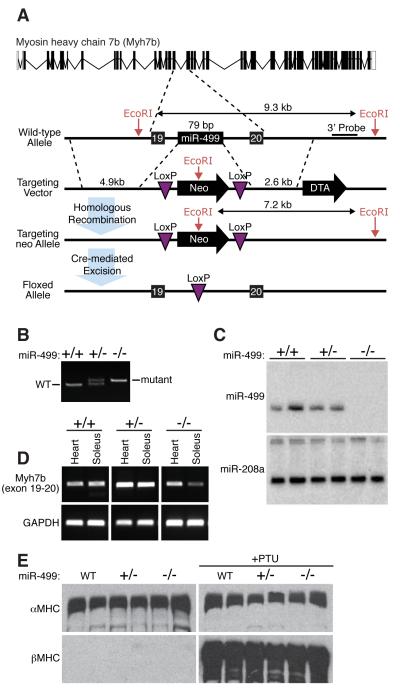
To further explore the functions of miR-499 and miR-208b in vivo, we generated mutant mice with germ line deletions of the pre-miR regions of the Myh7b and β-MHC introns encoding these miRNAs, respectively. LoxP sites were introduced into the corresponding introns at both ends of the miRNA-coding regions (Fig. 3A and 4A). Breeding of these mice to mice expressing a CAG-Cre transgene, which is expressed ubiquitously, allowed for deletion of the corresponding miRNA and its complete absence in homozygous mutant animals as shown by PCR using primers flanking the loxP site (Fig. 3B and 4B). miR-499−/− and miR-208b−/− mice were obtained at Mendelian ratios from heterozygous intercrosses and displayed no overt abnormalities (Supplemental Tables 1 and 2 and data not shown).
Figure 3. Generation of miR-499 null mice.
A) Strategy to generate miR-499 mutant mice by homologous recombination. The pre-miRNA sequence was replaced with a neomycin resistance cassette flanked by loxP sites. The neomycin cassette was removed in the mouse germline by breeding heterozygous mice to transgenic mice harboring the CAG-Cre transgene.
B) Detection of the miR-499 mutation by PCR. Primers flanking the loxP site in intron 19 of the Myh7b gene generate PCR products as indicated. The mutant band in the PCR reaction is larger than the wild type band due to replacement of the miRNA with a larger loxP site.
C) Detection of miR-499 and miR-208a transcripts by Northern analysis of hearts from wild-type, heterozygout and miR-499 mutant mice.
D) Detection of Myh7b expression by RT-PCR of RNA from heart and soleus of mice of the indicated genotypes using primers flanking intron 19. Deletion of miR-499 does not disrupt cardiac expression of Myh7b, while there is a slight decrease in expression in soleus.
E) Western analysis of α-MHC and β-MHC protein levels in hearts of neonatal mice of the indicated genotypes in the absence (left) and presence (right) of PTU. Two mice of each genotype were analyzed. Unlike miR-208, miR-499 is not required for up-regulation of β-MHC in response to PTU.
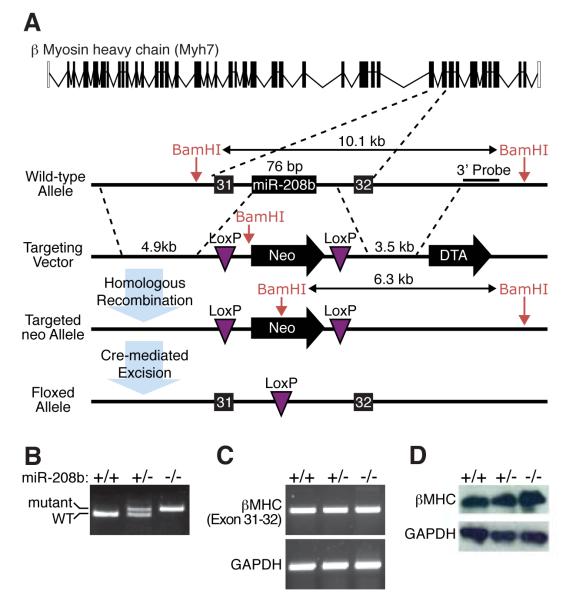
Figure 4. Generation of miR-208b null mice.
A) Strategy to generate miR-208b mutant mice by homologous recombination. The pre-miRNA sequence was replaced with a neomycin resistance cassette flanked by loxP sites. The neomycin cassette was removed in the mouse germline by breeding heterozygous mice to transgenic mice harboring the CAG-Cre transgene.
B) Detection of the miR-208b mutation by PCR. Primers flanking the loxP site in intron 31 of the β-MHC gene generate PCR products as indicated.
C) Detection of β-MHC expression by RT-PCR of RNA from hearts of mice of the indicated genotypes using primers flanking intron 31. Deletion of miR-208b does not disrupt expression of β-MHC.
D) Detection of β-MHC by western blot analysis of heart from P0 mice of the indicated genotypes, showing that β-MHC expression is not altered by deletion of miR-208b. GAPDH was detected as a loading control.
Northern blot analysis on adult heart tissue showed that miR-208a expression was unperturbed by the deletion of miR-499 (Fig. 3C). We confirmed that Myh7b mRNA expression was unaltered in hearts of miR-499−/− mice, while there was a decrease in the soleus as detected by RT-PCR using primers in exons flanking the deletion in intron 19 (Fig. 3D). Western blot analysis for both α-MHC and β-MHC, showed no difference between wild-type or miR-499−/− mice, and a comparable up-regulation in β-MHC expression in wild-type and miR-499−/− mice in response to PTU (Fig. 3E). These data, combined with the finding that over-expression of miR-499 in miR-208a−/− mice can restore β-MHC expression and repress fast skeletal muscle genes in the heart, indicate that both miR-208a and miR-499 must be absent for the blockade to β-MHC expression in response to stress and hypothyroidism.
Deletion of miR-208b also did not alter β-MHC expression as determined by both RT-PCR using primers in exons flanking the deletion in intron 31 (Fig. 4 C) and western blot analysis on neonatal cardiac tissue of miR-208b−/− mice (Fig. 4D). Thus, miR-208a appears to function as the most “upstream” and dominant regulatory miRNA to control expression of both slow myosins, β-MHC and myh7b, and their corresponding intronic miRNAs in the heart, whereas miR-499 and miR-208b are not individually required for expression of either myosin or other members of the MyomiR trio.
Control of skeletal muscle fiber type by miR-499 and -208b
The cardiac specificity of miR-208a led us to investigate the potential involvement of miR-208b and miR-499 in the control of skeletal muscle fiber types. Deletion of either miR-208b or miR-499 did not alter the expression of the other miRNA in the soleus (data not shown), and fiber type analysis showed little or no difference in the number of type I myofibers in either of these mutant mice compared to wild-type (Supplemental Fig. 3).
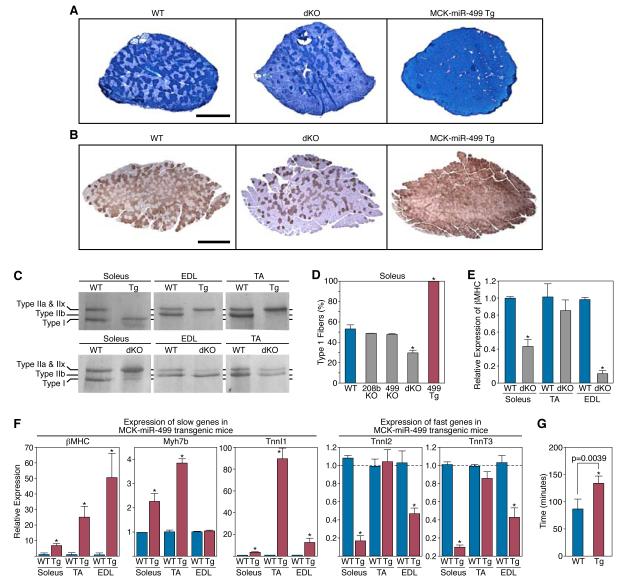
Wondering whether the homology of miR-208b and miR-499 and their coexpression in soleus might allow for redundant functions that were masked in mice with either single gene deletion, we generated miR-208b−/−; miR-499−/− double knockout (dKO) mice. These mice were obtained at predicted Mendelian ratios from transheterozygous intercrosses and displayed no overt abnormalities (Supplemental Table 3). However, staining of histological sections for myofiber type with metachromatic ATPase stain and immunohistochemistry against β-MHC showed a substantial loss of type I myofibers in the soleus of dKO mice (Fig. 5A and 5B). The loss of slow myofibers in dKO mice was also evidenced by the reduced expression of slow β-MHC at the protein and mRNA levels (Fig. 5C and 5E), and by a concomitant increase in the expression of faster type IIx/d and type IIb myosin isoforms (Fig. 5C and Supplemental Fig. 3).
Figure 5. Control of skeletal muscle fiber type by miR-208b and miR-499.
A) Detection of type I myofibers in soleus of wild-type, miR-208b−/−/miR-499−/− dKO, and MCK-miR-499 transgenic mice by metachromatic ATPase staining. Scale bar, 500 μm.
B) Immunohistochemistry for βMHC to identify type I myofibers in the soleus of wild-type, miR-208b−/−/miR-499−/− dKO, and MCK-miR-499 transgenic mice. Scale bar, 500 μm.
C) Identification of myosin isoform content of Soleus, EDL, and TA muscles from wild-type, miR-208b−/−/miR-499−/− dKO, and MCK-miR-499 transgenic mice by gel electrophoresis.
D) The percentage of type I myofibers within soleus muscles of wild type, MyomiR mutant, and MCK-miR-499 transgenic mice was determined as shown in Panel B.
E) βMHC mRNA levels in skeletal muscle of miR-208b−/−/miR-499−/− dKO adult mice were detected by real time PCR. Loss of MyomiRs in skeletal muscle results in repression of βMHC.
F) Expression of slow and fast myofiber genes in soleus, TA and EDL muscles of MCK-miR-499 transgenic mice compared to their level of expression in wild type muscles (set at a value of 1), as detected by real time PCR. Transgenic over-expression of miR-499 is sufficient to up-regulate slow myofiber genes and repress fast myofiber genes in soleus and EDL.
G) WT (n = 4) and MCK-miR-499 transgenic (n = 5) mice were subjected to a regimen of forced running to exhaustion on a treadmill. The time to run to exhaustion of each mouse is shown. p = 0.0039.
Conversely, forced expression of miR-499 under control of MCK regulatory elements, was sufficient to induce a complete conversion of all fast myofibers in soleus to a slow, type I phenotype (Fig. 5A, 5B, and 5C). Analysis of gene expression in the soleus by real-time PCR also demonstrated an induction of the slow fiber gene program in response to miR-499 overexpression (Fig. 5F). In the TA and EDL, which contain predominantly fast myofibers, we observed a pronounced induction of slow myofiber gene expression in MCK-miR-499 transgenic mice (Fig. 5F). Metachromatic ATPase staining of TA and EDL muscles from MCK-miR-499 transgenic animals also revealed conversion to a slower myofiber type(Supplimental Fig. 3). Separation of myosin isoforms by gel electrophoresis indicated a switch from fast type IIb fibers to slower type IIx/d and type IIa fibers in the EDL and TA of transgenic mice(Fig. 5C). This shift in fiber type was confirmed by real time PCR for fiber-type specific myosins (Supplemental Fig. 3). In contrast, fast myofiber genes were repressed in both soleus and EDL muscles from MCK-miR-499 transgenic mice (Fig. 5E). Even more remarkable, when mice were subjected to a regimen of forced treadmill running, the miR-499 transgenic animals ran more than 50% longer than wild-type littermates, indicative of enhanced endurance resulting from the reprogramming of fast myofibers to a slower fiber type (Fig. 5F). We conclude that miR-208b and miR-499 redundantly program skeletal myofibers to a slow phenotype at the expense of fast myofibers.
MyomiR targets and mechanism
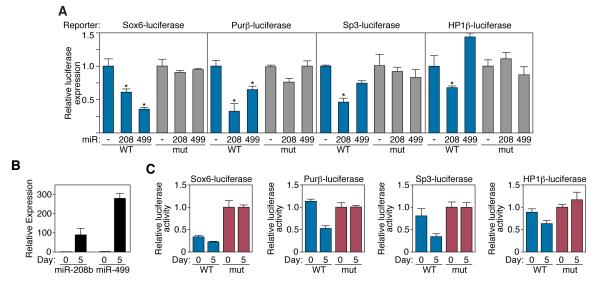
To begin to define the mRNA targets of MyomiRs that mediate their influence on myosin expression and myofiber phenotypes, we used targeting algorithms (TargetScan) to search for predicted targets that were evolutionarily conserved and encoded transcriptional regulators implicated in myofiber gene expression. In addition to the thyroid hormone receptor coregulator Thrap1, shown previously to be a target for repression by miR-208a in the heart (van Rooij et al., 2007; Callis et al., 2009), we identified several transcriptional repressors among the predicted targets of the MyomiRs (Supplemental Fig. 4). These include Sox6, Purβ, and Sp3, each of which has been reported to repress β-MHC expression (Adolph et al., 1993; Azakie et al., 2006; Gupta et al., 2003; Hagiwara et al., 2005; Hagiwara et al., 2007; Ji et al., 2007; Tsika et al., 2004; von Hofsten et al., 2008); and HP-1β, a corepressor of MEF2 (Zhang et al., 2002), which activates slow fiber gene expression (Wu et al., 2000).
Remarkably, the 3′ UTR of Sox6 mRNA contains four evolutionarily conserved target sites for miR-499, one of which is also a predicted site for miR-208 (Supplementary Fig. 4). The 3′ UTRs of Purβ and HP-1β each contain single conserved predicted target sequences for miR-499 and miR-208a, while Sp3 contains a conserved miR-208 targeting sequence. We cloned the 3′ UTRs of each of these predicted targets downstream of a luciferase reporter and assayed for repression by miR-208 and -499 in transfected COS cells. Expression of the MyomiRs repressed wild-type 3′ UTR-luciferase reporter constructs, but had little effect on reporters containing 2 nucleotide mutations in the conserved targeting sequence (Fig. 6A). Similarly, wild-type luciferase reporter constructs were repressed in C2C12 cells as MyomiR expression increased over the course of differentiation, while mutated constructs displayed no change in activity (Fig. 6B and 6C). Consistent with previous results, the 3′UTR of Thrap1 displayed repression by both miR-208a and miR-499 (data not shown).
Figure 6. MyomiRs directly repress predicted target genes.
A) Luciferase studies of predicted MyomiR target transcripts. Wild-type and mutant Sox6, Purβ, Sp3, and HP1β 3′UTRs were cloned into a luciferase reporter plasmid. These constructs were co-transfected into COS-1 cells with expression vectors for miR-208a or miR-499. Luminescence values were assayed forty-eight hours post transfection and were normalized to β-galactosidase activity.
B) Expression of MyomiRs increases following C2C12 myoblast differentiation. Relative levels of miR-208b and miR-499 expression were detected by real-time PCR at days 0 and 5 of C2C12 differentiation.
C) Wild-type, but not mutant luciferase reporter constructs are repressed by MyomiRs in C2C12 cells.
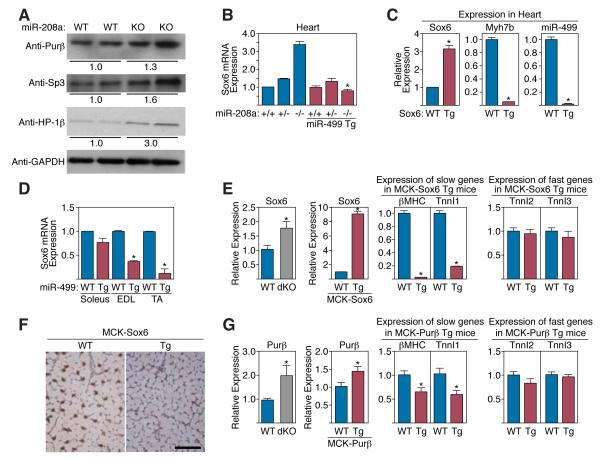
To further validate the above repressors as potential targets of MyomiRs, we compared the expression of the corresponding proteins in cardiac extracts from wild-type and miR-208a−/− mice (Fig. 7A). Consistent with a repressive effect of miR-208a on their expression Purβ, Sp3 and HP-1β proteins were elevated in expression in hearts from mutant mice, while their mRNA levels were unchanged (data not shown). Additionally, Sox6 mRNA was up-regulated in miR-208a−/− hearts, indicative of a destabilizing effect of miR-208a on Sox6 mRNA. Transgenic expression of miR-499 was sufficient to reduce Sox6 mRNA expression in miR-208a−/− hearts to wild-type levels (Fig. 7B). In addition, Sox6 mRNA levels were reduced in skeletal muscles of MCK-miR-499 transgenic mice (Fig. 7D). Such down-regulation of Sox6 suggests that Sox6 mediates, at least in part, the increase in slow and decrease in fast myofiber gene expression in MCK-miR-499 transgenic mice, as shown in Figure 5F.
Figure 7. Target proteins that mediate the actions of MyomiRs.
A) Western blot analysis of miR-208a/499 target expression in adult wild type (WT) and miR-208a−/− (KO) mouse heart lysate. Relative expression was calculated from comparison of band intensity following normalization to GAPDH loading control.
B) Sox6 mRNA levels are elevated in the miR-208a−/− heart as measured by real time PCR. Transgenic expression of miR-499 abolished the increase in Sox6 mRNA in hearts lacking miR-208.
C) Myh7b and miR-499 are repressed in the hearts of MCK-Sox6 transgenic mice. Transcript levels were measured in the hearts of P8 transgenic mice and wild type controls (WT) by real time PCR.
D) Real time PCR indicates Sox6 mRNA levels are depressed in skeletal muscle of MCK-miR-499 mice.
E) Sox6 mRNA transcript is overexpressed in MyomiR dKO skeletal muscle. MCK-Sox6 transgenic mice exhibit repression of slow skeletal muscle genes and unaltered levels of fast skeletal troponins in skeletal muscle. Expression was measured by real time PCR in total hind limb muscle of MCK-Sox6 transgenic mice and wild type (WT) controls.
F) Immunohistochemistry for Type I myosin (βMHC) in the hind limb of MCK-Sox6 transgenic mice indicates an absence of Type I fibers in the EDL of transgenic mice as compared to wild-type (WT) littermate controls. Scale bar, 100 μm.
G) Purβ mRNA transcript is overexpressed in MyomiR dKO skeletal muscle. MCK-Purβ transgenic mice demonstrate repression of slow skeletal muscle genes and unaltered fast skeletal gene expression. Expression was measured by real time PCR in total hind limb muscle of MCK-Purβ transgenic mice and wild type (WT) controls.
Transgenic expression of Sox6 and Purβ phenocopies MyomiR gene deletions
To further validate Sox6 and Purβ as potential mediators of MyomiR function, we generated MCK-Sox6 and MCK-Purβ transgenic mice. MCK-Sox6 mice appeared normal at birth, however they demonstrated retarded growth at postnatal day 8 and ultimately died by 28 days of age. Analysis of multiple F0 transgenic animals revealed that simply elevating the expression of Sox6 by 3-fold in the neonatal heart was sufficient to extinguish expression of Myh7b and miR-499 (Fig. 7C), and that over-expression in skeletal muscle strongly repressed β-MHC and slow TnnI transcript levels (Fig. 7E). Immunohistochemistry for β-MHC on skeletal muscle sections revealed a complete loss of type I fibers in MCK-Sox6 mice (Fig. 7F). MCK-Purβ transgenic mice demonstrated that a 1.4-fold increase in Purβ expression was sufficient to repress β-MHC expression in skeletal muscle at the mRNA level, but had little effect on the overall number of type I fibers (Fig. 7G and data not shown). Thus, over-expression of Sox6 at similar levels to the miR-208a KO heart, in addition to relatively modest elevation in expression of Sox6 and Purβ to levels comparable to those in MyomiR dKO mice (Fig. 7E and 7G) was sufficient to evoke myofiber phenotypes similar to those of miR-208a KO and MyomiR dKO mice. These results provide strong support for the conclusion that these transcriptional repressors mediate the actions of MyomiRs on slow myofiber gene expression.
In contrast, these targets did not activate the expression of fast TnnI2 or TnnI3 (Fig. 7E and 7G), as seen in miR-208b−/−/miR-499−/− dKO mice. The latter findings suggest that Sox6 and Purβ mediate the actions of miR-499 and miR-208 on the slow myofiber gene program, but other targets of these MyomiRs are required for the activation of fast myofiber genes.
Discussion
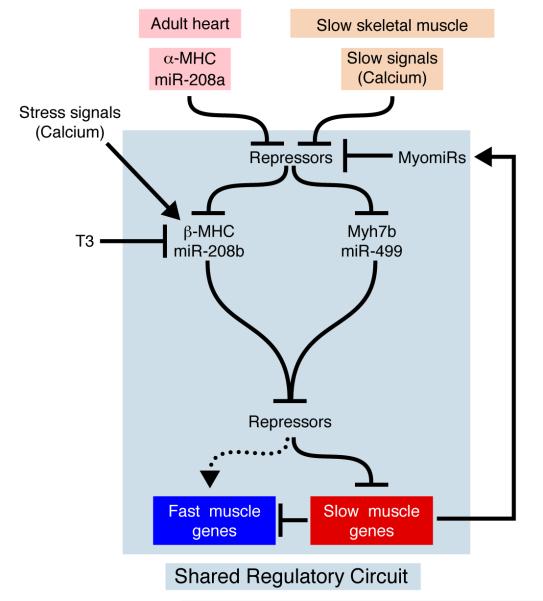
The results of this study reveal a network of miRNAs within myosin genes that regulate myosin expression, fiber type gene expression, and muscle performance. The functions of myosin genes thus extend far beyond the mere expression of myosin proteins to the control of myriad functions of striated muscles. A model to account for our results is shown in Figure 8.
Figure 8. Control of myosin and fast versus slow muscle gene expression by the MyomiR network.
In the adult heart, miR-208a encoded by the α-MHC gene is required for expression of β-MHC and Myh7b, which encode miR-208b and miR-499, respectively. Activation of Myh7b by miR-208a is constitutive, whereas activation of β-MHC also requires stress signals or absence of thyroid hormone. In slow skeletal muscle, Myh7b and β-MHC are regulated independently of miR-208a. miR-208a, -208b, and -499 repress the expression of a common set of transcriptional repressors that repress slow myosin and the slow myofiber gene program at the expense of fast muscle gene expression. Activation of the slow myofiber gene program also creates a positive feedback loop via the expression of miR-208b and miR-499, which further reinforce slow muscle gene program. Regulatory interactions within the blue box are shared by cardiac and slow skeletal muscle.
Functions of MyomiRs in the heart
As schematized in Figure 8, α-MHC and miR-208a sit atop a hierarchy of regulatory steps leading to myofiber diversification, stress-responsiveness and thyroid hormone sensitivity of the heart. In the mouse heart, miR-208a, encoded by α-MHC, is required for up-regulation of β-MHC and miR-208b in response to stress and hypothyroidism. miR-208a also regulates the expression of Myh7b and its intronic miRNA, miR-499; however, this regulatory step is distinct from the regulation of β-MHC/miR-208b in that Myh7b/miR-499 are highly expressed in the adult heart in the absence of stress, whereas β-MHC/miR-208b require stress or hypothyroidism for expression. In addition to the requisite role of miR-208a in the expression of β-MHC and Myh7b in the adult heart, miR-208a is also required for repression of fast muscle genes in the heart.
Within the adult mouse heart, α-MHC is the predominant myosin, accounting for greater than 90% of myosin expression (Lompre et al., 1984), and miR-208a is the dominant miRNA among the trio of MyomiRs. Although miRNAs are typically envisioned to function as fine-tuners of gene expression, miR-208a operates in a precise, stoichiometric manner to control the expression of Myh7b/miR-499, such that Myh7b/miR-499 expression is reduced by ~50% in hearts of miR-208a+/− mice and is extinguished in hearts of miR-208a−/− mice. Forced expression of miR-499 in miR-208a−/− hearts restores expression of β-MHC and represses ectopic expression of fast muscle genes, consistent with the conclusion that miR-499 functions downstream of miR-208a. Notably, transgenic expression of miR-499 in the hearts of miR-208a−/− mice also reactivates the expression of Myh7b, indicative of a positive autoregulatory loop (Fig. 7).
In large animals, β-MHC is the predominant myosin in the adult heart, so we presume that miR-208b, which shares an identical seed sequence with miR-208a, fulfills the functions of miR-208a in large animals. Whether miR-208b is required for pathological remodeling of hearts with predominantly β-MHC expression (Lompre et al., 1991), as is miR-208a in mice (van Rooij et al., 2007), is an important issue with respect to the potential therapeutic manipulation of MyomiRs in the setting of human heart disease.
There are several other noteworthy conclusions to be drawn about the regulation and functions of MyomiRs in the heart based on our findings with MyomiR mutant mice. First, the cardiac regulatory circuitry amongst the MyomiRs and their host myosins appears to be operative specifically in the adult but not in the fetal heart. We base this conclusion on the finding that β-MHC and Myh7b are expressed normally in neonatal hearts of miR-208a−/− mice, whereas these myosin genes are silenced in heart of adult mutant mice. Thus, there must be a regulatory switch that distinguishes neonatal and adult cardiac gene programs with respect to their sensitivity to MyomiR functions. Second, none of the MyomiRs are essential for function of the adult heart, as evidenced by the phenotype of miR-208a−/− mice, which are essentially MyomiR nulls, since the absence of miR-208a abolishes the expression of miR-208b and miR-499. Thus, the MyomiRs appear to function primarily to adapt adult cardiac gene expression to physiological and pathological signaling rather than to control cardiac homeostasis.
Functions of MyomiRs in skeletal muscle
Unlike α-MHC and miR-208a, which are expressed only in the heart, β-MHC/miR-208b and Myh7b/miR-499 are also expressed in slow skeletal muscle. The latter two myosins and their intronic miRNAs appear to function in a shared regulatory circuit in cardiac and skeletal muscle muscle; however, the upstream inputs into this myosin/miRNA circuit differ in cardiac and skeletal muscle in multiple ways (Fig. 8). For example, the expression of miR-208b and miR-499 in slow skeletal muscle does not require an “upstream” MyomiR, since both of these miRNAs are expressed normally in the absence of the other, as shown from our analysis of miR-208b and miR-499 null mice. The activation of slow myofiber genes in skeletal muscle depends on calcium signaling through calcineurin and various kinases (Bassel-Duby and Olson, 2006; Wu et al., 2000), leading us to propose that β-MHC and Myh7b are activated independently through such signals, resulting in the production of their encoded MyomiRs, which then reinforce the slow skeletal muscle gene program through the actions of their targets. Importantly, this model infers the existence of a positive feedback loop whereby the targets of miR-208b and -499 in slow skeletal muscle enhance their own expression via their targets, which act on their host myosins. Such a mechanism also enables MyomiRs to function as binary regulators of slow versus fast muscle gene programs – activating slow and repressing fast – through their downstream targets. Moreover, we have observed that manipulation of MyomiR levels in the soleus, TA, and EDL elicits a distinct shift towards the slow phenotype in each muscle. In addition to up-regulating β-MHC, transgenic expression of miR-499 primarily drives these muscles to shift one myosin isoform slower in the sequential transition towards a slow myofiber phenotype(Pette and Staron, 1997). We hypothesize that divergent gene expression profiles, as well as different frequencies of muscle use between the soleus, TA, and EDL may be responsible for the distinct changes we observe in response to gain of MyomiR function.
A collection of transcriptional repressors downstream of MyomiRs
It has been demonstrated that microRNAs function by finely modulating the expression of their downstream target genes at the protein level (Baek et al., 2008). Consistent with this idea, our results demonstrate that the MyomiRs target a cohort of transcriptional repressors to mediate their actions on striated muscle gene expression and function. Among their targets, Sox6, Purβ and Sp3 have previously been shown to repress the expression of slow skeletal muscle genes, but the mechanisms that control the expression of these repressors have not been investigated. In Sox6 mutant mice, slow fiber type-specific gene expression, including the expression of β-MHC, is increased, whereas fast myofiber genes are repressed (Hagiwara et al., 2005; Hagiwara et al., 2007; von Hofsten et al., 2008). It has been proposed that the repressive influence of Sox6 on β-MHC expression is mediated through direct binding of Sox6 to the promoter of the gene (Hagiwara et al., 2007). Purβ, a single-stranded DNA binding protein, cooperates with Sp3 to repress β-MHC expression (Ji et al., 2007). Thus, by dampening the expression of Sox6, Purβ, and Sp3, the MyomiRs enable the expression of β-MHC and other genes under the inhibitory influence of the repressors. Conversely, in the absence of MyomiRs, the expression of these repressors is enhanced, resulting in the inhibition of β-MHC and other downstream targets. Our finding that transgenic over-expression of Sox6 or Purβ represses slow genes, but does not activate fast genes suggests that another of the targets of MyomiRs likely mediates fast muscle gene expression downstream of MyomiRs or could require the combined actions of multiple targets.
In addition to the above targets, HP-1β, a transcriptional repressor shown previously to function as a class II HDAC co-repressor, is also a target for repression by the MyomiRs. Class II HDACs function as calcium-sensitive repressors of the MEF2 transcription factor, which is necessary and sufficient for activation of slow myofiber gene expression (Potthoff et al., 2007). The repressive influence of MyomiRs on HP-1 expression would therefore be expected to diminish the repressive influence of class II HDACs on MEF2, promoting the expression of slow muscle genes. Thrap1, another validated target of the MyomiRs, functions as a modulator of thyroid hormone signaling and likely plays a key role in regulating the response of β-MHC and myofiber gene programs to hypothyroidism.
Interestingly, our data indicates that the MyomiR family functions redundantly in vivo, while luciferase reporter assays suggest that these micoRNAs may exhibit differential effects on target gene repression. These differences in our reporter assays may be attributed to dissimilarity in sequence outside of the seed region of miR-208 and miR-499, which may have effects on the micoRNA:target interactions that dictate repression, or possibly to differences in microRNA associated machinery in vitro and in vivo.
Implications for gene regulatory networks
Sequence analysis of MHC genes indicates that Myh7b is the most ancient of this trio of myosins and gene duplications gave rise to α- and β-myosins. The MyomiRs are conserved from fish to human genomes, but do not exist in invertebrates, suggesting that the ancestral Myh7b gene contained the ancestral MyomiR in an intron and when that myosin gene duplicated, so too did its intronic MyomiR. It is intriguing, in this regard, that the existence of MyomiRs correlates with myofiber diversification and the acquisition of hormonal sensitivity and stress responsiveness. These findings argue strongly for selection pressure to maintain not only the conserved sequence of MyomiRs, and presumably of their targets, but also the integration of this family of miRNAs into myosin genes, so as to ensure their regulatory relationship. The incorporation of MyomiRs into the introns of myosin genes that they regulate also provides an efficient means of ensuring the co-regulation of the miRNA and the gene programs under its control, rather than creating separate sets of cis-regulatory elements to control expression of the miRNA and the myosin gene. The functions and regulation of the MyomiR network serve as a paradigm for phenotypic control. This elaborate regulatory network provides for communication between genes and reveals an unanticipated breadth of functions of myosin genes, previously recognized simply for their roles as protein-coding genes. We speculate that protein-coding genes with central roles in the functionality of other tissues will also be found to encode families of miRNAs that dictate the functions of those tissues, especially in settings of disease. Perhaps most provocative is the realization that miRNA functions can be perturbed without deleterious consequences on tissue function, but with dramatic effects on signal-dependent tissue remodeling responses. As such, miRNAs represent potentially powerful disease modifiers and therapeutic targets.
Materials and methods
Northern blot analysis
Total RNA was isolated from mouse, and human cardiac tissue samples by using Trizol reagent (Gibco/BRL). Northern blots to detect microRNAs were performed as described previously (van Rooij et al., 2006). U6 was detected as a loading control using the following primers in RT-PCR reactions:
(U6 forward: 5-GTGCTCGCTTCGGCAGC-3,
U6 reverse: 5-AAAATATGGAACGCTTCACGAATTTGCG-3).
RNA in situ hybridization
Tissues used for histology were incubated in Krebs-Henselheit solution, fixed in 4% paraformaldehyde, sectioned, and processed for in situ hybridization by standard techniques (Shelton et al., 2000). 35S-labeled RNA probes were generated using Maxiscript kit (Amersham). Signals were pseudocolored in red using Adobe Photoshop.
MicroRNA microarray analysis
Microarray assay was performed using a service provider (LC Sciences, Houston). The assay started from 10 μg total RNA from cardiac tissue of both wild-type and miR-208−/−mice, which was size fractionated and the small RNAs (< 300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining. Hybridization was performed overnight on a μParaflo microfluidic chip on which each detection probe consisted of a chemically modified nucleotide coding segment complementary to target microRNA (from miRBase, http://microrna.sanger.ac.uk/sequences/) After RNA hybridization, tag-conjugating Cy5 dyes were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Device) and digitized using Array-Pro image analysis software (Media Cybernetics). Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter (Locally-weighted Regression). The ratio of the two sets of detected signals (log2 transformed, balanced) and p-values of the t-test were calculated; differentially detected signals were those with less than 0.01 p-values.
RT-PCR and Realtime analysis
RT-PCR with random hexamer primers (Invitrogen) was performed on RNA samples, after which the expression of a subset of genes was analyzed by either PCR or quantitative real time PCR using Taqman probes purchased from ABI.
Generation of transgenic mice
A mouse genomic fragment flanking miR-499 was subcloned into a muscle-specific expression plasmid containing the MCK enhancer and human GH poly(A)+ signal (Sternberg et al., 1988). Genomic DNA was isolated from mouse tail biopsies and analyzed by PCR using primers specific for the human GH poly(A)+ signal.
Western blots analysis
Myosin was extracted from cardiac tissue as described (Hamalainen and Pette, 1995). MHC isoforms were separated by SDS PAGE and Western blotting was performed with mouse monoclonal αMHC (BA-G5) (ATCC, Rockville, MD) and mouse monoclonal anti-myosin (slow, skeletal M8421) (Sigma, MO), which is highly specific for βMHC. Total heart lysate was prepared by homogenizing tissue in RIPA buffer, was resolved by SDS-PAGE, and analyzed to detect HP1β, PURβ, and SP3 using mouse monoclonal anti-HP1β (Cell Signaling #2613) at a dilution of 1:1000, anti-PURβ (a gift from Robert J Kelm Jr at University of Vermont College of Medicine(Kelm et al., 1999)) at a dilution 1:1000, and anti-SP3 (Abcam ab18334) at a dilution of 1:200. Anti-mouse IgG conjugated to horseradish peroxidase (Biorad) at a dilution of 1:5000 was used as a secondary antibody, followed by detection with ECL detection kit (Amersham).
Generation of miR-499 and miR-208b mutant mice
To generate the miR-499 targeting vector, a 4.9kb region (5′ arm) extending upstream of the miR-499 coding region was ligated into the pGKneoF2L2dta targeting plasmid upstream of the loxP sites and the Frt-flanked neomycin cassette. A 2.6 kb fragment (3′ arm) was ligated into the vector between the neomycin resistance and Dta negative selection cassettes. Targeted ES-cells carrying the disrupted allele were identified by Southern blot analysis with 5′ and 3′ probes. Three miR-499 targeted ES clones were identified and used for blastocyst injection. A comparable approach was taken to generate the miR-208b targeting vector, with a 4.9kb region extending upstream of the miR-208b coding region functioning as the 5′ arm and a 3.5 kb fragment was used for the 3′arm. The resulting chimeric mice were bred to C57BL/6 to obtain germline transmission of the mutant allele. PCR primer sequences are available upon request.
Fiber-type staining
Soleus, GP, TA, and EDL muscles were isolated at 8 weeks of age and were embedded in a 3:1 ratio of Tissue Freezing Medium to gum tragacanth. Samples were flash frozen and sectioned on a cryostat-microtome. Metachromatic ATPase staining was performed as previously described (Ogilvie and Feeback, 1990).
Forced running
Forced treadmill running of 6 week old wild-type and MCK-miR-499 transgenic mice was performed as follows. Animals were trained on the treadmill (Colombus Instruments, 10% incline) for 5 minutes at 7m/min for two consecutive days. The following day mice were run at 8m/min for 30 minutes, then 9m/min for 15 minutes, followed by 10m/min for 15 minutes. Finally, speed was incrementally increased by 1m/min every 10 minutes until the mouse exhibited exhaustion. The end point was reached when a mouse received repeated electrical stimuli for greater than 5 seconds.
MHC electrophoresis
Myosin was isolated from skeletal muscle and run on glycerol-SDS-PAGE gels as previously described (Talmadge and Roy, 1993). Gels were stained with a silver nitrate staining kit(Biorad).
Cell culture, transfection and luciferase assays
A 300-500 bp genomic fragment encompassing the miR-208a, miR-499, miR-21 or miR-206 coding region, were amplified by PCR and ligated into pCMV6. Full length 3′ UTRs of Sox6, Sp3, HP1b, and PURβ were cloned into the pMiR-report vector (Ambion). Cell culture, transfection, and luciferase studies were performed as previously described (van Rooij et al., 2007), Mutations in the 3′UTRs alter the 2nd and 3rd nucleotides of the targeting sequence, and were genereated using QuickChange Lightning kit (Stratagene).
Immunohistochemistry
Tissue samples were fixed in 4% paraformaldehyde and embedded in paraffin for sectioning. Deparaffinized sections were permeabilized with 0.04% Pronase E, then blocked in 0.5% bovine serum albumin/5% normal goat serum in PBS. NOQ7.5.4D (1:16000) was used for primary detection of Type I myosin, and HRP-conjugated secondary (Sigma A8924) followed by DAB chromagen reaction were used for detection. Samples were then counterstained with hematoxylin.
Analysis of transgenic animals
2-3 F1 transgenic lines of MCK-miR-499 and MCK-PURβ, and 3 F0 transgenic MCK-Sox6 animals were analyzed in this study.
Statistical Analysis
All graphs represent mean values +/− SEM. Asterisk(*) represents p-value<0.05 as calculated by unpaired t-test.
Animal Care
All animal procedures were previously approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center.
Supplementary Material
Figure 1. Northern analysis for MyomiRs shows that their expression correlates with that of their host gene. Blockade of thyroid receptor signaling by treating animals with PTU represses Myh6 and induces Myh7 expression in cardiac tissue of rats. This effect can be reversed by subsequent supplementation of thyroid hormone (T3). While PTU induces both the expression of Myh7/miR-208b and Myh7b/miR-499, T3 can potently reverse the increase in expression. Label indicates the mature miRNA.
Figure 2. Microarray analysis for miRNA expression of cardiac tissue of wild-type and miR-208a mutant mice. miRNAs downregulated in hearts of miR-208a−/− mice are shown. The colors indicate the relative abundance of the miRNA; blue indicates highest expression and orange indicates the miRNA to be absent.
Figure 3. Analysis of myosin expression in skeletal muscle of MCK-miR-499 transgenic and dKO animals. (A) Metachromatic ATPase staining of miR-208KO soleus, miR-499 KO soleus, and MCK-miR-499 TA and EDL muscles and wild-type littermate controls. (B) Detection of fast skeletal myosin isoform expression in dKO skeletal muscle by real-time PCR. (C) Detection of fast skeletal myosin isoform expression in MCK-miR-499 skeletal muscle by real-time PCR.
Figure 4. MyomiR target sites Predicted targets of MyomiRs are shown along with the positions of target sequences in the 3′ UTRs of mouse mRNAs.
Supplemental Table 1 Genotyping of offspring from miR-499+/− intercrosses
Supplemental Table 2 Genotyping of offspring from miR-208b+/− intercrosses
Supplemental Table 3 Genotyping of offspring from miR-499+/−/miR-208b+/− intercrosses
Acknowledgements
We are grateful to John McAnally and John Shelton for experimental assistance. Work in Eric Olson’s laboratory was supported by grants from the National Institutes of Health, the Donald W. Reynolds Cardiovascular Clinical Research Center, the Leducq Foundation, the Robert A. Welch Foundation and the American Heart Association - Jon Holden DeHaan Foundation. Eva van Rooij was supported by grants from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolph EA, Subramaniam A, Cserjesi P, Olson EN, Robbins J. Role of myocyte-specific enhancer-binding factor (MEF-2) in transcriptional regulation of the alpha-cardiac myosin heavy chain gene. J Biol Chem. 1993;268:5349–5352. [PubMed] [Google Scholar]
- Azakie A, Fineman JR, He Y. Sp3 inhibits Sp1-mediated activation of the cardiac troponin T promoter and is downregulated during pathological cardiac hypertrophy in vivo. Am J Physiol Heart Circ Physiol. 2006;291:H600–611. doi: 10.1152/ajpheart.01305.2005. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin KM, Haddad F. Effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J Appl Physiol. 2001;90:345–357. doi: 10.1152/jappl.2001.90.1.345. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatkin D, McConnell BK, Mudd JO, Semsarian C, Moskowitz IG, Schoen FJ, Giewat M, Seidman CE, Seidman JG. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest. 2000;106:1351–1359. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Sueblinvong V, Raman J, Jeevanandam V, Gupta MP. Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. Implications in the repression of alpha-myosin heavy chain during heart failure. J Biol Chem. 2003;278:44935–44948. doi: 10.1074/jbc.M307696200. [DOI] [PubMed] [Google Scholar]
- Gupta MP. Factors controlling cardiac myosin-isoform shift during hypertrophy and heart failure. J Mol Cell Cardiol. 2007;43:388–403. doi: 10.1016/j.yjmcc.2007.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara N, Ma B, Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Dev Dyn. 2005;234:301–311. doi: 10.1002/dvdy.20535. [DOI] [PubMed] [Google Scholar]
- Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn. 2007;236:2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- Herron TJ, McDonald KS. Small amounts of alpha-myosin heavy chain isoform expression significantly increase power output of rat cardiac myocyte fragments. Circ Res. 2002;90:1150–1152. doi: 10.1161/01.res.0000022879.57270.11. [DOI] [PubMed] [Google Scholar]
- Ji J, Tsika GL, Rindt H, Schreiber KL, McCarthy JJ, Kelm RJ, Jr., Tsika R. Puralpha and Purbeta collaborate with Sp3 to negatively regulate beta-myosin heavy chain gene expression during skeletal muscle inactivity. Mol Cell Biol. 2007;27:1531–1543. doi: 10.1128/MCB.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm RJ, Jr., Cogan JG, Elder PK, Strauch AR, Getz MJ. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter. J Biol Chem. 1999;274:14238–14245. doi: 10.1074/jbc.274.20.14238. [DOI] [PubMed] [Google Scholar]
- Kiriazis H, Kranias EG. Genetically engineered models with alterations in cardiac membrane calcium-handling proteins. Annu Rev Physiol. 2000;62:321–351. doi: 10.1146/annurev.physiol.62.1.321. [DOI] [PubMed] [Google Scholar]
- Korte FS, Herron TJ, Rovetto MJ, McDonald KS. Power output is linearly related to MyHC content in rat skinned myocytes and isolated working hearts. Am J Physiol Heart Circ Physiol. 2005;289:H801–812. doi: 10.1152/ajpheart.01227.2004. [DOI] [PubMed] [Google Scholar]
- Krenz M, Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J Am Coll Cardiol. 2004;44:2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lompre AM, Mercadier JJ, Schwartz K. Changes in gene expression during cardiac growth. Int Rev Cytol. 1991;124:137–186. doi: 10.1016/s0074-7696(08)61526-0. [DOI] [PubMed] [Google Scholar]
- Lompre AM, Nadal-Ginard B, Mahdavi V. Expression of the cardiac ventricular alpha- and beta-myosin heavy chain genes is developmentally and hormonally regulated. J Biol Chem. 1984;259:6437–6446. [PubMed] [Google Scholar]
- Lowes BD, Minobe W, Abraham WT, Rizeq MN, Bohlmeyer TJ, Quaife RA, Roden RL, Dutcher DL, Robertson AD, Voelkel NF, et al. Changes in gene expression in the intact human heart. Downregulation of alpha-myosin heavy chain in hypertrophied, failing ventricular myocardium. J Clin Invest. 1997;100:2315–2324. doi: 10.1172/JCI119770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K, Phillips PC, Postlethwait JH. Evolution of sarcomeric myosin heavy chain genes: evidence from fish. Mol Biol Evol. 2004;21:1042–1056. doi: 10.1093/molbev/msh103. [DOI] [PubMed] [Google Scholar]
- Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res. 2000;86:386–390. doi: 10.1161/01.res.86.4.386. [DOI] [PubMed] [Google Scholar]
- Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie RW, Feeback DL. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 1990;65:231–241. doi: 10.3109/10520299009105613. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Mammalian skeletal muscle fiber type transitions. Int Rev Cytol. 1997;170:143–223. doi: 10.1016/s0074-7696(08)61622-8. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell VL, Manaves V, Martin AF, de Tombe PP. Impact of beta-myosin heavy chain isoform expression on cross-bridge cycling kinetics. Am J Physiol Heart Circ Physiol. 2005;288:H896–903. doi: 10.1152/ajpheart.00407.2004. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- Schuyler GT, Yarbrough LR. Changes in myosin and creatine kinase mRNA levels with cardiac hypertrophy and hypothyroidism. Basic Res Cardiol. 1990;85:481–494. doi: 10.1007/BF01931494. [DOI] [PubMed] [Google Scholar]
- Shelton JM, Lee MH, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Stelzer JE, Brickson SL, Locher MR, Moss RL. Role of myosin heavy chain composition in the stretch activation response of rat myocardium. J Physiol. 2007;579:161–173. doi: 10.1113/jphysiol.2006.119719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg EA, Spizz G, Perry WM, Vizard D, Weil T, Olson EN. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988;8:2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge RJ, Roy RR. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J Appl Physiol. 1993;75:2337–2340. doi: 10.1152/jappl.1993.75.5.2337. [DOI] [PubMed] [Google Scholar]
- Tsika G, Ji J, Tsika R. Sp3 proteins negatively regulate beta myosin heavy chain gene expression during skeletal muscle inactivity. Mol Cell Biol. 2004;24:10777–10791. doi: 10.1128/MCB.24.24.10777-10791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vanderheyden M, Mullens W, Delrue L, Goethals M, de Bruyne B, Wijns W, Geelen P, Verstreken S, Wellens F, Bartunek J. Myocardial gene expression in heart failure patients treated with cardiac resynchronization therapy responders versus nonresponders. J Am Coll Cardiol. 2008;51:129–136. doi: 10.1016/j.jacc.2007.07.087. [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Elworthy S, Gilchrist MJ, Smith JC, Wardle FC, Ingham PW. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 2008;9:683–689. doi: 10.1038/embor.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Leinwand LA. The mammalian myosin heavy chain gene family. Annu Rev Cell Dev Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1. Northern analysis for MyomiRs shows that their expression correlates with that of their host gene. Blockade of thyroid receptor signaling by treating animals with PTU represses Myh6 and induces Myh7 expression in cardiac tissue of rats. This effect can be reversed by subsequent supplementation of thyroid hormone (T3). While PTU induces both the expression of Myh7/miR-208b and Myh7b/miR-499, T3 can potently reverse the increase in expression. Label indicates the mature miRNA.
Figure 2. Microarray analysis for miRNA expression of cardiac tissue of wild-type and miR-208a mutant mice. miRNAs downregulated in hearts of miR-208a−/− mice are shown. The colors indicate the relative abundance of the miRNA; blue indicates highest expression and orange indicates the miRNA to be absent.
Figure 3. Analysis of myosin expression in skeletal muscle of MCK-miR-499 transgenic and dKO animals. (A) Metachromatic ATPase staining of miR-208KO soleus, miR-499 KO soleus, and MCK-miR-499 TA and EDL muscles and wild-type littermate controls. (B) Detection of fast skeletal myosin isoform expression in dKO skeletal muscle by real-time PCR. (C) Detection of fast skeletal myosin isoform expression in MCK-miR-499 skeletal muscle by real-time PCR.
Figure 4. MyomiR target sites Predicted targets of MyomiRs are shown along with the positions of target sequences in the 3′ UTRs of mouse mRNAs.
Supplemental Table 1 Genotyping of offspring from miR-499+/− intercrosses
Supplemental Table 2 Genotyping of offspring from miR-208b+/− intercrosses
Supplemental Table 3 Genotyping of offspring from miR-499+/−/miR-208b+/− intercrosses