Abstract
Aside from Myc-activating translocations characteristic of plasmacytomas (PCT), little is known about genetic factors and signaling pathways responsible for the development of spontaneous B-cell lineage lymphomas of mice. Here, we characterized the transcriptional profiles of PCT, centroblastic diffuse large B-cell lymphomas (CBL), and high-grade splenic marginal zone B-cell lymphoma (MZL++) using high-throughput quantitative reverse transcription-PCR. Expression profiles of CBL and MZL++ were strikingly similar and quite unlike that of PCT. Among the genes expressed at significantly higher levels by PCT were a number involved in NOTCH signaling, a finding supported by gene set enrichment analyses of microarray data. To investigate the importance of this pathway, NOTCH signaling was blocked in PCT cell lines by treatment with a γ-secretase inhibitor (GSI) or transduction of a dominant-negative mutant of MAML1. These treatments resulted in reduced expression of NOTCH transcriptional targets in association with impaired proliferation and increased apoptosis. GSI treatment of transformed plasma cells in a primary PCT also induced apoptosis. These results integrate NOTCH activation with oncogenic signaling pathways downstream of translocated Myc in the pathogenesis of mouse PCT, two signaling pathways also implicated in development of human multiple myeloma and T-cell lymphoblastic lymphoma.
Introduction
A spectrum of mature B-cell lineage lymphomas of different histologic types develops spontaneously in various strains of mice, including NFS.V+ congenic mice, which express ecotropic murine leukemia viruses (MuLV) at high levels (1). Many of these tumors have phenotypic similarities to subsets of human B-cell lymphomas (2), and a range of attendant studies has generated fundamental insights into related human diseases. Nonetheless, certain aspects of the mechanisms involved in the transformation of human and mouse lymphomas seem to be quite distinct. A hallmark feature of mature B-cell tumors in humans is the occurrence of balanced chromosomal translocations, most often involving immunoglobulin (Ig) genes and a wide range of partner genes. Some of these translocations are associated with specific subtypes of lymphomas, such as Ig/MYC translocations in Burkitt's lymphoma, Ig/BCL6 translocations in diffuse large B-cell lymphoma (DLBCL), and Ig/BCL10 or API2/MALT1 translocations in marginal zone lymphomas (MZL) of mucosa-associated lymphoid tissue (3). In most instances, expression of the partner genes is transcriptionally dysregulated with expression being directed by Ig gene or other regulatory sequences in the place of cognate elements. The consequences of altered expression of these proto-oncogenes can often be deduced from their normal biological functions.
Ig/oncogene translocations also occur in mice. However, they are regularly associated with a specific tumor type only for plasmacytomas (PCT), a neoplasm that features Ig/Myc translocations in over 95% of cases (4). Ig/BCL6 translocations have been described but are the exception rather than the rule in mouse DLBCL (5). Instead, the mode of oncogene activation in many spontaneous mouse B-cell tumors can often be ascribed to proviral insertional mutagenesis with proto-oncogenes being brought under the control of regulatory sequences in the MuLV long terminal repeats. Candidate cancer genes can be identified by cloning and sequencing proviral-host junction fragments (6). The advent of rapid PCR cloning methods and the availability of the entire mouse genome sequence have given this approach new life. However, this approach has been applied in only a few instances to specific lymphoma subsets (7) and its use is obviously dependent on tumors in mice that express ecotropic MuLV at high levels, either from endogenous loci or following inoculation.
Although it was anticipated that cDNA or oligonucleotide microarray-based transcriptional profiling would permit the association of aberrant oncogene expression with distinct lymphoma classes of humans and mice, the technique has proven to be much more useful in defining clinically distinct subsets of lymphomas belonging to single histologic classes (5, 8, 9).
As an alternative approach to addressing these issues in mouse lymphomagenesis, we have used a high-throughput quantitative real-time reverse transcription-PCR (qPCR) approach to simultaneously examine the expression of 384 genes selected because of their involvement in the pathogenesis of hematopoietic neoplasms or in signaling pathways governing the growth, survival, and differentiation of normal cells. We have applied this approach to studies of three histologically defined classes of mouse B-cell lineage tumors: splenic MZL, DLBCLs of centroblastic type (CBL), and PCT from interleukin-6 (IL-6) transgenic mice (10). These classes were chosen to sample lymphomas derived from different lineages of mature B cells (marginal zone versus follicular B cells) and dissimilar states of differentiation (germinal center centroblasts versus terminally differentiated plasma cells). The results of the study showed that patterns of gene expression for MZL++ and CBL were remarkably similar, in keeping with their cytologic similarities, and readily distinguishable from the transcriptional profile of PCT. Several genes that served to distinguish PCT from MZL++ and CBL were elements of the NOTCH signaling pathway. Studies on NOTCH activity in PCT cell lines showed that it was involved in regulating proliferation and promoting survival. These results suggest that in mouse PCT, NOTCH and MYC govern two overlapping transcriptional programs that promote plasma cell growth and transformation.
Materials and Methods
Mice, splenic B cells, plasma cells, and PCT cell lines
NFS.V+ mice that develop a range of B-cell lymphomas (1) and BALB/c-IL-6 transgenic mice that develop PCT in lymph nodes and spleen (10) were described previously. Controls included young BALB/cPt and NFS.V+ mice without tumors. All animal studies were performed in accordance with approved animal protocols of the National Institute of Allergy and Infectious Diseases (NIAID) and the National Cancer Institute (NCI), NIH. At necropsy, portions of spleens and lymph nodes of mice with enlarged spleens and nodes were fixed in formaldehyde for histologic studies or snap frozen for later preparation of RNA. Histologic diagnoses based on studies of sections stained with H&E were made according to the Bethesda classification of mouse lymphoid neoplasms (2) by a pathologist (Dr. Torgny N. Fredrickson) who contributed to the classification. Pooled spleen cells from three to four IL-6 transgenic mice were processed with antibody-coated magnetic-activated Dynal beads (Invitrogen) to negatively enrich B cells. The enriched cells were stained with antibodies to B220 and CD138 and then sorted using a fluorescence-activated cell sorting (FACS) Aria-Green cell sorter (Becton Dickinson) to generate highly enriched (>90%) populations of normal B cells and plasma cells, respectively. PCT cell lines were generously provided by Dr. Michael Potter (NCI, NIH).
Preparation of RNA and cDNA
RNA was prepared from frozen tumor tissues, normal B cells and plasma cells, or cell lines using the RNeasy Mini kit (Qiagen), and the quality of the RNA was examined by Bioanalyzer (Agilent). cDNA was synthesized according to the manufacturer's protocol (MessageSensor RT kit, Ambion).
Fabrication of real-time qPCR array and data analysis
Three hundred eighty-four primer pairs for cancer-related genes were selected using the Primer3 program and in some cases by manual design. Each primer was searched against Genbank via the National Center for Biotechnology Information BLAST to insure target specificity to each target gene. The primer pairs were then dried in the wells of 384-well plates. A list of the genes is given in Supplementary Table S1. cDNA and SYBR Green RT-PCR mastermix (Applied Biosystems) were added to the cells using a RoboGo (Aviso) and reactions were run using an ABI Prism 7900HT (Applied Biosystems). Differentially expressed genes were identified using a global pattern recognition (GPR) algorithm, including global normalization that compares the Ct in each gene with the Ct of every other gene and a two-tailed unpaired t test that compares Cts from the control and experimental group (11). Subsequently, hierarchical clustering and functional grouping with Fisher's exact test were conducted. ANOVA test was also performed to find significant genes among the three B-cell lymphoma classes and the results were shown in k-means clustering (k = 10).
Oligomicroarrays
Microarray chips printed by the NIAID Microarray Research Facility comprised ~18,000 genes represented by 70-mer oligonucleotides. Sample preparation and hybridization were as described (12). After the raw data were normalized with lowess smoothing function, the significant genes were identified with significance analysis of microarrays (13) and gene set enrichment analysis (GSEA) was performed (14).
Treatments with γ-secretase inhibitors
Four PCT cell lines (ABPC8298, SIPC8523, SIPC8195, and TEPC2372) and three Abelson-transformed early B-cell lines [Rag–/– (obtained from Dr. Jessica Jones, Georgetown University), 18-81, and 220-8 (obtained from Dr. Fred Alt, Harvard Medical School)] were treated with medium containing either the γ-secretase inhibitor (GSI) GSI-XII (Calbiochem) dissolved in DMSO or DMSO alone for 36 h. Primary PCTs were cultured in medium with DMSO or GSI for 24 h.
Transfection of dominant-negative MAML1
Cultured PCT cell lines were infected with supernatants of Pheonix-Eco cells that had been transfected with the dominant-negative MAML1 (DNMAML1)-EGFP fusion construct in a murine stem cell virus–based retroviral vector or a construct expressing EGFP alone (15). The vectors were generously provided by Dr. Warren Pear (University of Pennsylvania). Cells expressing EGFP were purified by FACS sorting.
Cell proliferation, bromodeoxyuridine incorporation, and apoptosis assays
Cell proliferation assays were carried out using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Roche) or the CellTiter-Glo assay (Promega). For bromodeoxyuridine (BrdUrd) incorporation assays, cells were pulsed with 10 μmol/L BrdUrd for 30 min and then fixed, permeabilized, stained with allophycocyanin (APC)-labeled anti-BrdUrd antibody (Becton Dickinson), and analyzed by FACS. Apoptosis assay were performed by FACS analysis of cells stained with APC-conjugated Annexin V and 7-amino-actinomycin D (7-AAD; Becton Dickinson). Caspase-3 activity in cell lysates was analyzed using a colorimetric Caspase-3 Assay kit according to the manufacturer's instruction (Sigma-Aldrich).
Western blotting
Western blotting was performed as described previously (12) using antibodies specific for CDK4, CDK6, phosphorylated Rb (pRb; Ser807/811), caspase-8, and cleaved caspase-3 (Cell Signaling Technology) and anti-tubulin (Sigma-Aldrich).
Results
Comparisons of gene expression profiles of MZL++, CBL, and PCT using qPCR arrays
A qPCR array platform composed of 384 cancer-related genes (Supplementary Table S1) was used to examine the features of three histologically defined classes of primary mouse B-cell neoplasms: CBL, MZL++, and PCT. For splenic MZL, we selected high-grade cases from NFS.V+ mice classified as MZL++ (16). MZL initially seem as low-grade tumors with the cytology of normal marginal zone B cells and exhibit little if any mitotic or apoptotic activity. Over time, the tumor cells have been shown to increase not only in number but also in grade, with cells from high-grade MZL++ cases having cytologic features indistinguishable from those of CBL but also in association with frequent mitotic and apoptotic figures (16). Splenic CBL were also from NFS.V+ mice, whereas PCT were from BALB/c-IL-6 transgenic mice (10). Although it would have been ideal to have a single strain as the source of all three tumor types, NFS.V+ mice rarely develop PCT and high-grade MZL are seen very rarely in strains other than the NFS congenics (2, 17). PCT are very uncommon in NFS.V+ mice (1, 5), and BALB/c-IL-6 transgenic mice do not develop MZL or CBL.
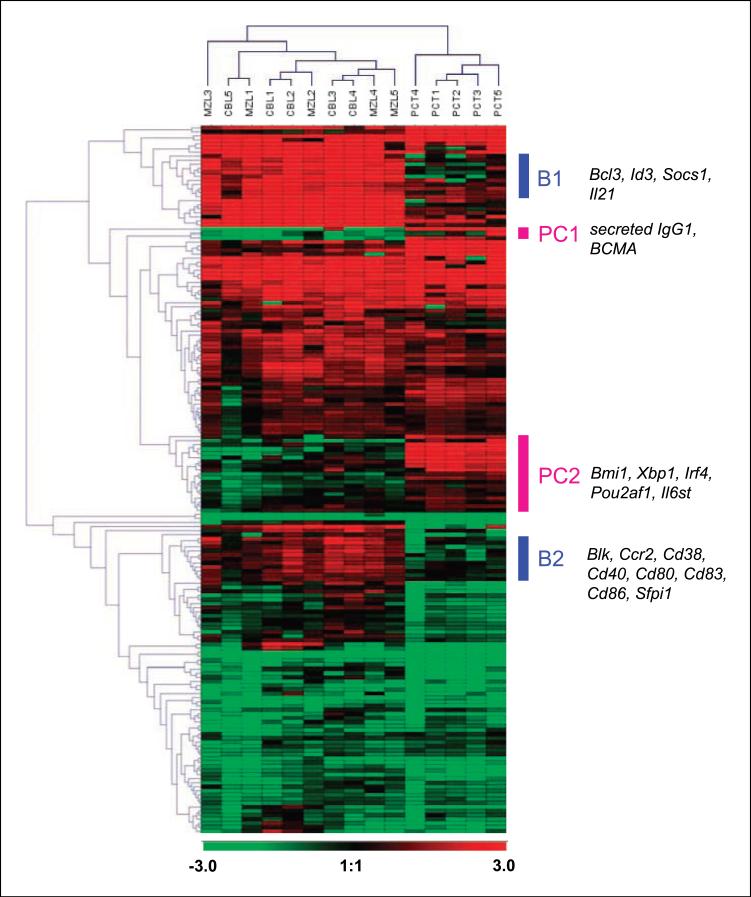
Tests of RNA prepared from spleens of five tumor-bearing mice of each histologic class were compared with analyses of RNA prepared from spleens of five normal NFS.V+ or BALB/c mice. We then used the GPR algorithm to identify genes that significantly distinguished each lymphoma type from the expression pattern of normal spleens. Hierarchical clustering of the 174 genes found to be differentially expressed by at least one class of lymphoma and the 15 individual cases is shown in Fig. 1. The clustering identified sets of genes with closely related expression patterns that are preferentially expressed in normal B cells (subsets designated B) or in normal plasma cells (subsets PC). As would be expected, B-cell–related genes, including Bcl3, Id3, Socs1, Il21, Blk, Ccr2, Cd38, Cd40, Cd80, Cd83, Cd86, and Sfpi1, were expressed at increased and comparable levels by MZL++ and CBL. Likewise, the plasma cell–related genes, including secreted IgG1, BCMA, Bmi1, Xbp1, Irf4, Pou2af1, and Il6st, exhibited heightened expression in the PCT.
Figure 1.
Clustering analysis of gene expression data from qPCR arrays. A hierarchical clustering algorithm was used to group the genes that significantly distinguished each lymphoma type from normal controls, as determined by the GPR algorithm–bearing t test, and the tumor samples. The dendrograms depict the relatedness of individual genes and cases. The colored bars to the right of the figure indicate clusters of coordinately expressed genes overexpressed in normal B cells compared with normal plasma cells (blue) or in normal plasma cells compared with normal B cells (red). Selected genes associated with the clusters are shown. The ratios of relative gene expression are depicted according to the color scale shown at the bottom.
A dendrogram showed the segregation of PCT cases on one branch with CBL and MZL++ on another but with the MZL++ and CBL cases intermingled (Fig. 1). This intermingling of MZL++ and CBL cases was quite unexpected because the B cells of origin (marginal zone and follicular, respectively) are anatomically, phenotypically, and functionally quite distinct. This suggests that the transition of MZL from low-grade to a high-grade disease featuring cells with the cytology of centroblasts is associated with the acquisition of a transcriptional program remarkably similar to that of CBL.
Identification of NOTCH pathway activation in PCT
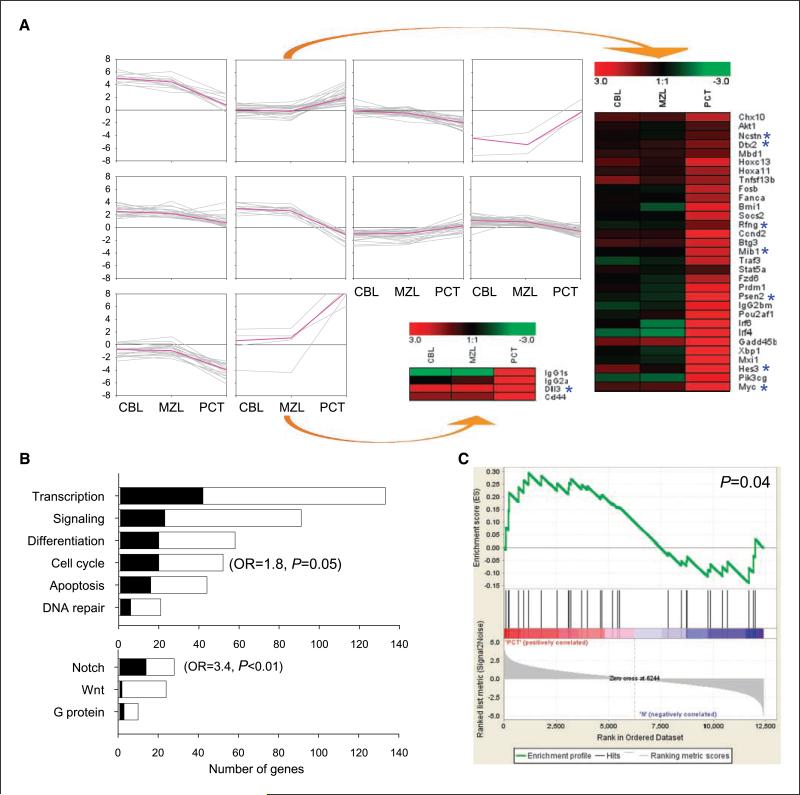
ANOVA analyses, performed as another statistical approach to identifying genes from the qPCR data set that distinguish one lymphoma class from the others, revealed 196 genes that were differentially expressed among the three classes (P < 0.05; Supplementary Table S2). Subsequently, k-means clustering was applied to the set of genes that showed significant group differences (k = 10; Fig. 2A). The second and tenth clusters, for which expression was similar for CBL and MZL++ but increased in PCT (Fig. 2A), comprised a series of genes known to be activated by translocation or other mechanisms in human multiple myeloma (MM), a plasma cell neoplasm with some similarities to PCT (5, 18, 19). These included Bmi1, Myc, Pou2af1, Ccnd2, Akt1, and Pik3gc (20–22). These two clusters also included genes involved in normal plasma cell proliferation and/or differentiation, such as Tnfsf13b, Prdm1, Irf4, and Xbp1. In contrast, genes in the ninth cluster, which were prominently down-regulated in PCT, predictably included several involved in normal B-cell differentiation and function, such as Pax5 and Irf8 (Supplementary Table S2; refs. 15, 23).
Figure 2.
Identification of NOTCH signaling as a significant pathway in PCT. A, k-means clustering for the significant genes revealed by qPCR that distinguish CBL, MZL++, and PCT. ANOVA analysis identified the genes with significant group differences (P < 0.05), which were clustered by k-means algorithm (k = 10). The pink line in each cluster connects the median values for each lymphoma type and the gray lines connect the value for each gene. Expression patterns for the genes comprising clusters 2 and 10 are shown. The ratios of relative gene expression are depicted as in Fig. 1. *, a NOTCH signaling pathway gene. B, functional categorization by Gene Ontology of genes differentially expressed by PCT. The number of genes in each of the six categories for which expression was unchanged is represented by the white portion of each bar, whereas the black portion depicts the number of genes that were significantly changed in expression. Bottom, breakdown of signaling genes involved in Notch, Wnt, and G protein signaling. The odds ratios (OR) for each class with a significant P value are shown. C, GSEA of oligonucleotide microarray data generated from the same PCT cases as used for qPCR arrays identified genes involved in Notch signaling as enriched among the set of differentially expressed genes in PCT.
Interestingly, a series of genes involved in the NOTCH signaling pathway, Ncstn, Dtx2, Rfng, Mib1, Psen2, Hes3, and Dll3, were enriched in the clusters in which gene expression was substantially increased in PCT (Fig. 2A). This observation was reinforced by the results of a functional categorization of differentially expressed genes based on Gene Ontology (Fig. 2B), which showed that genes involved in cell cycle regulation and NOTCH signaling were modulated more than those in other biological processes. In addition, GSEA of oligomicroarray data from the same tumors confirmed that components of the NOTCH signaling pathway were significantly enriched in PCT (Fig. 2C).
Comparative expression of NOTCH signaling components in normal plasma cells and PCT
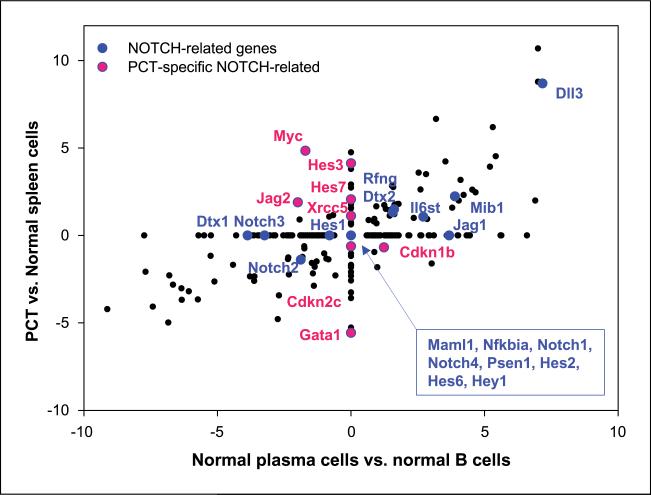
The recent observation that NOTCH signaling is involved in the later stages of B-cell maturation (24) raised the possibility that some of the NOTCH pathway molecules seen to be up-regulated in PCT might simply be reflective of the fully differentiated status of normal plasma cells. To examine this possibility, we used data from qPCR arrays to evaluate differences in gene expression among normal spleen cells, purified normal splenic B cells from BALB/c-IL-6 transgenic mice, purified premalignant plasma cells from BALB/c-IL-6 transgenic mice, and the PCT described previously. In Fig. 3, the relative expression of genes by normal plasma cells compared with normal B cells is plotted on the X axis and the relative expression of genes by PCT compared with normal spleen cells is plotted on the Y axis. Genes represented by points that fall on the diagonal from upper right to lower left are expressed at similar levels by normal plasma cells and PCT and are thus indicative of a state of differentiation rather than features of the transformed state. Points that lie on the vertical line, except those at the “0,0” intersection, represent genes that distinguish PCT from normal plasma cells but are uninformative for distinguishing normal plasma cells from normal B cells. Consequently, these genes are PCT specific. Points on the horizontal line are associated with genes that tend to distinguish normal plasma cells from normal B cells but do not distinguish PCT from normal plasma cells. Finally, points that fall into the upper left and lower right quadrants can also be viewed as PCT specific and thus potentially contributory to PCT development.
Figure 3.
Characterization of NOTCH signaling genes associated with normal plasma cell differentiation and PCT development. Data from gene expression profiling by qPCR arrays were used to compare normal plasma cells with normal B cells (X axis) and PCT with normal spleen cells (Y axis). NOTCH signaling genes associated with normal plasma cell differentiation are labeled in blue and PCT-specific NOTCH-related genes are labeled in red. The scale for each axis is fold change in log2.
By this reasoning, expression of 10 of the 26 genes recognized as components of the NOTCH signaling pathway and shown in blue is reflective of their expression in normal plasma cells and probable roles in late B-cell differentiation (24). Seven others, shown in red, are candidate contributors to PCT induction, progression, or maintenance. Five of these seven are direct transcriptional targets of NOTCH: Xrcc5 (Gpp21), Hes7, Myc, Cdkn2c (p18), and Cdkn1b (p27). Importantly, both p18 and p27 are negatively regulated by NOTCH (25). In addition, Hes3 is rapidly activated by NOTCH via a recently described pathway involving serial signaling through phosphatidylinositol 3-kinase/AKT, mammalian target of rapamycin, and signal transducer and activator of transcription 3 (26). In addition, transcription of Gata1 is suppressed by HES1, a direct target of NOTCH (27). Recent studies of human T-cell lymphomas showed that NOTCH is a direct transcriptional activator of MYC and that together they form a “feed-forward transcriptional loop” that promotes leukemia development (25). It is therefore of interest that six additional genes, Cdk4, Cdc20, Gapd, Set, Jak1, and Jak2, are direct transcriptional targets of MYC (28, 29). This suggests that a similar loop involving NOTCH and MYC could contribute to the development of PCT.
Effects of NOTCH signaling on proliferation and cell survival in PCT cell lines
NOTCH signaling is activated by binding of ligands to NOTCH receptors (NOTCH1-NOTCH4) and followed by two sequential cleavages of the receptors by the enzymes metalloproteinase and γ-secretase. The cleavages release intracellular NOTCH (ICN), which translocates to nucleus where it interacts with the transcription factor, CSL, and transcriptional coactivators, including MAML proteins (MAML1-MAML3). To determine if NOTCH signaling was of biological importance to PCT, we blocked the signaling at two different stages of this cascade: cleavage of NOTCH by γ-secretase at the cell membrane and interaction of ICN with MAML in the nucleus.
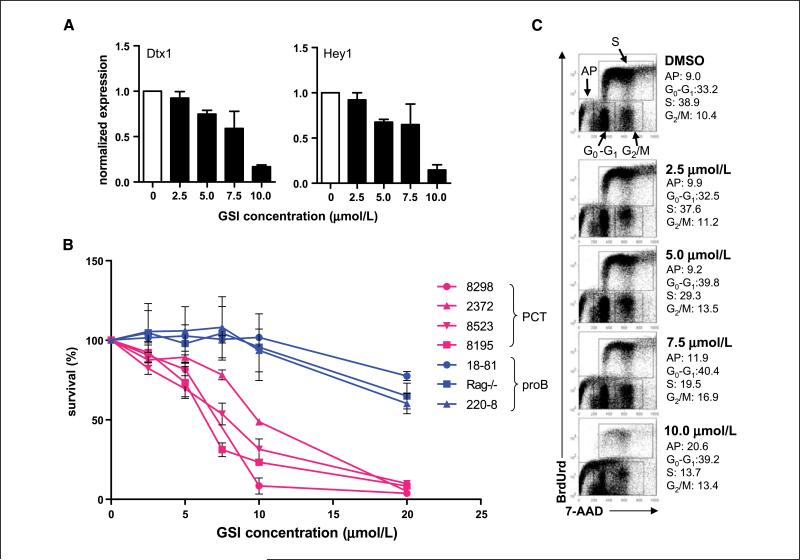
To block the cleavage of NOTCH, we used the GSI, GSI-XII (30), to treat four PCT cell lines in vitro. To evaluate the effectiveness of GSI treatment on NOTCH inhibition, we quantified expression of the NOTCH-specific targets genes, Dtx1 and Hey, by qPCR. The transcript levels or both genes were reduced by treatment with the GSI in a dose-dependent manner (Fig. 4A). We next investigated if inhibition of NOTCH signaling by GSI might affect proliferation of the PCT cell lines using three pro-B-cell lines as controls. GSI treatment remarkably inhibited growth of the PCT cell lines at concentrations that had much less effect on the early B-cell lines (Fig. 4B). To determine whether this growth inhibition was caused by apoptosis and/or cell cycle inhibition, the PCT cell lines were labeled with BrdUrd and 7-AAD and analyzed for incorporation of BrdUrd and DNA content by flow cytometry (Fig. 4C). These studies showed that the proportions of apoptotic cells (labeled AP) were correlated directly with increasing concentrations of GSI, whereas the proportions of S-phase cells (labeled S) were correlated inversely. Thus, GSI treatment inhibited cell proliferation in PCT cell lines by induction of apoptosis and suppression of cell cycle progression. To investigate whether primary PCT would show dependence on Notch signaling for growth, GSI treatment was carried out using ascites cells from pristine-induced PCT mice, which include primary PCT along with lymphocytes and inflammatory cells. After 24 hours of GSI treatment, the cells were harvested and examined by flow cytomery for Annexin V and 7-AAD to determine the extent of apoptosis in both PCT cell (CD138+) and non-PCT cell (CD138–) subsets. About 25% of CD138+ cells cultured in DMSO were apoptotic, whereas the frequencies in cultures with two different concentration of GSI-XII were increased to 45% and 56%. Only less than 1% of the CD138– cells were apoptotic (Supplementary Fig. S1). This result indicates that the proportion of apoptotic population in cultured PCT cells is significantly increased with GSI treatment.
Figure 4.
Inhibition of NOTCH signaling in PCT cell lines by GSI treatment. PCT cell lines and pro-B-cell lines were treated with different concentration of GSI-XII for 36 h. A, the expressions of NOTCH targets Dtx1 and Hey1 were measured by qPCR in the GSI-treated ABPC8298 PCT cell line. Columns, mean compared with DMSO-treated cells for three independent experiments; bars, SE. B, proliferation of GSI-treated PCT and pro-B-cell lines was examined by the MTT assay. Points, mean compared with DMSO-treated cells for three independent experiments; bars, SE. C, representative flow cytometric analysis of BrdUrd incorporation and DNA content in GSI-treated PCT cell lines. GSI-treated cells were pulsed with 10 μmol/L BrdUrd for 30 min and then fixed, permeabilized, and stained for BrdUrd and 7-AAD. Compartments containing the apoptotic population (AP) and cells in the G0-G1, S, and G2-M phases of the cell cycle are identified at the top. The proportions of cells in each compartment for cells treated with DMSO alone and increasing concentrations of the GSI are given to the right of each profile.
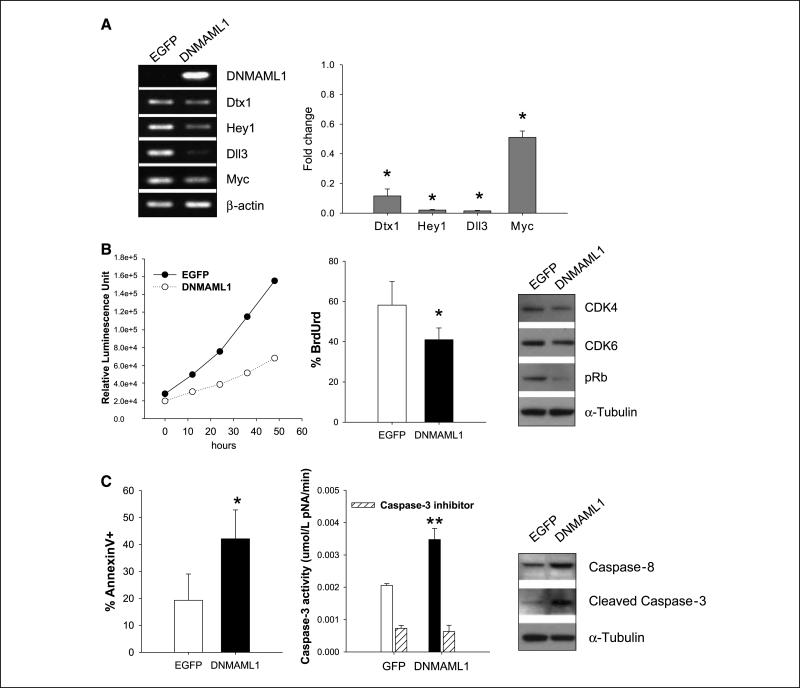
Although GSI-XII is quite specific for γ-secretase, it still has activity against targets other than NOTCH, such as β-amyloid precursor protein (31). This prompted us to take an alternative approach to assessing the contributions of NOTCH signaling to PCT growth and survival. To do this, we took advantage of the fact that transcriptional activation by NOTCH is dependent on its association with members of the MAML family of coactivators through their NH2-terminal sequences. Previous studies of a human MAML1 mutant containing only the ICN binding domain, termed DNMAML1, showed that it functioned as a potent dominant-negative inhibitor of signaling by each of the NOTCH receptors in mouse cells (15). We therefore infected PCT cell lines with retroviral vectors expressing DNMAML1 and EGFP or EGFP alone. RT-PCR showed that human DNMAML1 was expressed in PCT cell lines (Fig. 5A, left) and was associated with reduced expression of the NOTCH target genes Dtx1, Hey1, Dll3, and Myc. Quantitative analyses showed that expression of each of these targets was reduced by 50% to >95% in cells expressing DNMAML1.
Figure 5.
Inhibition of NOTCH signaling in PCT by DNMAML1. A, PCR analysis (left) of expression of DNMAML1; the NOTCH targets Dtx1, Hey1, Dll3, and Myc; and β-actin in the ABPC8298 PCT cell line infected with viruses expressing EGFP alone or human DNMAML1 and EGFP. The MAML1 primers are specific for the human transcript. Right, quantitative analyses of NOTCH target gene expression in ABPC8298 expressing DNMAML1. Data are normalized to level of expression in the PCT cell lines expressing only EGFP. Columns, mean compared with the expression in cells expressing EGFP only for three independent experiments; bars, SE. *, significantly different from expression in EGFP-only cells (P < 0.05). B, analyses of cell proliferation. Left, proliferation assay in PCT cell lines expressing DNMAML or EGFP only; middle, percent of cells incorporating BrdUrd after 30 min in culture (*, P < 0.05); right, Western blot analysis of CDK4, CDK6, pRb, and α-tubulin expression. C, analyses of cell death in cultured ABPC8298 cells: percent of cells staining with Annexin V. Left, columns, mean of three independent experiments; bars, SE. *, P < 0.05. Middle, capase-3 activity determined in the cell lysates (black columns) or cell lysates incubated with the caspase-3 inhibitor. **, P < 0.005. Right, Western blot analyses of caspase-8, cleaved caspase-3, and α-tubulin levels.
Previous studies of human T-cell acute lymphoblastic leukemias (T-ALL) showed that deregulated expression of NOTCH1 is responsible for activating transcriptional programs that promote the growth of leukemic cells (25). To reassess the importance of NOTCH signaling to growth of the PCT cell line, clones expressing DNMAML1 and EGFP or EGFP alone were cultured and viable cell numbers were determined at 12-hour intervals (Fig. 5B, left). These results showed that cell expansion was greatly reduced in cultures expressing DNMAML1. To determine if this phenotype could be ascribed to altered proliferation, we first tested cells for uptake of BrdUrd into DNA, which occurs during S phase, providing an estimate of cell growth fraction. In cells, expressing DNMAML1, the frequency of BrdUrd+ cells was significantly reduced (Fig. 5B, middle). To examine molecular mechanisms that might affect cell cycle regulation and proliferation, we performed Western blot analyses of CDK4, CDK6, and pRb expression in cells expressing DNMAML1 or EGFP only (Fig. 5B, right). Whereas the levels of both kinases were somewhat reduced in DNMAML1-expressing cell, levels of pRb were markedly lower. This indicated that the reduced expansion of DNMAML1+ cells was due, at least in part, to altered G1-S progression.
We also evaluated the possible contributions of enhanced cell death to the slower expansion of DNMAML1-expressing cells. Flow cytometric studies showed that the proportions of cells binding Annexin V were significantly increased in cells expressing DNMAML1 (Fig. 5C, left). A representative flow cytometry analysis of EGFP only and DNMAML1 cells stained with Annexin V and 7-AAD is shown in Supplementary Fig. S2. In addition, DNMAML1-positive cells were found to have increased levels of caspase-3 activity that was blocked by a specific inhibitor (Fig. 5C, middle). Finally, Western blot analyses showed that levels of caspase-8 and cleaved caspase-3 were significantly increased in cells expressing DNMAML1 (Fig. 5C, right). These results indicated that NOTCH signaling in the PCT cell line promoted survival as well as proliferation.
Discussion
It is well established that NOTCH signaling pathways can promote or inhibit the differentiation of lymphoid progenitors and their progeny in a lineage-dependent manner (32). Heightened expression of NOTCH1 in hematopoietic progenitors, for example, promotes T-cell differentiation while inhibiting B-cell development (33). In addition, DLL1, NOTCH2, CSL, and MAML1 are required for the development of marginal zone but not of follicular B cells (34–36) and activated NOTCH2 promotes the development of B1 B cells (37). Finally, DLL1-NOTCH1 interactions were shown to promote the terminal differentiation of mature B cells to antibody-secreting plasma cells (24).
Importantly, aberrant expression of various NOTCH signaling molecules has also been tied to several human and mouse lymphoid malignancies. The most clearly defined role for NOTCH as an oncogene is in human T-ALL in which NOTCH1 is activated by chromosomal translocations or, more frequently, by mutations that cause either ligand-independent activation of the receptor or increased stability. Notch1 is also a frequent target of retroviral integrations in mouse B-cell lymphomas (38).
Studies of possible contributions of disordered NOTCH signaling to the pathogenesis of human MM are of particular relevance to the current study of mouse PCT. In this regard, several investigations have shown that JAG1, JAG2, NOTCH1, NOTCH2, NOTCH3, NOTCH4, MIB2, and LFNG are expressed by primary tumor cells of patients with MM and/or MM cell lines and may contribute to disease initiation or progression (39–42). It should be noted that other conflicting studies showed that heightened expression of NOTCH induced growth inhibition of several MM cell lines (40, 43). However, the mechanisms contributing to growth inhibition differ between the reports. In one report (40), growth inhibition was associated with reduced apoptosis, whereas in the second (43), growth inhibition was associated with increased apoptosis. In a follow-up study by Nefedova and colleagues (30), it was shown that inhibition of NOTCH signaling was associated with increased apoptosis, but effect on cell cycle progression was not examined.
In the present study, data from qPCR array analyses of primary mouse B-cell lineage tumors established that several genes involved in NOTCH signaling were expressed at significantly higher levels by PCT than by CBL or MZL++, including Myc, Dll3, Rfng, Ncstn, Psen2, Mib1, and Dtx2. Earlier studies that identified contributions of NOTCH signaling to terminal B-cell differentiation allowed us to predict that some of these differences would be reflective of normal plasma cell differentiation but also to anticipate that others may have contributed to tumor induction, progression, or maintenance. Indeed, direct comparisons of data obtained from studies of normal plasma cells and PCT made it possible to distinguish a “differentiation” subset of genes from a candidate “cancer” subset.
Direct evidence for the involvement of NOTCH signaling in plasmacytomagenesis came from studies of cultured PCT cell lines treated with a GSI or expressing DNMAML1. Treatment with the GSI inhibited cell expansion by slowing G1-S progression and inducing apoptosis. DNMAML1 was previously shown to bind only CSL in the presence of ICN and to interact comparably with ICN1, ICN2, ICN3, and ICN4 (15). Consequently, DNMAML1 can be used to screen for the activities of all NOTCH receptors. We showed first that expression of a series of known NOTCH target genes was significantly reduced in cells expressing DNMAML1 and that the extent of reduction was inversely correlated with the levels of DNMAML1 transcripts. We also showed that the growth of cultured PCT cell lines expressing DNMAML1 was greatly reduced and that this was due to NOTCH promotion of cell cycle progression and survival.
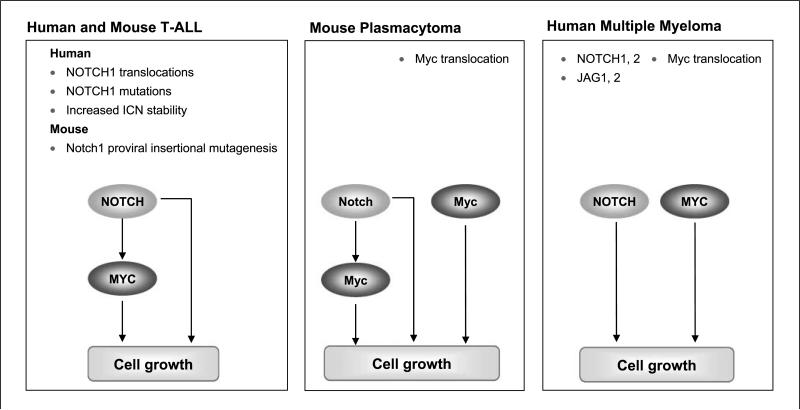
Based on combined data from qPCR as well as oligomicroarrays, it seems that the NOTCH signaling pathway is activated in PCT of differing origins: pristane-treated or Abelson MuLV-infected BALB/c mice or BALB/c-IL-6 transgenic mice. PCT from almost all these mice carry balanced chromosomal translocations that activate Myc (4). Myc is a transcription factor widely recognized as a master regulator of cell growth, proliferation, differentiation, metabolism, and death, which acts by occupying over 4,000 genomic loci (44, 45). Recent genome-wide studies designed to identify target genes regulated by NOTCH1 in T-ALL cells showed that NOTCH1 is not only a direct activator of biosynthetic pathways controlling cell growth and metabolism but also a direct regulator of MYC (25, 46). By reverse engineering of regulatory networks from expression profiles, it was found that NOTCH1 and MYC orchestrate two overlapping transcriptional programs with many common targets that synergize to regulate the growth of primary T-ALL cells (Fig. 6; ref. 25). We suggest that a similar synergy between MYC and NOTCH is also characteristic of mouse PCT, but with Myc being activated by translocations in addition to its role as a downstream target of NOTCH (Fig. 6). It is noteworthy that the levels of Myc transcripts were significantly reduced in cells expressing DNMAML1. This suggests that NOTCH continues to influence expression from the translocated Myc gene as the other Myc allele is silenced in tumors carrying an Ig/Myc translocation (47). The mechanisms responsible for NOTCH activation in primary PCT, like those of BALB/c-IL-6 transgenic mice described here, await clarification. Cultured cell lines are unlikely to provide clear insights into these issues as they have had to adapt to growth in vitro in the absence of stromal elements and soluble mediators known to influence the growth and survival of human MM and mouse PCT.
Figure 6.
Integration of NOTCH and MYC signaling pathways in T-ALL of humans and mice, mouse PCT, and human MM. The results of this study and other analyses have shown that signaling pathways activated by NOTCH and MYC are common to all three types of tumors but that the mechanisms responsible for their activation are different for each. In mouse PCT and human T-ALL, NOTCH and MYC form integrated feed-forward loops that were shown in T-ALL to direct transcriptional programs with extensive overlap of target genes that regulate the growth of tumor cells (26). PCT differ from T-ALL in that MYC is also activated almost universally by chromosomal translocations. MYC is almost always activated in MM as well and sometimes by chromosomal translocations but also by mechanisms that remain undefined. The mechanisms responsible for NOTCH activation in T-ALL of humans and mice are well defined but are poorly understood in PCT and MM. Regardless of the mechanisms responsible for their activation, the integration of NOTCH and MYC signaling pathways in these tumors may help model the structures of key transcriptional networks in a variety of mouse and human tumors.
Recent studies have drawn renewed attention to several similarities between mouse PCT and human MM (5, 18, 19, 48). These parallels have been extended by the demonstrations that NOTCH receptors and ligands can be identified in almost all MM cell lines and cells from primary cases (39, 40, 49) and that MYC is activated by translocation, amplification, or other mechanisms in nearly all MM cell lines and in cells from more than half of primary cases. Although Myc-activating translocations are considered to be early and perhaps initiating events in PCT, they are thought to occur during the later stages of MM progression (50). Nonetheless, the understanding that the NOTCH and MYC signaling pathways are active in both diseases suggests that PCT might be valuable for modeling late-stage MM as it occurs in a substantial subset of patients and examining therapeutics that target both pathways.
Supplementary Material
Acknowledgments
Grant support: Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases, and NIH grants DK56597 and RO1AI28802 (D.C. Roopenian).
We thank Drs. Siegfried Janz, Alexander Kovalchuk, Janet Hartley, and Torgny N. Fredrickson for providing materials, histologic studies of mouse lymphomas, and many helpful discussions and Drs. Zohreh Naghashfar and Chang Hoon Lee for microarray hybridization and technical advice.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
D.J. Shaffer: commercial research support and ownership interest, Bar Harbor BioTechnology. D.C. Roopenian: ownership interest, Bar Harbor BioTechnology. The other authors disclosed no potential conflicts of interest.
References
- 1.Hartley JW, Chattopadhyay SK, Lander MR, et al. Accelerated appearance of multiple B cell lymphoma types in NFS/N mice congenic for ecotropic murine leukemia viruses. Lab Invest. 2000;80:159–69. doi: 10.1038/labinvest.3780020. [DOI] [PubMed] [Google Scholar]
- 2.Morse HC, III, Anver MR, Fredrickson TN, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–58. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 3.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–62. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 4.Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–95. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 5.Qi C-F, Zhou JX, Lee CH, et al. Anaplastic, plasmablastic, and plasmacytic plasmacytomas of mice: relationships to human plasma cell neoplasms and late-stage differentiation of normal B cells. Cancer Res. 2007;67:2439–47. doi: 10.1158/0008-5472.CAN-06-1561. [DOI] [PubMed] [Google Scholar]
- 6.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 7.Shin MS, Fredrickson TN, Hartley JW, Suzuki T, Agaki K, Morse HC., III High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 2004;64:4419–27. doi: 10.1158/0008-5472.CAN-03-3885. [DOI] [PubMed] [Google Scholar]
- 8.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 9.Zhan F, Tian E, Bumm K, Smith R, Barlogie B, Shaughnessy J., Jr Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 2003;101:1128–40. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 10.Kovalchuk AL, Kim JS, Park SS, et al. IL-6 transgenic mouse model for extraosseous plasmacytoma. Proc Natl Acad Sci U S A. 2002;99:1509–14. doi: 10.1073/pnas.022643999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akilesh S, Shaffer DJ, Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res. 2003;13:1719–27. doi: 10.1101/gr.533003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CH, Melchers M, Wang H, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. From the Cover: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maillard I, Weng AP, Carpenter AC, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 16.Fredrickson TN, Lennert K, Chattopadhyay SK, Morse HC, III, Hartley JW. Splenic marginal zone lymphomas of mice. Am J Pathol. 1999;154:805–12. doi: 10.1016/S0002-9440(10)65327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morse HC., III . Classification and characteristics of mouse B cell-lineage lymphoma. In: Honjo T, Alt F, Neuberger M, editors. Molecular biology of B cells. Elsevier Academic Press; San Diego: 2004. pp. 365–75. [Google Scholar]
- 18.Janz S, Morse HC, III, Teitell M. Mouse models of human mature B cells and plasma cell neoplasm. In: Li S, editor. Mouse models of human blood cancers. Springer Science; Berlin (Germany): 2008. pp. 179–226. [Google Scholar]
- 19.Park ES, Shaughnessy J, Gupta S, et al. Gene expression profiling reveals different pathways related to Abl and other genes that cooperate with c-Myc in a model of plasma cell neoplasia. BMC Genomics. 2007;8:302. doi: 10.1186/1471-2164-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Vos J, Thykjaer T, Tarte K. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene. 2002;21:6848–57. doi: 10.1038/sj.onc.1205868. [DOI] [PubMed] [Google Scholar]
- 21.Hanamura I, Huang Y, Zhan F, Barlogie B, Shaughnessy J. Prognostic value of cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia. 2006;20:1288–90. doi: 10.1038/sj.leu.2404253. [DOI] [PubMed] [Google Scholar]
- 22.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–98. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 23.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 24.Santos MA, Sarmento LM, Rebelo M, et al. Notch1 engagement by Delta-like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proc Natl Acad Sci U S A. 2007;104:15454–9. doi: 10.1073/pnas.0702891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Androutsellis-Theotokis A, Leker RR, Soldner F, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–6. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 27.Ishiko E, Matsumura I, Ezoe S, et al. Notch signals inhibit the development of erythroid/megakaryocytic cells by suppressing GATA-1 activity through the induction of HES1. J Biol Chem. 2005;280:4929–39. doi: 10.1074/jbc.M406788200. [DOI] [PubMed] [Google Scholar]
- 28.Menssen A, Hermeking H. From the Cover: Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A. 2002;99:6274–9. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006;66:4591–601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- 30.Nefedova Y, Sullivan DM, Bolick SC, Dalton WS, Gabrilovich DI. Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood. 2008;111:2220–9. doi: 10.1182/blood-2007-07-102632. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Nadeau P, Song W, et al. Presenilins are required for γ-secretase cleavage of β-APP and transmembrane cleavage of Notch-1. Nat Cell Biol. 2000;2:463–5. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- 32.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–6. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 33.Pui JC, Allman D, Xu L, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 34.Hozumi K, Negishi N, Suzuki D, et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–44. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 35.Tanigaki K, Han H, Yamamoto N, et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–50. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Maillard I, Nakamura M, Pear WS, Griffin JD. The transcriptional coactivator Maml1 is required for Notch2-mediated marginal zone B-cell development. Blood. 2007;110:3618–23. doi: 10.1182/blood-2007-06-097030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witt CM, Won W-J, Hurez V, Klug CA. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J Immunol. 2003;171:2783–8. doi: 10.4049/jimmunol.171.6.2783. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–74. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 39.Houde C, Li Y, Song L, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104:3697–704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- 40.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–10. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 41.De Vos J, Couderc G, Tarte K, et al. Identifying intercellular signaling genes expressed in malignant plasma cells by using complementary DNA arrays. Blood. 2001;98:771–80. doi: 10.1182/blood.v98.3.771. [DOI] [PubMed] [Google Scholar]
- 42.Jundt F, Probsting KS, Anagnostopoulos I, et al. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–5. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 43.Zweidler-McKay PA, He Y, Xu L, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106:3898–906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levens D. Disentangling the MYC web. Proc Natl Acad Sci U S A. 2002;99:5757–9. doi: 10.1073/pnas.102173199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–9. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ar-Rushdi A, Nishikura K, Erikson J, Watt R, Rovera G, Croce C. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983;222:390–3. doi: 10.1126/science.6414084. [DOI] [PubMed] [Google Scholar]
- 48.Boylan KLM, Gosse MA, Staggs SE, et al. A transgenic mouse model of plasma cell malignancy shows phenotypic, cytogenetic, and gene expression heterogeneity similar to human multiple myeloma. Cancer Res. 2007;67:4069–78. doi: 10.1158/0008-5472.CAN-06-3699. [DOI] [PubMed] [Google Scholar]
- 49.Jundt F, Anagnostopoulos I, Forster R, Mathas S, Stein H, Dorken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99:3398–403. doi: 10.1182/blood.v99.9.3398. [DOI] [PubMed] [Google Scholar]
- 50.Shou Y, Martelli ML, Gabrea A, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000;97:228–33. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.