Abstract
G protein-coupled receptors (GPCRs) are a large superfamily of signaling proteins expressed on the plasma membrane. They are involved in a wide range of physiological processes and, therefore, are exploited as drug targets in a multitude of therapeutic areas. In this extent, knowledge of structural and functional properties of GPCRs may greatly facilitate rational design of modulator compounds. Solution and solid-state nuclear magnetic resonance (NMR) spectroscopy represents a powerful method to gather atomistic insights into protein structure and dynamics. In spite of the difficulties inherent the solution of the structure of membrane proteins through NMR, these methods have been successfully applied, sometimes in combination with molecular modeling, to the determination of the structure of GPCR fragments, the mapping of receptor-ligand interactions, and the study of the conformational changes associated with the activation of the receptors. In this review, we provide a summary of the NMR contributions to the study of the structure and function of GPCRs, also in light of the published crystal structures.
Keywords: G protein-coupled receptors (GPCRs), NMR, three-dimensional structures, ligand recognition, receptor activation
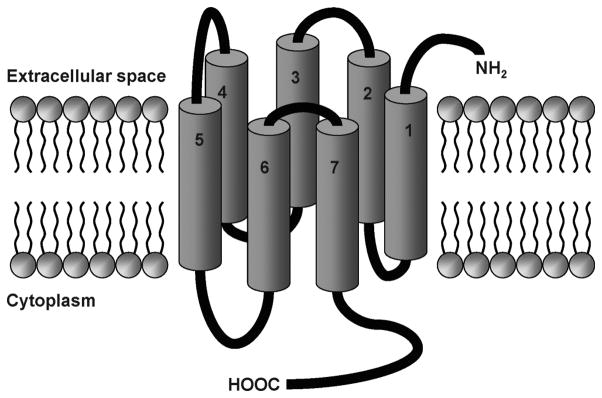
G protein-coupled receptors (GPCRs), also known as seven transmembrane-spanning receptors (7TMRs), are a large superfamily of signaling proteins expressed on the plasma membrane that function as receivers for extracellular stimuli [1]. About 1000 GPCRs have been identified in the human genome [2]. Since their signaling is involved in numerous physiological functions and pathological conditions, GPCRs constitute the molecular target of a significant percentage of the currently marketed drugs. Moreover, many additional members of the superfamily have been identified as potential targets for the treatment of a variety of diseases and are the object of substantial drug discovery efforts. From the molecular point of view, all GPCRs share a common molecular architecture, being composed of a single polypeptide chain folded into a bundle of seven α-helical transmembrane domains (TMs) connected by three extracellular and three intracellular loops (ELs and ILs). The N-terminus is located in the extracellular space, while the C-terminus is in the cytosol (Fig. (1)).
Figure 1.
The topology of G protein coupled receptors.
Given that structure-based drug discovery is an efficient method to rationally design novel drugs and improve the properties of old drugs, the scientific community has been striving for a long time to shed light onto the elusive structure-function relationships of GPCRs employing a variety of direct biophysical and indirect biochemical methods [3]. The most direct method that has been used is X-ray crystallography. However, up until now, this technique has been applied successfully only to a limited number of receptors, namely rhodopsin, the unliganded opsin, the beta-adrenergic receptors (β-ARs), and the adenosine A2A receptor [4–13]. In particular, the low resolution projection map of bovine rhodopsin published by Schertler in 1993 [14] and the subsequent 2.8 Å resolution structure published by Paczewski in 2000 [15] provided the first static three-dimensional (3D) pictures of a GPCR, and have been widely used as templates for the construction of homology models. The subsequent recent publication of the X-ray structure of the β-ARs and the A2A receptor [6;7;10;11] proved conclusively that GPCRs share a common arrangement of the TMs, while suggesting a greater variability for the extracellular and intracellular regions. This includes the second extracellular loop (EL2), which, together with the portions of the TMs facing the extracellular side, is thought to line the ligand binding pocket for the great majority of GPCRs.
On the basis of the available crystallographic information, various studies reported insights into the structure and functions of GPCRs obtained through indirect methods such as site-directed mutagenesis, zinc crosslinking of histidines, photoaffinity labeling and site-specific chemical labeling (see as examples [16–22]). In this context, in silico molecular modeling conducted in an iterative manner with the gathering of experimental data became a widespread tool to infer the 3D structure of the receptors [23–25]. Like other researchers, we have successfully applied this approach to the identification of modulators of several GPCRs (see as examples [26–29]). Despite the relatively low sequence identity between rhodopsin and most other GPCRs, which instilled many doubts on the reliability and the scope of rhodopsin-based modeling, a recent comparison between in silico models and X-ray structure of theβ2-AR firmly supported the applicability of homology modeling and molecular docking to the study of GPCR/ligand complexes [30].
Besides crystallography, other direct methods of investigation aimed at gathering atomic-resolution information have been also applied, among which electron crystallography, electron paramagnetic resonance, UV absorbance and fluorescence spectroscopy, and, as we will discuss at length in this review, nuclear magnetic resonance (NMR) spectroscopy (see as examples [14;31–38]).
NMR spectroscopy, conducted in solution or in solid-state, is a powerful method to study protein structure, protein dynamics, and protein-ligand interactions. NMR techniques are routinely used in drug discovery for soluble protein targets [39;40]. Although their application to GPCRs is hindered by difficulties related to the preparation of receptor samples and the interpretation of the complex spectra, many studies directed towards a structural characterization of GPCRs through NMR have been reported, providing useful information for structure-based drug discovery.
The methods and technical challenges underlying the application of NMR spectroscopy to membrane proteins and GPCRs have been covered in recently published reviews [41–43]. Here, we will focus on the advances in the understanding of the structure and function of GPCRs that have been made through NMR studies. In particular, the review is divided in three sections: in the first one, we will discuss attempts to determine 3D structures of GPCRs; in the second one, we will illustrate studies on receptor-ligand interactions conducted by labeling ligands and/or selected residues in the binding pocket; in the third section we will review studies intended to elucidate the motions and the structural changes consequent to the activation of the receptor, also conducted by labeling selected receptor residues.
1. Determination of the 3D structure of GPCRs
Many obstacles interfere with the determination of the complete 3D structure of a GPCR. Structural studies by NMR require the assignment of all resonance picks in the spectra. Since 1H is the only NMR active isotope abundantly present in proteins, a complete assignment of the complex NMR spectra for a whole protein the size of a GPCR is contingent to the preparation of protein samples uniformly labeled with NMR active C and/or N isotopes.
NMR spectra are complicated by anisotropic nuclear spin interactions, such as dipole-dipole coupling, chemical shielding anisotropy, and quadrupole coupling. Solution-state NMR, which takes advantage of the averaging effect on anisotropic interactions due to the fast tumbling of the samples, is routinely applied to the analysis of proteins with a molecular weight between 30 and 50 kDa. Solid-state NMR spectra, ideal for membrane proteins such as GPCRs, are complicated by the restricted molecular movements that prevent the averaging of the anisotropic effects, inducing a peak-broadening that lowers significantly the resolution of the spectra [42;43].
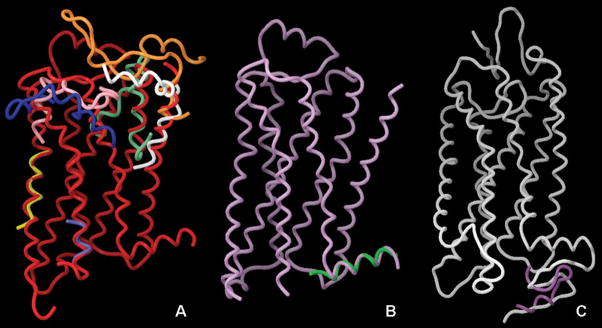
The introduction of the magic-angle spinning and oriented-sample techniques substantially addressed these problems. However, the complexity of the solid-state NMR spectra and difficulties related to sample preparation have prevented, so far, the solution of the structure of whole GPCRs through NMR. For these reasons, most of the studies have been directed toward the solution of individual portions of the receptor structure either in aqueous solution or in different lipid-mimetic solvents [41]. In Table 1, we report a synoptic view of the available 3D structural data. Additionally, for the NMR-derived GPCR fragments with 3D coordinates available in the Protein Data Bank, we provide, in Fig. (2) and (3), a structural alignment with the X-ray structure of rhodopsin and the β-ARs.
Table 1.
A synoptic view of the available NMR structural data for GPCRs. A list of the abbreviations is provided in footnotea.
| Receptor | Source | Domain | Sequence | Constraints | Solvent | Optimization | Summary of Results | Ref. |
|---|---|---|---|---|---|---|---|---|
| α2 adrenergic receptor | human | TM3-IL2-TM4 | 121–155 | TMA | DPC | DG/EM | 120–126: α-helix. 127–131: flexible α-helix. 132–143: α-helix. |
[65] |
| β2 adrenergic receptor | turkey | IL3 | 284–295 | TMA | LPC TFE AS |
290–294: α-helix and flexible Nt in LPC; α-helix in TFE; unstructured in AS; random coil in TFE/AS. | [67] | |
| human | Helix 8 | 324–357 | AS DMSO DPC |
SA/MD | Unstructured in AS. α-helix in DMSO and DPC. |
[72] | ||
| β1 adrenergic receptor | turkey | Helix 8 | 345–359 | LMPC DMPC |
EM | α-helix (PDB code:1dep). | [73] | |
| AT1A angiotensin receptor | rat | Helix 8 | 300–320 | AS/TFE | SA/MD | 306–320: α-helix. | [74] | |
| IL2 | 123–156 | TMA | DPC-d38 | DG/MD | 128–139, 147–156: α-helices. 141–145: “U-shape” form. |
[66] | ||
| B2 bradykinin receptor | rat | Ct | 309–366 | DPC | 311–326, 333–345, and 348–363: α-helices. | [75] | ||
| CB1 cannabinoid receptor | human | IL3 | 338–346 | AS AS/Gαi1 |
DG | Unstructured in AS; α-helix with Gαi1 (PDB code: 1lvq). | [68] | |
| IL3 | 300–343 | TMA | SDS | DG/MD | 300–310, 312–319 and 332–346: α-helices. 321–327 and 329–331: turns. |
[124] | ||
| Helix 8 | 397–418 | AS DPC SDS |
SA/MD | Unstructured in AS; α-helix in DPC/SDS. | [71] | |||
| CB2 cannabinoid receptor | human | TM1-IL1-TM2 | 27–101 | DMSO | Preliminary results | [50] | ||
| Helix 8 | 298–319 | AS DMSO DPC |
SA/MD | Unstructured in AS; α-helix in DMSO and DPC. | [70] | |||
| CCK1 cholecystokini n receptor | human | Nt EL3 |
1–47, 329–357 352–379 |
TMA | DPC/AS | DG/MD MD-wd |
1–47: α-helical structures at N- and C-termini and β-sheet stabilized by a disulfide bridge between C18 and C29 (PDB code: 1d6g). 333–337, 341–345, 353–356, 364–367, 372–378: contain helical regions (PDB code: 1hzn). |
[54–56] |
| CCK2 cholecystokini n receptor | human | EL3 | 352–379 | TMA | DPC/AS | DG/MD-wd | 332–339, 341–346, and 351–356: contain helical regions (PDB code: 1l4t). | [56] |
| α-Factor receptor (Ste2pR) | S. cerevisiae | TM6 | 252–269, C252A | TFE/AS | DG/EM | α-helix with a proline kink. | [44] | |
| TM6 | 252–269, C252A | DMPC | α-helix with a proline kink (PDB code: 1pjd). | [49] | ||||
| TM7, EL3, Cs | 267–339 | TFE/AS CDCl3/CD3 H/AS LPC DPC |
DG/SA | High helical content of TM7 and Ct in TFE/AS and CDCl3/CD3OH/AS. Sample stability in detergents was not insufficient for NMR experiments. |
[48] | |||
| NK1 Neurokinin receptor | rat | Ns, EL3 | 1–39, 264–290 | TMA | DPC/AS | DG/MD-wd | 3–10: α-helix. 264–270: α-helix. 276–289: α-helix. |
[58] |
| EL2 | 162–198 | TMA | DPC/AS | MD-wd | 176–182: α-helix. 187–189: turn. |
[125] | ||
| PTH1 parathyroid hormone receptor | human | Nt | 168–198 | TMA | AS DPC |
DG/MD | Low solubility in AS. 170–174, 178–185, and 189–195: contain α-helical structures. 175–177 and 186–188: contain turns (PDB code: 1bl1.pdb). |
[69] |
| EL1 | 241–285 | TMA | DPC/AS | DG/MD-wd | 256–264: α-helix. 275–284: three α-helical loops. |
[126] | ||
| IL3 | 382–408 | LF, CF2 | SDS AS |
DG | 382–393: α-helix with LF and CF2. 394–398: flexible with CF2. 394–406: flexible with LF. 402–408: ordered with CF2. Unstructured in AS. |
[53] | ||
| Rhodopsin | bovine | TM3-IL2-TM4 | 125–158 | TMA | DPC | DG/EM | 125–137: α-helix. 143–146: turn-like structure. |
[65] |
| Ct | 330–348 | AS | Unstructured peptide. | [33] | ||||
| Phosphorilated Ct | 330–348 | AS AS/arrestin |
DG/MD | Unstructured peptide in AS. The N-terminal portion is flexible and the C-terminal portion assumes a stable helix- loop structure in the presence of arrestin (PDB code: 1nzs). |
[77;78;127] | |||
| V1a Vaspressin receptor | human | IL2 | 146–166 | LF, CF1 | AS TFE/AS |
Random coil conformation in AS; α-helix followed by a β-turn in TFE/AS. | [52] | |
| S1P4 Sphingosine 1-phosphate receptor | human | EL1 | 95–125 | DB TMA |
AS/TFE | DG/MD | Partially helical structure (PDB code: 2dco). | [61] |
| NK1 tachykinin receptor | TM7 | AMSSTM YNPIISS L |
DMSO DMSO/CD Cl3 PFTB/CD3 OD |
EM/MD | Kinked α-helix as a minor conformer in DMSO; increased folded content in DMSO/CDCl3; twoα-helical regions linked by a hinge around the NP motif in PFTB/CD3OD. | [45] | ||
| A2 tromboxane receptor | human | EL1 | 88–104 | DB | AS | DG/MD | β-turns. | [59] |
| EL2 | 173–193 | DG/MD | A two-turns loop in correspondence of 183 and 188. | [60] | ||||
| IL2–3 | 129–149, 220–246 | DB | AS | EM/MD | IL2: large turn. IL3: helical-like conformation. |
[51] |
AS–aqueous solution; BS–biosynthesis in Escherichia coli; CD–circular dichroism; CF1: cyclized form of peptide with termini substituted by S-carboxymethylcysteine; CF2: cyclized form of the peptide with a linker of 8 methylenes; Ct–C-terminus; DB disulfide bridge; DG–distance geometry; DMPC–deuterated dimyristoyl- phosphatidylcholine; DMSO–dimethyl sulfoxide; DPC dodecylphosphocholine micelles; DQF-COSY–double-quantum filtered correlation spectroscopy; EL–extracellular loop EM–energy minimization; HSQC–Heteronuclear single quantum coherence; IL–intracellular loop; LF–linear form of the peptide; LMPC–deuterated lysomyristoyl-phosphatidylcholine; LPC–1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine; MD–molecular dynamics; MD -wd- molecular dynamics in water-decane environment; MS–manual synthesis; NOESY–nuclear overhauser enhancement spectroscopy; Nt–N-terminus; PFTB–perfluoro-tert-butanol; PC–Peptide Constrains; SA–simulated annealing; SDS sodium dodecyl sulfate; SF–solid fase automatic synthesis; ssNMR–solid-state NMR; TFE–trifluoroethanol; TM–transmembrane helix; TMA–transmembrane helix anchor; TOCSY–total correlation spectroscopy; TROSY–transverse relaxation optimized spectroscopy.
Figure 2.
Structural alignment of the NMR coordinates of GPCR portions deposited in the PDB and the crystal structures of the and bovine rhodopsin. Panel A: superimposition of the β2-AR crystal structure (2RH1, red) with the NMR-derived structures of the N-termini of PTH1 (1BL1–white) and CCK1 (1D6G, orange), EL1 of S1P4, (2DCO, aquamarine), IL3 of CB1 (1LVQ, light blue), TM6 of Ste2pR (1PJD, yellow), and EL3 of CCK1 (1HZN, pink) and CCK2 (1L4T, dark blue). Panel B: superimposition of the crystal structure of the β1-AR (2VT4, plum) with the NMR-derived structure of H8 of the same receptor (1DEP, green). Panel C: superimposition of the crystal structure of rhodopsin (2HPY, white) and the NMR-derived structure of the C-terminus of the same receptor (1NZS, blue purple). Pictures prepared with Maestro 8.0.308, Schrodinger.
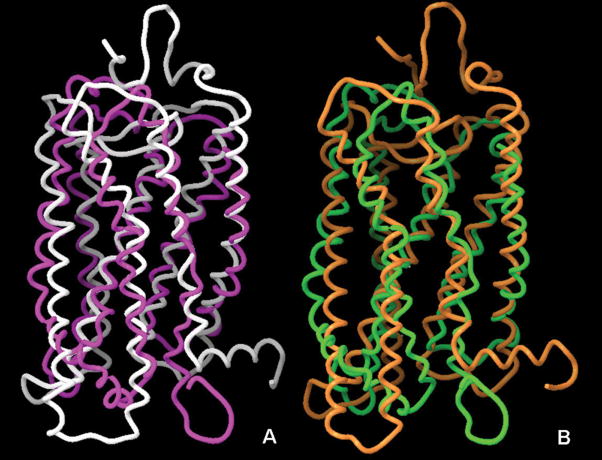
Figure 3.
Superimposition of the crystal structures and the NMR-based three-dimensional models of bovine rhodopsin. Panel A: Superimposition of the X-ray structure (1GZM, white) and the NMR-based model of the ground state (1LN6, purple) receptor. Panel B: Superimposition of the X-ray structure of opsin in its G-protein-interacting conformation (3DQB, orange) and NMR-based model of Meta II rhodopsin (1JFP, green). Picture prepared with Maestro 8.0.308, Schrodinger.
Transmembrane domains
We now know from crystallographic data that some of the TMs present kinks in their α-helical structures, particularly pronounces in TM6 and TM7. These data emerged clear from NMR studies, even before the publication of GPCR X-ray structures. In particular, kinks were detected studying portions of TM6 of the Saccharomyces cerevisiae α-factor receptor (Ste2pR) [44] and TM7 of the tachykinin receptor NK1 [45] (Table 1). Solution and solid-state NMR studies of the TMs of the Ste2pR continued after the disclosure of the X-ray structure of rhodopsin and, among other findings, confirmed the existence of the helical kink of TM6 previously detected through solution NMR with the a more sophisticated solid-state NMR of the 15N labeled TM6 peptide in 1,2-dimyristoyl-sn-glycero-phosphocholine bilayers [46–49]. As mentioned, the kinks of TM6 and TM7 postulated on the basis of the NMR results are consistent with the conformation of the homologous regions of the receptors that have now been crystallized (Fig. (2), panel A) [12;15], supporting the idea that even structural data gathered for isolated domains can be very informative. Probably, even greater insights could be obtained from the analyses of larger substructures or even whole receptors. At this purpose, Zheng and coworkers biosynthesized a quite large segment of the cannabinoid CB2 receptor, including TM1, IL1 and TM2, using a fusion protein overexpression strategy [50]. The preliminary results gathered by the authors for the 13C/15N labeled peptide argue in favor of the applicability of this strategy to the synthesis of GPCR helical bundles for NMR studies.
Extracellular loops
While homology modeling based on the available GPCR X-ray structures allows the construction of reliable models of the TMs, modeling the 3D structure of extracellular and intracellular regions remains a difficult task [30]. Low sequence similarity as well as numerous insertions and deletions, hamper an effective application of homology modeling to these regions. NMR spectroscopy could prove particularly useful in solving this problem: the structures of loops and termini of the receptor of interest could be derived through NMR and subsequently combined with homology models to build complete 3D structures.
To obtain structures of ILs and ELs close to the naturally occurring ones, many experimentalists turned to the application of structural constraints meant to keep the termini at a distance compatible with their role of connecting two TMs. In some cases, portions of the adjacent TMs have been added to act as anchors for shackling loops or termini to the lipid environment. Alternatively, the loops have been constrained with disulfide bridges between cysteines [51] or S-carboxymethylcysteines [52] added to the peptide termini, with or without methylene linkers [53].
Since for most of the GPCRs activated by peptides ligand binding is thought to occur at the level of the extracellular domains, NMR analyses have been extensively applied to study receptor-peptide interactions. For example, the contact interactions of CCK8–the natural cholecystokinin peptide, composed of eight amino acids–with the N-terminus and EL3 of the CCK1 and with EL3 of the CCK2 subtypes of the cholecystokinin receptors have been studied by NMR by monitoring chemical shift perturbations and intermolecular NOEs [54–56] (Table 1). For the CCK2 the analysis has been complemented by integrating NMR-derived structural information with rhodopsin-based homology modeling. With a similar strategy, the interactions between the CCK1 and synthetic agonists have also been studied [57]. The integration of homology modeling and NMR-derived information provided also a structural hypothesis for the binding of substance P to the NK1 receptor, confirming and rationalizing the results previously obtained through a variety of indirect biochemical methods [58]. A similar approach also corroborated the postulated existence of a disulfide bridge between EL1 and EL2 in the thromboxane A2 receptor (TP), highlighting also the involvement in ligand binding of the WCF motif of EL2 [59;60].
Lastly, Pham and coworkers developed an alternative methodology for studying GPCR loops based on the computational design of a peptide containing segments that mimic the self assembling of the ends of two TMs connected by the loop under examination [61]. When applying their strategy to the analyses of EL1 of the sphingosine 1-phosphate receptor S1P4, the authors obtained the best results with the coiled-coil strategy, i.e. by substituting the TM2 and TM3 segments with sequences from structurally characterized water-soluble proteins with a pair of antiparallel helices oriented in a way consistent with the S1P4 homology model. The addition of a disulfide bridge appeared also necessary to keep the helices together. The ability of the peptide to bind an analog of the agonist’s headgroup suggested that EL1 adopted a biologically relevant conformation. Here, we superimposed this NMR-derived structure to the homologous regions in the crystal structure of the β2 receptor (Fig. (2), panel A) and, in agreement with the conclusions of the authors, revealed a sensible conformation and orientation for the TM2 and TM3 mimetics.
Intracellular loops
Since the ILs, and IL2 in particular, are directly involved in the coupling of the receptors with the G proteins, [62–64] the experimental elucidation of the structure of these binding interfaces would provide insights into the molecular mechanisms of the selectivity toward the various G protein subtypes.
NMR studies of IL2 in the adrenergic α2A receptor, rhodopsin, the bradykinin B2 receptor, and the vaspressin V1a receptor have been performed (Table 1) [52;65;66]. In particular, NMR studies of the peptide corresponding to the IL2 of theα2A receptor revealed the presence of a cytoplasmic helix not detected in the analogous rhodopsin peptides [65] or in the crystal structure of rhodopsin. X-ray crystallography revealed that this helix, although not detected in the β2, is indeed present in the β1 subtype [7;11].
Conversely, the NMR-derived structure of IL2 of the B2 receptor revealed a “U” shape [66] similar to that detected in rhodopsin. Also in the case of the vasopressin V1A receptor, the NMR analysis of a linear and a cyclic IL2 peptide revealed a conformation in TFE/H2O solvent similar to that shown in rhodopsin, particularly with the cyclic peptide [52].
The structure of IL3 has also been determined for several receptors, including the cannabinoid CB1 receptor, the β2 receptor, and the parathyroid hormone PTH1 receptor (Table 1) [53;67;68]. Of note, the IL3 peptide of the CB1 receptor (Fig. (2), panel A) assumed a helical conformation in aqueous solution when Gαi1 was added, while a mutated IL3 formed just a single helixturn [68]. The authors proposed the importance of this portion of IL3 in the formation of the G protein binding interface.
Also for the ILs, hybrid solutions composed of molecular modeling and NMR studies have been applied. In particular, Wu and coworkers derived by NMR the structure of all three ILs of the thromboxane A2 receptor and subsequently incorporated them into a rhodopsin-based homology model [51]. They consequently proposed a list of amino acid residues potentially involved in the formation of the G protein-binding interface that proved consistent with available mutagenesis data.
N-/C- termini
In addition to the mentioned NMR structure of the CCK1 N-terminus, the structure of the N-terminus of PTH1 receptor has also been solved [69], revealing to be different from that of rhodopsin (Fig. (2), panel A). These data confirm the idea that each GPCR may be characterized by unique extracellular regions that contribute to the selectivity of the receptors for specific ligands.
All the available GPCR X-ray structures are characterized by the presence of aα-helix, known as helix 8 (H8), in the portion of the C-terminus proximal to TM7. NMR in conjunction with CD spectroscopy confirmed the presence of an H8 for the CB1 andCB2 receptors, the Ste2p receptor, the β1and β2 receptors, the angiotensin AT1A receptor, and the bradykinin B2 receptor in detergent [70–75]. In Fig. (2), panel B, we superimposed the NMR-derived the β1 receptor H8 to the corresponding segment of the crystal structure of the same receptor, revealing a very good agreement between crystallographic and NMR data. Conversely, a random coil conformation was detected in water for this region [71–73]. These conformational changes of H8 have been speculated to be indicative of possible structural rearrangements consequent the activation of the receptors. For instance, partial unfolding of H8 upon light activation has been detected in rhodopsin by Fourier transform infrared (FTIR) and fluorescence spectroscopy [76].
Several NMR studies of the rhodopsin C-terminus have been published [33;77;78]. Beyond H8, this region resulted unstructured in aqueous solution, both in unphosphorylated or phosphorylated form, while has adopted a defined 3D structure when phosphorylated and in the presence of arrestin (Fig. (2), panel C) [78]. Here we could only compare this NMR-derived structure (pdb code: 1nzs) with the crystal structures of lumirhodopsin and batorhodopsin (pdb code: 2hpy and 2g87), for which this distal portion of the C-terminus has been determined. The detected fairly high root mean square deviations of 5.5–6 Å suggest that the C-terminus of rhodopsin undergoes a conformational change when bound to arrestin.
Yeagle’s approach to derive the structure of rhodopsin through NMR
Before the obtainment of high resolution X-ray structures of GPCRs, Yeagle and coworkers devised a methodology intended to solve the 3D structure of portions of the receptors and subsequently assemble them together. Through two dimensional homonuclear 1H NMR analyses in solution, the authors determined the structure of overlapping peptides spanning the entire sequence of the rhodopsin and subsequently computationally assembled the fragments into a single structure [79–81]. The information about the packing of the helical bundle, necessary to put the pieces together, was gathered from electron paramagnetic spectroscopy (EPS) [36;82;83], zinc crosslinking of histidines [16], and electron cryo-microscopy data [84]. In the form of distance constraints, this was then applied to the NMR-derived fragments through simulated annealing. Using different sets of experimental data collected for the ground-state and activated (META II) rhodopsin, the authors intended to generate models for both states (PDB codes: 1jfp and 1ln6). After the publication of the crystal structure of rhodopsin, the authors discovered that the largest TM peptides yielded the best superimposition with the corresponding segments of the crystal structure [85].
Here we superimposed Yeagle’s ground state rhodopsin to the crystal structures of the ground state rhodopsin (PDB code: gzm), detecting a RMSD of the backbone of about 6 Å (Fig. (3), panel A). The packing of the TM helical bundle is similar in the two structures, however, Yeagle’s model predicts the presence of several non helical portions, in particular in TM7, not confirmed by the crystallography. The intracellular and extracellular regions appear significantly divergent in the Yeagle’s model and the crystal structure. We also superimposed Yeagle’s Meta II rhodopsin to the recently solved crystal structure of opsin in its G protein interacting conformation [8], detecting, also in this case, an RMSD of the backbone of about 6 Å (Fig. (3), panel B).
Whole GPCRs
Dispite the difficulties, several attempts to solve the structure of whole GPCRs by NMR are currently being undertaken. Solution NMR spectroscopy experiments, conducted for the uniformly 15N-labelled vasopressin V2 receptor, solubilized with lyso-myristoylphosphatidylcholine, indicated the potential suitability of the technique for structural studies of GPCRs. Notably, the authors observed over 250 amide peaks out of the expected 349 in their 1H,15N-TROSY spectrum [86]. Additionally, solid-state NMR studies of a uniformly 15N-labeled and selectively 15N-Ile-labeled chemokine CXCR1 receptor in magnetically aligned bicelles provided spectra that suggested the potential applicability of NMR to structural determinations and the elucidation of structure-activity relationships (vide infra) [32].
2. Mapping receptor-ligand interactions
Direct atomic resolution information on the interactions of GPCR with their ligands is available through X-ray crystallography only for a limited number of receptors, while, for the great majority of them, indirect methods of analysis have been used. A synergistic application of mutagenesis studies, photoaffinity labeling, chemical modifications of the ligands, and computational modeling has been the most common way of studying the determinants of ligand recognition. Taken together, these results have contributed to the general definition of a common binding pocket for GPCRs located towards the extracellular opening of the helical bundle [87;88]. Also for receptors naturally stimulated by ligands that bind to their N-terminal ectodomains, such as the calcium sensing or the glycoprotein hormone receptors, the possibility of modulation through ligands that bind within their helical bundle has been demonstrated [89–93].
In this context, structure-activity relationships (SAR) analyses and drug-discovery studies would greatly benefit from direct experimental proofs of receptor-ligand interactions, such as those that NMR could provide. Solid-state NMR spectroscopy has been applied successfully to membrane proteins to detect ligand binding and to analyze protein-ligand interactions, especially when coupled to selective isotopic labeling of specific residues located in the binding pocket and/or of the ligand [94]. Through this expedient, significantly simplified spectra with a manageable amount of signals have been obtained, thus rendering NMR applicable to the analysis of bioactive ligand conformations, protonation states, receptor/ligand interactions and residue/residue interactions. Moreover, although not yet for GPCRs, NMR coupled to selective isotopic labeling has successfully been applied to rational fragment-based drug discovery. In particular, ligands or fragments that bind to specific pockets of a protein have been identified by monitoring the changes in the chemical shifts of 15N or 1H-amide atoms in 15N-labelled protein, an approach known as SAR by NMR [95;96].
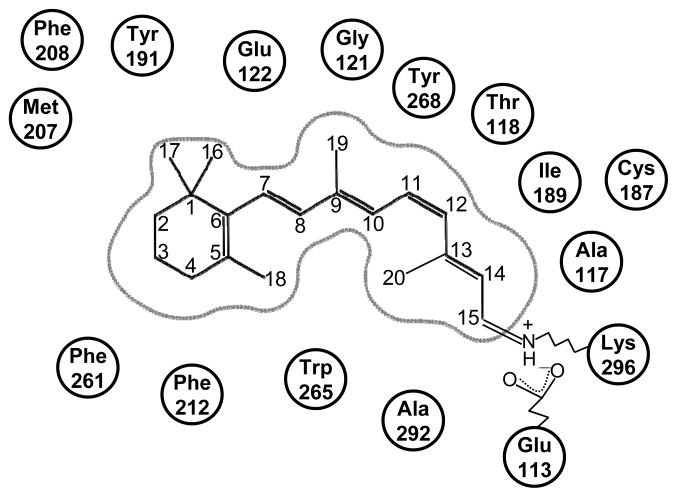
Rhodopsin has been extensively studied by solid-state NMR coupled to selective labeling. Before any high-resolution crystal structures were released, selective 15N-labeling of Lys residues led to the conclusion that the distance between the protonated Schiff base by which the retinylidene chromophore is covalently bound to K296(7.43 according to the Ballesteros and Weinstein residue indexing [97]) and the E113(3.28) counterion (Fig. (4)) was greater than 4 Å [98]. The subsequent publication of crystal structures, revealed that the distance is, in fact, about 3.5 Å.
Figure 4.
Two dimensional schematic representation of the retinal binding site in ground state rhodopsin. Picture prepared with MOE 2008.10, Chemical Computing Group.
The conformation of retinal and its interactions with rhodopsin have also been studied extensively through NMR coupled to selective labeling. For example, high-resolution solid state deuterium (2H) NMR experiments provided detailed information on the orientation of retinal in the binding pocket and on the conformational changes that occur with the transition from the ground state to the Meta I intermediate of the activation cycle. The study applied three retinal analogues labeled with deuterium at three different pairs of adjacent carbons [99]. Ultra high field solid-state magic angle spinning 2D homonuclear and 2D heteronuclear NMR spectroscopy has also been used to study rhodopsin reconstituted with a uniformly 13C-labeled 11-cis-retinal. Complete assignment of the 13C and 1H chemical shifts for retinal highlighted nonbonding interactions between the protons of the methyl groups of its ionone ring and the nearby aromatic acid residues F208(5.43), F212(5.47), and W265(6.48) (Fig. (4)). Furthermore, it was shown that binding of retinal involves a chiral selection of the ring conformation, resulting in equatorial and axial positions for CH3-16 and CH3-17 [100]. Additional evidences of the interactions between retinal and rhodopsin in the ground and Meta I states have also been obtained through NMR experiments conducted variously labeling the ligand with 2H [101–104]. These data support the hypothesis that a strain in the polyen around the cis bond assists the photoisomerization of retinal. A 13C-labelled 9-cis retinal isomer, typical of isorhodopsin, has also been investigated, leading to similar conclusions [105].
Application to rhodopsin of an NMR technique named by the authors “selective interface detection spectroscopy” (SIDY), based on the detection of the correlations between 13C atoms of labeled ligands and 1H atoms of unlabelled receptors, led to the detection of several contacts between the aliphatic carbons of the 11-cis-retinal ionone ringand residues in the binding pocket. Although, SIDY data do not provide sequence-specific assignments of the contacts, these could be identified by means of additional data, and, in the case of rhodopsin, resulted in good agreement with the available crystallographic structures [106].
NMR spectroscopy provides also a very effective way of studying the conformational changes that a ligand undergoes upon binding to a receptor. In this context, the analysis of the unbound and receptor-bound 13C,15N-labelled neurotensin, a 13-residue peptide, revealed that the ligand is in a disordered state in the absence of the receptor, while adopts a beta-strand conformation when bound to the NTS1 receptor [107]. Similarly, the structure of the bradykinin (BK) peptide bound to the bradykinin B2 receptor has been studied through solid-state NMR, revealing a double S-shape structure [108]. These type of studies can be also conducted to rationalize the biological activities of compounds on the basis of their bioactive conformations, as it has been done in the case of two natural peptides active at the neuropeptide NPR-1 receptor one [109].
In addition to conformational changes, NMR spectroscopy can also detect changes in the protonation state of a ligand as a consequence of binding to a receptor. In this context, solid state NMR experiments conducted with uniformly labeled 13C,15N-histamine bound to the human histamine H1 receptor revealed that the ligand can bind either in a monocationic or a dicationic form. On the basis of their data, the authors hypothesized that, in analogy with rhodopsin, also in the case of the H1 receptor a protonation switch might be part of the activation mechanism [110].
3. Detection of motions and conformational changes associated with receptor activation
Upon binding of agonists, which typically occurs in proximity of the extracellular opening of the helical bundle, GPCRs undergo a series of structural changes that cascade from the extracellular to the intracellular part of the receptor and ultimately lead to G protein activation. Detailed structural knowledge of the ground and activated state of the receptor and mechanistic insights into the activation process would significantly assist drug discovery, allowing a more rational design of compounds capable of stimulating or blocking the receptor.
A wealth of information has been derived by the analysis of mutations that affect the basal activity of GPCR, either naturally occurring or generated through site-directed mutagenesis. Mutations that cause an increase of the basal activity are likely to stabilize the activated state of the receptor and are referred to as constitutively active mutants. Those that prevent the activation of the receptor are, instead, called uncoupling mutants and are likely to stabilize its inactive state or to disrupt the cascade of conformational changes that lead to signaling. Through such mutational analyses, various molecular switches for GPCR activation have been proposed [111;112]. Alternatively, the structural changes that occur upon receptor activation have been monitored through electron paramagnetic resonance spectroscopy (EPR) [34;36;83;113], UV absorbance spectroscopy [35], engineering of metal-ion-binding sites [16], as well as site-specific chemical labeling coupled to fluorescence spectroscopy [37], and, as we will see in the coming paragraphs, NMR spectroscopy. These data shed light onto a number of intramolecular distances that are characteristic of the activated receptors. We recently used these biophysically measured distances to construct a computational structure of the activated rhodopsin based on coarse-grained and all atom simulations, and subsequently study the dynamics of the activation process [114]. The substantial conformational changes predicted by these indirect methods were not detected in the crystal structure of a photoactivated deprotonated intermediate (PDI) of rhodopsin that shows absorption maxima consistent with the META II state [13]. The crystal lattice may have prevented large-scale structural rearrangements. However, a significant opening of the intracellular surface of the receptor and a furthering of the intracellular ends of TM3 and TM6 have been captured in the crystal structure of opsin with a fragment of the C-terminus of transducin. At least relatively to the cytosolic half, this may represent the first detailed structure of an activated GPCR [8].
Compared to crystallography, NMR spectroscopy offers the possibility of conducting structural analyses in a less constrained membrane-like environment. In this context, NMR is well suited to the study of receptor activation through isotopic labeling of specific residues located in areas affected by the structural changes. In this context, the specific labeling of rhodopsin led to observance of the transition between its inactive and activated states.
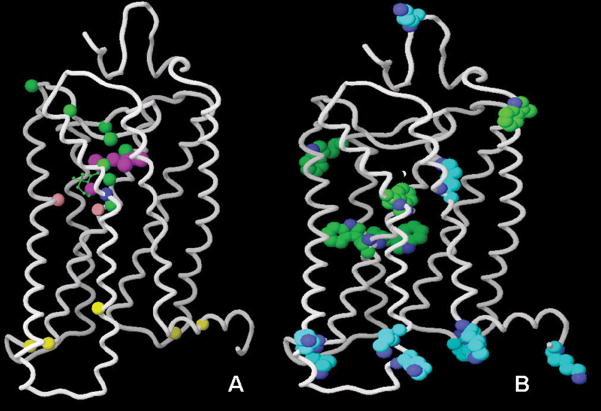
2D-dipolar-assisted rotational resonance NMR measurements between 13C-labels on the C14, C15, C19, C20 atoms of retinal and 13C-labeled G114(3.29), T118(3.33), G121(3.36), Y178(4.68), G188(EL2), Y191(EL2), S196(EL2), and Y268(6.51), all located in the retinal binding pocket, was performed for the ground state and META II rhodopsin (Fig. (5), panel A, purple and green spheres indicate retinal and rhodopsin atoms, respectively) [115]. The results highlighted that the extracellular portion of the receptor undergoes conformational changes upon activation. In particular, a large rotation of the C20 methyl group of retinal toward EL2 and also a translation of about 4–5 Å of the retinal chromophore toward TM5 were observed. This displacement of retinal has been associated with the motions of the TM5, TM6, and TM7 predicted by different biophysical and biochemical methods [115]. Subsequent measurements between the C19 and C20-methyl groups of retinal and 13C-labeled W265(6.48) confirmed that, in agreement with what suggested by atomic microscopy [38], the movement of the beta-ionon ring causes a reorientation of the side chain of W265(6.48) upon activation (Fig. (5), panel A, a blue sphere indicates W265). In agreement with what seen in the crystal structure of opsin [8], the study also suggested that TM6, but not TM3, undergoes a significant rearrangement [116]. Most likely, the local movements triggered by the isomerization of retinal cause a perturbation of the complex network of hydrogen bonds that stabilizes the helical bundle of the ground state rhodopsin. This is suggested also by the comparison of the chemical shifts of 15N and 13C-labeled wild type and mutant rhodopsin in the ground and META II states [117], which, among other observations, led to the detection of the disruption of the strong hydrogen bond between Glu 122 in TM3 and H211 in TM5 (Fig. (5), pink spheres indicate Glu122 and H211), thus indicating a motion of TM5. A more recent study demonstrated that the transition to META II is accompanied also by a displacement of EL2 from the retinal binding site, and suggested that this displacement is coupled to the rotation of TM5 and the breakage of the ionic lock connecting TM3 and TM6 [118]. As mentioned, all these interconnected conformational changes caused by the photoisomerization of retinal eventually cascade down towards the cytosolic portion of the TMs, thus leading to G protein activation. To specifically monitor the changes that occur in this region, NMR spectroscopy of 19F labeled rhodopsin has been applied after mutation of specific residues to Cys and subsequent attachment of trifluoroethylthio groups via disulfide bridges (C65, C139, C140, C251, C316, indicated by yellow spheres in Fig. (5), panel A). The results support the idea that the tertiary structure of the cytoplasmic face of the receptor changes significantly upon light activation [119;120].
Figure 5.
Panel A: Positions of the ligand atoms and residues labeled in order to study the activation-related conformational changes of rhodopsin. The retinal atoms (C5, C9, C11, C13, C14, C15, C19, C20) are in purple; residues 114, 118, 121, 178, 188, 191, 196, and 268 are in green; residue 265 is in blue; residues 122 and 211 are in pink; the residues mutated to Cys to be labeled with trifluoroethylthio groups are in yellow. For simplicity, only one carbon of the residue backbone is shown. Panel B: All Trp and Lys residues of rhodopsin. Picture prepared with Maestro 8.0.308, Schrodinger.
Beyond the activation process, NMR analyses of 15N labeled rhodopsin have been applied also to the study of the dynamics of the dark adapted receptor. In particular, a study on α-15N-Lys-labeled rhodopsin revealed that, while the single Lys residue located in the C-terminus is endowed with nanosecond scale movements, those located in different regions of the receptors present micro- to millisecond timescale motions (Fig. (5), panel B). In contrast with Lys residues, which in rhodopsin are located mostly in the cytosolic domains, Trp residues are, for the most part, present in the TMs (Fig. (5), panel B). A subsequent study of α and ε-15N-Trp-labeled rhodopsin, confirmed that there are micro- to millisecond timescale backbone motions in the inactive dark state, while suggesting a substantial restriction of the Trp side chains to a single specific conformation [121;122].
Conclusion
Despite the numerous technical challenges, NMR spectroscopy has the potential of becoming a leading technique in the study of the structure-function relationships of GPCRs and their interactions with ligands. In contrast to crystallographic methods, which provide high-resolution but static molecular snapshots, NMR techniques can provide dynamic pictures of receptor structures and receptor-ligand interactions, thus offering insights into the molecular mechanisms of ligand recognition and receptor activation.
Through this article, we reviewed many of the contributions that NMR studies have given to the understanding of the molecular structure and functioning of GPCRs. In particular, NMR spectroscopy has been extensively applied to the study of individual portions of GPCRs, also in combination with homology modeling. Through selective labeling of ligands and/or specific receptor residues, NMR studies have also been applied to the study of GPCR-ligand interactions and to the investigations of the molecular changes coupled to activation of the receptor.
Recently, dynamic nuclear polarization (DNP), a technique intended to increase the NMR sensitivity by transferring the polarization of electron spins to nuclei, has been successfully applied to the study the photocycle of bacteriorhodopsin, a 7TM light-driven ion pump with some structural analogies to rhodopsin [123]. Due to the enhanced sensitivity of the experiments, DNP allows the analysis of low concentration samples in a lower timeframe, and the detection of low populated conformations.
The introduction of DNP, as well as other advances in solution and solid-state NMR technology, together with progresses in the preparation of GPCR samples, are expected to lead in the future to the determination of the 3D structure of whole GPCRs, and to provide further mechanistic insights into the complex cascade of motions and rearrangements characteristic of the activation process. Furthermore, these methodological advances are also likely to foster the application of SAR by NMR techniques to the discovery of lead compounds for GPCRs through NMR-based high throughput screenings and their subsequent structure-based optimization.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nature Reviews Molecular Cell Biology. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. Febs Letters. 2002;520:97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 3.Yeagle PL, Albert AD. G-protein coupled receptor structure. Biochimica et Biophysica Acta-Biomembranes. 2007;1768:808–824. doi: 10.1016/j.bbamem.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 5.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 6.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, et al. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, et al. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–4U2. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–U30. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 9.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–U33. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 10.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 13.Salom D, Lodowski DT, Stenkamp RE, Le TI, Golczak M, Jastrzebska B, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schertler GFX, Villa C, Henderson R. Projection Structure of Rhodopsin. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 15.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 16.Sheikh SP, Zvyaga TA, Lichtarge O, Sakmar TP, Bourne HR. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 17.Marco E, Foucaud M, Langer I, Escrieut C, Tikhonova IG, Fourmy D. Mechanism of activation of a G protein-coupled receptor, the human cholecystokinin-2 receptor. J Biol Chem. 2007;282:28779–28790. doi: 10.1074/jbc.M700349200. [DOI] [PubMed] [Google Scholar]
- 18.Tan YV, Couvineau A, Murail S, Ceraudo E, Neumann JM, Lacapere JJ, et al. Peptide agonist docking in the N-terminal ectodomain of a class II G protein-coupled receptor, the VPAC1 receptor - Photoaffinity, NMR, and molecular modeling. Journal of Biological Chemistry. 2006;281:12792–12798. doi: 10.1074/jbc.M513305200. [DOI] [PubMed] [Google Scholar]
- 19.Dawson ES, Henne RM, Miller LJ, Lybrand TP. Molecular models for cholecystokinin-A receptor. Pharmacology & Toxicology. 2002;91:290–296. doi: 10.1034/j.1600-0773.2002.910605.x. [DOI] [PubMed] [Google Scholar]
- 20.Turek JW, Halmos T, Sullivan NL, Antonakis K, Le Breton GC. Mapping of a ligand-binding site for the human thromboxane A(2) receptor protein. Journal of Biological Chemistry. 2002;277:16791–16797. doi: 10.1074/jbc.M105872200. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann C, Gaietta G, Bunemann M, Adams SR, Oberdorff-Maass S, Behr B, et al. A FlAsH-based FRET approach to determine G protein - coupled receptor activation in living cells. Nature Methods. 2005;2:171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- 22.Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacology & Therapeutics. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Moro S, Spalluto G, Jacobson KA. Techniques: Recent developments in computer-aided engineering of GPCR ligands using the human adenosine A(3) receptor as an example. Trends in Pharmacological Sciences. 2005;26:44–51. doi: 10.1016/j.tips.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Moro S, Deflorian F, Bacilieri M, Spalluto G. Ligand-based homology modeling as attractive tool to inspect GPCR structural plasticity. Current Pharmaceutical Design. 2006;12:2175–2185. doi: 10.2174/138161206777585265. [DOI] [PubMed] [Google Scholar]
- 25.Patny A, Desai PV, Avery MA. Homology modeling of G-protein-coupled receptors and implications in drug design. Current Medicinal Chemistry. 2006;13:1667–1691. doi: 10.2174/092986706777442002. [DOI] [PubMed] [Google Scholar]
- 26.Tikhonova IG, Sum CS, Neumann S, Thomas CJ, Raaka BM, Costanzi S, et al. Bidirectional, Iterative Approach to the Structural Delineation of the Functional “Chemoprint” in GPR40 for Agonist Recognition. J Med Chem. 2007;50:2981–2989. doi: 10.1021/jm0614782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel S, Skoumbourdis AP, Childress J, Neumann S, Deschamps JR, Thomas CJ, et al. A virtual screen for diverse ligands: discovery of selective g protein-coupled receptor antagonists. Journal of the American Chemical Society. 2008;130:5115–5123. doi: 10.1021/ja077620l. [DOI] [PubMed] [Google Scholar]
- 28.Kellenberger E, Springael JY, Parmentier M, Hachet-Haas M, Galzi JL, Rognan D. Identification of nonpeptide CCR5 receptor agonists by structure-based virtual screening. Journal of Medicinal Chemistry. 2007;50:1294–1303. doi: 10.1021/jm061389p. [DOI] [PubMed] [Google Scholar]
- 29.Tikhonova IG, Sum CS, Neumann S, Engel S, Raaka BM, Costanzi S, et al. Discovery of novel Agonists and antagonists of the free fatty acid receptor 1 (FFAR1) using virtual screening. Journal of Medicinal Chemistry. 2008;51:625–633. doi: 10.1021/jm7012425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costanzi S. On the Applicability of GPCR Homology Models to Computer-Aided Drug Discovery: A Comparison between In Silico and Crystal Structures of the beta2-Adrenergic Receptor. J Med Chem. 2008;51:2907–2914. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peleg G, Ghanouni P, Kobilka BK, Zare RN. Single-molecule spectroscopy of the beta(2) adrenergic receptor: Observation of conformational substates in a membrane protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8469–8474. doi: 10.1073/pnas.151239698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. Journal of the American Chemical Society. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner K, Richter C, Klein-Seetharaman J, Schwalbe H. Isotope labeling of mammalian GPCRs in HEK293 cells and characterization of the C-terminus of bovine rhodopsin by high resolution liquid NMR spectroscopy. Journal of Biomolecular Nmr. 2008;40:49–53. doi: 10.1007/s10858-007-9205-3. [DOI] [PubMed] [Google Scholar]
- 34.Altenbach C, Cai KW, Klein-Seetharaman J, Khorana FG, Hubbell WL. Structure and function in rhodopsin: Mapping light-dependent changes in distance between residue 65 in helix TM1 and residues in the sequence 306–319 at the cytoplasmic end of helix TM7 and in helix H8. Biochemistry. 2001;40:15483–15492. doi: 10.1021/bi011546g. [DOI] [PubMed] [Google Scholar]
- 35.Lin SW, Sakmar TP. Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996;35:11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- 36.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 37.Dunham TD, Farrens DL. Conformational changes in rhodopsin. Movement of helix f detected by site-specific chemical labeling and fluorescence spectroscopy. J Biol Chem. 1999;274:1683–1690. doi: 10.1074/jbc.274.3.1683. [DOI] [PubMed] [Google Scholar]
- 38.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GF. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogtherr M, Fiebig K. Modern Methods of Drug Discovery. In: Hillish A, Hilgenfeld R, editors. Birkhauser. 2003. pp. 183–202. [Google Scholar]
- 40.Homans SW. NMR spectroscopy tools for structure-aided drug design. Angewandte Chemie-International Edition. 2004;43:290–300. doi: 10.1002/anie.200300581. [DOI] [PubMed] [Google Scholar]
- 41.Ratnala VRP. New tools for G-protein coupled receptor (GPCR) drug discovery: combination of baculoviral expression system and solid state NMR. Biotechnology Letters. 2006;28:767–778. doi: 10.1007/s10529-006-9005-y. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen NC, Malmendal A, Vosegaard T. Techniques and applications of NMR to membrane proteins (Review) Molecular Membrane Biology. 2004;21:129–141. doi: 10.1080/09687680410001693679. [DOI] [PubMed] [Google Scholar]
- 43.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magnetic Resonance in Chemistry. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 44.Arshava B, Liu SF, Jiang HL, Breslav M, Becker JM, Naider F. Structure of segments of a G protein-coupled receptor: CD and NMR analysis of the Saccharomyces cerevisiae tridecapeptide pheromone receptor. Biopolymers. 1998;46:343–357. doi: 10.1002/(SICI)1097-0282(199811)46:6<343::AID-BIP1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 45.Berlose JP, Convert O, Brunissen A, Chassaing G, Lavielle S. 3-Dimensional Structure of the Highly Conserved 7Th Transmembrane Domain of G-Protein-Coupled Receptors. European Journal of Biochemistry. 1994;225:827–843. doi: 10.1111/j.1432-1033.1994.0827b.x. [DOI] [PubMed] [Google Scholar]
- 46.Arevalo E, Estephan R, Madeo J, Arshava B, Dumont M, Becker JM, et al. Biosynthesis and biophysical analysis of domains of a yeast G protein-coupled receptor. Biopolymers. 2003;71:516–531. doi: 10.1002/bip.10491. [DOI] [PubMed] [Google Scholar]
- 47.Naider F, Arshava B, Ding FX, Arevalo E, Becker JM. Peptide fragments as models to study the structure of a G-protein coupled receptor: The alpha-factor receptor of Saccharomyces cerevisiae. Biopolymers. 2001;60:334–350. doi: 10.1002/1097-0282(2001)60:5<334::AID-BIP10175>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 48.Estephan R, Englander J, Arshava B, Samples KL, Becker JM, Naider F. Biosynthesis and NMR analysis of a 73-residue domain of a Saccharomyces cerevisiae G protein-coupled receptor. Biochemistry. 2005;44:11795–11810. doi: 10.1021/bi0507231. [DOI] [PubMed] [Google Scholar]
- 49.Valentine KG, Liu SF, Marassi FM, Veglia G, Opella SJ, Ding FX, et al. Structure and topology of a peptide segment of the 6th transmembrane domain of the Saccharomyces cerevisae alpha-factor receptor in phospholipid bilayers. Biopolymers. 2001;59:243–256. doi: 10.1002/1097-0282(20011005)59:4<243::AID-BIP1021>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng HA, Zhao J, Sheng WY, Xie XQ. A transmembrane helix-bundle from G-protein coupled receptor CB2: Biosynthesis, purification, and NMR characterization. Biopolymers. 2006;83:46–61. doi: 10.1002/bip.20526. [DOI] [PubMed] [Google Scholar]
- 51.Wu JX, Feng M, Ruan KH. Assembling NMR structures for the intracellular loops of the human thromboxane A(2) receptor: Implication of the G protein-coupling pocket. Archives of Biochemistry and Biophysics. 2008;470:73–82. doi: 10.1016/j.abb.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demene H, Granier S, Muller D, Guillon G, Dufour MN, Delsuc MA, et al. Active peptidic mimics of the second intracellular loop of the V-1A vasopressin receptor are structurally related to the second intracellular rhodopsin loop: A combined H-1 NMR and biochemical study. Biochemistry. 2003;42:8204–8213. doi: 10.1021/bi027358n. [DOI] [PubMed] [Google Scholar]
- 53.Mierke DF, Royo M, Pellegrini M, Sun HM, Chorev M. Peptide mimetic of the third cytoplasmic loop of the PTH/PTHrP receptor. Journal of the American Chemical Society. 1996;118:8998–9004. [Google Scholar]
- 54.Pellegrini M, Mierke DF. Molecular complex of cholecystokinin-8 and N-terminus of the cholecystokinin A receptor by NMR spectroscopy. Biochemistry. 1999;38:14775–14783. doi: 10.1021/bi991272l. [DOI] [PubMed] [Google Scholar]
- 55.Giragossian C, Mierke DF. Intermolecular interactions between cholecystokinin-8 and the third extracellular loop of the cholecystokinin A receptor. Biochemistry. 2001;40:3804–3809. doi: 10.1021/bi002659n. [DOI] [PubMed] [Google Scholar]
- 56.Giragossian C, Mierke DF. Intermolecular interactions between cholecystokinin-8 and the third extracellular loop of the cholecystokinin-2 receptor. Biochemistry. 2002;41:4560–4566. doi: 10.1021/bi0160009. [DOI] [PubMed] [Google Scholar]
- 57.Giragossian C, Sugg EE, Szewczyk JR, Mierke DF. Intermolecular interactions between peptidic and nonpeptidic agonists and the third extracellular loop of the cholecystokinin 1 receptor. Journal of Medicinal Chemistry. 2003;46:3476–3482. doi: 10.1021/jm030144z. [DOI] [PubMed] [Google Scholar]
- 58.Ulfers AL, Piserchio A, Mierke DF. Extracellular domains of the neurokinin-1 receptor: Structural characterization and interactions with substance P. Biopolymers. 2002;66:339–349. doi: 10.1002/bip.10312. [DOI] [PubMed] [Google Scholar]
- 59.Ruan KH, Wu JX, So SP, Jenkins LA, Ruan CH. NMR structure of the thromboxane A(2) receptor ligand recognition pocket. European Journal of Biochemistry. 2004;271:3006–3016. doi: 10.1111/j.1432-1033.2004.04232.x. [DOI] [PubMed] [Google Scholar]
- 60.Ruan KH, So SP, Wu JX, Li DW, Huang AM, Kung J. Solution structure of the second extracellular loop of human thromboxane A(2) receptor. Biochemistry. 2001;40:275–280. doi: 10.1021/bi001867c. [DOI] [PubMed] [Google Scholar]
- 61.Pham TCT, Kriwacki RW, Parrill AL. Peptide design and structural characterization of a GPCR loop mimetic. Biopolymers. 2007;86:298–310. doi: 10.1002/bip.20745. [DOI] [PubMed] [Google Scholar]
- 62.Kushwaha N, Harwood SC, Wilson AM, Berger M, Tecott LH, Roth BL, et al. Molecular determinants in the second intracellular loop of the 5-hydroxytryptamine-1A receptor for G-protein coupling. Molecular Pharmacology. 2006;69:1518–1526. doi: 10.1124/mol.105.019844. [DOI] [PubMed] [Google Scholar]
- 63.Neumann S, Krause G, Claus M, Paschke R. Structural determinants for G protein activation and selectivity in the second intracellular loop of the thyrotropin receptor. Endocrinology. 2005;146:477–485. doi: 10.1210/en.2004-1045. [DOI] [PubMed] [Google Scholar]
- 64.Havlickova M, Blahos J, Brabet I, Liu JF, Hruskova B, Prezeau L, et al. The second intracellular loop of metabotropic glutamate receptors recognizes C termini of G-protein alpha-subunits. Journal of Biological Chemistry. 2003;278:35063–35070. doi: 10.1074/jbc.M306555200. [DOI] [PubMed] [Google Scholar]
- 65.Chung DA, Zuiderweg ERP, Fowler CB, Soyer OS, Mosberg HI, Neubig RR. NMR structure of the second intracellular loop of the alpha 2A adrenergic receptor: Evidence for a novel cytoplasmic helix. Biochemistry. 2002;41:3596–3604. doi: 10.1021/bi015811+. [DOI] [PubMed] [Google Scholar]
- 66.Piserchio A, Prado GN, Zhang R, Yu J, Taylor L, Polgar P, et al. Structural insight into the role of the second intracellular loop of the bradykinin 2 receptor in signaling and internalization. Biopolymers. 2002;63:239–246. doi: 10.1002/bip.10072. [DOI] [PubMed] [Google Scholar]
- 67.Jung H, Windhaber R, Palm D, Schnackerz KD. Nmr and Circular-Dichroism Studies of Synthetic Peptides Derived from the 3Rd Intracellular Loop of the Beta-Adrenoceptor. Febs Letters. 1995;358:133–136. doi: 10.1016/0014-5793(94)01409-t. [DOI] [PubMed] [Google Scholar]
- 68.Ulfers AL, McMurry JL, Miller A, Wang LG, Kendall DA, Mierke DF. Cannabinoid receptor-G protein interactions: G(alpha i1)-bound structures of IC3 and a mutant with altered G protein specificity. Protein Science. 2002;11:2526–2531. doi: 10.1110/ps.0218402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pellegrini M, Bisello A, Rosenblatt M, Chorev M, Mierke DF. Binding domain of human parathyroid hormone receptor: From conformation to function. Biochemistry. 1998;37:12737–12743. doi: 10.1021/bi981265h. [DOI] [PubMed] [Google Scholar]
- 70.Choi G, Landin J, Xie XQ. The cytoplasmic helix of cannabinoid receptor CB2, a conformational study by circular dichroism and H-1 NMR spectroscopy in aqueous and membrane-like environments. Journal of Peptide Research. 2002;60:169–177. doi: 10.1034/j.1399-3011.2002.21012.x. [DOI] [PubMed] [Google Scholar]
- 71.Choi G, Guo JX, Makriyannis A. The conformation of the cytoplasmic helix 8 of the CB1 cannabinoid receptor using NMR and circular dichroism. Biochimica et Biophysica Acta-Biomembranes. 2005;1668:1–9. doi: 10.1016/j.bbamem.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 72.Katragadda M, Maciejewski MW, Yeagle PL. Structural studies of the putative helix 8 in the human beta(2) adrenergic receptor: an NMR study. Biochimica et Biophysica Acta-Biomembranes. 2004;1663:74–81. doi: 10.1016/j.bbamem.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 73.Jung H, Windhaber R, Palm D, Schnackerz KD. Conformation of a beta-adrenoceptor-derived signal transducing peptide as inferred by circular dichroism and H-1 NMR spectroscopy. Biochemistry. 1996;35:6399–6405. doi: 10.1021/bi952575s. [DOI] [PubMed] [Google Scholar]
- 74.Franzoni L, Nicastro G, Pertinhez TA, Tato M, Nakaie CR, Paiva ACM, et al. Structure of the C-terminal fragment 300–320 of the rat angiotensin II AT(1A) receptor and its relevance with respect to G-protein coupling. Journal of Biological Chemistry. 1997;272:9734–9741. doi: 10.1074/jbc.272.15.9734. [DOI] [PubMed] [Google Scholar]
- 75.Piserchio A, Zelesky V, Yu J, Taylor L, Polgar P, Mierke DF. Bradykinin B2 receptor signaling: Structural and functional characterization of the C-terminus. Biopolymers. 2005;80:367–373. doi: 10.1002/bip.20220. [DOI] [PubMed] [Google Scholar]
- 76.Lehmann N, Alexiev U, Fahmy K. Linkage between the intramembrane H-bond network around aspartic acid 83 and the cytosolic environment of helix 8 in photoactivated rhodopsin. Journal of Molecular Biology. 2007;366:1129–1141. doi: 10.1016/j.jmb.2006.11.098. [DOI] [PubMed] [Google Scholar]
- 77.Getmanova E, Patel AB, Klein-Seetharaman J, Loewen MC, Reeves PJ, Friedman N, et al. NMR spectroscopy of phosphorylated wild-type rhodopsin: Mobility of the phosphorylated C-terminus of rhodopsin in the dark and upon light activation. Biochemistry. 2004;43:1126–1133. doi: 10.1021/bi030120u. [DOI] [PubMed] [Google Scholar]
- 78.Kisselev OG, McDowell JH, Hargrave PA. The arrestin-bound conformation and dynamics of the phosphorylated carboxy-terminal region of rhodopsin. Febs Letters. 2004;564:307–311. doi: 10.1016/S0014-5793(04)00226-1. [DOI] [PubMed] [Google Scholar]
- 79.Choi G, Landin J, Galan JF, Birge RR, Albert AD, Yeagle PL. Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin. Biochemistry. 2002;41:7318–7324. doi: 10.1021/bi025507w. [DOI] [PubMed] [Google Scholar]
- 80.Yeagle PL, Choi G, Albert AD. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms. Biochemistry. 2001;40:11932–11937. doi: 10.1021/bi015543f. [DOI] [PubMed] [Google Scholar]
- 81.Yeagle PL, Alderfer JL, Albert AD. Structure of the third cytoplasmic loop of bovine rhodopsin. Biochemistry. 1995;34:14621–14625. doi: 10.1021/bi00045a002. [DOI] [PubMed] [Google Scholar]
- 82.Cai K, Itoh Y, Khorana HG. Mapping of contact sites in complex formation between transducin and light-activated rhodopsin by covalent crosslinking: use of a photoactivatable reagent. Proc Natl Acad Sci U S A. 2001;98:4877–4882. doi: 10.1073/pnas.051632898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai K, Klein-Seetharaman J, Hwa J, Hubbell WL, Khorana HG. Structure and function in rhodopsin: Effects of disulfide cross-links in the cytoplasmic face of rhodopsin on transducin activation and phosphorylation by rhodopsin kinase. Biochemistry. 1999;38:12893–12898. doi: 10.1021/bi9912443. [DOI] [PubMed] [Google Scholar]
- 84.Unger VM, Schertler GFX. Low-Resolution Structure of Bovine Rhodopsin Determined by Electron Cryomicroscopy. Biophysical Journal. 1995;68:1776–1786. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katragadda M, Chopra A, Bennett M, Alderfer JL, Yeagle PL, Albert AD. Structures of the transmembrane helices of the G-protein coupled receptor, rhodopsin. Journal of Peptide Research. 2001;58:79–89. doi: 10.1034/j.1399-3011.2001.00904.x. [DOI] [PubMed] [Google Scholar]
- 86.Tian CL, Breyer RM, Kim HJ, Karra MD, Friedman DB, Karpay A, et al. Solution NMR spectroscopy of the human vasopressin V2 receptor, a G protein-coupled receptor. Journal of the American Chemical Society. 2005;127:8010–8011. doi: 10.1021/ja051161b. [DOI] [PubMed] [Google Scholar]
- 87.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:929–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 88.Surgand JS, Rodrigo J, Kellenberger E, Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins. 2006;62:509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- 89.Jaschke H, Neumann S, Moore S, Thomas CJ, Colson AO, Costanzi S, et al. A low molecular weight agonist signals by binding to the transmembrane domain of thyroid-stimulating hormone receptor (TSHR) and luteinizing hormone/chorionic gonadotropin receptor (LHCGR) Journal of Biological Chemistry. 2006;281:9841–9844. doi: 10.1074/jbc.C600014200. [DOI] [PubMed] [Google Scholar]
- 90.Moore S, Jaeschke H, Kleinau G, Neumann S, Costanzi S, Jiang JK, et al. Evaluation of small-molecule modulators of the luteinizing hormone/choriogonadotropin and thyroid stimulating hormone receptors: Structure-activity relationships and selective binding patterns. Journal of Medicinal Chemistry. 2006;49:3888–3896. doi: 10.1021/jm060247s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu JX, Jiang JK, Costanzi S, Thomas C, Yang W, Feyen JHM, et al. A missense mutation in the seven-transmembrane domain of the human Ca2+ receptor converts a negative allosteric modulator into a positive allosteric modulator. Journal of Biological Chemistry. 2006;281:21558–21565. doi: 10.1074/jbc.M603682200. [DOI] [PubMed] [Google Scholar]
- 92.Heitman LH, Oosterom J, Bonger KM, Timmers CM, Wiegerinck PHG, Ijzerman AP. [H-3]Org 43553, the first low-molecular-weight agonistic and allosteric Radioligand for the human luteinizing hormone receptor. Molecular Pharmacology. 2008;73:518–524. doi: 10.1124/mol.107.039875. [DOI] [PubMed] [Google Scholar]
- 93.Neumann S, Kleinau G, Costanzi S, Moore S, Jiang JK, Raaka BM, et al. A Low Molecular Weight Antagonist for the Human Thyrotropin Receptor with Therapeutic Potential for Hyperthyroidism. Endocrinology. 2008;149:5945–5950. doi: 10.1210/en.2008-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Middleton DA. Solid-state NMR spectroscopy as a tool for drug design: from membrane-embedded targets to amyloid fibrils. Biochemical Society Transactions. 2007;35:985–990. doi: 10.1042/BST0350985. [DOI] [PubMed] [Google Scholar]
- 95.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 96.Hajduk PJ. SAR by NMR: Putting the pieces together. Molecular Interventions. 2006;6:266–272. doi: 10.1124/mi.6.5.8. [DOI] [PubMed] [Google Scholar]
- 97.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 98.Eilers M, Reeves PJ, Ying WW, Khorana HG, Smith SO. Magic angle spinning NMR of the protonated retinylidene Schiff base nitrogen in rhodopsin: Expression of N-15-lysine-and C-13-glycine-labeled opsin in a stable cell line. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:487–492. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grobner G, Burnett IJ, Glaubitz C, Choi G, Mason AJ, Watts A. Observations of light-induced structural changes of retinal within rhodopsin. Nature. 2000;405:810–813. doi: 10.1038/35015604. [DOI] [PubMed] [Google Scholar]
- 100.Creemers AFL, Kiihne S, Bovee-Geurts PHM, Degrip WJ, Lugtenburg J, de Groot HJM. H-1 and C-13 MAS NMR evidence for pronounced ligand-protein interactions involving the ionone ring of the retinylidene chromophore in rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9101–9106. doi: 10.1073/pnas.112677599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown MF, Heyn MP, Job C, Kim S, Moltke S, Nakanishi K, et al. Solid-State H-2 NMR spectroscopy of retinal proteins in aligned membranes. Biochimica et Biophysica Acta-Biomembranes. 2007;1768:2979–3000. doi: 10.1016/j.bbamem.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Salgado GFJ, Struts AV, Tanaka K, Fujioka N, Nakanishi K, Brown MF. Deuterium NMR structure of retinal in the ground state of rhodopsin. Biochemistry. 2004;43:12819–12828. doi: 10.1021/bi0491191. [DOI] [PubMed] [Google Scholar]
- 103.Salgado GFJ, Struts AV, Tanaka K, Krane S, Nakanishi K, Brown MF. Solid-state H-2 NMR structure of retinal in metarhodopsin I. Journal of the American Chemical Society. 2006;128:11067–11071. doi: 10.1021/ja058738+. [DOI] [PubMed] [Google Scholar]
- 104.Struts AV, Salgado GFJ, Tanaka K, Krane S, Nakanishi K, Brown MF. Structural analysis and dynamics of retinal chromophore in dark and metal states of rhodopsin from H-2 NMR of aligned membranes. J Mol Biol. 2007;372:50–66. doi: 10.1016/j.jmb.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Creemers AFL, Bovee-Geurts PHM, Degrip WJ, Lugtenburg J, de Groot HJM. Solid-state NMR analysis of ligand-receptor interactions reveals an induced misfit in the binding site of isorhodopsin. Biochemistry. 2004;43:16011–16018. doi: 10.1021/bi048541e. [DOI] [PubMed] [Google Scholar]
- 106.Kiihne SR, Creemers AFL, de Grip WJ, Bovee-Geurts PHM, Lugtenburg J, de Groot HJM. Selective interface detection: Mapping binding site contacts in membrane proteins by NMR spectroscopy. Journal of the American Chemical Society. 2005;127:5734–5735. doi: 10.1021/ja045677r. [DOI] [PubMed] [Google Scholar]
- 107.Luca S, White JF, Sohal AK, Filippov DV, van Boom JH, Grisshammer R, et al. The conformation of neurotensin bound to its G protein-coupled receptor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10706–10711. doi: 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lopez JJ, Shukla AK, Reinhart C, Schwalbe H, Michel H, Glaubitz C. The structure of the neuropeptide bradykinin bound to the human G-protein coupled receptor bradykinin B2 as determined by solid-state NMR spectroscopy. Angewandte Chemie-International Edition. 2008;47:1668–1671. doi: 10.1002/anie.200704282. [DOI] [PubMed] [Google Scholar]
- 109.Dossey AT, Reale V, Chatwin H, Zachariah C, Debono M, Evans PD, et al. NMR analysis of Caenorhabditis elegans FLP-18 neuropeptides: Implications for NPR-1 activation. Biochemistry. 2006;45:7586–7597. doi: 10.1021/bi0603928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ratnala VRP, Kiihne SR, Buda F, Leurs R, de Groot HJM, Degrip WJ. Solid-state NMR evidence for a protonation switch in the binding pocket of the H1 receptor upon binding of the agonist histamine. Journal of the American Chemical Society. 2007;129:867–872. doi: 10.1021/ja0652262. [DOI] [PubMed] [Google Scholar]
- 111.Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SGF, Shi L, Gether U, et al. Activation of the beta(2)-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. Journal of Biological Chemistry. 2001;276:29171–29177. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- 112.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. beta2 Adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch Journal of Biological Chemistry. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 113.Altenbach C, Klein-Seetharaman J, Cai K, Khorana HG, Hubbell WL. Structure and function in rhodopsin: mapping light-dependent changes in distance between residue 316 in helix 8 and residues in the sequence 60–75, covering the cytoplasmic end of helices TM1 and TM2 and their connection loop CL1. Biochemistry. 2001;40:15493–15500. doi: 10.1021/bi011545o. [DOI] [PubMed] [Google Scholar]
- 114.Tikhonova IG, Best RB, Engel S, Gershengorn MC, Hummer G, Costanzi S. Atomistic insights into rhodopsin activation from a dynamic model. Journal of the American Chemical Society. 2008;130:10141–10149. doi: 10.1021/ja0765520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Patel AB, Crocker E, Eilers M, Hirshfeld A, Sheves M, Smith SO. Coupling of retinal isomerization to the activation of rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Crocker E, Eilers M, Ahuja S, Hornak V, Hirshfeld A, Sheves M, et al. Location of Trp265 in metarhodopsin II: Implications for the activation mechanism of the visual receptor rhodopsin. Journal of Molecular Biology. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 117.Patel AB, Crocker E, Reeves PJ, Getmanova EV, Eilers M, Khorana HG, et al. Changes in interhelical hydrogen bonding upon rhodopsin activation. J Mol Biol. 2005;347:803–812. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 118.Ahuja S, Hornak V, Yan ECY, Syrett N, Goncalves JA, Hirshfeld A, et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat Struct Mol Biol. 2009;16:168–175. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loewen MC, Klein-Seetharaman J, Getmanova EV, Reeves PJ, Schwalbe H, Khorana HG. Solution F-19 nuclear Overhauser effects in structural studies of the cytoplasmic domain of mammalian rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4888–4892. doi: 10.1073/pnas.051633098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Klein-Seetharaman J, Getmanova EV, Loewen MC, Reeves PJ, Khorana HG. NMR spectroscopy in studies of light-induced structural changes in mammalian rhodopsin: Applicability of solution F-19 NMR. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Klein-Seetharaman J, Yanamala NVK, Javeed F, Reeves PJ, Getmanova EV, Loewen MC, et al. Differential dynamics in the G protein-coupled receptor rhodopsin revealed by solution NMR. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3409–3413. doi: 10.1073/pnas.0308713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung L, Schwalbe H, et al. Solution NMR spectroscopy of [alpha-N-15]lysine-labeled rhodopsin: The single peak observed in both conventional and TROSY-type HSQC spectra is ascribed to Lys-339 in the carboxyl-terminal peptide sequence. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3452–3457. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mak-Jurkauskas ML, Bajaj VS, Hornstein MK, Belenky M, Griffin RG, Herzfeld J. Energy transformations early in the bacteriorhodopsin photocycle revealed by DNP-enhanced solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:883–888. doi: 10.1073/pnas.0706156105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ulfers AL, McMurry JL, Kendall DA, Mierke DF. Structure of the third intracellular loop of the human cannabinoid 1 receptor. Biochemistry. 2002;41:11344–11350. doi: 10.1021/bi0259610. [DOI] [PubMed] [Google Scholar]
- 125.Macdonald D, Mierke DF, Li HZ, Pellegrini M, Sachais B, Krause JE, et al. Photoaffinity labeling of mutant neurokinin-1 receptors reveals additional structural features of the substance P/NK-1 receptor complex. Biochemistry. 2001;40:2530–2539. doi: 10.1021/bi001880x. [DOI] [PubMed] [Google Scholar]
- 126.Piserchio A, Bisello A, Rosenblatt M, Chorev M, Mierke DF. Characterization of parathyroid hormone/receptor interactions: Structure of the first extracellular loop. Biochemistry. 2000;39:8153–8160. doi: 10.1021/bi000196f. [DOI] [PubMed] [Google Scholar]
- 127.Dorey M, Hargrave PA, McDowell JH, Arendt A, Vogt T, Bhawsar N, et al. Effects of phosphorylation on the structure of the G-protein receptor rhodopsin. Biochimica et Biophysica Acta-Biomembranes. 1999;1416:217–224. doi: 10.1016/s0005-2736(98)00224-7. [DOI] [PubMed] [Google Scholar]