Abstract
Although gonadotropins and androgen are required for normal spermatogenesis and both testosterone and follicle-stimulating hormone (FSH) are responsible for the inhibition of spermatogonial differentiation that occurs in irradiated rats, it has been difficult to identify the specific genes involved. To study specific hormonally regulated changes in somatic cell gene expression in the testis that may be involved in these processes, without the complication of changing populations of germ cells, we used irradiated LBNF1 rats, the testes of which contain almost exclusively somatic cells except for a few type A spermatogonia. Three different groups of these rats were treated with various combinations of gonadotropin-releasing hormone antagonist, an androgen receptor antagonist (flutamide), testosterone, and FSH, and we compared the gene expression levels 2 wk later to those of irradiated-only rats by microarray analysis. By dividing the gene expression patterns into three major patterns and 11 subpatterns, we successfully distinguished, in a single study, the genes that were specifically regulated by testosterone, by luteinizing hormone (LH), and by FSH from the large number of genes that were not hormonally regulated in the testis. We found that hormones produced more dramatic upregulation than downregulation of gene expression: Testosterone had the strongest upregulatory effect, LH had a modest but appreciable upregulatory effect, and FSH had a minor upregulatory effect. We also separately identified the somatic cell genes that were chronically upregulated by irradiation. Thus, the present study identified gene expression changes that may be responsible for hormonal action on somatic cells to support normal spermatogenesis and the hormone-mediated block in spermatogonial development after irradiation.
Keywords: follicle-stimulating hormone, irradiation, luteinizing hormone, spermatogenesis, spermatogonial differentiation, testis, testosterone
Dissection of the effects of testosterone, LH, and FSH on irradiated rat testes somatic cells identifies genes that may be responsible for hormonal actions on somatic cells that affect spermatogenesis.
INTRODUCTION
The completion of the meiotic and postmeiotic stages of spermatogenesis in normal mammals is dependent primarily on testosterone, which is produced by the Leydig cells through the action of luteinizing hormone (LH). Secondarily, these stages are also dependent on the action of follicle-stimulating hormone (FSH), which stimulates premeiotic germ cell development as well [1]. Paradoxically, in irradiated and other toxicant-treated rats, testosterone and, to a lesser extent, FSH inhibit spermatogonial differentiation [2, 3].
To study the mechanisms of action by these hormones in the rat, scientists most often have altered their levels by pharmacological or surgical means or have administered receptor antagonists [1, 4, 5], and in the mouse, they have produced null mutations for the hormones or their receptors [6] (for review, see [7]). Receptor localization and cell-specific knockouts of the hormone receptors in the testis have shown that the stimulatory effect of gonadotropins and testosterone on germ cells is an indirect result of action on the somatic cells. Likewise, spermatogonial transplantation studies have shown that the hormone-mediated block in spermatogonial differentiation in irradiated rats is also an indirect result of hormone action on the somatic cells [8].
This hormonal regulation of genes in specific somatic cells of the testis had been studied previously using RNA and protein analysis of individual genes. Recently, microarrays have been employed to obtain global analyses of hormone-regulated gene expression in these cells. To our knowledge, however, the only such studies in the rat have examined the effects of FSH on the acute responses of Sertoli cells in vitro [9] or on the short-term responses of immature rat testes in vivo [10]. No microarray studies regarding the effects of testosterone on rat testicular cells have been performed; all of these studies have been done in the mouse. Three studies, using immature mice that overexpress androgen-binding protein [11], androgen receptor (AR)-null immature mice [12], or AR-hypomorphic adult mice [13], have characterized the chronic effects of androgen reduction in the testis. The chronic effects of elimination of androgen action in Sertoli cells have been examined in Sertoli-cell AR knockout immature [14] or adult [13] mice. Other studies have examined the acute effects of testosterone or FSH in hypogonadal (hpg) [15, 16] or neonatal [17] mice. The effects of FSH also have been examined in mouse Sertoli cell lines by overexpressing a constitutively active FSH receptor [18].
These studies have provided valuable information, but the results have been discordant, both because of differences between systems and because of limitations in the methodology. Some have used cell lines or primary cells, which differ in fundamental ways from cells in vivo. In most of the in vivo studies, hormone treatment or receptor modulation altered the germ cell composition. Studies employing immature animals were influenced by differences in gene expression between immature and adult animals. Those in vivo studies that examined only acute changes upon addition of the hormones may have missed some important functional changes that result from chronic treatment, such as indirect regulation of targets. Finally, knockout or overexpression of a gene in vivo may have altered earlier differentiation pathways in the cells.
We used the irradiated adult LBNF1 rat as a novel in vivo model to identify genes for which expression in the somatic cells were altered by chronic hormonal changes without most of the complications that arise from changing germ cell populations. A single dose of 6 Gy removes all differentiated germ cells but does not eliminate any of the somatic cell types; however, it alters somatic cell function to block the spermatogonial differentiation [8], resulting in undifferentiated type A spermatogonia (1.4 per 100 Sertoli cells) as the only germ cells remaining [19]. In this model, most of the hormonally regulated changes in gene expression that are identified should be in the somatic cells, because nearly all of the germ cells, which constitute most of the testis mass, are absent and the remaining type A spermatogonia do not possess AR [20] or gonadotropin receptors. In support of the relationship of this model to the normal testes, we noted that the effect of hormone suppression on extent of progression of spermatogenesis in irradiated rats, which was to the spermatocyte stage with occasional spermatids [21], was similar in that in nonirradiated rats [22].
Specifically, we analyzed the changes induced by 2-wk treatments with gonadotropin-releasing hormone antagonist (GnRH-ant) given along with various combinations of an AR antagonist (flutamide), testosterone, or FSH to dissect out the chronic effects of testosterone, LH, and FSH on gene expression in the somatic cells of testes. The 2-wk treatment was chosen because it produces minimal changes in the cellular composition of the testis [3, 23]. However, based on the initiation of spermatogonial differentiation after 4 wk of GnRH-ant treatment [3, 24], the primary and secondary changes in gene expression could be expected to have occurred by 2 wk. The genes for which expression is regulated by testosterone or FSH will be considered to be specifically involved in the reversal of the hormone-mediated inhibition of spermatogonial differentiation in toxicant-treated rats and, possibly, in the support of normal spermatogenesis. Furthermore, comparisons between nonirradiated control and irradiated rats would show which genes are expressed primarily in germ cells and which primarily in the somatic cells and, of those localized to the somatic cells, the ones for which expression is increased by irradiation.
MATERIALS AND METHODS
Animals
The LBNF1 male rats were obtained from Harlan Sprague-Dawley and irradiated at approximately 8 wk of age. Animals were housed under a 12L:12D photoperiod and allowed food and water ad libitum. Animal facilities were approved by the American Association for the Accreditation of Laboratory Animal Care, and all procedures were approved by the Institutional Animal Care and Use Committee.
Irradiation
Rats were anesthetized, and all except nonirradiated controls were irradiated to the lower part of the body with a single dose of 6 Gy of 60Co gamma radiation as described previously [21, 24].
Animal Groups and Treatments
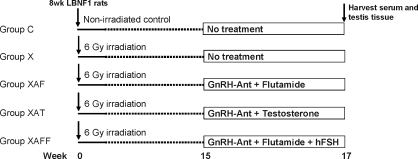
The schematic of the experimental design is illustrated in Figure 1. Five groups of animals were used. The nonirradiated control group (group C) received only sham or placebo treatments. One group received only irradiation (group X). The other three irradiated groups were treated, starting 15 wk later, with the GnRH-ant acyline (National Institute of Child Health and Human Development) and either the AR-antagonist flutamide (group XAF), testosterone (group XAT), or flutamide plus human FSH (hFSH; group XAFF). Acyline was dissolved in water and administered as weekly subcutaneous injections of 1.5 mg/kg [24]. Testosterone was administered via three 8-cm-log Silastic capsules (inner diameter, 2 mm; outer diameter, 3.2 mm; Dow Corning Corp.) [25]. Flutamide was administered either by subcutaneous implantation of a pellet (Innovative Research of America) calculated to deliver 20 mg kg−1 day−1 for 2 wk [25] or in four 5-cm-long Silastic capsules, which gave results equivalent to the pellets [26]. Recombinant hFSH (5 U/μg, based on bioactivity; National Hormone and Peptide Program) was dissolved in saline and placed in Alzet (model 2001; Durect Corp.) osmotic pumps and implanted subcutaneously to produce a dose of 4 U/day [3]. Pumps were replaced after 1 wk. Rats were killed after 2 wk of hormonal treatment for RNA extraction, hormone measurements, and histological analysis. Blood was collected by terminal cardiac puncture. The testes were weighed, and one was processed for RNA extraction. A portion of the other testis was fixed in Bouin solution for histological examination, and the remainder was reweighed, homogenized in water, and stored at −20°C for intratesticular testosterone (ITT) measurements.
FIG. 1.
Schematic of experimental design.
Assessment of Spermatogenesis
Tubule differentiation index was determined by counting the number of tubules with differentiating germ cells (at least three cells that had reached the type B spermatogonial stage or later) in sections from methacrylate-embedded tissue stained with periodic acid-Schiff reagent and hematoxylin [27].
Hormone Measurements
Serum testosterone and ITT concentrations were measured using coated-tube radioimmunoassay kits (DSL 4000; Diagnostic Systems Laboratories) as described previously [25]. Rat serum FSH and LH levels were measured using immunofluorometric assays (Delfia; Perkin Elmer Wallac) as described previously [3, 25, 28, 29]. Rat serum FSH measurements were adjusted for the cross-reaction contribution of hFSH (for details, see Supplemental Methods; all Supplemental Data are available online at www.biolreprod.org).
RNA Preparation and Microarray Hybridization
Testis tissue samples were immediately placed in RNAlater (Qiagen) and stored at −20°C until total RNA was extracted using the RNeasy Midi kit (Qiagen). RNA quality was determined by electrophoresis and by 230-, 260-, and 280-nm absorption readings (NanoDrop). Ten micrograms of total RNA from each sample were submitted to the Genomics Facility at M.D. Anderson Cancer Center for microarray hybridization using the Rat 230 2.0 GeneChips (Affymetrix).
Two separate sets of microarray runs, using a relatively large number of samples for each, were performed. The final data set (after exclusion of outliers in cluster plots) involved comparison of 10 nonirradiated control (group C) and five irradiated (group X) samples for the first microarray run and comparison among the irradiated group (14 group X samples) and the hormonal treatment groups (11 group XAF, 5 group XAT, and 5 group XAFF samples) for the second run.
Data Analysis
After normalization among the arrays, the expression levels within each chip were divided by the median value of all the probes for which expression was regarded as present on the array and were then log base 2 transformed. Only probes that had a present call based on the Affymetrix criteria in more than 70% of the chips for at least one treatment group were used in subsequent analysis. The statistical significance of changes in expression level for each probe between two treatment groups was determined with the Student t-test, with P < 0.05 being considered as significant. In addition to having statistical significance, probes were considered to be hormonally regulated only if the changes in expression met fold-change criteria described in the Supplemental Methods. Briefly, only differences of 1.41-fold or greater between the X and XAF groups and of 1.26-fold or greater between the other hormonal treatment groups, with P < 0.05 as determined by a t-test, were considered to be potentially statistically and biologically significant. Array data sets were deposited in Gene Expression Omnibus as GSE15223 and GSE15243.
The probes were related to specific genes according to the Affymetrix annotation of the GeneChips on November 5, 2007, using the UniGene Build 165. When multiple probes were associated with a specific UniGene entry, they were ranked according to the quality of the probe set, the presence in the treatment groups, and the level of expression (Supplemental Table S1).
For calculation of the minimum fold-induction of expression level after irradiation (group X vs. group C) in Table 1, expression levels of these genes were so low in nonirradiated control testes that they were classified as absent in the control testes by the Affymetrix criteria. In these cases, fold-induction in expression was calculated using the average of the expression values of the 100 most weakly expressed present probes (−3.73) as baseline. For the listed genes, the average SEMs were 5% of the mean fold-induction.
TABLE 1.
Somatic cell genes that are maximally upregulated after irradiation.
For calculation of the minimum fold-induction of expression level by irradiation after maximal hormone effect correction (group XAF vs. group C) in Table 1, the maximal hormone effect correction of the fold-induction in expression was calculated by taking the fold-induction of expression resulting from irradiation [2(X−C)], which is associated with moderate increases in hormone levels, and multiplying it by the fold-change resulting from testosterone/LH/FSH suppression [2(XAF−X)], which is associated with even larger decreases in the elevated hormone levels in the irradiated testis to below the levels observed in nonirradiated control testes. In these expressions, C, X, and XAF are the log2-transformed values of the normalized expression levels of the probe in the control, irradiated, and irradiated plus GnRH-ant plus flutamide groups, respectively.
The canonical pathways and functional analysis were generated through the use of Ingenuity Pathway Analysis (IPA; Ingenuity Systems) as instructed by the manufacturer.
Quantitative Real-Time PCR
Total RNA (3 μg) was used to generate the template cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Sciences). Quantitative real-time PCR was performed using the Rotor-Gene 3000 thermocycler (Corbett Research) with the primer sequences and conditions listed in Supplemental Table S2. Relative levels of mRNA concentrations were calculated using Rotor-Gene 6.0 software and normalized using ribosomal protein S2 (Rps2). All samples were run in triplicate. The rat testis samples used in the real-time PCR were partially overlapping with the samples used in the microarray hybridization.
RESULTS
We first examined changes in gene expression after irradiation. We then identified genes that were regulated by testosterone, FSH, and LH, respectively, in irradiated testes.
Germ Cell-Specific and Somatic Cell-Expressed Genes
Whereas the nonirradiated control rat testis contains largely germ cells (20 per Sertoli cell) [10, 30], the irradiated rat testis contains very few germ cells (0.015 per Sertoli cell) (Supplemental Fig. S1, A and B), making the comparison of genes expressed useful for identifying germ and somatic cell genes. All somatic elements, including Sertoli cells, peritubular myoid cells, Leydig cells, macrophages, and vascular cells, were present (Supplemental Fig. S1Bi). Microarray analysis of control and irradiated rat testes revealed a total of 14 021 genes (distinct UniGene entries) expressed in either normal or irradiated rat testis (see Supplemental Table S1 for results of all probes). Among them, 1489 genes were scored as present only in normal testes, and 4808 were scored as present only in irradiated testes. The large number of genes that appeared to be present only in irradiated testes is mostly a consequence of the increased sensitivity for detection of somatic cell genes that resulted from the germ cell depletion and, hence, a greater concentration of somatic cell RNA. We then used cutoff values to identify whether these genes are primarily expressed in the germ or somatic cells. Because the microarray data were not compensated for changes in cellular composition, the apparent levels of RNA from somatic cell-enriched genes that were unchanged in expression per cell would be increased in the irradiated testes. Indeed, the average increase in expression in the microarray of three Sertoli cell-specific genes for which the expression was not hormonally regulated (Wt1, Clu, and Cldn11) was 4.3-fold. We therefore used a cutoff of at least a fourfold increase to define genes that were highly likely to be somatic cell-enriched. In addition, we chose an arbitrary 10-fold or greater increase as a cutoff for somatic genes that were upregulated as a consequence of irradiation. Because irradiation depleted all differentiated germ cells in the testis, we initially chose a 10-fold or greater decrease as a cutoff for germ cell-specific genes. Based on these restrictive criteria, 674 genes were expressed primarily in germ cells and 2534 mainly in somatic cells, but the majority (9718 genes) could not be localized (Supplemental Table S1, column “Somatic or Germ”). The most highly expressed germ cell-specific and somatic cell-enriched genes are listed in Supplemental Table S3.
A total of 597 somatic cell-enriched genes showed more than a 10-fold increase in expression after irradiation. Ten of these genes were even upregulated by more than 45-fold after irradiation (Table 1).
Hormone Levels and Cellular Complement
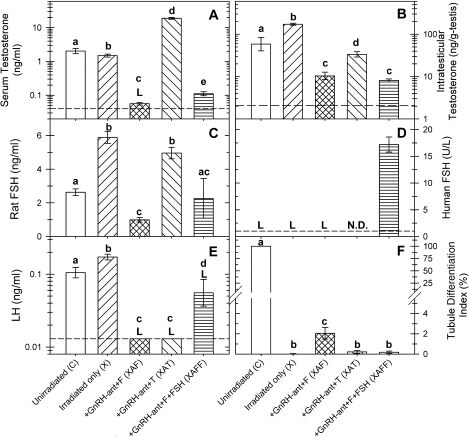
As shown previously [21], irradiation increased ITT, FSH, and LH levels (Fig. 2). Treatment of irradiated rats with GnRH-ant along with flutamide (group XAF) markedly reduced serum testosterone to near baseline, but substitution of testosterone (group XAT) for flutamide raised serum testosterone to supraphysiological levels (Fig. 2A). In contrast, ITT concentrations, which were also markedly reduced by GnRH-ant treatment, were only partially restored by exogenous testosterone treatment (Fig. 2B). Endogenous rat FSH was also reduced in the XAF group, but exogenous testosterone treatment largely reversed this suppression (Fig. 2C). Treatment of rats with exogenous hFSH (group XAFF) raised the serum level of hFSH in the rats (Fig. 2D) to well above the median FSH level in normal human males (3.3 IU/L) [31]. LH levels were decreased to below the limit of detection in both the XAF and XAT groups (Fig. 2E). Detectible LH levels, averaging 33% of those in irradiated-only rats, were observed in most group-XAFF rats treated with exogenous hFSH, the reasons for which are not known. We have ruled out cross-reaction of hFSH in the rat LH assay, but it is possible that some metabolites of hFSH do cross-react in this assay. However, as will be shown later, the exogenous hFSH stimulated expression of a different set of genes than were stimulated by LH, indicating that this increased LH level detected by the antibody is not of biological significance.
FIG. 2.
Hormone levels (A–E) and spermatogonial differentiation as assessed by tubule differentiation indices (TDI; F) of nonirradiated control and irradiated rats receiving different hormonal treatments for 2 wk. The data for hormone levels are presented as the mean ± SEM calculated from either untransformed data (FSH, TDI) or from log-transformed data (serum testosterone [T], ITT, LH) obtained from individual rats. Except for the hFSH assay, all other assays had 5–19 animals in each group. For the hFSH assay, seven animals were used in group XAFF. Only one animal from groups C, X, and XAF were assayed, and hFSH was not determined (N.D.) for group XAT, because in our experience, rats not treated with hFSH do not show any detectable levels in the hFSH assay. ANOVA was used for the statistical analysis. Lowercase letters a, b, c, and d indicate the values in different groups are significantly different (P < 0.05). An uppercase letter L indicates that the concentration was below the detection limit in some or all samples of the group. Detectable levels of LH were observed in most of the rats in group XAFF, but the reason is unknown. Please note that the levels of the serum testosterone, ITT, and LH are expressed at log scales.
The histological changes in the irradiated rat testes after 2 wk of hormonal treatment were minimal (Supplemental Fig. S1, B–E). The only consistent qualitative changes we observed in the somatic component was a reduction in nuclear size and apparent cytoplasmic size of Leydig cells when treated with GnRH-ant (Supplemental Fig. S1, compare Ci, Di, and Ei with Bi) [32]. With respect to the germ cells, GnRH-ant treatment induced only a doubling of the number of type A spermatogonia [23]. A few differentiating germ cells (type B spermatogonia or later) were observed in only 2% of the tubules in group XAF (amounting to <1 type B spermatogonium per 100 Sertoli cells) and in less than 0.22% of the tubules in other irradiated groups (Fig. 2F). Therefore, we deemed the 2-wk time point to be suitable for microarray analysis.
Relationship of Patterns of Gene Expression to Hormone Levels
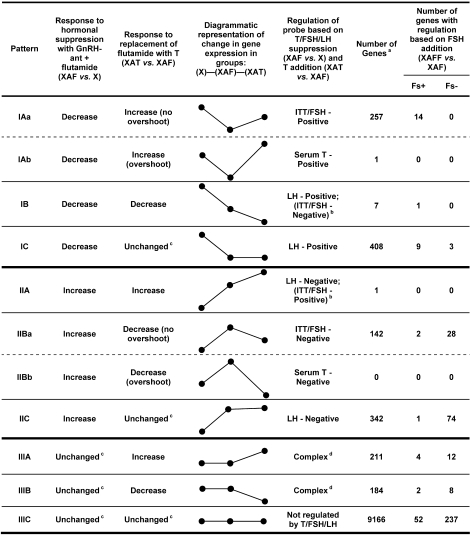
To determine the roles of testosterone, FSH, and LH in their expression, genes were first divided into three patterns of change in response to total hormonal suppression with GnRH-ant plus flutamide (group XAF): pattern I, decreased; pattern II, increased; and pattern III, small or nonsignificant changes (Fig. 3).
FIG. 3.
Definition of patterns of changes in gene expression and numbers of genes in different patterns. Comparisons were first made among changes in expression of genes in testes of irradiated rats (group X); after suppression of FSH, LH, and testosterone (T; group XAF); and after addition of exogenous testosterone (group XAT) to determine the regulation of the probe based on testosterone/FSH/LH suppression and testosterone addition. Then, the genes in each pattern were subdivided according to their response to addition of exogenous FSH (group XAFF) as being either increased (Fs+) or decreased (Fs−). a Genes were counted only if they could be placed in the indicated categories by being present in the groups that were being compared or, if present in only one group, the values were significantly higher than those in the other group. b If the levels of LH, which were below the limit of detection of the assay, were actually further reduced in group XAT compared to group XAF, then no need exists to invoke regulation by ITT/FSH. c Any difference is either small or not significant. d Pattern not simply explained by hormonal changes and may involve complex, counterbalancing regulation by LH and ITT/FSH.
To distinguish genes specifically regulated by testosterone/FSH from those regulated by LH, these patterns were further subdivided depending on whether the expression increased (A), decreased (B), or remained unchanged (C) upon the replacement of flutamide with testosterone (group XAT). This substitution increased the levels of testosterone and FSH, whereas no detectable changes occurred in the level of LH (Fig. 2). The genes that showed changes in expression upon testosterone/FSH/LH suppression that were not even partially reversed by testosterone substitution were considered to be LH regulated. Because LH levels were at the limit of detection in the XAF and XAT groups (Fig. 2), the actual LH levels in these two groups were uncertain. If the LH levels were further reduced in group XAT, we would expect LH-regulated genes to show patterns IB or IIA (Fig. 3). However, if LH levels were unchanged between the XAF and XAT groups, we expect that LH-regulated genes would show patterns IC or IIC, but could show patterns IB or IIA if the genes were regulated in one direction by LH and in the opposite direction by testosterone or FSH. Hence, genes with the IB, IC, IIA, and IIC patterns of gene expression changes will be considered to be LH-regulated.
The genes with expression patterns IA and IIB were subdivided further into patterns in which this change upon treatment with testosterone (group XAT) was less than or equal to that induced by hormonal suppression (patterns IAa and IIBa) and those in which the change in expression was greater (patterns IAb and IIBb) (Fig. 3). Because this treatment only partially restored ITT and FSH but greatly elevated serum testosterone (Fig. 2), the hormonal regulation of genes with patterns IAa and IIBa should be through ITT or FSH levels, whereas the regulation of genes in groups IAb and IIBb should correlate with the serum levels of testosterone.
Comparison of the expression levels in the rats treated with the XAF regimen versus the XAFF regimen was used to identify genes in the various subgroups that were positively (Fs+) or negatively (Fs−) regulated by FSH, although they may also be regulated by other hormones besides FSH. In particular the hormonal regulation of the genes with patterns IAa and IIBa, which were already shown to be regulated by ITT or FSH, was further defined by the changes in gene expression when FSH was added (group XAFF) to the treatment with GnRH-ant plus flutamide (group XAF). Genes with patterns IAa or IIBa for which expression was changed by FSH addition in a manner similar to that of testosterone replacement were considered to be mainly regulated by FSH; all others were considered to be regulated by ITT.
Finally, some genes (pattern III) did not have their expression altered by testosterone/FSH/LH suppression. The expression of some of these was changed by the addition of testosterone (patterns IIIA and IIIB); however, their expression patterns cannot be explained simply by hormone changes. All the other genes (pattern IIIC after excluding FSH-regulated genes) were considered to be unaffected by all of these hormone manipulations.
Assignment of Genes to Different Hormonally Regulated Expression Patterns
The assignment of all genes to the various patterns is given in Supplemental Table S1.
When the genes with patterns IAa (positively regulated by ITT/FSH) or IIBa (negatively regulated by ITT/FSH) were further subdivided according to their regulation by FSH (Fig. 3), most were shown not to be regulated by the FSH levels and, hence, must be regulated by the ITT levels. Among the FSH-regulated genes with these two expression patterns, the direction of actual FSH regulation was nearly always consistent with the predicted assignment based on comparing groups X, XAF, and XAT. For instance, all of the 14 FSH-regulated genes in group IAa were positively regulated by FSH (Fs+), and of the 30 FSH-regulated genes in group IIBa, 28 were negatively regulated (Fs−).
In contrast to the large number of genes with patterns IAa and IIBa, only one gene had pattern IAb, and none had pattern IIBb. This emphasizes that the testosterone regulation of gene expression was not correlated with the serum testosterone levels but, rather, with the ITT levels, as indicated earlier.
Among the putative LH-regulated genes, most showed no significant changes when testosterone was substituted for flutamide in the treatment regimen, and 408 were assigned to pattern IC (upregulated by LH) and 342 to IIC (downregulated by LH). A small number showed further changes in the same direction when testosterone was restored as when all hormones were suppressed (patterns IB and IIA).
A total of 104 positively FSH-regulated and 391 negatively FSH-regulated genes were identified by comparing the expression levels in the rats in group XAFF with those in group XAF (Supplemental Table S1, column “Exp. Change Fs”). Most of these genes fell into seven of the 11 patterns described in Figure 3. However, some could not fit into the patterns described above, because they were not expressed in one or more treatment group.
Appreciable numbers of genes showed some weak but complex hormonal regulation according to patterns IIIA (211 genes) and IIIB (184 genes), but the changes in expression were only, at most, twofold. However, the major portion (9166 genes) of the gene entries was not regulated at all by hormones.
Hormonally Regulated Genes
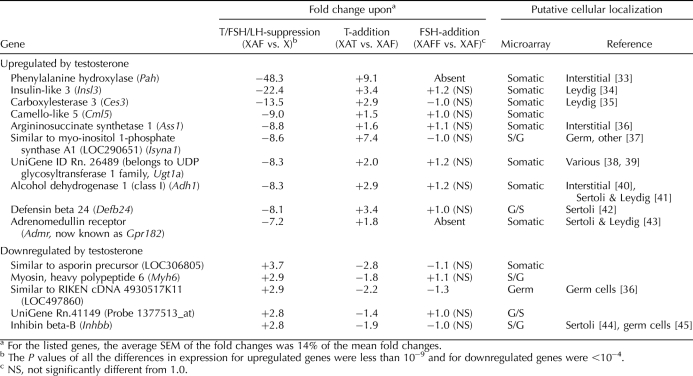
Among the genes regulated by the intratesticular levels of testosterone, not only were more genes positively (>240 genes) regulated than negatively (<120 genes) regulated (Fig. 3), the expression-fold changes were more dramatic for the positively regulated ones (Table 2) [33–45]. The 10 genes that were most strongly upregulated by ITT had at least a sevenfold decrease in expression upon testosterone/FSH/LH suppression, whereas none of the genes downregulated by ITT showed more than a 3.7-fold change. Unfortunately, we were unable to assay for the well-known testosterone-regulated gene, Rhox5 [46], because no probe is available for it on the rat microarray and, furthermore, its androgen-dependent proximal promoter is inactive in irradiated rat testes [47].
TABLE 2.
Genes that are maximally regulated by testosterone.
Among those genes that were the most strongly downregulated by testosterone levels, the only one that has been well characterized in the testis is Inhbb, which codes for the βB chain of inhibin B and activin B and is expressed in Sertoli cells. The RIKEN cDNA 4930517K11 is a not-yet-characterized transcribed sequence, which is highly expressed primarily in germ cells. Three additional germ cell-specific genes—Ret, testis-specific cytochrome c (Cyct), and synaptonemal complex protein 3 (Sycp3)—also show a twofold or greater downregulation by testosterone (Supplemental Table S1). We propose that it was the increased number and differentiation of these germ cells after 2 wk of suppression of ITT levels that made it appear these genes had been downregulated by the testosterone levels in the irradiated testes. This phenomenon further reduces the number of genes actually downregulated by testosterone.
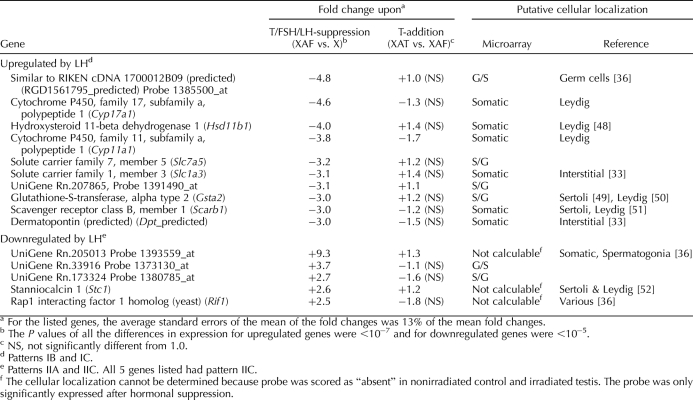
The changes in gene expression resulting from LH (less than fivefold upregulation) (Table 3) [33, 36, 48–52] were less dramatic than the changes resulting from testosterone (Table 2). In contrast to the many well-known genes, including Cyp17a1, Hsd11b1, and Cyp11a1, that were upregulated by LH, the LH-downregulated genes were more weakly expressed, and many were not identifiable.
TABLE 3.
Genes that are maximally regulated by LH.
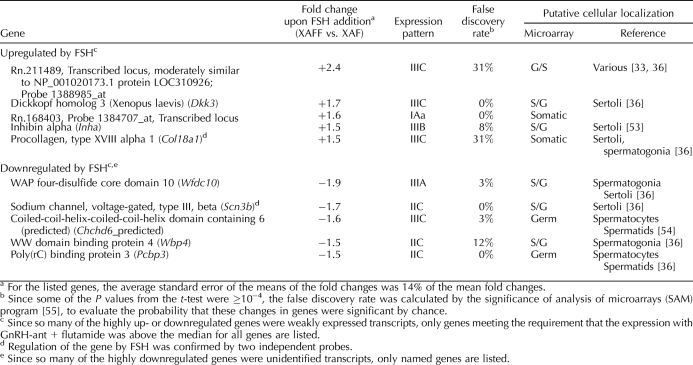
The changes in expression of genes attributable to FSH were even more modest than the regulation observed with LH, with no genes showing upregulation by more than 2.5-fold or downregulation by more than 1.9-fold (Table 4 and Supplemental Table S1, column “Fs/F or −F/Fs”) [33, 36, 53–55]. Several of these (Dkk3, Inha, Col18a1, Wfdc10, and Scn3b) were primarily expressed in Sertoli cells (Table 4). Among the FSH-downregulated genes were also an appreciable number of germ cell genes, such as Chchd6, Pcbp3 (Table 4), Ret, Cyct, and Sycp3. As was the case with the apparent testosterone downregulation of germ-cell genes described earlier, the apparent FSH downregulation of such genes may also have been a result of the enhancement of the block in spermatogonial differentiation caused by addition of FSH. To demonstrate that the apparent LH levels in the FSH-treated rats were not biologically significant, we examined the 10 most strongly LH-upregulated genes for changes in expression after FSH treatment. Just one, Dpt, showed even a marginal increase, and that occurred only with one of the two probes for this gene on the microarray.
TABLE 4.
Moderate–strongly expressed genes that are maximally regulated by FSH.
In contradistinction to the hormonally regulated genes described above, the majority of the genes were quite stable across the different treatment groups (pattern IIIC excluding Fs+ and Fs–) (Fig. 3 and Supplemental Table S1, columns “Exp. Pattern” and “Exp. Change Fs”). Among the most highly expressed of these genes were the housekeeping genes beta-actin (Actb) and the ribosomal proteins, including Rps2; hence, these are appropriate for use as the internal controls in real-time RT-PCR comparisons of the gene expression changes with the different hormonal treatments. In addition, several strongly expressed Sertoli cell-specific genes, clusterin (Clu) and claudin 11 (Cldn11), cathepsin L (Ctsl), and Wilms tumor 1 (Wt1), were not hormonally regulated.
Confirmation of Array Data by Real-Time RT-PCR
Up to three genes (most of which were chosen because they have reported roles in testis functions) from each hormone-regulated expression pattern from the microarrays, except for patterns IAb, IIA, and IIBb, which contain no genes or only one gene, were tested by real-time PCR (Supplemental Fig. S2). In general, the real-time PCR results agreed well with the array data. In only two of these instances did the PCR results put a gene into a different category than the microarray did. The real-time PCR results for Igfbp3 showed that it was increased significantly with GnRH-ant plus flutamide treatment and decreased significantly by testosterone addition and, therefore, should have pattern IIB, whereas array data did not detect these differences as being significant, thus assigning it to pattern IIIC. On the other hand, the microarray results indicated that Timp1 was increased significantly with the addition of FSH, but the real-time data did not confirm that the increase was significant.
The microarrays also identified a set of genes known to be activated by liver X receptors (LXR) [56] that were found to have more than a 10-fold increase in expression after irradiation (for details, see Discussion). Real-time PCR (Supplemental Table S4) confirmed the microarray observations of these increases.
DISCUSSION
In the present study, we used irradiated rat testes as a model to investigate the changes in gene expression profiles upon hormonal modulation and radiation-induced germ cell depletion. By comparing normal rat testes with irradiated rat testes, many germ cell-specific and somatic cell-enriched genes could be identified. Also, somatic genes that were highly upregulated as a result of irradiation could be distinguished. Most importantly, genes in somatic cells that were regulated by testosterone, LH or FSH, respectively, could be identified in this unique model, which contains almost exclusively somatic cells. In particular, we analyzed the genes that were strongly regulated by testosterone (patterns IAa and IIBa) to identify factors critical for the recovery of spermatogonial differentiation after irradiation. It should be noted, however, that differences in somatic cell gene expression exist between the irradiated and the nonirradiated testis (Table 1) [47], and that these differences may affect the response of specific genes to hormonal regulation. Therefore, the possibility that some genes identified in this model may display different hormonal regulation in the normal testis needs to be considered.
Distinguishing Germ Cell-Specific and Somatic Cell-Enriched Genes
This method of distinguishing germ cell-specific and somatic cell-enriched genes appeared to be quite useful and was generally accurate. Of the 10 most highly expressed germ-cell genes listed in Supplemental Table S3, eight (Gsg1, Oaz3, Odf2, Smcp, Odf1, Tnp1, Crisp2, and Tnp2) are all well-known male germ cell-specific genes, and Hspb9 has also been shown to be germ-cell specific [57]. Cmtm2a (previously known as Cklfsf1) is highly expressed in spermatocytes [58], and the data of the current study now indicate it to be germ-cell specific.
We further compared our array results with published data using the same microarray system that was previously used to distinguish genes expressed in Sertoli cells, spermatogonia plus early spermatocytes, pachytene spermatocytes, and round spermatids [59]. Among the 797 probe sets (probe sets instead of UniGene entries are counted for easier comparison with other arrays) that were defined as germ-cell specific based on the 10-fold reduction after irradiation (Supplemental Table S1), 749 (94%) of them had at least fourfold higher expression levels in one of the four germ cell types than in Sertoli cells [59] and, hence, appear to be germ-cell genes. The genes with lower fold-decreases after irradiation (4- to 10-fold) were also mostly germ-cell genes. Of the 812 probe sets within this range, 614 (76%) were considered to be germ-cell genes based on the enrichment of expression compared to Sertoli cells.
Originally, one limitation of this analysis was that it could not, on its own, distinguish between somatic cell-enriched genes downregulated by irradiation [47] and germ cell-specific genes. However, we can now determine some somatic cell-enriched genes that were downregulated by combining the present results with the above-mentioned database on gene expression in separated seminiferous tubule cells [59]. The cell separation studies show that two genes, Prnd (prion protein 2 probe set 1384924_at) and MGC114529 (similar to melanoma antigen family A, 10, probe 1382906_at) are highly expressed in Sertoli cells but absent in germ cells; therefore, these should be somatic cell-enriched genes that were downregulated upon irradiation. Among the 10 most strongly expressed somatic cell-enriched genes (Supplemental Table S3), our localization of most of them is supported by published reports using immunohistochemistry, in situ hybridization, separated cells, or other array data using different separated testicular cell types [33, 36, 49, 54, 60].
In addition, the somatic genes that were upregulated by irradiation were identified (Table 1). The three most strongly upregulated ones—RGD:708447 (previously known as Testin, GenBank accession no. NM_173132), Wnt4, and Tnfrsf1a—are all expressed in Sertoli cells [49, 61, 62], with RGD:708447 being specifically expressed in these cells. They have important roles in the testis, because RGD:708447 is a component of Sertoli-cell junctional complexes [63], Wnt4 is important for sex-specific vasculature formation and steroidogenic cell recruitment during gonad development [64], and Tnfrsf1a, the receptor for tumor necrosis factor alpha, is involved in Sertoli-cell mediation of toxicant-induced germ-cell apoptosis [65].
The greatly increased expression of these somatic genes after irradiation cannot be explained simply by cellular composition changes and loss of mass of irradiated rat testis. Rather, it must be caused by additional effects induced by irradiation, such as 1) germ cell depletion or interruption of germ cell-Sertoli cell junctions; 2) changes in levels of hormones, such as ITT, LH, and FSH, which are all elevated in irradiated rats (Fig. 2); or 3) a direct radiation effect. Germ cell depletion or interruption of germ cell-Sertoli cell junctions is, for example, known to upregulate RGD:708447 [66]. Although others have shown that testosterone upregulates RGD:708447 [67] and FSH upregulates Tnrfsf1a expression [68] in Sertoli cells, we showed here that even after maximal correction for the upregulation from increased hormone levels (Table 1), these genes were still highly upregulated after irradiation, indicating nonhormonal mechanisms must also be involved. In contrast, the increase in hormone levels may have contributed to a significant part of the increase in Ogn expression (Table 1).
Our microarray data also identified possible cases of coregulation of genes that may be more direct results of irradiation. For example, Ctsa (also known as Ppgb) and Pltp, which were both upregulated more than 40-fold (Table 1), are adjacent on rat chromosome 3q42. Further investigation of the upregulation of Pltp, which is a direct target gene of LXRs [56], by IPA and real-time RT-PCR, revealed that multiple LXR-target genes, including Apoe, Abca1, Lpl, Cd36, and Abcg1 [69], were upregulated more than 10-fold by irradiation of rat testes (Supplemental Fig. S3 and Supplemental Table S4). This indicates that the LXR/retinoid X receptor (RXR) pathway is activated in irradiated rat tests. IPA analysis also showed the upregulation of genes that are involved in complement system initiation (Supplemental Fig. S4), indicating the activation of immune function in irradiated rat testes.
Hormonal Regulation of Gene Expression
We were indeed able to dissect the distinct effects of testosterone, LH, and FSH on gene expression in our irradiated rat model. It should be noted, however, that because the tissue was harvested after 2 wk of continuous hormonal changes, the observed regulation of genes could have resulted from both direct and indirect actions, which is likely to mimic the in vivo situation more than acute hormone modulation. These results revealed that ITT was the most important regulator of somatic cell gene expression, that LH was also an appreciable gene regulator, but that FSH produced minimal changes. This sequence correlates with the severity of reproductive phenotypes, which were greatest in AR-null mice, intermediate in LH receptor-null mice, and minimal in FSH receptor-null mice [6, 70, 71].
Three genes (Pah, Insl3, and Ces3) showed very strong upregulation by testosterone (Table 2). One of these genes, Insl3, is known to have an important role in the testis and controls testis descent to the inguinal area during male embryonic development [72]. Among the genes that were strongly upregulated by testosterone, most were enriched in testis interstitial cells, and at least two are already known to be localized in Leydig cells (Table 2). This suggests that the Leydig cell is quantitatively the main target of androgen regulation within the testis. However, except for Gpr182 (previously known as Admr), which has several potential androgen-response elements in the 5′ promoter [73], no androgen-response elements have been reported in the promoters of the other genes; hence, it is likely that they are indirectly upregulated by testosterone.
Evidence in the literature supports some of the changes in gene expression induced by testosterone (Table 2). Androgen upregulates the expression of Ces3 in adipose tissues [74] and some members of UDP glycosyltransferase 1 family, such as Ugt1a1, in Sertoli cells [38]. Similarly, the presence of AR in peritubular myoid cells is reported to maintain the expression levels of Gpr182 [73]. Furthermore, the increased levels of Inhbb mRNA when Leydig cells are eliminated with ethane dimethane sulfonate [75] are consistent with our observation that these levels were downregulated by testosterone. Two of the genes (Pex11a and Ctgf) that have been reported to be acutely regulated by testosterone in hpg mouse testes [15] were regulated in the same directions in irradiated rats (Supplemental Table S1). In addition, several genes, including Spinlw1 (previously known as Eppin) and Cdo1, that were upregulated by testosterone in irradiated rats (Supplemental Table S1) were also chronically upregulated by the presence of AR in control as opposed to Sertoli-cell AR knockout mice [14]; thus, the testosterone regulation of these genes most likely occurred in the Sertoli cell.
Our results also showed some changes in gene expression that were at variance with some published reports or their interpretations. Several studies have drawn the conclusion that Insl3 is under the control of LH because of its significantly reduced expression levels after treatments that included GnRH-ant [76, 77]. However, these treatments also decreased ITT. Our experiments, which include separate treatment groups (XAF and XAT), showed that Insl3 was quite clearly regulated by testosterone (Table 2) and not by LH, which was undetectable in both treatment groups (Fig. 2) [78]. Recently it has been confirmed that testosterone does increase Insl3 mRNA levels in Leydig cells [79]. Similarly, it has been reported that LH restores the Isyna1 activity in testes of hypophysectomized rats [80]; our data further clarify that it is actually testosterone, not LH, that regulates Isyna1. Whereas others have reported that Adh1 expression was lower in wild-type than in either AR-null or hypomorphic mice, and that testosterone treatment did not induce any changes in Adh1 expression levels in hpg mice [12, 13], our array data (Table 2), using two different two-probe sets, strongly indicated that in rats, Adh1 was upregulated by testosterone, in turn indicating a very significant interspecies difference.
The LH-upregulated genes listed in Table 3 showed a bias toward those involved in steroidogenic pathways. Cyp17α1 and Cyp11α1 are in the testosterone biosynthesis pathway, Hsd11b1 is involved in corticosteroid metabolism [48], and Scarb1 mediates the uptake of cholesterol esters into the cells [81]. In addition, our assay showed other steroidogenic-related genes, including Star, which is responsible for cholesterol intake in mitochondria; Alas1, which catalyzes the first step of the heme biosynthesis pathway for mitochondrial cytochromes [82]; and Fdxr, which transfers electrons to CYP11A1 [83], to be LH upregulated (Supplemental Table S1). Previous studies have also shown that LH can upregulate Cyp17a1 [84], Cyp11a1 [85], Scarb1 [86], Star [87], and Alas1 [82] and downregulate Stc1 [52], supporting the validity of our LH-regulated gene list. Furthermore, our LH-upregulated genes were predominantly ones expressed in the Leydig cells, or at least localized to the interstitial compartment, which is consistent with localization of the LH receptor on the Leydig cells. IPA analysis revealed that of 68 LH-regulated genes involved in lipid and steroid metabolism, 61 were upregulated by LH, and only seven were downregulated by LH (data not shown), supporting the qualitative observation that these genes were almost exclusively in the LH-upregulated category.
Although 495 genes were apparently regulated by FSH in our array analysis using a t-test, the false-discovery rates indicated that some were most likely valid but that others could have been identified by chance (Table 4). We have confirmed the upregulation of Star by real-time RT-PCR, and the indications of FSH-upregulation of Star, Inha, Pde4b, Wfdc1, and Sod3 and downregulation of Foxq1 and Map3k1 (Table 4 and Supplemental Table S1) were supported by independent microarray analyses reported in the literature [9, 10, 16]. We note that in the present study, the fold-changes of their expression were smaller than previously reported. This is likely a result of our 2-wk treatment compared to shorter-term exposures in the other studies, because even in those shorter-term assays, many of the genes that were acutely regulated by FSH already showed declines in their changes in expression within 24 h [9]. It is also possible that the failure to produce larger changes in gene expression in our study was a result of the remaining levels of FSH in the GnRH-ant-treated rats.
In conclusion, using the irradiated rat testis, the present study successfully identified distinct genes that are regulated by testosterone, by LH, and by FSH in a single experimental model. The results clearly show that the hormones induce much greater upregulation than downregulation of gene expression. Future studies of separated cells and histological material are needed to determine the cell types responsible for the gene profile changes. The data shown here and in the Supplemental Materials also will be useful for documenting the in vivo hormonal regulation of other known genes and for further gene and pathway discovery. Many of these genes and pathways will likely be involved in the hormonal regulation of normal spermatogenesis. In this particular model system, the knowledge of radiation-induced and testosterone-dependent changes in gene expression will help us to elucidate the genes responsible for the spermatogonial block in irradiated rat testis and its reversal by hormone manipulation [2].
Supplementary Material
Acknowledgments
We thank Mr. Kuriakose Abraham for the histological preparations and Mr. Walter Pagel for editorial advice. Special thanks to Dr. Miles Wilkinson for comments on the manuscript. We also thank Ms. Tarja Laiho for the skillful assistance in performing gonadotropin assays and Dr. Mini Kapoor and Ms. Sarah Rizvi for performing the microarray procedures. We sincerely thank Drs. R.P. Blye and Hyun K. Kim (Contraceptive Development Branch, National Institute of Child Health and Human Development) and Dr. Sheri Hild (Bioqual, Inc.) for providing the acyline and Dr. A.F. Parlow (National Hormone and Peptide Program) for providing recombinant hFSH.
Footnotes
Supported by NIH research grants ES-08075 (to M.L.M.) and HD-16229 (to Joanne Richards, Baylor College of Medicine, Houston, TX), training grant T32 HD-07324, Cancer Center Support Grant CA-16672, and Cooperative Centers Program in Reproduction Grant HD-07495.
These authors contributed equally to this work.
REFERENCES
- Meachem SJ, McLachlan RI, Stanton PG, Robertson DM, Wreford NG.FSH immunoneutralization acutely impairs spermatogonial development in normal adult rats. J Androl 1999; 20: 756–762. [PubMed] [Google Scholar]
- Meistrich ML, Shetty G.Inhibition of spermatogonial differentiation by testosterone. J Androl 2003; 24: 135–148. [DOI] [PubMed] [Google Scholar]
- Shetty G, Weng CC, Meachem SJ, Bolden-Tiller OU, Zhang Z, Pakarinen P, Huhtaniemi I, Meistrich ML.Both testosterone and FSH independently inhibit spermatogonial differentiation in irradiated rats. Endocrinology 2006; 147: 472–482. [DOI] [PubMed] [Google Scholar]
- Ultee-van Gessel AM, Timmerman MA, de Jong FH.Effects of treatment of neonatal rats with highly purified FSH alone and in combination with LH on testicular function and endogenous hormone levels at various ages. J Endocrinol 1988; 116: 413–420. [DOI] [PubMed] [Google Scholar]
- Duckett RJ, Wreford NG, Meachem SJ, McLachlan RI, Hedger MP.Effect of chorionic gonadotropin and flutamide on Leydig cell and macrophage populations in the testosterone-estradiol-implanted adult rat. J Androl 1997; 18: 656–662. [PubMed] [Google Scholar]
- Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, Xing L, Boyce BF, et al. Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci U S A 2002; 99: 13498–13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhtaniemi I.Mutations along the pituitary-gonadal axis affecting sexual maturation: novel information from transgenic and knockout mice. Mol Cell Endocrinol 2006; 7: 254–255. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shao S, Meistrich M.The radiation-induced block in spermatogonial differentiation is due to damage to the somatic environment, not the germ cells. J Cell Physiol 2007; 211: 149–158. [DOI] [PubMed] [Google Scholar]
- McLean DJ, Friel PJ, Pouchnik D, Griswold MD.Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone-treated rat Sertoli cells. Mol Endocrinol 2002; 16: 2780–2792. [DOI] [PubMed] [Google Scholar]
- Meachem SJ, Ruwanpura SM, Ziolkowski J, Ague JM, Skinner MK, Loveland KL.Developmentally distinct in vivo effects of FSH on proliferation and apoptosis during testis maturation. J Endocrinol 2005; 186: 429–446. [DOI] [PubMed] [Google Scholar]
- Petrusz P, Jeyaraj DA, Grossman G.Microarray analysis of androgen-regulated gene expression in testis: the use of the androgen-binding protein (ABP)-transgenic mouse as a model. Reprod Biol Endocrinol 2005; 3: 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Abel M, Charlton HM, Hu B, Johnston H, Baker PJ.Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive Tfm mouse testis. Endocrinology 2007; 148: 2914–2924. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Shima JE, Connolly CM, Sharma M, Holdcraft RW, Griswold MD, Braun RE.Transcriptional profiling of androgen receptor (AR) mutants suggests instructive and permissive roles of AR signaling in germ cell development. Mol Endocrinol 2007; 21: 895–907. [DOI] [PubMed] [Google Scholar]
- Denolet E, De Gendt K, Allemeersch J, Engelen K, Marchal K, Van Hummelen P, Tan KA, Sharpe RM, Saunders PT, Swinnen JV, Verhoeven G.The effect of a Sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Mol Endocrinol 2006; 20: 321–334. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD.Identification of testosterone-regulated genes in testes of hypogonadal mice using oligonucleotide microarray. Mol Endocrinol 2004; 18: 422–433. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD.Follicle-stimulating hormone induced changes in gene expression of murine testis. Mol Endocrinol 2004; 18: 2805–2816. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Shima JE, Nie R, Friel PJ, Griswold MD.Androgen-regulated transcripts in the neonatal mouse testis as determined through microarray analysis. Biol Reprod 2005; 72: 1010–1019. [DOI] [PubMed] [Google Scholar]
- Strothmann K, Simoni M, Mathur P, Siakhamary S, Nieschlag E, Gromoll J.Gene expression profiling of mouse Sertoli cell lines. Cell Tissue Res 2004; 315: 249–257. [DOI] [PubMed] [Google Scholar]
- Kangasniemi M, Huhtaniemi I, Meistrich ML.Failure of spermatogenesis to recover despite the presence of A spermatogonia in the irradiated LBNF1 rat. Biol Reprod 1996; 54: 1200–1208. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Shetty G, Bolden-Tiller OU, Porter KL.Hormones and spermatogonial development. Skinner MK, Griswold MD.Sertoli Cell Biology San Diego:Elsevier Academic Press;2005: 437–448. [Google Scholar]
- Meistrich ML, Kangasniemi M.Hormone treatment after irradiation stimulates recovery of rat spermatogenesis from surviving spermatogonia. J Androl 1997; 18: 80–87. [PubMed] [Google Scholar]
- Sinha Hikim AP, Swerdloff RS.Temporal and stage-specific changes in spermatogenesis of rat after gonadotropin deprivation by a potent gonadotropin-releasing hormone antagonist treatment. Endocrinology 1993; 133: 2161–2170. [DOI] [PubMed] [Google Scholar]
- Shuttlesworth GA, de Rooij DG, Huhtaniemi I, Reissmann T, Russell LD, Shetty G, Wilson G, Meistrich ML.Enhancement of A spermatogonial proliferation and differentiation in irradiated rats by GnRH antagonist administration. Endocrinology 2000; 141: 37–49. [DOI] [PubMed] [Google Scholar]
- Porter KL, Shetty G, Meistrich ML.Testicular edema is associated with spermatogonial arrest in irradiated rats. Endocrinology 2006; 147: 1297–1305. [DOI] [PubMed] [Google Scholar]
- Shetty G, Wilson G, Huhtaniemi I, Shuttlesworth GA, Reissmann T, Meistrich ML.Gonadotropin-releasing hormone analogs stimulate and testosterone inhibits the recovery of spermatogenesis in irradiated rats. Endocrinology 2000; 141: 1735–1745. [DOI] [PubMed] [Google Scholar]
- Porter KL, Shetty G, Shuttlesworth G, Weng CY, Huhtaniemi I, Pakarinen P, Meistrich ML.Estrogen enhances recovery from radiation-induced spermatogonial arrest in rat testes. J Androl 2009; 30: 440–451. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, van Beek MEAB.Spermatogonial stem cells: Assessing their survival and ability to produce differentiated cells. Chapin RE, Heindel J.Methods in Toxicology, vol. 3A New York:Academic Press;1993: 106–123. [Google Scholar]
- Haavisto A, Pettersson K, Bergendahl M, Perheentupa A, Roser JF, Huhtaniemi I.A supersensitive immunofluorometric assay for rat luteinizing hormone. Endocrinology 1993; 132: 1687–1691. [DOI] [PubMed] [Google Scholar]
- Hakola K, Van der Boogaart P, Mulders J, de Leeuw R, Schoonen W, Van Heyst J, Swolfs A, Van Casteren J, Huhtaniemi I, Kloosterboer H.Recombinant rat follicle-stimulating hormone: production by Chinese hamster ovary cells, purification and functional characterization. Mol Cell Endocrinol 1997; 127: 59–69. [DOI] [PubMed] [Google Scholar]
- Meachem SJ, McLachlan RI, de Kretser DM, Robertson DM, Wreford NG.Neonatal exposure of rats to recombinant follicle stimulating hormone increases adult Sertoli and spermatogenic cell numbers. Biol Reprod 1996; 54: 36–44. [DOI] [PubMed] [Google Scholar]
- Simoni M, Tuttelmann F, Michel C, Bockenfeld Y, Nieschlag E, Gromoll J.Polymorphisms of the luteinizing hormone/chorionic gonadotropin receptor gene: association with maldescended testes and male infertility. Pharmacogenet Genomics 2008; 18: 193–200. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Wilson G, Shuttlesworth G, Huhtaniemi I, Reissmann T.GnRH agonists and antagonists stimulate recovery of fertility in irradiated LBNF1 rats. J Androl 2001; 22: 809–817. [PubMed] [Google Scholar]
- Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, Garbers DL.Defining the spermatogonial stem cell. Dev Biol 2004; 269: 393–410. [DOI] [PubMed] [Google Scholar]
- Adham IM, Burkhardt E, Benahmed M, Engel W.Cloning of a cDNA for a novel insulin-like peptide of the testicular Leydig cells. J Biol Chem 1993; 268: 26668–26672. [PubMed] [Google Scholar]
- von Deimling O, Ronai A, de Looze S.Nonspecific esterases of mammalian testis. Comparative studies on the mouse (Mus musculus) and rat (Rattus norvegicus). Histochemistry 1985; 82: 547–555. [DOI] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jegou B, Primig M.The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci U S A 2007; 104: 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin TR, Griswold MD.Characterization of the expression and regulation of genes necessary for myo-inositol biosynthesis and transport in the seminiferous epithelium. Biol Reprod 2004; 70: 744–751. [DOI] [PubMed] [Google Scholar]
- Magnanti M, Giuliani L, Gandini O, Gazzaniga P, Santiemma V, Ciotti M, Saccani G, Frati L, Agliano AM.Follicle-stimulating hormone, testosterone, and hypoxia differentially regulate UDP-glucuronosyltransferase 1 isoforms expression in rat Sertoli and peritubular myoid cells. J Biochem Biophys Methods 2000; 74: 149–155. [DOI] [PubMed] [Google Scholar]
- Brands A, Munzel PA, Bock KW.In situ hybridization studies of UDP-glucuronosyltransferase UGT1A6 expression in rat testis and brain. Biochem Pharmacol 2000; 59: 1441–1444. [DOI] [PubMed] [Google Scholar]
- Vonesch JL, Nakshatri H, Philippe M, Chambon P, Dolle P.Stage and tissue-specific expression of the alcohol dehydrogenase 1 (Adh-1) gene during mouse development. Dev Dyn 1994; 199: 199–213. [DOI] [PubMed] [Google Scholar]
- Deltour L, Haselbeck RJ, Ang HL, Duester G.Localization of class I and class IV alcohol dehydrogenases in mouse testis and epididymis: potential retinol dehydrogenases for endogenous retinoic acid synthesis. Biol Reprod 1997; 56: 102–109. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Matsui Y.Testis-specific expression of a novel mouse defensin-like gene, Tdl. Mech Dev 2002; 116: 217–221. [DOI] [PubMed] [Google Scholar]
- Li YY, Hwang IS, O WS, Tang F.Adrenomedullin peptide: gene expression of adrenomedullin, its receptors and receptor activity modifying proteins, and receptor binding in rat testis—actions on testosterone secretion. Biol Reprod 2006; 75: 183–188. [DOI] [PubMed] [Google Scholar]
- Kaipia A, Penttila TL, Shimasaki S, Ling N, Parvinen M, Toppari J.Expression of inhibin bA and bB, follistatin and activin-A receptor messenger ribonucleic acids in the rat seminiferous epithelium. Endocrinology 1992; 131: 2703–2710. [DOI] [PubMed] [Google Scholar]
- Buzzard JJ, Loveland KL, O'Bryan MK, O'Connor AE, Bakker M, Hayashi T, Wreford NG, Morrison JR, de Kretser DM.Changes in circulating and testicular levels of inhibin A and B and activin A during postnatal development in the rat. Endocrinology 2004; 145: 3532–3541. [DOI] [PubMed] [Google Scholar]
- Lindsey SJ, Wilkinson M.A testosterone- and LH-regulated homeobox gene expressed in mouse Sertoli cells and epididymis. Dev Biol 1996; 179: 471–484. [DOI] [PubMed] [Google Scholar]
- Maiti S, Meistrich ML, Wilson G, Shetty G, Marcelli M, McPhaul MJ, Morris PL, Wilkinson MF.Irradiation selectively inhibits expression from the androgen-dependent Pem homeobox gene promoter in Sertoli cells. Endocrinology 2001; 142: 1567–1577. [DOI] [PubMed] [Google Scholar]
- Gomez-Sanchez EP, Romero DG, de Rodriguez AF, Warden MP, Krozowski Z, Gomez-Sanchez CE.Hexose-6-phosphate dehydrogenase and 11beta-hydroxysteroid dehydrogenase-1 tissue distribution in the rat. Endocrinology 2008; 149: 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD.The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319–330. [DOI] [PubMed] [Google Scholar]
- Rabahi F, Brule S, Sirois J, Beckers JF, Silversides DW, Lussier JG.High expression of bovine alpha glutathione S-transferase (GSTA1, GSTA2) subunits is mainly associated with steroidogenically active cells and regulated by gonadotropins in bovine ovarian follicles. Endocrinology 1999; 140: 3507–3517. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Nagaosa K, Hirose T, Tsuda K, Hasegawa K, Shiratsuchi A, Nakanishi Y.Expression and function of class B scavenger receptor type I on both apical and basolateral sides of the plasma membrane of polarized testicular Sertoli cells of the rat. Dev Growth Differ 2004; 46: 283–298. [DOI] [PubMed] [Google Scholar]
- Li L, Wong CK.Effects of dexamethasone and dibutyryl cAMP on stanniocalcin-1 mRNA expression in rat primary Sertoli and Leydig cells. Mol Cell Endocrinol 2008; 283: 96–103. [DOI] [PubMed] [Google Scholar]
- Cuevas P, Ying SY, Ling N, Ueno N, Esch F, Guillemin R, Healy D, Ta S.Immunohistochemical detection of inhibin in the gonad. Biochem Biophys Res Commun 1987; 142: 23–30. [DOI] [PubMed] [Google Scholar]
- Schlecht U, Demougin P, Koch R, Hermida L, Wiederkehr C, Descombes P, Pineau C, Jegou B, Primig M.Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol Biol Cell 2004; 15: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G.Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Beyer TP, Yang XP, Schmidt RJ, Zhang Y, Bensch WR, Kauffman RF, Gao H, Ryan TP, Liang Y, Eacho PI, Jiang XC.Phospholipid transfer protein is regulated by liver X receptors in vivo. J Biol Chem 2002; 277: 39561–39565. [DOI] [PubMed] [Google Scholar]
- Kappe G, Verschuure P, Philipsen RL, Staalduinen AA, Van de Boogaart P, Boelens WC, De Jong WW.Characterization of two novel human small heat shock proteins: protein kinase-related HspB8 and testis-specific HspB9. Biochim Biophys Acta 2001; 1520: 1–6. [DOI] [PubMed] [Google Scholar]
- Wang L, Wu C, Zheng Y, Qiu X, Wang L, Fan H, Han W, Lv B, Wang Y, Zhu X, Xu M, Ding P, et al. Molecular cloning and characterization of chemokine-like factor superfamily member 1 (CKLFSF1), a novel human gene with at least 23 alternative splicing isoforms in testis tissue. Int J Biochem Cell Biol 2004; 36: 1492–1501. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Wright WW, Dicandeloro P, Wilson E, Kopf GS, Jelinsky SA.Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc Natl Acad Sci U S A 2008; 105: 8315–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Zhi Y, Finger JN, Kopf GS, Wright WW.Identification of testis-specific male contraceptive targets: insights from transcriptional profiling of the cycle of the rat seminiferous epithelium and purified testicular cells. Ann N Y Acad Sci 2007; 1120: 36–46. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Grima J, Stahler MS, Lockshin RA, Bardin CW.Testins are structurally related Sertoli cell proteins whose secretion is tightly coupled to the presence of germ cells. J Biol Chem 1989; 264: 21386–21393. [PubMed] [Google Scholar]
- De SK, Chen HL, Pace JL, Hunt JS, Terranova PF, Enders GC.Expression of tumor necrosis factor-alpha in mouse spermatogenic cells. Endocrinology 1993; 133: 389–396. [DOI] [PubMed] [Google Scholar]
- Zong SD, Bardin CW, Phillips D, Cheng CY.Testins are localized to the junctional complexes of rat Sertoli and epididymal cells. Biol Reprod 1992; 47: 568–572. [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K, Hoyle C, Brennan J, Dandonneau M, Alldus G, Capel B, Swain A.Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development 2003; 130: 3663–3670. [DOI] [PubMed] [Google Scholar]
- Yao PL, Lin YC, Sawhney P, Richburg JH.Transcriptional regulation of FasL expression and participation of sTNF-alpha in response to Sertoli cell injury. J Biol Chem 2007; 282: 5420–5431. [DOI] [PubMed] [Google Scholar]
- Grima J, Cheng CY.Testin induction: the role of cyclic 3′,5′-adenosine monophosphate/protein kinase A signaling in the regulation of basal and lonidamine-induced testin expression by rat Sertoli cells. Biol Reprod 2000; 63: 1648–1660. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Bardin CW.Identification of two testosterone-responsive testicular proteins in Sertoli-cell enriched culture medium whose secretion is suppressed by cells of the intact seminiferous tubule. J Biol Chem 1987; 262: 12768–12779. [PubMed] [Google Scholar]
- Mauduit C, Besset V, Caussanel V, Benahmed M.Tumor necrosis factor alpha receptor p55 is under hormonal (follicle-stimulating hormone) control in testicular Sertoli cells. Biochem Biophys Res Commun 1996; 224: 631–637. [DOI] [PubMed] [Google Scholar]
- Lundholm L, Moverare S, Steffensen KR, Nilsson M, Otsuki M, Ohlsson C, Gustafsson JA, Dahlman-Wright K.Gene expression profiling identifies liver X receptor alpha as an estrogen-regulated gene in mouse adipose tissue. J Mol Endocrinol 2004; 32: 879–892. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Poutanen M, Wilbertz J, Huhtaniemi I.Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice. Mol Endocrinol 2001; 15: 172–183. [DOI] [PubMed] [Google Scholar]
- Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, Sassone-Corsi P.Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc Natl Acad Sci U S A 1998; 95: 13612–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nef S, Parada LF.Cryptorchidism in mice mutant for Insl3. Nat Genet 1999; 22: 295–299. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yeh S, Chen YT, Wu CC, Chuang KH, Lin HY, Wang RS, Chang YJ, Mendis-Handagama C, Hu L, Lardy H, Chang C.Oligozoospermia with normal fertility in male mice lacking the androgen receptor in testis peritubular myoid cells. Proc Natl Acad Sci U S A 2006; 103: 17718–17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc C, Yoshioka M, St-Amand J.Transcriptomic characterization of the long-term dihydrotestosterone effects in adipose tissue. Obesity (Silver Spring)2007; 15: 1107–1132. [DOI] [PubMed] [Google Scholar]
- Tena-Sempere M, Kero J, Rannikko A, Yan W, Huhtaniemi I.The pattern of inhibin/activin alpha- and betaB-subunit messenger ribonucleic acid expression in rat testis after selective Leydig cell destruction by ethylene dimethane sulfonate. Endocrinology 1999; 140: 5761–5770. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Kumagai J, Sudo S, Chun SY, Pisarska M, Morita H, Toppari J, Fu P, Wade JD, Bathgate RA, Hsueh AJ.Paracrine regulation of mammalian oocyte maturation and male germ cell survival. Proc Natl Acad Sci U S A 2004; 101: 7323–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne RA, Forster T, Burgess ST, Craigon M, Walton MJ, Baird DT, Ghazal P, Anderson RA.Molecular profiling of the human testis reveals stringent pathway-specific regulation of RNA expression following gonadotropin suppression and progestogen treatment. J Androl 2008; 29: 389–403. [DOI] [PubMed] [Google Scholar]
- Bolden-Tiller OU, Shetty G, Weng CC, Stivers DN, Meistich ML.Hormone-regulated somatic testicular cell genes correlating with blocked spermatogonial differentiation in irradiated rats. Biol Reprod 2005; Special Issue: 88 [Google Scholar]
- Lague E, Tremblay JJ.Antagonistic effects of testosterone and the endocrine disruptor mono-(2-ethylhexyl) phthalate on INSL3 transcription in Leydig cells. Endocrinology 2008; 149: 4688–4694. [DOI] [PubMed] [Google Scholar]
- Hasegawa R, Eisenberg F., JrSelective hormonal control of myo-inositol biosynthesis in reproductive organs and liver of the male rat. Proc Natl Acad Sci U S A 1981; 78: 4863–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RM, Jo Y, Leers-Sucheta S, Bose HS, Miller WL, Azhar S, Stocco DM.Differential regulation of steroid hormone biosynthesis in R2C and MA-10 Leydig tumor cells: role of SR-B1-mediated selective cholesteryl ester transport. Biol Reprod 2003; 68: 114–121. [DOI] [PubMed] [Google Scholar]
- Tofilon PJ, Piper WN.Inhibition of rat testicular heme synthesis and depression of microsomal cytochrome P-450 by estradiol benzoate. Biochem Pharmacol 1982; 31: 3667–3671. [DOI] [PubMed] [Google Scholar]
- Payne AH, Hales DB.Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev 2004; 25: 947–970. [DOI] [PubMed] [Google Scholar]
- Shan L, Hardy DO, Catterall JF, Hardy MP.Effects of luteinizing hormone (LH) and androgen on steady-state levels of messenger ribonucleic acid for LH receptors, androgen receptors, and steroidogenic enzymes in rat Leydig cell progenitors in vivo. Endocrinology 1995; 136: 1686–1693. [DOI] [PubMed] [Google Scholar]
- Pakarinen P, Vihko KK, Voutilainen R, Huhtaniemi I.Differential response of luteinizing hormone receptor and steroidogenic enzyme gene expression to human chorionic gonadotropin stimulation in the neonatal and adult rat testis. Endocrinology 1990; 127: 2469–2474. [DOI] [PubMed] [Google Scholar]
- Landschulz KT, Pathak RK, Rigotti A, Krieger M, Hobbs HH.Regulation of scavenger receptor, class B, type I, a high-density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J Clin Invest 1996; 98: 984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM.The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR). J Biol Chem 1994; 269: 28314–28322. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.