Abstract
• Background and Aims Sulfate assimilation is a pathway used by prokaryotes, fungi and photosynthetic organisms to convert inorganic sulfate to sulfide, which is further incorporated into carbon skeletons of amino acids to form cysteine or homocysteine. The pathway is highly regulated in a demand-driven manner; however, this regulation is not necessarily identical in various plant species. Therefore, our knowledge of the regulation of sulfate assimilation is reviewed here in detail with emphasis on different plant species.
• Scope Although demand-driven control plays an essential role in regulation of sulfate assimilation in all plants, the molecular mechanisms of the regulation and the effects of various treatments on the individual enzymes and metabolites are often different. This review summarizes (1) the molecular regulation of sulfate assimilation in Arabidopsis thaliana, especially recent data derived from platform technologies and functional genomics, (2) the co-ordination of sulfate, nitrate and carbon assimilations in Lemna minor, (3) the role of sulfate assimilation and glutathione in plant–Rhizobia symbiosis, (4) the cell-specific distribution of sulfate reduction and glutathione synthesis in C4 plants, (5) the regulation of glutathione biosynthesis in poplar, (6) the knock-out of the adenosine 5′phosphosulfate reductase gene in Physcomitrella patens and identification of 3′-phosphoadenosyl 5′-phosphosulfate reductase in plants, and (7) the sulfur sensing mechanism in green algae.
• Conclusions As the molecular mechanisms of regulation of the sulfate assimilation pathway are not known, the role of Arabidopsis as a model plant will be further strengthened. However, this review demonstrates that investigations of other plant species will still be necessary to address specific questions of regulation of sulfur nutrition.
Keywords: Sulfate assimilation, plant nutrition, glutathione, cysteine, C4 photosynthesis, Arabidopsis thaliana, Physcomitrella patens, poplar, Lemna minor
INTRODUCTION
Sulfur is found in nature in different oxidation states in inorganic, organic and bioorganic forms. For living organisms sulfur is an essential element with many different functions. It is found in reduced form in amino acids, peptides and proteins, in iron–sulfur clusters, lipoic acid and other co-factors, and in oxidized form as sulfonate group modifying proteins, polysaccharides and lipids. In addition, reduced sulfur compounds, such as hydrogen sulfide, serve as electron donors for chemotrophic or phototrophic growth in a large and diverse group of bacteria and archae, including purple and green sulfur bacteria (Trüper and Fischer, 1982). By contrast, oxidized sulfur compounds such as sulfate can function as terminal electron acceptor in respiration to support growth of sulfate-reducing bacteria (Postgate, 1984). Plants, yeast and most prokaryotes cover their demand for reduced sulfur by reduction of inorganic sulfate to sulfide, which is then incorporated into organic compounds. Sulfur is the least abundant of the six macronutrients required by plants, and perhaps therefore its metabolism has been least studied. In the last decade, however, significant progress in understanding the pathway of sulfate assimilation in plants has been made, and has been summarized in several reviews (Hawkesford and Wray, 2000; Leustek et al., 2000; Kopriva and Koprivova, 2004; Kopriva and Rennenberg, 2004; Saito, 2004). A combination of biochemical and molecular methods have revealed that adenosine 5′phosphosulfate reductase is the key enzyme of this pathway (Koprivova et al., 2000; Suter et al., 2000; Bick et al., 2001; Tsakraklides et al., 2002; Vauclare et al., 2002). Recently, a combination of transcriptome and metabolome analyses has significantly increased our knowledge of the cellular processes affected by sulfur starvation, which is becoming a serious problem in agriculture (Hirai et al., 2003, 2004, 2005; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003, 2005). Furthermore, the first reports on molecular mechanisms of regulation of sulfate uptake and assimilation have emerged very recently, opening a new era of our understanding of the pathway (Maruyama-Nakashita et al., 2004, 2005). These new developments were enabled by the availability of numerous platform resources for the model plant Arabidopsis thaliana. However, it is also becoming increasingly apparent that not all biological questions on sulfur metabolism can be addressed in Arabidopsis and that not all knowledge gained on this model is transferable to other species. In this review, therefore, the newest developments in the field of plant sulfur nutrition will be summarized with special emphasis on choice of different plant species for particular experiments and on differences in regulation of the pathway in different plants.
PLANT SULFUR ASSIMILATION
The major source of sulfur for plants is inorganic sulfate, although they are also able to use reduced sulfur compounds from the atmosphere, such as sulfur dioxide or hydrogen sulfide (Leustek et al., 2000). Sulfate is taken up and distributed within cells and the plant as a whole by sulfate transporters. Plants possess multiple sulfate transporters with different properties and functions (reviewed in Buchner et al., 2004). The first plant sulfate transporters were cloned from a tropical legume, Stylosanthes hamata, by Smith et al. (1995), who utilized functional complementation screening of a yeast sulfate transporter mutant. Arabidopsis possess 14 genes for sulfate transporters, divided into five groups according to sequence similarity and function. All these genes have significant sequence similarities with the originally identified cDNA clone, SHST1 (Smith et al., 1995). However, an exception appears with the group 5 members, which show limited sequence identity and structural similarity with the remaining sulfate transporter family members, and their transport activity has yet been unverified. High-affinity sulfate transporters, group 1, are responsible for uptake of sulfate from soil solution into the root cells (Shibagaki et al., 2002; Yoshimoto et al., 2002). Low-affinity transporters, group 2, are required for translocation of sulfate within the plant; therefore, they are localized in xylem parenchyma and phloem cells of roots and leaves (Takahashi et al., 2000). Group 4 transporters are localized in tonoplast and are responsible for sulfate efflux from the vacuole (Kataoka et al., 2004a). Very little is known about the function of sulfate transporters of groups 3 and 5, except a demonstrated ability of SULTR3;5 to increase the rate of root-to-shoot sulfate translocation in Arabidopsis (Kataoka et al., 2004b). Unexpectedly, group 3 sulfate transporter was identified as essential for nitrogen fixation in legume nodules (Krusell et al., 2005; see also below). The identity of the plastidic sulfate uptake system remains one of the greatest challenges for the sulfate transport community.
Intracellular sulfate is further metabolized into a large variety of primary and secondary metabolites. For assimilation into cysteine, sulfate has to be transported into plastids and activated by adenylation to adenosine 5′ phosphosulfate (APS) in a reaction catalysed by ATP sulfurylase (ATPS; EC 2.7.7.4). APS is reduced to sulfite by APS reductase (APR; EC 1.8.4.9); the electrons are derived from glutathione. Sulfite is further reduced by a ferredoxin-dependent sulfite reductase (SiR; EC 1.8.7.1) to sulfide, which is incorporated by O-acetylserine (thiol)lyase (OASTL; 2.5.1.47) into the amino acid skeleton of O-acetylserine (OAS) to form cysteine. OAS is synthesized by acetylation of serine with acetyl-CoenzymeA catalysed by serine acetyltransferase (SAT; EC 2.3.1.30) (Leustek et al., 2000; Suter et al., 2000; Kopriva and Koprivova, 2003; Fig. 1). SAT and OASTL form a multi-enzyme complex of cysteine synthase (Hell et al., 2002). Cysteine can be directly incorporated into proteins or peptides, such as glutathione (GSH), the most abundant low-molecular-weight thiol with a plethora of functions in plant stress defence, redox regulation, and sulfur storage and transport. Alternatively, cysteine is further metabolized and serves as a donor of reduced sulfur for synthesis of methionine, iron–sulfur centres, and various coenzymes and secondary metabolites. An intermediate of the sulfate assimilation pathway, sulfite, is metabolized into sulfolipids, essential components of chloroplast membranes (Sanda et al., 2001). Whereas sulfate reduction is localized in plastids only, cysteine is synthesized in all three compartments capable of protein synthesis: plastids, mitochondria and the cytosol (Leustek et al., 2000).
Fig. 1.
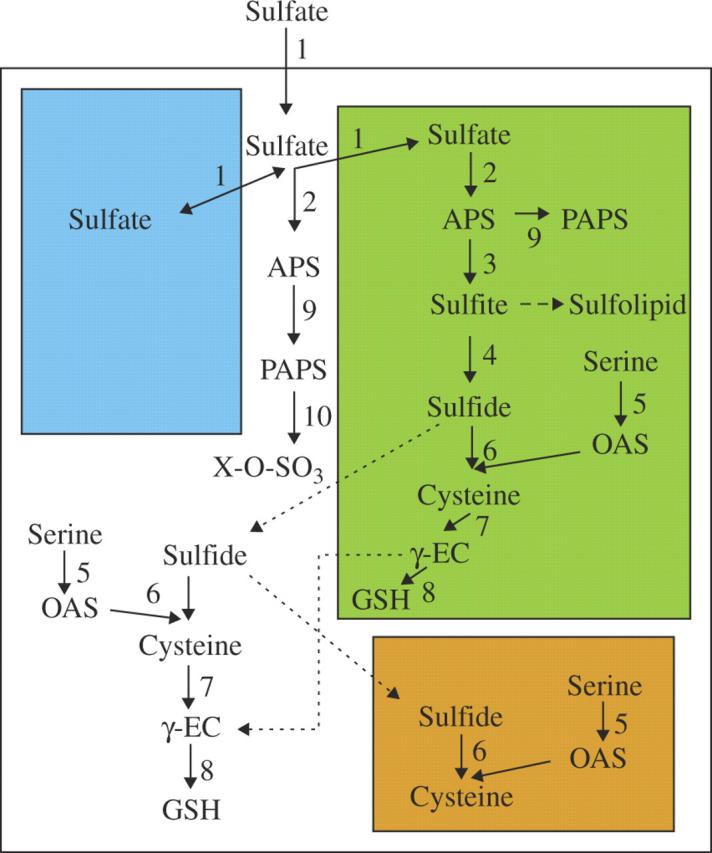
Schema of plant sulfur assimilation and subcellular localization of its major steps. Green colour represents plastids, brown mitochondria and blue vacuoles. Numbers represent enzymes as follows: 1, sulfate transporter; 2, ATP sulfurylase; 3, APS reductase; 4, sulfite reductase; 5, serine acetyltransferase; 6, O-acetylserine thiollyase; 7, γ-glutamylcysteine synthetase; 8, glutathione synthetase; 9, APS kinase; 10, sulfotransferase. Dashed lines represent multiple reaction steps; dotted lines indicate unconfirmed transport steps.
Sulfur is, however, also present in plant metabolites in the oxidized state as a sulfo-group modifying carbohydrates, proteins and many natural products. A large proportion of the known sulfated metabolites play various roles in plant defence against biotic and abiotic stress. A well-studied example of such compounds is glucosinolates, which participate in defence against herbivores and pathogens in Brassicales (Mikkelsen et al., 2002). They are also responsible for taste and flavour of cruciferous vegetables, and some of their degradation products, isothiocyanates, possess an anti-cancer activity (Mithen, 2001; Fahey et al., 2001). Another large group of sulfated compounds important for medicine are sulfated flavonoids, present in more than 250 species of 32 families (Barron et al., 1988), where they are involved in detoxification of reactive oxygen species and regulation of plant growth (Varin et al., 1997). Several other sulfated compounds were shown to participate directly in plant defence against pathogens, such as a sulfated derivate of jasmonic acid identified in Arabidopsis (Gidda et al., 2003) or sulphated β-1,3 glucan (Menard et al., 2004). The transfer of the sulfo-group, i.e. sulfation, is catalysed by sulfotransferases (SOT) (Klein and Papenbrock, 2004). The SOT reaction requires 3′-phosphoadenosyl 5′-phosphosulfate (PAPS) as the sulfate donor and a compound with a free hydroxyl group as an acceptor. Multiple SOT isoforms are found in higher eukaryots because of the structural diversity of the biological acceptors of the sulfate group (Klein and Papenbrock, 2004; Table 1). PAPS is synthesized by phosphorylation of APS by APS kinase (Fig. 1). APS thus can be withdrawn from the primary sulfate assimilation pathway in plastids. In addition, there seems to be a sulfation-dedicated PAPS synthesis in the cytosol, as ATPS and APS kinase activity are present both in plastids and in the cytosol, in contrast to APR and SiR, which are strictly plastidic (Leustek et al., 2000; Rotte and Leustek, 2000; Koprivova et al., 2001).
Table 1.
Genomic organization of transporters and enzymes for sulfate assimilation, GSH synthesis and sulfation reactions in sequenced plant and algal genomes
| Arabidopsis thaliana | Oryza sativa | Populus trichocarpa | Chlamydomonas reinhardtii | |
|---|---|---|---|---|
| SULTR | 14 | 13 | 14 | 5 |
| ATPS | 4 | 2 | 4 | 1 |
| APS kinase | 4 | 3 | 3 | 1 |
| APR | 3 | 2 | 2 | 1 |
| SiR | 1 | 2 | 2 | 1 |
| OASTL | 9 | 9 | 10 | 4 |
| SAT | 5 | 5 | 5 | 2 |
| γ-ECS | 1 | 2 | 2 | 1 |
| GSHS | 1 | 3 | 3 | 1 |
| SOT | 22 | 27 | 22 | 0 |
The number of genes was determined by tblastn against genomic sequences available in GenBank (Arabidopsis thaliana, Oryza sativa) or from the DOE Joint Genome Institute website (http://genome.jgi-psf.org/).
ATPS, ATP sulfurylase; APS, adenosine 5′ phosphosulfate; APR, APS reductase; SiR, sulfite reductase; OASTL, O-acetylserine (thiol)lyase; SAT, serine acetyltransferase; γ-ECS, γ-glutamylcysteine synthetase; GSHS, GSH synthetase; SOT, sulfotransferase.
Very recently, a further complexity was added to the sulfate assimilation pathway. First, sulfite oxidase, an enzyme oxidizing sulfite to sulfate, was detected in plants (Eilers et al., 2001). This enzyme is responsible for catabolism of sulfur-containing metabolites in animal mitochondria (Kisker et al., 1997). In plants it is localized in peroxisomes, i.e. spatially separated from sulfate assimilation, but its function is not yet known (Nowak et al., 2004). Secondly, PAPS reductase, an enzyme reducing PAPS to sulfite in yeast, fungi and some enteric bacteria, was found in the moss Physcomitrella patens (Koprivova et al., 2002a; see also below). The greatest progress in understanding the regulation of sulfate assimilation has been achieved in Arabidopsis thaliana. This recent development is therefore summarized in the next section, before the species-specific variations of sulfur metabolism are discussed.
SULFATE ASSIMILATION IN ARABIDOPSIS THALIANA
Arabidopsis thaliana became the model plant for studies of sulfur assimilation only recently, when the Arabidopsis sequencing project was initiated. This plant was long used predominantly for molecular cloning (Hell et al., 1994; Leustek et al., 1994; Brühl et al., 1996; Gutierrez-Marcos et al., 1996; Setya et al., 1996; Noji et al., 1998). These experiments revealed a great complexity of the sulfate assimilation pathway in Arabidopsis. With the exception of SiR and the enzymes of GSH synthesis, all other enzymes were encoded by small gene families (Table 1). It is not yet clear whether this is a functional redundancy or if all genes have a specific function. The sulfate transporter family is the best understood in this respect (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002, 2003; Kataoka et al., 2004a, b). Each analysed isoform has a specific localization, affinity to sulfate or regulation, indicating different biological functions. Differences in subcellular localizations and in kinetic parameters were found also among the five members of the Arabidopsis SAT gene family (Kawashima et al., 2005); however, their different functions remain to be demonstrated in vivo, e.g. by analysis of knock-out lines. By contrast, very little is known about the individual members of the ATPS, APR and APS kinase families. In the case of ATPS, all four genes code for proteins with putative organelle targeting peptides, so that it is not known which isoform is responsible for the cytosolic ATPS activity. This contrasts with other plant species, e.g. rice or potato, for which a cytosolic and plastidic ATPS were clearly identified (Klonus et al., 1994; Table 1). GSH biosynthesis is also differently organized in Arabidopsis and other plant species. Whereas in Arabidopsis and possibly other Brassicales the first enzyme of GSH synthesis, γ-glutamylcysteine synthetase (γ-ECS), seems to be exclusively localized in plastids and the second one, GSH synthetase (GSHS), is dually targeted to plastids and the cytosol from a single gene (Wachter et al., 2005), other plant species contain multiple copies of both genes (Table 1) and possess γ-ECS activity also in the cytosol (Hell and Bergmann, 1990).
The availability of cDNA sequences allowed using Arabidopsis for investigation of regulation of sulfate metabolism at the molecular level. Many reports primarily addressed changes in accumulation of mRNA of sulfate assimilation genes; for example, Takahashi et al. (1997) showed that a low-affinity sulfate transporter, SULTR2;1, and one isoform of APR and SAT were strongly induced by 2 d of sulfate starvation, and Harada et al. (2000) demonstrated regulation of sulfate assimilation by jasmonate. However, it was also possible to conduct more complex studies of the level of enzyme activities, mRNA and protein accumulation, metabolites (Cys and GSH), and even fluxes through the pathway. The first among such studies was the investigation of the control of sulfate assimilation by light (Kopriva et al., 1999). APR activity was shown to undergo a diurnal rhythm with maximum activity 4 h after light onset and minimum activity at the beginning of the night. Furthermore, sucrose was able to mimic the effect of light, revealing an interaction of sulfate assimilation with carbon metabolism (Kopriva et al., 1999). Sulfate assimilation is also strongly interconnected with the assimilation of nitrogen (Reuveny et al., 1980; Brunold and Suter, 1984). In A. thaliana withdrawal of nitrogen from nutrition for 3 d led to a specific decrease of APR activity, whereas OASTL and thiol contents were not affected (Koprivova et al., 2000). In all these experiments, the changes in APR activity corresponded to changes in mRNA levels of all three APR isoforms and APR protein accumulation, showing that APR is primarily regulated on the level of transcription. In addition, however, Bick et al. (2001) revealed a further, post-translational level of APR regulation by redox processes. These and other experiments concentrated on APR, as this enzyme was long known to be strongly regulated by various environmental factors, nutrient availability and stress (Brunold, 1990). The finding that treatment of Arabidopsis root cultures with thiols reduced solely APR, but not ATPS, SiR or OASTL, enabled use of feeding experiments with 35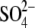 for control flux analysis to quantify the contribution of APR in the control of sulfate assimilation (Vauclare et al., 2002). Starting from internal sulfate the flux control coefficient of APR was calculated to be between 0·7 and 0·9 (equivalent to 70 and 90 % of the total control), indicating strong control of the pathway by APR. However, because sulfate uptake was even more inhibited by GSH than APS reduction, APR shares control of the flux with the sulfate uptake system. APR is thus indeed a key enzyme of the sulfate reduction pathway (Vauclare et al., 2002). This conclusion was further corroborated by Tsakraklides et al. (2002), who demonstrated that expression of bacterial APR in Arabidopsis results in deregulation of the pathway and accumulation of thiols and other reduced sulfur compounds, such as thiosulfate.
for control flux analysis to quantify the contribution of APR in the control of sulfate assimilation (Vauclare et al., 2002). Starting from internal sulfate the flux control coefficient of APR was calculated to be between 0·7 and 0·9 (equivalent to 70 and 90 % of the total control), indicating strong control of the pathway by APR. However, because sulfate uptake was even more inhibited by GSH than APS reduction, APR shares control of the flux with the sulfate uptake system. APR is thus indeed a key enzyme of the sulfate reduction pathway (Vauclare et al., 2002). This conclusion was further corroborated by Tsakraklides et al. (2002), who demonstrated that expression of bacterial APR in Arabidopsis results in deregulation of the pathway and accumulation of thiols and other reduced sulfur compounds, such as thiosulfate.
In the search for molecular mechanisms of regulation of sulfate assimilation, several potential signals were identified (Fig. 2). The most prominent signal is the precursor of cysteine, OAS. OAS has been identified as a limiting factor for cysteine synthesis (Rennenberg, 1983) and was shown to induce APR activity and rate of thiol synthesis in Lemna minor (Neuenschwander et al., 1991). In Arabidopsis, OAS induced mRNA accumulation of all genes of sulfate assimilation and dramatically increased flux through sulfate assimilation (Koprivova et al., 2000). OAS also strongly affects the cysteine synthase complex: even a less than two-fold increase in OAS concentration results in dissociation of the complex and inactivation of SAT (Berkowitz et al., 2002). Because OAS accumulates during sulfur deficiency, and because of its effects on cysteine synthase and expression of sulfate assimilation genes, OAS was proposed to act as a mediator of plant sulfur status (Hell et al., 2002). Accordingly, the activity of a βSR S-responsive region of the β-subunit of soybean conglycinin (Awazuhara et al., 2002) was up-regulated by OAS (Ohkama-Ohtsu et al., 2005). In the search for mutants with altered sulfur deficiency response using the βSR-driven green fluorescent protein (GFP) expression as a tool, a mutant accumulating OAS was identified, again indicating OAS as a signal of sulfur deficiency (Ohkama-Ohtsu et al., 2005). This conclusion was further strengthened by a transcriptome analysis suggesting the role of OAS as general regulator of gene expression (Hirai et al., 2003). The mRNA levels of more than 850 genes were affected by treatment of Arabidopsis with 1 mm OAS for 48 h. However, the correlation of changes in expression levels after sulfur deficiency and OAS treatment was significant only in the leaves. Further analysis of metabolite-to-gene networks revealed that expression of several genes of sulfate assimilation, such as ATPS3, APR2 and 3, SULTR1;1, 1;2 and 2;1, was indeed tightly linked to OAS level (Hirai et al., 2005). Transcript levels of other genes also regulated by sulfur deficiency, such as SiR, APS kinase or group 3 sulfate transporters, by contrast, were not correlated with OAS, showing that OAS cannot be the sole sensor of sulfur deficiency (Hirai et al., 2005). It is thus evident that OAS is an important player in regulation of plant sulfur homeostasis but its exact molecular function and the other components of the regulatory circuit are unresolved.
Fig. 2.
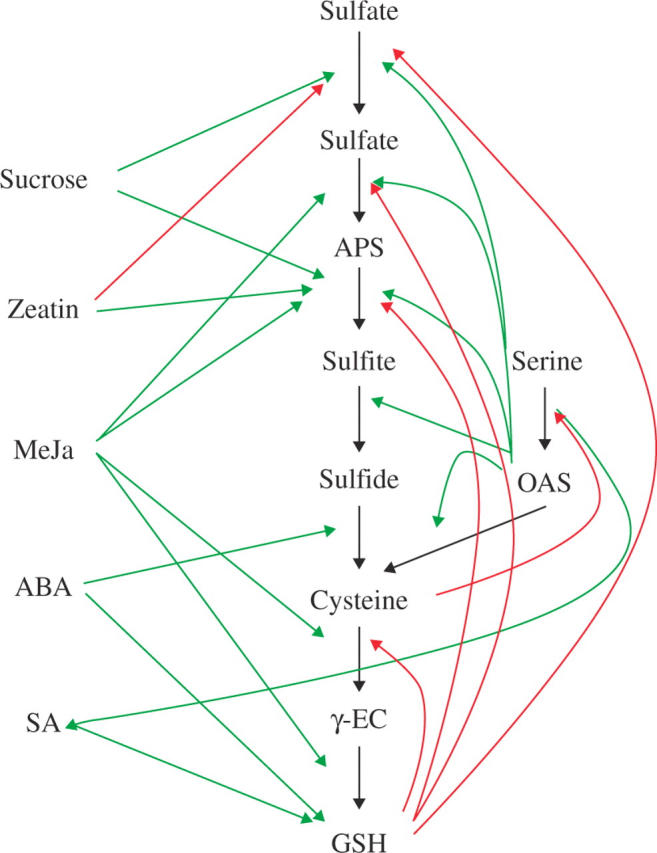
Schematic representation of putative signals involved in regulation of sulfate assimilation and GSH synthesis. Red arrows represent repression of the metabolic step; green arrows indicate an increase in metabolite accumulation (pointed towards metabolite) or induction of the reaction.
The general role of OAS as signal in the sulfur deficiency response was further questioned in experiments with potato, in which induction of sulfate uptake and mRNA levels of sulfate transporter and APR by sulfur starvation preceded accumulation of OAS (Hopkins et al., 2005). Whether this is due to different regulatory circuits in Arabidopsis and potato remains to be elucidated. This explanation is possible, as it is not the first major variation to be described in the regulation of sulfur metabolism between the two species. The cystathionine γ-synthase (CgS), first step in synthesis of methionine from cysteine and O-phosphohomoserine, is strongly down-regulated by Met. In Arabidopsis this is achieved by S-adenosylmethionine-dependent degradation of CgS mRNA (Chiba et al., 2003). The CgS protein plays an important role in this autoregulation, as mto1 mutants accumulating Met and CgS transcript have alterations in amino acid sequence of the N-terminal part of the protein (Chiba et al., 1999). Met levels in Arabidopsis can be modulated by changes in CgS mRNA level using a transgenic approach (Hesse and Hoefgen, 2003). However, in potato, neither overexpression nor antisense downregulation of CgS affected Met levels (Kreft et al., 2003). In addition, potato CgS mRNA expressed in Arabidopsis does not undergo the same degradation as the endogenous transcript (Kreft et al., 2003). It is thus possible that different molecular mechanisms of regulation of sulfate assimilation exist in Arabidopsis and potato (and potentially in other plants) so that the knowledge obtained from the model species may not always be simply transferred to other, for example, crop species.
Another compound possibly regulating sulfate assimilation directly is glutathione (Fig. 2). Reduced forms of sulfur, such as H2S, cysteine or GSH, trigger a strong decrease in sulfate uptake and assimilation (Brunold and Schmidt, 1978; Lappartient et al., 1999; Westerman et al., 2001). In A. thaliana root cultures APR activity and transcript levels were decreased by feeding either cysteine or GSH (Vauclare et al., 2002). Because external GSH supply also increases the accumulation of cysteine, both Cys and GSH might be responsible for the control of 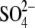 uptake and assimilation. Blocking GSH synthesis by l-buthionine [S, R] sulfoximine, an inhibitor of γ-ECS, relieved the repression APR, indicating that GSH is most probably the acting molecular signal (Vauclare et al., 2002).
uptake and assimilation. Blocking GSH synthesis by l-buthionine [S, R] sulfoximine, an inhibitor of γ-ECS, relieved the repression APR, indicating that GSH is most probably the acting molecular signal (Vauclare et al., 2002).
Carbohydrates induce APR mRNA accumulation and activity in the dark (Kopriva et al., 1999, 2002a; Hesse et al., 2003); the mechanism involved is, however, distinct from the carbohydrate regulation of the ADP-glucose pyrophosphorylase subunit ApL3 gene used to decipher sugar sensing in Arabidopsis (Rook et al., 2001; S. Kopriva, unpubl. res.).
Little is known about the role of phytohormones in the control of sulfur assimilation, but recent developments indicate that this group of compounds is very important for regulation of S nutrition (Ohkama et al., 2002; Maruyama-Nakashita et al., 2004, 2005). Ohkama et al. (2002) used transgenic Arabidopsis plants, expressing GFP under control of a chimeric promoter containing a sulfur responsive element of β-conglycinin (Awazuhara et al., 2002), to test the influence of phytohormones on the sulfur deficiency response. Whereas abscisic acid (ABA), indole-3-acetic acid (IAA), aminocyclopropane carboxylic acid (ACC, precursor of ethylene), gibberelic acid (GA3) and jasmonic acid (JA) were not able to induce expression of GFP derived from the sulfur responsive element and, thus, mimic the sulfur starvation response, trans-zeatin caused an increase in GFP synthesis both in sulfur-sufficient and in sulfur-deficient conditions. In addition, zeatin treatment resulted in an increased accumulation of mRNA for APR and a low-affinity sulfate transporter (Ohkama et al., 2002). By contrast, cytokinins repressed the expression of high-affinity sulfate transporters and sulfate uptake capacity of Arabidopsis roots. The CRE1/WOL/AHK4 cytokinin receptor was found to be important for regulation given that the effect of cytokinins was attenuated in cre1-1 mutants (Maruyama-Nakashita et al., 2004). Cytokinins are known to be closely related to the nitrogen status of the plant and to be involved in regulation of nitrogen assimilation (Samuelson et al., 1995; Takei et al., 2001; Collier et al., 2003). Recently, they were shown to be involved also in regulation of phosphate uptake (Martin et al., 2000). It therefore seems likely that cytokinins may play a more general role in co-ordination of uptake and assimilation of nutrients, including sulfur.
Although auxin was not able to induce expression of GFP from the sulfur responsive element, its role in regulation of sulfur assimilation cannot be excluded. A NIT3 nitrilase, involved in synthesis of IAA, belongs to genes strongly induced by sulfur deficiency (Kutz et al., 2002). In addition, the cis-acting element conferring sulfur starvation response recently identified in Arabidopsis SULTR1;2 promoter contains an auxin response factor (ARF) binding sequence (Maruyama-Nakashita et al., 2005). In contrast to typical ARF sites the motif was found as a monomer in the sulfur responsive element and the promoter activity was not affected by naphthalene acetic acid (Maruyama-Nakashita et al., 2005). However, more information is needed to evaluate comprehensively the regulatory interactions of auxin with sulfur metabolism.
JA did not affect the expression of the sulfur responsive promotor element (Ohkama et al., 2002), but is nevertheless involved in regulation of sulfate assimilation. Treatment of Arabidopsis with methyljasmonate resulted in a fast but transient increase in mRNA levels of many genes involved in sulfate assimilation and GSH synthesis, but without affecting sulfur metabolite levels (Xiang and Oliver, 1998; Harada et al., 2000; Jost et al., 2005). The mRNA for sulfate transporters was not affected, confirming that JA does not participate in the regulation by sulfur nutrition although genes of jasmonate biosynthesis are among those induced by sulfur starvation (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003). The induction of sulfate assimilation by JA is not surprising as JA is known to participate in the transduction of stress responses (Reymond and Farmer, 1998) and sulfur compounds often play an important role in plant stress defence (Foyer and Rennenberg, 2000).
The interaction of sulfate assimilation and GSH synthesis with stress defence is further corroborated by the finding that the level of GSH increased in plants treated with abscisic acid (Jiang and Zhang, 2001) and salicylic acid (SA) (Fodor et al., 1997). ABA plays an important role in adaptive responses to environmental stresses (Chandler and Robertson, 1994) and leads to increased production of reactive oxygen species (Guan et al., 2000). It is therefore not clear whether GSH synthesis is regulated by ABA itself or by the oxidative stress resulting from ABA treatment. Because ABA induces mRNA accumulation of cytosolic OASTL (Barroso et al., 1999) it seems that this compound may have a more profound effect on control of sulfur metabolism. SA plays a central role in plant defence against pathogens. SA accumulates upon pathogen attack, induces expression of pathogenesis-related genes and is a necessary component of systemic acquired resistance (Kunkel and Brooks, 2002). Treatment of tobacco leaves with SA as well as infection with tobacco mosaic virus resulted in an increase in GSH content in inoculated but not in systemic leaves (Fodor et al., 1997). In addition, treatment with the biologically active SA analogue 2,6-dichloroisonicotinic acid increased the GSH level leading to a reduction of NPR1, a regulator of systemic acquired resistance, and expression of the PR1 gene for a pathogenesis-related protein (Mou et al., 2003). SA was also implicated in the mechanism of nickel tolerance in hyperaccumulator Thlaspi species. Elevated SA levels engineered in Arabidopsis resulted in an increase in SAT activity and GSH content and, consequently, in increased tolerance to Ni (Freeman et al., 2005). Whether SA regulates the expression of γECS and GSHS, if and how it affects SAT, or if it utilizes another mechanism to increase GSH synthesis remain to be elucidated.
Arabidopsis is the plant model of choice for global investigations of transcriptome and metabolome. As the major physiological problem connected with sulfur metabolism is sulfur deficiency, the extent to which this condition affects plant metabolism was investigated by expression profiling (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003) and combined transcriptome and metabolome analysis (Hirai et al., 2005; Nikiforova et al., 2005). More than 2700 genes were found to be affected by sulfur starvation. As expected, the genes induced by sulfur deficiency included those coding for sulfate transporters and APR; other genes of sulfate assimilation were not significantly and/or consistently affected. However, the genes involved in jasmonate and auxin biosynthesis were induced irrespective from the experimental set-up, as well as a gene for NADPH oxidoreductase, which is involved in oxidative stress defence. Several genes were strongly regulated in one experiment but not others. Examples of such genes are thioglucosidase, which was strongly induced in the roots after 24 h of sulfur deficiency (Maruyama-Nakashita et al., 2003) and was suggested to be involved in providing additional sulfur via degradation of glucosinolates (Wittstock and Halkier, 2002), or phenylalanine ammonia-lyase, strongly reduced after 6 and 10 d of sulfur starvation (Nikiforova et al., 2003). Metabolome analysis revealed that from approx. 6000 analysed metabolites, 11·5 % were significantly affected by 13 d of sulfur starvation (Nikiforova et al., 2005). Examples are tryptophan, concentration of which increased up to 28-fold, other amino acids and flavonoids, whereas levels of thiols, lipids and chlorophyll decreased (Nikiforova et al., 2005). The power of the global study of metabolite and transcript networks was demonstrated by Hirai et al. (2005), who used a batch-learning self-organizing mapping analysis to reveal clusters of genes and metabolites regulated by the same mechanism. This led, for example, to prediction of function of three sulfotransferase genes in glucosinolate metabolism that were confirmed after biochemical analysis of corresponding recombinant proteins. Altogether, transcriptome and metabolome analyses revealed the complexity of the interactions between S, N and C metabolism and opened new perspectives for dissecting the molecular mechanisms of regulation of sulfate assimilation. In this context, the finding of 49 transcription factor genes that specifically responded to sulfur deficiency is of greatest importance (Nikiforova et al., 2003).
Compared with the number for other metabolic pathways, only few recent reports have described the use of genetic approaches for studies of sulfate assimilation (Shibagaki et al., 2002; Ohkama-Ohtsu et al., 2005). Analysis of selenate-resistant mutants resulted in functional characterization of SULTR1;2 and its identification as the major uptake system for sulfate into the roots (Shibagaki et al., 2002). GFP expression from the well-characterized βSR promoter fragment from conglycinin (Awazuhara et al., 2002) was used as a tool in the search for mutants with altered sulfur deficiency response. Seeds from plants harvesting the βSR::GFP construct were mutagenized and mutants were selected with increased GFP expression at normal sulfur supply (Ohkama-Ohtsu et al., 2005). In one of these mutants, the level of OAS was increased and, in addition to GFP expression, the mRNA levels of several other genes responsive to sulfate starvation were increased even at normal sulfur concentration. Map-based cloning and sequence analysis identified a thiol reductase to be responsible for the elevated OAS levels (Ohkama-Ohtsu et al., 2005). This result, although not easy to interpret, indicates the power of the genetic approach, which allows identification of unexpected connections in plant metabolism. Surely, this approach, together with the use of other genetic resources, such as T-DNA or transposon mutants, will deliver the much needed identity of molecular signals and transcription factors regulating sulfur metabolism. Arabidopsis thaliana will certainly be the major player in this process.
SULFATE ASSIMILATION IN LEMNA MINOR
Traditionally, the water plant Lemna minor was used as a model to study the regulation of sulfate assimilation in higher plants. The advantages of Lemna compared with other plant systems, such as tobacco cell cultures or spinach chloroplasts, were (1) the use of an intact plant, (2) direct contact of the fronds with nutrient solutions, (3) rapid increase in biomass and (4) the possibility of growth in aseptic liquid cultures under steady-state conditions. L. minor was used predominantly to decipher the regulation of sulfate assimilation by sulfur compounds (Brunold and Schmidt, 1978; Brunold et al., 1987) and the co-ordination of sulfate reduction with the assimilation of nitrogen (Brunold and Suter, 1984; Suter et al., 1986). Sulfur deficiency induced activity of APR; other enzymes of the sulfate reduction pathway were much less influenced (Brunold et al., 1987). An increase in sulfate concentration in the nutrition or addition of cysteine or H2S led to decreased APR activity, but activity could be quickly re-established on a return to control conditions (Brunold and Schmidt, 1978; Brunold et al., 1987). The activities of ATPS and APR decreased under nitrogen-deficient conditions (Brunold and Suter, 1984). At the same time, addition of nitrate or ammonia to the medium quickly restored the activity of these two enzymes. By contrast, addition of ammonia or amino acids (Arg, Asn, Gln) to the nutrient solution caused a 50–110 % increase in extractable APR activity in Lemna (Brunold and Suter, 1984; Suter et al., 1986). Addition of ammonia increased the flux through the sulfate assimilation pathway, measured as incorporation of 35S in proteins after feeding [35S]sulfate (Brunold and Suter, 1984). Finally, APR was shown to be repressed by dark, a process attenuated by OAS (Neuenschwander et al., 1991). Addition of OAS also led to an increase in thiol concentration both in light and in dark and to an increased flux through the sulfate assimilation pathway. From these results, Neuenschwander et al. (1991) for the first time suggested a role for OAS in regulation of APR and sulfate assimilation.
Lemna proved to be useful also in molecular studies. The purification of plant APR was successful only from L. minor, which together with cloning the corresponding mRNA led to the conclusion that APS sulfotransferase and APS reductase are identical enzymes (Suter et al., 2000). In order to study the interconnection of sulfate, nitrogen and carbon assimilation, L. minor was used to analyse the effects of CO2 omission from the atmosphere and simultaneous application of alternative carbon sources on APR and nitrate reductase (NR) (Kopriva et al., 2002a). Incubation in a CO2-free atmosphere led to a severe decrease in APR and NR activities and mRNA levels, but Rubisco was not affected to any great degree. Simultaneous addition of sucrose prevented the reduction in enzyme activities but not in mRNA levels. OAS could also attenuate the effect of missing CO2 on APR, but did not affect NR. 35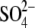 feeding showed that withdrawal of CO2 severely inhibited sulfate uptake and the flux through the sulfate assimilation pathway. After re-supply of normal air or addition of sucrose, incorporation of 35S into proteins and glutathione greatly increased. OAS treatment resulted in high-level labelling of cysteine; incorporation of 35S in proteins and glutathione was increased to a much lower extent (Kopriva et al., 2002a). These results corroborated the close link between sulfate, nitrate and carbon assimilation and indicated that OAS might signal the plant N status towards S assimilation, but most probably not vice versa.
feeding showed that withdrawal of CO2 severely inhibited sulfate uptake and the flux through the sulfate assimilation pathway. After re-supply of normal air or addition of sucrose, incorporation of 35S into proteins and glutathione greatly increased. OAS treatment resulted in high-level labelling of cysteine; incorporation of 35S in proteins and glutathione was increased to a much lower extent (Kopriva et al., 2002a). These results corroborated the close link between sulfate, nitrate and carbon assimilation and indicated that OAS might signal the plant N status towards S assimilation, but most probably not vice versa.
SULFATE ASSIMILATION IN LEGUMES
Legumes are unique among plants because they are able to form symbiosis with rhizobial bacteria. These bacteria are able to reduce atmospheric nitrogen to ammonium, which is incorporated into carbohydrate skeletons provided by the plant to form amino acids (Udvardi and Day, 1997). Infection of plants with Rhizobia causes development of nodules, where the bacteria in the cytoplasm of plant cells differentiate into bacteroids (Van Rhijn and Vanderleyden, 1995). Nodulation is very important for plant nitrogen nutrition, especially in soils with low nitrogen content. Sulfur and sulfur compounds were shown to play an important role in establishing the symbiosis and its function (Zhao et al., 1999; Frendo et al., 2005; Harrison et al., 2005; Krusell et al., 2005). Growth and nitrogen fixation of pea were greatly enhanced by providing additional sulfur to the soil (Zhao et al., 1999). Legumes are the only plant family with significant amounts of a GSH analogue homoglutathione, which contains a β-alanine instead of glycine (Klapheck, 1988). Homoglutathione (hGSH) is synthesized by an hGSH synthase, which is distinct from GSHS. The presence of the two genes in legumes is the result of relatively recent gene duplication after divergence of the Fabales (Frendo et al., 2001). No functional differences between GSH and hGSH are apparent as GSH to hGSH ratios vary largely between different legume species (Matamoros et al., 1999). Nodules are organs with the highest GSH and/or hGSH content in legumes because of their role in defence of the nitrogenase against reactive oxygen species (Matamoros et al., 2003). However, GSH and hGSH are important also for establishing the symbiosis. Reducing the tripeptide contents either by antisense expression of the corresponding biosynthetic genes or by inhibition of the GSH/hGSH synthetase with buthionine sulfoximine dramatically inhibited the formation of nodules (Frendo et al., 2005). In addition, the bacteria have a requirement for GSH. Sinorhizobium mutants in γ-ECS were not able to nodulate plant host whereas disruption of GSHS led to slow growth, delayed and deformed development of nodules, and a reduced capacity to fix nitrogen (Harrison et al., 2005). By contrast, Rhizobium mutants unable to reduce sulfate and nitrate due to disruption of siroheme synthase and thus with non-functional sulfite and nitrite reductase were unable to grow in the soil but produced functional nodules (Tate et al., 1997). It therefore seems that Rhizobia are able to take up and utilize cysteine but not GSH.
The importance of sulfur for nodule function was further corroborated by analysis of Lotus sym mutants with non-functional nodules (Krusell et al., 2005). They cloned the gene responsible for the sym13 and sym81 mutations, which display nitrogen deficiency syndromes under symbiotic but not non-symbiotic growth conditions and form smaller nodules with reduced nitrogenase content and nitrogen fixing capacity. In both cases, an SST1 sulfate transporter was defective. SST1 is a group 3 sulfate transporter expressed in a nodule-specific manner and located in the symbiosome membrane (Krusell et al., 2005). Thus, interestingly, the first sulfate transporter with clearly a unique function is essential not for sulfur nutrition but for metabolism of nitrogen. However, the strong sym phenotype of the sst1 mutants is surprising as at least one additional sulfate transporter is expressed specifically in Lotus nodules (Colebatch et al., 2002). In addition, the reduction of total sulfur content of 20–25 % in the mutants compared with wild-type nodules can hardly explain the severe disruption in nitrogen fixation as plants can reduce sulfur content by up to 70 % without phenotypic changes (Nikiforova et al., 2003). The mechanism by which the loss of SST1 aborts nitrogen fixation thus needs to be addressed in more detail.
SULFATE ASSIMILATION IN C4 PLANTS
An interesting variation in sulfate assimilation was observed in plants with C4 photosynthesis (reviewed in Kopriva and Koprivova, 2005). C4 photosynthesis is characterized by spatial separation of a primary CO2 fixation step into C4 acids from their decarboxylation and refixation of the released CO2 by Rubisco. This separation of the photosynthetic reactions is usually linked with occurrence of two distinct cell types: the bundle sheath cells (BSCs) and mesophyll cells (MCs) arranged around vascular tissue in a radial pattern known as Kranz anatomy (Laetsch, 1974). CO2 is initially fixed by phosphoenolpyruvate carboxylase (PEPCase) in the MCs to form a C4 compound oxaloacetate, which is subsequently converted to malate and/or aspartate. These C4 acids diffuse to BSCs, where they become decarboxylated and the released CO2 is refixed by Rubisco in the Calvin cycle, as in C3 plants (Edwards and Huber, 1981; Hatch, 1987). The CO2 concentration is therefore increased at the site of its photosynthetic fixation and thus eliminates the oxygenation reaction of Rubisco and photorespiration. A characteristic feature of C4 plants is a cell-specific localization of many enzymes of primary metabolism in BSCs or MCs. Clearly, the enzymes involved in the primary CO2 fixation and malate and/or aspartate synthesis, such as cytosolic carbonic anhydrase, PEPCase, pyruvate phosphate dikinase and NADP-malate dehydrogenase, are localized predominantly or exclusively in the MCs, whereas NAD(P)-malic enzyme, Rubisco, Rubisco activase and some enzymes of the Calvin cycle are found exclusively in BSCs (reviewed in Sheen, 1999; Edwards et al., 2001). In addition, glycine decarboxylase, a key enzyme of photorespiration, is localized exclusively in BSCs of C4 and C3–C4 intermediate plants (Hylton et al., 1988). C3–C4 intermediate plants were originally identified by having a CO2 compensation point intermediate between that of C3 and C4 species and can be considered as evolutionary intermediates in the path from C3 to C4 photosynthesis (Monson and Moore, 1989; Kopriva et al., 1996). Interestingly, enzymes participating in the assimilation of nitrogen are also localized in a cell-specific manner in various C4 plants; nitrate reductase and nitrite reductase are specifically localized in MCs, whereas glutamine synthetase is equally distributed between MCs and BSCs, and glutamate synthetase and glutamate dehydrogenase are predominantly but not exclusively localized within BSCs (Rathnam and Edwards, 1976; Moore and Black, 1979).
Given the number of cellular processes spatially distributed in C4 plants, it was not a great surprise that also enzymes of sulfate assimilation were found to be differentially localized (Gerwick and Black, 1979; Gerwick et al., 1980; Passera and Ghisi, 1982; Schmutz and Brunold, 1984). Several groups reported that 75–100 % of total leaf ATP sulfurylase activity in maize was confined to BSCs (Gerwick and Black, 1979; Passera and Ghisi, 1982; Burnell, 1984; Schmutz and Brunold, 1984). These findings were extended to 17 other C4 species, in which 95–100 % of total leaf ATPS activity was found in chloroplasts of BScs (Gerwick et al., 1980). In addition, APR activity was found almost exclusively in BSCs of maize (Schmutz and Brunold, 1984; Burgener et al., 1998), whereas activities of SiR and OASTL were found in MCs and BSCs at comparable levels (Passera and Ghisi, 1982; Burnell, 1984; Schmutz and Brunold, 1985). The mRNA for APR, ATPS and SiR accumulated in BSCs only, whereas OASTL transcript was detected in both MCs and BSCs (Kopriva et al., 2001).
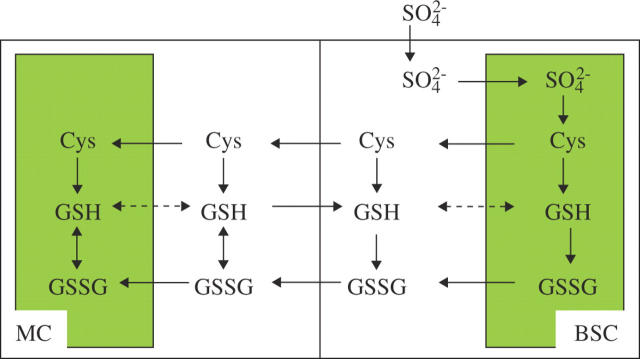
Not only sulfate assimilation but also the synthesis and reduction of glutathione seem to be differently localized in C4 plants than in, for example, Arabidopsis. GSH is particularly important in maize and other low-temperature-sensitive C4 plants, because it protects against chilling stress by detoxification of H2O2. The oxidized glutathione (GSSG) formed in this reaction is subsequently reduced by NADPH-dependent glutathione reductase (GR). At low temperature, GSH content and reduction state are higher in chilling-tolerant genotypes of maize than in chilling-sensitive genotypes (Kocsy et al., 1996). When GSH content was increased in chilling-sensitive maize by treatment with herbicide safeners, chilling-induced injury was significantly reduced (Kocsy et al., 2001), whereas reduction of GSH by inhibiting its synthesis in a chilling-tolerant genotype resulted in increased leaf injury at low temperature (Kocsy et al., 2000). Consequently, in maize chilling induces foliar thiol levels and activities of APR, γ-ECS and GSHS (Brunner et al., 1995), and total GSH content and the activities of APR and GR are increased in chilling-tolerant maize compared with a sensitive genotype even at standard growth conditions (Kocsy et al., 1997). Although GSH is essential for protection against chilling injury and other stress, it is not equally distributed between MCs and BSCs in maize. GSHS activity is greater in MCs than in BSCs, leading to GSH synthesis predominantly in the MCs (Burgener et al., 1998) and higher GSH levels in this cell type (Doulis et al., 1997; Burgener et al., 1998; Kopriva et al., 2001). Cysteine was shown to be exported from BSC protoplast as a transport form of reduced sulfur to the MCs (Burgener et al., 1998; Fig. 3). The enzymes of GSH synthesis and corresponding mRNAs were, however, found to be localized in both MCs and BSCs by immunohistochemistry (Gómez et al., 2004). Both enzymes were detected in chloroplasts and in the cytosol (Gómez et al., 2004); this contrasts with results for the Brassicaceae, where γ-ECS is localized in plastids and GSHS is prevalently cytosolic (Wachter et al., 2005). Probably as a result of the low capacity for NADPH formation in BSCs, GR was found exclusively in MCs of maize (Doulis et al., 1997; Pastori et al., 2000). These results thus demonstrate that there are species-specific differences in the intercellular localization of GSH biosynthetic enzymes, which are dependent on increased capacity for transport of various sulfur compounds and possibly result in different regulatory mechanisms for sulfur assimilation in C4 plants.
Fig. 3.
Schematic representation of the distribution of the major steps in sulfate assimilation and GSH synthesis and reduction between mesophyll (MC) and bundle sheath (BSC) of maize. Dashed lines represent unconfirmed transport steps.
Most results on the regulation of sulfate assimilation obtained with maize fitted well to the general hypothesis of demand-driven control of the pathway (Lappartient and Touraine, 1996). A co-ordinate increase in mRNA levels for sulfate transporters, ATPS and APR was observed in maize roots and leaves upon sulfate starvation (Bolchi et al., 1999; Hopkins et al., 2004) and the ATPS mRNA level was repressed in the presence of reduced sulfur compounds (Bolchi et al., 1999). Accordingly, ATPS and APR activities were increased upon treatments of maize with cadmium or chilling, which resulted in higher demand for reduced sulfur (Nussbaum et al., 1988; Brunner et al., 1995). In these and other reports the regulation of sulfate assimilation in maize was not distinguishable from that in other plants. Bolchi et al. (1999), however, described an interesting result which differentiates maize from other plant species analysed to date. Whereas in Brassicaceae GSH exerts the feedback repression of sulfate assimilation (Lappartient and Touraine, 1996; Vauclare et al., 2002), in maize cysteine is acting directly without conversion to GSH (Bolchi et al., 1999). This variation might well be a consequence of the BSC localization of sulfate assimilation (discussed in Kopriva and Koprivova, 2005).
Because the biological significance of the cell-specific distribution of sulfate assimilation and GSH synthesis is not known, the evolution of this trait is of particular interest. To address the question of whether the BSC localization of sulfate assimilation was a prerequisite or a consequence of C4 photosynthesis, the distribution of APR was studied in Flaveria species with different types of photosynthesis (Koprivova et al., 2001). The dicot genus Flaveria (Flaveriinae—Asteraceae) is an excellent model to study the evolution of C4 photosynthesis because, in addition to C3 and C4 species, a relatively large number of C3–C4 intermediates occur in this genus (Ku et al., 1991) and a continuous gradation both in the physiology and the biochemistry of photosynthesis exists among Flaveria species (Monson and Moore, 1989). APR activity, and cysteine and glutathione levels were significantly higher in C4-like and C4 species than in C3 and C3–C4 species. However, surprisingly, by northern analysis of cell-specific RNA and in situ hybridization ATPS and APR mRNA were present at comparable levels in both MCs and BSCs of the C4 species Flaveria trinervia. In addition, immunogold electron microscopy confirmed the presence of APR protein in chloroplasts of both cell types (Koprivova et al., 2001). Apparently, the localization of assimilatory sulfate reduction in the BSCs is not ubiquitous among C4 plants but occurs most probably only in C4 monocots and is, therefore, neither a prerequisite nor a consequence of C4 photosynthesis. The finding of different compartmentation of ATPS and APR in maize and Flaveria clearly revealed the dangers involved in generalization, and that species-specific variations in regulation and compartmentation might be far greater than are usually assumed.
SULFATE ASSIMILATION IN TREES
In trees the regulation of sulfate assimilation is necessarily more complex than in herbaceous plants because of their long life cycles connected with seasonal processes. Trees possess several specific physiological characteristics such as wood production, periods of dormancy combined with storage and mobilization processes that require transport between the shoot and the roots over extremely long distances. In conifers GSH accumulates in winter in the needles as storage of reduced sulfur (Schupp and Rennenberg, 1992). Deciduous trees also seem to store reduced sulfur in the form of GSH; the storage organ is the bark (Siller-Cepeda et al., 1991; Herschbach and Rennenberg, 2001). Under ambient conditions, sulfate is a major sulfur source. It is taken up by the roots, loaded to xylem and transported to the leaves where its reduction takes place. Uptake of sulfate in trees is facilitated by association with mycorrhiza. Although the association with ectomycorrhiza has no impact on the rate of sulfate uptake in trees, it does affect sulfate loading into the xylem (Seegmüller et al., 1996; Kreuzwieser and Rennenberg, 1998). In addition, the de-repression of sulfate transport by sulfur deficiency observed in non-mycorrhizal beech roots is attenuated in ectomycorrhizal roots, indicating a significant contribution of the fungal partner to the regulation of sulfate uptake (Kreuzwieser et al., 1996; Kreuzwieser and Rennenberg, 1998). From these and other studies (Herschbach et al., 2000) it is apparent that sulfate uptake and xylem loading are regulated differently. However, only little is known about the molecular mechanisms of regulation of sulfate reduction in trees. Poplar has proved to be an excellent model plant for molecular studies, as demonstrated by the fact that it was the third plant species for which the genome was sequenced (Tuskan et al., 2004). The genomic organization of the pathway in poplar is similar to that in herbaceous plants; ATPS and APR are present in several isoforms, SiR and γ-ECS are single copy genes (Table 1; Kopriva et al., 2004). In a first attempt to characterize the regulation of sulfur nutrition in poplar, the effects of sulfur and nitrogen deficiency were monitored (Kopriva et al., 2004). Although distinct differences were found, such as no effect of sulfate deficiency on APR in leaves, the regulation of sulfate assimilation by nutrient availability observed in poplar showed strong similarities to the regulation described for herbaceous plants (Kopriva et al., 2004).
In sulfur research, poplars over-expressing bacterial genes for GSH biosynthesis in the cytosol or in the chloroplast are the best characterized plants with manipulated GSH content. Indeed, our current understanding of the regulation of GSH synthesis in plants is mostly derived from experiments with transgenic poplars. GSH synthesis is regulated by the supply of the constituent amino acids and by feedback inhibition of γ-ECS by GSH (Hell and Bergmann, 1990; Noctor et al., 1998b). To address this regulation in more detail a poplar hybrid Populus tremula × P. alba (INRA clone no. 717-1-B4, Versailles, France) was transformed to express bacterial γ-ECS or GSHS either in the cytosol (Noctor et al., 1996; Arisi et al., 1997) or in the chloroplast (Noctor et al., 1998a). Over-expression of γ-ECS, but not of GSHS, increased foliar and root GSH concentration (Strohm et al., 1995; Noctor et al., 1996, 1998a; Arisi et al., 1997; Herschbach et al., 2000), thus confirming the major role of γ-ECS in the control of GSH synthesis. Experiments with poplar leaf discs revealed that feeding of γ-EC dramatically enhanced GSH synthesis compared with feeding of Cys and Glu, this effect being more profound in the GSHS-overexpressing plants, whereas Cys was more effective in the γ-ECS-overexpressing poplar (Strohm et al., 1995; Noctor et al., 1996). Interestingly, over-expression of γ-ECS either in the cytosol or in the chloroplast did not decrease cysteine and methionine concentrations (Herschbach et al., 2000). Presumably, sulfate reduction and Cys formation are adjusted to the higher demand for GSH synthesis in γ-ECS transgenic trees by a still unknown mechanism. The third amino acid constituting GSH, glycine, is produced through photorespiration and restricts GSH synthesis in the dark in both the cytosol and the chloroplast (Noctor et al., 1997a, b, 1999). Furthermore, over-expression of γ-ECS in the chloroplast, but not in the cytosol, was accompanied by higher foliar concentrations of valine, leucine, isoleucine, tyrosine and lysine (Noctor et al., 1998a). As synthesis of these amino acids is predominantly localized in the chloroplasts, γ-ECS over-expression seems to interact with general amino acid synthesis in this compartment.
GSH concentration was increased in phloem exudates from stem bark tissues of poplars over-expressing γ-ECS in the cytosol, but not in GSHS-expressing plants (Herschbach et al., 1998). The higher GSH concentration in the phloem correlated linearly with the GSH concentrations in the leaves and roots. In deciduous trees both reduced sulfur, mainly as GSH, and sulfate are transported in the phloem to the roots (Herschbach and Rennenberg, 2001). Indeed, Herschbach et al. (2000) concluded, based on correlation analysis, that sulfate uptake and xylem loading under both enhanced and decreased sulfur demand might be controlled by the sulfate-to-glutathione ratio. How such regulation could be achieved on molecular level is, however, not clear.
GSH is an important molecule in the protection against reactive oxygen species and, therefore, increasing its foliar concentration might be a good strategy to increase the tolerance of plants to oxidative stress. However, over-expression of γ-ECS or GSHS in transgenic poplar did not increase resistance against ozone or the herbicide paraquat, although this was enhanced in wild-type poplar upon feeding GSH (Will et al., 1997). In addition, exposure to heavy metals represents a stress that can be resisted by increased production of GSH and other sulfur-containing metabolites. Sulfur is important for chelating heavy metals through metalothioneins, i.e. Cys-rich proteins, and phytochelatins (PCs), small polypeptides with repeating γ-EC units (Cobbett, 2000). PCs are synthesized from GSH by phytochelatin synthase (Grill et al., 1989). The sulfhydryl groups of Cys residues bind the heavy metal ions and the resulting complexes are excreted to the vacuole. Heavy metals induce the synthesis of PCs via activation of phytochelatin synthase and induction of γ-ECS (Cobbett, 2000). Apparently, GSH synthesis is the rate-limiting step in PC synthesis as exposure of the γ-ECS-overexpressing poplars to Cd enhanced PC production (Rennenberg and Will, 2000). Although enhanced PC concentration by over-expression of genes for GSH synthesis increased Cd tolerance in Brassica juncea (Zhu et al., 1999a, b), the same effect was not observed in poplar (Arisi et al., 2000; Rennenberg and Will, 2000; Koprivova et al., 2002b). Cd accumulation correlated with PC levels; poplars over-expressing γ-ECS accumulated more Cd, but without increase in Cd tolerance (Koprivova et al., 2002b). GSH is also important for resistance against herbicides. Differences in herbicide toxicity are often based on the capacity of the plants to detoxify the herbicide, e.g. through the glutathione-S-transferase (GST) reaction and subsequent excretion of the conjugate into the vacuole (Edwards et al., 2000). Consequently, growth of transgenic poplars over-expressing γ-ECS in the cytosol or in the chloroplast was less reduced upon treatment with chloroacetanilide herbicides than that of the wild-type (Gullner et al., 2001). It seems, however, that a simple increase in cellular GSH level per se is not able significantly to increase plant tolerance to oxidative stress, but a more precise fine-tuning, e.g. by tissue- or compartment-specific manipulation, might be necessary.
The poplars over-expressing γ-ECS offered an opportunity to address the effects of increased GSH synthesis on the sulfate assimilation pathway because cysteine availability is most critical for the rate of GSH synthesis (Strohm et al., 1995). Although foliar GSH levels were three- to four-fold higher in transgenic poplars over-expressing γ- ECSin the cytosol, foliar activities of enzymes of sulfate assimilation, ATPS, APR, SiR, SAT and OASTL, and their mRNA levels were not different from those of wild-type poplars (Hartmann et al., 2004). The results show that sulfate reduction in poplar is sufficient to provide the additional cysteine necessary to accommodate the enhanced GSH synthesis. By contrast, the increased GSH level in transgenic poplars did not down-regulate ATPS and APR as described for several herbaceous plant species (Lappartient et al., 1999; Vauclare et al., 2002). Exogenous feeding of GSH to the roots, however, caused the APR activity and mRNA accumulation to decrease (Hartmann et al., 2004). The poplars thus reacted differently to increased GSH levels due to feeding and endogenous synthesis. The lack of regulation of APR and ATPS in the transgenic poplar lines must be caused by a second signal that positively influences APR mRNA accumulation and activity and overrides the negative signal of GSH. Note that γ-ECS was over-expressed also in other plant species. However, the effects of the over-expression were either detrimental in tobacco, which ironically suffered from a highly increased oxidative stress (Creissen et al., 1999), or too moderate in Arabidopsis, where GSH levels were increased only by approx. 50 % (Xiang et al., 2001). The physiology of poplar was thus especially suitable for this kind of manipulation. Poplar might therefore serve as an alternative model for genetic engineering if the more generally used herbaceous plants are affected too severely as a result of the manipulations.
SULFATE ASSIMILATION IN PHYSCOMITRELLA PATENS
One of the remaining questions regarding sulfate assimilation is the presence of the PAPS-dependent pathway in plants. PAPS is not only a source of activated sulfate for sulfation reactions but an intermediate of sulfate assimilation in fungi, yeast and some bacteria (Kopriva et al., 2002b). In these organisms APS cannot be reduced directly but a second activation step to PAPS is necessary. The reduction is achieved in a thioredoxin-dependent reaction catalysed by PAPS reductase. As the sulfate assimilation pathway was first resolved in yeast and in PAPS-reducing enteric bacteria Escherichia coli and Salmonella typhimurium (Jones-Mortimer, 1968; Kredich, 1971) and PAPS had been detected in plants (Schiff, 1959), the same sequence of reactions including PAPS reductase was proposed for plant sulfate assimilation (Kopriva and Koprivova, 2004). However, studies with the green alga Chlorella revealed that APS rather than PAPS was reduced (Tsang et al., 1971; Schmidt, 1972). APS reductase activity was detected in a variety of plants and photosynthetic bacteria, and therefore it was considered to represent the major sulfate-reducing enzyme in photosynthetic organisms (Schmidt, 1975; Schmidt and Trüper, 1977). Three isoforms of APR were then cloned from Arabidopsis by complementation of E. coli mutants deficient in PAPS reductase (Gutierrez-Marcos et al., 1996; Setya et al., 1996). The N-terminal part of the mature APR was homologous to E. coli PAPS reductase, with a C-terminal thioredoxin-like extension. Nevertheless, a PAPS-dependent pathway of plant sulfate assimilation was never excluded, in particular when the purification of PAPS reductase from spinach was reported (Schwenn 1989).
Analysis of the Arabidopsis genome did not identify any gene homologous to E. coli PAPS reductase other than APR. As PAPS reductase in plants may have a completely different structure, only analysis of plants lacking APR activity may prove or exclude the PAPS-dependent sulfate assimilation. However, no such plants have yet been described, probably because APR is encoded by small multigene families of 2–3 isoforms in most plant species analysed to date (Gutierrez-Marcos et al., 1996; Setya et al., 1996; Koprivova et al., 2001; Kopriva et al., 2004). This problem was overcome by the use of the moss Physcomitrella patens, which in the last few years has become an increasingly used model system to study the function of plant genes (Schaefer, 2002). The single-copy APR gene in P. patens was disrupted by homologous recombination, resulting in complete loss of the correct transcript and enzymatic activity (Koprivova et al., 2002a). Surprisingly, however, the knockout plants grew on sulfate as the sole sulfur source. The knockouts showed a Cd-sensitive phenotype and reduced flux through sulfate reduction, as measured by incorporation of [35S]sulfate (Koprivova et al., 2002a). Although PAPS reductase activity could not be measured in moss extracts, cDNA and the gene coding for this enzyme were isolated from P. patens. The corresponding recombinant protein possessed PAPS reductase activity, confirming the identity of the gene. The moss P. patens is thus the first plant species for which PAPS reductase was confirmed at the molecular level and also the first organism for which both APS- and PAPS-dependent sulfate assimilation were found to coexist (Koprivova et al., 2002a). Very recently, PAPS reductase was identified in the expressed sequence tag (EST) collection from the spike moss Selaginella lepidophylla, a vascular plant (Kopriva and Koprivova, 2004). PAPS reductase thus must have been present in the common ancestor of bryophytes and vascular plants. This result opens many interesting questions on the role of the two parallel sulfate-reducing systems, their co-ordination and regulation, and the evolution of sulfate assimilation in plants. Because of the rapid and simple method used to obtain gene knockouts, P. patens is an interesting alternative to Arabidopsis for functional genomics. Furthermore, being a lower plant makes P. patens an important species for evolutionary studies and a potential source of new genes.
In addition to possessing a PAPS reductase, bryophytes differ from higher plants in defence mechanisms against heavy metals. Similar to higher plants, exposure to heavy metals induces accumulation of GSH. Phytochelatins were, however, not detected in bryophytes but the heavy metals were chelated and detoxified by GSH (Bruns et al., 2001).
SULFUR METABOLISM IN ALGAE
Green algae have contributed substantially to our understanding of plant sulfur metabolism. Experiments with Chlorella led to the discovery of APS-dependent reduction (Tsang et al., 1971; Schmidt, 1972). Most information on the biochemistry of APS reductase before its cloning from Arabidopsis was derived from studies of various algae, such as Chlorella, Euglena and Porphyra (Schmidt, 1972; Li and Schiff, 1991; Kanno et al., 1996). Euglena is unique among all photosynthesizing organisms as its sulfate assimilation is localized in mitochondria (Brunold and Schiff, 1976). Algae are also an excellent system to study the effects of nutrient variations. For example, in the green alga Dunaliella salina PEPCase activity decreased 11-fold upon sulfur starvation, as did accumulation of Rubisco and light-harvesting proteins, suggesting significant reduction in photosynthetic capacity and changes in the allocation of carbon (Giordano et al., 2000). In the freshwater green alga Chlamydomonas reinhardtii sulfur starvation resulted in changes in cell-wall composition by induction of proteins containing very low cysteine and methionine (Takahashi et al., 2001). Sulfur starvation in Chlamydomonas results in induction of periplasmic arylsulfatase, which enables the algae to utilize alternative sources of sulfur (de Hostos et al., 1988). This specific response to nutritional stress was utilized to screen for Chlamydomonas mutants in response to sulfur starvation (Davies et al., 1994). Although some information about the signal transduction of sulfur starvation in plants has begun to appear (see above), Chlamydomonas is still the only organism in which this sulfur sensing is at least partially understood. Mutations in sac genes (for sulfur acclimation) result in a lack of response to sulfur limitation (Davies et al., 1994). The sac1 mutants are not able to respond to sulfur starvation and induce arylsulfatase and sulfate uptake and assimilation, as well as down-regulation of photosynthesis, whereas in sac2 and sac3 mutants only sulfate uptake and assimilation are disturbed. The Sac1 gene encodes a protein homologous to ion channels that seems to function as a sulfate sensor (Davies et al., 1996). The Sac3 encodes an Snf1-like kinase; its disruption leads to de-repression of the arylsulfatases even at normal sulfur concentrations but, by contrast, to the inability to induce fully the sulfate uptake system (Davies et al., 1999; Ravina et al., 2002). Sac2 is involved in post-transcriptional regulation of APR, but has not yet been identified (Ravina et al., 2002).
In contrast to reports on freshwater algae, there have been few studies of the basic sulfur metabolism of marine algae because, contrary to the other major inorganic nutrients nitrogen, phosphate and silicate, sulphur is assumed not to limit the growth of marine phytoplankton (Giordano et al., 2005). On the other hand, the production of dimethyl sulfide (DMS) has attracted considerable interest since Lovelock et al. (1978) showed it to be the dominant sulfur gas in the sea. Research interest was further stimulated when Charlson et al. (1987) highlighted the connection between marine micro-organisms, the sulfur aerosol particles that form when DMS oxidizes in air and global climate. The major precursor for DMS is the tertiary sulfonium compound dimethylsulfonio-propionate (DMSP), which is found in various marine phytoplankton, some seaweeds and a few saltmarsh and terrestrial plants. In marine phytoplankton DMSP has some of the properties of a compatible solute in that it accumulates in response to changes in salinity and low temperature but may also represent an overflow mechanism for excess reduced compounds and energy or to counter oxidative stress (Stefels, 2000; Sunda et al., 2002). The pathway of DMSP synthesis differs in higher plants and the green macroalga Ulva intestinalis (Hanson et al., 1994; Gage et al., 1997). Whereas in higher plants S-methylmethionine is metabolized to DMSP-aldehyde and than oxidized to DMSP, in the marine algae Met is first transaminated to 4-methylthio-2-oxobutyrate, reduced to methylthio hydroxybutyrate, and methylated and decarboxylated to DMSP (Gage et al., 1997). However, nothing is known about the control of DMSP synthesis and its interaction with sulfate uptake and assimilation.
CONCLUSIONS
Great progress has been achieved recently in linking sulfur metabolism to a plethora of cell processes and has made sulfur deficiency one of the best characterized environmental conditions. The molecular mechanisms controlling the sulfate assimilation pathway are still far from being understood. The role of Arabidopsis as a model plant in deciphering these mechanisms will thus be further strengthened due to the wealth in genetic resources and available genomics data. This review, however, demonstrates that not all data from Arabidopsis are simply transferable to other species and that investigations of other plant species will be necessary to address specific questions of sulfur metabolism.
Acknowledgments
Research in my laboratory at JIC is supported by the Biotechnology and Biological Sciences Research Council (BBSRC). Funding from German Research Council DFG and Swiss National Science Foundation is also acknowledged. I would like to thank Dr A. Koprivova for critical reading of the manuscript.
LITERATURE CITED
- Arisi AC, Noctor G, Foyer CH, Jouanin L. 1997. Modification of thiol contents in poplars (Populus tremula×P. alba) overexpressing enzymes involved in glutathione synthesis. Planta 203: 362–372. [DOI] [PubMed] [Google Scholar]
- Arisi A-CM, Mocquot B, Lagriffoul A, Mench M, Foyer CH, Jouanin L. 2000. Responses to cadmium in leaves of transformed poplars overexpressing γ-glutamylcysteine synthetase. Physiologia Plantarum 109: 143–149. [Google Scholar]
- Awazuhara M, Kim H, Goto DB, Matsui A, Hayashi H, Chino M, Kim S-G, et al. 2002. A 235-bp region from a nutritionally regulated soybean seed-specific gene promoter can confer its sulfur and nitrogen response to a constitutive promoter in aerial tissues of Arabidopsis thaliana. Plant Science 163: 75–82. [Google Scholar]
- Barron D, Varin L, Ibrahim RK, Harborne JB, Williams CA. 1988. Sulfated flavonoids—An update. Phytochemistry 27: 2375–2395. [Google Scholar]
- Barroso C, Romero LC, Cejudo FJ, Vega JM, Gotor C. 1999. Salt-specific regulation of the cytosolic O-acetylserine(thiol)lyase gene from Arabidopsis thaliana is dependent on abscisic acid. Plant Molecular Biology 40: 729–736. [DOI] [PubMed] [Google Scholar]
- Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R. 2002. Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. Journal of Biological Chemistry 277: 30629–30634. [DOI] [PubMed] [Google Scholar]
- Bick JA, Setterdahl AT, Knaff DB, Chen Y, Pitcher LH, Zilinskas BA, et al. 2001. Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry 40: 9040–9048. [DOI] [PubMed] [Google Scholar]
- Bolchi A, Petrucco S, Tenca PL, Foroni C, Ottonello S. 1999. Coordinate modulation of maize sulphate permease and ATP sulphurylase mRNAs in response to variations in sulphur nutritional status: stereospecific down-regulation by L-cysteine. Plant Molecular Biology 39: 527–537. [DOI] [PubMed] [Google Scholar]
- Brühl A, Haverkamp T, Gisselmann G, Schwenn JD. 1996. A cDNA clone from Arabidopsis thaliana encoding plastidic ferredoxin:sulfite reductase. Biochemica et Biophysica Acta 1295: 119–124. [DOI] [PubMed] [Google Scholar]
- Brunner M, Kocsy G, Rüegsegger A, Schmutz D, Brunold C. 1995. Effect of chilling on assimilatory sulfate reduction and glutathione synthesis in maize. Journal of Plant Physiology 146: 743–747. [Google Scholar]
- Brunold C. 1990. Reduction of sulfate to sulfide. In: Rennenberg H, Brunold C, De Kok LJ, Stulen I, eds. Sulfur nutrition and sulfur assimilation in higher plants. The Hague: SPB Academic Publishing, 13–31.
- Brunold C, Schiff JA. 1976. Studies of sulfate utilization by algae. 15. Enzymes of assimilatory sulfate reduction in Euglena and their cellular localization. Plant Physiology 57: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunold C, Schmidt A. 1978. Regulation of sulfate assimilation in plants. 7. Cysteine inactivation of adenosine 5′-phosphosulfate sulfotransferase in Lemna minor L. Plant Physiology 61: 342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunold C, Suter M. 1984. Regulation of sulfate assimilation by nitrogen nutrition in the duckweed Lemna minor L. Plant Physiology 76: 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunold C, Suter M, Lavanchy P. 1987. Effect of high and low sulfate concentrations on adenosine 5′-phosphosulfate sulfotransferase from Lemna minor L. Physiologia Plantarum 70: 168–174. [Google Scholar]
- Bruns I, Sutter K, Menge S, Neumann D, Krauss GJ. 2001. Cadmium lets increase the glutathione pool in bryophytes. Journal of Plant Physiology 158: 79–89. [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ. 2004. Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. Journal of Experimental Botany 55: 1765–1773. [DOI] [PubMed] [Google Scholar]
- Burgener M, Suter M, Jones S, Brunold C. 1998. Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiology 116: 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN. 1984. Sulfate assimilation in C4 plants. Plant Physiology 75: 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. 1994. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annual Review of Plant Physiology and Plant Molecular Biology 45: 113–141. [Google Scholar]
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. 1987. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature 326: 655–661. [Google Scholar]
- Chiba Y, Ishikawa M, Kijima F, Tyson RH, Kim J, Yamamoto A, et al. 1999. Evidence for autoregulation of cystathionine gamma-synthase mRNA stability in Arabidopsis. Science 286: 1371–1374. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Sakurai R, Yoshino M, Ominato K, Ishikawa M, Onouchi H, et al. 2003. S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 100: 10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett CS. 2000. Phytochelatins and their roles in heavy metal detoxification. Plant Physiology 123: 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK. 2002. Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Molecular Plant Microbe Interactions 15: 411–420. [DOI] [PubMed] [Google Scholar]
- Collier M, Fotelli M, Nahm M, Kopriva S, Rennenberg H, Hanke D, et al. 2003. Regulation of nitrogen uptake by Fagus sylvatica on a whole plant level—Interactions between cytokinins and soluble N compounds. Plant Cell Environment 26: 1549–1560. [Google Scholar]
- Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, et al.1999. Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11: 1277–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. 1994. Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell 6: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. 1996. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO Journal 1: 2150–2159. [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. 1999. Sac3, a Snf1-like serine threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulis AG, Debian N, Kingston-Smith AH, Foyer CH. 1997. Differential localization of antioxidants in maize leaves. Plant Physiology 114: 1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards GE, Huber SC. 1981. The C4 Pathway. In: Hatch MD, Boardman NK, eds. Biochemistry of plants. New York: Academic Press, 238–281.
- Edwards GE, Franceschi VR, Ku MS, Voznesenskaya EV, Pyankov VI, Andreo CS. 2001. Compartmentation of photosynthesis in cells and tissues of C4 plants. Journal of Experimental Botany 52: 577–590. [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. 2000. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends in Plant Science 5: 193–198. [DOI] [PubMed] [Google Scholar]
- Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, et al. 2001. Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. Journal of Biological Chemistry 276: 46989–46994. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P. 2001. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56: 5–51. [DOI] [PubMed] [Google Scholar]
- Fodor J, Gullner G, Adam AL, Barna B, Komives T, Kiraly Z. 1997. Local and systemic responses of antioxidants to tobacco mosaic virus infection and to salicylic acid in tobacco—Role in systemic acquired resistance. Plant Physiology 114: 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Rennenberg H. 2000. Regulation of glutathione synthesis and its role in abiotic and biotic stress defence. In: Brunold C, Rennenberg H, De Kok LJ, Stulen I, Davidian JC, eds. Sulfur nutrition and sulfur assimilation in higher plants: molecular, biochemical and physiological aspects. Berne: Paul Haupt, 127–153.
- Freeman JL, Garcia D, Kim D, Hopf A, Salt DE. 2005. Constitutively elevated salicylic acid signals glutathione-mediated nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Physiology 137: 1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo P, Jimenez MJ, Mathieu C, Duret L, Gallesi D, Van de Sype G, et al. 2001. A Medicago truncatula homoglutathione synthetase is derived from glutathione synthetase by gene duplication. Plant Physiology 126: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo P, Harrison J, Norman C, Hernandez Jimenez MJ, Van de Sype G, et al. 2005. Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Molecular Plant Microbe Interactions 18: 254–259. [DOI] [PubMed] [Google Scholar]
- Gage DA, Rhodes D, Nolte KD, Hicks WA, Leustek T, Cooper AJ, et al. 1997. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 387: 891–894. [DOI] [PubMed] [Google Scholar]
- Gerwick BC, Black CC. 1979. Sulfur assimilation in C4 plants. Plant Physiology 64: 590–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick BC, Ku SB, Black CC. 1980. Initiation of sulfate activation: a variation in C4 photosynthesis plants. Science 209: 513–515. [DOI] [PubMed] [Google Scholar]
- Gidda SK, Miersch O, Levitin A, Schmidt J, Wasternack C, Varin L. 2003. Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. Journal of Biological Chemistry 278: 17895–17900. [DOI] [PubMed] [Google Scholar]
- Giordano M, Pezzoni V, Hell R. 2000. Strategies for the allocation of resources under sulfur limitation in the green alga Dunaliella salina. Plant Physiology 124: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Norici A, Hell R. 2005. Sulfur and phytoplankton: acquisition, metabolism and impact on the environment. New Phytologist 166: 371–382. [DOI] [PubMed] [Google Scholar]
- Gómez LD, Vanacker H, Buchner P, Noctor G, Foyer CH. 2004. Intercellular distribution of glutathione synthesis in maize leaves and its response to short-term chilling. Plant Physiology 134: 1662–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Löffler S, Winnacker E-L, Zenk MH. 1989. Phytochelatins, the heavy-metal binding peptides of plants, are synthesised from glutathione by a specific γ-glutamylcysteine dipeptidyl transpeptidase (phytochelatine synthase). Proceedings of the National Academy of Sciences of the USA 86: 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan LM, Zhao J, Scandalios JG. 2000. Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant Journal 22: 87–95. [DOI] [PubMed] [Google Scholar]
- Gullner G, Kömives T, Rennenberg H. 2001. Enhanced tolerance of transgenic poplar plants overexpressing γ-glutamylcysteine synthetase towards chloroacetanilide herbicides. Journal of Experimental Botany 52: 971–979. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL. 1996. Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proceedings of the National Academy of Sciences of the USA 93: 13377–13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Rivoal J, Paquet L, Gage DA. 1994. Biosynthesis of 3-dimethylsulfonio-propionatein Wollastonia biflora (L.) DC. Evidence that S-methylmethionine is an intermediate. Plant Physiology 105: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada E, Kusano T, Sano H. 2000. Differential expression of genes encoding enzymes involved in sulfur assimilation pathways in response to wounding and jasmonate in Arabidopsis thaliana. Journal of Plant Physiology 156: 272–276. [Google Scholar]
- Harrison J, Jamet A, Muglia CI, Van de Sype G, Aguilar OM, et al. 2005. Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. Journal of Bacteriology 187: 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T, Hönicke P, Wirtz M, Hell R, Rennenberg H, Kopriva S. 2004. Sulfate assimilation in poplars (Populus tremula×P. alba) overexpressing γ-glutamylcysteine synthetase in the cytosol. Journal of Experimental Botany 55: 837–845. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy, and ultrastructure. Biochimica et Biophysica Acta 895: 81–106. [Google Scholar]
- Hawkesford M, Wray J. 2000. Molecular genetics of sulphate assimilation. Advances in Botanical Research 33: 159–223. [Google Scholar]
- Hell R, Bergmann L. 1990. γ-glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localisation. Planta 180: 603–612. [DOI] [PubMed] [Google Scholar]
- Hell R, Bork C, Bogdanova N, Frolov I, Hauschild R. 1994. Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine (thiol) lyase from Arabidopsis thaliana. FEBS Letters 351: 257–262. [DOI] [PubMed] [Google Scholar]
- Hell R, Jost R, Berkowitz O, Wirtz M. 2002. Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22: 245–257. [DOI] [PubMed] [Google Scholar]
- Herschbach C, Rennenberg H. 2001. Significance of phloem-translocated organic sulfur compounds for the regulation of sulfur nutrition. Progress in Botany 62: 177–192. [Google Scholar]
- Herschbach C, Jouanin L, Rennenberg H. 1998. Overexpression of gamma-glutamylcysteine synthetase, but not of glutathione synthetase, elevates glutathione allocation in the phloem of transgenic poplar trees. Plant and Cell Physiology 39: 447–451. [Google Scholar]
- Herschbach C, van Der Zalm E, Schneider A, Jouanin L, De Kok LJ, Rennenberg H. 2000. Regulation of sulfur nutrition in wild-type and transgenic poplar over-expressing gamma-glutamylcysteine synthetase in the cytosol as affected by atmospheric H2S. Plant Physiology 124: 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R. 2003. Molecular aspects of methionine biosynthesis. Trends in Plant Science 8: 259–262. [DOI] [PubMed] [Google Scholar]
- Hesse H, Trachsel N, Suter M, Kopriva S, von Ballmoos P, Rennenberg H, et al. 2003. Effect of glucose on assimilatory sulfate reduction in roots of Arabidopsis thaliana. Journal of Experimental Botany 54: 1701–1709. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. 2003. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant Journal 33: 651–663. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. 2004. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA 101: 10205–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, et al. 2005. Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. Journal of Biological Chemistry 280: 25590–25595. [DOI] [PubMed] [Google Scholar]
- Hopkins L, Parmar S, Bouranis DL, Howarth JR, Hawkesford MJ. 2004. Coordinated expression of sulfate uptake and components of the sulfate assimilatory pathway in maize. Plant Biology 6: 408–414. [DOI] [PubMed] [Google Scholar]
- Hopkins L, Parmar S, Blaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ. 2005. O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiology 138: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Togasaki RK, Grossman AR. 1988. Purification and biosynthesis of derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. Journal of Cell Biology 106: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylton CM, Rawsthorne S, Smith AM, Jones DA, Woolhouse HW. 1988. Glycine decarboxylase is confined to the bundle-sheath cells of leaves of C3–C4 intermediate species. Planta 175: 452–459. [DOI] [PubMed] [Google Scholar]
- Jiang M, Zhang J. 2001. Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant and Cell Physiology 42: 1265–1273. [DOI] [PubMed] [Google Scholar]
- Jones-Mortimer MC. 1968. Positive control of sulphate reduction in Escherichia coli. Biochemical Journal 110: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost R, Altschmied L, Bloem E, Bogs J, Gershenzon J, Hähnel U, et al. 2005. Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynthesis Research 86: 491–508. [DOI] [PubMed]
- Kanno N, Nagahisa E, Sato M, Sato Y. 1996. Adenosine 5′-phosphosulfate sulfotransferase from the marine macroalga Porphyra yezoensis Ueda (Rhodophyta): stabilization, purification, and properties. Planta 198: 440–446. [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, et al. 2004a. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H. 2004b. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiology 136: 4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Berkowitz O, Hell R, Noji M, Saito K. 2005. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiology 137: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Rees DC. 1997. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annual Review of Biochemistry 66: 233–267. [DOI] [PubMed] [Google Scholar]
- Klapheck S. 1988. Homoglutathione—Isolation, quantification and occurrence in legumes. Physiologia Plantarum 74: 727–732. [Google Scholar]
- Klein M, Papenbrock J. 2004. The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. Journal of Experimental Botany 55: 1809–1820. [DOI] [PubMed] [Google Scholar]
- Klonus D, Hofgen R, Willmitzer L, Riesmeier JW. 1994. Isolation and characterization of two cDNA clones encoding ATP-sulfurylases from potato by complementation of a yeast mutant. Plant Journal 6: 105–112. [DOI] [PubMed] [Google Scholar]
- Kocsy G, Brunner M, Rüegsegger A, Stamp P, Brunold C. 1996. Glutathione synthesis in maize genotypes with different sensitivities to chilling. Planta 198: 365–370. [Google Scholar]
- Kocsy G, Owttrim G, Brander K, Brunold C. 1997. Effect of chilling on the diurnal rhythm of enzymes involved in protection against oxidative stress in a chilling-tolerant and a chilling-sensitive maize genotype. Physiologia Plantarum 99: 249–254. [Google Scholar]
- Kocsy G, von Ballmoos P, Suter M, Ruegsegger A, Galli U, Szalai G, et al. 2000. Inhibition of glutathione synthesis reduces chilling tolerance in maize. Planta 211: 528–536. [DOI] [PubMed] [Google Scholar]
- Kocsy G, Galiba G, Brunold C. 2001. Role of glutathione in adaptation and signalling during chilling and cold acclimation in plants. Physiologia Plantarum 113: 158–164. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Koprivova A. 2003. Sulphate assimilation: a pathway which likes to surprise. In: Abrol YP, Ahmad A, eds. Sulphur in higher plants. Dordrecht: Kluwer, 87–112.
- Kopriva S, Koprivova A. 2004. Plant adenosine 5′-phosphosulphate reductase: the past, the present, and the future. Journal of Experimental Botany 55: 1775–1783. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Koprivova A. 2005. Sulfate assimilation and glutathione synthesis in C4 plants. Photosynthesis Research 86: 363–372. [DOI] [PubMed]
- Kopriva S, Rennenberg H. 2004. Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. Journal of Experimental Botany 55: 1831–1842. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Chu C-C, Bauwe H. 1996. Molecular phylogeny of Flaveria as deduced from the analysis of H-protein nucleotide sequences. Plant Cell and Environment 19: 1028–1036. [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsel N, Catalano C, Suter M, et al. 1999. Light regulation of assimilatory sulfate reduction in Arabidopsis thaliana. Plant Journal 20: 37–44. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Jones S, Koprivova A, Suter M, von Ballmoos P, Brander K, et al. 2001. Influence of chilling stress on the intercellular distribution of assimilatory sulfate reduction and thiols in Zea mays. Plant Biology 3: 24–31. [Google Scholar]
- Kopriva S, Suter M, von Ballmoos P, Hesse H, Krähenbühl U, Rennenberg H, et al. 2002a. Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiology 130: 1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Büchert T, Fritz G, Suter M, Benda R, Schünemann V, et al. 2002b. The presence of an iron–sulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadenosine 5′-phosphosulfate for sulfate assimilation. Journal of Biological Chemistry 277: 21786–21791. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Hartmann T, Massaro G, Hönicke P, Rennenberg H. 2004. Regulation of sulfate assimilation by nitrogen and sulfur nutrition in poplar trees. Trees 18: 320–326. [Google Scholar]
- Koprivova A, Suter M, Op den Camp R, Brunold C, Kopriva S. 2000. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiology 122: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Melzer M, von Ballmoos P, Mandel T, Brunold C, Kopriva S. 2001. Assimilatory sulfate reduction in C3, C3–C4, and C4 species of Flaveria. Plant Physiology 127: 543–550. [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Meyer A, Schween G, Herschbach C, Reski R, Kopriva S. 2002a. Functional knockout of the adenosine 5′ phosphosulfate reductase gene in Physcomitrella patens revives an old route of sulfate assimilation. Journal of Biological Chemistry 277: 32195–32201. [DOI] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S, Jäger D, Will B, Jouanin L, Rennenberg H. 2002b. Evaluation of poplar lines overexpressing enzymes of glutathione synthesis for phytoremediation of cadmium. Plant Biology 4: 664–670. [Google Scholar]
- Kredich NM. 1971. Regulation of L-cysteine biosynthesis in Salmonella typhimurium. Journal of Biological Chemistry 246: 3474–3484. [PubMed] [Google Scholar]
- Kreft O, Hoefgen R, Hesse H. 2003. Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiology 131: 1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzwieser J, Rennenberg H. 1998. Sulphate uptake and xylem loading of mycorrhizal beech roots. New Phytologist 140: 319–329. [DOI] [PubMed] [Google Scholar]
- Kreuzwieser J, Herschbach C, Rennenberg H. 1996. Sulphate uptake and xylem loading of non-mycorrhizal excised roots of young beech (Fagus sylvatica) trees. Plant Physiology and Biochemistry 34: 409–416. [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Kramer U, Sato S, et al. 2005. The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku MSB, Wu JR, Dai ZY, Scott RA, Chu C, Edwards GE. 1991. Photosynthetic and photorespiratory characteristics of Flaveria species. Plant Physiology 96: 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. 2002. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology 5: 325–331. [DOI] [PubMed] [Google Scholar]
- Kutz A, Muller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW. 2002. A role for nitrilase3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant Journal 30: 95–106. [DOI] [PubMed] [Google Scholar]
- Laetsch WM. 1974. The C4 syndrom: a structural analysis. Annual Review of Plant Physiology 25: 27–52. [Google Scholar]
-
Lappartient AG, Touraine B. 1996. Demand-driven control of root ATP sulphurylase activity and
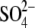 uptake in intact canola. The role of phloem-translocated glutathione. Plant Physiology 111: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
uptake in intact canola. The role of phloem-translocated glutathione. Plant Physiology 111: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar] - Lappartient AG, Vidmar JJ, Leustek T, Glass ADM, Touraine B. 1999. Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant Journal 18: 89–95. [DOI] [PubMed] [Google Scholar]
- Leustek T, Murillo M, Cervantes M. 1994. Cloning of a cDNA encoding ATP sulfurylase from Arabidopsis thaliana by functional expression in Saccharomyces cerevisiae. Plant Physiology 105: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP. 2000. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annual Review of Plant Physiology and Plant Molecular Biology 51: 141–165. [DOI] [PubMed] [Google Scholar]
- Li JY, Schiff JA. 1991. Purification and properties of adenosine 5′-phosphosulfate sulfotransferase from Euglena. Biochemical Journal 274: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock JE, Maggs RJ, Rasmussen RA. 1978. Atmospheric sulphur and the natural sulphur cycle. Nature 237: 452–453. [Google Scholar]
- Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Pena A, et al. 2000. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant Journal 24: 559–567. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yarnaya T, Takahashi H. 2003. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways Plant Physiology 132: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. 2004. A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant Journal 38: 779–789. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. 2005. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant Journal 42: 305–314. [DOI] [PubMed] [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. 1999. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiology 121: 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Dalton DA, Ramos J, Clemente MR, Rubio MC, Becana M. 2003. Biochemistry and molecular biology of antioxidants in the rhizobia–legume symbiosis. Plant Physiology 133: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard R, Alban S, de Ruffray P, Jamois F, Franz G, Fritig B, et al. 2004. Beta-1,3 glucan sulfate, but not beta-1,3 glucan, induces the salicylic acid signaling pathway in tobacco and Arabidopsis. Plant Cell 16: 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Olsen CE, Halkier BA. 2002. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids 22: 279–295. [DOI] [PubMed] [Google Scholar]
- Mithen R. 2001. Glucosinolates—biochemistry, genetics and biological activity. Plant Growth Regulation 34: 91–103. [Google Scholar]
- Monson RK, Moore BD. 1989. On the significance of C3–C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant Cell and Environment 12: 689–699. [Google Scholar]
- Moore R, Black CC. 1979. Nitrogen assimilation pathways in leaf mesophyll and bundle sheath cells of C4 photosynthesis plants formulated from comparative studies with Digitaria sanguinalis (L.) Scop. Plant Physiology 64: 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan WH, Dong XN. 2003. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Neuenschwander U, Suter M, Brunold C. 1991. Regulation of sulfate assimilation by light and O-acetyl-L-serine in Lemna minor L. Plant Physiology 97: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. 2003. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant Journal 33: 633–650. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, et al. 2005. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiology 138: 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Strohm M, Jouanin L, Kunert K-J, Foyer CH, Rennenberg H. 1996. Synthesis of glutathione in leaves of transgenic poplar overexpressing γ-glutamylcysteine synthetase. Plant Physiology 112: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Jouanin L, Arisi A-CM, Valadier M-H, Roux Y, Foyer CH. 1997a. Light-dependent modulation of foliar glutathione synthesis and associated amino acid metabolism in transformed poplar. Planta 202: 357–369. [Google Scholar]
- Noctor G, Arisi A-CM, Jouanin L, Valadier M-H, Roux Y, Foyer CH. 1997b. The role of glycine in determining the rate of glutathione synthesis in poplar. Possible implications for glutathione production during stress. Physiologia Plantarum 100: 255–263. [Google Scholar]
- Noctor G, Arisi A-CM, Jouanin L, Foyer CH. 1998a. Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiology 118: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Arisi A-CM, Jouanin L, Kunert KJ, Rennenberg H, Foyer CH. 1998b. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. Journal of Experimental Botany 49: 623–647. [Google Scholar]
- Noctor G, Arisi A-CM, Jouanin L, Foyer C. 1999. Photorespiratory glycine enhances glutathione accumulation in both the chloroplastic and cytosolic compartments. Journal of Experimental Botany 50: 1157–1167. [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K. 1998. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. Journal of Biological Chemistry 273: 32739–32745. [DOI] [PubMed] [Google Scholar]
- Nowak K, Luniak N, Witt C, Wustefeld Y, Wachter A, Mendel RR, et al. 2004. Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant and Cell Physiology 45: 1889–1894. [DOI] [PubMed] [Google Scholar]
- Nussbaum S, Schmutz D, Brunold C. 1988. Regulation of assimilatory sulfate reduction by cadmium in Zea-mays-L. Plant Physiology 88: 1407–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama N, Takei K, Sakakibara H, Hayashi H, Yoneyama T, Fujiwara T. 2002. Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant and Cell Physiology 43: 1493–1501. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Kasajima I, Fujiwara T, Naito S. 2005. Isolation and characterization of an Arabidopsis mutant that overaccumulates O-Acetyl-L-Ser. Plant Physiology 136: 3209–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passera C, Ghisi R. 1982. ATP sulphurylase and O-acetylserine sulphydrylase in isolated mesophyll protoplasts and bundle sheath strands of S-deprived maize leaves. Journal of Experimental Botany 33: 432–438. [Google Scholar]
- Pastori GM, Mullineaux PM, Foyer CH. 2000. Post-transcriptional regulation prevents accumulation of glutathione reductase protein and activity in the bundle sheath cells of maize. Plant Physiology 122: 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postgate JR. 1984. The sulphate-reducing bacteria. Cambridge: Cambridge University Press.
- Rathnam CKM, Edwards GE. 1976. Distribution of nitrate-assimilating enzymes between mesophyll protoplasts and bundle sheath cells in leaves of three groups of C-4 plants. Plant Physiology 57: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravina CG, Chang CI, Tsakraklides GP, McDermott JP, Vega JM, Leustek T, et al. 2002. The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Physiology 130: 2076–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennenberg H. 1983. Role of O-acetylserine in hydrogen-sulfide emission from pumpkin leaves in response to sulfate. Plant Physiology 73: 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennenberg H, Will B. 2000. Phytochelatin production and cadmium accumulation in transgenic poplar (Populus tremula×P. alba). In: Brunold C, Rennenberg H, De Kok LJ, Stulen I, Davidian J-C, eds. Sulfur nutrition and sulfur assimilation in higher plants. Berne: Paul Haupt, 393–398.
- Reuveny Z, Dougall DK, Trinity PM. 1980. Regulatory coupling of nitrate and sulfate assimilation pathways in cultured tobacco cells. Proceedings of the National Academy of Sciences of the USA 77: 6670–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. 1998. Jasmonate and salicylate as global signals for defense gene expression. Current Opinion in Plant Biology 1: 404–411. [DOI] [PubMed] [Google Scholar]
- van Rhijn P, Vanderleyden J. 1995. The Rhizobium–plant symbiosis. Microbiological Reviews 59: 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. 2001. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant Journal 26: 421–433. [DOI] [PubMed] [Google Scholar]
- Rotte C, Leustek T. 2000. Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiology 124: 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. 2004. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiology 136: 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson ME, Campbell WH, Larsson CM. 1995. The influence of cytokinins in nitrate regulation of nitrate reductase-activity and expression in barley. Physiologia Plantarum 93: 533–539. [Google Scholar]
- Sanda S, Leustek T, Theisen MJ, Garavito RM, Benning C. 2001. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. Journal of Biological Chemistry 276: 3941–3946. [DOI] [PubMed] [Google Scholar]
- Schaefer DG. 2002. A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annual Review of Plant Biology 53: 477–501. [DOI] [PubMed] [Google Scholar]
- Schiff JA. 1959. Studies on sulfate utilization by Chlorella pyrenoidosa using sulfate-S-35—The occurrence of S-adenosyl methionine. Plant Physiology 34: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. 1972. On the mechanism of photosynthetic sulfate reduction. An APS-sulfotransferase from Chlorella. Archives of Microbiology 84: 77–86. [DOI] [PubMed] [Google Scholar]
- Schmidt A. 1975. Distribution of the APS-sulfotransferase activity among higher plants. Plant Science Letters 5: 407–415. [Google Scholar]
- Schmidt A, Trüper HG. 1977. Reduction of adenylylsulfate and 3′-phosphoadenylylsulfate in phototrophic bacteria. Experientia 33: 1008–1009. [DOI] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. 1984. Intercellular localization of assimilatory sulfate reduction in leaves of Zea mays and Triticum aestivum. Plant Physiology 74: 866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. 1985. Localization of nitrite and sulfite reductase in bundle sheath and mesophyll cells of maize leaves. Physiologia Plantarum 64: 523–528. [Google Scholar]
- Schwenn JD. 1989. Sulfate assimilation in higher plants: a thioredoxin-dependent PAPS-reductase from spinach leaves. Zeitschrift für Naturforschung 44c: 504–508. [Google Scholar]
- Schupp R, Rennenberg H. 1992. Changes in sulfur metabolism during needle development of Norway spruce. Botanica Acta 105: 180–189. [Google Scholar]
- Seegmüller S, Schulte M, Herschbach C, Rennenberg H. 1996. Interactive effects of mycorrhization and elevated atmospheric CO2 on sulphur nutrition of young pedunculate oak (Quercus robur L.) trees. Plant Cell and Environment 19: 418–426. [Google Scholar]
- Setya A, Murillo M, Leustek T. 1996. Sulfate reduction in higher plants—molecular evidence for a novel 5′-adenylylsulfate reductase. Proceedings of the National Academy of Sciences of the USA 93: 13383–13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller-Cepeda JH, Chen THH, Fuchigami LH. 1991. High performance liquid chromatography analysis of reduced and oxidised glutathione in woody plant tissues. Plant and Cell Physiology 32: 1179–1185. [Google Scholar]
- Sheen J. 1999. C4 gene expression. Annual Review of Plant Physiology and Plant Molecular Biology 50: 187–217. [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, et al. 2002. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant Journal 29: 475–486. [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. 1995. Plant members of a family of sulfate transporters reveal functional subtypes. Proceedings of the National Academy of Sciences of the USA 92: 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefels J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants Journal of Sea Research 43: 183–197. [Google Scholar]
- Strohm M, Jouanin L, Kunert KJ, Pruvost C, Polle A, Foyer CH, et al. 1995. Regulation of glutathione synthesis in leaves of transgenic poplar (Populus tremula×P. alba) overexpressing glutathione synthetase. Plant Journal 7: 141–145. [Google Scholar]
- Sunda W, Kieber DJ, Kiene RP, Huntsman S. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418: 317–320. [DOI] [PubMed] [Google Scholar]
- Suter M, Lavanchy P, von Arb C, Brunold C. 1986. Regulation of sulfate assimilation by amino acids in Lemna minor L. Plant Science 44: 125–132. [Google Scholar]
- Suter M, von Ballmoos P, Kopriva S, Op den Camp R, Schaller J, Kuhlemeier C, et al. 2000. Adenosine 5′-phosphosulfate sulfotransferase and adenosine 5′-phosphosulfate reductase are identical enzymes. Journal of Biological Chemistry 275: 930–936. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, de Almeida Engler J, et al. 1997. Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA 94: 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K. 2000. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant Journal 23: 171–182. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Braby CE, Grossman AR. 2001. Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiology 127: 665–673. [PMC free article] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. 2001. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant and Cell Physiology 42: 85–93. [DOI] [PubMed] [Google Scholar]
- Tate R, Riccio A, Iaccarino M, Patriarca EJ. 1997. A cysG mutant strain of Rhizobium etli pleiotropically defective in sulfate and nitrate assimilation. Journal of Bacteriology 179: 7343–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trüper HG, Fischer U. 1982. Anaerobic oxidation of sulphur compounds as electron donors for bacterial photosynthesis. Philosophical Transactions of the Royal Society B: Biological Sciences 298: 529–542. [Google Scholar]
- Tsang ML-S, Goldschmidt EE, Schiff JA. 1971. Adenosine-5′-phosphosulfate (APS35) as an intermediate in the conversion of adenosine-3′-phosphate-5′-phosphosulfate (PAPS35) to acid volatile radioactivity. Plant Physiology 47 (Suppl.): 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides G, Martin M, Chalam R, Tarczynski MC, Schmidt A, Leustek T. 2002. Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant Journal 32: 879–889. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio SP, Teichmann T. 2004. Poplar genomics is getting popular: the impact of the poplar genome project on tree research. Plant Biology 6: 2–4. [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Day DA. 1997. Metabolite transport across symbiotic membranes of legume nodules. Annual Review of Plant Physiology and Plant Molecular Biology 48: 493–523. [DOI] [PubMed] [Google Scholar]
- Varin L, Marsolais F, Richard M, Rouleau M. 1997. Sulfation and sulfotransferases 6: Biochemistry and molecular biology of plant sulfotransferases. FASEB Journal 11: 517–525. [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, et al. 2002. Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible to negative control by thiols than ATP sulphurylase. Plant Journal 31: 729–740. [DOI] [PubMed] [Google Scholar]
- Wachter A, Wolf S, Steininger H, Bogs J, Rausch T. 2005. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant Journal 41: 15–30. [DOI] [PubMed] [Google Scholar]
- Westerman S, Stulen I, Suter M, Brunold C, De Kok LJ. 2001. Atmospheric H2S as sulphur source for Brassica oleracea: consequences for the activity of the enzymes of the assimilatory sulphate reduction pathway. Plant Physiology and Biochemistry 39: 425–432. [Google Scholar]
- Will B, Eiblmeier M, Langebartels C, Rennenberg H. 1997. Consequences of chronic ozone exposure in transgenic poplars overexpressing enzymes of the glutathione metabolism. In: Cram WJ, De Kok LJ, Stulen I, Brunold C, Rennenberg H, eds. Sulfur metabolism in higher plants. Leiden: Backhuys Publishers, 257–259.
- Wittstock U, Halkier BA. 2002. Glucosinolate research in the Arabidopsis era. Trends in Plant Science 7: 263–270. [DOI] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ. 1998. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10: 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Werner BL, Christensen EM, Oliver DJ. 2001. The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiology 126: 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K. 2002. Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant Journal 29: 465–473. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H. 2003. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiology 131: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FJ, Wood AP, McGrath SP. 1999. Effects of sulphur nutrition on growth and nitrogen fixation of pea (Pisum sativum L.). Plant and Soil 212: 209–219. [Google Scholar]
- Zhu YL, Pilon-Smits EAH, Jouanin L, Terry N. 1999a. Overexpression of glutathione synthetase in Indian mustard enhanced cadmium accumulation and tolerance. Plant Physiology 119: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Pilon-Smits EAH, Tarun AS, Weber SU, Jouanin L, Terry N. 1999b. Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing γ-glutamylcysteine synthetase. Plant Physiology 121: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]