Abstract
Protein kinase Cδ (PKCδ) is an essential component of the intrinsic apoptotic program. Following DNA damage, such as exposure to UV radiation, PKCδ is cleaved in a caspase-dependent manner, generating a constitutively active catalytic fragment (PKCδ-cat), which is necessary and sufficient for keratinocyte apoptosis. We found that in addition to inducing apoptosis, expression of PKCδ-cat caused a pronounced G2/M cell cycle arrest in both primary human keratinocytes and immortalized HaCaT cells. Consistent with a G2/M arrest, PKCδ-cat induced phosphorylation of Cdk1 (Tyr15), a critical event in the G2/M checkpoint. Treatment with the ATM/ATR inhibitor caffeine was unable to prevent PKCδ-cat-induced G2/M arrest, suggesting that PKCδ-cat is functioning downstream of ATM/ATR in the G2/M checkpoint. To better understand the role of PKCδ and PKCδ-cat in the cell cycle response to DNA damage, we exposed wild-type and PKCδ null mouse embryonic fibroblasts (MEFs) to UV radiation. Wild-type MEFs underwent a pronounced G2/M arrest, Cdk1 phosphorylation, and induction of apoptosis following UV exposure, whereas PKCδ null MEFs were resistant to these effects. Expression of PKCδ-green fluorescent protein, but not caspase-resistant or kinase-inactive PKCδ, was able to restore G2/M checkpoint integrity in PKCδ null MEFs. The function of PKCδ in the DNA damage-induced G2/M cell cycle checkpoint may be a critical component of its tumor suppressor function.
Keywords: Cell/Checkpoint, Cell/Cycle, DNA/Damage, Signal Transduction/Protein Kinases/Serine/Threonine, Tissue/Organ Systems/Skin, Tumor/Suppressor, Ultraviolet Radiation
Introduction
Cells frequently encounter both internal and environmental stresses that cause genomic damage. This damage can be fatal if present at sufficient levels, but may also introduce genomic mutation. To preserve genomic integrity, eukaryotic cells have developed mechanisms to cope with DNA damage, which include the detection of DNA damage, engagement of cell cycle checkpoints, and repair of the damage prior to cell cycle progression.
Cell cycle checkpoints are present at several stages of the eukaryotic cell cycle. The major cell cycle checkpoints responsible for maintaining the genomic integrity of a cell include the G1/S checkpoint, the G2/M DNA damage checkpoint, and the metaphase spindle attachment checkpoint (1, 2). These checkpoints ensure that DNA replication and segregation do not proceed until DNA damage is repaired and thus perform the vital function of preserving genomic integrity. Non-functional cell cycle checkpoints lead to an increased rate of mutation as well as aneuploidy and therefore promote tumorigenesis. The importance of cell cycle checkpoints in the maintenance of genomic integrity and prevention of tumor formation is indicated by the frequent loss or mutation of key cell cycle regulators such as p53 in cancer (3).
The G2/M checkpoint is activated by the presence of DNA damage and prevents entry into mitosis until the damage is repaired. Key proteins in this pathway include the apical kinases ATM and ATR, which are activated following recognition of DNA damage, the checkpoint kinases Chk1 and Chk2, as well as the tumor suppressor p53 (4–7). The G2/M checkpoint culminates with the inhibition of the mitosis-promoting factor Cdk1/cyclin B. Cdk1/cyclin B is inhibited both by phosphorylation catalyzed by Wee1 and Myt1 and cytoplasmic sequestration by the 14-3-3 chaperone proteins (8–10). The Cdc25 phosphatases counteract G2/M checkpoint activation by removing inhibitory phosphate groups from Cdk1 and are themselves negatively regulated by Chk1 (11).
Protein kinase Cδ (PKCδ)2 is a calcium-independent member of the protein kinase C family of serine/threonine protein kinases (12). PKCδ is expressed ubiquitously in human cells and has been shown to play important roles in both cell cycle signaling and apoptosis. The structure of the PKCδ protein consists of a regulatory domain, which contains a pseudosubstrate region connected to the catalytic domain by a short hinge region (13). When in an inactive state, the pseudosubstrate region of the regulatory domain binds to the active site of the catalytic domain and thus represses PKCδ catalytic activity (14). Diacylglycerol or the phorbol ester 12-O-tetradecanoylphorbol-13-acetate can relieve the repressive pseudosubstrate binding and activate PKCδ.
PKCδ plays a well established role in the apoptotic cascade and has been shown to phosphorylate many targets, including p53, Mcl-1, and lamin B, in a manner that promotes apoptosis (15–18). It has been demonstrated that during UV radiation-induced apoptosis, PKCδ is cleaved in its hinge region by activated caspase 3 (19). This cleavage frees the active site from pseudosubstrate inhibition, generating a constitutively active catalytic fragment, termed PKCδ-cat, which phosphorylates Mcl-1 to accelerate apoptosis (16, 19). This cleavage event is vital to the apoptotic cascade because the overexpression of a mutant PKCδ that cannot be cleaved by caspase 3 suppresses UV radiation-induced apoptosis in human keratinocytes (KCs) (20). It has also been reported that PKCδ contains a nuclear localization sequence and that nuclear localization of PKCδ is of critical importance to successful completion of apoptosis (21, 22).
PKCδ expression is lost in both human squamous cell carcinomas and chemically induced mouse skin tumors, supporting its function as a cutaneous squamous cell carcinoma tumor suppressor gene (23, 24). The vital role of PKCδ-mediated apoptosis in tumor suppression is bolstered by work demonstrating that transgenic mice overexpressing PKCδ are resistant to chemically induced squamous cell carcinomas and have elevated 12-O-tetradecanoylphorbol-13-acetate-induced apoptosis (24, 25). Furthermore, re-expression of PKCδ in human squamous carcinoma cells induces spontaneous apoptosis and inhibits tumorigenesis (23).
In addition to the well studied pro-apoptotic effects of PKCδ, several studies have reported effects of PKCδ on the cell cycle. For example, PKCδ stimulates apoptosis by initiating an S phase arrest in rat thyroid cells (26). Other studies have tied PKCδ to the G2/M phases of the cell cycle by demonstrating that PKCδ can localize to the nucleus, where it is associated with chromatin condensation as well as inhibition of cytokinesis (27, 28). Furthermore, research on the effects of PKCδ overexpression in HCT116 cells revealed that PKCδ induced many of the morphological features of mitotic catastrophe, including multimicronucleation and centrosomal amplification (29). In this study, we report that PKCδ-cat plays a critically important role in enforcing the G2/M checkpoint in response to UV radiation.
EXPERIMENTAL PROCEDURES
Retroviral Constructs, Packaging, and Infection
PKCδ-green fluorescent protein (GFP), PKCδ(D327A)-GFP, and PKCδ(K376R)-GFP constructs were graciously provided by Dr. Mary E. Reyland (University of Colorado Health Sciences Center) and were previously described (21). These constructs were subcloned in XhoI/NotI sites of the LZRS-Linker retroviral expression vector for infection of cultured KCs and mouse embryonic fibroblasts (MEFs). LZRS-PKCδ-cat-FLAG and the 4-hydroxytamoxifen-activatable LZRS-PKCδ-cat-estrogen receptor (ER) were previously described (18). Retroviral supernatant was generated by calcium phosphate-mediated transfection of Phoenix-Ampho packaging cells as described previously (30). Retroviral infection was done for 1 h at 1200 rpm and 32 °C.
Cell Culture and UV Treatment
Primary human KCs were isolated from neonatal foreskins with Loyola Institutional Review Board approval as described previously (31, 32). KCs and HaCaT cells were cultured in Medium 154CF with 0.07 mm calcium and human keratinocyte growth supplement (Cascade Biologics). Spontaneously immortalized MEFs from wild-type or PKCδ null mice were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum and were kindly provided by Dr. Anning Lin (University of Chicago). PKCδ-cat-ER was activated by treatment with 10 μm 4-hydroxytamoxifen (Alexis Biochemicals) (18). In all experiments using PKCδ-cat-ER, control (Linker) transduced cells were also treated with 4-hydroxytamoxifen. ATM/ATR inhibition was achieved by the addition of 2 mm caffeine in water to the culture medium (33). UV irradiation was done using a UV Panelite unit with ∼65% emission in the UVB spectrum as previously described (23).
Antibodies and Western Blotting
Cell lysates were collected by scraping cells in radioimmune precipitation assay buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate). Lysates were briefly sonicated and centrifuged at 14,000 rpm for 5 min to remove cellular debris. Protein concentrations were determined using standard Bradford reagent methodology.
Antibodies used for Western blotting include actin (69100, MP Biomedical); Cdk1 (sc-747), phosphorylated Cdk1 (Tyr15) (sc-7989R), and PKCδ (sc-937) (Santa Cruz Biotechnology); γH2A.X (UBI 05-636, Upstate); histone H3 (ab1791) and phosphorylated histone H3 (Ser10) (UBI 06-570) (Abcam); p-p53 (Ser15) (9284S, Cell Signaling); PKCδC terminus (610397, BD Transduction Laboratories); and vinculin (V4505, Sigma).
Flow Cytometry
Cells were collected by trypsinization, pelleted, and washed in fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline, 5% fetal bovine serum). Cells were then pelleted and resuspended in 100 μl of fetal bovine serum. Cells were fixed by addition of ice-cold 100% ethanol for at least 30 min. RNA was digested by treatment with 10 μg/ml RNase for 15 min at 37 °C. Propidium iodide was added to a final concentration of 50 μg/ml, and samples were incubated on ice for at least 1 h. Cell cycle profiles were analyzed using a Beckman Coulter EPICS XL-MCL flow cytometer. Histogram overlays were generated using FlowJo software.
Mitotic Index
For mitotic index measurements in MEFs, the cells were treated with or without UV radiation, followed by 10 ng/ml nocodazole to trap any cells that had overcome UV radiation-induced G2/M arrest and entered mitosis. Mitotic index measurements in HaCaT cells were performed 3 days after PKCδ-cat-ER transduction and thus were not treated with nocodazole. Cells were collected and washed in FACS buffer before being fixed in 3.7% formaldehyde for 10 min. After fixation, cells were permeabilized in 70% EtOH at 4 °C for 30 min. After washing in FACS buffer, cells were incubated for 2 h in 100 μl of phosphorylated histone H3 (Ser10) antibody. Cells were washed and incubated on ice in 100 μl of diluted Alexa Fluor 488-conjugated anti-rabbit secondary antibody for 30 min. Following secondary antibody incubation, cells were washed twice and incubated in 500 μl of solution containing 10 μg/ml RNase and 50 μg/ml propidium iodide for 30 min prior to FACS analysis.
Confocal Microscopy
Cells were cultured on glass coverslips, fixed in 3.7% formaldehyde, and permeabilized with 0.1% Triton X-100. To stain chromatin, coverslips were incubated in 100 ng/ml 4′,6-diamidino-2-phenylindole for 5 min. Coverslips were mounted onto slides using Gelvatol. Images were generated using a Carl Zeiss LSM-510 confocal microscope with 1-μm optical slice at ×40 magnification.
Nuclear/Cytoplasmic Fractionation
Cells were collected in phosphate-buffered saline and incubated in hypotonic buffer (10 mm KCl, 50 mm HEPES, pH 7.9, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, Complete protease inhibitor (Roche Applied Science), 40 mm glycerophosphate, 2 mm sodium fluoride, and 1 mm sodium orthovanadate) for 15 min on ice. Triton X-100 was added to a final concentration of 0.5% to complete lysis. Samples were centrifuged, and the resulting supernatant contained enriched cytoplasmic proteins. The pellet (containing nuclei) was washed in hypotonic buffer plus 1 m sucrose and lysed in high-salt buffer (400 mm KCl, 50 mm HEPES, pH 7.0, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1 mm dithiothreitol, Complete Protease Inhibitor, and phosphatase inhibitors). Samples were centrifuged, and the resulting supernatant contained enriched nuclear proteins.
RESULTS
PKCδ-cat Induces G2/M Cell Cycle Arrest
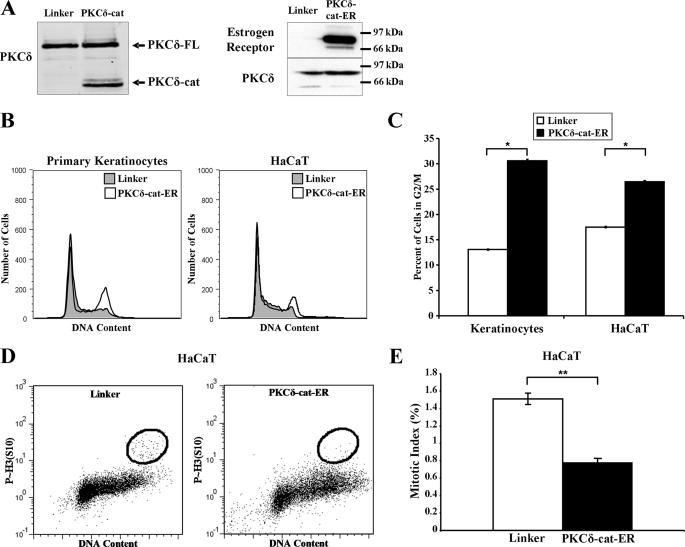
To determine the effect of PKCδ-cat on the cell cycle, we retrovirally expressed PKCδ-cat or an inducible PKCδ-cat-ER fusion protein in cultured primary human KCs, HaCaT cells (Fig. 1A), and MEFs (supplemental Fig. 1). Propidium iodide staining revealed that PKCδ-cat expression increased G2/M KCs ∼2-fold (p < 0.005) compared with the control Linker virus population (Fig. 1, B and C). PKCδ-cat expression had a similar effect on the immortalized HaCaT cell line, which harbors mutant p53, although the effect was slightly diminished (34). To distinguish between arrest in the G2 or M phase, mitotic index measurements were performed on PKCδ-cat-ER-transduced HaCaT cells. Fig. 1, D and E, shows that PKCδ-cat-ER caused a significant (p < 0.001) reduction in mitotic cells, signifying that the cells arrested with 4N DNA were in the G2 phase. Because PKCδ-cat can also induce apoptosis, we inhibited apoptosis with the caspase 3 inhibitor benzyloxycarbonyl-VAD to determine whether the cell cycle effects were independent of apoptosis. Benzyloxycarbonyl-VAD slightly accentuated the G2/M accumulation induced by PKCδ-cat, although the effect was not statistically significant, suggesting that G2/M arrest may precede apoptosis (data not shown).
FIGURE 1.
PKCδ-cat expression induces G2/M cell cycle arrest. A, Western blots displaying relative levels of full-length PKCδ protein (PKCδ-FL) and PKCδ-cat are shown in control Linker-, PKCδ-cat-FLAG-, and PKCδ-cat-ER-transduced KCs. PKCδ-cat-ER-transduced cells display a strong ER-positive band at the same position as the PKCδ protein, indicating successful expression of the ER fusion protein. B, representative DNA content histograms of primary KCs and HaCaT cells transduced with either Linker or PKCδ-cat-ER retrovirus are shown. Propidium iodide staining was performed 2–3 days after infection. Similar results were obtained in at least three independent experiments. C, quantitation of the percentage of cells containing G2/M DNA content in KCs and HaCaT cells 2–3 days following infection with either Linker control or PKCδ-cat-ER retrovirus. Graphs represent experiments done in triplicate. *, Student's t test value of p < 0.005. D, HaCaT cells transduced with either Linker or PKCδ-cat-ER retrovirus were harvested and stained for phosphorylated histone H3 (Ser10) and propidium iodide after 3 days and analyzed by flow cytometry. E, quantitation of mitotic indices from HaCaT cells treated as described for D. **, Student's t test value of p < 0.001. Error bars denote the S.D.
PKCδ-cat Induces G2/M Checkpoint Pathway
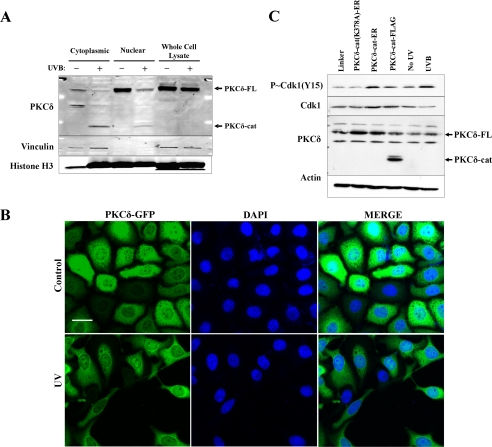
To have a direct role in cell cycle regulation, it is likely that PKCδ-cat would need to have at least some nuclear localization. To examine this, we performed nuclear/cytoplasmic fractionation on HaCaT cells before and 18 h after exposure to 30 mJ/cm2 UV radiation. Fractionation results revealed that both the full-length PKCδ and PKCδ-cat were present in the nucleus, with PKCδ-cat detected only after UV exposure (Fig. 2A). UV radiation caused a significant fraction of histone H3 to be detected in the cytoplasmic extract, suggesting that apoptotic degradation of the nuclear membrane occurred in the cells exposed to UV radiation. Confocal microscopy using a carboxyl-terminal GFP fusion of PKCδ (PKCδ-GFP) and 4′,6-diamidino-2-phenylindole staining was used to confirm the subcellular fractionation data. PKCδ-GFP was localized to both the cytoplasm and nucleus before and after UV irradiation (Fig. 2B). This localization provides a means by which PKCδ may be interacting with important components of the G2/M cell cycle checkpoint pathway.
FIGURE 2.
PKCδ-cat expression induces G2/M checkpoint activation in KCs. A, Western blot showing levels of PKCδ protein in cytoplasmic and nuclear extracts of untreated or 30 mJ/cm2 UV radiation-exposed KCs after 18 h. Full-length PKCδ (PKCδ-FL) and PKCδ-cat are indicated by arrows. Vinculin and histone H3 are presented as cytoplasmic and nuclear markers, respectively. B, confocal microscope images were taken showing cytoplasmic and nuclear staining of PKCδ-GFP fusion protein both before and 18 h after exposure to 30 mJ/cm2 UV radiation. 4′,6-Diamidino-2-phenylindole (DAPI) staining is displayed to demonstrate nuclear localization. Images were taken using a ×40 objective and 1-μm optical slice. Scale bar denotes 25 μm. C, Western blot showing levels of p-Cdk1 (Tyr15), total Cdk1, and PKCδ after retroviral transduction of PKCδ-cat-ER, PKCδ-cat, or the kinase-dead PKCδ(K378A)-cat-ER. The PKCδ-cat-ER fusion proteins run to the same position in the gel as the full-length PKCδ. KC lysate harvested 18 h after exposure to 30 mJ/cm2 UV radiation is shown as a positive control for p-Cdk1 (Tyr15) induction. Actin protein levels are displayed as a loading control.
We next tested whether PKCδ-cat was sufficient to induce the G2/M DNA damage checkpoint. One key aspect of this checkpoint involves the phosphorylation of Cdk1 on Tyr15 by Wee1 and Myt1 kinases, an event that has been shown to inhibit Cdk1 activity and to prevent entry into mitosis (8, 9). We measured relative levels of p-Cdk1 (Tyr15) in PKCδ-cat-expressing KCs and found that PKCδ-cat expression induced elevated levels of p-Cdk1 (Tyr15) compared with the control Linker-transduced cells. The kinase-dead mutant (K378A) did not induce p-Cdk1 (Tyr15) (Fig. 2C). As a positive control, UV radiation also induced p-Cdk1 (Tyr15).
UV Radiation-induced G2/M Cell Cycle Arrest Requires PKCδ
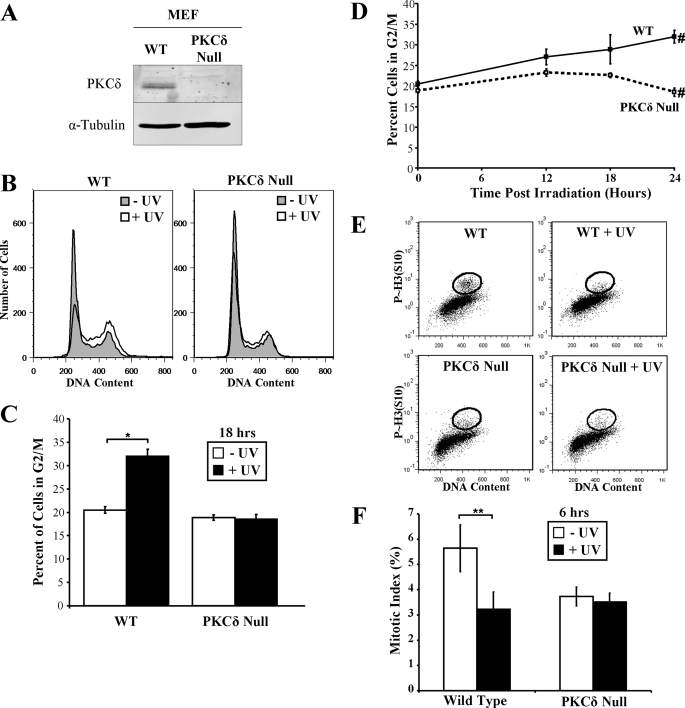
Because expression of PKCδ-cat was capable of inducing a pronounced G2/M arrest and checkpoint activation in KCs and HaCaT cells, we next determined the requirement for PKCδ in DNA damage-induced cell cycle arrest. To address this issue, wild-type and PKCδ null MEFs were exposed to UV radiation (Fig. 3A). We found that exposure of the wild-type MEFs to 30 mJ/cm2 UV radiation induced a pronounced G2/M arrest, which persisted up to 24 h after exposure (Fig. 3, B–D). Strikingly, UV exposure failed to induce G2/M arrest in PKCδ null MEFs, although there was a slight increase in the percentage of cells in S phase. PKCδ null MEFs did undergo a G2/M arrest when transduced with PKCδ-cat-ER virus (supplemental Fig. 1). Mitotic index measurements revealed that UV caused a significant (p < 0.05) decrease in mitotic wild-type MEFs, but not PKCδ null MEFs, at 6 h, indicating that the UV radiation-induced arrest was in the G2 phase (Fig. 3, E and F). These results suggest that PKCδ is critical for G2/M checkpoint integrity after DNA damage.
FIGURE 3.
PKCδ null MEFs fail to arrest in G2/M phase following UV irradiation. A, Western blot displaying PKCδ protein levels in wild-type (WT) and PKCδ null MEF whole cell lysates. α-Tubulin levels are shown as a loading control. B, representative DNA content histograms of wild-type and PKCδ null MEFs before and 18 h after exposure to 30 mJ/cm2 UV radiation. C, the percentage of wild-type and PKCδ null MEFs in G2/M phase of the cell cycle before and after exposure to 30 mJ/cm2 UV radiation is displayed. Error bars denote the S.D. from experiments performed in triplicate. *, Student's t test value of p < 0.005. D, the percentage of wild-type and PKCδ null MEFs with G2/M DNA content at various times following exposure to 30 mJ/cm2 UV radiation is displayed. Error bars denote the S.D. from experiments performed in triplicate. #, Student's t test value of p < 0.005. E, wild-type and PKCδ null MEFs were exposed to 10 mJ/cm2 UV radiation, treated with 100 ng/ml nocodazole, and stained for phosphorylated histone H3 (Ser10) and propidium iodide 6 h after UV exposure. Flow cytometry analysis is shown, with the phosphorylated histone H3 (Ser10)-positive G2/M DNA content cells circled as the mitotic cells. F, quantitation of mitotic indices from HaCaT cells treated as described for D. **, Student's t test value of p < 0.05.
Interestingly, G2/M checkpoint override by the ATM/ATR inhibitor caffeine drove UV radiation-irradiated wild-type MEFs into apoptosis but did not induce apoptosis in PKCδ null MEFs (supplemental Fig. 2). The lack of apoptosis after caffeine-mediated inhibition of checkpoint activation in PKCδ null MEFs is likely a reflection of the crucial pro-apoptotic functions of PKCδ. Interestingly, PKCδ null cells treated with UV radiation plus caffeine accumulated in S phase (p < 10−4), indicating a possible S phase checkpoint independent of PKCδ and ATM/ATR function.
G2/M Checkpoint Integrity Requires PKCδ Cleavage and Kinase Activity
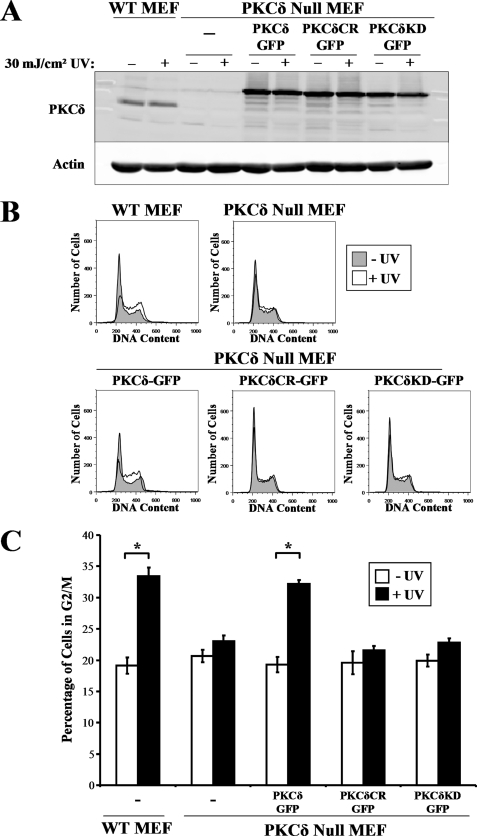
We next attempted to rescue G2/M checkpoint integrity in PKCδ null MEFs by retrovirally transducing them with the catalytically competent PKCδ-GFP (Fig. 4A). To address whether PKCδ kinase activity or caspase cleavage is required to restore the G2/M checkpoint response, we also transduced a kinase-dead mutant, PKCδ(K376R)-GFP, or a kinase competent mutant that possesses a mutated caspase cleavage site, PKCδ(D327A)-GFP. All PKCδ-GFP constructs were expressed at similar levels (Fig. 4A). Expression of PKCδ-GFP in PKCδ null MEFs had little effect on the cell cycle distribution of non-damaged cells (Fig. 4, B and C). Expression of PKCδ-GFP in PKCδ null MEFs restored the G2/M checkpoint response to UV radiation (Fig. 4, B and C). The kinase-dead PKCδ(K376R)-GFP mutant was not able to restore the G2/M checkpoint in the PKCδ null MEF background, indicating that kinase activity is necessary for this PKCδ checkpoint function. Strikingly, expression of the cleavage site mutant PKCδ was also unable to restore UV radiation-induced G2/M arrest in the PKCδ null MEFs (Fig. 4, B and C). The dose of UV radiation (30 mJ/cm2) used in these experiments was sufficient to induce caspase 2 and 3 activities, both of which are capable of cleaving PKCδ (data not shown). This suggests that PKCδ cleavage is a critically important event in the enforcement of the G2/M checkpoint after exposure to UV radiation.
FIGURE 4.
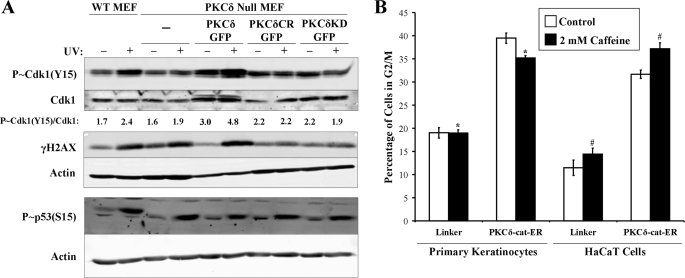
G2/M checkpoint induction requires PKCδ kinase activity. A, wild-type (WT) and PKCδ null MEFs were untransduced or transduced with wild-type PKCδ-GFP, cleavage-resistant (CR) PKCδ(D327A)-GFP, or kinase-dead (KD) PKCδ(K376R)-GFP and exposed to 30 mJ/cm2 UV radiation. Protein lysates were analyzed for expression of PKCδ and actin. B, representative DNA content histograms of wild-type and PKCδ null MEFs before and 18 h after exposure to 30 mJ/cm2 UV radiation. C, the percentage of wild-type and PKCδ null MEFs in G2/M after transduction with the indicated PKCδ-GFP fusion protein before and after exposure to 30 mJ/cm2 UV radiation is displayed. Error bars denote the S.D. from experiments performed in triplicate. *, Student's t test value of p < 0.005.
UV Radiation-induced G2/M Checkpoint Activation in Wild-type and PKCδ Null MEFs
We next examined components of the G2/M checkpoint pathway before and after UV exposure in wild-type and PKCδ null MEFs. In agreement with the cell cycle analysis in Figs. 3 and 4, we found that UV exposure induced G2/M checkpoint activation as assessed by increased levels of p-Cdk1 (Tyr15) in wild-type but not PKCδ null MEFs (Fig. 5A). Despite lacking an intact G2/M checkpoint response, PKCδ null MEFs still exhibited UV radiation-induced γH2A.X and p-p53 (Ser15), both substrates of ATM and ATR kinases (35–37). The induction of γH2A.X and p-p53 (Ser15), but not p-Cdk1, suggests that PKCδ may be acting downstream of ATM/ATR in the DNA damage. To examine this, we treated primary human KCs and HaCaT cells ectopically expressing the constitutively active PKCδ-cat with 2 mm caffeine (Fig. 5B). We found that inhibition of ATM/ATR activity by caffeine treatment did not block PKCδ-cat-induced G2/M arrest in primary KCs or HaCaT cells, indicating a role for PKCδ-cat in the G2/M checkpoint downstream of ATM/ATR activation.
FIGURE 5.
UV radiation-induced G2/M DNA damage checkpoint activation requires PKCδ function. A, wild-type (WT) and PKCδ null MEFs expressing PKCδ-GFP, cleavage-resistant (CR) PKCδ(D327A)-GFP, or kinase-dead (KD) PKCδ(K376R)-GFP were exposed to 10 mJ/cm2 UV radiation. Protein lysates were collected 18 h after irradiation and analyzed for levels of p-Cdk1 (Tyr15), total Cdk1, γH2A.X, and actin. p-Cdk1 (Tyr15)/Cdk1 densitometry ratios are displayed and were similar in multiple experiments. B, propidium iodide cell cycle analysis of LZRS-Linker- or LZRS-PKCδ-cat-ER-transduced primary KCs and HaCaT cells incubated with 2 mm caffeine for 48 h. The percentage of cells in the G2/M phase of the cell cycle is displayed from experiments performed in triplicate. *, Student's t test value of p < 10−5; #, Student's t test value of p < 10−4.
In agreement with the cell cycle data presented in Fig. 4, re-expression of PKCδ-GFP, but not either kinase-dead PKCδ or caspase cleavage mutant PKCδ, restored phosphorylation of p-Cdk1 (Tyr15) after UV exposure (Fig. 5A). This supports the idea that both kinase activity and caspase cleavage of PKCδ are important events in the activation of the G2/M checkpoint after DNA damage.
DISCUSSION
The majority of studies on PKCδ function have focused on the pro-apoptotic role that the kinase plays both in vivo and in vitro. We have now demonstrated that in addition to these pro-apoptotic effects, PKCδ also regulates cell cycle progression by participating in the G2/M DNA damage checkpoint. We have shown that retroviral expression of PKCδ-cat in primary KCs, immortalized KCs with mutant p53, and MEFs is sufficient to induce G2/M checkpoint activation and cell cycle arrest (34). PKCδ-cat-induced G2/M checkpoint activation requires kinase competent PKCδ because a kinase-dead PKCδ mutant failed to activate the G2/M checkpoint (Fig. 2C).
The cleavage of PKCδ appears to be critical to checkpoint integrity because the expression of the caspase-resistant PKCδ(D327A)-GFP mutant was unable to restore the G2/M checkpoint in PKCδ null MEFs (Figs. 4 and 5). PKCδ is cleavable by numerous proteases including caspases 2 and 3, but the protease responsible for cleaving PKCδ in this circumstance remains unclear (19, 38). It is possible that PKCδ may be a substrate for the caspase 2 containing DNA-PKcs-PIDDosome complex, which was recently demonstrated to be integral to proper G2/M checkpoint maintenance after ionizing radiation exposure (39). PKCδ appears to be required for G2/M checkpoint maintenance rather than induction because a decrease in mitotic index was observed in both wild-type and PKCδ null MEFs 1 h after UV exposure (data not shown) but not after 6 h (Fig. 3, E and F). Proteins such as p53, p21, and the DNA-PKcs-PIDDosome complex have been similarly implicated in G2/M checkpoint maintenance rather than initial checkpoint activation (39, 40).
Several PKCδ substrates involved in the DNA damage response have been identified. These include both p53 and the p53 family member p73β (15, 41, 42). PKCδ has been demonstrated to phosphorylate and activate the DNA repair protein Rad9 after exposure to the DNA-damaging agent 5-azacytidine in an ATM-dependent manner (43). It is possible that ATR, a major kinase activated by UV radiation, is also capable of activating PKCδ in a similar fashion. Other components of the DNA damage response that have been identified as PKCδ substrates include DNA-PKcs and topoisomerase IIα (44, 45). Together, these studies reveal the importance of PKCδ in the DNA damage repair pathway and indicate that it may be functioning at several levels. Nuclear localization of PKCδ is important for DNA damage-induced apoptosis and may be required during multiple processes following DNA damage, including checkpoint activation (21, 22).
The novel cell cycle regulatory role for PKCδ described in this study has important implications for the role of this protein as a tumor suppressor. Based on these results, the loss of PKCδ during tumor development would allow for the cell to progress through the cell cycle, even in the presence of DNA damage, while at the same time evading apoptosis. This is illustrated by wild-type MEFs being driven into apoptosis by caffeine-induced G2/M checkpoint override after UV irradiation, whereas PKCδ null MEFs do not arrest or undergo apoptosis even after treatment with caffeine (supplemental Fig. 2). Similar to wild-type MEFs, KCs are highly sensitive to caffeine treatment after UV irradiation (46, 47). Thus, KCs lacking PKCδ, such as in squamous papillomas and carcinomas, are also likely to have this dual defect in the response to DNA damage, resulting in both reduced cell cycle arrest and reduced apoptosis (23, 24, 48).
Furthermore, concomitant treatment of LZRS-PKCδ-cat-ER-transduced cells with the ATM/ATR inhibitor caffeine was unable to prevent PKCδ-cat-ER-induced G2/M arrest. This suggests that PKCδ-cat is inducing G2/M checkpoint activation independent of the ATM/ATR kinases and may actually function internally to the G2/M checkpoint signal cascade. Consistent with this, UV induced γH2A.X and p-p53 (Ser15) in PKCδ null MEFs but did not induce p-Cdk1 (Tyr15) (Fig. 5A). It will be important in the future to identify substrates for PKCδ within the G2/M checkpoint signaling hierarchy. Possible targets include, but are not limited to, the Cdc25 phosphatases, the checkpoint kinases Chk1 and Chk2, and the Cdk1 kinase Wee1 (8, 49). The importance of cell cycle checkpoint function to cancer cells has prompted intensive study of small molecule inhibitors targeted toward components of the G2/M checkpoint such as ATM/ATR, Chk1, Chk2, and the Aurora family for potential use as cancer therapeutics (50–52). Better understanding of how PKCδ participates in this cell cycle checkpoint is important for developing better cancer treatments in the future.
Supplementary Material
Acknowledgments
We thank all members of the Skin Cancer Research Program of Loyola University Medical Center for helpful discussions and the Molecular and Cellular Biochemistry Division. Reagents were graciously provided by Drs. Anning Lin and Mary Reyland.
This work was supported, in whole or in part, by National Institutes of Health Grant CA083784 (to M. F. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- PKCδ
- protein kinase Cδ
- KC
- keratinocyte
- PKCδ-cat
- PKCδ catalytic fragment
- GFP
- green fluorescent protein
- MEF
- mouse embryonic fibroblast
- ER
- estrogen receptor
- FACS
- fluorescence-activated cell sorter.
REFERENCES
- 1.Bartek J., Lukas J. (2007) Curr. Opin. Cell Biol. 19, 238–245 [DOI] [PubMed] [Google Scholar]
- 2.Musacchio A., Salmon E. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M., Sidransky D., Vogelstein B., Harris C. C. (1991) Science 253, 49–53 [DOI] [PubMed] [Google Scholar]
- 4.Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 5.Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 6.Hirao A., Kong Y. Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S. J., Mak T. W. (2000) Science 287, 1824–1827 [DOI] [PubMed] [Google Scholar]
- 7.Agarwal M. L., Agarwal A., Taylor W. R., Stark G. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8493–8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell P., Nurse P. (1987) Cell 49, 559–567 [DOI] [PubMed] [Google Scholar]
- 9.Mueller P. R., Coleman T. R., Kumagai A., Dunphy W. G. (1995) Science 270, 86–90 [DOI] [PubMed] [Google Scholar]
- 10.Chan T. A., Hermeking H., Lengauer C., Kinzler K. W., Vogelstein B. (1999) Nature 401, 616–620 [DOI] [PubMed] [Google Scholar]
- 11.Chen M. S., Ryan C. E., Piwnica-Worms H. (2003) Mol. Cell. Biol. 23, 7488–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappa H., Murray-Rust J., Dekker L. V., Parker P. J., McDonald N. Q. (1998) Structure 6, 885–894 [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K. (2007) Cell Signal. 19, 892–901 [DOI] [PubMed] [Google Scholar]
- 14.Pears C. J., Kour G., House C., Kemp B. E., Parker P. J. (1990) Eur. J. Biochem. 194, 89–94 [DOI] [PubMed] [Google Scholar]
- 15.Yoshida K., Liu H., Miki Y. (2006) J. Biol. Chem. 281, 5734–5740 [DOI] [PubMed] [Google Scholar]
- 16.Sitailo L. A., Tibudan S. S., Denning M. F. (2006) J. Biol. Chem. 281, 29703–29710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross T., Griffiths G., Deacon E., Sallis R., Gough M., Watters D., Lord J. M. (2000) Oncogene 19, 2331–2337 [DOI] [PubMed] [Google Scholar]
- 18.Sitailo L. A., Tibudan S. S., Denning M. F. (2004) J. Invest. Dermatol. 123, 434–443 [DOI] [PubMed] [Google Scholar]
- 19.Denning M. F., Wang Y., Nickoloff B. J., Wrone-Smith T. (1998) J. Biol. Chem. 273, 29995–30002 [DOI] [PubMed] [Google Scholar]
- 20.D'Costa A. M., Denning M. F. (2005) Cell Death Differ. 12, 224–232 [DOI] [PubMed] [Google Scholar]
- 21.DeVries T. A., Neville M. C., Reyland M. E. (2002) EMBO J. 21, 6050–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeVries-Seimon T. A., Ohm A. M., Humphries M. J., Reyland M. E. (2007) J. Biol. Chem. 282, 22307–22314 [DOI] [PubMed] [Google Scholar]
- 23.D'Costa A. M., Robinson J. K., Maududi T., Chaturvedi V., Nickoloff B. J., Denning M. F. (2006) Oncogene 25, 378–386 [DOI] [PubMed] [Google Scholar]
- 24.Reddig P. J., Dreckschmidt N. E., Ahrens H., Simsiman R., Tseng C. P., Zou J., Oberley T. D., Verma A. K. (1999) Cancer Res. 59, 5710–5718 [PubMed] [Google Scholar]
- 25.Humphries M. J., Limesand K. H., Schneider J. C., Nakayama K. I., Anderson S. M., Reyland M. E. (2006) J. Biol. Chem. 281, 9728–9737 [DOI] [PubMed] [Google Scholar]
- 26.Santiago-Walker A. E., Fikaris A. J., Kao G. D., Brown E. J., Kazanietz M. G., Meinkoth J. L. (2005) J. Biol. Chem. 280, 32107–32114 [DOI] [PubMed] [Google Scholar]
- 27.Watanabe T., Ono Y., Taniyama Y., Hazama K., Igarashi K., Ogita K., Kikkawa U., Nishizuka Y. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10159–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi A., Ohtani N., Yamakoshi K., Iida S., Tahara H., Nakayama K., Nakayama K. I., Ide T., Saya H., Hara E. (2006) Nat. Cell Biol. 8, 1291–1297 [DOI] [PubMed] [Google Scholar]
- 29.Perletti G., Marras E., Dondi D., Osti D., Congiu T., Ferrarese R., de Eguileor M., Tashjian A. H., Jr. (2005) Int. J. Cancer 113, 42–53 [DOI] [PubMed] [Google Scholar]
- 30.Sitailo L. A., Tibudan S. S., Denning M. F. (2002) J. Biol. Chem. 277, 19346–19352 [DOI] [PubMed] [Google Scholar]
- 31.Mitra R., Nickoloff B. (1994) in Keratinocyte Methods (Leigh I., Watt F. eds) Cambridge University Press, Cambridge, UK [Google Scholar]
- 32.Tibudan S. S., Wang Y., Denning M. F. (2002) J. Invest. Dermatol. 119, 1282–1289 [DOI] [PubMed] [Google Scholar]
- 33.Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. (1999) Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 34.Lehman T. A., Modali R., Boukamp P., Stanek J., Bennett W. P., Welsh J. A., Metcalf R. A., Stampfer M. R., Fusenig N., Rogan E. M., Harris C. C. (1993) Carcinogenesis 14, 833–839 [DOI] [PubMed] [Google Scholar]
- 35.Ward I. M., Chen J. (2001) J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 36.Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001) J. Biol. Chem. 276, 42462–42467 [DOI] [PubMed] [Google Scholar]
- 37.Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., Abraham R. T. (1999) Genes Dev. 13, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emoto Y., Manome Y., Meinhardt G., Kisaki H., Kharbanda S., Robertson M., Ghayur T., Wong W. W., Kamen R., Weichselbaum R., Kufe D. (1995) EMBO J. 14, 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi M., Vivian C. J., Lee K. J., Ge C., Morotomi-Yano K., Manzl C., Bock F., Sato S., Tomomori-Sato C., Zhu R., Haug J. S., Swanson S. K., Washburn M. P., Chen D. J., Chen B. P., Villunger A., Florens L., Du C. (2009) Cell 136, 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 41.Liu H., Lu Z. G., Miki Y., Yoshida K. (2007) Mol. Cell. Biol. 27, 8480–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren J., Datta R., Shioya H., Li Y., Oki E., Biedermann V., Bharti A., Kufe D. (2002) J. Biol. Chem. 277, 33758–33765 [DOI] [PubMed] [Google Scholar]
- 43.Yoshida K., Wang H. G., Miki Y., Kufe D. (2003) EMBO J. 22, 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bharti A., Kraeft S. K., Gounder M., Pandey P., Jin S., Yuan Z. M., Lees-Miller S. P., Weichselbaum R., Weaver D., Chen L. B., Kufe D., Kharbanda S. (1998) Mol. Cell. Biol. 18, 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida K., Yamaguchi T., Shinagawa H., Taira N., Nakayama K. I., Miki Y. (2006) Mol. Cell. Biol. 26, 3414–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Y. P., Lou Y. R., Peng Q. Y., Xie J. G., Nghiem P., Conney A. H. (2008) Cancer Res. 68, 2523–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heffernan T. P., Kawasumi M., Blasina A., Anderes K., Conney A. H., Nghiem P. (2009) J. Invest. Dermatol. 129, 1805–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aziz M. H., Wheeler D. L., Bhamb B., Verma A. K. (2006) Cancer Res. 66, 713–722 [DOI] [PubMed] [Google Scholar]
- 49.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 50.Schmidt M., Bastians H. (2007) Drug Resist. Updates 10, 162–181 [DOI] [PubMed] [Google Scholar]
- 51.Bucher N., Britten C. D. (2008) Br. J. Cancer 98, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hose D., Rème T., Meissner T., Moreaux J., Seckinger A., Lewis J., Benes V., Benner A., Hundemer M., Hielscher T., Shaughnessy J. D., Jr., Barlogie B., Neben K., Krämer A., Hillengass J., Bertsch U., Jauch A., De Vos J., Rossi J. F., Möhler T., Blake J., Zimmermann J., Klein B., Goldschmidt H. (2009) Blood 113, 4331–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.