Abstract
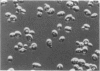
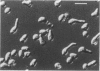
The CDC28 gene of Saccharomyces cerevisiae encodes a protein kinase that is required for passage through the G1 phase of the cell cycle. We have used an inducible promoter fused to the CDC28 coding sequence to isolate conditionally dominant mutant alleles of CDC28. Overexpression of these dominant alleles causes arrest in the G1 phase of the cell cycle but permits the distinctive asymmetric growth that is characteristic of recessive temperature-sensitive cdc28 mutants. The dominant alleles encode products with no detectable protein kinase activity, and their phenotypic effects can be suppressed by simultaneous overproduction of the wild-type protein. DNA sequence analysis showed that the mutant site in at least one of the dominant alleles is in a residue that is highly conserved among protein kinases. These properties are best understood if the dominant mutation results in the catalytic inactivation of the protein kinase but still allows the binding of another component needed for CDC28 function. By this model, high levels of the mutant protein arrest cell division by denying the wild-type protein access to this other component. Suppressors that may encode this other component have been isolated on high-copy-number plasmids.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beach D., Durkacz B., Nurse P. Functionally homologous cell cycle control genes in budding and fission yeast. Nature. 1982 Dec 23;300(5894):706–709. doi: 10.1038/300706a0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Bücking-Throm E., Duntze W., Hartwell L. H., Manney T. R. Reversible arrest of haploid yeast cells in the initiation of DNA synthesis by a diffusible sex factor. Exp Cell Res. 1973 Jan;76(1):99–110. doi: 10.1016/0014-4827(73)90424-2. [DOI] [PubMed] [Google Scholar]
- DeFeo-Jones D., Scolnick E. M., Koller R., Dhar R. ras-Related gene sequences identified and isolated from Saccharomyces cerevisiae. Nature. 1983 Dec 15;306(5944):707–709. doi: 10.1038/306707a0. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Ellis R. W., Scolnick E. M. Autophosphorylation of v-Ha-ras p21 is modulated by amino acid residue 12. Proc Natl Acad Sci U S A. 1984 May;81(9):2674–2678. doi: 10.1073/pnas.81.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974 Jun;38(2):164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley J., Phear G. A. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe; patterns of splicing and homology to protein kinases. Gene. 1984 Nov;31(1-3):129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hunter T. A thousand and one protein kinases. Cell. 1987 Sep 11;50(6):823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- Hutter K. J., Eipel H. E. Microbial determinations by flow cytometry. J Gen Microbiol. 1979 Aug;113(2):369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. G., Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987 May 7;327(6117):31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- Lörincz A. T., Reed S. I. Primary structure homology between the product of yeast cell division control gene CDC28 and vertebrate oncogenes. Nature. 1984 Jan 12;307(5947):183–185. doi: 10.1038/307183a0. [DOI] [PubMed] [Google Scholar]
- Lörincz A. T., Reed S. I. Sequence analysis of temperature-sensitive mutations in the Saccharomyces cerevisiae gene CDC28. Mol Cell Biol. 1986 Nov;6(11):4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Dobson M. J., Roberts N. A., Tuite M. F., Emtage J. S., White S., Lowe P. A., Patel T., Kingsman A. J., Kingsman S. M. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene. 1983 Sep;24(1):1–14. doi: 10.1016/0378-1119(83)90126-9. [DOI] [PubMed] [Google Scholar]
- Mendenhall M. D., Jones C. A., Reed S. I. Dual regulation of the yeast CDC28-p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell. 1987 Sep 11;50(6):927–935. doi: 10.1016/0092-8674(87)90519-8. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983 Oct;34(3):961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Peden K. W., Nathans D. Local mutagenesis within deletion loops of DNA heteroduplexes. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7214–7217. doi: 10.1073/pnas.79.23.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A., Yochem J., Byers B., Nunn M. F., Duesberg P. H., Doolittle R. F., Reed S. I. A relationship between the yeast cell cycle genes CDC4 and CDC36 and the ets sequence of oncogenic virus E26. Nature. 1984 Jun 7;309(5968):556–558. doi: 10.1038/309556a0. [DOI] [PubMed] [Google Scholar]
- Reed S. I., Hadwiger J. A., Lörincz A. T. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B. J., Hartwell L. H. Regulation of mating in the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1977 Nov;75(2 Pt 1):355–365. doi: 10.1083/jcb.75.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Seraphin B., Faye G. KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986 Oct;5(10):2697–2701. doi: 10.1002/j.1460-2075.1986.tb04553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt P. K., Bos T. J., Doolittle R. F. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci U S A. 1987 May;84(10):3316–3319. doi: 10.1073/pnas.84.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C., Richardson S. L., Reed S. I. Subcellular localization of a protein kinase required for cell cycle initiation in Saccharomyces cerevisiae: evidence for an association between the CDC28 gene product and the insoluble cytoplasmic matrix. J Cell Biol. 1987 Oct;105(4):1527–1538. doi: 10.1083/jcb.105.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]