Abstract
Francisella tularensis, the zoonotic cause of tularemia, can infect numerous mammals and other eukaryotes. Although studying F. tularensis pathogenesis is essential to comprehending disease, mammalian infection is just one step in the ecology of Francisella species. F. tularensis has been isolated from aquatic environments and arthropod vectors, environments in which chitin could serve as a potential carbon source and as a surface for attachment and growth. We show that F. tularensis subsp. novicida forms biofilms during the colonization of chitin surfaces. The ability of F. tularensis to persist using chitin as a sole carbon source is dependent on chitinases, since mutants lacking chiA or chiB are attenuated for chitin colonization and biofilm formation in the absence of exogenous sugar. A genetic screen for biofilm mutants identified the Sec translocon export pathway and 14 secreted proteins. We show that these genes are important for initial attachment during biofilm formation. We generated defined deletion mutants by targeting two chaperone genes (secB1 and secB2) involved in Sec-dependent secretion and four genes that encode putative secreted proteins. All of the mutants were deficient in attachment to polystyrene and chitin surfaces and for biofilm formation compared to wild-type F. novicida. In contrast, mutations in the Sec translocon and secreted factors did not affect virulence. Our data suggest that biofilm formation by F. tularensis promotes persistence on chitin surfaces. Further study of the interaction of F. tularensis with the chitin microenvironment may provide insight into the environmental survival and transmission mechanisms of this pathogen.
Francisella tularensis is a Gram-negative facultative intracellular pathogen that causes the zoonotic disease tularemia (50). Although researchers have focused on various aspects of F. tularensis infections in mammalian hosts, this organism can survive and grow in one of the widest environmental ranges of any studied pathogen. Indeed, F. tularensis has been isolated from a variety of sources, including lagomorphs, arthropods, amoebae, and freshwater (54, 59, 60, 68). Mammals either succumb to infection or clear the bacterium (68), suggesting that mammals may not support prolonged persistence of F. tularensis in nature. Understanding the environmental lifestyle of F. tularensis will help elucidate the survival mechanisms of this pathogen outside of a host and identify risks for human exposure. Recently, outbreaks of tularemia were associated with freshwater, particularly outbreaks of F. tularensis subsp. holarctica (type B) in Eurasia (10, 78). While the most virulent subspecies, F. tularensis subsp. tularensis (type A), was historically linked with the arid climates of North America, a recent epidemiological study found that 100% of the deaths due to tularemia were associated with type A1 strains found in moist climates of the United States (37), suggesting that water may serve as an environmental reservoir for F. tularensis.
The survival of some bacteria in an aquatic environment is associated with their ability to utilize chitin as a carbon source. Chitin is the second most abundant biopolymer in nature and provides structure to many organisms, including the cell wall of fungi (4) and the exoskeleton of arthropods and insects (48). This oligomer of N-acetyl-d-glucosamine (GlcNAc) is hydrolyzed by a family of enzymes termed chitinases (5). These enzymes serve a variety of roles and are conserved from bacteria to mammals. Bacterial chitinases provide environmental organisms the ability to acquire carbon under otherwise nutrient-limiting conditions (34). For example, Vibrio cholerae, the etiological agent of cholera, utilizes chitinases to persist in marine environments on copepod molts (51). The interaction of V. cholerae with chitin influences various metabolic and physiologic responses in this microorganism. For example, Meibom et al. demonstrated that association with chitin and chitin derivatives led to a specific expression profile in V. cholerae that included two chitinase genes and the pilus genes required for colonization and subsequent biofilm formation on nutritive and nonnutritive surfaces (46). Environmental studies have clearly shown that attachment to chitin surfaces is an integral part of the aquatic lifestyle of V. cholerae, and these surface-attached bacterial communities constitute a successful survival mechanism (63).
Formation of biofilms is associated with enhanced survival during environmental stress (1) and increased resistance to antibiotics (12). Biofilms formed by many pathogenic bacteria play an important role in environmental persistence and disease transmission. For instance, Yersinia pestis biofilms are reported to function in the transmission of plague bacteria via colonization of the proventriculus of fleas and the mouth of nematodes (14, 30). We hypothesized that biofilm formation by F. tularensis may represent a mechanism of persistence and transmission as well.
A review by Hassett et al. (28) indicated that the F. tularensis subsp. holarctica live vaccine strain (LVS) can form biofilms on glass coverslips (28). However, the environmental relevance and molecular mechanisms of F. tularensis biofilm formation were not characterized. F. tularensis subspecies encode two conserved putative chitinases, chiA and chiB (http://www.biohealthbase.org/). Various F. tularensis subspecies have been isolated from arthropods with a chitin exoskeleton (54) and from freshwater, where outbreaks have been associated with chitinous crustaceans (2, 16). We therefore investigated the interaction of F. tularensis subsp. novicida (F. novicida) with chitin. We show that F. novicida forms biofilms on natural and synthetic chitin surfaces. Formation of these bacterial communities was dependent on two chitinase genes when exogenous sugar was not present. Attachment to chitin was dependent on factors that are secreted by the Sec translocon protein export system. This mechanism of colonization is specific for environmental surfaces because deletion of genes that facilitate attachment to chitin did not result in defects in virulence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
F. novicida strain U112 and the F. tularensis subsp. holarctica LVS (17, 58), as well as F. novicida Francisella pathogenicity island (FPI) and hspX deletion mutants (77), have been previously described. F. tularensis subsp. tularensis strains SchuS4 and AS2058 (FT-10) were provided by Jean Celli and the New Mexico Department of Health, respectively, and handled under biosafety level 3 precautions in accordance with the Centers for Disease Control and Prevention protocol. Unless otherwise noted, strains were grown in modified Mueller-Hinton medium (MMH; Difco, Corpus Christi, TX) supplemented with 0.025% ferric pyrophosphate, 0.02% IsoVitaleX (Becton Dickinson, Franklin Lakes, NJ) as a cysteine source, and 0.1% glucose as a carbon source. For some experiments, F. tularensis strains were grown in Chamberlain's defined medium (CDM) (44) with or without glucose. For enumeration studies, bacteria were grown on MMH agar plates.
Imaging of F. novicida colonization on chitin films and sterile crab shell pieces.
Wild-type F. novicida was allowed to attach to either synthetic chitin films (79) or sterile crab shell pieces for 1 h. After 1 h, surfaces were washed three times with phosphate-buffered saline to remove nonadherent bacteria and samples were incubated at 30°C in CDM without glucose. After 1 h and 1 week of incubation, respectively, crab shell and chitin film samples were processed for scanning electron microscopy (SEM) investigation. Substrates with attached cells were fixed for 3 days at 4°C in 4% paraformaldehyde with 2% glutaraldehyde in 0.1 M Na cacodylate buffer (pH 7.3, electron microscopy grade; EMS, Hatfield, PA). After primary fixation, samples were rinsed in the same buffer, postfixed in 1% aqueous OsO4 for 1 h, and dehydrated in an ascending ethanol series (30, 50, 70, 80, 90, and 100% for 20 min each), followed by critical-point drying with liquid CO2 using a Tousimis SAMDRI-VT-3B apparatus (Tousimis, Rockville, MD). Samples were mounted on adhesive carbon film on 15-mm aluminum stubs and sputter coated with 100 Å gold/palladium using a Denton Desk 11 TSC sputter coater. Visualization was carried out with a Hitachi S-3400N VP scanning electron microscope (Hitachi Ltd., Pleasanton, CA) operated at 10 to 15 kV at a working distance of 8 to 10 mm with a secondary electron detector. Images were capture in TIF format.
Growth in CDM broth.
F. novicida was grown overnight in CDM at 37° with aeration. The culture was then diluted to an optical density at 600 nm (OD600) of 0.05 using an Ultropec 2100 Pro spectrophotometer (Amersham Biosciences, Pittsburgh, PA) in CDM with no sugar or in CDM with 10 mM glucose or 10 mM GlcNAc. OD and CFU were monitored over time for each medium condition. The doubling time of each culture was calculated.
Imaging of flow cell-grown biofilms.
Flow cells were assembled as previously described (11, 73). The flow system apparatus was sterilized and preconditioned with MMH plus 5 μg/ml tetracycline (Tet5) overnight at ambient temperature (20 to 22°C). F. novicida harboring the pKK219-GFP plasmid (24, 38) was grown overnight at 26°C in MMH plus Tet5 with aeration. Overnight-grown bacteria were diluted 1:50 in fresh medium and grown to an OD600 of 1.0. The culture was then diluted to an OD600 of 0.1. The flow on the flow system was stopped, and 1 ml of culture was inoculated into each channel of the flow cell. Flow cells were inverted for 1 h to allow the bacteria to adhere. Flow cells were then uprighted, and flow was initiated at 0.1 ml/min. Biofilm progression at ambient temperature was imaged by confocal microscopy (Bio-Rad, Hercules, CA) every 24 h over the course of 5 days. z sections were taken at 0.1-μm steps, and three-dimensional (3D) renderings of the z stacks were generated using Volocity imaging software (Improvision, Lexington, MA).
Crystal violet assaying for biofilm formation.
Crystal violet assaying for biofilm formation was performed as previously described (57). Briefly, Francisella strains were grown overnight at the appropriate temperature. Cultures were diluted into fresh medium to an OD600 of 0.05, and 200 μl was aliquoted per well in a 96-well polystyrene plate in at least triplicate. The bacteria were allowed to grow statically and were sampled at various time points. The OD570 was read in a 96-well microplate reader (BioTek, Winooski, VT). At each time point, nonadherent bacteria were removed from the well and 30 μl of 0.1% crystal violet was added to each well for 15 min of incubation. Wells were washed three times with distilled water, and the remaining biomass-absorbed crystal violet was solubilized with 95% ethanol. Staining was then quantified by determining the OD570 in a 96-well microplate reader (labeled CV570). All OD readings for the assay comparing relative crystal violet staining between laboratory Francisella strains and type A Francisella were obtained at 600 nm (CV600) using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Transposon library screen for biofilm-deficient mutants.
A sequenced two-allele transposon mutant library was used to test for F. novicida transposon mutants that were deficient in biofilm formation (the following reagent was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: F. tularensis subsp. novicida “two-allele” transposon mutant library plates 1 to 14 and 16 to 32). The library represents two or more transposon insertions in all nonessential genes. At the time of screening, plate 15 of the library was unavailable due to quality control issues, resulting in a library size of 2,954 mutants. The two-allele library was received frozen in a 96-well format. MMH was inoculated into the wells of 96-well plates with the library, and mutants were grown overnight to stationary phase at 37°C with shaking at 200 rpm. Overnight cultures were diluted 1:50 in 200 μl of fresh MMH in 96-well plates. The plates were grown statically for 10 h in a 37°C incubator, and the ability of each transposon mutant to form a biofilm was assessed as described above. Mutants exhibiting a lower potential for biofilm formation were classified by crystal violet staining more than 2 standard deviations lower than the plate average. Wild-type F. novicida was included on each plate as a positive control, and a well of MMH only was used as a blank. To account for small differences in culture growth, crystal violet staining was normalized to each mutant culture on the basis of the OD570. Wells where significant growth defects were observed were excluded. Biofilm-deficient transposon mutants were retested in triplicate.
Secondary screening for attachment.
Overnight cultures of biofilm mutants identified in our screen were grown in triplicate with shaking (200 rpm) at 37°C. Stationary-phase cultures (200 μl) were transferred to new 96-well plates and allowed to adhere statically for 1 h at 37°C. Crystal violet staining was assayed as before. Attachment deficiency was defined as crystal violet staining 2 standard deviations below that of the wild type.
Bacterial mutagenesis.
Targeted deletions were generated in the U112 strain as previously described (8); for the primers used, see Table S1 in the supplemental material. Briefly, the regions of the chromosome 5′ and 3′ to the gene of interest were amplified by PCR. Using splicing by overlap extension (SOE) PCR (41), a kanamycin resistance cassette expressed by the groEL promoter was introduced between these regions of homology. Briefly, ∼500-bp sequences flanking the targeted gene were amplified and spliced to either end of the gro promoter-resistance cassette construct. The resulting PCR product was transformed into F. novicida strain U112 by chemical transformation, and transformants were selected on MMH agar with 30 μg/ml kanamycin. Gene deletions were confirmed by sequencing. ΔsecB1, ΔFTN_1750, and chiA targeted deletion strains were subsequently complemented in cis by reintroducing the wild-type gene into the chromosome at the original locus, along with the chloramphenicol acetyltransferase (CAT) cassette chloramphenicol resistance marker, again by SOE and homologous recombination of a spliced PCR construct. ΔsecB2, ΔostA2, ΔFTN_0308, and chiB deletion mutants were complemented in trans by introducing the wild-type gene, as well as the CAT cassette, into gro-gfp pFNLTP6 (43). Approximately 500-bp regions flanking the gfp gene of pFNTLTP were amplified and spliced to the wild-type copy of the gene for complementation with the CAT cassette on the 3′ end. SOE PCR complementation constructs were introduced by homologous recombination with pFNLTP6 at the NdeI and BamHI sites, removing the gfp gene. The resulting plasmid expressed the complementing gene under the regulation of the constitutive groEL promoter. Complemented strains were selected for growth on 3 μg/ml chloramphenicol and also confirmed by sequencing. Complementation plasmids were then chemically transformed into deletion strains. For the complementation primers used, see Table S1 in the supplemental material. The ΔchiA ΔchiB double mutant was constructed by the same method as that used for the single-deletion strains, except that the chiB gene was replaced with the CAT cassette instead of the kanamycin resistance cassette.
RAW264.7 macrophage infections.
RAW264.7 macrophages were seeded at 2.5 × 105 cells per well in 24-well tissue culture plates (Becton Dickinson, Franklin Lakes, NJ) and incubated overnight at 37°C with 5% CO2. Wild-type and mutant U112 strains were grown overnight to stationary phase at 37°C with aeration and diluted to 5 × 106 CFU/ml in Dulbecco's modified Eagle medium (Gibco, Carlsbad, CA) with 10% fetal bovine serum. For each strain, 1-ml inocula were added to triplicate wells and centrifuged at 730 × g for 15 min to mediate attachment. Infected plates were incubated at 37°C with 5% CO2 (time zero) for 0.5 h and washed three times with warm medium. Three wells per strain were harvested at this time using 0.1% saponin to lyse the cells. CFU were enumerated by serial dilution, and the percentage recovered was calculated by normalizing to the inocula. Fresh warm medium was added to the remaining wells, and the wells were harvested in triplicate, as described above, at 8 h and 24 h postinfection.
Mouse infections.
Competitive-index (CI) infections of 6- to 8-week-old female C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were performed as previously described (77). Mice were infected intradermally (i.d.) or intraperitoneally (i.p.) with equal amounts (5 × 103 CFU) of wild-type and mutant F. novicida in 0.05 ml. Mice were monitored for morbidity and death during the course of infection. Mice were sacrificed at 2 days postinfection, and their skin and spleens were removed and homogenized for CFU enumeration. CIs were calculated as the ratio of mutant to wild-type bacteria in the output, normalized for the input, and significance was calculated by comparing the CI to 1 (CI of a gene with no role in virulence) using one-sample t tests. All animal infection experiments were approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of Stanford University. Mutants with deletions of the entire FPI and the negative control hspX chaperone gene were described previously (77).
Crab shell attachment.
Overnight cultures were grown at 30°C in MMH. Approximately 1-cm2 pieces of sterile crab shell were inoculated with 2 ml of stationary-phase cultures in 12-well plates. After 1 h at 30°C, the shells were washed to remove unattached bacteria. Attached bacteria were recovered by vortexing, and CFU were enumerated. All of the strains were tested in triplicate. Unpaired t tests were used to determine statistical differences between wild-type and mutant counts.
Statistical analysis.
Statistical analysis was performed using Prism4 software (GraphPad, La Jolla, CA). Unless otherwise stated, unpaired Student t tests were applied, and two-tailed P values are shown. For mouse CI data, a one-sample t test was used to compare mutant to wild-type bacterial ratios to an expected value of 1.
RESULTS AND DISCUSSION
F. novicida forms a biofilm on chitin surfaces.
We hypothesized that chitin may be an environmentally relevant surface for the persistence of F. tularensis in nature based on the presence of two well-conserved chitinase genes in the sequenced F. tularensis genomes (Table 1). Maintenance of the chiA and chiB genes in F. tularensis subspecies and the related but divergent fish pathogen Francisella philomiragia suggested that chitinases provide a selective advantage for Francisella species in nature. F. novicida is a close relative of the highly virulent type A bacterium F. tularensis subsp. tularensis and encodes both chitinase enzymes. Because F. novicida is genetically tractable, we use this subspecies here as a model to study the molecular aspects of F. tularensis ecology.
TABLE 1.
Francisella species chitinase genes
| Strain | Homolog (E value)a |
|
|---|---|---|
| chiA | chiB | |
| F. tularensis subsp. tularensis SchuS4 | FTT0715 (9e−66) | FTT_1768c (2e−15) |
| F. tularensis subsp. tularensis FSC198 | FTF0715 (3e−70) | FTF_1768c (2e−15) |
| F. tularensis subsp. holarctica LVS | FTL_1521 (9e−66) | FTL_0093 (1e−15) |
| F. tularensis subsp. holarctica OSU18 | FTH_1471 (2e−68) | FTH_0088 (1e−15) |
| F. tularensis subsp. novicida U112 | FTN_0627 (4e−69) | FTN_1744 (4e−14) |
| F. philomiragia | Fphi_0215 (1e−66) | Fphi_0864 (1e−15) |
E value based on comparison to glycosyl hydrolase 18 family chitinases.
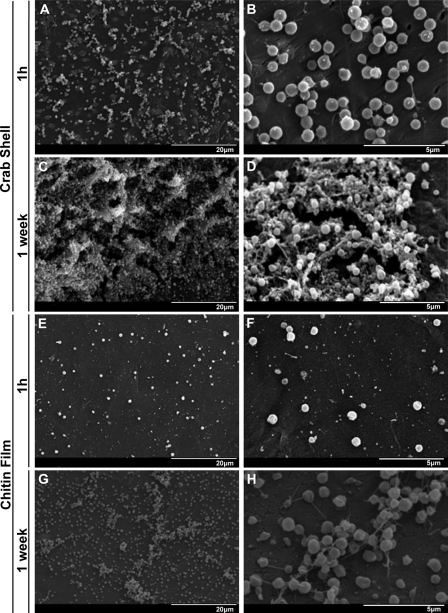
To test the ability of F. tularensis species to adhere to a chitin-containing surface, we incubated F. novicida with crab shell pieces. Crab shells are rich in chitin, a constituent of various surfaces Francisella species may encounter and subsequently colonize in their natural habitats. These surfaces include copepod and zooplankton shells in freshwater environments and the exoskeletons of arthropod vectors. After 1 h at 30°C, individual and small groups of adherent bacteria were present on the shell surface as visualized by SEM (Fig. 1A and B). After 1 week on the crab shells in the presence of minimal CDM without exogenous sugar, 3D bacterial communities were present on the chitin-based surface (Fig. 1C). At higher magnification (Fig. 1D), we saw microcolonies consisting of individual bacteria surrounded by a matrix of extracellular polymeric substance (EPS). The observed community structure suggests that F. novicida can attach to and proliferate as biofilms on the environmentally relevant surface chitin.
FIG. 1.
F. novicida biofilm formation on chitin surfaces. Images display SEM visualization of F. novicida colonization of crab shell pieces (A to D) and synthetic chitin films (E to H). Individual attached bacteria and small attached microcolonies were observed on crab shell pieces at 1 h (A and B). After 1 week, typical 3D biofilm architecture was observed, consisting of bacteria surrounded by an EPS matrix (C and D). Similar results were obtained after 1 h (E and F) and 1 week (G and H) on synthetic chitin.
Although crab shells consist mainly of chitin, they contain additional components, such as other carbohydrates and protein (53). To test if chitin is sufficient to support F. novicida colonization and proliferation, we visualized bacterial attachment and biofilm formation on synthetic chitin films (79). F. novicida attached at lower levels to smooth chitin films compared to the topographically varied crab shells after 1 h (Fig. 1A and E). At 1 week after a shift to minimal medium, the surface of the chitin films contained F. novicida microcolonies and EPS extensions (Fig. 1G and H), indicating the initiation of biofilm formation. However, the architecture of the bacterial communities on chitin films was not as developed as that of the communities on the crab shell pieces (Fig. 1A to D), which may be explained by the smaller starting population on this surface (Fig. 1C and G). More likely, additional components of the crab shell, like protein, may allow more rapid expansion of the adherent population. We conclude that chitin is necessary, but not necessarily sufficient, for wild-type levels of F. novicida biofilm maturation in the absence of exogenous sugar.
F. novicida can utilize GlcNAc as a carbon source for growth.
F. novicida persistence and proliferation on chitin surfaces in the absence of exogenous sugar suggested that this pathogen was able to utilize the chitin component of the surface as a nutrient source. To test this, we grew F. novicida in CDM without added sugar or supplemented with 10 mM glucose (a known metabolic substrate for Francisella species) or with 10 mM GlcNAc (the monosaccharide end product of chitin hydrolysis) in aerated batch culture. F. novicida growth was negligible in CDM in the absence of added sugar (doubling time, 11.25 h). In contrast, F. novicida grew in CDM supplemented with 10 mM glucose with a doubling time of 63 min. Similarly, F. novicida grew in CDM supplemented with 10 mM GlcNAc (doubling time, 76 min). The high proliferation of F. novicida on chitin surfaces (Fig. 1) may therefore be explained by the ∼11-fold increase in the growth rate between F. novicida grown in CDM with GlcNAc and F. novicida grown in CDM without sugar. We conclude that F. novicida can metabolize GlcNAc, suggesting that hydrolysis of chitin by chitinases to generate GlcNAc (34) may provide a local nutrient source for persistence and growth.
Chitinase genes facilitate F. novicida growth on chitin surfaces.
To further address the importance of chitin as a nonhost niche for Francisella in nature, we constructed F. novicida mutants lacking either chitinase gene chiA or chiB and a mutant lacking both chitinase genes. Hager et al. demonstrated that the F. novicida homologs of these enzymes that contain chitin-binding domains are secreted and bind to chitin beads (27). We expected these deletion mutant strains to be attenuated for persistence and biofilm formation on chitin surfaces if F. tularensis species have evolved to form biofilms on chitin surfaces to scavenge carbon. Indeed, the ΔchiA and ΔchiB deletion mutants were attenuated for colonization of crab shells when incubated in CDM without sugar. Although the chitinase mutant bacteria attached to chitin to the same extent as wild-type F. novicida at 1 h (data not shown), we recovered 16- and 15-fold fewer ΔchiA and ΔchiB mutant bacteria than wild-type F. novicida bacteria (P < 0.001), respectively, after 2 days of colonization of crab shells (Fig. 2A). Furthermore, we recovered the same number of ΔchiA ΔchiB double chitinase mutant bacteria as ΔchiA and ΔchiB single chitinase mutant bacteria (Fig. 2A), suggesting that the two chitinase genes act in the same metabolic pathway, as predicted by KEGG pathway analysis (32). The abilities of the ΔchiA and ΔchiB mutant bacterial strains to grow on chitin was restored by the reintroduction of a wild-type copy of each chitinase gene into the coinciding mutant strain, as measured by increased crab shell colonization to levels near that of wild-type F. novicida (Fig. 2A). The ability of the chitinase mutant strains to persist at low levels could be due to the utilization of the amino acids present in the CDM. Alternatively, natural degradation of the crab shell during the experiment could liberate enough free GlcNAc to enable the bacteria to persist but not replicate. Regardless, the highly significant difference between wild-type and chitinase mutant bacteria suggests that chitinase activity strongly contributes to F. novicida persistence on chitin under otherwise carbon-limiting conditions.
FIG. 2.
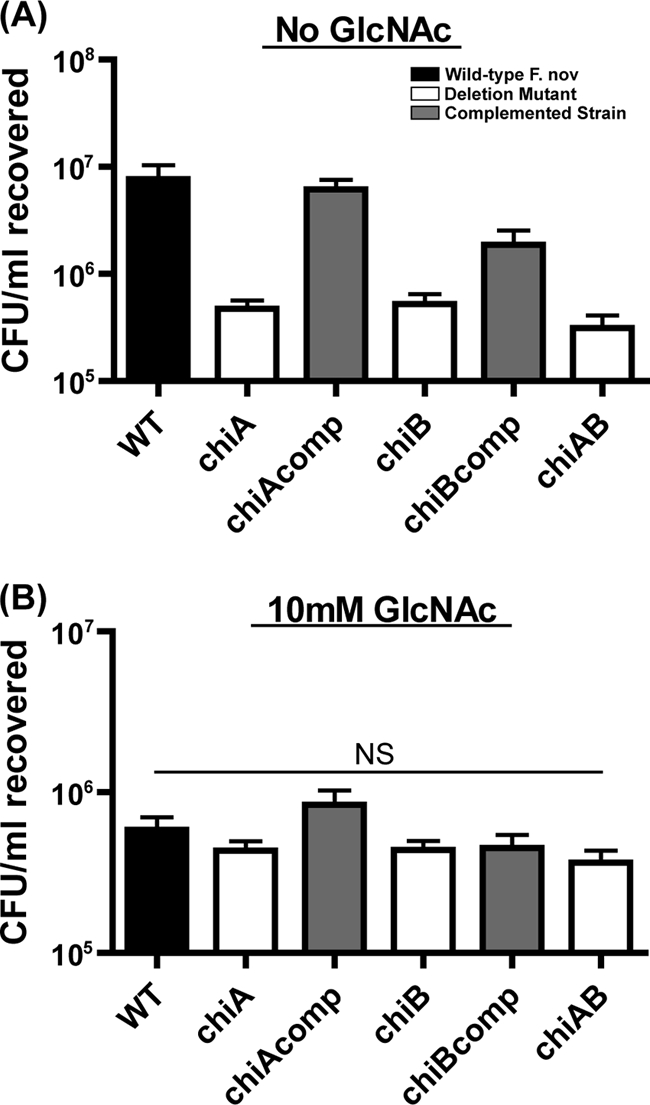
Chitinase mutants are attenuated for chitin colonization in the absence of exogenous sugar. Stationary-phase wild-type (WT) and chitinase mutant bacteria were allowed to adhere for 1 h to crab shell pieces. Equivalently adherent strains were allowed to colonize these chitin surfaces in CDM with or without GlcNAc at 30°C. Triplicate samples were harvested 2 days postinoculation, and CFU were enumerated. F. novicida chitinase mutants (white) were recovered at statistically significantly lower levels than wild-type bacteria (black) (P < 0.001) when incubated in CDM (A) but in equivalent numbers in CDM with GlcNAc (B). Addition of the wild-type chiA and chiB genes to deletion mutant strains (grey; chiAcomp and chiBcomp) complemented the chitin colonization defects observed during colonization in CDM without GlcNAc (A).
We postulated that the inability of chitinase mutant bacteria to convert chitin to the useable metabolite GlcNAc explains their attenuated colonization of chitin. Indeed, the inability of the chitinase mutants to colonize crab shells was alleviated by the addition of 10 mM GlcNAc to the exogenous medium (Fig. 2B), indicating that the chitinase mutant bacteria possess the determinants required to colonize a chitin surface but lack the ability to generate a useable carbon source in order to proliferate. The 13-fold decrease in the recovered wild-type F. novicida when GlcNAc was added (Fig. 2) is consistent with microarray data published for V. cholerae demonstrating that when this pathogen was grown in the presence of excess GlcNAc, the pili and chitinases required to colonize this surface were repressed (46).
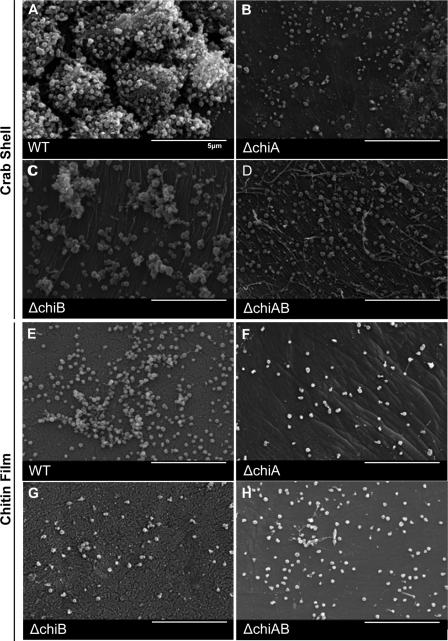
We next compared the architecture of the communities formed by the chitinase mutants on crab shells or chitin films in the absence of exogenous sugar for 1 week by SEM. In contrast to wild-type F. novicida, the chitinase mutants were present as single bacteria or small, mostly monolayer, clusters of bacteria (Fig. 3). We conclude that F. novicida biofilm formation on chitin in the absence of exogenous sugar requires functional chitinase enzymes. Unlike motile V. cholerae that show chemotaxis toward nutrients, F. tularensis species are nonflagellated and nonmotile under laboratory conditions (13). Therefore, the ability of Francisella species to adhere to and colonize chitin may represent a single mechanism for survival in nutrient-poor nonhost environments. Growth on chitin may trigger a specific biofilm program of genes that promote the retention of scavenged GlcNAc in the local microenvironment for use by F. tularensis.
FIG. 3.
Chitinase genes are required for biofilm architecture on chitin surfaces during nutrient stress. Images show representative colonization by wild-type and chitinase mutant strains on crab shells (A to D) or synthetic chitin films (E to H). Bacteria were allowed to attach for 1 h and then incubated for 1 week at 30°C before being processed for SEM. In contrast to the extensive 3D biofilm development in wild-type F. novicida, the chitinase mutants were present as single bacteria or small clusters of bacteria on both natural and synthetic chitin.
Beyond scavenging carbon in the environment, the secreted chitinases that are vital for biofilm formation on chitin could be important for the establishment of arthropod infection, similar to the malaria parasite Plasmodium falciparum (76). The P. falciparum chitinase allows the parasite to penetrate the chitin-containing peritrophic matrix surrounding the blood meal in the mosquito midgut and establish the infection. Efforts to target this chitinase to block the transmission of malaria are ongoing (66, 70). We are currently working to discern the role(s) of F. tularensis chitinases in arthropod vectors.
Characterization of biofilm development by Francisella species.
F. tularensis chitin utilization provides insight into potential persistence mechanisms of this highly virulent pathogen. The missing piece of our model was the proteins that promote attachment to chitin surfaces. We established in vitro systems for studying F. tularensis biofilm formation to aid in identifying attachment determinants. In vitro biofilms on abiotic surfaces provided a model system to characterize and genetically dissect F. novicida biofilm formation and test the ability of other pathogenic F. tularensis strains to similarly attach to and proliferate on a surface.
We incubated green fluorescent protein (GFP)-labeled F. novicida in the flow cell system (11) to confirm the in vitro formation of these bacterial communities under flow conditions. Bacterial attachment and surface growth at ambient temperature (20 to 22°C) and a flow rate of 0.1 ml/min were analyzed by confocal laser scanning microscopy (CLSM) at various time points (24 h, 48 h, 72 h, 96 h, and 120 h) (Fig. 4). We observed the formation of a mat-like biofilm with an average depth of approximately 15 μm. This architecture of flow cell-grown F. novicida biofilms was similar to that reported for other Gram-negative species, including the related gammaproteobacterium Shewanella oneidensis (73) and alphaproteobacterium Caulobacter crescentus (19). Our results indicate that F. novicida is able to form biofilms on an abiotic surface such as glass with architecture similar to that observed on chitin (Fig. 1). These results are consistent with the report by Hassett et al. indicating that LVS can form biofilms on glass coverslips in the absence of flowing medium (28).
FIG. 4.
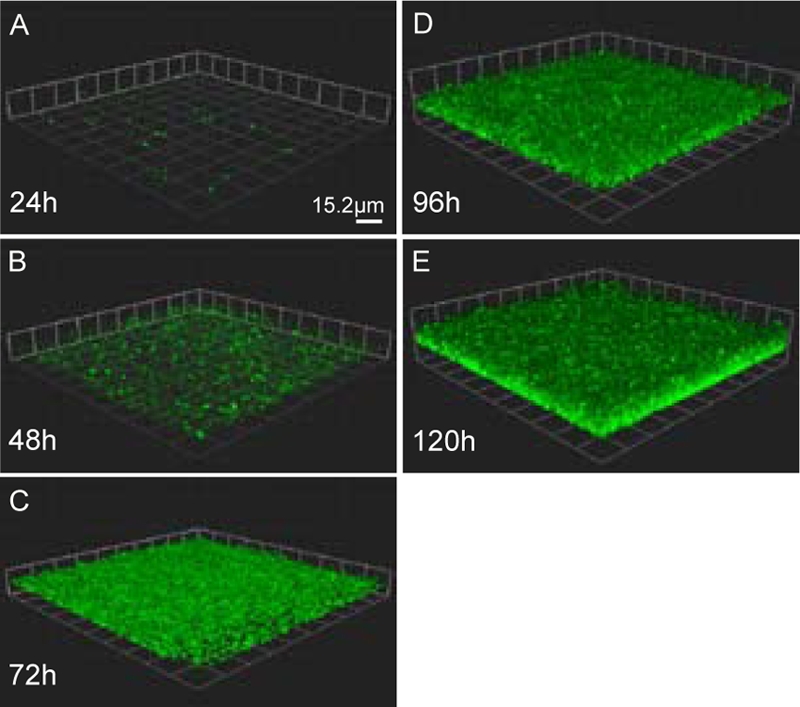
Francisella forms a mat-like biofilm under flow conditions. GFP-expressing F. novicida grown at room temperature (20 to 22°C) was imaged daily in flow cells run at 0.1 ml/min using CLSM. Representative images from triplicate experiments are shown. At 24 h (A), small groups of bacteria were present. Over the next 48 h (B and C), a uniform monolayer of bacteria was observed on the surface. By 96 h (D), depth in the biofilm was observed, and at 120 h (E), the biofilm reached an average thickness of 15 μm.
A modified O'Toole and Kolter microtiter assay (57) was utilized to establish a high-throughput model for F. tularensis biofilm formation. This assay measures adherent biomass under static conditions by crystal violet staining. F. novicida and F. tularensis subsp. holarctica LVS were grown at 26°C and 37°C in 96-well microtiter plates. Recent work by Horzempa et al. found that LVS demonstrated different expression profiles at these two temperatures (29). The OD570 (Fig. 5A and B) and crystal violet staining (CV570) (Fig. 5C and D) were measured over 152 h. Both F. novicida and LVS showed increased crystal violet staining over time when grown at 26°C and 37°C, indicating increased accumulation of adherent biomass. This result is consistent with our finding that F. novicida forms biofilms when adhering to an abiotic surface (Fig. 4). At both of the temperatures assayed, we observed a decrease in crystal violet staining (Fig. 5C and D) concurrent with F. novicida and LVS entering stationary phase (Fig. 5A and B). This result suggested that the biofilms were undergoing dispersion (72), a process of biofilm dissolution and reseeding that occurs during decreased oxygen tension and nutrient deprivation. Similar dispersal did not occur in the flow cell system-grown F. novicida biofilms (Fig. 4), presumably because the population was constantly provided an undepleted carbon and oxygen source under flow conditions.
FIG. 5.
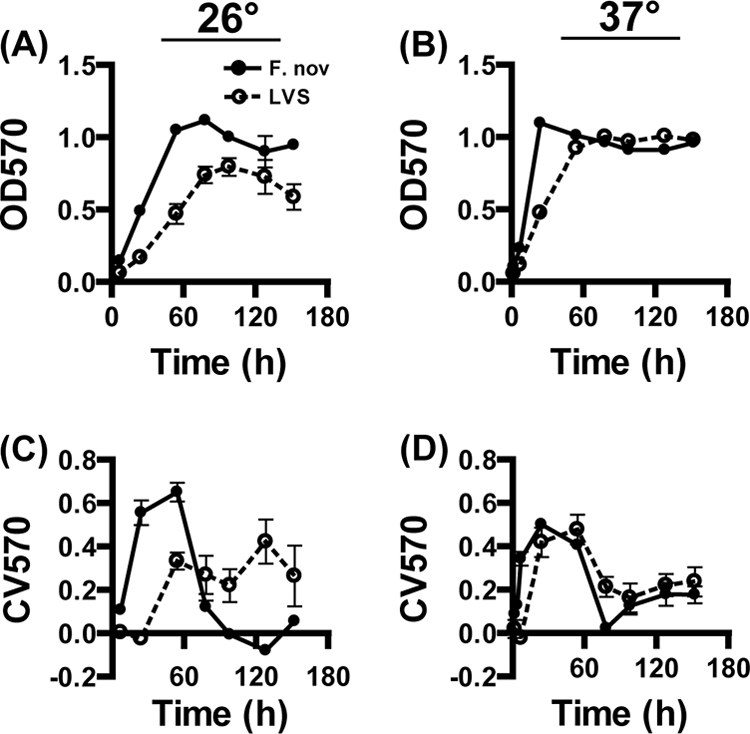
Kinetics of F. tularensis biofilm formation under static conditions. A modified O'Toole and Kolter assay was performed to compare the kinetics and relative levels of biofilm formation of F. novicida (solid circles) and an LVS (open circles). Bacterial growth (A and B) and crystal violet staining (C and D) were determined over time at 26°C (A and C) and 37°C (B and D) by OD570 readings. Both F. tularensis strains were found to acquire crystal violet stain at both temperatures. Growth and crystal violet staining were faster at 37°C for both strains.
Type A Francisella strains form biofilms in the microtiter plate assay.
A high percentage of tularemia morbidity and mortality is caused by infection with F. tularensis subsp. tularensis (type A) strains (21). These strains have a very low infectious dose, and as few as 10 organisms can cause a lethal infection in humans (18). Molecular subtyping of type A strains has identified two distinct subtypes (A1 and A2) with specific geographic distributions (37). Type A1 strains are primarily found in the eastern United States, while type A2 strains are almost exclusively isolated in the western United States. The O'Toole and Kolter assay demonstrated that these highly virulent strains were able to form biofilms to levels similar to those of F. novicida and LVS (Fig. 6). F. tularensis subsp. tularensis strains SchuS4 (type A1) and FT-10 (type A2) reached ODs similar to that of an LVS (type B) when grown under static conditions (Fig. 6), while SchuS4 and FT-10 exhibited higher crystal violet staining at 24 h (P < 0.05), implying increased biofilm formation of type A strains (Fig. 6). F. novicida CV600 staining was approximately twofold higher than that of the other strains tested (P < 0.001). However, the OD of the F. novicida culture was 2.5-fold higher than that of the other strains at 24 h. Similar crystal violet staining by type A1 and type A2 strains compared to the type B LVS suggests that biofilm formation may be pertinent to the survival of pathogenic F. tularensis strains in the environment.
FIG. 6.
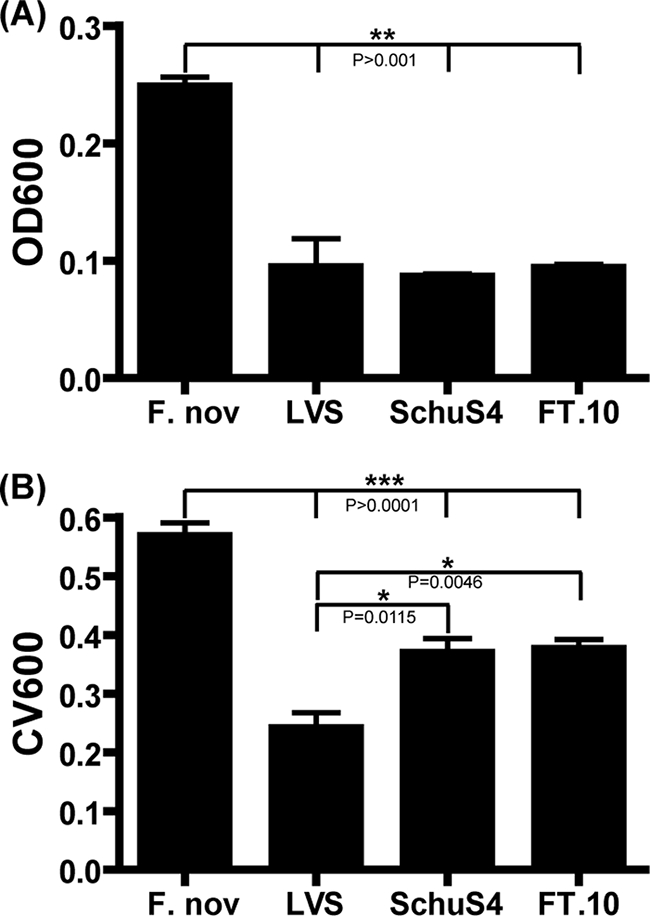
Biofilm formation by virulent F. tularensis subsp. tularensis strains. F. novicida, an LVS, and type A strains SchuS4 and FT-10 were assayed for growth and crystal violet staining at 24 h postinoculation. Culture OD600 (A) and crystal violet staining (B) were determined after static growth at 37°C. F. novicida demonstrated increased growth kinetics and crystal violet staining compared to that of the other strains (P > 0.001). Virulent strains SchuS4 and FT-10 exhibited significantly higher crystal violet staining than the LVS.
Screen for biofilm-deficient mutants identifies novel genes important for F. novicida biofilm formation.
We screened a two-allele transposon library (BEI Resources, Manassas, VA) that represented two or more transposon insertion mutants per nonessential gene in the F. novicida genome to elucidate the genetic determinants of F. novicida interaction with abiotic and biotic surfaces. To facilitate high-throughput screening, individual insertion mutants were assayed for biofilm formation in the microtiter assay established above rather than on chitin. We defined biofilm-deficient mutants as strains where crystal violet staining was 2 standard deviations below the mean of the plate. We eliminated mutant strains that exhibited a significant growth defect from further characterization. In total, we identified 98 F. novicida transposon insertion mutants, representing 88 genes that were attenuated for biofilm formation (see Table S2 in the supplemental material). To elucidate pathways important for F. novicida surface attachment and growth, we assigned gene ontology classifications to the genes identified in the biofilm screen (www.biohealthbase.org) (see Fig. S1 in the supplemental material). Roles for the 64 annotated genes included protein secretion, various metabolic pathways, signal transduction, protein transport, and cell envelope biogenesis. Although many Gram-negative and Gram-positive bacteria can form biofilms, the bacterial mechanisms utilized to facilitate these communities vary (3, 42, 52, 57, 75). For example, the role of type IV pili and flagella in biofilm formation is well documented (56, 62). However, little is known about attachment and surface growth during the biofilm maturation of nonmotile bacteria. By characterizing the roles of the genes we identified in this study, including the approximately 25% with no annotated function, we aim to elucidate alternate methods of environmental persistence by nonmotile bacteria.
Sec-dependent secretion functions in initial attachment during F. novicida biofilm formation on abiotic and biotic surfaces.
We were particularly interested in the four transposon insertion mutants we identified in the Sec translocon complex involved in protein export from the cytoplasm (23). The core components of the Sec translocon in Escherichia coli are the SecYEG protein channel and the SecA ATPase motor protein (9). Due to the pleiotropic roles of general protein secretion in bacteria, components of this apparatus are considered essential in other Gram-negative organisms (23, 40). The Sec translocon in F. novicida is composed of 13 proteins, but only the four genes we identified in our screen were represented in the two-allele library, the secA motor ATPase and secG pore genes, as well as the secB1 and secB2 genes that encode chaperones which specifically target preproteins to SecA (74). Additionally, we identified 18 transposon biofilm mutant clones, representing 14 genes that are predicted to encode proteins with secretion signals based on the signal sequence detection algorithm SignalP (6) (Table 2).
TABLE 2.
Sec translocon and Sec-dependent secreted proteins involved in biofilm formation
| Designation | Well IDa | Gene | Gene product | Biological process(es) | Sec secretionb |
|---|---|---|---|---|---|
| FTN_0090 | 4F07 | acpA | Acid phosphatase | Fatty acid and lipid metabolism | Secreted |
| FTN_0100 | 20C12 | Hypothetical membrane protein | Hypothetical, novel | Secreted | |
| FTN_0109 | 14G06 | Protein of unknown function | Unknown function, novel | Secreted | |
| FTN_0121 | 26G09 | secB1 | Preprotein translocase, subunit B | Motility, attachment and secretion, structure | Translocon |
| FTN_0121 | 4F06 | secB1 | Preprotein translocase, subunit B | Motility, attachment and secretion, structure | Translocon |
| FTN_0191 | 19E06 | Polar amino acid uptake transporter | Transport of amino acids | Secreted | |
| FTN_0304 | 20C11 | Pilus assembly protein | Motility, attachment and secretion, structure | Secreted | |
| FTN_0308 | 19H06 | Membrane protein of unknown function | Unknown function, novel | Secreted | |
| FTN_0357 | 21B08 | pal | Peptidoglycan-associated lipoprotein, OmpA family | Transport of drugs, antibacterial compounds | Secreted |
| FTN_0429 | 14G12 | Conserved protein of unknown function | Unknown function, conserved | Secreted | |
| FTN_0635 | 25C04 | Serine-type d-Ala-d-Ala carboxypeptidase | Cell wall, LPS, capsule | Secreted | |
| FTN_0672 | 12G03 | secA | Preprotein translocase, subunit A (ATPase, RNA helicase) | Motility, attachment and secretion, structure | Translocon |
| FTN_0713 | 14C04 | ostA2 | Organic solvent tolerance protein OstA | Cell wall, LPS, capsule | Secreted |
| FTN_0713 | 21H10 | ostA2 | Organic solvent tolerance protein OstA | Cell wall, LPS, capsule | Secreted |
| FTN_0713 | 26E07 | ostA2 | Organic solvent tolerance protein OstA | Cell wall, LPS, capsule | Secreted |
| FTN_0714 | 12G01 | Protein of unknown function | Unknown function, novel | Secreted | |
| FTN_0714 | 27C09 | Protein of unknown function | Unknown function, novel | Secreted | |
| FTN_1093 | 18A05 | Protein of unknown function | Unknown function, novel | Secreted | |
| FTN_1476 | 26A03 | Protein of unknown function | Unknown function, novel | Secreted | |
| FTN_1503 | 26A08 | Protein of unknown function | Unknown function, novel | Secreted | |
| FTN_1510 | 1E01 | secB2 | Preprotein translocase, subunit B | Motility, attachment and secretion, structure | Translocon |
| FTN_1630 | 13C11 | secG | Preprotein translocase, subunit G, membrane protein | Motility, attachment and secretion, structure | Translocon |
| FTN_1750 | 19H02 | Acyltransferase | Fatty acid and lipid metabolism | Secreted | |
| FTN_1750 | 23D04 | Acyltransferase | Fatty acid and lipid metabolism | Secreted |
Well identity (ID) annotation from the BEI Resources F. novicida two-allele transposon library.
Genes labeled translocon are structural components of Sec-dependent secretion. Secreted proteins were predicted using the SignalP algorithm.
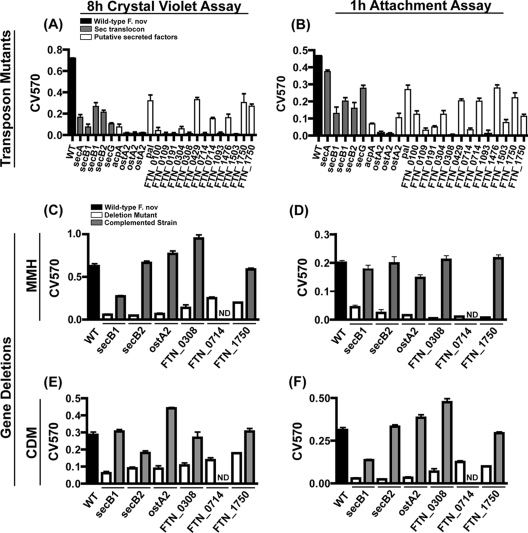
We hypothesized from the results of our genetic biofilm screen and a secondary attachment assay that proteins secreted by the Sec translocon may represent novel mediators of F. novicida adhesion, a process that has not been characterized. We confirmed that transposon mutants in the secretion apparatus were deficient in biofilm formation (Fig. 7A) and attachment (Fig. 7B). secB1 and secB2 deletion mutants were constructed, while secA and secG deletions could not be generated, suggesting that these genes are essential and the transposon insertion mutations present in the library represent an incomplete loss of gene function. Additionally, attempts to construct a double deletion of secB1 and secB2 did not yield viable colonies. Growth curves obtained with the ΔsecB1 and ΔsecB2 mutants showed no growth defect in batch culture compared to wild-type F. novicida, and microscopic analysis of cell morphology revealed no alterations in bacterial shape (data not shown). Both the ΔsecB1 and ΔsecB2 mutants were deficient in biofilm formation (Fig. 7C) and attachment (Fig. 7D) when grown in MMH. The ΔsecB1 and ΔsecB2 mutant phenotypes were restored to wild-type attachment and biofilm formation levels when wild-type copies of secB1 and secB2 were added back to the deletion mutants (Fig. 7C and D). These experiments were also performed with CDM to confirm that the role of Sec-dependent secreted factors in biofilm formation is not limited to growth in a nutrient-rich environment (Fig. 7E and F). Our data indicate that Sec-dependent secretion is important for F. novicida attachment to abiotic surfaces and biofilm formation. We therefore postulated that Sec-secreted proteins represent novel mediators of F. novicida adherence.
FIG. 7.
Sec-secreted factors mediate initial attachment during biofilm formation. Five transposon insertions representing mutants defective in four genes in the Sec translocon (gray) and 18 transposon insertions representing mutants defective in 14 genes in putative secreted factors (white) identified in the forward genetic screen were tested in triplicate compared to wild-type (WT) F. novicida (black) for 8 h of biomass accumulation (A) and 1 h of initial attachment (B). Multiple transposon mutants were tested for genes identified more than once in the screen. Adherence of biomass at 8 h was used to measure biofilm formation. Attachment was assessed by crystal violet staining 1 h postinoculation of stationary-phase cultures. Targeted mutants defective in selected representative genes (white) showed similar defects in biofilm formation (C and E) and attachment (D and F) compared to wild-type F. novicida (black) when grown in MMH and CDM, respectively, based on crystal violet staining. Complementation of the deleted genes (gray) restored mutants to wild-type levels in all cases. Bars represent the means, and the lines indicate standard deviations calculated from triplicate samples of a representative experiment. Each experiment was repeated in triplicate. No data (ND) were obtained for FTN_0714 complementation due to technical difficulties.
The 18 Sec-dependent transposon insertion mutants (Table 2) were all defective in biofilm formation (Fig. 7A) and initial attachment (Fig. 7B) based on crystal violet staining, confirming our screen results. Surprisingly, type IV pilus genes, known mediators of biofilm formation in Gram-negative bacteria, were not among the Sec-secreted factors identified and were found to be dispensable for F. novicida biofilm formation upon further study (data not shown).
We focused on four of the secreted factors that have homologs in all F. tularensis subspecies and cause bacteria to be highly attenuated for biofilm formation when deleted, i.e., FTN_0308, FTN_0713, FTN_0714, and FTN_1750. FTN_0713 (ostA2), FTN_0714, and FTN_1750 were all identified at least twice in the biofilm screen. We selected FTN_0308 due to the strong biofilm phenotype of the one transposon insertion mutant that was identified in the genetic screen (Fig. 7A and B). We constructed a deletion mutant for each of these genes and tested it for attachment and biofilm formation. All four mutants were defective in initial attachment and biofilm formation in both rich and defined media (Fig. 7C to F). The ΔostA2, ΔFTN_1750, and ΔFTN_0308 mutants were complemented for attachment and biofilm attenuation by reintroduction of the deleted genes in cis into the chromosome or in trans by expressing the gene in pFNLTP6 using the constitutive gro promoter. The ΔFTN_0714 mutant could not be complemented for technical reasons, likely due to the length of the complementation PCR product (∼8 kb). Taken together, our data indicate that initial attachment during Francisella biofilm formation is facilitated by proteins secreted by the Sec-dependent secretion system.
The protein encoded by FTN_0713 (ostA2) has significant homology (E value, 6e−64) to organic solvent tolerance proteins involved in lipopolysaccharide (LPS) modification (7). Although ostA2 homologs have not been implicated in biofilm formation, LPS chemistry has been shown to influence attachment during biofilm formation by other bacteria (3, 15, 22). The unique structure of Francisella species LPS (26) could contribute to adhesion of F. tularensis to nonmammalian surfaces. FTN_1750 encodes a putative acyltransferase with strong homology (E value, 4e−27) to acylhomoserine lactone biosynthesis enzyme HdtS, suggesting that this protein may function in quorum sensing, a cell-cell communications process that regulates biofilm formation under certain conditions (35).
While F. novicida biofilm genes were identified by screening for mutants defective in adherence and biofilm formation on polystyrene, we identified two novel putative chitin-binding proteins encoded by FTN_0308 and FTN_0714. The FTN_0714-encoded protein is annotated as a hypothetical lipoprotein (BioHealthBase). The SMART domain prediction algorithm (39, 65) indicates that FTN_0714 contains repeating polycystic kidney disease family domains conserved from archaea through mammals that facilitate adhesion (31). This domain family plays a role in the binding and hydrolysis of chitin by the chiA chitinase of the aquatic bacterial strain Alteromonas sp. strain 0-7 (55). FTN_0308 is annotated to encode a hypothetical protein with unknown function (BioHealthBase). However, the Phyre protein-folding prediction algorithm (33) indicates a structural homology to the Streptomyces chitinase C chitin-binding domain and the C terminus contains homology to F17c family bacterial adhesins. We are currently determining the specific roles that these two gene products may have in attachment to both abiotic and chitin surfaces.
F. tularensis species genomes contain an annotated chitin-binding protein, encoded by cbpA, that was not identified by our biofilm screen. This gene product may specifically mediate association with chitin. Additionally, we did not identify chiA and chiB in our screen despite their conserved Sec-dependent secretion signals and role in biofilm formation on chitin. We would not expect chitinases to mediate biofilm formation on polystyrene, however. This result was confirmed by using clean deletion mutant stains (data not shown).
Sec secretion mutants are not attenuated in murine models of infection.
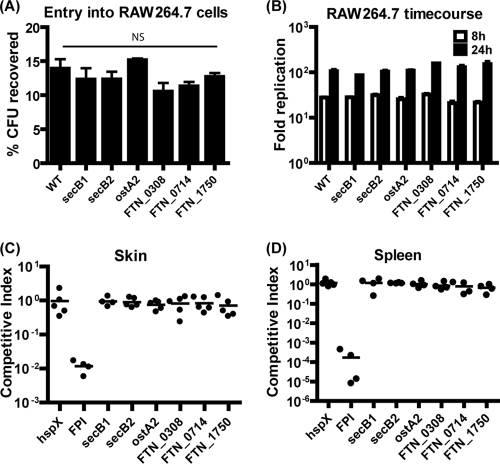
While no evidence of F. tularensis biofilm formation inside mammalian hosts exists, bacteria often utilize proteins to attach to both environmental and host surfaces (62, 64, 67). Attachment of F. tularensis in any context is poorly understood. We therefore tested if the biofilm attachment factors we describe also mediate host tissue association by using in vitro and in vivo infection models. F. tularensis is primarily found within macrophages in a mammalian host (25). Therefore, RAW264.7 macrophage-like cells were infected at a multiplicity of infection of 20:1 with wild-type F. novicida or the Sec-dependent secretion mutants. At 0.5 h postinfection, non-cell-associated F. novicida bacteria were washed away and the remaining bacteria were recovered and enumerated. No statistically significant differences in CFU counts were observed (Fig. 8A), suggesting that the mutants that are defective in attachment to polystyrene and chitin were still able to efficiently associate with eukaryotic cells. Intracellular replication was monitored in the presence of extracellular gentamicin for 8 and 24 h (Fig. 8B). Wild-type F. novicida and all of the mutants showed approximately 100-fold replication at 24 h compared to the initial 0.5-h counts. Thus, the mutants successfully entered and replicated within macrophages, demonstrating that the Sec secretion biofilm mutants that we characterized are not deficient in attachment to or replication within macrophages.
FIG. 8.
The Sec translocon and secreted factors do not influence F. novicida virulence. Sec secretion-targeted F. tularensis deletion mutants were assessed for virulence in in vitro and in vivo models. Entry efficiency of F. novicida strains into RAW264.7 macrophage-like cells was measured as the percentage of the inoculum recovered from inside the cells at 30 min postinfection (A). Intracellular replication of wild-type (WT) and mutant bacteria was assessed as fold replication compared to 30-min counts at 8 h and 24 h postinfection (B). The ability of mutants to colonize the skin after i.d. inoculation (C) and the spleen after i.p. inoculation (D) was determined by determining CIs in C57BL/6J mice at 2 days postinfection. For all of the virulence assays, no difference was observed between the Sec secretion biofilm mutants and wild-type F. novicida.
To test the potential roles of secB1, secB2, ostA2, FTN_0308, FTN_0714, and FTN_1750 during a systemic mouse infection, we infected C57BL/6J mice with a 1:1 mixture of 5 × 103 CFU of wild-type and deletion mutant bacteria. CIs for each wild-type-mutant combination were obtained for both the i.d. and i.p. routes of infection. Mutants that are not attenuated in mice should have a CI of 1; i.e., equal numbers of wild-type and mutant bacteria should be recovered in the tissue at the time of harvest, as was observed for the previously described ΔhspX mutant strain (77). As a positive control, we included an F. novicida mutant that lacks the entire FPI (77). As expected, this mutant was severely attenuated in mice (Fig. 8C and D). However, none of the Sec secretion biofilm mutants demonstrated a CI value statistically significantly different from 1 via either route of infection (Fig. 8C and D). Additionally, no defect was observed in the spread of Sec secretion mutants to systemic tissues such as the liver and spleen after i.d. inoculation (data not shown). Our data indicate that these genes that are crucial for association with nonmammalian surfaces do not contribute to local or systemic colonization of mammalian hosts. FTN_0713 (ostA2), the putative LPS modification gene, was identified by Kraemer et al. in a negative selection screen for F. novicida mutants attenuated for infection via intranasal inoculation of mice, indicating that this mutant may be more sensitive to the innate immune response in the lung (e.g., antimicrobial peptides) due to an altered LPS (36, 77). secA and secE transposon mutants were identified by Su et al. as being involved in lung colonization (69). These attenuated phenotypes for the nonredundant Sec translocon genes imply that Sec-secreted proteins other than those characterized here do influence host colonization. The lack of attenuation of the secB1 and secB2 deletion mutants in the virulence assays tested here supports the idea that these two genes encode redundant functions.
F. novicida biofilm determinants also play a role in attachment to chitin-based surfaces.
Given that Francisella species induce biofilm formation on both abiotic and chitin surfaces, we hypothesized that the attachment determinants we identified for association with polystyrene may also facilitate attachment to chitin. After a 1 h of incubation at 30°C, an average of 3.33 × 107 CFU/ml wild-type F. novicida were attached to the crab shell pieces (Fig. 9). Sec secretion mutants were 5.6- to 16.2-fold attenuated for attachment to this chitin-based surface compared to wild-type bacteria (P < 0.01), confirming that Sec-secreted proteins contribute to attachment to chitin-based surfaces. The specificity of these adherence factors for nonmammalian surfaces further supports our suggestion that F. tularensis biofilm formation in nature has evolved to promote this pathogen's survival outside of a host, potentially by facilitating chitin utilization.
FIG. 9.
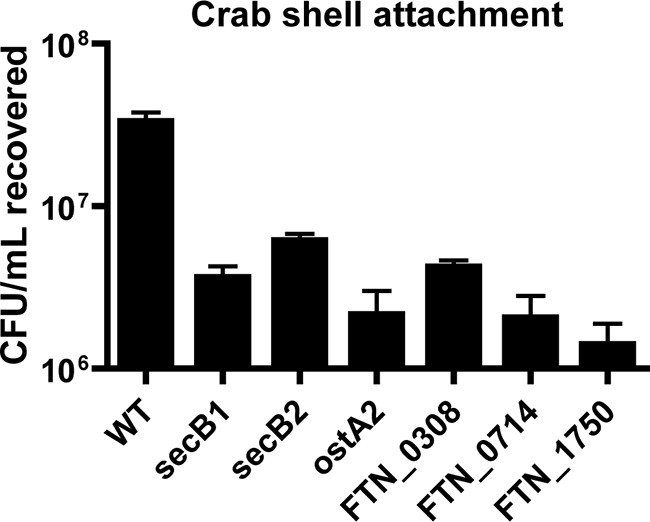
Biofilm mutants are attenuated for attachment to chitin-based crab shell pieces. Stationary-phase cultures of secB1, secB2, ostA2, FTN_0308, FTN_0714, and FTN_1750 deletion mutants were allowed to attach for 1 h to sterile crab shell pieces. CFU of attached bacteria were enumerated in triplicate samples. Sec secretion biofilm mutants were found to attach statistically significantly less than wild-type (WT) F. novicida by unpaired t test (P > 0.01).
From our collective data, we propose a model in which these early determinants of biofilm formation allow association with chitin surfaces in nature. Through this interaction, F. tularensis chitinases have access to this substrate and provide bacteria with GlcNAc, which is utilized for growth in nutrient-limiting environments. Biofilm maturation on chitin would then create a local microenvironment enriched in this carbon source, providing a nonhost niche for this zoonotic pathogen.
The ability of F. tularensis to form biofilms on chitin may also provide the bacterium resistance to grazing by freshwater protozoa. For the chitin colonizer V. cholerae, biofilm formation was shown to reduce grazing by flagellate organisms compared to planktonic bacteria (45). Thelaus et al. found that F. tularensis subsp. holarctica had increased resistance to both ciliate and flagellate protozoa compared to that of E. coli (71). Although the role of biofilm formation in resistance to predation was not addressed, this observation suggests that F. tularensis may actively prevent protozoal grazing in nature. Coupled with the ability to survive in nutrient-limited aquatic environments, biofilm-mediated resistance to predation could contribute to F. tularensis persistence in the environment and allow prolonged transmission of this pathogen.
We provide here the first extensive characterization of F. tularensis biofilm formation and explore how these communities may promote environmental persistence and transmission on chitin surfaces. The F. novicida biofilm genes we describe contribute to the ability of this pathogen to colonize a surface it may encounter in nature. Very little is known about how and where Francisella species persist in nature when not replicating within a host. Our findings may help explain how tularemia outbreaks that have been attributed to freshwater crustaceans (2, 16) occur. Additionally, chitin utilization may support F. tularensis persistence on other arthropods, such as zooplankton, copepods, and biting arthropod vectors. A study of F. tularensis survival in artificial water found that the presence of chitinous freshwater shrimps, mollusks, diatoms, or zooplankton promoted the sustained viability of this pathogen for an additional week to 1 month in nutrient-poor water (49). Survival on environmental chitin may therefore serve as a reservoir for disease transmission during seasonal tularemia outbreaks. Palo et al. identified a strong epidemiological correlation between areas with low rates of water turnover and human cases of tularemia (61). These researchers postulated low water turnover as an environmental cue for a burst of F. tularensis replication. As with cholera outbreaks (20), conditions that promote interaction of F. tularensis with chitin surfaces on which the bacteria can replicate may seed infection. Further study of F. tularensis biofilm formation and the role of these communities in chitin colonization could clarify the open question of the location of the F. tularensis environmental reservoir.
Supplementary Material
Acknowledgments
We thank Melanie Blokesch for her generous gifts of reagents and technical assistance, Gary K. Schoolnik for his thoughtful discussions and for providing the chitin films, Jonathan W. Jones and Thomas Henry for help with mouse experiments, and Carmen D. Cordova for assistance with the biofilm CLSM imaging. Jean Celli graciously provided the SchuS4 strain.
J.J.M. was supported by National Science Foundation and Department of Homeland Security graduate fellowships, as well as a National Institutes of Health Cell and Molecular Biology training grant. This work was supported by grants AI063302 and AI065359 from the NIH-NIAID to D.M.M.
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, J. L., and R. J. C. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anda, P., J. Segura del Pozo, J. M. Díaz García, R. Escudero, F. J. García Peña, M. C. López Velasco, R. E. Sellek, M. R. Jiménez Chillarón, L. P. Sánchez Serrano, and J. F. Martínez Navarro. 2001. Waterborne outbreak of tularemia associated with crayfish fishing. Emerg. Infect. Dis. 7:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balestrino, D., J. M. Ghigo, N. Charbonnel, J. A. Haagensen, and C. Forestier. 2008. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10:685-701. [DOI] [PubMed] [Google Scholar]
- 4.Bartnicki-Garcia, S. 1968. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu. Rev. Microbiol. 22:87-108. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276-24286. [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Braun, M., and T. J. Silhavy. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45:1289-1302. [DOI] [PubMed] [Google Scholar]
- 8.Brotcke, A., and D. M. Monack. 2008. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect. Immun. 76:3473-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brundage, L., J. P. Hendrick, E. Schiebel, A. J. Driessen, and W. Wickner. 1990. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62:649-657. [DOI] [PubMed] [Google Scholar]
- 10.Celebi, S., M. Hacimustafaoglu, and S. Gedikoglu. 2008. Tularemia in children. Indian J. Pediatr. 75:1129-1132. [DOI] [PubMed] [Google Scholar]
- 11.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. Toftgaard Nielsen, M. Givskov, S. Molin, and J. D. Ron. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Cowan, S. T., and R. K. A. Feltham (ed.). 2004. Cowan and Steel's manual for the identification of medical bacteria, 3rd ed. Cambridge Press, Cambridge, United Kingdom.
- 14.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 15.de Lima Pimenta, A., P. Di Martino, E. Le Bouder, C. Hulen, and M. A. Blight. 2003. In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microbes Infect. 5:1177-1187. [DOI] [PubMed] [Google Scholar]
- 16.Díaz de Tuesta, A. M., Chow-Quan, M. P. Geijo Martínez, J. Dimas Núñez, F. J. Díaz de Tuesta, C. R. Herranz, and E. Val Pérez. 2001. Tularemia outbreak in the province of Cuenca associated with crab handling. Rev. Clin. Esp. 201:385-389. [DOI] [PubMed] [Google Scholar]
- 17.Eigelsbach, H. T., and C. M. Downs. 1961. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87:415-425. [PubMed] [Google Scholar]
- 18.Ellis, J., P. C. F. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Entcheva-Dimitrov, P., and A. M. Spormann. 2004. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J. Bacteriol. 186:8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epstein, P. R. 1993. Algal blooms in the spread and persistence of cholera. Biosystems 31:209-221. [DOI] [PubMed] [Google Scholar]
- 21.Farlow, J., D. M. Wagner, M. Dukerich, M. Stanley, M. Chu, K. Kubota, J. Petersen, and P. Keim. 2005. Francisella tularensis in the United States. Emerg. Infect. Dis. 11:1835-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujise, O., Y. Wang, W. Chen, and C. Chen. 2008. Adherence of Aggregatibacter actinomycetemcomitans via serotype-specific polysaccharide antigens in lipopolysaccharides. Oral Microbiol. Immunol. 23:226-233. [DOI] [PubMed] [Google Scholar]
- 23.Gold, V. A. M., F. Duong, and I. Collinson. 2007. Structure and function of the bacterial Sec translocon. Mol. Membr. Biol. 24:387-394. [DOI] [PubMed] [Google Scholar]
- 24.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71:5940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golovliov, I., M. Ericsson, G. Sandstrom, A. Tarnvik, and A. Sjostedt. 1997. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65:2183-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunn, J. S., and R. K. Ernst. 2007. The structure and function of Francisella lipopolysaccharide. Ann. N. Y. Acad. Sci. 1105:202-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hager, A. J., D. L. Bolton, M. R. Pelletier, M. J. Brittnacher, L. A. Gallagher, R. Kaul, S. J. Skerrett, S. I. Miller, and T. Guina. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227-237. [DOI] [PubMed] [Google Scholar]
- 28.Hassett, D. J., P. A. Limbach, R. F. Hennigan, K. E. Klose, R. E. W. Hancock, M. D. Platt, and D. F. Hunt. 2003. Bacterial biofilms of importance to medicine and bioterrorism: proteomic techniques to identify novel vaccine components and drug targets. Expert Opin. Biol. Ther. 3:1201-1207. [DOI] [PubMed] [Google Scholar]
- 29.Horzempa, J., P. Carlson, D. O'Dee, R. Shanks, and G. Nau. 2008. Global transcriptional response to mammalian temperature provides new insight into Francisella tularensis pathogenesis. BMC Microbiol. 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 31.Jing, H., J. Takagi, J.-H. Liu, S. Lindgren, R.-G. Zhang, A. Joachimiak, J.-H. Wang, and T. A. Springer. 2002. Archaeal surface layer proteins contain [beta] propeller, PKD, and [beta] helix domains and are related to metazoan cell surface proteins. Structure 10:1453-1464. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa, M., M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu, and Y. Yamanishi. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480-D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 34.Keyhani, N. O., and S. Roseman. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108-122. [DOI] [PubMed] [Google Scholar]
- 35.Kirisits, M. J., and M. R. Parsek. 2006. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 8:1841-1849. [DOI] [PubMed] [Google Scholar]
- 36.Kraemer, P. S., A. Mitchell, M. R. Pelletier, L. A. Gallagher, M. Wasnick, L. Rohmer, M. J. Brittnacher, C. Manoil, S. J. Skerett, and N. R. Salama. 2009. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect. Immun. 77:232-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kugeler, K. J., P. S. Mead, A. M. Janusz, J. E. Staples, K. A. Kubota, L. G. Chalcraft, and J. M. Petersen. 2009. Molecular epidemiology of Francisella tularensis in the United States. Clin. Infect. Dis. 48:863-870. [DOI] [PubMed] [Google Scholar]
- 38.Kuoppa, K., Å. Forsberg, and A. Norqvist. 2001. Construction of a reporter plasmid for screening in vivo promoter activity in Francisella tularensis. FEMS Microbiol. Lett. 205:77-81. [DOI] [PubMed] [Google Scholar]
- 39.Letunic, I., T. Doerks, and P. Bork. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lill, R., K. Cunningham, L. A. Brundage, K. Ito, D. Oliver, and W. Wickner. 1989. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 8:961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, J., X. Zogaj, J. R. Barker, and K. E. Klose. 2007. Construction of targeted insertion mutations in Francisella tularensis subsp. novicida. Biotechniques 43:487-490. [DOI] [PubMed] [Google Scholar]
- 42.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 70:7511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maier, T. M., R. Pechous, M. Casey, T. C. Zahrt, and D. W. Frank. 2006. In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl. Environ. Microbiol. 72:1878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matz, C., D. McDougald, A. M. Moreno, P. Y. Yung, F. H. Yildiz, and S. Kjelleberg. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 102:16819-16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meibom, K. L., X. B. Li, A. T. Nielsen, C.-Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melillo, A., D. D. Sledjeski, S. Lipski, R. M. Wooten, V. Basrur, and E. R. Lafontaine. 2006. Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263:102-108. [DOI] [PubMed] [Google Scholar]
- 48.Merzendorfer, H., and L. Zimoch. 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 206:4393-4412. [DOI] [PubMed] [Google Scholar]
- 49.Mironchuk, I. U. V., and A. V. Mazepa. 2002. Viability and virulence of Francisella tularensis subsp. holarctica in water ecosystems (experimental study). Zh. Mikrobiol. Epidemiol. Immunobiol. March-April:9-13. [PubMed]
- 50.Mörner, T. 1992. The ecology of tularaemia. Rev. Sci. Tech. 11:1123-1130. [PubMed] [Google Scholar]
- 51.Mueller, R. S., D. McDougald, D. Cusumano, N. Sodhi, S. Kjelleberg, F. Azam, and D. H. Bartlett. 2007. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J. Bacteriol. 189:5348-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muñoz-Elías, E. J., J. Marcano, and A. Camilli. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 76:5049-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naczk, M., J. Williams, K. Brennan, C. Liyanapathirana, and F. Shahidi. 2004. Compositional characteristics of green crab (Carcinus maenas). Food Chem. 88:429-434. [Google Scholar]
- 54.Nigrovic, L. E., and S. L. Wingerter. 2008. Tularemia. Infect. Dis. Clin. North Am. 22:489-504. [DOI] [PubMed] [Google Scholar]
- 55.Orikoshi, H., S. Nakayama, C. Hanato, K. Miyamoto, and H. Tsujibo. 2005. Role of the N-terminal polycystic kidney disease domain in chitin degradation by chitinase A from a marine bacterium, Alteromonas sp. strain O-7. J. Appl. Microbiol. 99:551-557. [DOI] [PubMed] [Google Scholar]
- 56.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 57.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 58.Owen, C. R., E. O. Buker, W. L. Jellison, D. B. Lackman, and J. F. Bell. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J. Bacteriol. 87:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967-978. [DOI] [PubMed] [Google Scholar]
- 60.Oyston, P. C. F. 2008. Francisella tularensis: unravelling the secrets of an intracellular pathogen. J. Med. Microbiol. 57:921-930. [DOI] [PubMed] [Google Scholar]
- 61.Palo, R., C. Ahlm, and A. Tärnvik. 2005. Climate variability reveals complex events for tularemia dynamics in man and mammals. Ecol. Soc. 10:22. [Google Scholar]
- 62.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pruzzo, C., L. Vezzulli, and R. R. Colwell. 2008. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 10:1400-1410. [DOI] [PubMed] [Google Scholar]
- 64.Rosen, D. A., J. S. Pinkner, J. M. Jones, J. N. Walker, S. Clegg, and S. J. Hultgren. 2008. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 76:3337-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schultz, J. R., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shahabuddin, M., and J. M. Vinetz. 1999. Chitinases of human parasites and their implications as antiparasitic targets. EXS 87:223-234. [DOI] [PubMed] [Google Scholar]
- 67.Shanks, R. M. Q., N. A. Stella, E. J. Kalivoda, M. R. Doe, D. M. O'Dee, K. L. Lathrop, F. L. Guo, and G. J. Nau. 2007. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 189:7262-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sjöstedt, A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1-29. [DOI] [PubMed] [Google Scholar]
- 69.Su, J., J. Yang, D. Zhao, T. H. Kawula, J. A. Banas, and J.-R. Zhang. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75:3089-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takeo, S., D. Hisamori, S. Matsuda, J. Vinetz, J. Sattabongkot, and T. Tsuboi. 2009. Enzymatic characterization of the Plasmodium vivax chitinase, a potential malaria transmission-blocking target. Parasitol. Int. 58:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thelaus, J., A. Andersson, P. Mathisen, A. L. Forslund, L. Noppa, and M. Forsman. 2009. Influence of nutrient status and grazing pressure on the fate of Francisella tularensis in lake water. FEMS Microbiol. Ecol. 67:69-80. [DOI] [PubMed] [Google Scholar]
- 72.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thormann, K. M., R. M. Saville, S. Shukla, D. A. Pelletier, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valent, Q. A., P. A. Scotti, S. High, J. W. de Gier, G. von Heijne, G. Lentzen, W. Wintermeyer, B. Oudega, and J. Luirink. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17:2504-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vilain, S., J. M. Pretorius, J. Theron, and V. S. Brozel. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilm. Appl. Environ. Microbiol. 75:2861-2868. [DOI] [PMC free article] [PubMed]
- 76.Vinetz, J. M., S. K. Dave, C. A. Specht, K. A. Brameld, B. Xu, R. Hayward, and D. A. Fidock. 1999. The chitinase PfCHT1 from the human malaria parasite Plasmodium falciparum lacks proenzyme and chitin-binding domains and displays unique substrate preferences. Proc. Natl. Acad. Sci. U. S. A. 96:14061-14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104:6037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willke, A., M. Meric, R. Grunow, M. Sayan, E. J. Finke, W. Splettstosser, E. Seibold, S. Erdogan, O. Ergonul, Z. Yumuk, and S. Gedikoglu. 2009. An outbreak of oropharyngeal tularaemia linked to natural spring water. J. Med. Microbiol. 58:112-116. [DOI] [PubMed] [Google Scholar]
- 79.Yusof, N. L., L. Y. Lim, and E. Khor. 2004. Flexible chitin films: structural studies. Carbohydr. Res. 339:2701-2711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.