Abstract
Iron is required by almost all bacteria, but concentrations above physiological levels are toxic. In bacteria, intracellular iron is regulated mostly by the ferric uptake regulator, Fur, or a similar functional protein. Iron limitation results in the regulation of a number of genes, especially those involved in iron uptake. A subset of these genes is the Fur regulon under the control of Fur. In the present study, we have identified Fur- and iron-regulated genes in Listeria monocytogenes by DNA microarray analysis using a fur mutant and its isogenic parent. To identify genes regulated exclusively in response to iron limitation, the whole-genome transcriptional responses to the iron limitation of a fur mutant and its isogenic parent were compared. Fur-regulated genes were identified by comparing the transcriptional profile of the parent with the transcriptional profile of the isogenic fur mutant. Our studies have identified genes regulated exclusively in response to iron and those that are negatively regulated by Fur. We have identified at least 14 genes that were negatively regulated directly by Fur. Under iron-limited conditions, these genes were upregulated, while the expression of fur was found to be downregulated. To further investigate the regulation of fur in response to iron, an ectopic fur promoter-lacZ transcriptional fusion strain was constructed, and its isogenic fur and perR mutant derivatives were generated in L. monocytogenes 10403S. Analysis of the iron limitation of the perR mutant indicated that the regulation of genes under the negative control of Fur was significantly inhibited. Our results indicate that Fur and PerR proteins negatively regulate fur and that under iron-limited conditions, PerR is required for the negative regulation of genes controlled by Fur.
Iron is an important nutrient required by almost all bacteria and is an abundant element in nature. However, it is not readily available for bacteria to use, especially under aerobic conditions because iron is present in the insoluble Fe3+ form. In the human host, free iron is present in almost negligible concentrations, as it is sequestered by high-affinity proteins such as albumin, transferrin, and lactoferrin, which are also potential sources of iron for many pathogenic bacteria (5, 25). It has been repeatedly demonstrated that the presence of Fur as well as the iron acquisition ability of bacteria are important requirements for survival in the host (23, 48). In order to acquire iron from diverse sources, bacteria have evolved various mechanisms (26). However, iron uptake needs to be tightly controlled since excess levels of iron can generate toxic hydroxyl radicals by the Fenton reaction.
In most bacteria, iron uptake regulation is under the control of Fur or a similar protein of equivalent function (1). When iron is available to the bacteria in adequate levels, Fur complexes with iron and represses several genes by binding to their promoters at a consensus nucleotide sequence, the Fur box (6, 21). Some studies have shown that Fur can function as a repressor even in the absence of iron (20, 33). Moreover, for some bacteria, the Fur protein was identified as being a positive regulator (14, 16).
In order to identify genes under the control of Fur, methods such as systematic evolution of ligands by exponential enrichment (SELEX) and Fur titration assay (FURTA) have been developed. The recent use of DNA microarrays has greatly improved our understanding of the role of Fur and iron in bacteria (2, 12, 17). However, little is known about iron- and Fur-regulated genes in Listeria monocytogenes.
L. monocytogenes is a Gram-positive, rod-shaped, facultative, intracellular pathogen that can cause serious food-borne illness in humans (10, 13, 29, 46). In the host environment, the availability and acquisition of iron are important for the survival of this bacterium (11, 52). In L. monocytogenes, Fur is understood to be the regulator of iron acquisition systems that help to utilize diverse iron sources, such as transferrin, ferritin, lactoferrin, hemin, catecholamine, and siderophores, produced by other bacteria (28). We have recently cloned, purified, and characterized the Fur protein from L. monocytogenes. Our studies showed that, in vitro, the purified Fur protein binds to Fur box-containing promoters (Pfur and PfhuC) without iron, and fur is expressed in the presence of iron (33). Studies from other laboratories have shown that fur regulates virulence genes and almost all iron acquisition genes in this bacterium, while the expression of fur seems to be negatively regulated by PerR (41, 47). Although few roles for Fur and iron in L. monocytogenes have been described, the independent functions of these two factors are not completely known.
In this study, L. monocytogenes EGDe and its isogenic fur deletion mutant were compared for genome-wide expression changes in response to iron limitation. The implementation of this approach helped to identify genes that were regulated exclusively by either iron or Fur. We found 7 new genes with Fur boxes in their upstream putative promoter regions. Among the Fur-regulated genes, iron limitation resulted in a significant upregulation of many genes except fur. To confirm the microarray data, Northern blotting and quantitative reverse transcription-PCR (qRT-PCR) analysis were performed with cells grown in iron-limiting KRM medium (41) and iron-chelated (2,2′-dipyridyl [DPD]) brain heart infusion (BHI) medium, and the expressions of fur, feoB, lmo0514, and lmo2185 (previously named svpA) were analyzed. Furthermore, the regulation of fur gene expression and the role of PerR were investigated by using a fur::lacZ fusion strain of L. monocytogenes 10403S and its isogenic fur and perR mutant derivatives.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains of Escherichia coli and L. monocytogenes used in this study are listed in Table 1. E. coli was grown at 37°C on LB agar or in LB broth (Difco, Sparks, MD). L. monocytogenes EGDe and its isogenic fur deletion mutant were grown at 30°C or 37°C in BHI broth, BHI agar (Difco, Sparks, MD), or KRM medium. The composition and preparation of KRM medium was previously described by Newton et al. (41). To prepare KRM medium, all ingredients except metal ions were dissolved in water and treated with 10 g of Chelex per liter of medium for 5 h at room temperature, and Chelex was removed by using a 0.22-μm filter (Nalgene, Rochester, NY). A total of 10.4 g liter−1 of RPMI 1640 medium (Sigma, St. Louis, MO) and all the required metals except iron were added to this solution, and filter sterilization was performed by using 0.22-μm filter units. Iron-replete conditions were achieved by adding 20 μM sterile FeSO4 to KRM minimal medium. To reduce the iron in the cultures, the bacteria were grown overnight in BHI broth, diluted 1:100 in KRM medium, and grown to stationary phase. This culture was diluted once again (1:100) in fresh KRM medium and grown to the exponential phase. Iron contamination was minimized by washing and then rinsing all the glassware with concentrated HCl. In addition, all the required metals were dissolved in Chelex-treated water.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or primer sequence (5′→3′)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 supE44 hsdR17 gyrA96 relA1 thi (lac-proAB) F′ (traD36 proAB+lacIqZ M15) | This study |
| L. monocytogenes | ||
| 10403S | Serotype 1/2a | This study |
| Pfl | 10403S Pfur::lacZ fur+ | This study |
| Pf-1 | PFL fur::pAUL-A fur mutant | This study |
| Pfl-2 | PFL perR::pAUL-A perR mutant | This study |
| EGDe | Serotype 1/2a | 47 |
| EGDeΔfur | fur deletion mutant of EGDe | 47 |
| Plasmids | ||
| pHK77 | Integratable lacZ fusion vector | 31 |
| pHK77-F | 720-bp upstream region of fur cloned into pHK77 | This study |
| pIntFM | pAUL-A containing 275-bp fur internal PCR fragment | This study |
| pIntPM | pAUL-A containing 275-bp perR internal PCR fragment | This study |
| Primers | ||
| 514F | TATGCGCAAAACTCCATGAA | |
| 514R | CTTTGAGTCCTGCAGCATTTT | |
| FB1F | CGCTTGTTTTACCATACGGAG | |
| FB1R | CCATAAAGTTTTAAGCGATGGC | |
| HK091 | GTTTCCCGCTCGAATTC | |
| HKlacZ | TTCTGGTGCCGGAAACCAG | |
| HKFF | GCTAGCGTTGTGCTTTAATGCGTCCA | |
| HKFR | GGTACCGGATGGCAGGAAATATTTGG | |
| Paf-F | GCAAATGGTACCTGCACCTGACACTGGCCT | |
| Paf-R | TGAATCGGAGCTCCCACTTCGATTCGACGATTT | |
| PaP-F | CCTGAAAGCTTTTAACTCACATACACACCCAACC | |
| pap-R | TGAATGGATCCATATCCGGTCATATGTGCTGC | |
| Svpa1F | AGATGCAATTAGTTATGATCACTGGT | |
| Svpa1R | AGATGCAATTAGTTATGATCACTGGT | |
| qRT-PCR primers | ||
| 16S rRNART-F | CCCTTATGACCTGGGCTACA | |
| 16S rRNART-R | CCTACCGACTTCGGGTGTTA | |
| lmo0485RT-F | CGTATTGCACACACTTGCTG | |
| lmo0485RT-R | CCTGCTTTTAAATCCGCAAT | |
| lmo1956RT-F | TGGCCTTGCAACCGTTTATAG | |
| lmo1956RT-R | TTATCAACCACGCGCAATTC | |
| lmo1960RT-F | CCAACTAGAGACGTTGCCAAA | |
| lmo1960RT-R | ACTTTGATGCGGAGAACGTC | |
| lmo2185RT-F | GATGCAATTAGTTATGATCACTGGT | |
| lmo2185RT-R | CCATCCGAAAGAGTCACAGG |
Underlining indicates restriction endonuclease sites.
For the preparation of iron-depleted BHI medium, cultures grown overnight in BHI medium were diluted 1:100 in BHI broth and grown to an optical density at 600 nm (OD600) of ∼0.1, 0.3 mM 2,2′-dipyridyl (DPD) (Sigma, St. Louis, MO) was then added, and the cultures were grown to an OD600 of ∼0.7 as described previously by Jin et al. (28).
When antibiotic selection pressure was required, ampicillin (50 μg ml−1), erythromycin (300 μg ml−1), or kanamycin (50 μg ml−1) for E. coli or chloramphenicol (10 μg ml−1), erythromycin (3 μg ml−1), or neomycin (10 μg ml−1) for L. monocytogenes was added to the medium.
Construction of a fur::lacZ fusion in L. monocytogenes.
To construct the transcriptional fusion consisting of the fur promoter and lacZ, two primers, hkf-F and hkf-R, with NheI and KpnI restriction sites, respectively, were used to amplify a 720-bp region upstream of the fur coding region from L. monocytogenes chromosomal DNA. The PCR product was digested with the enzymes KpnI and NheI (Promega, Madison, WI), ligated into NheI- and KpnI-digested plasmid pHK77, and transformed into E. coli JM109 cells. The resulting clone was designated pHK77-F.
To create a fur::lacZ fusion in an ectopic region of the chromosome, pHK77-F was transformed by electroporation into L. monocytogenes competent cells. The resulting transformants were selected at 30°C on BHI agar plates containing chloramphenicol. The resultant colonies were transferred into BHI broth without antibiotics and grown overnight. This culture was then diluted 1:1,000 in BHI broth with neomycin (10 μg ml−1) and grown overnight at 41°C without shaking to facilitate the integration of the plasmid. This process was sequentially repeated 5 times. Later, the culture was diluted 1:1,000 with fresh BHI medium, and 50 μl was plated onto BHI agar containing neomycin. Several colonies were inoculated into BHI broth containing neomycin and grown at 30°C overnight to facilitate plasmid excision. Diluted cultures were plated onto BHI agar containing neomycin, and the resultant colonies were tested for chloramphenicol sensitivity to confirm the double-crossover recombination and integration of Pfur-lacZ as described previously (31). The recombinants, which were neomycin resistant and chloramphenicol sensitive, were tested for β-galactosidase activity on BHI agar plates containing 50 μg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Invitrogen, Carlsbad, CA).
Construction of fur and perR mutants of the L. monocytogenes 10403S fur::lacZ strain.
The procedure for the construction of the mutants by using plasmid pAUL-A was previously described by Chakraborty et al. (9). To construct mutations in the fur and perR regions of the fur::lacZ fusion strain, an internal fragment from each respective coding region was generated from the chromosomal DNA of L. monocytogenes 10403S by PCR amplification. To create a mutation in the fur region, primers paf-F and paf-R with KpnI and SacI restriction sites, respectively, were used to amplify a 275-bp fragment internal to the fur coding region. Similarly for the construction of the perR mutation, primers pap-F and pap-R with HindIII and BamHI restriction sites, respectively, were used to generate a 275-bp PCR product internal to the perR coding region. The PCR fragment corresponding to the regions of fur or perR were cloned separately into pAUL-A, a temperature-sensitive integratable plasmid, by appropriate restriction enzyme digestions, ligated using T4 DNA ligase (Promega, Madison, WI), and transformed into E. coli JM109 cells. The resultant clones were designated pintFM, for pAUL-A:fur internal fragment, and pintPM, for pAUL-A:perR internal fragment. These plasmids were purified and transformed separately into the fur::lacZ strain of L. monocytogenes 10403S and selected by incubation at 30°C on BHI agar plate containing erythromycin. For the integration of the plasmids into either fur or perR coding regions, colonies were transferred twice by streaking onto BHI agar containing erythromycin and incubated at 42°C. Mutants of fur (Pfl-1) or perR (Pfl-2) were confirmed by PCR, sequencing, and transcript analysis.
Construction of an L. monocytogenes EGDe perR mutant.
Plasmid pAUL-A, containing the perR internal fragment constructed as described above, was transformed into competent EGDe cells. Integration of the plasmid and mutant confirmation were performed as described above for L. monocytogenes 10403S.
RNA purification and analysis.
For the extraction of total RNA, 3 ml of culture was mixed with 6 ml of bacterial RNA Protect solution (Qiagen, Valencia, CA), and the mixture was centrifuged for 20 min at 3,000 × g in a swinging-bucket rotor centrifuge to pellet the cell. These pellets were resuspended in 1 ml of Trizol (Invitrogen, Carlsbad, CA), and the cells were broken using the FastPrep system (Qbiogene, Irvine, CA) at a speed of 6.0 for 40 s. RNA was extracted from the broken-cell lysate according to the manufacturer's instructions. Extracted RNA was treated with RNase-free DNase I (Qiagen, Valencia, CA) and purified by using the RNeasy minikit (Qiagen, Valencia, CA). The concentration of RNA was measured by using a spectrophotometer, and its quality was assessed by agarose gel electrophoresis before being used for microarray, Northern blot, or qRT-PCR analysis. Three independent bacterial cultures for each test and controls were prepared as biological replicates for RNA isolation.
Probe synthesis and microarray hybridization.
cDNA was generated by reverse transcription using random hexamers (Invitrogen, Carlsbad, CA) as primers. The primers were annealed (70°C for 10 min, followed by snap-freezing in ice for 1 min) to 5 μg of total RNA, extended with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) in a buffer containing 0.1 M dithiothreitol-12.5 mM deoxynucleoside triphosphate (dNTP)/5-(3-aminoallyl)-dUTP (Ambion, Austin, TX), and incubated overnight at 42°C. Residual RNA was removed from these samples by alkaline treatment and neutralized, and cDNA was purified by using a QIAquick PCR purification kit (Qiagen, Valencia, CA). Purified aminoallyl-modified cDNA was labeled with Cy3 or Cy5 monofunctional N-hydroxysuccinimide (NHS) ester cyanogen dyes (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions. Labeled cDNA was purified by using a QIAquick PCR purification kit.
The full genome of the L. monocytogenes array consists of single-stranded (70-mer) epoxy-coated oligonucleotides representing 6,347 open reading frames (ORFs) from L. monocytogenes strains EGDe, 4b F2365, 1/2a F6854, and 4b H7858 (http://pfgrc.tigr.org). Purified labeled cDNA was hybridized with L. monocytogenes genome microarrays as previously described (37, 40). Briefly, the slides were prehybridized in a buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, and 0.1% bovine serum albumin (BSA) and then washed and blown to dryness. The two different labeled cDNA samples were dried and resuspended in hybridization solution (40% deionized formamide, 5× SSC, 0.1% SDS, 0.6 μg μl−1 sheared salmon sperm DNA). The labeled cDNA samples were hybridized with slides at 42°C for 18 to 20 h. After hybridization, the slides were washed in low-stringency buffer (2× SSC, 0.1% SDS) for 2 min, in medium-stringency buffer (0.1× SSC, 0.1% SDS) for 5 min, and, finally, in high-stringency buffer (0.1× SSC) for 10 min. The slides were dried and scanned by use of a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA).
Data analysis.
Scanned tagged image file format (TIFF) images obtained from hybridized microarray slides were analyzed by using TIGR-Spotfinder (http://www.tm4.org/spotfinder.html). Spots were analyzed by adaptive quantification, and the local background was subsequently subtracted. Spots with background-corrected signal intensities (median) in both channels with less-than-twofold background intensity (median) were rejected from further analyses. Data normalization was performed on the remaining spots by use of the LOWESS algorithm (block mode; smooth parameter, 0.33) using TIGR-MIDAS software (http://www.tm4.org/midas.html), and the normalized log2 ratio of test/reference signal for each spot was recorded. Genes with less than three data points were considered unreliable and were excluded from further analysis. For the remaining genes, the averaged log2 ratio was calculated, and the change of significance in expression for a gene was determined with Significance Analysis of Microarrays (SAM) software (www.stat.standford.edu/∼tibs/SAM/index.html) (56) using one class mode (Δ = 1.0) [the measurement is the log(test/reference)2 ratio from two labeled samples hybridized to an array]. In this analysis, SAM assigns a score to each gene on the basis of the change in gene expression relative to the standard deviation of repeated measurements. For genes with scores greater than an adjustable threshold, SAM uses permutations of repeated measurements to estimate the percentage of genes identified by chance, the false discovery rate (FDR). A “q value” is assigned to each gene and corresponds to the lowest false discovery rate at which the gene is considered significant. The differentially expressed genes identified by SAM were further filtered on the basis of previously defined criteria (29, 35, 38). Several controls were employed to minimize the technical and biological variations. To ensure the quality of the data, the following steps were performed: (i) each ORF was present as a 4× replicate on each array; (ii) array slides were prepared in duplicate for each experiment, and fluorophore dyes were swapped between replicates to account for dye bias; and (iii) the experiment was repeated twice.
To examine changes in the levels of transcripts among different functional categories, we classified these genes using in-house Gene Sorter software according to the categories described by the comprehensive microbial resource of TIGR (http://cmr.tigr.org/tigr-scripts/CMR/shared/Genomes.cgi).
Fur box analysis.
To identify putative Fur boxes, intergenic regions of the L. monocytogenes EGDe genome were analyzed by using a custom position weight matrix (CPWM) sequence generated from a previously identified Fur box nucleotide sequence (31, 36). The generation of CPWM and whole-genome analysis were done using an online bioinformatics software tool, Virtual Footprint Analysis (http://www.prodoric.de/vfp) (39).
Northern blot analysis.
Ten micrograms of purified total RNA samples from wild-type L. monocytogenes EGDe and its isogenic fur mutant, obtained from cultures grown in KRM medium with or without added iron, was separated on a 1.0% agarose-0.66 M formaldehyde gel and transferred onto a nylon membrane. RNA isolation, hybridization, and detection were performed as described previously (36). To confirm the microarray data, regulations of lmo0514, fur (lmo1956), lmo2185, and feoB (lmo2105) transcripts were analyzed by Northern blotting. Total RNA was also isolated from L. monocytogenes 10403S cultures grown in BHI medium or BHI medium containing 0.3 mM DPD and analyzed for changes of fur and lmo2185 gene expression. The PCR primers used for the preparation of radioactive probes are shown in Table 1.
Quantitative real-time PCR analysis.
To analyze the effects of iron limitation on the regulation of various Fur repression control (FRC) genes, 0.3 mM 2,2′-dipyridyl (DPD) was added to cultures growing in BHI medium. In order to titrate out the DPD, 0.1 mM ferrous sulfate was added to the samples. Cells were grown in biological triplicates as follows: cultures in BHI medium grown overnight were diluted 1:100 in 10 ml BHI broth and grown to an OD600 of 0.1, 0.3 mM DPD was added to the first set, 0.3 mM DPD and 0.1 mM FeSO4 were added to the second set, and the third set had no DPD and FeSO4 and thus served as a control. In addition, the effect of excess iron on gene expression was quantified for samples treated with 0.1 mM, 0.5 mM, or no ferric citrate and grown in biological triplicates. Cultures grown overnight were diluted 1:100 in 10 ml BHI medium supplemented with (i) no ferric citrate (control), (ii) 0.1 mM ferric citrate, and (iii) 0.5 mM ferric citrate. All the cultures were grown until they reached an OD600 of 0.7 before total RNA was isolated.
To investigate gene expression, 0.5 μg of total RNA was reverse transcribed to cDNA using a high-capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) was performed by using Power SYBR green PCR master mix (Applied Biosystems) in a 7300 Fast system instrument (Applied Biosystems) according to the manufacturer's instructions. The levels of gene expression in the treated cells and the nontreated controls were calculated by relative quantification using the 16S rRNA gene as the endogenous reference gene (30). All the samples were amplified in triplicates, and data analysis was carried out using 7300 Fast system software (Applied Biosystems). Primers were designed specifically to amplify 100-nucleotide stretches from the genes being analyzed (Table 1).
β-Galactosidase assay for fur expression analysis.
Cultures of the fur::lacZ fusion strain and its isogenic fur or perR mutants of L. monocytogenes were grown overnight in BHI medium. These cultures were diluted 1:100 and grown to an OD600 of 0.1. At this culture density, 0.3 mM DPD was added, and the culture was grown until it reached an OD600 of 0.7. For β-galactosidase assays, cells were permeabilized with 0.5% toluene and 4.5% ethanol, and the activity was measured as previously described (40). The activity for each sample was calculated as follows: 1,000 × (OD420 − 1.75 × OD550)/(OD600 × reaction time × volume).
Microarray data accession number.
The data set discussed in this publication is shown in Table S1 in the supplemental material and was deposited in the Gene Expression Omnibus (GEO) (19), accessible through GEO series accession number GSE12735 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127355).
RESULTS AND DISCUSSION
Bacterial growth in KRM medium.
KRM medium, which was used to grow the bacteria, was previously described to be very low in iron (46). In this medium, EGDe and the fur mutant reached an approximate final-density OD600 of 0.9 (see Fig. S1 in the supplemental material). Although a growth defect for the fur mutant in iron-limited tropolone-containing medium was reported previously (48), we did not observe any defect in growth for the mutant in KRM medium. Reasons for the difference in the growth of the mutant in different media reported by the two studies are not completely clear. Experiments described previously by Jin et al. (28) showed that L. monocytogenes can efficiently grow in RPMI-based medium and that this medium has about 1 μM iron. From our experience with various media, we have observed that the minimum concentration of iron required to support growth varies from one medium to another. For example, the minimum iron concentration required for medium containing or treated with tropolone would not be the same as that required for RPMI-based medium. Hence, the minimum amount of iron is dependent on the medium, growth conditions, and, specifically, the presence of the iron-chelating agent added or used. Furthermore, the iron toxicity for a bacterium is an intrinsic physiological phenomenon and is determined by its state of growth, oxygen requirement, and efficiency in regulating iron transport. Researchers have reported that the concentration of iron in BHI medium is about 25 μM, and in our experiments, we used up to 1,000 μM iron, but this did not result in any growth defect. However, we did not test whether 1,000 μM or any other concentration of iron was toxic to the fur mutant.
To perform our experiments, total RNA was isolated from strain EGDe and its fur mutant grown to exponential phase (OD600 = 0.55). We chose this growth phase because it is considered to be the metabolically active phase of growth for bacteria. Recent studies have shown that the transcriptomic response of exponential-phase-grown cultures is closely related to the response elicited by Listeria cells growing in vivo (7).
Microarray experiments.
Microarray experiments were aimed at determining significant transcriptional changes of the fur mutant compared to EGDe when iron is available for both strains (experiment a), the fur mutant compared to EGDe when iron is limited for both strains (experiment b), when iron is limited for the fur mutant only (experiment c), and when iron is limited for the parent only (experiment d). A similar experimental design was used to study iron acquisition and regulation in Campylobacter jejuni (45) and Fur- and iron-regulated gene expression in Helicobacter pylori (20).
In our study, genes that were significantly regulated were identified by estimating the false discovery rate (FDR) using SAM software (Δ = 1.01; FDR of 5%; q < 0.05). The false discovery rate has been well described to be a suitable method for microarray analysis and has become one of the most important standards by which genes showing significant changes in transcription are listed and sorted (40, 42, 51). In a recent proteomic study, researchers compared t statistics and SAM methods to analyze the liquid chromatography-mass spectrometry (LC/MS) proteomic data, and the results indicated that SAM identified a large number of different expressed proteins while overcoming the false-positive results that one would encounter with a t test (50).
In our experiments, iron deficiency resulted in transcriptional changes of a variety of genes in the parent and the fur mutant (see Table S1 in the supplemental material). In the parent, these changes are likely mediated through Fur, which is known as an iron-responsive transcriptional regulator for almost all bacteria. Thus, the genes that are repressed by Fur are commonly observed to be upregulated when iron is limited or when fur is mutated, as was observed in experiments a, b, and d. However, bacteria have several genes that are regulated directly by iron and not by Fur. In our study, genes that were exclusively iron dependent were identified by comparing data from experiment c to data from experiment d.
L. monocytogenes response to iron limitation.
Iron limitation resulted in significant changes in the expression of several genes in L. monocytogenes EGDe and the fur mutant. From the microarray experiments, we obtained genes that responded especially to iron and those that were negatively regulated by Fur.
Genes regulated exclusively in response to iron limitation.
Comparison of gene lists from experiments c and d showed that iron limitation resulted in the upregulation of 10 genes (Table 2) and the downregulation of 15 genes (Table 3).
TABLE 2.
Genes upregulated exclusively in response to iron limitation
| Locus tag | Predicted function (gene) | Fold valuea |
|||
|---|---|---|---|---|---|
| Δfur+Fe/WT+Fe | Δfur−Fe/WT−Fe | Δfur−Fe/Δfur+Fe | WT−Fe/WT+Fe | ||
| Amino acid biosynthesis | |||||
| lmo2090 | Argininosuccinate synthase (argG) | — | — | 2.43 | 7.47 |
| lmo2091 | Argininosuccinate lyase (argH) | — | −1.42 | 3.41 | 3.2 |
| lmo2252 | Aspartate aminotransferase (aspB) | −1.38 | −1.31 | 2.27 | 2.47 |
| Energy metabolism | |||||
| lmo0014 | Similar to quinol oxidase AA3, subunit I (qoxB) | — | −1.46 | 2.02 | 2.54 |
| lmo0016 | Similar to quinol oxidase AA3-600 chain IV (qoxD) | 1.37 | — | 2.77 | 2.81 |
| Protein fate | |||||
| lmo2068 | Chaperone protein, GroEL | 1.6 | — | 2.31 | 2.28 |
| Protein synthesis | |||||
| lmo0569 | Histidyl-tRNA synthetase (hisZ) | — | — | 2.07 | 2.1 |
| Transport and binding proteins | |||||
| lmo1738 | Similar to amino acid ABC transporter (binding protein) | — | −1.52 | 3.88 | 4.68 |
| lmo1739 | Similar to amino acid (glutamine) ABC transporter (ATP-binding protein) | — | −1.34 | 2.58 | 8.31 |
| lmo2251 | Similar to amino acid ABC transporter (ATP-binding protein) (GlnQ) | — | — | 2.45 | 6.01 |
Numbers in the fold value column are ratios of expression levels in the fur mutant (Δfur) and in the L. monocytogenes wild type (WT) in the presence of iron (+Fe) or in iron-limiting conditions (−Fe). —, no change or data not available.
TABLE 3.
Genes downregulated exclusively in response to iron limitation
| Locus tag | Predicted function (gene) | Fold valuea |
|||
|---|---|---|---|---|---|
| Δfur+Fe/WT+Fe | Δfur−Fe/WT−Fe | Δfur−Fe/Δfur+Fe | WT−Fe/WT+Fe | ||
| Amino acid biosynthesis | |||||
| lmo1986 | Similar to ketol-acid reductoisomerase (ilvC) | 1.45 | 1.32 | −2.38 | −4.38 |
| lmo1988 | 3-Isopropylmalate dehydrogenase (leuB) | 1.4 | — | −2.63 | −3.36 |
| lmo1989 | 3-Isopropylmalate dehydratase (large subunit) (leuC) | — | — | −2.86 | −2.57 |
| lmo1990 | 3-Isopropylmalate dehydratase (small subunit) (leuD) | 1.47 | — | −3.49 | −2.05 |
| Cellular processes | |||||
| lmo0515 | Universal stress protein, UspA-like | — | — | −2.32 | −3.53 |
| Hypothetical proteins | |||||
| lmo0584 | Similar to membrane protein YubA | −1.6 | −1.8 | −3.03 | −4.03 |
| lmo0585 | Unknown, CscB-like | — | 1.27 | −2.68 | −4.96 |
| lmo0587 | Unknown, CscC-like | −1.37 | −1.84 | −2.79 | −3.68 |
| lmo0576 | Cell wall-associated MucBP | — | — | −2.33 | −2.97 |
| lmo0850 | Transcriptional regulator | — | — | −2.45 | −4.17 |
| Regulatory functions | |||||
| lmo0733 | Putative transcriptional regulator | — | −1.28 | −2.01 | −3.36 |
| Transport and binding proteins | |||||
| lmo2254 | Xanthine/uracil permease family protein | — | — | −2.08 | −2.19 |
| Unclassified | |||||
| lmo0160 | Putative peptidoglycan-bound protein (LPXTG motif) | −1.49 | −1.68 | −2.53 | −3.86 |
| lmo0732 | Putative peptidoglycan bound protein (LPXTG motif) | −1.47 | −1.28 | −2.25 | −7 |
| lmo2522 | Cell wall-binding protein, unknown | — | −1.74 | −2.28 | −3.94 |
Numbers in the fold value column are ratios of expression levels in the fur mutant (Δfur) and in the L. monocytogenes wild type (WT) in the presence of iron (+Fe) or in iron-limiting conditions (−Fe). —, no change or data not available.
(i) Genes induced in response to iron limitation.
Iron limitation resulted in the induction of genes involved in arginine biosynthesis, argH and argG. Bioinformatic analysis of these two genes revealed that they encode arginosuccinate synthase and arginosuccinate lyase, respectively, from a single operon and are involved in a two-step process to enzymatically convert citrulline to l-arginine. In our experiments, various other genes, lmo0569 (hisZ), lmo2252 (aspartate aminotransferase), and amino acid transport genes of the ABC type, lmo1738, lmo1739, and lmo2251, were induced under low-iron conditions. lmo2251 belongs to an operon consisting of two additional structural genes, arpJ and arginine ABC transporter. Researchers reported previously that arpJ is highly induced in L. monocytogenes-infected mammalian cells and that the mutant for this gene was defective in intracellular growth (32). The induction of these two genes was also observed for a Vibrio cholerae mouse infection study (8).
In our study, the induction of two genes, lmo0014 and lmo0016, known to encode subunits for the enzyme quinol oxidase, indicates that oxygen is still a preferred electron acceptor under iron-deprived growth conditions. This condition seems to be causing damage to some proteins, as shown by the upregulation of lmo2068 (chaperone protein GroEL) in our experiments (Table 2). In bacteria, GroEL is an important heat shock chaperone that works together with GroES to help cells cope with damaged or improperly folded proteins. In L. monocytogenes, these genes are present in a single operon and are known to be induced in response to various environmental stress stimuli (36).
(ii) Genes repressed in response to iron limitation.
In the amino acid biosynthesis group, the most downregulated genes were found to be involved in leucine (lmo1988 to lmo1990) and isoleucine (lmo1986) biosynthesis. It appears that in the absence of iron, bacteria try to carefully regulate branched-amino-acid biosynthesis to ensure that iron is properly utilized since many enzymes involved in amino acid biosynthesis are iron dependent: the adaptive “iron-sparing” response (24). We also found that the lmo0515, lmo0576, lmo0585, lmo0587, and lmo0850 genes, encoding hypothetical proteins, were downregulated. Bioinformatic analysis of the corresponding representative proteins, Lmo0850 and Lmo0515, showed that they are similar to a transcriptional regulator and universal stress protein (UspA), respectively. In E. coli, UspA is known to protect the bacteria from different stress conditions generated during growth, such as hydrogen peroxide stress. Hence, the repression of this gene in our experiments may suggest lower oxidative stress generated under low-iron growth conditions (43). Other genes encoding several putative cell wall- or membrane protein-related genes, such as lmo0160, lmo0576, lmo0584, lmo0732, and lmo2522, were downregulated in the absence of iron. Protein sequence analysis for Lmo0160 and Lmo0732 showed that they have LPXTG wall-anchoring motifs and could serve as substrates for sortase A (SrtA), whose function is to recognize the motif and display the protein on the bacterial cell surface (3). Analysis of the other sequences showed that Lmo0584 is similar to YubA, a membrane protein of unknown function, whereas Lmo0576 is similar to mucin binding protein (MucBP). In Lactobacillus reuteri 1063, mucBP was shown to be involved in adhesion to mucin (49). Downstream of the lmo0584 gene, there is a Csc-like cluster present, comprised of the genes lmo585 to lmo587. Among the proteins encoded in this cluster, we found that Lmo0585 (CscB-like) and Lmo0587 (CscC-like) have a WxL domain on their C termini, which may help in associating with the bacterial surface (4). The function of Lmo2522 is not completely known; experiments have shown that this protein has two LysM domains and is released into the supernatant of a bacterial culture (55). We found that lmo2254 (xanthine/uracil transport system permease protein) and lmo0733, a transcriptional factor, were downregulated (Table 3). However, we do not understand the significance of the regulation of these two genes.
Genes regulated by Fur.
To identify the genes under the direct control of Fur, the gene expression profiles from experiments a, b, and d were compared. In this approach, the genes that were observed to be simultaneously regulated either as a consequence of fur mutation (experiments a and b) or as a response to low-iron conditions in the medium (experiment d) can be considered to be regulated by Fur. The difference in the responses of the Fur-regulated genes in experiments a and b is due to the different regulation of fur in the parent; fur is significantly repressed in the absence of iron. Our analysis showed several genes to be negatively regulated by Fur, and only one gene, lmo0514 (contains the LPXTG sorting signal) appeared to be positively regulated in the presence of iron. However, we did not experimentally determine whether lmo0514 is directly or indirectly regulated by Fur. Also, we did not find any consensus Fur box sequence in the upstream putative promoter region of this gene.
Genes negatively regulated by Fur under iron limitation conditions.
Analysis of the iron limitation response showed that several genes under FRC were upregulated (Table 4). Northern blot analysis showed that the regulation of fur, which is also under Fur regulation, was significantly repressed. Based on these preliminary results, the response of the FRC genes to iron limitation seems to mimic the response observed when fur is mutated in this bacterium.
TABLE 4.
Genes regulated by Fur under iron limitation conditions
| Locus tag | Predicted function (gene) | Fold valuea |
|||
|---|---|---|---|---|---|
| Δfur+Fe/WT+Fe | Δfur−Fe/WT−Fe | Δfur−Fe/Δfur+Fe | WT−Fe/WT+Fe | ||
| Cell envelope | |||||
| lmo0366 | Similar to putative lipoprotein involved in iron transport | 27.61 | 2.1 | — | 5.47 |
| Energy metabolism | |||||
| lmo0485 | Nitroreductase family protein | 3.83 | 1.65 | 1.61 | 2.17 |
| lmo0943 | Nonheme iron-binding ferritin (fri) | 2.4 | 1.89 | 1.18 | 2.35 |
| lmo1131 | Unknown, similar to ABC transporter, ATP-binding protein | 9.93 | 1.39 | — | 5.97 |
| Hypothetical proteins | |||||
| lmo0365 | Similar to high-affinity iron transporter | 25.03 | 2.43 | — | 5.58 |
| lmo1007 | Unknown | 5.61 | — | — | 2.48 |
| lmo2132 | Unknown | 2.04 | 1.4 | 1.26 | 2 |
| lmo2181 | Sortase B (srtB) | 12.83 | 1.34 | −1.21 | 4.6 |
| lmo2104 | Ferrous iron transport protein (A) | 15.17 | — | 1.22 | 7 |
| lmo2105 | Ferrous iron transport protein (B) | 7.76 | 1.27 | — | 4.34 |
| Regulatory functions | |||||
| lmo1956 | Ferric uptake regulator | −12.75 | −3.25 | — | 1.44b |
| Transport and binding proteins | |||||
| lmo0541 | Similar to iron compound binding lipoprotein, ABC Transporter | 7.01 | — | — | 2.62 |
| lmo1957 | Ferrichrome ABC transporter (permease) | 5.86 | 1.79 | −1.27 | 2.22 |
| lmo1958 | Iron compound ABC transporter, permease protein (fhuB) | 4.7 | 1.44 | — | 2.01 |
| lmo1960 | Iron compound ABC transporter (ATP binding) (fhuC) | 2.44 | — | — | 2.02 |
| lmo2182 | Ferrichrome ABC transporter (ATP binding) | 13.3 | 1.34 | — | 3.97 |
| lmo2183 | Ferrichrome ABC Transporter (permease) | 15.76 | 1.41 | — | 4.48 |
| Unclassified | |||||
| lmo0361 | Twin-arginine-targeting protein translocase (tatC) | 22.75 | 2.85 | −1.17 | 3.61 |
| lmo0362 | Twin-arginine-targeting protein translocase (tatA) | 19.75 | 3.21 | −1.16 | 4.29 |
| lmo2180 | Unknown, similar to GP157 family of protein | 4.09 | — | — | 4.71 |
| lmo2185 | (svpA) | 8.91 | 1.58 | — | 2.99 |
| lmo2186 | Unknown | 9.48 | — | — | 4.24 |
| lmo0367 | Putative iron-dependent peroxidase | 25.83 | 1.57 | — | 4.07 |
| Unknown function | |||||
| lmo1961 | Similar to oxidoreductase | 3.58 | — | — | 2.4 |
Numbers in the fold value column are ratios of expression levels in the fur mutant (Δfur) and in the L. monocytogenes wild type (WT) in the presence of iron (+Fe) or in iron-limiting conditions (−Fe). —, no change or data not available.
Downregulated.
Comparative analysis of the FRC genes in the fur mutant showed that their response (fold value) was much higher in medium containing iron (Δfur+Fe/WT+Fe) than when this iron was limited (Δfur−Fe/WT−Fe). For example, the fold value for the expression of fur changed from −12.75-fold (Δfur+Fe/WT+Fe) to −3.25-fold (Δfur−Fe/WT−Fe), and that of lmo2185 changed from 8.91-fold (Δfur+Fe/WT+Fe) to 1.58-fold. Based on these results, we hypothesized that (i) the presence of an optimal concentration of iron is required for fur to be maximally induced, and any significant limitation of iron would result in the repression of fur, and (ii) it is the repression of fur rather than the availability of iron that is important for the regulation of genes under Fur repression control (FRC).
Some of the FRC genes identified by microarray analysis were previously studied for L. monocytogenes EGDe (18, 22, 28). It was previously shown that that deletion of fhuD, fhuC, or hupC reduced the uptake of ferric hydroxamate and hemin/hemoglobin, respectively (28). Moreover, the authors of that study also observed that iron transport was impaired when fri was mutated, but the deletion of fur, feoB, lmo2183, lmo2184, or lmo2185 did not result in the same impairment (28). Analysis of FhuD from L. monocytogenes showed that it is similar to FhuD from E. coli and FhuD2 from Staphylococcus aureus, but unlike some S. aureus strains, no gene duplication was observed. Studies of other organisms have shown that Fur regulates fhuA in E. coli and fhuD in Bacillus subtilis (6, 25). As expected, other iron uptake genes, feoAB (lmo2104 and lmo2105), fhuGB operon genes (lmo1957 and lmo1958), fhuD (lmo1959), fhuC (lmo1960), lmo2183, and lmo2431, were found to be significantly upregulated in response to iron limitation.
Studies have shown that in bacteria, the Feo transport system, Fe2+ type, is used for the uptake of the Fe2+ form of iron and is under the negative control of Fur (25). In E. coli, the proteins involved in this system are encoded from the feoABC operon. Among the Gram-positive Bacillus species, the feoAB operon is present in Bacillus cereus but not in Bacillus subtilis. In L. monocytogenes, both the feoA and feoB genes are present.
In our study, we have found that the tatC (lmo0361), tatA (lmo0362), lmo0365 to lmo0367, lmo0485, lmo0541, and lmo1007 genes are under the regulation of Fur. Extensive studies of the Tat system of E. coli showed that this machinery translocates proteins that have twin-arginine motifs on their N termini and also posses hydrophobic H domains. In E. coli, this machinery is made of several Tat proteins, encoded by tatABCD, but in L. monocytogenes, only tatA and tatC are present. It seems that one of the substrates translocated by this system is an iron-dependent peroxidase, encoded by lmo0367 (15), and we found a significant induction of this gene in our experiments. Nucleotide sequence analysis of this gene showed that it is part of the operon consisting of lmo0365 to lmo0367. Analysis of the protein products encoded by this operon and Lmo0541 indicated that they are all probably involved in iron transport. We found that Lmo0365 was 40% identical to YwbL, Lmo0366 was 43% identical to YwbM, and Lmo0367 was 51% identical to YwbN of B. subtilis. It is known that all these proteins in B. subtilis are involved in iron transport and are regulated by Fur in response to iron.
In our microarray analysis, we also identified lmo0485, encoding a nitroreductase family protein, to be upregulated. Protein sequence alignment showed that it is 28% identical to B. subtilis YfhC. Experiments done with B. subtilis showed that yfhC is regulated by peroxide stress and iron limitation (2, 38). Furthermore, analysis of Lmo0541 showed that it is about 30% identical to FhuD in B. subtilis, and when Lmo1131 was analyzed, it was found to be 26% identical to B. subtilis CydC (44). However, more experiments are needed to fully understand and confirm their function in L. monocytogenes.
Identification of Fur boxes.
For the identification of Fur boxes, all the intergenic regions within the whole-genome sequence of L. monocytogenes EGDe were analyzed using a custom position weight matrix generated from Fur box sequences. Fur boxes in the upstream putative promoter sequences were identified in 14 genes, and the locations of these consensus sequences in the genome are listed in Table S2 in the supplemental material.
While several genes were observed to be regulated by fur mutation or due to iron limitation, we found Fur boxes in only a few of them, such as the ones involved in iron acquisition and virulence. We performed an electrophoretic mobility shift assay (EMSA) for these putative Fur box sequence-containing promoter regions and confirmed that the purified Fur protein can efficiently shift the mobility of Fur box-containing DNA upon binding (see Fig. S2 in the supplemental material). Comparison of all the identified Fur box sequences showed that they have a small number of nucleotide differences from one another; such differences were also observed previously for B. subtilis (2). In our present study, we have identified Fur boxes in 7 new genes. Among the 14 Fur-regulated genes, fur was downregulated only in KRM medium or when DPD was added. We hypothesized that this mode of regulation may be a way to effectively reduce Fur protein synthesis to relax the genes under its direct control. This mechanism seems plausible, as there are only a few genes under the direct control of Fur. Moreover, no genes except lmo0514, which would require the presence of this protein for their expression, were identified. To understand whether iron availability or fur regulation is important for controlling FRC genes, we performed experiments to observe the regulation of fur in the presence of excess and limited concentrations of iron.
Regulation of fur in L. monocytogenes in response to iron limitation.
In most bacteria, fur is regulated positively or negatively in response to iron in an autoregulatory manner. In autoregulatory control, fur has been shown to be iron dependent where Fur complexes with metals and binds to a putative Fur box sequence present in its promoter region. In addition to this mode of regulation, fur is also understood to be regulated by other proteins such as PerR. In L. monocytogenes, fur has a Fur box and PerR box in the upstream promoter region. We have previously shown that the purified Fur protein can bind a putative Fur box in the absence of iron and proposed an autoregulatory mode of regulation for this gene (33). In our microarray analysis experiment, fur was observed to be downregulated in response to iron limitation, and in our previous report, we showed that the addition of excess iron also results in the repression of fur. PerR was previously shown to be a negative regulator of fur in L. monocytogenes (47). From all these studies, fur appears to be under the dual control of the Fur and PerR proteins, and knowledge of the regulatory mechanism is important for an understanding of its global regulatory control.
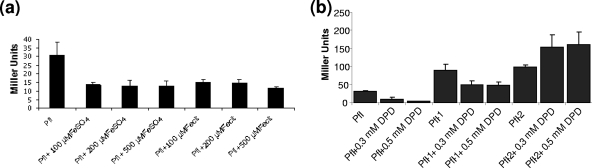
In order to understand the regulation of fur, we constructed a fur::lacZ promoter fusion in the ectopic chromosomal region of the parent and generated its derivatives with mutations in fur and perR regions. Analysis of β-galactosidase expression showed that the addition of either iron or DPD in increasing concentrations for the parent growing in BHI medium resulted in a decrease of β-galactosidase activity (Fig. 1a and b). To understand the mechanism behind the repression of fur under iron-limiting conditions, DPD was added to fur or perR mutant derivatives. In this assay, we found high levels of β-galactosidase activity for fur or perR mutants growing in BHI medium (Fig. 1b). The addition of DPD at 0.3 mM or 0.5 mM concentrations resulted in a significant repression of the activity in the fur mutant and its parent fusion strain, but no significant change in the activity was observed for the fusion strain with the perR mutation. For these studies, we could not include complementation studies for either the fur or perR mutant. Several attempts to complement the fur mutant or make a perR and fur double mutant were unsuccessful, and we believe that it was due partly to the significant downregulation of comK in the fur mutant, as observed for microarray analyses (experiment a). We encountered a different problem in the case of complementing the perR mutant, where we were unable to transform pCU1 containing the perR structural gene. We concluded that it was not a transformation problem since we were able to successfully transform plasmid pCU1 alone into the perR mutant.
FIG. 1.
(a) Effect of iron concentration on β-galactosidase expression of L. monocytogenes fur::lacZ fusion strain Pfl (10403S). Error bars represent standard deviations for triplicate experiments. (b) Effect of iron limitation caused by 2,2′-dipyridyl on β-galactosidase expression of Pfl. Shown are data for the effect of DPD on the β-galactosidase expression of L. monocytogenes fur::lacZ fusion strains, Pfl (10403S), Pfl-1(fur mutant), and Pfl-2 (perR mutant). Errors bar represent standard deviations for triplicate experiments.
However, we have taken several steps as a quality control measure to asses the stability of plasmid pAUL-A integration. We checked the mutants by growing them in medium with or without antibiotics for 24 h and 48 h. Cultures from these two time points were then serially diluted and plated onto appropriate antibiotic-containing BHI agar medium. Simultaneously, genomic DNA was also isolated from several colonies growing on agar medium and checked by Southern blotting. For both of these analyses, we found that the plasmid integration was stable. For β-galactosidase assays or Northern blot analysis (pAUL-A insertion mutants), we grew the bacteria overnight in the presence of antibiotics, but antibiotics were eliminated from the diluted cultures. We did not make any attempt to understand the response of our strains or the regulation of the FRC genes when iron was limited at the stationary phase of growth. Although an understanding of the regulation in this phase is important, we believe that this study should be carried out separately because the stationary phase would add significant metabolic and other nutrient limitation stresses to the already subsisting iron limitation stress.
Our results suggest that the regulation of fur is autoregulatory and negatively regulated by PerR. When iron is limited, fur is repressed, and this is mediated through the change in activity of PerR. Previously, PerR from B. subtilis was biochemically characterized, and it was proposed that this protein associates with Fe(II) as a cofactor that is sensitive to peroxide (27). When intracellular iron levels are low, PerR binds Mn(II) and represses several genes under its regulon, which include fur (27, 34, 54). Based on our data and data from previous studies of PerR, we hypothesize that in the absence of iron, PerR changes from a less active (complexed to Fe) to a more active (uses Mn as cofactor) form, which can cause the repression of fur, resulting in the upregulation of the Fur repressor control genes. However, it is not known if PerR mediates any factors that may target fur mRNA for degradation or reduce the repression caused by Fur.
Northern analysis.
To confirm microarray results, expression levels of four genes, lmo0514, fur, feoB, and lmo2185, were analyzed with the fur deletion mutant and its isogenic parent grown in minimal medium (KRM medium) with or without added iron (see Fig. S3 and S4 in the supplemental material). Under low-iron conditions (KRM medium) or because fur was deleted, both fur and lmo0514 were found to be repressed, while feoB and lmo2185 were induced. It should be noted that the absence of fur mRNA in the mutant is due to the complete deletion of the structural gene.
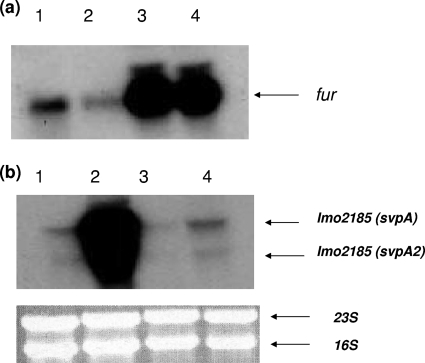
In order to confirm that the repression of fur in KRM medium was due to iron limitation and that this regulation requires PerR, total RNA isolated from the cultures grown in BHI medium with DPD was analyzed by Northern blotting (see Fig. S5 in the supplemental material). These samples were also analyzed to confirm the induction of lmo2185 in the presence of DPD. We probed for lmo2185 transcripts for control purposes in these experiments, as the regulation of this gene is a good indication of iron limitation in the environment (41). In the case of the perR mutant, fur expression was upregulated. However, when the same mutant was grown in the presence of DPD, no significant change in lmo2185 expression was observed (Fig. 2).
FIG. 2.
Northern blot analysis to determine the expression of fur and lmo2185 from L. monocytogenes in response to 2,2′-dipyridyl. (a) Northern blot analysis for fur in the perR mutant in response to iron limitation by DPD. Ten micrograms of total RNA was isolated from parent cells grown in BHI medium (lane 1) or 0.3 mM DPD (lane 2) or the perR mutant grown in BHI medium (lane 3) or 0.3 mM DPD (lane 4). (b) Northern blot analysis of lmo2185 expression. The amounts of RNA and lanes are similar to those described above (a).
qRT-PCR analysis.
To demonstrate that fur is regulated in the presence of excess iron and under conditions of iron limitation, we performed qRT-PCR using fur (lmo1956), lmo0485, lmo1960, and lmo2185 in wild-type L. monocytogenes EGDe (see Tables S3 and S4 in the supplemental material). The data generated by the qRT-PCR analysis supported data from the Northern blot analysis. The presence of excess iron, in the form of ferric citrate, showed a direct proportional downregulation of the fur transcript. However, the level of repression was less than that observed when iron was limited. Among other genes, lmo0485 showed an upregulation as in the microarray analysis. lmo0485 is a member of the nitroreductase family and is similar to yfhC of B. subtilis. In B. subtilis, yfhC is induced upon exposure to hydrogen peroxide and paraquat (38). The iron compound ABC transporter gene fhuC (lmo1960) is also upregulated initially in the presence of iron, but increasing concentrations of iron resulted in a decreased level of expression. lmo1960 is responsible for the uptake of iron, but excess iron is detrimental to the cell, hence the reduced levels of expression. In this analysis, the downregulation of fur and lmo2185 could also be a result of the sensing of excess iron by some unknown factor. Work is under way in our laboratory to understand the mechanism of regulation of fur and FRC genes under excess-iron conditions.
qRT-PCR data obtained from the experiments under iron-depleted conditions, i.e., in the presence of DPD, also support the microarray and Northern blot results. A downregulation of fur and lmo1956 in the absence of iron was observed. However, the level of repression was reduced in the presence of 0.1 mM FeSO4, thus demonstrating that the DPD added was effectively chelating iron in our experiments. On the other hand, lmo0485, lmo1960, and lmo2185 showed markedly decreased levels of expression in the presence of 0.1 mM FeSO4 compared to those under iron-depleted conditions.
The results observed from fur::lacZ fusions, Northern blot analysis, and qRT-PCR clearly indicated that fur is repressed when iron availability is limited. Several studies demonstrated that the regulation of genes under the control of Fur changes as a consequence, resulting from the deliberate deletion of fur, repression of fur, degradation of fur mRNA, or intracellular inactivation of Fur repression activity. Although any of the above-mentioned processes could be operating in Listeria, our studies support the hypothesis that the repression of fur is the probable way of regulating FRC genes when iron is limited. In this bacterium, fur is under the control of its own protein and PerR. Based on all our studies, we believe that fur repression is essential for regulating FRC genes because a characterization of the Fur protein in Listeria indicated that this protein can actively bind in the absence of metal ions (33). fur is repressed under iron limitation conditions, but repression was significantly inhibited when perR was mutated. Using microarray analysis, we have also observed that the addition of DPD to the perR mutant does not result in significant changes in fur or FRC genes (data not shown). We hypothesize that iron limitation and the regulation of iron acquisition genes depend on the activity of PerR but not on the availability of iron as a cofactor for the Fur protein. Fur complexing with iron is a generally observed mechanism of regulation seen for many bacteria. In B. subtilis, PerR functions as a transcriptional repressor of fur, but no role in the degradation of fur mRNA or inactivation of Fur has been demonstrated. However, we do not have sufficient evidence to rule out the involvement of PerR-mediated molecules in the regulation of Fur or iron acquisition genes. Based on our current understanding of fur and FRC gene expression in L. monocytogenes, we have proposed a model for regulation, as shown in Fig. 3.
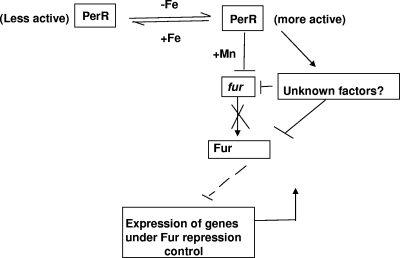
FIG. 3.
Proposed model of PerR-mediated iron acquisition in L. monocytogenes. PerR is constitutively expressed, and it is in a less active state (associated with Fe). When iron is limited, PerR binds to manganese (more active state; complexed with Mn) and represses fur, which consequently results in a reduction of Fur expression and upregulation of FRC genes. Broken lines indicate a change in control over FRC genes by Fur due to a decrease in expression. Alternatively, PerR in the active form may be interacting with unknown factors to cause the same effect on fur and FRC gene regulation.
Supplementary Material
Acknowledgments
We thank Colin Hill for providing the fur deletion mutant of L. monocytogenes EGDe and Abraham L. Sonenshein for generously providing plasmid pHK77. The L. monocytogenes microarrays were obtained through the NIAID Pathogen Functional Genomics Resource Center, managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by The Institute for Genomic Research (TIGR). We appreciate Anthony J. Otsuka for critical reading of the manuscript.
This work was partially supported by the Illinois State University Research Program, a grant from the NIH to R.K.J., and award 2006-35201-17356 from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 20 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 3.Bierne, H., and P. Cossart. 2007. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol. Mol. Biol. Rev. 71:377-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinster, S., S. Furlan, and P. Serror. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 189:1244-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 6.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 7.Camejo, A., C. Buchrieser, E. Couve, F. Carvalho, O. Reis, P. Ferreira, S. Sousa, P. Cossart, and D. Cabanes. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty, T. 1999. Molecular and cell biological aspects of infection by Listeria monocytogenes. Immunobiology 201:155-163. [DOI] [PubMed] [Google Scholar]
- 11.Conte, M. P., C. Longhi, M. Polidoro, G. Petrone, V. Buonfiglio, S. Di Santo, E. Papi, L. Seganti, P. Visca, and P. Valenti. 1996. Iron availability affects entry of Listeria monocytogenes into the enterocyte like cell line Caco-2. Infect. Immun. 64:3925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 13.Cossart, P. 2002. Molecular and cellular basis of the infection by Listeria monocytogenes: an overview. Int. J. Med. Microbiol. 291:401-409. [DOI] [PubMed] [Google Scholar]
- 14.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 15.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dussurget, O., E. Dumas, C. Archambaud, I. Chafsey, C. Chambon, M. Hebraud, and P. Cossart. 2005. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol. Lett. 250:253-261. [DOI] [PubMed] [Google Scholar]
- 19.Edgar, R., and T. Barrett. 2006. NCBI GEO standards and services for microarray data. Nat. Biotechnol. 24:1471-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst, F. D., S. Bereswill, B. Waidner, J. Stoof, U. Mader, J. G. Kusters, E. J. Kuipers, M. Kist, A. H. van Vliet, and G. Homuth. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151:533-546. [DOI] [PubMed] [Google Scholar]
- 21.Escolar, L., J. Perez-Martin, and. V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiorini, F., S. Stefanini, P. Valenti, E. Chiancone, and D. De Biase. 2008. Transcription of the Listeria monocytogenes fri gene is growth-phase dependent and is repressed directly by Fur, the ferric uptake regulator. Gene 410:113-121. [DOI] [PubMed] [Google Scholar]
- 23.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaballa, A., H. Antelmann, C. Aguilar, S. K. Khakh, K. B. Song, G. T. Smaldone, and J. D. Helmann. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U. S. A. 105:11927-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 26.Heinrichs, D. V., A. Rahn, E. Dale, and M. T. Sebulsky. 2004. Staphylococcus, Streptococcus, and Bacillus, p. 387-401. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 27.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849-859. [DOI] [PubMed] [Google Scholar]
- 28.Jin, B., S. M. Newton, Y. Shao, X. Jiang, A. Charbit, and P. E. Klebba. 2006. Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol. Microbiol. 59:1185-1198. [DOI] [PubMed] [Google Scholar]
- 29.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 30.Kenny, J. G., D. Ward, E. Josefsson, I. M. Jonsson, J. Hinds, H. H. Rees, J. A. Lindsay, A. Tarkowski, and M. J. Horsburgh. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, H. J., M. Mittal, and A. L. Sonenshein. 2006. CcpC-dependent regulation of citB and lmo0847 in Listeria monocytogenes. J. Bacteriol. 188:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klarsfeld, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13:585-597. [DOI] [PubMed] [Google Scholar]
- 33.Ledala, N., S. L. Pearson, B. J. Wilkinson, and R. K. Jayaswal. 2007. Molecular characterization of the Fur protein of Listeria monocytogenes. Microbiology 153:1103-1111. [DOI] [PubMed] [Google Scholar]
- 34.Lee, J. W., and J. D. Helmann. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485-499. [DOI] [PubMed] [Google Scholar]
- 35.Lin, J. T., M. B. Connelly, C. Amolo, S. Otani, and D. S. Yaver. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob. Agents Chemother. 49:1915-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 39.Munch, R., K. Hiller, A. Grote, M. Scheer, J. Klein, M. Schobert, and D. Jahn. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187-4189. [DOI] [PubMed] [Google Scholar]
- 40.Muthaiyan, A., J. A. Silverman, R. K. Jayaswal, and B. J. Wilkinson. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newton, S. M., P. E. Klebba, C. Raynaud, Y. Shao, X. Jiang, I. Dubail, C. Archer, C. Frehel, and A. Charbit. 2005. The svpA-srtB locus of Listeria monocytogenes: Fur-mediated iron regulation and effect on virulence. Mol. Microbiol. 55:927-940. [DOI] [PubMed] [Google Scholar]
- 42.Norris, A. W., and C. R. Kahn. 2006. Analysis of gene expression in pathophysiological states: balancing false discovery and false negative rates. Proc. Natl. Acad. Sci. U. S. A. 103:649-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nystrom, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11:537-544. [DOI] [PubMed] [Google Scholar]
- 44.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Portnoy, D. A., V. Auerbuch, and I. J. Glomski. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rea, R., C. Hill, and C. G. Gahan. 2005. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71:8314-8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rea, R. B., C. G. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 50.Roxas, B. A., and Q. Li. 2008. Significance analysis of microarray for relative quantitation of LC/MS data in proteomics. BMC Bioinformatics 9:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schafer, E. R., M. J. Oneal, M. L. Madsen, and F. C. Minion. 2007. Global transcriptional analysis of Mycoplasma hyopneumoniae following exposure to hydrogen peroxide. Microbiology 153:3785-3790. [DOI] [PubMed] [Google Scholar]
- 52.Sword, C. P. 1966. Mechanisms of pathogenesis in Listeria monocytogenes infection. I. Influence of iron. J. Bacteriol. 92:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reference deleted.
- 54.Traore, D. A., A. El Ghazouani, L. Jacquamet, F. Borel, J. L. Ferrer, D. Lascoux, J. L. Ravanat, M. Jaquinod, G. Blondin, C. Caux-Thang, V. Duarte, and J. M. Latour. 2009. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 5:53-59. [DOI] [PubMed] [Google Scholar]
- 55.Trost, M., D. Wehmhoner, U. Karst, G. Dieterich, J. Wehland, and L. Jansch. 2005. Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5:1544-1557. [DOI] [PubMed] [Google Scholar]
- 56.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.