Abstract
The wide variety of RNAs produced in the nucleus must be localized correctly to perform their functions. However, the mechanism of this localization is poorly understood. We report here the differential subnuclear localization of RNA strands of opposite polarity derived from the replicating Potato spindle tuber viroid (PSTVd). During replication, (+)- and (−)-strand viroid RNAs are produced. We found that in infected cultured cells and plants, the (−)-strand RNA was localized in the nucleoplasm, whereas the (+)-strand RNA was localized in the nucleolus as well as in the nucleoplasm with distinct spatial patterns. Furthermore, the presence of the (+)-PSTVd in the nucleolus caused the redistribution of a small nucleolar RNA. Our results support a model in which (1) the synthesis of the (−)- and (+)-strands of PSTVd RNAs occurs in the nucleoplasm, (2) the (−)-strand RNA is anchored in the nucleoplasm, and (3) the (+)-strand RNA is transported selectively into the nucleolus. Our results imply that the eukaryotic cell has a machinery that recognizes and localizes the opposite strands of an RNA, which may have broad ramifications in the RNA regulation of gene expression and the infection cycle of pathogenic RNAs and in the development of RNA-based methods to control gene expression as well as pathogen infection.
INTRODUCTION
The eukaryotic cell has evolved highly elaborate subcellular organelles/structures that specialize in distinct biochemical/biophysical processes. The biogenesis of organelles/structures, the progression of localized biochemical reactions, and the cellular integration of these processes involve extensive subcellular traffic and localization of a wide variety of molecules. Within an organelle, the spatially clustered biochemical reactions also require the trafficking and precise localization of all of the molecules involved. Thus, organized trafficking and localization of molecules are hallmarks of a living cell. Elucidating the underlying pathways and regulatory mechanisms of these processes has vast importance to understanding the basic principles of life.
Once considered merely the bridge for genetic information flow from DNA to protein, RNA is rapidly emerging as a key player in the regulation of a wide variety of cellular processes (Grosshans and Slack, 2002; Storz, 2002; Andersen and Panning, 2003; Greenspan, 2003; Lai, 2003; Masse et al., 2003). One mechanism involves complementary base-pairing of an antisense RNA, such as a micro-RNA, with its target RNA to regulate gene expression key to development (Reinhart et al., 2000; Llave et al., 2002; Kawasaki and Taira, 2003; Masse et al., 2003). The abnormal production and function of an antisense RNA has been reported as a novel mechanism of genetic diseases (Tufarelli et al., 2003). Various functions require precise subcellular localizations of the RNAs, the mechanism of which is poorly understood. In particular, whether cellular machinery exists to deliberately regulate the localization of RNAs of opposite polarity is virtually unknown. Elucidating the mechanisms of subcellular RNA localization has significance beyond understanding the RNA regulation of growth and development as well as understanding the basic principles of traffic regulation within a cell. The knowledge gained from studies of this nature is essential for the development of RNA-based gene technologies such as gene therapy, the success of which depends ultimately on the delivery of a designed RNA to the subcellular compartment where the target is localized (Samarsky et al., 1999).
Pathogens that partially or wholly depend on cellular factors to replicate themselves must be localized to the proper subcellular compartments to access these factors. Therefore, studying the infection patterns of pathogens can contribute greatly to our understanding of the basic mechanisms of traffic. The replication of viroids in the Pospiviroidae family offers a simple model system to investigate the mechanisms of the subcellular localization of RNAs. Viroids are self-replicating RNAs that infect plants. They can alter the expression of selective plant genes important to growth and development (Itaya et al., 2002; Qi and Ding, 2003). A viroid genome consists of a single-stranded, covalently closed circular, noncoding, and nonencapsidated RNA, with genome sizes ranging from 275 to 400 nucleotides (Hull, 2002). Depending on viroid-host combinations, these small RNAs may be highly pathogenic (Schnölzer et al., 1985; Owens et al., 1991; De la Peña et al., 1999; S̆korić et al., 2001; Qi and Ding, 2003). Viroids in the Pospiviroidae replicate in the nucleus, and viroids in the Avsunviroidae replicate in the chloroplasts (Flores et al., 2000). Without encoding proteins, viroid RNAs must interact directly with cellular factors to achieve the correct subcellular localization to accomplish replication functions.
Replication of Potato spindle tuber viroid (PSTVd), the type species of the Pospiviroidae, follows an asymmetric rolling-circle pathway (Owens and Diener, 1982; Branch and Robertson, 1984). The circular (+)-strand RNA is used as the initial template to synthesize multimeric and linear (−)-strand RNA. The latter then is used as a template to synthesize numerous copies of the multimeric linear (+)-strand RNA. These RNAs then are processed into unit-length monomers and circularized.
Inhibition of PSTVd replication by α-amanitin, but not by actinomycin D, suggests that the transcription of both (+)- and (−)-strands of PSTVd involves the nucleoplasm-localized DNA-dependent RNA polymerase II (Mühlbach and Sanger, 1979; Schindler and Mühlbach, 1992; Fels et al., 2001). However, in situ hybridization localized both the (+)- and (−)-strands of PSTVd in the nucleoli of nuclei isolated from infected tomato (Harders et al., 1989). Such localization patterns raise the question of how the nucleoplasm-localized RNA polymerase II could transcribe the nucleolus-localized (−)- and (+)-strands of PSTVd into each other. It is possible that the (−)- and (+)-strand PSTVd RNAs traffic from the nucleoplasm to the nucleolus (Harders et al., 1989), but there was no direct evidence to support this notion. The subcellular localization of two other members of the Pospiviroidae also has been investigated. In tissue sections of infected plants, in situ hybridization revealed that Citrus exocortis viroid (CEVd) was localized throughout the nucleus, whereas Coconut cadang cadang viroid was distributed mostly in the nucleolus and to a lesser extent in the nucleoplasm (Bonfiglioli et al., 1996). Both the (+)- and (−)-RNAs of these viroids exhibited similar distribution patterns, although the hybridization signal for the (−)-RNA was much weaker (Bonfiglioli et al., 1996). These data invoked speculation about whether the nucleolus-localized DNA-dependent RNA polymerase I was involved in some aspects of replication in addition to the possible nucleolar import of the (−)- and (+)-strand RNAs (Bonfiglioli et al., 1996). How the reported subnuclear localization patterns of the (−)- and (+)-strands of viroids would reconcile with the subnuclear localization of enzymes known to be involved in replication remains to be determined.
Using improved sample preparation and in situ hybridization protocols, we have studied the subnuclear localization of PSTVd in infected cultured cells of Nicotiana benthamiana and in infected tomato and N. benthamiana plants. We found that the (−)-PSTVd was localized in the nucleoplasm but not in the nucleolus. By contrast, the (+)-PSTVd was localized in the nucleolus as well as in the nucleoplasm, with various distinct spatial patterns. These findings resolved a major conundrum in the cell biology of viroid replication and support a model for PSTVd replication that includes intranuclear trafficking of the (+)-strand but not the (−)-strand RNA. These findings also suggest strongly that cellular machinery exists to control the trafficking and localization of RNAs of opposite polarity, which may have important implications in gene regulation and pathogen infection.
RESULTS
Localization of the (+)- and (−)-Strands of PSTVd in Infected Cultured Cells of N. benthamiana
PSTVd (intermediate strain) (Schnölzer et al., 1985) replicates in cultured cells (Qi and Ding, 2002) as well as in whole N. benthamiana plants (Hu et al., 1997; Zhu et al., 2001). We used electroporation (Qi and Ding, 2002) to deliver in vitro transcripts of PSTVd into protoplasts prepared from the cultured cells of this plant. At 2 days after inoculation, protoplasts were processed for fluorescence in situ hybridization (FISH) using Alexa Fluor 488–labeled riboprobes specific for the (+)- and (−)-strands of PSTVd. These riboprobes did not cross-hybridize between strands under our experimental conditions (Qi and Ding, 2002). In each experiment, the identity of the nucleolus was established by four means: (1) differential interference contrast microscopy that revealed the optically dense nucleolus within a nucleus (Figures 1A, 1F, and 1K, arrows); (2) 4′,6-diamidino-2-phenylindole (DAPI) staining specifically of the nucleoplasm but not the nucleolus (Beven et al., 1996) (Figures 1B, 1G, and 1L); (3) nucleolar localization of the U3 small nucleolar RNA (snoRNA) (Beven et al., 1996); and (4) nucleolar localization of the U14 snoRNA (Shaw et al., 1998). Both U3 and U14 were detected by hybridization with biotin-labeled riboprobes that were further detected by Alexa Fluor 594–conjugated streptavidin (red fluorescence).
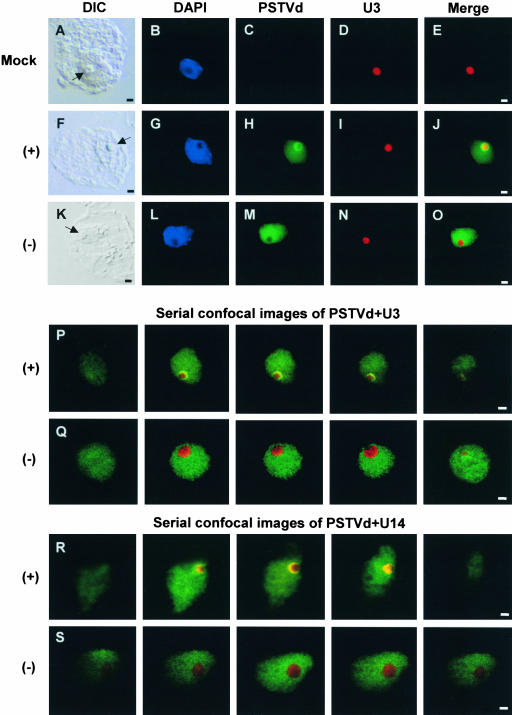
Figure 1.
Differential Subnuclear Localization of the (+)- and (−)-Strands of PSTVd in Infected N. benthamiana Cells.
The (+)- and (−)-strands of PSTVd were detected by Alexa Fluor 488–labeled riboprobes (green fluorescence). The U3 and U14 snoRNAs were detected by biotin-labeled antisense riboprobes that were further detected by streptavidin conjugated to Alexa Flour 594 (red fluorescence). The differential interference contrast (DIC) images ([A], [F], and [K]) show the nucleoli (arrows) in the cells. DAPI stains chromatin in the nucleoplasm (blue fluorescence in [B], [G], and [L]). Unstained nucleoli appear as dark spots in the nuclei. Bars = 2 μm.
(A) to (E) Mock-inoculated cell. No PSTVd hybridization signals were detected.
(F) to (J) Nucleolar as well as nucleoplasmic localization of the (+)-strand of PSTVd. The yellowish ring in the merged image ([J]) indicates overlap between (+)-PSTVd and U3 in the nucleolus.
(K) to (O) Nucleoplasmic localization of the (−)-strand of PSTVd. The merged image ([O]) shows no overlap between (−)-PSTVd and U3 snoRNA localization signals (see text for details).
(P) Serial confocal images showing localization of the (+)-PSTVd RNA in the nucleolus as well as in the nucleoplasm. The viroid signal (green) overlaps with U3 snoRNA signal (red) to produce a yellowish ring within the nucleolus.
(Q) Serial confocal images showing localization of the (−)-PSTVd RNA in the nucleoplasm. Note that there is no overlap between (−)-PSTVd (green) and U3 snoRNA (red) localizations throughout the serial sections.
(R) Serial confocal images showing localization of the (+)-PSTVd RNA in the nucleolus as well as in the nucleoplasm. The viroid signal (green) overlaps with U14 snoRNA signal (red) to produce a yellowish ring within the nucleolus.
(S) Serial confocal images showing localization of the (−)-PSTVd RNA in the nucleoplasm. Note that there is no overlap between (−)-PSTVd (green) and U14 snoRNA (red) localizations throughout the serial sections.
Fluorescence microscopic examination showed no PSTVd hybridization signals on cells that were mock-inoculated with water alone (Figure 1C). The (+)-strand of PSTVd was localized in the nucleolus as well as in the nucleoplasm (Figure 1H, green fluorescence). Within the nucleolus, the viroid signal often was concentrated as a highly visible ring-like structure in the periphery. In merged images, the green fluorescent signal of the (+)-PSTVd overlapped with the red fluorescent signal of U3 (Figure 1I) to produce the characteristic yellowish color in the nucleolus (Figure 1J). By contrast, the (−)-strand of PSTVd was localized only in the nucleoplasm (Figure 1M, green fluorescence). The lack of viroid signal in the nucleolus created the dark appearance of this subnuclear organelle (Figure 1M), which mirrored the dark nucleolar space in the DAPI-stained nucleus (Figure 1L). The localization of U3 further established the nucleolar identity in the same cell (Figure 1N). The localization patterns of the (+)- and (−)-strands of PSTVd were consistent in >300 cells examined.
To verify the results from fluorescence microscopic observations at higher resolutions, we examined cells with optical sectioning by laser scanning confocal microscopy. Serial confocal images presented in Figure 1P show that the (+)-PSTVd was localized in the nucleoplasm as well as in the nucleolus. Localization in the latter was revealed by a yellowish fluorescent signal produced by the physical colocalization of green viroid signal and red U3 signal. By contrast, the (−)-PSTVd was present only in the nucleoplasm (green signal). Its absence from the nucleolus was demonstrated by the nonoverlapping of the green viroid signal with the nucleolus-localized red U3 signal (Figure 1Q). Identical results were obtained from FISH experiments with U14 as a nucleolar marker (Figures 1R and 1S).
In summary, FISH experiments using a variety of nucleolar markers and the correlative use of fluorescence and confocal microscopy established unequivocally that the (+)-PSTVd was localized in the nucleoplasm as well as in the nucleolus. However, the (−)-PSTVd was localized in the nucleoplasm and absent from the nucleolus.
Localization of the (+)- and (−)-Strands of PSTVd in Infected Plant Tissues
To determine whether the localization patterns of the (−)- and (+)-strand PSTVd RNAs observed in cultured N. benthamiana cells would mirror the situation in infected plants, we used FISH to detect the subcellular localization of PSTVd on cryosections of systemically infected leaves of tomato and N. benthamiana collected 4 to 5 weeks after inoculation. The experiments on tomato were particularly important because we wanted to determine whether both the (+)- and (−)-strands of PSTVd would be localized in the nucleolus, as reported previously in studies with nuclei isolated from infected tomato plants (Harders et al., 1989). Initial analyses yielded similar results for infected N. benthamiana and tomato plants, and the latter were chosen for detailed analyses.
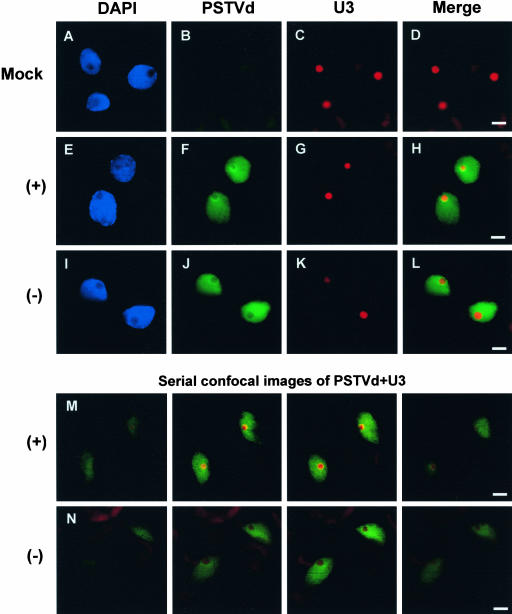
In mock-inoculated tomato leaves, there was no viroid hybridization signal, in contrast to the strong U3 snoRNA hybridization signal (Figures 2A to 2D). In infected tomato leaves, the (+)-PSTVd was localized in the nucleoplasm as well as in the nucleolus, with a higher concentration at the nucleolar periphery (Figures 2E to 2H; see further details below). The (−)-PSTVd, on the other hand, was localized in the nucleoplasm and was absent from the nucleolus (Figures 2I to 2L). Examination of >500 cells yielded the same results for both RNAs. FISH experiments with U14 as a nucleolar marker yielded the same results (see supplemental data online). These fluorescence microscopic observations were confirmed with serial confocal optical sectioning examinations. As shown in Figure 2M, the (+)-PSTVd was colocalized with U3 within the nucleolus (the yellowish fluorescent ring). By contrast, the (−)-PSTVd was detected only in the nucleoplasm and showed no overlapping with U3 localization, indicating absence from the nucleolus (Figure 2N).
Figure 2.
Differential Subnuclear Localization of the (+)- and (−)-Strands of PSTVd in Infected Tomato Tissues.
(A) to (D) Mock-inoculated cell. No PSTVd hybridization signals were detected.
(E) to (H) Nucleolar as well as nucleoplasmic localization of the (+)-strand of PSTVd. The yellowish ring in the merged image ([H]) indicates overlap between (+)-PSTVd and U3 in the nucleolus.
(I) to (L) Nucleoplasmic localization of the (−)-strand of PSTVd. The merged image ([L]) shows no overlap between (−)-PSTVd and U3 snoRNA localization signals (see text for details).
(M) Serial confocal images showing localization of the (+)-PSTVd RNA in the nucleolus as well as in the nucleoplasm. The viroid signal (green) overlaps with U3 snoRNA signal (red) to produce a yellowish ring within the nucleolus.
(N) Serial confocal images showing localization of the (−)-PSTVd RNA in the nucleoplasm. Note that there is no overlap between (−)-PSTVd (green) and U3 snoRNA (red) localizations throughout the serial sections.
Bars = 4 μm.
Together, the localization patterns of the (+)- and (−)-strands of PSTVd in the infected cultured cells and plants suggest that viroid transcription occurs in the nucleoplasm, consistent with the nucleoplasmic localization of the purported RNA polymerase II. Although the (−)-strand viroid RNAs were retained in the nucleoplasm, the (+)-strand RNAs were transported selectively into the nucleolus.
Potential Stages of (+)-Strand PSTVd RNA Traffic into the Nucleolus
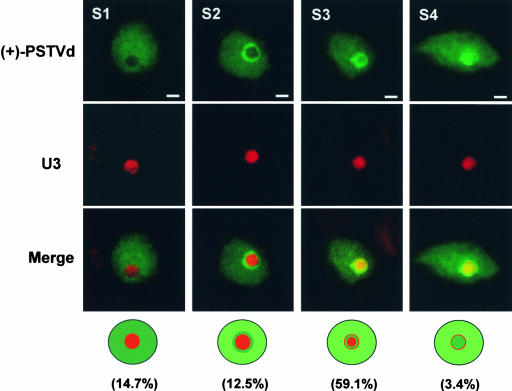
Detailed analyses of the (+)-strand PSTVd RNA localization in 806 cells from six infected tomato leaves revealed four distinct spatial patterns, which we interpreted as possibly representing successive stages of migration of the RNA from the nucleoplasm to the nucleolus. Single-plane confocal images of the localization patterns are presented in Figure 3 (for serial images obtained from optical sectioning of the same cells, see the supplemental data online). As shown in Figure 3, in 14.7% of the cells, the (+)-strand viroid RNA was localized only in the nucleoplasm (stage 1 or S1). In 12.5% of the cells, some viroid RNA appeared in a ring-like structure around the nucleolus (S2). The ring was found mostly in the nucleoplasm, because its green signal showed limited overlap with the nucleolus-localized red signal of U3. In 59.1% of the cells, some viroid RNA was present in a ring-like structure within the periphery of the nucleolus (S3). Its localization within the nucleolus was indicated by the distinct yellowish color when merged with the U3 signal. In 3.4% of the cells, some viroid RNA showed colocalization with U3 in the entire nucleolus (S4). These putative stages of (+)-PSTVd traffic from the nucleoplasm into the nucleolus are illustrated at the bottom of Figure 3. The localization pattern of (+)-PSTVd in the remaining cells is discussed below. Fluorescence microscopic images showing these putative stages of (+)-PSTVd traffic are presented in the supplemental data online.
Figure 3.
Confocal Images Showing the Spatial Localization Patterns of the (+)-Strand of PSTVd That Are Suggestive of Various Stages of Nucleolus-Bound Trafficking of the RNA from the Nucleoplasm.
Serial images of the same cells are presented in the supplemental data online. At stage 1 (S1), the viroid RNA is localized only in the nucleoplasm, as indicated by the nonoverlapping of PSTVd and U3 signals. At S2, there is concentration of the viroid RNA as a nucleoplasmic ring around the nucleolus. At S3, a ring of concentrated RNA appears within the nucleolus. At S4, the RNA is fully within the nucleolus. The diagrams at bottom summarize these patterns. The percentages of cells showing each RNA localization pattern are shown in parentheses. Bars = 2 μm.
The Presence of (+)-PSTVd in the Nucleolus Caused the Redistribution of U3 snoRNA
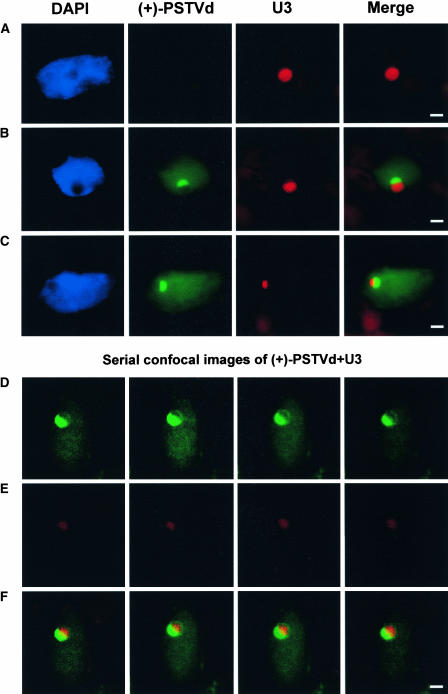
Previous studies showed that U3 snoRNA was localized in subnucleolar compartments in a symmetrical manner (Beven et al., 1996). Our examination of >1000 cells from the mock-inoculated plants confirmed this symmetrical distribution of U3 in the tomato nucleoli (Figure 4A). Strikingly, in >10% of the infected cells, the (+)-PSTVd and U3 were localized asymmetrically in separate domains within the nucleolus (Figures 4B and 4C). As shown in Figure 4B, DAPI staining of the nucleoplasm revealed the spherical nucleolus, which was not stained. Within this nucleolus, the green (+)-PSTVd signal was localized in the top half, with the lower half of the nucleolus appearing as a “black” hole. The red U3 signal filled this lower half of the nucleolus. The merged image shows that the (+)-PSTVd and U3 both were localized in the nucleolus but showed little overlapping of signals. The asymmetrical distribution of U3 and (+)-PSTVd within the nucleolus was further confirmed with confocal optical sectioning (Figures 4D to 4F). The spherical contour of the nucleolus can be discerned vividly in Figure 4D. Here, the (+)-PSTVd occupied approximately half of the nucleolus. The other half of the nucleolus contained no viroid green fluorescent signals and was encircled by a thin green fluorescent line. This viroid-devoid half of the nucleolus was occupied by U3 (Figures 4E and 4F). Importantly, redistribution of U3 did not occur in cells in which the (+)-PSTVd remained outside the nucleolus (see S1 and S2 in Figure 3). Thus, the presence of the viroid RNA within the nucleolus was correlated directly with the redistribution of U3. Interestingly, U14 also exhibited similar redistribution within the nucleolus in the presence of the (+)-PSTVd in a small proportion of the cells examined (data not shown).
Figure 4.
Asymmetrical Distribution of U3 snoRNA in the Presence of the (+)-PSTVd within the Nucleolus.
(A) Symmetrical distribution of U3 in the nucleolus of a mock-inoculated plant.
(B) and (C) Two infected cells showing the asymmetrical distribution of U3 (red) and PSTVd (green) in the nucleoli. Note that in each case the two RNAs occupy separate domains within the nucleolus with little overlapping.
(D) to (F) Serial confocal images showing the asymmetrical distribution of PSTVd (green in [D]) and U3 (red in [E]). The merged images ([F]) show little overlapping of the two RNAs.
Bars = 2 μm.
DISCUSSION
A Model of Intranuclear Traffic–Coupled Replication of Viroids in the Pospiviroidae
The results from this study provide novel information about the subnuclear localization of PSTVd replication intermediates in infected plant cells. First, the (−)-strand of PSTVd is localized in the nucleoplasm but not in the nucleolus. Second, the (+)-strand of PSTVd is localized in both the nucleoplasm and the nucleolus with distinct spatial patterns. The localization of the (−)-strand of PSTVd in the nucleoplasm is in sharp contrast with the localization of the same RNA in the nucleoli of nuclei isolated from infected tomato plants, as reported by Harders et al. (1989). The precise reason for this discrepancy is unclear. Our sample preparation protocols, especially the use of cryosections, should have preserved the subcellular distribution of the viroid RNAs more faithfully. Furthermore, the PSTVd RNA was detected directly by hybridization with fluorescent riboprobes with the highest possible spatial resolution. It is possible that in the previous study (Harders et al., 1989), the (−)-strand of PSTVd was redistributed within the nuclei during nuclear isolation from tomato. Most importantly, the nucleoplasmic localization of the (−)- and (+)-strands of PSTVd observed in the present study is fully consistent with molecular studies demonstrating that the nucleoplasm-localized DNA-dependent RNA polymerase II is involved in PSTVd transcription (Schindler and Mühlbach, 1992; Fels et al., 2001).
Our experiments have revealed high-resolution subnuclear localization patterns of the (+)-strand of PSTVd that are suggestive of successive stages of (+)-strand RNA migration from the nucleoplasm into the nucleolus: (1) nucleoplasm; (2) nucleoplasmic ring around the nucleolus; (3) peripheral ring inside the nucleolus; and (4) more central location in the nucleolus. Interestingly, Bonfiglioli et al. (1996) also reported concentration of Coconut cadang cadang viroid in the inner periphery of the nucleolus in infected oil palms. The prominent accumulation of the viroid RNA at a particular site suggests its interaction with a distinct cellular factor(s) to accomplish a specific function en route to the nucleolus.
The significance of (+)-strand PSTVd RNA trafficking into the nucleolus remains to be determined. Harders et al. (1989) discussed the possibility that the linear, multimeric (+)-strands of PSTVd synthesized in the nucleoplasm traffic into the nucleolus for processing into circular monomeric molecules. Processing of rRNAs and tRNAs occurs in the nucleolus (Lewis and Tollervey, 2000). In addition, some plant snoRNA genes are clustered and expressed polycistronically in the nucleoplasm (Leader et al., 1997); the precursor snoRNA appears to be transported into the nucleolus for processing into individual snoRNAs (Shaw et al., 1998). Thus, the nucleolus is an attractive candidate site for PSTVd processing. Our localization data support this hypothesis. The fibrillar center and dense fibrillar component are organized within the periphery of the nucleolus (Brown and Shaw, 1998). These are likely the specific sites where the processing of rRNA (Beven et al., 1996; Koberna et al., 2002) and some snoRNA (Shaw et al., 1998) occurs. U3 snoRNA, which is involved in rRNA processing in animal and yeast systems (Kass et al., 1990; Savino and Gerbi, 1990; Hughes and Ares, 1991; Beltrame and Tollervey, 1992), is localized in these structures in plant cells (Beven et al., 1996). Notably, in 59.1% of the infected tomato cells examined, the (+)-strand of PSTVd was colocalized with U3 as a ring in the nucleolar periphery, which corresponds to the fibrillar center and dense fibrillar component region. Before experimental data are available to verify this model, we also need to consider an alternative possibility, also discussed by Harders et al. (1989), that processing of the (+)-strand, multimeric PSTVd RNAs occurs in the nucleoplasm and the circular viroids then migrate into the nucleolus.
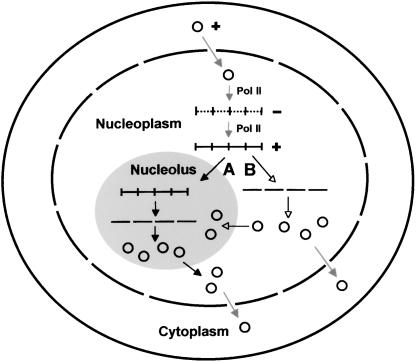
Based on these discussions, we present a model that places the rolling-circle replication of PSTVd in a cellular context (Figure 5). The key features of this model are as follows: (1) synthesis of the (−)- and (+)-strands of PSTVd occurs in the nucleoplasm; (2) the (−)-strand RNA is anchored in the nucleoplasm; (3) the (+)-strand RNA is transported selectively into the nucleolus; and (4) some (+)-strand RNA traffics from the nucleolus back into the nucleoplasm and further into the cytoplasm for spreading into neighboring cells. The two alternative mechanisms for the processing and trafficking of the (+)-strand PSTVd, which remain to be tested experimentally, are indicated by arrows A and B in Figure 5.
Figure 5.
Model of PSTVd Replication Featuring Intranuclear Trafficking of the (+)-Strand PSTVd.
The circular monomeric (+)-PSTVd traffics from the cytoplasm into the nucleoplasm to be used as the initial template to synthesize linear, multimeric (−)-PSTVd by DNA-dependent RNA polymerase II (Pol II). The latter then serves as the template to synthesize linear, multimeric (+)-PSTVd by the same enzyme. Two alternative pathways for the processing and trafficking of the (+)-PSTVd are illustrated in the model. In path A, the linear, multimeric (+)-PSTVd traffics into the nucleolus, where it is cleaved into monomers and then circularized. In path B, the linear, multimeric (+)-PSTVd is cleaved into monomers and circularized in the nucleoplasm. Some circular monomers then traffic into the nucleolus. In either pathway, some circular monomers traffic into the cytoplasm through the nuclear pore complex and further into neighboring cells through the plasmodesmata (not shown).
The inhibition by α-amanitin of the replication of Cucumber pale fruit viroid (Mühlbach and Sanger, 1979), Hop stunt viroid (Yoshikawa and Takahashi, 1986), and CEVd (Flores and Semancik, 1982; Semancik and Harper, 1984; Flores, 1989; Rivera-Bustamante and Semancik, 1989) in various experimental systems suggests the involvement of the DNA-dependent RNA polymerase II in the replication of these viroids, all of which are members of the Pospiviroidae. Significantly, both the (−)- and (+)-strands of CEVd RNA were associated with the largest subunit of RNA polymerase II in tomato in vivo, consistent with the role of this enzyme in CEVd replication (Warrilow and Symons, 1999). Thus, the model presented in Figure 5 may have general relevance to the subcellular trafficking and localization of replication intermediates of viroids in the Pospiviroidae.
Implications of the Intranuclear Localization of Viroid RNAs in Pathogenicity
The differential subnuclear localization of the (+)- and (−)-strands of PSTVd may be valuable in investigating the mechanisms of viroid diseases, which remain poorly understood. PSTVd may interact with a double-stranded RNA-activated protein kinase to disturb cellular functions leading to diseases (Diener et al., 1993). Recent studies have demonstrated comprehensive alteration in host gene expression during PSTVd infection (Itaya et al., 2002). Altered expression of certain genes can be linked to specific defects in cellular processes that lead to a particular symptom (Qi and Ding, 2003). Assuming that the (+)- and (−)-strands of PSTVd each can interact with specific cellular factors to disrupt normal cell functions to cause symptoms, the differential subnuclear localization of the (+)- and (−)-strands of PSTVd suggest different cellular targets for these RNAs.
Several animal and plant viruses target to the nucleolus and can cause the redistribution of nucleolar proteins with functions that are unclear (Hiscox, 2002). Our results provide evidence that a pathogenic RNA can cause the redistribution of a nucleolar RNA. An important control for the nucleolar identity is DAPI staining, which highlighted the nucleoplasm but not the nucleolus (Beven et al., 1996; Shaw et al., 1998; this study). It is intriguing that in each case, when the (+)-PSTVd was localized in one part of the nucleolus, the U3 was localized in the remaining part. Our interpretation of this finding is that the (+)-PSTVd competes with U3 for certain nucleolar factors, with the consequence that the U3 is displaced from part of its normal nucleolar domain. In a simple scenario, this redistribution of U3 could negatively affect rRNA processing and subsequent biological processes. Our preliminary observations showed that U14 also exhibited similar redistribution within the nucleolus in the presence of (+)-PSTVd in some cells. Further studies are needed to determine how widespread this redistribution is among the numerous snoRNAs, the dynamics of this redistribution, the underlying mechanisms, and its biological significance during viroid infection.
Nucleolar Traffic of PSTVd May Be Mediated by a Novel Mechanism
Besides its role in ribosome biosynthesis, the nucleolus is now implicated in many other important functions, including nuclear export, processing of small RNAs, tRNAs, and even some mRNAs, assembly of ribonucleoprotein, cell aging control, and cell cycle regulation (reviewed by Lewis and Tollervey, 2000; Olson et al., 2000; Gerbi et al., 2003). These functions necessitate heavy trafficking of many types of RNAs and proteins between the nucleolus and the nucleoplasm/cytoplasm. Most mechanistic insights on nucleolar RNA trafficking have come from recent studies on the nucleolar import of snoRNAs, which function as guide RNAs to mediate the cleavage of rRNA precursors and modifications of rRNAs (Kiss, 2002; Terns and Terns, 2002).
snoRNAs are synthesized in the nucleoplasm and imported into the nucleolus. The majority of snoRNAs fall into two categories: the C/D box snoRNAs that contain a C box (UGAUGA) and a D box (CUGA), and the hinge-hairpin-hinge-ACA (H/ACA) box snoRNAs (Kiss, 2002; Terns and Terns, 2002). Recent studies showed that the C/D and H/ACA boxes function as nucleolar localization elements (Lange et al., 1998, 1999; Samarsky et al., 1998; Narayanan et al., 1999a, 1999b). The nucleolar localization of a third family of snoRNAs, which consists of two species, the 7-2/MRP snoRNA and the RNA component of RNase P, is mediated by their To antigen binding domain (Jacobson et al., 1995, 1997). Other RNAs use different motifs for trafficking into the nucleolus. Several small nuclear RNAs that function in splicing in the nucleoplasm transiently pass through the nucleoli before reaching their nucleoplasmic destinations (Gerbi et al., 2003). The nucleotide sequence at the 3′ end of U6 small nuclear RNA is required for localization to the nucleolus (Gerbi and Lange, 2002).
Comparative sequence analysis did not reveal the presence of currently known nucleolar localization elements in PSTVd. Thus, the (+)-strand PSTVd RNA likely contains a novel motif(s) for nucleolar targeting. This motif(s) may bind a nuclear factor that is dedicated to transporting certain endogenous RNAs into the nucleolus. Alternatively, it may bind a nuclear factor that is not a dedicated RNA transporter but instead is a factor that relocates itself within the nucleus to perform specific functions in association with the life cycle of a cell. Resolving these issues should provide new insights into the dynamics of macromolecular movement within the nucleus.
Strand Polarity–Based Subcellular Localization of RNAs Has General Biological Significance
The localization of the (+)- and (−)-strands of PSTVd provides an excellent example of how specific subnuclear localization of an RNA is closely associated with its biological function. Furthermore, our results suggest strongly that the eukaryotic cell has mechanisms to recognize and localize the opposite strands of an RNA to specific subnuclear locations. It is unlikely that this strand polarity–based RNA trafficking/localization is a unique intranuclear phenomenon. Hepatitis delta virus, which contains a viroid-like circular RNA, replicates via a double-rolling circular mechanism in the nucleus by using host enzymes, including the DNA-dependent RNA polymerase II (Moraleda and Taylor, 2001; Macnaughton et al., 2002). Immediately after synthesis and processing, the genomic but not the antigenomic Hepatitis delta virus RNAs are exported out of the nucleus in infected human hepatoma cultured cells (Macnaughton and Lai, 2002). In an infected tobacco and cucumber cell, the Cucumber mosaic virus (+)-strand RNA is localized throughout the cytoplasm, whereas the (−)-strand RNA is localized predominantly to the tonoplast (Cillo et al., 2002). Although the role of virus-encoded proteins in the differential subcellular localization of viral RNAs of opposite polarity remains to be elucidated, it is possible that cellular factors also are involved. Together, the available data support the hypothesis that cellular mechanisms exist to localize RNAs of opposite polarity to distinct subcellular locations for specific functions. The strand polarity–based localization of viroid and viral RNAs may represent an evolutionary adaptation to the host mechanisms to achieve optimal replication, accumulation, and/or protection against cellular degradation.
Because the polarity of an RNA can influence its subcellular localization, an important question arises regarding how the increasing numbers of antisense, regulatory RNAs are colocalized with their sense targets within a cell at both the suborganellar and subcellular levels. This question has direct relevance to our understanding of the mechanisms of RNA regulation of gene expression. Practically, ectopic expression of antisense RNA has been used to study the function of a gene and to engineer organisms for a desirable trait. Such approaches have had varying results (Jen and Gewirtz, 2000). It will be interesting to determine whether the differential subcellular localization of sense and antisense RNAs can account for some failures of the antisense method.
METHODS
Plant Materials and Growth Conditions
Tomato (Lycopersicon esculentum cv Rutgers) and Nicotiana benthamiana plants were grown in a growth chamber with a 27/22°C day/night temperature regime and a 14-h-light/10-h-dark cycle. N. benthamiana suspension cells were cultured as described (Qi and Ding, 2002; Sunter and Bisaro, 2003).
DNA Constructs
Plasmid pRZ6-2, in which the cDNA of the intermediate strain of Potato spindle tuber viroid (PSTVdInt) (Gross et al., 1978) is flanked by ribozymes, was constructed by Hu et al. (1997) and was kindly provided by Robert Owens (U.S. Department of Agriculture/Agricultural Research Service, Beltsville, MD). The construction of plasmids pInter(+) and pInter(−) was described previously (Qi and Ding, 2002).
The full-length cDNA of tomato U3 small nucleolar RNA was amplified by reverse transcriptase–mediated PCR using primers LeU3-F (5′-ACG- ACCTTACTTGAACAG-3′) and LeU3-R (5′-TCTGTCAGACAGCCATGA-3′), which were designed based on the published U3 sequence (Kiss and Solymosy, 1990). The PCR fragment then was cloned into pGEM-T vector (Promega, Madison, WI) to give rise to plasmid pLeU3(−), which was used as a template to synthesize the antisense U3 riboprobes.
Partial cDNA of tomato U14 small nucleolar RNA was amplified by reverse transcriptase-mediated PCR using primers StU14-F (5′-AATTGAAGGCTTGTTTCT-3′) and StU14-R (5′-TCAGAGATCCAAGGAAGG-3′), which were designed based on the published potato U14 sequence (Leader et al., 1994). The PCR fragment then was cloned into pGEM-T vector (Promega) to give rise to plasmid pLeU14(−), which was used as a template to synthesize the antisense U14 riboprobes.
In Vitro Transcription
To prepare in vitro transcripts of PSTVdInt for inoculation, the plasmid pRZ6-2 was linearized by HindIII and used as a template for in vitro transcription using the T7 MEGAscript kit (Ambion, Austin, TX) according to the manufacturer's directions.
For in situ hybridization, riboprobes were prepared by in vitro transcription using the T7 Maxiscript kit (Ambion) according to the methods recommended by the manufacturer. SpeI-linearized pInter(+) and pInter(−) were used as templates to synthesize Alexa Fluor 488–5-UTP (Molecular Probes, Eugene, OR)–labeled sense and antisense PSTVd riboprobes, respectively. SpeI-linearized pLeU3(−) and pLeU14(−) were used as templates to produce Biotin–16-UTP (Roche, Indianapolis, IN)–labeled antisense U3 and U14 probes, respectively.
After transcription, DNA templates were removed by digestion with RNase-free DNase I, and RNA transcripts were purified using the MEGAclear kit (Ambion) and quantified by UV spectrometry.
Plant and Protoplast Inoculation
Cotyledons of 1-week-old tomato seedlings and young leaves of 3-week-old N. benthamiana were dusted with carborundum and mechanically inoculated with PSTVdInt in vitro transcripts (100 ng/μL). Water alone was used as a mock inoculum. At 4 to 5 weeks after inoculation, systemic leaves were collected and processed for in situ hybridization.
Protoplasts were prepared from N. benthamiana cultured cells and inoculated with PSTVdInt transcripts as described (Qi and Ding, 2002). At 2 days after inoculation, the protoplasts were collected and processed for in situ hybridization.
Tissue and Cell Processing
Leaf tissues were fixed to obtain cryosections of 20 μm thickness as described (Zhu et al., 2002). Protoplasts were fixed and processed as described previously (Qi and Ding, 2002). Before in situ hybridization, the fixed protoplasts were incubated in PBS containing 2% BSA, 0.1% cellulase, 0.05% macerozyme, and 0.5% Nonidet P-40 at room temperature for 30 min and then in prechilled methyl alcohol with 2% hydrogen peroxide at −10°C for 30 min.
Fluorescence in Situ Hybridization
In situ hybridization was performed essentially as described (Zhu et al., 2002) except that Alexa Fluor 488– or biotin-labeled probes were used instead of digoxigenin-labeled riboprobes. Alexa Fluor 488–labeled sense or antisense PSTVd probes each were mixed with biotin-labeled antisense U3 or U14 probes and added to the hybridization buffer. Samples were treated further with 2.5 μg/mL streptavidin conjugated to Alexa Fluor 594 (Molecular Probes). All samples were stained with the DNA fluorochrome 4′,6-diamidino-2-phenylindole (DAPI) for 2 min at 1 μg/mL in PBS and mounted in Fluoromount G (Electron Microscopy Sciences, Fort Washington, PA) before microscopic examination.
Microscopy
Samples were examined with an E600 fluorescence microscope (Nikon, Tokyo, Japan). DAPI fluorescence was visualized with a filter set consisting of an excitation filter of 330 to 380 nm, a dichroic mirror of 400 nm, and a barrier filter of 435 to 485 nm. Alexa Fluor 488 fluorescence was visualized with a filter set consisting of an excitation filter of 450 to 490 nm, a dichroic mirror of 510 nm, and a barrier filter of 520 to 560 nm. Alexa Fluor 594 fluorescence was visualized with a filter set consisting of an excitation filter of 540 to 580 nm, a dichroic mirror of 595 nm, and a barrier filter of 600 to 660 nm. Images were captured and processed with a SPOT 2 Slider charge-coupled device camera and the associated software (Diagnostics Instruments, Sterling Heights, MI). Samples also were examined with a Nikon PCM-2000 confocal laser scanning microscope equipped with argon and green HeNe lasers.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Biao Ding, ding.35@osu.edu.
Supplementary Material
Acknowledgments
We express our gratitude to David Bisaro for making his N. benthamiana cultured cells available to us and to Robert Owens for providing the PSTVd cDNA. We are indebted to Xuehua Zhong for assisting with part of this work and for helpful discussions. We thank Asuka Itaya for assistance with confocal microscopy, insightful discussions, and critical reading of the manuscript. This work was supported in part by a grant from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (2001-35303-11073) and in part by a grant from the National Science Foundation (IBN-0238412).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016576.
Footnotes
Online version contains Web-only data.
References
- Andersen, A.A., and Panning, B. (2003). Epigenetic gene regulation by noncoding RNAs. Curr. Opin. Cell Biol. 15, 281–289. [DOI] [PubMed] [Google Scholar]
- Beltrame, M., and Tollervey, D. (1992). Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven, A.F., Lee, R., Razaz, M., Leader, D.J., Brown, J.W., and Shaw, P.J. (1996). The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J. Cell Sci. 109, 1241–1251. [DOI] [PubMed] [Google Scholar]
- Bonfiglioli, R.G., Webb, D.R., and Symons, R.H. (1996). Tissue and intra-cellular distribution of coconut cadang cadang viroid and citrus exocortis viroid determined by in situ hybridization and confocal laser scanning and transmission electron microscopy. Plant J. 9, 457–465. [Google Scholar]
- Branch, A.D., and Robertson, H.D. (1984). A replication cycle for viroids and other small infectious RNAs. Science 223, 450–455. [DOI] [PubMed] [Google Scholar]
- Brown, J.W., and Shaw, P.J. (1998). Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo, F., Roberts, I.M., and Palukaitis, P. (2002). In situ localization and tissue distribution of the replication-associated proteins of Cucumber mosaic virus in tobacco and cucumber. J. Virol. 76, 10654–10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña, M., Navarro, B., and Flores, R. (1999). Mapping the molecular determinant of pathogenicity in a hammerhead viroid: A tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. USA 96, 9960–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, T.O., Hammond, R.W., Black, T., and Katze, M.G. (1993). Mechanism of viroid pathogenesis: Differential activation of the interferon-induced, double-stranded RNA-activated, Mr 68,000 protein kinase by viroid strains of varying pathogenicity. Biochimie 75, 533–538. [DOI] [PubMed] [Google Scholar]
- Fels, A., Hu, K., and Riesner, D. (2001). Transcription of potato spindle tuber viroid by RNA polymerase II starts predominantly at two specific sites. Nucleic Acids Res. 29, 4589–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R. (1989). Synthesis of RNAs specific to citrus exocortis viroid by a fraction rich in nuclei from infected Gynura aurantiaca: Examination of the nature of the products and solubilisation of the polymerase-template complex. J. Gen. Virol. 70, 2695–2706. [Google Scholar]
- Flores, R., Randles, J.W., Bar-Josef, M., and Diener, T.O. (2000). Subviral agents: Viroids. In Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses, M.H.V. van Regenmortel, C.M. Fauquet, D.H.L. Bishop, E.B. Carstens, M.K. Estes, S.M. Lemon, J. Manilof, M.A. Mayo, D.J. McGeoch, C.R. Pringle, and R.B. Wickner, eds (San Diego, CA: Academic Press), pp. 1009–1024.
- Flores, R., and Semancik, J.S. (1982). Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc. Natl. Acad. Sci. USA 79, 6285–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi, S.A., Borovjagin, A.V., and Lange, T.S. (2003). The nucleolus: A site of ribonucleoprotein maturation. Curr. Opin. Cell Biol. 15, 318–325. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A., and Lange, T.S. (2002). All small nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP localize to nucleoli: Identification of the nucleolar localization element of U6 snRNA. Mol. Biol. Cell 13, 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan, R.J. (2003). RNA and memory: From feeding to localization. Curr. Biol. 13, R126–R127. [DOI] [PubMed] [Google Scholar]
- Gross, H.J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H., and Sanger, H.L. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273, 203–208. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., and Slack, F.J. (2002). Micro-RNAs: Small is plentiful. J. Cell Biol. 156, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harders, J., Lukacs, N., Robert-Nicoud, M., Jovin, T.M., and Riesner, D. (1989). Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8, 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox, J.A. (2002). The nucleolus: A gateway to viral infection? Arch. Virol. 147, 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Feldstein, P.A., Hammond, J., Hammond, R.W., Bottino, P.J., and Owens, R.A. (1997). Destabilization of potato spindle tuber viroid by mutations in the left terminal loop. J. Gen. Virol. 78, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Hughes, J.M., and Ares, M., Jr. (1991). Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2002). Matthew's Plant Virology, 4th ed. (San Diego, CA: Academic Press).
- Itaya, A., Matsuda, Y., Gonzales, R.A., Nelson, R.S., and Ding, B. (2002). Potato spindle tuber viroid strains of different pathogenicity induce and suppress expression of common and unique genes in infected tomato. Mol. Plant-Microbe Interact. 15, 990–999. [DOI] [PubMed] [Google Scholar]
- Jacobson, M.R., Cao, L.G., Taneja, K., Singer, R.H., Wang, Y.L., and Pederson, T. (1997). Nuclear domains of the RNA subunit of RNase P. J. Cell Sci. 110, 829–837. [DOI] [PubMed] [Google Scholar]
- Jacobson, M.R., Cao, L.G., Wang, Y.L., and Pederson, T. (1995). Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell Biol. 131, 1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen, K.Y., and Gewirtz, A.M. (2000). Suppression of gene expression by targeted disruption of messenger RNA: Available options and current strategies. Stem Cells 18, 307–319. [DOI] [PubMed] [Google Scholar]
- Kass, S., Tyc, K., Steitz, J.A., and Sollner-Webb, B. (1990). The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60, 897–908. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H., and Taira, K. (2003). Hes1 is a target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2 cells. Nature 423, 838–842. [DOI] [PubMed] [Google Scholar]
- Kiss, T. (2002). Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Kiss, T., and Solymosy, F. (1990). Molecular analysis of a U3 RNA gene locus in tomato: Transcription signals, the coding region, expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res. 18, 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna, K., Malinsky, J., Pliss, A., Masata, M., Vecerova, J., Fialova, M., Bednar, J., and Raska, I. (2002). Ribosomal genes in focus: New transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J. Cell Biol. 157, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.C. (2003). RNA sensors and riboswitches: Self-regulating messages. Curr. Biol. 13, R285–R291. [DOI] [PubMed] [Google Scholar]
- Lange, T.S., Borovjagin, A., Maxwell, E.S., and Gerbi, S.A. (1998). Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 17, 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, T.S., Ezrokhi, M., Amaldi, F., and Gerbi, S.A. (1999). Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 10, 3877–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader, D.J., Clark, G.P., Watters, J., Beven, A.F., Shaw, P.J., and Brown, J.W. (1997). Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader, D.J., Sanders, J.F., Waugh, R., Shaw, P., and Brown, J.W. (1994). Molecular characterisation of plant U14 small nucleolar RNA genes: Closely linked genes are transcribed as polycistronic U14 transcripts. Nucleic Acids Res. 22, 5196–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D., and Tollervey, D. (2000). Like attracts like: Getting RNA processing together in the nucleus. Science 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Macnaughton, T.B., and Lai, M.M. (2002). Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76, 3928–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnaughton, T.B., Shi, S.T., Modahl, L.E., and Lai, M.M. (2002). Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76, 3920–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse, E., Majdalani, N., and Gottesman, S. (2003). Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6, 120–124. [DOI] [PubMed] [Google Scholar]
- Moraleda, G., and Taylor, J. (2001). Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75, 10161–10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbach, H.P., and Sanger, H.L. (1979). Viroid replication is inhibited by α-amanitin. Nature 278, 185–188. [DOI] [PubMed] [Google Scholar]
- Narayanan, A., Lukowiak, A., Jady, B.E., Dragon, F., Kiss, T., Terns, R.M., and Terns, M.P. (1999. a). Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 18, 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, A., Speckmann, W., Terns, R., and Terns, M.P. (1999. b). Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.O., Dundr, M., and Szebeni, A. (2000). The nucleolus: An old factory with unexpected capabilities. Trends Cell Biol. 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., and Diener, T.O. (1982). RNA intermediates in potato spindle tuber viroid replication. Proc. Natl. Acad. Sci. USA 79, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, R.A., Thompson, S.M., and Steger, G. (1991). Effects of random mutagenesis upon potato spindle tuber viroid replication and symptom expression. Virology 185, 18–31. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2002). Replication of Potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: The role of specific nucleotides in determining replication levels for host adaptation. Virology 302, 445–456. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2003). Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15, 1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Rivera-Bustamante, R.F., and Semancik, J.S. (1989). Properties of a viroid-replicating complex solubilized from nuclei. J. Gen. Virol. 70, 2707–2716. [Google Scholar]
- Samarsky, D.A., Ferbeyre, G., Bertrand, E., Singer, R.H., Cedergren, R., and Fournier, M.J. (1999). A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. USA 96, 6609–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A., Fournier, M.J., Singer, R.H., and Bertrand, E. (1998). The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino, R., and Gerbi, S.A. (1990). In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, I.M., and Mühlbach, H.P. (1992). Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: A reevaluation. Plant Sci. 84, 221–229. [Google Scholar]
- Schnölzer, M., Haas, B., Ramm, K., Hofmann, H., and Sänger, H.L. (1985). Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 4, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semancik, J.S., and Harper, K.L. (1984). Optimal conditions for cell-free synthesis of citrus exocortis viroid and the question of specificity of RNA polymerase activity. Proc. Natl. Acad. Sci. USA 81, 4429–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P.J., Beven, A.F., Leader, D.J., and Brown, J.W. (1998). Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J. Cell Sci. 111, 2121–2128. [DOI] [PubMed] [Google Scholar]
- S̆korić, D., Conerly, M., Szychowski, J.A., and Semancik, J.S. (2001). CEVd-induced symptom modification as a response to a host-specific temperature-sensitive reaction. Virology 280, 115–123. [DOI] [PubMed] [Google Scholar]
- Storz, G. (2002). An expanding universe of noncoding RNAs. Science 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- Sunter, G., and Bisaro, D.M. (2003). Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305, 452–462. [DOI] [PubMed] [Google Scholar]
- Terns, M.P., and Terns, R.M. (2002). Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- Tufarelli, C., Stanley, J.A., Garrick, D., Sharpe, J.A., Ayyub, H., Wood, W.G., and Higgs, D.R. (2003). Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34, 157–165. [DOI] [PubMed] [Google Scholar]
- Warrilow, D., and Symons, R.H. (1999). Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch. Virol. 144, 2367–2375. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, N., and Takahashi, T. (1986). Inhibition of hop stunt viroid replication by α-amanitin. Z. Pflanzenkr. Pflanzenschutz 93, 62–71. [Google Scholar]
- Zhu, Y., Green, L., Woo, Y.-M., Owens, R., and Ding, B. (2001). Cellular basis of potato spindle tuber viroid systemic movement. Virology 279, 69–77. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.