Abstract
In the classic paradigm of mammalian cell cycle control, Rb functions to restrict cells from entering S phase by sequestering E2F activators (E2f1, E2f2 and E2f3), which are invariably portrayed as the ultimate effectors of a transcriptional program that commit cells to enter and progress through S phase1, 2. Using a panel of tissue-specific cre-transgenic mice and conditional E2f alleles we examine the effects of E2f1, E2f2 and E2f3 triple deficiency in murine ES cells, embryos and small intestines. We show that in normal dividing progenitor cells E2F1-3 function as transcriptional activators, but contrary to current dogma, are dispensable for cell division and instead are necessary for cell survival. In differentiating cells they function in complex with Rb as repressors to silence E2F targets and facilitate exit from the cell cycle. The inactivation of Rb in differentiating cells resulted in a switch of E2F1-3 from repressors to activators, leading to the superactivation of E2F responsive targets and ectopic cell divisions, and loss of E2f1-3 completely suppressed these phenotypes. This work contextualizes the activator versus repressor functions of E2F1-3 in vivo, revealing distinct roles in dividing versus differentiating cells and in normal versus cancer-like cell cycles in vivo.
Keywords: Small intestine, cell cycle, E2F, retinoblastoma, tumor suppressor
E2Fs function as transcription factors, with E2F1-3 as activators and E2F4-8 as repressors3–8. Although it is a maxim of mammalian cell cycle regulation that the E2F1-3 activator subclass is required for cell proliferation, the evidence for this is based almost exclusively on in vitro studies using cells derived from murine and human tissues or on the in vivo analysis of Rb mutant mice1, 2. Other experiments, however, suggest that these E2Fs can also function as repressors in complex with Rb9–11, yet the relative contribution of activation version repression and the physiological contexts in which these contrary E2F functions are employed remain unclear.
To explore the functions of the E2F activator subclass, we derived E2f1−/−;E2f2−/−;E2f3LoxP/− ES cells (Supplementary Fig. 1a, 1b) and compared the consequences of inactivating the conditional E2f3LoxP allele in these cells versus in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs. The expression of E2f1, E2f2 and E2f3 in wild type ES cells was generally higher than in MEFs and the loading of E2F3 protein on classic E2F target promoters was comparable between the two proliferating cell types (Supplementary Fig. 2a-c). Consistent with previous observations, the ablation of E2f1-3 in MEFs with standard cre-expressing vectors led to the induction of p53 activity, the loading of E2F4-p130 repressor complexes on E2F target promoters and a marked decrease in E2F target expression (Fig. 1a, Supplementary Fig. 3a–c)5–7. Consequently, triply deficient MEFs underwent a complete cell cycle arrest (Fig. 1b)5–7. In contrast, E2f1−/−;E2f2−/−;E2f3Δ/−(TKO) ES cells failed to activate p53 or form E2F4/p130 repressive complexes, and as a result, E2F target expression was unaffected and cells proliferated equally well as E2f1−/−;E2f2−/−;E2f3LoxP/− (DKO) control cells (Supplementary Fig. 3a-c).
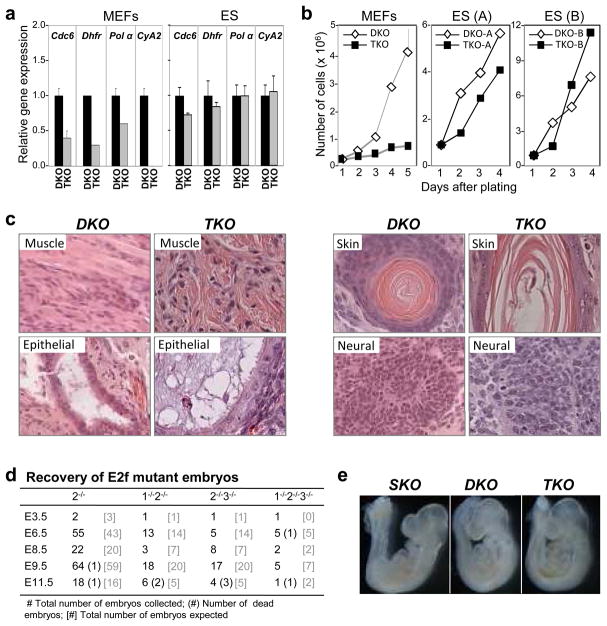
Figure 1. Cell proliferation in the absence of E2f1-3.
a Expression of E2F-regulated genes was measured by real-time RT-PCR in proliferating ES and MEFs cells with the indicated genotypes (primer information is provided in Supplementary Fig. 19). b. Growth curves of two sets of DKO and TKO ES cell clones (A and B) and DKO and TKO MEFs. c. DKO and TKO ES cells were injected underneath the skin of athymic nude mice and teratomas were harvested, sectioned and stained with H&E. Representative tissues of DKO and TKO teratomas include muscle (mesoderm), respiratory epithelium (endoderm), skin and neural cells (ectoderm). d. Embryos derived from intercrosses between E2f1+/−;E2f2−/−;E2f3+/− mice were collected at various timepoints during pregnancies. e. Representative E9.5 embryos were photographed immediately upon collection; E2f2−/− (SKO), E2f2−/−;E2f3−/− (DKO), and E2f1−/−E2f2−/−E2f3−/− (TKO) embryos.
We then evaluated whether triply-deficient ES cells could proliferate in vivo. Subcutaneous injection of TKO ES cells into athymic nude mice yielded efficient teratoma formation, producing mesoderm, endoderm, and ectoderm at a rate similar to DKO ES lines (Fig. 1c, Supplementary Fig. 4a, 4b). Moreover, from E2f1+/−;E2f2−/−;E2f3+/− intercrosses we recovered the expected number of live TKO embryos as late as E9.5, but none were recovered past E11.5 (Fig. 1d, and data not shown). The live E9.5 TKO embryos appeared morphologically normal by gross and histological examination (Fig. 1e and data not shown). While cell proliferation was normal in most tissues, there was evidence of decreased proliferation and increased apoptosis in the myocardium and the first branchial arch of TKO embryos (Supplementary Fig. 5a–d). These latter observations are consistent with heart defects found in E2f3 singly-deleted adult mice12.
To explore whether E2F1-3 might have cell cycle-related functions in tissues that arise later in embryonic and postnatal development, we exploited the highly organized cellular architecture of the small intestine. Maintenance of structural and functional integrity of the small intestine requires continuous epithelial regeneration13. Intestinal stem cells are housed at the base of crypts of Lieberkühn and give rise to transit-amplifying cells. As these cells migrate up from the base and into the finger-like extensions called villi, they exit the cell cycle and differentiate13. Western blot assays showed that E2f1, E2f2 and both isoforms of E2f3 (E2F3a and E2F3b) are expressed in the crypt and villus (Supplementary Fig. 6). We used Ah-cre mice14 to ablate E2f1-3 in the small intestine in utero or in adult mice (Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP, TKO). Induction of Ah-cre expression by intraperitoneal injection of β-napthoflavone (β-NF) led to the efficient deletion of E2f3LoxP in crypt stem cells and transit-amplifying cells by one day post-injection, and in the entire intestinal epithelium within 3–4 days (crypt and villus; Supplementary Fig. 7a–c). Loss of E2f1-3 did not result in a compensatory increase of other E2F family members, except for a modest increase in E2f8 (Supplementary Fig. 7d). Whether E2f3LoxP was deleted in utero at E15.5 or in the adult at 2 months of age, the architecture of TKO small intestines remained relatively intact and animals were asymptomatic for 90 days following β-NF administration (Fig. 2a, Supplementary Fig. 8a, 8b). Cell-type specific marker analysis demonstrated that all differentiated epithelial cell-types were appropriately represented in TKO small intestines (Fig. 2b, Supplementary Fig. 9). Remarkably, cell proliferation was identical in TKO and control intestines (Fig. 2c), however, we noted a marked increase in γ-H2AX and P-ATM1981 staining in TKO crypts and villi (Fig. 2d, 2e, Supplementary Fig. 10a). A parallel analysis of retinal (Chen et al, accompanying manuscript) and lens (P.W. unpublished observations) progenitors also revealed increased γ-H2AX staining in TKO samples (Supplementary Fig. 10b, 10c). Together, these observations suggest that counter to current dogma, E2F1-3 are dispensable for the proliferation of embryonic stem cells and their mesodermal, endodermal, and ectodermal derivatives, and cells in at least some adult tissues.
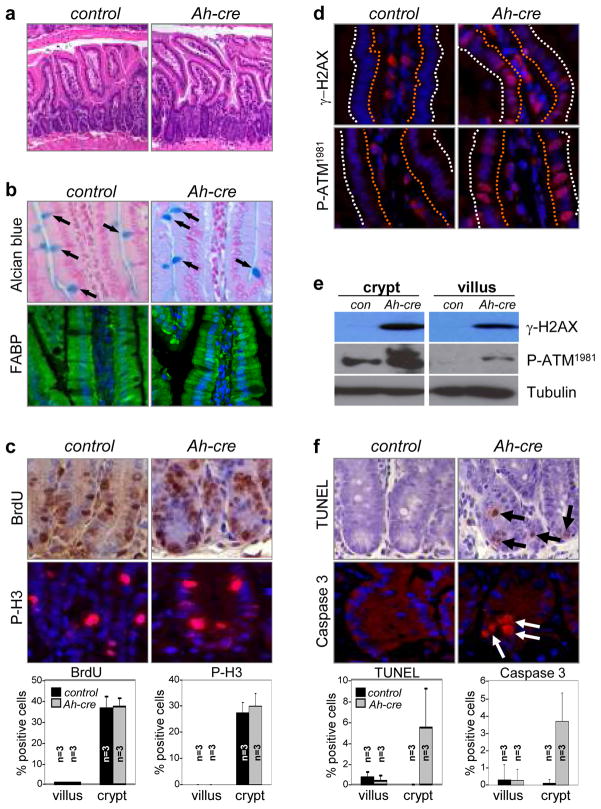
Figure 2. Apoptosis of crypt intestinal cells in the absence of E2f1, E2f2, and E2f3.
a H&E stained sections from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) intestines after 90 days of β-NF administration. b. Analysis of cell differentiation in control and Ah-cre small intestines. Goblet cells were identified by Alcian blue staining (arrows point to positive-stained goblet cells); absorptive cells were identified by anti-Fatty acid binding protein (FABP, green) antibodies; DAPI (blue) was used for staining nuclei. c. BrdU (brown) and phosphorylated histone H3 (P-H3, red) immunohistochemical staining was performed on small intestine sections from β-NF injected control and Ah-cre mice. Quantification of BrdU- and phosphorylated histone H3-positive cells in crypts and villi. n=3, 3 different animals with the indicated genotypes were analyzed (bottom panels); error bars indicate standard deviation. d. Immunohistochemical staining for γ-H2AX, P-ATM1981 in control and Ah-cre intestinal crypts and villi. The orange dotted line outlines the luminal side of the villus; the white dotted line outlines the outer side of the villus. DAPI (blue) was used for staining nuclei. e. Examination of γ-H2AX and P-ATM1981 in cell extracts from control and Ah-cre intestinal crypts and villi by Western blot assays. f. Sections of small intestines from β-NF injected control and Ah-cre mice were processed for TUNEL (brown) and cleaved caspase-3 (red) assays. DAPI (blue) or hematoxylin was used for staining nuclei. Quantification of TUNEL and cleaved caspase-3 positive cells in crypts and villi (bottom panels). n=3, 3 different animals with the indicated genotypes were analyzed; error bars indicate standard deviation.
Close examination of H&E-stained slides revealed increased numbers of pyknotic nuclei in TKO crypts (data not shown). TUNEL and cleaved caspase-3 assays confirmed the presence of apoptotic cells in crypts of TKO intestines (Fig. 2f). We also observed increased p53 immunoreactivity in TKO crypts (Supplementary Fig. 11a), which was reminiscent of previous work showing exquisite sensitivity of this cellular compartment to oncogene- and radiation-induced p53 responses15. While p53 was elevated in TKO crypts, we failed to detect any significant increase in the expression of p53-responsive genes and moreover, the conditional ablation of p53 (Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP;p53LoxP/LoxP) did not suppress the apoptosis caused by E2f1-3 deficiency (Supplementary Fig. 11b, 11c). Together, these surprising observations suggest that E2F1-3 are dispensable for cell division in the adult and that at least in the small intestine, they function in a p53-independent manner to maintain DNA integrity and cell survival.
To understand the underlying mechanism for these unexpected results, we isolated crypt and villus cell populations from control and E2f1-3-deficient (TKO) small intestines and analyzed global gene expression profiles. Sample preparation and processing of the Affymetrix oligo-arrays are described in the Methods section. We utilized an unbiased method similar to Gene Set Enrichment Analysis to identify genes that were differentially expressed16. Two variables contributed to the observed gene expression changes, cell compartment (crypt vs. villus) and genotype (TKO vs. control). The cell compartment analysis compared gene expression in crypts and villi of the same genotype (Fig. 3a). For control small intestines, this revealed that among the ~45K genes queried, 1207 genes were upregulated and 2363 genes were downregulated as progenitor cells in the crypt migrated up into the villus and exited the cell cycle (>1.5-fold, p<0.0001; Supplementary Fig. 12a, Supplementary Table 1). As expected, the expression of most known E2F targets, as defined by previous gene expression17, reporter and chromatin immunoprecipitation assays18 (Supplementary Fig. 12b), was markedly higher in control crypts than in associated villi (Fig. 3a, left panel), consistent with the proliferative status of crypts. For TKO small intestines, the expression of E2F targets in crypts was only marginally higher than in their associated villi (Fig. 3a, right panel), suggesting that expression of these genes were either reduced in crypts, elevated in villi, or both.
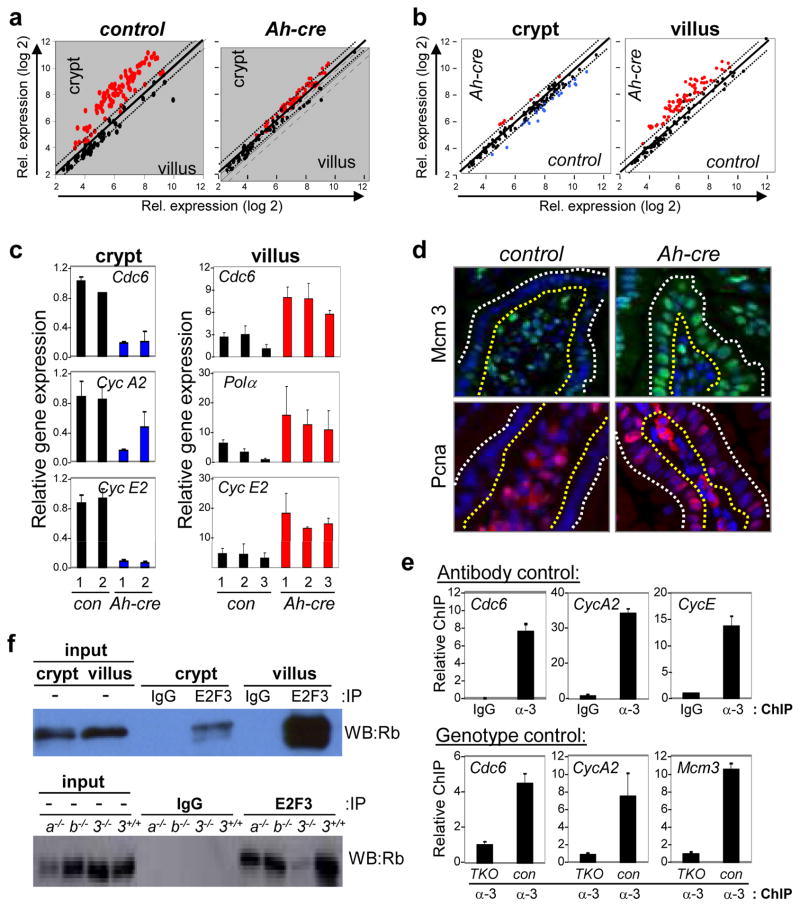
Figure 3. Repression of E2F-target genes in E2f1-3 deficient villi.
a Scatter plots comparing expression of known E2F-target genes (see Supplementary Fig. 16b) between cell compartments (crypt and villus); E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre). Genes with >1.5-fold increase in expression are depicted as red dots. b. Scatter plots comparing expression of known E2F-target genes between genotypes (control and Ah-cre samples); n=3 for each of the four samples. Red dots indicate genes whose expression increased >1.5-fold and blue dots indicate genes that decreased >1.5-fold. c. Quantitative real-time PCR was performed to compare the relative expression of selected E2F-target genes in control and Ah-cre crypts (left panels) and villi (right panels) using specific primers (Supplementary Fig. 20). d. Immunohistochemical staining of Mcm3 (green) and Pcna (red) in control and Ah-cre villi. DAPI (blue) was used for staining nuclei. Yellow dotted line outlines the luminal side of the villus; white dotted line outlines the outer side of the villus. Note that staining of blood cells in lumens of villi is non-specific. e. Chromatin immunoprecipitation (ChIP) assays using IgG or anti-E2F3 (α-3) antibodies with lysates from wild-type villi (Antibody control; top panels). ChIP assays using anti-E2F3 (α-3) antibodies with lysates from wild type (con) and Ah-cre (TKO) villi (Genotype control: bottom panels). Primers flanking known E2F-binding elements were used to detect the indicated gene promoters (Supplementary Fig. 19). f. Co-immunoprecipitation assays of cell extracts prepared from control villi and crypts. Imunoprecipitations (IP) used anti-E2F3 antibody or IgG. Anti-Rb antibody was used to probe Western blot (WB; left panel). The specificity of the anti-E2F3 antibody used in the left panel was evaluated in intestinal lysates derived from Ah-cre (3−/−), E2f3a−/− (3a−/−), E2f3b−/− (3b−/−) and E2f3+/+ mice. Anti-Rb antibody was used to probe Western blot (WB; right panel).
The genotype analysis compared gene expression in TKO vs. control samples of the same cell compartment. This comparison revealed a modest but significant downregulation of E2F targets in progenitor cells of TKO crypts, which included many but not all known classic targets such as Cdc6, Cyclin A2, Cyclin E2, Top2, and Hmgb2 (Fig. 3b–c, left panels, Supplementary Fig. 13a). We suspect that continued proliferation of TKO progenitors in the small intestine when E2F targets are limiting likely contributes to replicative stress, DNA damage and the observed increase in γ-H2AX labeling in these cells. Whether these aberrant processes are linked to the death of TKO progenitor cells remains to be rigorously evaluated. The genotype comparison also revealed a remarkable upregulation of a large number of E2F targets in differentiated cells of the TKO villus (Fig. 3b–c, right panels, Supplementary Fig. 13a). Western blot assays and IF staining showed that the accumulation of two of these E2F target gene products, Mcm3 and Pcna, was widespread throughout the TKO villus (Fig. 3d, Supplementary Fig. 13b). Similarly, there was increased expression of E2F targets in differentiated TKO cells of the retina and lens (Supplementary Fig. 13c; PW unpublished observations), suggesting a general role for E2F1-3 in transcriptional repression in post-mitotic cells in vivo. Chromatin immunoprecipitation (ChIP) assays using villus-enriched lysates derived from control and TKO small intestines showed that E2F3 occupies E2F binding sites on classic E2F-target promoters (Fig. 3e). Importantly, coimmunoprecipitation assays using intestinal epithelial cells derived from E2f3−/−, E2f3a−/− and E2f3b−/− villi showed that both E2F3a/b isoforms19–21 participate in a complex with the Rb protein (Fig. 3f). Consistent with this, Rb was found to be hypophosphorylated in the villus (Supplementary Fig. 14a). Together, these data suggest that E2F1-3 act as transcription activators in dividing progenitors, and as repressors (in complex with Rb) in differentiating cells of the small intestine.
On the surface, the observation that E2F1-3 repress E2F targets and are dispensable for cell proliferation contradict previous findings from the analysis of Rb/E2f double knockout animals1, 2, 8. Therefore, to thoroughly explore the mechanistic relationship between Rb and E2F1-3, we used the small intestine as an in vivo system where results could be uniformly compared across different genetic configurations. The Ah-cre mediated inactivation of Rb in utero or in adult mice resulted in increased proliferation of cells in the villus compartment but not in the crypt (Fig. 4a, 4b, Supplementary Fig. 14b–e), indicating that Rb-deleted transit-amplifying cells failed to appropriately exit the cell cycle. There was, however, no concomitant increase in apoptosis or defect in cell differentiation (Supplementary Fig. 15a, 15b), and as a result, Rb-deficient villi appeared uniformly hyperplastic. The combined ablation of the three E2fs completely suppressed the unscheduled proliferation and hyperplasia caused by Rb deficiency (QKO; Fig. 4a, 4b, Supplementary Fig. 16a). Importantly, the basal levels of proliferation in QKO crypts were indistinguishable from control or TKO samples (Supplementary Fig. 16b), consistent with the rather normal development of E2f1-3 deficient small intestines containing an intact Rb gene.
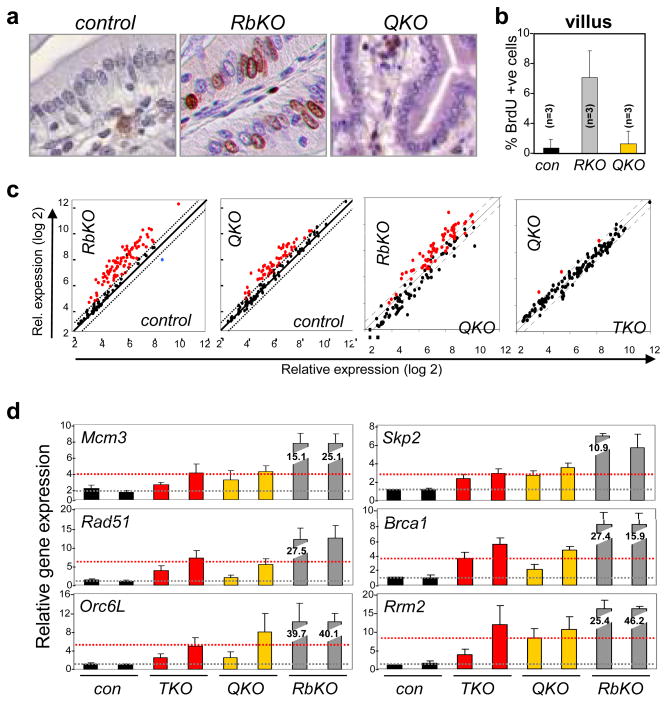
Figure 4. E2F1-3 contribute to the ectopic cell proliferation caused by Rb-deficiency.
a BrdU analysis was performed in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control), Ah-cre;RbLoxP/LoxP (RbKO) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP;RbLoxP/LoxP (QKO) small intestines. b. Quantification of BrdU incorporation. n=3, 3 different animals with the indicated genotypes were analyzed; error bars indicate standard deviation. c. Scatter plot analysis comparing differentially expressed E2F target genes in control, RbKO, Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (TKO) and QKO villi; n=3 for each of the eight samples. Red dots indicate genes whose expression increased >1.5-fold and blue dots indicate gene that decreased >1.5-fold. d. Quantitative RT-PCR analysis of selected E2F-target genes in control (con), RbKO, TKO, and QKO villi. The normal basal level of E2F target expression is illustrated as a grey dotted line and the threshold level of E2F target expression required for ectopic proliferation is illustrated as a red dotted line.
The selective requirement for E2f1-3 in the proliferation of Rb-deficient cells provided an opportunity to dissect possible cancer-specific mechanisms of E2F in cell cycle control. We therefore compared global gene expression programs in control, RbKO, TKO and QKO intestinal epithelia. Several important insights came from this analysis. First, there were expansive gene expression differences between control and RbKO villi (1290 upregulated and 487 downregulated genes; Fig. 4c, Supplementary Table 2), but relatively minor differences in their associated crypts (Supplementary Fig. 17a, 17b). Gene Ontology algorithms22 identified a bias for differentially expressed genes involved in the regulation of transcription, DNA metabolic processes and cell cycle (Supplementary Table 3). IF and quantitative RT-PCR assays confirmed the dramatic accumulation of most E2F-target genes in RbKO villi (Supplementary Fig. 17c–d). From these data we conclude that Rb is critical for the repression of E2F targets at a time when progenitor cells commit to exit the cell cycle and terminally differentiate. Second, hierarchical clustering of all data sets showed that TKO and QKO tissues clustered together in a separate group from control and RbKO tissues (Supplementary Fig. 18), suggesting that some functions coordinated by E2F1-3 may be Rb-independent. Finally and most importantly, the expression levels of E2F targets in TKO and QKO villi were equivalent, and while higher than in control villi, they were substantially lower than in RbKO villi (Fig. 4c). Quantitative RT PCR assays confirmed the relative expression of E2F targets to be: control < TKO = QKO ≪ RbKO (Fig. 4d). From these data, we conclude that the supra-elevated expression of E2F targets observed in RbKO villi is due to both ‘derepression’ (lacking intact Rb/E2F1-3 repressor complexes) and E2F1-3 mediated ‘hyper-activation’. In the absence of E2F1-3 mediated hyper-activation, cells in QKO villi fail to hyper-activate and thus do not accumulate sufficient levels of E2F targets to undergo ‘ectopic’ cell proliferation (this threshold level of expression is illustrated as a red dotted line in Fig. 4d).
We provide overwhelming evidence showing that normal cell proliferation in mice can be maintained in the absence of activator E2Fs. We conclude that E2F1-3, like G1 Cdk’s23–25, are not as critical for normal cell proliferation in mammals as original studies implied3, 4, 26–29. However, not all is well in the absence of E2F1-3, since TKO dividing progenitors in the small intestine undergo apoptosis. A prosurvival role for E2F1-3 was also evident in retinal progenitor cells of the mouse (Chen et al., accompanying paper), however, in the retina cell death was p53 dependent whereas in the small intestine it was p53 independent. Thus, the sensitivity of cells to p53 activation varies considerably across tissue types.
The findings presented here also expose dual functions for E2F1-3 in transcription activation and repression in vivo. In dividing progenitor cells, when Rb is inactive (hyperphosphorylated), free E2F1-3 are employed to optimally activate the expression of target genes. The inability to do so in E2f1-3 deficient tissues still permits cells to replicate their DNA and divide, but at the cost of increased DNA damage and cell death. As cells commit to a terminally differentiated fate, P-Rb is dephosphorylated and forms a physical complex with E2F1-3 proteins. We propose that this is not to just sequester E2F activators but rather, to form the first repressive complex that is necessary to downregulate E2F targets and usher transit-amplifying cells out of the cell cycle. Once cells exit the cell cycle, other professional repressor complexes accumulate, including p130/E2F4 and p107/E2F4, to more permanently enforce the repression of E2F targets. Given that inactivation of Rb, but not p107 or p13030, induces ectopic cell divisions in the small intestine, we suggest that Rb has a unique role in transit-amplifying cells that is dependent on its ability to associate with E2F1-3. Maintenance of quiescence in terminally differentiated cells of the villus, however, is a function that is shared among all members of the Rb family30. In summary, this work challenges the current paradigm of cell cycle control and provides, for the first time, a unified molecular view of how the dual functions of E2F1-3 in transcriptional activation and repression are employed in vivo to control normal versus Rb-mutant or cancer cell cycles.
METHODS SUMMARY
Mice (E2f1−/−, E2f2−/−, E2f3f/f, Ah-cre and Rbf/f) used for the studies were in mixed background (129SvEv, C57BL/6NTac and FVB/NTac). β-napthoflavone (sigma; N3633-5G) was administered into 2 month old Ah-cre mice three times within 24 hours as described previously14 and mice were harvested 7 or 90 days later. β-napthoflavone was also injected into pregnant female mice at 15.5 days postcoitum for analysis of embryos at E18.5. Villus and crypt fractions were isolated as previously described8. Three independent samples from each genetic group were used for gene expression analysis by Affymetrix microarray. Analysis of gene expression data were performed using BRB-array tools developed by Dr. Richard Simon and Amy Peng Lam of the National Cancer Institute. Gene Ontologies were predicted by DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources at the National Institute of Allergy and Infectious diseases, NIH. X-gal staining, real-time RT-PCR, BrdU, ChIP and TUNEL assays were performed as previously described8, 19. Primers for ChIP, real-time RT-PCR and genotyping are listed in Supplementary Fig. 19a-b. Antibodies used for Western blot or immunohistochemical staining are listed in Supplementary Fig. 19c.
Supplementary Material
Supplementary Figure 1. Deletion of E2f1-3 does not impair ES cell growth or differentiation. a. PCR genotyping of multiple ES cell clones after electroporation with a cre-expressing plasmid driven by the elongation factor-1 promoter (EF1-cre). b. Expression levels of nanog, oct4, eomes and cdx2 were measured by quantitative RT-PCR in trophoblast stem (TS) cells and ES cell clones. Consistent with the pluripotency of ES cells derived from E2f1−/−;E2f2−/−;E2f3LoxP/− embryos, these cells expressed high levels of oct4 and nanog but low levels of cdx2 and eomes (TS cell markers). ES cells were derived from blastocysts at E3.5 from crosses between E2f1+/−; E2f2−/−; E2f3+/− and E2f1+/−; E2f2−/−; E2f3LoxP/LoxP mice. Colonies that grew from disaggregated blastocysts with ES cell-like morphology were subcultured for expansion and were genotyped by PCR for E2f1, E2f2, and E2f3. E2f1+/−; E2f2−/−; E2f3LoxP/− clones derived from two different embryos were expanded and electroporated with EF1-Cre. Cells were plated at low density after electroporation and colonies were collected for genotyping and further culture. Approximately 55% to 70% of the resulting clones underwent recombination of the conditional E2f3 allele without any form of selection. Finally, ES cells were checked for expression of ES cell-specific and trophoblast stem (TS) cell-specific markers to verify their identities.
Supplementary Figure 2. E2F3 expression and promoter-binding activity in ES cells. a. Real-time RT-PCR analysis of E2f1, E2f2, E2f3a, and E2f3b expression in wild type MEFs and ES cells (TC-1 line). b. The TC-1 wild type ES cell line expresses equivalent levels of E2F3 protein as detected by Western blots probed with anti-E2F3. E2f1-3 triply-deleted (TKO) ES cells are used as a negative control. c. Chromatin immunoprecipitation was done with anti-E2F3 antibodies, using control (−) or cre-treated (+) E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs (left panel) as well as DKO and TKO ES cells (right panel).
Supplementary Figure 3. Gene expression and E2F4/p130 mediated repression in DKO and TKO ES cells. a. Real-time RT-PCR analysis of p53 target gene expression in control (−) or cre-treated (+) E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs (left panel) as well as DKO and TKO ES cells (right panel). The binding of E2F4 (b) and p130 (c) to E2F target promoters in control (−) or cre-treated (+) E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs (left panel) as well as DKO and TKO ES cells (right panel) was detected by chromatin immunoprecipitation with anti-E2F4 and anti-p130 antibodies, respectively.
Supplementary Figure 4. E2f1, E2f2, and E2f3 are dispensable for teratoma formation. a. Pluripotency of ES cell lines was measured by scoring the number of teratomas containing defined cell lineages (neural, muscle, epithelial, skin and inflammatory) from a total number of teratomas that developed in mice injected with one wild type ES cell line (TC-1), two DKO ES cell lines, and two TKO ES cell lines. b. Growth rates of teratomas in (a) were calculated as the number of days required for teratomas to reach a volume of 200 mm3.
Supplementary Figure 5. Cell proliferation and apoptosis in TKO embryos. Ki67 (a) and TUNEL (c) staining of various organs from E9.5 E2f1+/+;E2f2−/−;E2f3+/+ (SKO), E2f1−/−;E2f2−/−;E2f3+/+ (DKO) and E2f1−/−;E2f2−/−;E2f3−/− (TKO) embryos. Quantification of Ki-67-positive (b) and TUNEL-positive (d) cells from a minimum of three embryos of each genotype (SKO, DKO and TKO); at least three sections for each embryo were analyzed. Error bars represent standard deviation.
Supplementary Figure 6. Detection of E2F1, E2F2 and E2F3 proteins in the small intestine. Western blots of protein lysates derived from wild type crypts and villi were probed with antibodies specific for E2F1, E2F2, and E2F3. Villi lysates derived from Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP small intestines (TKO) were used as a negative control; note that a small amount of spill-over occurred from the adjacent lane. Tubulin was used as a loading control.
Supplementary Figure 7. Expression of Ah-cre and inactivation of E2f3LoxP/LoxP in the small intestine. a. In situ X-Gal staining was used to examine the expression of cre-recombinase in the small intestines of Rosa26LoxP/LoxP (control) and Ah-cre;Rosa26LoxP/LoxP (Ah-cre) mice after one, two and three days following β-napthoflavone (β-NF) injection. Deletion of E2f3LoxP was measured by Southern blot (b) and PCR (c) of DNA isolated from small intestines with the indicated genotypes. d. Real-time RT-PCR was used to measure E2f4-8 expression in villi from control and TKO small intestines.
Supplementary Figure 8. Normal architecture of E2f1-3 deficient small intestines. a. H&E stained sections of small intestines from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) pups that were harvested 3 and 24 days following β-NF administration into pregnant females (at 15.5 d.p.c.). b. H&E stained sections of small intestines from control and Ah-cre mice harvested 7 and 90 days following the administration of β-NF into 2 month-old mice.
Supplementary Figure 9. Normal cell differentiation in E2f1-3 deficient small intestines. Antibodies specific against cholecystokinin (CCK, red), somatostatin (red), serotonin (red) and lysozyme (red) were used for identifying various differentiated endocrine cells in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) small intestines. DAPI (blue) is used for staining nuclei. Arrows point to positively-stained cells.
Supplementary Figure 10. Increased γ-H2AX staining in E2f1-3 deficient small intestines a. Immunohistochemical staining of γ-H2AX (red) and P-ATM1981 (red) in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) intestinal crypts. DAPI (blue) was used for staining nuclei. The arrows indicate positively-stained cells. b. Immunohistochemical staining of γ-H2AX (red) in wild type (control) and cre-treated E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (E2f1-3 TKO) retinas at various developmental stages indicated (E14, P0 and P18). c. Quantification of the γ-H2AX positive cells stained in (b).
Supplementary Figure 11. Loss of E2F1-3 in the small intestine causes cell death in crypts. a. Small intestine sections from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre (TKO)) mice injected with β-NF and harvested 7 days later were stained with antibodies for p53. b. TUNEL assays were performed on small intestine sections from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP;p53LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP;p53LoxP/LoxP (Ah-cre (QKO)) mice treated as in (a). c. Quantification of TUNEL-positive cells from (b). Three mice were analyzed for each genetic group. Error bars represent standard deviation.
Supplementary Figure 12. Heirarchical clustering analysis of differentially expressed genes (>1.5-fold and p<0.0001) in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) villi and crypts is presented as a heat map (a). Samples from three animals of each genetic group were used for the analysis. b. List of E2F target genes as defined by previous gene expression, reporter and ChIP assays17, 18.
Supplementary Figure 13. Expression of E2F-responsive genes is upregulated in E2f1-3 deficient small intestines and retinas. a. Quantitative real-time PCR was used to examine the relative expression of selected E2F-target genes in control and Ah-cre crypts (left panels) and villi (right panels) using specific primers (Supplementary Fig. 20). b. Examination of Mcm3 and Pcna protein levels in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) villi by Western blot. c. Relative quantitative real-time PCR of a panel of genes in wild type and cre-treated P14 E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (TKO) retinas. Expression in TKO samples was relative to control (wild type) samples.
Supplementary Figure 14. Loss of Rb in differentiating intestinal crypt cells (crypt) results in ectopic proliferation in villi. a. Cell extracts isolated from control crypts and villi were used to detect Rb protein in Western blots. RbKO, Rb knockout crypt; Rb-P, hyperphosphorylated Rb; Rb, hyporphosphorylated Rb. b. PCR based assay used to examine deletion of the RbLoxP allele from genomic DNA purified from small intestines of Rb+/+, RbLoxP/LoxP (−), Ah-cre;RbLoxP/LoxP (+) mice after 7 days of β-napthoflavone (β-NF) administration. c. Small intestines from β-NF-injected 2 month-old RbLoxP/LoxP (control) and Ah-cre;RbLoxP/LoxP (RbKO) mice were harvested 7 or 90 days later, sectioned and stained with H&E. d. BrdU analysis was performed in control and RbKO intestines harvested at embryonic day 18.5 (E18.5) and at 2 months of age (P60). For the embryonic analysis of the small intestine (E18.5), β-NF was injected into pregnant females at 15.5 d.p.c. Other details of β-NF injection and tissue harvesting are provided in the Methods section. e. Quantification of BrdU incorporation in villi and crypts of animals with the indicated genotypes and ages. n=3, 3 different animals with the indicated genotypes were analyzed. Error bars indicate standard deviation.
Supplementary Figure 15. Normal differentiation of Rb-deficient small intestines. a. Small intestine sections from RbLoxP/LoxP (control) and Ah-cre;RbLoxP/LoxP (RbKO) mice, which were injected with β-NF and harvested 7 days after, were used for TUNEL staining. b. Analysis of cell differentiation in control and RbKO small intestines. Alcian blue, goblet cells were identified by Alcian blue staining; other antibody-based markers for cholecystokinin (CCK, red), serotonin (red), somatostatin (red), lysozyme (red) and fatty acid binding protein (FABP, green) were used to identify endocrine cells, paneth cells, and absorptive cells, respectively. DAPI (blue) was used for staining nuclei. Arrows indicate positive-stained differentiated cells.
Supplementary Figure 16. Normal cell proliferation in Rb-deficient crypts of the small intestine. a. Sections of small intestines from RbLoxP/LoxP (control), Ah-cre;RbLoxP/LoxP (RbKO) and Ah-cre;RbLoxP/LoxP;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (QKO) mice that were harvested 7 days post-β-NF-injection were stained with H&E. b. BrdU analysis was performed in control, RbKO and QKO small intestines from mice treated as in (a). Quantification of BrdU incorporation in crypts of three different animals with the indicated genotypes. Error bars indicate standard deviation.
Supplementary Figure 17. Global gene expression analysis in the Rb-deficient small intestine. a. Heatmap representation of heirarchical clustering analysis of differentially expressed genes in crypts and villi of RbLoxP/LoxP (control) and Ah-cre;RbLoxP/LoxP (RbKO) mice (p<0.0001). Three independent samples from each genetic group and tissue (crypt/villus) were analyzed. b. Scatter plots comparing the expression of known E2F-target genes in control and RbKO crypt and villus. c. Expression of E2F-target genes in control and RbKO villi was examined by Real-time RT-PCR. The two bars for each set represents two independent samples of that genotype. Error bars indicate standard deviation from triplicate samples. d. Immunohistochemical staining of Mcm3 (green) and Pcna (red) in control, RbKO and QKO villi. DAPI (blue) was used for staining nuclei. Yellow dotted line outlines the luminal side of the villus; white dotted line outlines the outer side of the villus. Note that staining of blood cells in lumens of villi is non-specific.
Supplementary Figure 18. Hierarchical clustering analysis of differentially expressed genes in control, RbKO (RKO), TKO, QKO villi (p<0.0001). Note that wild type (CON) and RKO samples segregate from TKO and QKO villi samples; a similar separation of genetic groups was achieved when the associated crypts were analyzed in the same manner.
Supplementary Figure 19. List of Primers used for genotyping (a) and ChIP assays (b). c. List of antibodies used in this study.
Supplementary Figure 20. List of Primers used for real-time RT-PCR analysis of gene expression.
Acknowledgments
We thank L. Rawahneh and J. Moffitt and R. Rajmohan for excellent technical assistance with histology. We also thank A. de Bruin and S. Naidu for assistance in analyzing histological slides. We are thankful to Drs. Joanna Groden, Mandy Simcox and Denis Guttridge for their critical comments. This work was funded by NIH grants to G.L. (R01CA85619, R01CA82259, R01HD04470, P01CA097189) and NIH grant to J.M.P. (CA098956); J.-L.C. is the recipient of a DoD award (BC061730). P.L.W. was supported by NIH training grant 5 T32 CA106196-04.
Footnotes
Author Contributions. M.L.R., J.M.P. and G.L. designed and supervised this study, analyzed data, and helped write and edit the manuscript. J-L.C., P.L.W. and M.T.S. designed and performed experiments, collected and analyzed data, and co-wrote the paper. V.N., A.F., Y.M.G., N.S., H-Z.C., M.O., S-H.W., P.T., B.C. and L.M. technically assisted with experiments and collected and analyzed data. D.C. and R.B. performed and analyzed gene expression of retina. J.P.H. and P.G.C. contributed to the analysis and comparison of gene microarray data. D.J.W. and O.J.S. contributed to the generation of key reagents.
Author information. All microarray data has been deposited to the Gene Expression Omnibus at the National Center for Biotechnology Information under accession numbers GSE16454. Reprints and permissions information is available at ww.nature.com/reprints.
References
- 1.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 3.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 6.Timmers C, et al. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27:65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma N, et al. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281:36124–36131. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 8.Saenz-Robles MT, et al. Intestinal hyperplasia induced by simian virus 40 large tumor antigen requires E2F2. J Virol. 2007;81:13191–13199. doi: 10.1128/JVI.01658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell. 2006;127:871–874. doi: 10.1016/j.cell.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Murga M, et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- 11.Iglesias A, et al. Diabetes and exocrine pancreatic insufficiency in E2F1/E2F2 double-mutant mice. J Clin Invest. 2004;113:1398–1407. doi: 10.1172/JCI18879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloud JE, et al. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol Cell Biol. 2002;22:2663–2672. doi: 10.1128/MCB.22.8.2663-2672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Flier LG, Clevers H. Stem Cells, Self-Renewal, and Differentiation in the Intestinal Epithelium. Annu Rev Physiol. 2008 doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 14.Ireland H, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–1246. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Coopersmith CM, Gordon J. I gamma-Ray-induced apoptosis in transgenic mice with proliferative abnormalities in their intestinal epithelium: re-entry of villus enterocytes into the cell cycle does not affect their radioresistance but enhances the radiosensitivity of the crypt by inducing p53. Oncogene. 1997;15:131–141. doi: 10.1038/sj.onc.1201176. [DOI] [PubMed] [Google Scholar]
- 16.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 17.Kong LJ, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, et al. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong JL, et al. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol Cell Biol. 2009;29:414–424. doi: 10.1128/MCB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone G, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leone G, et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Martin A, et al. Cdk2 is dispensable for cell cycle inhibition and tumor suppression mediated by p27(Kip1) and p21(Cip1) Cancer Cell. 2005;7:591–598. doi: 10.1016/j.ccr.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Malumbres M, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Russell P, Nurse P. Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell. 1986;45:781–782. doi: 10.1016/0092-8674(86)90550-7. [DOI] [PubMed] [Google Scholar]
- 27.Helin K, et al. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 28.Kaelin WG, Jr, et al. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 29.Nevins JR. Transcriptional regulation. A closer look at E2F. Nature. 1992;358:375–376. doi: 10.1038/358375a0. [DOI] [PubMed] [Google Scholar]
- 30.Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Deletion of E2f1-3 does not impair ES cell growth or differentiation. a. PCR genotyping of multiple ES cell clones after electroporation with a cre-expressing plasmid driven by the elongation factor-1 promoter (EF1-cre). b. Expression levels of nanog, oct4, eomes and cdx2 were measured by quantitative RT-PCR in trophoblast stem (TS) cells and ES cell clones. Consistent with the pluripotency of ES cells derived from E2f1−/−;E2f2−/−;E2f3LoxP/− embryos, these cells expressed high levels of oct4 and nanog but low levels of cdx2 and eomes (TS cell markers). ES cells were derived from blastocysts at E3.5 from crosses between E2f1+/−; E2f2−/−; E2f3+/− and E2f1+/−; E2f2−/−; E2f3LoxP/LoxP mice. Colonies that grew from disaggregated blastocysts with ES cell-like morphology were subcultured for expansion and were genotyped by PCR for E2f1, E2f2, and E2f3. E2f1+/−; E2f2−/−; E2f3LoxP/− clones derived from two different embryos were expanded and electroporated with EF1-Cre. Cells were plated at low density after electroporation and colonies were collected for genotyping and further culture. Approximately 55% to 70% of the resulting clones underwent recombination of the conditional E2f3 allele without any form of selection. Finally, ES cells were checked for expression of ES cell-specific and trophoblast stem (TS) cell-specific markers to verify their identities.
Supplementary Figure 2. E2F3 expression and promoter-binding activity in ES cells. a. Real-time RT-PCR analysis of E2f1, E2f2, E2f3a, and E2f3b expression in wild type MEFs and ES cells (TC-1 line). b. The TC-1 wild type ES cell line expresses equivalent levels of E2F3 protein as detected by Western blots probed with anti-E2F3. E2f1-3 triply-deleted (TKO) ES cells are used as a negative control. c. Chromatin immunoprecipitation was done with anti-E2F3 antibodies, using control (−) or cre-treated (+) E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs (left panel) as well as DKO and TKO ES cells (right panel).
Supplementary Figure 3. Gene expression and E2F4/p130 mediated repression in DKO and TKO ES cells. a. Real-time RT-PCR analysis of p53 target gene expression in control (−) or cre-treated (+) E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs (left panel) as well as DKO and TKO ES cells (right panel). The binding of E2F4 (b) and p130 (c) to E2F target promoters in control (−) or cre-treated (+) E2f1−/−;E2f2−/−;E2f3LoxP/LoxP MEFs (left panel) as well as DKO and TKO ES cells (right panel) was detected by chromatin immunoprecipitation with anti-E2F4 and anti-p130 antibodies, respectively.
Supplementary Figure 4. E2f1, E2f2, and E2f3 are dispensable for teratoma formation. a. Pluripotency of ES cell lines was measured by scoring the number of teratomas containing defined cell lineages (neural, muscle, epithelial, skin and inflammatory) from a total number of teratomas that developed in mice injected with one wild type ES cell line (TC-1), two DKO ES cell lines, and two TKO ES cell lines. b. Growth rates of teratomas in (a) were calculated as the number of days required for teratomas to reach a volume of 200 mm3.
Supplementary Figure 5. Cell proliferation and apoptosis in TKO embryos. Ki67 (a) and TUNEL (c) staining of various organs from E9.5 E2f1+/+;E2f2−/−;E2f3+/+ (SKO), E2f1−/−;E2f2−/−;E2f3+/+ (DKO) and E2f1−/−;E2f2−/−;E2f3−/− (TKO) embryos. Quantification of Ki-67-positive (b) and TUNEL-positive (d) cells from a minimum of three embryos of each genotype (SKO, DKO and TKO); at least three sections for each embryo were analyzed. Error bars represent standard deviation.
Supplementary Figure 6. Detection of E2F1, E2F2 and E2F3 proteins in the small intestine. Western blots of protein lysates derived from wild type crypts and villi were probed with antibodies specific for E2F1, E2F2, and E2F3. Villi lysates derived from Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP small intestines (TKO) were used as a negative control; note that a small amount of spill-over occurred from the adjacent lane. Tubulin was used as a loading control.
Supplementary Figure 7. Expression of Ah-cre and inactivation of E2f3LoxP/LoxP in the small intestine. a. In situ X-Gal staining was used to examine the expression of cre-recombinase in the small intestines of Rosa26LoxP/LoxP (control) and Ah-cre;Rosa26LoxP/LoxP (Ah-cre) mice after one, two and three days following β-napthoflavone (β-NF) injection. Deletion of E2f3LoxP was measured by Southern blot (b) and PCR (c) of DNA isolated from small intestines with the indicated genotypes. d. Real-time RT-PCR was used to measure E2f4-8 expression in villi from control and TKO small intestines.
Supplementary Figure 8. Normal architecture of E2f1-3 deficient small intestines. a. H&E stained sections of small intestines from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) pups that were harvested 3 and 24 days following β-NF administration into pregnant females (at 15.5 d.p.c.). b. H&E stained sections of small intestines from control and Ah-cre mice harvested 7 and 90 days following the administration of β-NF into 2 month-old mice.
Supplementary Figure 9. Normal cell differentiation in E2f1-3 deficient small intestines. Antibodies specific against cholecystokinin (CCK, red), somatostatin (red), serotonin (red) and lysozyme (red) were used for identifying various differentiated endocrine cells in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) small intestines. DAPI (blue) is used for staining nuclei. Arrows point to positively-stained cells.
Supplementary Figure 10. Increased γ-H2AX staining in E2f1-3 deficient small intestines a. Immunohistochemical staining of γ-H2AX (red) and P-ATM1981 (red) in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) intestinal crypts. DAPI (blue) was used for staining nuclei. The arrows indicate positively-stained cells. b. Immunohistochemical staining of γ-H2AX (red) in wild type (control) and cre-treated E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (E2f1-3 TKO) retinas at various developmental stages indicated (E14, P0 and P18). c. Quantification of the γ-H2AX positive cells stained in (b).
Supplementary Figure 11. Loss of E2F1-3 in the small intestine causes cell death in crypts. a. Small intestine sections from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre (TKO)) mice injected with β-NF and harvested 7 days later were stained with antibodies for p53. b. TUNEL assays were performed on small intestine sections from E2f1−/−;E2f2−/−;E2f3LoxP/LoxP;p53LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP;p53LoxP/LoxP (Ah-cre (QKO)) mice treated as in (a). c. Quantification of TUNEL-positive cells from (b). Three mice were analyzed for each genetic group. Error bars represent standard deviation.
Supplementary Figure 12. Heirarchical clustering analysis of differentially expressed genes (>1.5-fold and p<0.0001) in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) villi and crypts is presented as a heat map (a). Samples from three animals of each genetic group were used for the analysis. b. List of E2F target genes as defined by previous gene expression, reporter and ChIP assays17, 18.
Supplementary Figure 13. Expression of E2F-responsive genes is upregulated in E2f1-3 deficient small intestines and retinas. a. Quantitative real-time PCR was used to examine the relative expression of selected E2F-target genes in control and Ah-cre crypts (left panels) and villi (right panels) using specific primers (Supplementary Fig. 20). b. Examination of Mcm3 and Pcna protein levels in E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (control) and Ah-cre;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (Ah-cre) villi by Western blot. c. Relative quantitative real-time PCR of a panel of genes in wild type and cre-treated P14 E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (TKO) retinas. Expression in TKO samples was relative to control (wild type) samples.
Supplementary Figure 14. Loss of Rb in differentiating intestinal crypt cells (crypt) results in ectopic proliferation in villi. a. Cell extracts isolated from control crypts and villi were used to detect Rb protein in Western blots. RbKO, Rb knockout crypt; Rb-P, hyperphosphorylated Rb; Rb, hyporphosphorylated Rb. b. PCR based assay used to examine deletion of the RbLoxP allele from genomic DNA purified from small intestines of Rb+/+, RbLoxP/LoxP (−), Ah-cre;RbLoxP/LoxP (+) mice after 7 days of β-napthoflavone (β-NF) administration. c. Small intestines from β-NF-injected 2 month-old RbLoxP/LoxP (control) and Ah-cre;RbLoxP/LoxP (RbKO) mice were harvested 7 or 90 days later, sectioned and stained with H&E. d. BrdU analysis was performed in control and RbKO intestines harvested at embryonic day 18.5 (E18.5) and at 2 months of age (P60). For the embryonic analysis of the small intestine (E18.5), β-NF was injected into pregnant females at 15.5 d.p.c. Other details of β-NF injection and tissue harvesting are provided in the Methods section. e. Quantification of BrdU incorporation in villi and crypts of animals with the indicated genotypes and ages. n=3, 3 different animals with the indicated genotypes were analyzed. Error bars indicate standard deviation.
Supplementary Figure 15. Normal differentiation of Rb-deficient small intestines. a. Small intestine sections from RbLoxP/LoxP (control) and Ah-cre;RbLoxP/LoxP (RbKO) mice, which were injected with β-NF and harvested 7 days after, were used for TUNEL staining. b. Analysis of cell differentiation in control and RbKO small intestines. Alcian blue, goblet cells were identified by Alcian blue staining; other antibody-based markers for cholecystokinin (CCK, red), serotonin (red), somatostatin (red), lysozyme (red) and fatty acid binding protein (FABP, green) were used to identify endocrine cells, paneth cells, and absorptive cells, respectively. DAPI (blue) was used for staining nuclei. Arrows indicate positive-stained differentiated cells.
Supplementary Figure 16. Normal cell proliferation in Rb-deficient crypts of the small intestine. a. Sections of small intestines from RbLoxP/LoxP (control), Ah-cre;RbLoxP/LoxP (RbKO) and Ah-cre;RbLoxP/LoxP;E2f1−/−;E2f2−/−;E2f3LoxP/LoxP (QKO) mice that were harvested 7 days post-β-NF-injection were stained with H&E. b. BrdU analysis was performed in control, RbKO and QKO small intestines from mice treated as in (a). Quantification of BrdU incorporation in crypts of three different animals with the indicated genotypes. Error bars indicate standard deviation.
Supplementary Figure 17. Global gene expression analysis in the Rb-deficient small intestine. a. Heatmap representation of heirarchical clustering analysis of differentially expressed genes in crypts and villi of RbLoxP/LoxP (control) and Ah-cre;RbLoxP/LoxP (RbKO) mice (p<0.0001). Three independent samples from each genetic group and tissue (crypt/villus) were analyzed. b. Scatter plots comparing the expression of known E2F-target genes in control and RbKO crypt and villus. c. Expression of E2F-target genes in control and RbKO villi was examined by Real-time RT-PCR. The two bars for each set represents two independent samples of that genotype. Error bars indicate standard deviation from triplicate samples. d. Immunohistochemical staining of Mcm3 (green) and Pcna (red) in control, RbKO and QKO villi. DAPI (blue) was used for staining nuclei. Yellow dotted line outlines the luminal side of the villus; white dotted line outlines the outer side of the villus. Note that staining of blood cells in lumens of villi is non-specific.
Supplementary Figure 18. Hierarchical clustering analysis of differentially expressed genes in control, RbKO (RKO), TKO, QKO villi (p<0.0001). Note that wild type (CON) and RKO samples segregate from TKO and QKO villi samples; a similar separation of genetic groups was achieved when the associated crypts were analyzed in the same manner.
Supplementary Figure 19. List of Primers used for genotyping (a) and ChIP assays (b). c. List of antibodies used in this study.
Supplementary Figure 20. List of Primers used for real-time RT-PCR analysis of gene expression.