Abstract
Summary: The emergence of vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) over the past decade has provided a challenge to diagnostic microbiologists to detect these strains, clinicians treating patients with infections due to these strains, and researchers attempting to understand the resistance mechanisms. Recent data show that these strains have been detected globally and in many cases are associated with glycopeptide treatment failure; however, more rigorous clinical studies are required to clearly define the contribution of hVISA to glycopeptide treatment outcomes. It is now becoming clear that sequential point mutations in key global regulatory genes contribute to the hVISA and VISA phenotypes, which are associated predominately with cell wall thickening and restricted vancomycin access to its site of activity in the division septum; however, the phenotypic features of these strains can vary because the mutations leading to resistance can vary. Interestingly, changes in the staphylococcal surface and expression of agr are likely to impact host-pathogen interactions in hVISA and VISA infections. Given the subtleties of vancomycin susceptibility testing against S. aureus, it is imperative that diagnostic laboratories use well-standardized methods and have a framework for detecting reduced vancomycin susceptibility in S. aureus.
INTRODUCTION
Staphylococcus aureus has been recognized as an important cause of human disease for more than 100 years (186). Alexander Ogston first isolated Staphylococcus aureus from a surgical abscess in 1880 and described the role of S. aureus in localized infection and septicemia, including the use of animal models of infection (233, 325). Staphylococcus aureus is recognized as a cause of a wide range of infections, from minor skin infections and chronic bone infections to devastating septicemia and endocarditis (44, 45, 48, 63, 99, 124, 179, 207, 218, 277, 308-311). The history of S. aureus is one of evolution and change. The acquisition of antimicrobial resistance and changing patterns of staphylococcal disease have been common themes in the staphylococcal literature over the past 50 years (25). Significant events in the evolution of S. aureus have included the development of methicillin resistance, now a problem for many hospitals around the world, and the recent emergence of community strains of S. aureus that are methicillin resistant but also harbor genes associated with increased virulence (42, 43, 365). Methicillin-resistant S. aureus (MRSA) alone (which probably accounts for fewer than one-third of all S. aureus infections) caused more deaths in the United States in 2005 than human immunodeficiency virus infection (estimated MRSA mortality rate in 2005 of 6.3 per 100,000 individuals) and caused more invasive infections (estimated MRSA incidence in 2005 of 31.8 per 100,000 individuals) than other important bacterial pathogens such as Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis (36, 159).
The glycopeptide antibiotic vancomycin was first released in 1958. Subsequently, vancomycin has been the treatment of choice for serious infections caused by MRSA, which are becoming increasingly common globally. For many years there was no indication that vancomycin resistance in S. aureus was likely to be a problem. Therefore, initial reports of reduced vancomycin susceptibility in clinical isolates of S. aureus from Japan in 1997 generated significant concern in the medical community (114, 115). Since that time there has been uncertainty regarding optimal laboratory detection and the clinical relevance of reduced vancomycin susceptibility in S. aureus, changes in Clinical and Laboratory Standards Institute (CLSI) breakpoints for vancomycin against S. aureus, and increasing concern regarding the efficacy of vancomycin for the treatment of S. aureus infections. Reduced teicoplanin susceptibility in S. aureus was reported prior to the first reports of clinical S. aureus isolates from Japan with reduced vancomycin susceptibility (189). Both the terms glycopeptide-intermediate S. aureus (GISA) and vancomycin-intermediate S. aureus (VISA) have been used in the literature and in essence are interchangeable. However, it is clear that reduced teicoplanin susceptibility can be present in S. aureus without a clearly demonstrated reduction in vancomycin susceptibility (34, 113), whereas generally, VISA strains have demonstrated reduced teicoplanin susceptibility (183). Because the majority of in vitro susceptibility testing uses vancomycin, and much of the literature uses the term vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA), this review will use this terminology.
Significant controversy still exists regarding the current and future roles of vancomycin in the treatment of serious MRSA infections. A resolution of this controversy requires a detailed understanding of the mechanisms and clinical impact of changes in vancomycin susceptibility in S. aureus. This review will summarize current knowledge regarding the mechanisms and clinical impact of reduced vancomycin susceptibility in S. aureus.
Vancomycin-Resistant Staphylococcus aureus
After the emergence of vancomycin-resistant enterococci in the 1980s, significant concern existed with regard the potential for large outbreaks of vancomycin-resistant S. aureus (VRSA) due to the acquisition of the vanA gene from enterococci (172, 363). Fully vancomycin-resistant strains of S. aureus (VRSA) due to the acquisition of the vanA gene from vancomycin-resistant enterococci were first reported from the United States in 2002 (38, 47). However, to date, only nine cases of VRSA have been reported from the United States, with two additional cases, one from India and one from Iran; however, the genetics of resistance have not been verified in a second laboratory in these cases (3, 84, 247, 292, 323). This indicates that although this mechanism of resistance is significant, it is not evolving or spreading rapidly. Therefore, this review will focus on the more common and controversial area of reduced vancomycin susceptibility in S. aureus not related to the acquisition of vanA.
Understanding Vancomycin Resistance: the Staphylococcal Cell Wall
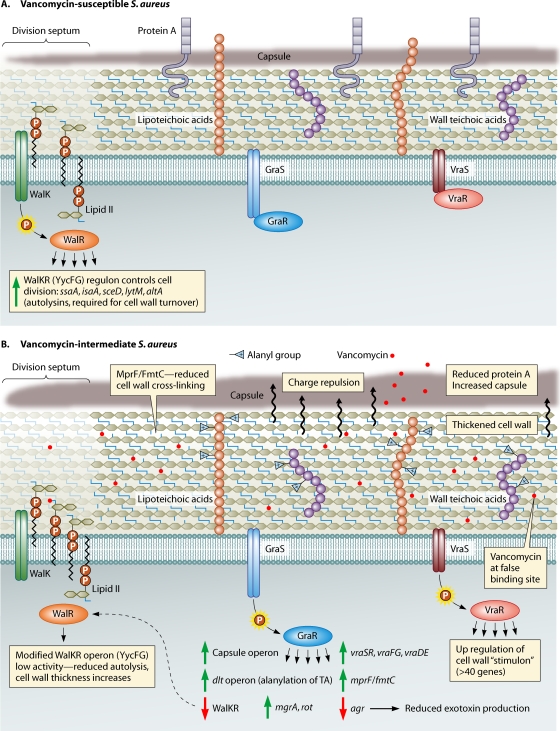
To understand the mechanisms and potential impacts of vancomycin resistance in S. aureus, a clear understanding of the organism's cell wall is required. The staphylococcal cell wall is a dynamic structure important for maintaining cell integrity and critical in host-pathogen interactions (351). The outermost surface of S. aureus is usually covered by a polysaccharide capsule (Fig. 1). Under the capsule lies the cell wall, a structure composed of highly cross-linked peptidoglycan (PG) (a complex structure composed of sugars and amino acids, also called murein), teichoic acids, and cell wall-associated proteins (72). The peptidoglycan is composed of glycan chains made up of the alternating amino sugars N-acetylglucosamine and N-acetylmuramic acid. Stem pentapeptides (l-Ala-d-iso-Gln-l-Lys-d-Ala-d-Ala) are attached to the carboxyl group of each N-acetylmuramic acid, and interpeptide bridges (pentaglycines, made up of glycine residues) connect the lysine component of one stem peptide to the penultimate d-alanine of a neighboring stem peptide (351). Teichoic acid chains are attached to the 6-hydroxyl groups of some of the N-acetylmuramic acid residues of the glycan chains and, together with the peptidoglycan, form a multilayered network that surrounds the S. aureus cell (161, 351). The stress-bearing murein therefore represents a continuous macromolecule encasing the sacculus (72). Typically, the degree of murein cross-linking in the S. aureus cell wall is high, with bridged peptides as a ratio of all peptide ends in the order of 80 to 90% (72, 328). The peptidoglycan composition from different S. aureus strains is highly conserved, with almost identical high-performance liquid chromatography (HPLC) muropeptide patterns across strains, suggesting that the composition is species specific (351).
FIG. 1.
Overview of some general cell wall characteristics of VSSA and VISA strains showing the key regulatory elements linked to intermediate-level vancomycin resistance, as uncovered by comparative genomics and genetic studies. (A) VSSA strain in the absence of vancomycin showing a normal peptidoglycan (PG) layer, with the production of protein A and normal capsular polysaccharide expression. Also shown is the division septum, where cell wall growth occurs. The lipid II-linked PG precursors assemble at the division septa, and the dipeptide moiety of lipid II is the lethal target of vancomycin. (B) VISA strain with mutations in either the graRS, vraSR, or walKR operon (or all) that might lead to their respective regulons remaining in an activated “locked-on” or otherwise modified state. The consequence of this modification includes cell wall thickening, decreased autolysis, reduced protein A production, increased capsule expression, increased d-alanylation of teichoic acids, and reduced agr activity.
By electron microscopy, the cell wall of S. aureus appears as a thick (20- to 40-nm-thick) homogeneous structure (72). The actual orientation of the glycan chains within the cell wall is uncertain. Early models suggested that the glycan strands were arranged in shell-like, parallel structures around the cell (72, 170). A recent model suggests that the glycan and oligopeptide chains are in fact perpendicular to the plasma membrane, with oligopeptide chains adopting a zigzag conformation to connect adjacent glycan strands (72).
The staphylococcal cell wall also contains teichoic acids, which represent up to 50% of the dry weight of the purified staphylococcal cell wall (214). Ribitol teichoic acids (or wall teichoic acids [WTAs]) are covalently linked to peptidoglycan and decorated with d-alanine and N-acetylglucosamine residues (379). Lipoteichoic acids (LTAs) are glycerol phosphate polymers linked to a glycolipid terminus in the cytoplasmic membrane (186). The functions of WTAs and LTAs are still being elucidated, with the recent generation of defined mutants, strains producing reduced amounts of teichoic acids, and strains producing altered teichoic acids providing significant insight into their functional roles (68, 78, 163, 248, 377, 381). It appears that many teichoic acid functions may be nonessential and may possibly involve indirect interactions with other cell wall components (379); however, the complete loss of LTA leads to cell death in S. aureus (104). Lipoteichoic acids appear to be involved in cell division (379). Some data suggest that the teichoic acids have a role to help protect the cell envelope as a mechanical barrier to host defense molecules and antibiotics, and also, the positive charge of d-alanine residues repels positively charged molecules such as defensins (54, 248, 249). Wall teichoic acids also contribute to lysozyme resistance in S. aureus by preventing lysozyme binding to peptidoglycan (19). The dltABCD operon is controlled by the regulator GraRS (also called ApsRS), which senses and responds to defensins and other antimicrobial peptides and regulates the alanylation of teichoic acids in response to the presence of antimicrobial compounds, indicating that the structure of teichoic acids can change in response to challenges (111, 180, 181). Wall teichoic acids also have a role in attachment to host cells, with studies demonstrating reduced nasal colonization and reduced binding to endothelial cells in strains deficient in WTA and in strains with reduced dltABCD-mediated alanylation of teichoic acids (378, 380, 381).
An understanding of the genetic determinants and enzymatic control of cell wall biosynthesis in S. aureus has been difficult. Many studies have used randomly generated mutants of methicillin-resistant S. aureus (MRSA) strains that demonstrate a reduction in the methicillin MIC, followed by biochemical analysis, to define the genes linked to cell wall biosynthesis (65, 66, 164, 217). A large number of genes appear to be involved in staphylococcal cell wall precursor production (66). Important genes include the femA, femB, femC, and femX genes; genes encoding the penicillin binding proteins (PBPs) (pbpA, pbpB, pbpC, and pbpD); and regulatory genes involved in cell wall biosynthesis, such as vraSR (351). The femA, femB, femC, and femX genes are involved in the stepwise synthesis of the pentaglycine bridge that attaches to the lysine residue of the stem peptide and are essential for bacterial survival (20, 278-280). The PBPs also have an important role in cell wall synthesis. In particular, high-molecular-weight PBPs (PBP1, PBP2, and PBP3) have a transglycosidase function (to link N-acetylglucosamine to N-acetylmuramic acid) and a transpeptidase function to link the penultimate d-Ala to a glycine acceptor in the nascent cell wall (214, 256). There is significant interest in the PBPs because of their relevance to antimicrobial therapy (they are the target site for beta-lactam antibiotics) and to antimicrobial resistance (PBP2a, encoded by mecA, is responsible for methicillin resistance in S. aureus) (256). Despite this interest, it has been difficult to assign specific functions to each PBP. PBP1 is essential but does not appear to play an important role in the cross-linking of peptidoglycan. It does, however, appear to be important for cell division (246, 371). PBP2 also plays an essential role in bacterial growth and survival, and the protein has a transpeptidase (TPase) and transglycosylase (TGase) domain (351). PBP3 does not appear to be essential; however, mutants demonstrate altered autolytic activity (254). PBP4 is the only low-molecular-weight PBP in S. aureus; however, it does possess transpeptidase and carboxypeptidase activities (205). Although initial studies showed a minimal impact of the deletion of the gene encoding PBP4 on cell growth and methicillin resistance (351), it was recently demonstrated that PBP4 may play an important role in the expression of methicillin resistance in community-associated MRSA strains (205).
Recently, a core and accessory set of cell wall-associated genes (the “cell wall stimulon”) has been described based on the results of microarray transcriptional analysis experiments after the exposure of S. aureus to cell wall-active agents (92, 168, 329, 361). These genes are predominately under the control of the two-component regulatory (2CR) system vraSR. These cell wall synthesis genes are summarized in Table 1 .
TABLE 1.
Genes involved in cell wall biosynthesis in S. aureusa
| ORF | Gene | Function |
|---|---|---|
| Core genes | ||
| SACOL1066 | fmt | Autolysis and methicillin resistance-related protein |
| SACOL1777 | htrA | Putative serine protease |
| SACOL1897 | prsA | Putative protein export protein |
| SACOL1932 | sgtB | Transglycosylase domain protein |
| SACOL1943 | vraS | Sensor histidine kinase |
| SACOL1944 | Conserved hypothetical protein | |
| SACOL1945 | Conserved hypothetical protein | |
| SACOL1956 | Conserved hypothetical protein | |
| SACOL2116 | murZ | UDP-N-Acetylglucosamine 1-carboxylvinyl transferase 2 |
| SACOL2302 | lytR | Transcriptional regulator, putative |
| SACOL2352 | tcaA | Teicoplanin resistance-associated protein |
| SACOL2435 | Conserved hypothetical protein | |
| SACOL2436 | Conserved hypothetical protein | |
| SACOL2518 | Conserved hypothetical protein | |
| SACOL2571 | Conserved hypothetical protein | |
| Additional genes | ||
| SACOL0033 | mecA | Penicillin binding protein 2A |
| SACOL0636 | mvaK1 | Melavonate kinase |
| SACOL0693 | tagA | Putative teichoic acid biosynthesis protein |
| SACOL0743 | bacA | Bacitracin resistance protein |
| SACOL1161 | murI | Glutamate racemase |
| SACOL1279 | upps | Undecaprenyl pyrophosphatase synthetase |
| SACOL1396 | fmtC | Autolysis and methicillin-resistant-related protein |
| SACOL1490 | pbpB | Penicillin binding protein 2 |
| SACOL2116 | murZ | UDP-N-Acetylglucosamineenolpyruvate transferase |
| SACOL2540 | srtA | Sortase A |
| SACOL1194 | pbpA | Penicillin binding protein 1 |
| SACOL1932 | sgtB | Monofunctional glycosyltransferase |
Mechanism of Vancomycin Action
After the emergence of methicillin resistance in S. aureus in the 1960s, the glycopeptides, particularly vancomycin, became the mainstay of therapy for serious MRSA infections. Vancomycin was discovered by Eli Lilly in the 1950s, after a missionary who was visiting Borneo sent a sample of dirt to a colleague who subsequently isolated the organism Amycolatopsis orientalis (previously designated Streptomyces orientalis and Nocardia orientalis), which was found to produce a substance that inhibited gram-positive organisms (compound 05865) (178). “Mississippi mud,” as it was affectionately known because of its brown color, was used in clinical trials in the mid-1950s and was approved for use by the U.S. Food and Drug Administration in 1958 (178).
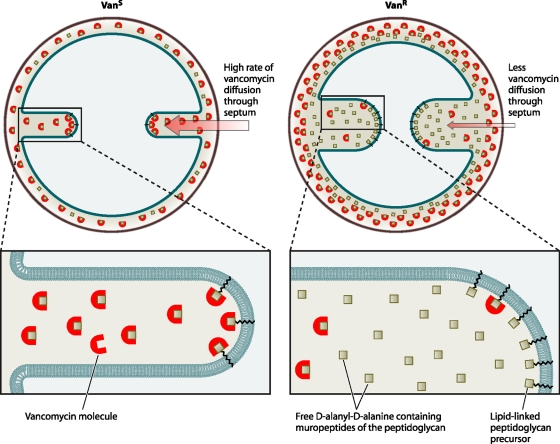
Vancomycin is an inhibitor of cell wall synthesis in S. aureus and other gram-positive organisms. While beta-lactam antibiotics inhibit cell wall synthesis by binding to the transpeptidase active site of penicillin binding proteins, vancomycin acts by a completely different mechanism. It binds to the C-terminal d-Ala-d-Ala residue of the peptidoglycan precursor and forms a stable, noncovalent complex, which prevents the use of the precursor for cell wall synthesis (Fig. 1) (259). Vancomycin inhibits late-stage peptidoglycan biosynthesis and acts outside the cytoplasmic membrane, which results in the intracellular accumulation of UDP-linked MurNAc-pentapeptide precursors (272, 273). The vancomycin complex involves a number of hydrogen bonds between the peptide component of vancomycin and the d-Ala-d-Ala residue (5, 15). Any process that interferes with vancomycin binding to d-Ala-d-Ala residues in the cell wall will decrease the potency of the drug. The addition of “false” binding sites (e.g., a d-Ala-d-Ala-containing ligand) to a bacterial culture containing vancomycin leads to competition between binding sites and a reduction of vancomycin activity (4). The main location for cell wall synthesis in S. aureus is the division septum and not the whole-cell membrane (245, 255). This means that vancomycin has to diffuse to the tip of the division septum to bind to peptidoglycan precursors at this location, and the distance of this diffusion varies depending on the cell cycle, where a longer septum exists later in the cycle (Fig. 2) (245).
FIG. 2.
Model depicting the site of vancomycin activity in the division septum and the changes associated with the VISA phenotype. The path of vancomycin to its lethal target (lipid II) should be through the division septum. In vancomycin-intermediate cells (Vanr), the rate of diffusion of vancomycin molecules to the septal tip is decreased, lowering the effective concentration of antibiotic that reaches the lipid-linked peptidoglycan precursor (lipid II) at the site of cell wall synthesis, per unit time, and therefore tilting the balance in favor of continued cell wall synthesis. This model implies that vancomycin efficiency varies during the cell cycle, as the path from the outside of the cell to the lethal targets is shorter when the septum starts to be formed and longer when septum synthesis approaches completion. (Adapted from reference 245 with permission.)
DEFINITIONS
A number of methods are available to determine vancomycin susceptibility in S. aureus, including methods to determine vancomycin MICs that use a relatively low inoculum and techniques to detect heteroresistance, which tend to rely on a higher inoculum and prolonged incubation. The standard method for hVISA detection is the population analysis profile (PAP) (see below); however, a number of surrogates for the detection of hVISA are also available, such as the macromethod Etest (MET). The definition of VISA is more straightforward, as it is defined based on a standard vancomycin MIC, while the definition of hVISA is more difficult and not well standardized.
Vancomycin-Intermediate Staphylococcus aureus (VISA)
The Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) defined resistance breakpoints for MIC and disc diffusion testing of vancomycin against S. aureus over 20 years ago (346). Initial breakpoints were as follows: susceptible at a vancomycin broth MIC of ≤4 μg per ml, intermediate at a vancomycin broth MIC of 8 to 16 μg per ml, and resistant at a vancomycin broth MIC of ≥32 μg per ml. Subsequently, in 2006, the CLSI redefined vancomycin breakpoints as follows: susceptible at a vancomycin broth MIC of ≤2 μg per ml, intermediate at a vancomycin broth MIC of 4 to 8 μg per ml, and resistant at a vancomycin broth MIC of ≥16 μg per ml (Table 2) (53). Hence, the current definition for VISA is an S. aureus isolate with a vancomycin broth MIC of 4 to 8 μg per ml. Other terminology, such as S. aureus with reduced vancomycin susceptibility (SA-RVS), has also been used to describe these strains and is summarized in Table 2. The rationale for changing breakpoints was an increasing association between a vancomycin MIC of 4 μg per ml and vancomycin treatment failure and also the increased detection of heteroresistant strains (346). However, the change in breakpoints will not help detect heteroresistant strains with a vancomycin broth MIC of ≤2 μg per ml. Additionally, given the potential for differences in the vancomycin MIC results based on the methodology used (263, 291), a vancomycin broth MIC using reference methodology such as CLSI broth microdilution should be used as the definitive test for the definition of VISA.
TABLE 2.
Summary of terminology for strains of S. aureus with reduced vancomycin susceptibility referred to in clinical case reportsd
| Glycopeptidea susceptibility classification | Broth microdilution (μg/ml) |
||
|---|---|---|---|
| CLSIb prior to 2006 | CLSI after 2006 | EUCASTc | |
| Susceptible (VSSA) | ≤4 | ≤2 | ≤2 |
| Intermediate (VISA) | 8-16 | 4-8 | No longer included in definition |
| Resistant (VRSA) | ≥32 | ≥16 | ≥4 |
Most often defined with reference to vancomycin, but some authors use “VISA” and “GISA” to indicate the presence of a class effect.
NCCLS before 2005.
New EUCAST breakpoints (released 20 May 2009) no longer define a VISA category and have reduced the resistant breakpoint to ≥4 μg per ml.
S. aureus with reduced susceptibility (SA-RVS) is defined as follows: (i) MIC of ≥4 μg per ml (91, 306) or (ii) area under vancomycin concentration-kill curve of ≥0.9 of the AUC of type strain Mu3 (124). hVISA is defined as follows: (i) VSSA strain that upon subculture stably produces subcolonies with MICs in the VISA/VRSA range at a frequency of ≥1 × 106 according to the population analysis profile (PAP) (114); (ii) AUC ratio of ≥0.9 of the AUC of type strain Mu3, referred to as PAP/AUC (48, 118, 155); or (iii) modified high-inoculum Etest read at 48 h (193, 194).
Heterogeneous Vancomycin-Intermediate Staphylococcus aureus (hVISA)
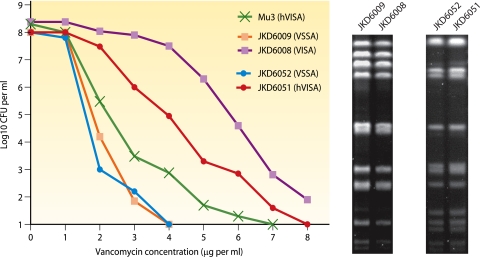
The definition and optimal laboratory detection of hVISA remain uncertain. Essentially, an hVISA isolate is an S. aureus isolate with a vancomycin MIC within the susceptible range when tested by routine methods (previously a vancomycin broth MIC of ≤4 μg per ml and now a vancomycin broth MIC of ≤2 μg per ml) but where a proportion of the population of cells are in the vancomycin-intermediate range (113). Typically, the resistant population is present at a frequency of ≤10−5 to 10−6, hence the difficulty in the detection of this resistance phenotype using CLSI methods where an inoculum of 5 × 104 CFU per well (broth MIC) or 1 × 104 CFU per spot (agar dilution) is used (53). The relative proportion of the population of cells that are resistant to vancomycin at 4 μg per ml can vary from strain to strain so that there is a spectrum from vancomycin-susceptible S. aureus (VSSA) to VISA (Fig. 3). The accurate detection of this phenotype requires a vancomycin population analysis profile (PAP), which is described below. In the modified vancomycin PAP described by Wootton et al., prototype hVISA strain Mu3 is used as a standard reference for the detection of hVISA isolates (387). By using PAP as a reference method, the hVISA phenotype can be detected for strains of S. aureus with vancomycin MICs as low as 0.5 to 1 μg per ml (174, 346). In a recent clinical study, the hVISA phenotype was detected in 50% of clinical MRSA isolates with a vancomycin broth MIC of 2 μg per ml (118). Therefore, if heteroresistance to vancomycin is clinically important for S. aureus, the current CLSI guidelines for the testing of vancomycin against S. aureus will not detect potentially important resistance in many isolates.
FIG. 3.
Example of population analysis profile curves for vancomycin-susceptible and heterogeneous vancomycin-intermediate S. aureus strains. The in vivo evolution of the resistant phenotype is depicted with a shift in the PAP curve to the right, with SmaI-digested pulsed-field gel electrophoresis patterns being identical for paired isolates from the same patient. (Adapted from reference 121.)
HISTORY AND EPIDEMIOLOGY
First Reports of hVISA and VISA
Reports of clinical S. aureus isolates that demonstrated reduced teicoplanin susceptibility and the in vivo emergence of resistance during teicoplanin therapy came from Europe in the early 1990s, although these strains remained susceptible to vancomycin (153, 189, 192). In 1997, strains of S. aureus with reduced susceptibility to vancomycin were reported from Japan. This included a strain with a vancomycin MIC of 8 μg per ml (strain Mu50, vancomycin-intermediate S. aureus [VISA] [ATCC 700699]) isolated from a surgical wound infection from a 4-month-old individual who had undergone cardiac surgery, where vancomycin failed to cure the infection (115), and a strain with a vancomycin MIC of 4 μg per ml that harbored subpopulations with a higher MIC at a rate of greater than 1 in 106 cells (strain Mu3, designated heterogeneous VISA [hVISA] [ATCC 700698]) (114). Mu3 was isolated in 1996 from the sputum of a 64-year-old patient with MRSA pneumonia who failed vancomycin therapy (114). Using the latest CLSI breakpoints, the original description of Mu3 would now classify the strain as being a VISA strain (53); however, subsequent studies where Mu3 has been tested reported vancomycin MICs of 2 μg per ml (57, 58). The subsequent screening of seven university hospitals in Japan found no additional VISA strains but a rate of isolation of hVISA strains of between 9 and 20% of MRSA isolates by using a simplified “mini-PAP” procedure (114).
Global Epidemiology and Associated Features of hVISA and VISA
A clear difficulty in interpreting the literature in this area is the lack of standardized criteria for the definition of hVISA and the use of different methodologies to detect VISA. The prevalence of hVISA has varied significantly. Some of these differences may well be explained by differences in laboratory definitions and testing strategies; however, it appears that rates of hVISA vary globally. For example, Hiramatsu et al. detected the hVISA phenotype in up to 20% of MRSA isolates from their hospital (114), but a subsequent study from Japan recently challenged this finding by detecting no hVISA strains among 6,625 strains tested (9, 137). In the original description of hVISA epidemiology in Japan by Hiramatsu et al., a brain heart infusion (BHI) agar (BHIA) plate with 4 μg vancomycin per ml was used for the screening of isolates (114). In a more recent study, vancomycin heteroresistance was detected after the selection of strains from screening agar containing 2 μg vancomycin per ml (257). In many other studies, hVISA has been defined by the use of a modified vancomycin population analysis profile (PAP) described previously by Wootton et al. (387).
After the first reports of VISA and hVISA from Japan, it did not take long for this resistance phenotype to be recognized around the world. Strains of S. aureus (predominately MRSA) demonstrating the hVISA or VISA phenotype have now been reported for many countries including the United States, Japan, Australia, France, Scotland, Brazil, South Korea, Hong Kong, South Africa, Thailand, Israel, and others (22, 67, 69, 77, 79, 82, 94, 158, 167, 194, 195, 206, 258, 284, 300, 326, 327, 331, 352, 354, 366, 373). A retrospective analysis of stored isolates detected previously unrecognized hVISA/VISA strains at least back to 1987 in the United States and back to a similar period in France, Spain, and Germany (11, 22, 39, 276, 288); however, there has been some concern about the potential loss of resistance during prolonged storage, which could impact results of such retrospective analyses. Although reported predominately for MRSA, the hVISA phenotype can be detected among methicillin-sensitive S. aureus (MSSA) strains (6, 26, 93, 187, 271). Additionally, some strains of MRSA that also express the VISA phenotype have been shown to have a deletion of the mecA gene or demonstrate reduced methicillin resistance despite the presence of mecA (107, 223).
After the first report of hVISA from Australia in 2001, hVISA and VISA have been increasingly reported from around Australia and New Zealand (100, 103, 119, 219, 375). At our institution in Australia, 9.4% of blood culture isolates of MRSA were found to be hVISA strains by modified vancomycin PAP in a 12-month period from July 2001 (48). In a more recent study at our institution, we found a similar percentage of MRSA blood culture isolates to be hVISA isolates (13%) but found a remarkably high rate of hVISA (approximately 50%) when all clinical MRSA isolates were tested by PAP analysis (118). Studies in the United States have generally detected very low rates of hVISA (70, 133, 165, 341); however, a range of methods were used for detection in these studies. A recent analysis of vancomycin susceptibility in S. aureus over a 22-year period from the Detroit, MI, region demonstrated an increasing rate of hVISA over the period (from 2.2 to 8.3%) among clinical MRSA isolates by using MET screening and confirmation by PAP (289).
In Israel in 2003 and 2004, 6% of patients with MRSA bacteremia had hVISA when blood culture isolates were screened using the MET, many of which would have been missed with routine testing (194). A study of MRSA isolates from 63 French hospitals found that only 0.7% of isolates were hVISA isolates after screening with BHI agar containing 6 μg teicoplanin per ml and confirmation by PAP (37), with similarly low rates found by some other French studies and in Belgium (69, 230, 271). However, a subsequent study at one French institution, which screened 2,300 S. aureus isolates using BHIA with 4 μg teicoplanin per ml, followed by MET and then PAP confirmation, found that 11% of the isolates were hVISA isolates, which were found to be clonal by pulsed-field gel electrophoresis (93). Seven of the 255 hVISA isolates detected were MSSA isolates.
A variability in rates of hVISA strains in other countries has also been demonstrated. After the early detection of hVISA and VISA in South Korea (157, 158), a follow-up study found no evidence of hVISA (156). In a Turkish hospital, the rate of hVISA among MRSA strains increased from 1.6% in 1998 to 32% in 2001 (300). In a review by Liu and Chambers in 2003 (183), data from 14 previous studies were combined, and hVISA rates were 2.16% in 6,052 MRSA isolates and 0.05% in 1,868 MSSA isolates in the literature at that time. Rates of hVISA varied from 0 to 8.24% in studies where at least 50 isolates were included. The true prevalence of hVISA is unclear and may have been significantly underestimated by many studies. To better understand hVISA epidemiology and clinical relevance, it is important that standard criteria for investigating hVISA are developed and used in studies of prevalence and clinical impact.
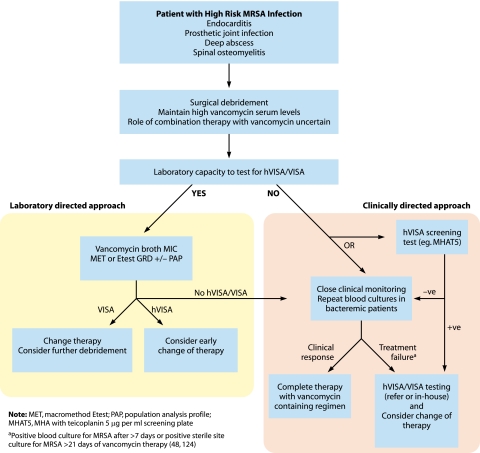
Risk factors for hVISA and VISA.
Not surprisingly, the main risk factors for infection with VISA and hVISA appear to be prior MRSA infection or colonization and exposure to vancomycin (48, 55, 91, 124). Additionally, most VISA and hVISA infections occur in patients with serious underlying disease such as malignancy, diabetes, renal failure, or recent major surgery (48). A “high bacterial load,” which occurs with infections such as endocarditis, deep abscess, or infection of a prosthetic joint, may also predispose an individual to the development of hVISA infection during failed glycopeptide therapy, probably because large numbers of organisms are present and the penetration of antibiotics into such infected areas may be limited (48). Related to this, some data suggest that low serum levels of vancomycin early in the treatment course of MRSA infections may also be associated with the emergence of VISA and hVISA (48). In studies to determine factors associated with higher vancomycin MICs and the reduced in vitro bactericidal activity of vancomycin against S. aureus (rather than hVISA or VISA per se), prior vancomycin exposure and residence in an intensive care unit (ICU) were independent predictors (185, 209).
For many patients, the hVISA or VISA phenotype was detected in bacterial isolates only after a prolonged period of infection associated with the failure of glycopeptide therapy (48, 121, 124, 151, 213, 313, 316, 326, 337, 340, 376). In many of these cases, a detailed analysis of the earlier clinical isolate failed to detect any vancomycin heteroresistance, and the phenotype appears to have emerged from a vancomycin-susceptible strain during therapy. Pulsed-field gel electrophoresis patterns have been used to demonstrate the clonality of vancomycin-susceptible and subsequent vancomycin-resistant strains, suggesting the emergence of resistance from the earlier vancomycin-susceptible isolate (Fig. 3).
In other cases, a nosocomial spread of hVISA or VISA has been suggested, with a number of outbreaks reported, mostly in France (64, 105, 190, 253). An outbreak reported by de Lassence et al. was described as 21 patients in a French ICU who had isolates of S. aureus with reduced glycopeptide susceptibility (64). Although that report described an outbreak of glycopeptide-intermediate S. aureus, the isolates in that study were all susceptible to vancomycin and teicoplanin according to routine Etest MIC determinations and were positive for reduced susceptibility only by a MET, suggesting that the strains were in fact heteroresistant. This is an example of difficulties that arise when trying to interpret the literature in this area and an example of an overinterpretation of the MET result. In addition to patient isolates, a number of environmental surfaces were positive for the resistant outbreak strain, demonstrating that hVISA/VISA can contaminate the environment. A comment in the discussion of that report stated that all isolates were of the same strain, suggesting nosocomial spread rather than the in vivo generation of resistant isolates.
Vancomycin MIC creep and hVISA/VISA.
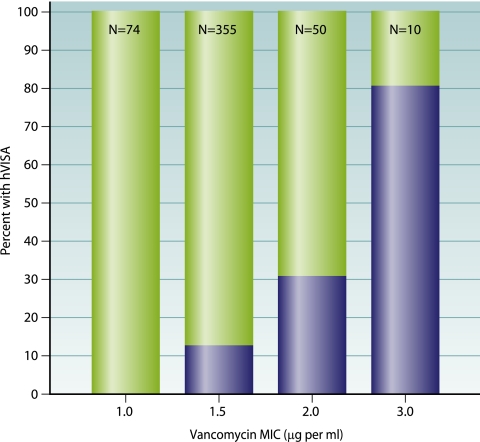
In addition to increasing numbers of reports of hVISA and VISA, there has been significant interest regarding the changing patterns of vancomycin MICs within the S. aureus population. This has been driven partly by studies demonstrating poorer outcomes for vancomycin treatment of MRSA infections with higher vancomycin MICs, even when these MICs are within the susceptible range (112, 184, 298, 332). Changing vancomycin MICs will directly impact the rates of hVISA, as it was clearly demonstrated that the proportion of isolates that are hVISA isolates increases with increasing vancomycin MICs within the susceptible range (Fig. 4) (289, 346). The term vancomycin “MIC creep” is now frequently used (101, 276, 333, 374). This phenomenon has been demonstrated at a number of centers in the United States over recent years by using Etest or vancomycin broth MIC (289, 333, 374), while at another location in the United States, no change in vancomycin MIC was detected between 1999 and 2006 (117). In France, increases in glycopeptide MICs (predominately teicoplanin) were observed over a 20-year period (276), while in Spain, the testing of over 3,000 clinical isolates by broth microdilution between 2002 and 2006 found no change in vancomycin MICs at one institution (6). Interestingly, an analysis of SENTRY data for the years 1998 to 2003, which included 35,458 S. aureus isolates, detected no change in vancomycin MICs over this time by using standard CLSI methods (147). Therefore, changes in S. aureus vancomycin MICs over time can occur within certain institutions; however, a limitation of previous studies is the lack of molecular typing of strains to determine if the MIC creep is actually due to the emergence of a new clone rather than the gradual reduction in vancomycin susceptibility within the clonal population of the institution. Ultimately, given the SENTRY results, it does not appear that changes in vancomycin MICs are rapidly occurring on a global scale but may be shifting within some institutions.
FIG. 4.
Correlation between vancomycin Etest MIC and heteroresistance (defined by macromethod Etest) for MRSA blood culture isolates collected between 1996 and 2006. The shaded area represents the percentage of isolates that are hVISA isolates at a given MIC and demonstrates that strains with an Etest MIC as low as 1.5 μg per ml also demonstrated heteroresistance. (Adapted from reference 220 with permission.)
Molecular epidemiology of hVISA and VISA.
Initially, studies of VISA strains using staphylococcal cassette chromosome mec (SCCmec) analysis and multilocus sequence typing (MLST) suggested that the phenotype was present predominately in one pandemic clone of MRSA; however, it has become clear that resistance has emerged in many major epidemic clones of MRSA (113, 126a).
Community-acquired S. aureus, including PVL-positive strains and hVISA/VISA.
A laboratory-based study of in vitro resistance development demonstrated a limited vancomycin resistance potential in community-acquired S. aureus clones from Australia (216). Recently, however, the community MRSA clone USA300 (Panton-Valentine leukocidin [PVL] positive) with a VISA phenotype found in San Francisco, CA, and Kansas has been described (102, 106). Therefore, the expression of the hVISA and VISA phenotypes can occur in many S. aureus lineages and is not limited to typical “hospital” clones of S. aureus. The recent emergence of multidrug resistance in USA300 (71), in addition to the recent description of the VISA phenotype in this clone, is concerning given the rapid spread of this strain across the United States. There is no reason to believe that PVL per se would affect vancomycin activity against S. aureus.
agr group and hVISA/VISA.
There initially appeared to be a link between the agr group II locus and VISA (296); however, subsequent studies have demonstrated the VISA phenotype in isolates from other agr groups (369). There does, however, appear to be a link between hVISA/VISA and the expression of agr in S. aureus and a potential association between the agr group and the response to vancomycin therapy (see below).
SCV S. aureus and hVISA/VISA.
The S. aureus small-colony-variant (SCV) phenotype is a slow-growing variant with distinct characteristics including reduced pigmentation and hemolytic activity as well as altered host-pathogen interactions favoring persistent and recurrent infection (265, 266). Small-colony variants of S. aureus have demonstrated reduced susceptibility to a number of antimicrobials including gentamicin, fluoroquinolones, and linezolid (16, 52, 264). Recently, the in vitro bactericidal activity of vancomycin against an SCV-defined hemB mutant was tested. The vancomycin killing activity was reduced in the SCV compared to the parental strain (359). It is uncertain if reduced vancomycin susceptibility is a common feature of clinical SCV S. aureus isolates; however, some of the typical phenotypic features found in hVISA and VISA strains are reminiscent of the SCV, such as reduced growth rate, altered pigmentation, and reduced hemolytic activity, and in VISA isolates demonstrating a heterogeneous colony morphology, the small-colony forms tend to display greater vancomycin resistance.
PHENOTYPIC FEATURES AND MECHANISMS OF RESISTANCE
In the late 1990s, Sieradzki et al. suggested that alterations in cell wall structure inhibit vancomycin access to its active site in a laboratory-induced strain with a vancomycin MIC of 100 μg per ml (315, 320). Over recent years, a model of the resistance mechanism of hVISA and VISA has evolved, where VISA emerges by sequential mutations from VSSA, with hVISA as an intermediary between VSSA and VISA. Reduced cell wall turnover and reduced autolytic activity, and in some cases activated cell wall synthesis, are thought to lead to cell wall thickening and reduced vancomycin access to its active site, which is localized to the division septum (Fig. 2). The key phenotypic features of hVISA and VISA are summarized in Table 3.
TABLE 3.
Phenotypic characteristics of clinical and laboratory-induced hVISA and VISA strains compared to related vancomycin-susceptible S. aureus strainsa
| Isolate studied | Phenotype (vancomycin MIC [μg/ml]) | Isolate description | Phenotypic feature(s) of hVISA or VISA isolate compared to VSSA | Reference(s) |
|---|---|---|---|---|
| MRSA COL | VSSA | VRSA (VM50) induced from MRSA COL by vancomycin selective pressure | Gradual alterations in cell wall with increasing resistance; removed vancomycin from broth (vancomycin absorbed into cell wall); reduced growth rate; reduced X linking of cell wall PG; inactivated PBP4; reduced methicillin MIC due to inactivation of mecA; delayed access of vancomycin to active site in division septum | 245, 315, 317, 319, 320, 322 |
| VM (VM50) | VRSA (100) | |||
| VM3 | hVISA/VISA | |||
| VM6 | hVISA/VISA | |||
| VM12 | hVISA/VISA | |||
| VM25 | hVISA/VISA | |||
| 523 | VSSA | Laboratory-induced VISA and VRSA strains; 523teico induced by teicoplanin (teicoplanin MIC of 128 μg per ml) | Slower growth, smaller colony size; increased lysostaphin resistance; decreased coagulase activity; increased cell diam; increased cell wall thickness; forms clumps in liquid culture; changes in PG X linking; increased production of PBPs; reduced Triton X-induced autolysis | 31, 62, 108, 215 |
| 523a | hVISA/VISA (3) | |||
| 523c | VISA (4) | |||
| 523k | VISA (8) | |||
| 523teico | VISA (4) | |||
| 1714 | VSSA | |||
| 1714s | VRSA (32) | |||
| 1725 | VSSA | |||
| 1725w | VISA (16) | |||
| Mu3 | hVISA (2-4) | First reported hVISA and VISA strains | vanA, vanB, and vanC1 to vanC3 negative; increased production of PBP2 and PBP2′; activated cell wall synthesis; increased glutamate-containing muropeptides; correlation between cell wall thickness and vancomycin resistance; Mu50 reduced resistance after 15 days of serial passage; reduced whole-cell autolytic activity; vancomycin clogging of cell wall with anomalous diffusion | 29, 56, 58, 59, 108, 109, 362 |
| Mu50 | VISA (8) | |||
| Mu50ω | VSSA/hVISA | Mu50ω isolated 1 year later from same patient as Mu50 | ||
| SA137/93A | VISA (8) | Germany; SA137/93A clinical isolate; other spontaneous mutants with increased and decreased resistances | Increased cell wall thickness; reduced beta-lactam resistance; increased cell wall X linking in SA137/93G | 267 |
| SA137/93G | VISA (12) | |||
| SA137/93G1 | VSSA | |||
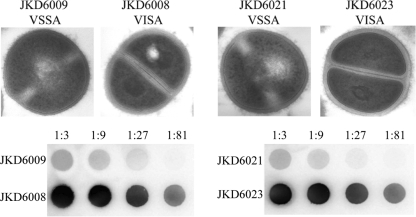
| JKD6000 | VSSA (2) | Paired isolates from 5 patients with persistent MRSA infections and vancomycin treatment failure | vanA, vanB, and vanC1 to vanC3 negative; thickened cell wall; reduced agr activity; reduced autolytic activity (4/5); reduced in vitro biofilm formation | 121 |
| JKD6001 | VISA (4) | |||
| JKD6004 | VSSA (1) | |||
| JKD6005 | hVISA (2) | |||
| JKD6009 | VSSA (1) | |||
| JKD6008 | VISA (4) | |||
| JKD6021 | VSSA (1) | |||
| JKD6023 | VISA (4) | |||
| JKD6052 | VSSA (1) | |||
| JKD6051 | hVISA (2) | |||
| PC-1 | hVISA (2) | Paired isolates from patient with vancomycin treatment failure | Reduced vancomycin concn in broth culture of PC-3, recovered from staphylococcal cell wall; cell wall thickening; reduced resistance after 15 days of serial passage; no increase in glutamate-containing muropeptides; reduced Triton X-induced autolysis | 29, 31, 32, 58, 284, 316 |
| PC-3 | VISA (8) | |||
| BB225 | VSSA (1) | Series of laboratory-induced VISA strains | No change in oxacillin MIC; increased cell wall thickness (with vancomycin); increased doubling time; reduced lysostaphin susceptibility of whole cells but increased susceptibility of purified cell walls; reduced autolytic activity in part due to lower activity of VISA autolysin extracts; reduced alt expression; removed vancomycin from broth culture; no mutS mutations | 58, 160, 221, 250 |
| BB225V3 | VISA (4) | |||
| BB270 | VSSA (1) | |||
| BB270V15 | VISA (12) | |||
| 13136p−m− | VSSA (1) | |||
| 13136p−m−V5 | VISA (4) | |||
| 13136p−m+ | VSSA (1.5) | |||
| 13136p−m+V20 | VISA (16) | |||
| SH108 | VSSA (2) | |||
| SH108V5 | VISA (6) | |||
| BB399 | VSSA (2) | |||
| BB399V12 | VISA (12) | |||
| BB568 | VSSA (1) | |||
| BB568V15 | VISA (12) | |||
| COL | VSSA (2) | |||
| COLV10 | VISA (8) | |||
| Others | ||||
| MI | VISA (6) | Michigan VISA isolate, July 1997, CAPD-associated peritonitis | Reduced resistance after 15 days of serial passage; cell wall thickening; increased extracellular matrix; increased glutamate-containing muropeptides; reduced Triton X-induced autolysis | 29, 31, 32, 58, 326 |
| NJ | VISA (5) | New Jersey VISA isolate, August 1997, bacteremia | Reduced resistance after 15 days of serial passage; cell wall thickening; increased extracellular matrix; increased glutamate-containing muropeptides; reduced Triton X-induced autolysis | 29, 31, 32, 58, 326 |
| IL-A | hVISA | VISA emerged from hVISA during 13 days of persistent bacteremia | Cell wall thickening; no increase in glutamate-containing muropeptides; increased cell wall thickness; reduced lysostaphin susceptibility; reduced Triton X-induced autolysis; increased cell wall X linking; reduced Triton X-induced autolysis | 30-32, 58 |
| IL-F | VISA (8) | |||
| AMC11094 | VISA (8) | South Korea, clinical isolate | Cell wall thickening | 58, 158 |
| 99/3759-V | VISA (8) | Scotland, UK | Cell wall thickening | 58 |
| 99/3700-W | VISA (8) | |||
| LIM-1 | VSSA (2) | France; isolated from patient with persistent infection and failed teicoplanin therapy; all blood culture isolates except LIM-3 (purulent discharge) | Cell wall thickening; vanA, vanB, and vanC1 to vanC3 negative | 58, 258 |
| LIM-2 | VISA (8) | |||
| LIM-3 | VISA | |||
| LIM-4 | VISA | |||
| 28160 | VISA (8) | South Africa | Cell wall thickening | 58, 79 |
| JH1 | VSSA (1) | Baltimore, MD; series of blood culture isolates from patient with endocarditis who failed vancomycin therapy; JH14, valve isolate | Cell wall thickening; decreased wall X linking; reduced PBP4; decreased cell wall turnover and autolysis; changes in wall teichoic acids; delayed access of vancomycin to active site in division septum | 245, 314, 318 |
| JH2 | VISA (4) | |||
| JH3 | VISA (4) | |||
| JH5 | VISA (6) | |||
| JH6 | VISA (8) | |||
| JH9 | VISA (8) | |||
| JH14 | VISA (8) | |||
| BR1 | VISA (8) | Clinical isolates from Brazil | Cell wall thickening; vanA, vanB, and vanC1 to vanC3 negative | 58, 235 |
| BR2 | VISA (8) | |||
| BR3 | VISA (8) | |||
| BR4 | VISA (8) | |||
| BR5 | VISA (8) | |||
| 98141 | VISA (8) | France, clinical isolate | Cell wall thickening | 49, 58 |
| SF1 | VSSA (1) | San Francisco, CA, paired clinical isolates; endocarditis, failed vancomycin therapy | Significant reduction in efficacy of vancomycin against SF2 in rabbit endocarditis model | 213 |
| SF2 | hVISA (2) | |||
| MRGR3 | VSSA (1), TSSA (0.5) | Teicoplanin-resistant subclones emerged in rat model of foreign-body infection without antibiotic exposure; stable clones selected on teicoplanin-containing medium (14-4 and 17-2); 14-4rev, spontaneous revertant | Unstable resistance in rat model without antibiotic exposure; increased fibronectin-mediated adherence; reduced autolytic activity; reduced extracellular hydrolase activity; reduced agr | 268, 269, 367 |
| 14-4 | VISA (4; teicoplanin, 16) | |||
| 14-4rev | VSSA (1) | |||
| 17-2 | VISA (4; teicoplanin, 16) | |||
| RN6607 | VSSA (1) | RN strains laboratory induced | Reduced agr activity in VISA and hVISA; reduced in vitro bactericidal activity of vancomycin in agr-null vs parent strains; agr-null increased propensity for hVISA; reduced agr activity associated with reduced autolysis and resistance to tPMP | 294, 295 |
| RN6607V | hVISA (2) | A5937 and A5940 clinical isolate pair | ||
| RN9120 | VSSA (1) | |||
| RN9120V | hVISA (2) | |||
| RN9120V-GISA | VISA (8) | |||
| A5937 | VSSA (2) | |||
| A5940 | VISA (4) | |||
| SA113 | TSSA (teicoplanin, 3) | In vitro-derived strains, teicoplanin selected | Slower growth; thickened cell wall; increased N-acetylglucosamine incorporation; reduced fitness in resistant strain | 201 |
| NM18 | Teicoplanin MIC, 16 | |||
| NM30 | Teicoplanin MIC, 48 | |||
| NM67 | Teicoplanin MIC, 64 (vancomycin, 24) | |||
| Hershey MC | Series of isolates from one patient | Reduced muropeptide cross-linking; reduced O-acetylation of muramic acid | 151 | |
| 1 | VSSA | |||
| 3 | VSSA | |||
| 10 | VISA (daptomycin resistant) | |||
| 25 | VISA |
Note that some laboratory-induced isolates had MICs of vancomycin in the resistant range despite the absence of the vanA operon. CAPD, continuous ambulatory peritoneal dialysis; tPMP, thrombin-induced platelet microbicidal protein; TSSA, teicoplanin-sensitive S. aureus.
Most of the work assessing the mechanisms of resistance in S. aureus strains with low-level vancomycin resistance has been performed using VISA isolates with vancomycin MICs of 8 μg per ml. hVISA strains appear to be the precursors of VISA strains and appear to be induced to homogenous resistance (VISA) after exposure to cell wall-active antibiotics (29, 30). One of the difficulties in working with these strains in the laboratory is the tendency for some resistant isolates to revert to a more susceptible phenotype. For example, in one study of four VISA strains (NJ, MI, PC, and Mu50), all strains reverted to a more susceptible phenotype after 15 days of passage on nonselective medium; however, three of the strains maintained a subpopulation that would grow on 4 μg vancomycin per ml (250). Cui et al. (58) also demonstrated a reduction in resistance levels of VISA isolates upon serial passage on drug-free medium; however, strains typically maintained an hVISA phenotype (58). Other studies demonstrated a more stable resistance phenotype. For example, 6 laboratory-derived VISA strains demonstrated a stable phenotype after 20 passages on nonselective medium (250). Many of the earlier studies of the features of VISA were performed with laboratory-derived strains after stepwise selection on glycopeptide-containing medium (62, 215, 250, 315, 319, 320, 322); however, more recent studies have utilized clinical isolates (32, 56, 121, 151, 314, 318), and in many cases, the phenotypic features have been similar. The ability to induce the selection of VISA from vancomycin-susceptible parent isolates varies between strains in terms of both the level of resistance attained and the rate at which resistance develops (58, 250, 267). After early studies of the effects of glycopeptides on S. aureus and investigation of laboratory-derived vancomycin-resistant strains of S. aureus and a clinical isolate of teicoplanin-resistant S. aureus revealed an increased level of production of PBP2 and cell wall thickening (62, 80, 313), much of the early investigation of hVISA and VISA was focused on cell wall changes.
Cell Wall Changes
Common biochemical and morphological changes can be found in many S. aureus isolates that demonstrate low-level vancomycin resistance (hVISA and VISA), either laboratory-induced or clinical isolates; however, when looked at in detail, the cell wall rearrangements that occur in VISA strains can vary between strains (32). Cell wall thickening is a consistent feature and was recognized well before the first description of clinical VISA isolates (Fig. 5) (30, 59, 62, 80, 108, 121, 187, 229, 267). It may be associated with activated cell wall synthesis (108), and the cell wall thickening is reduced when isolates are serially passaged and resistance levels drop (58). In some isolates, cell wall thickening may not be obvious by electron microscopy without vancomycin, but after vancomycin exposure, it becomes more obvious (59, 250, 316). Other common features include an increased level of production of abnormal muropeptides (319); an overexpression of PBP2 and PBP2′ (108, 215), although reduced levels of expression of PBP2′ were found in a laboratory-derived mutant due to the inactivation of mecA (322); reduced PBP4 expression levels (319); increased levels of d-Ala-d-Ala residues; and reduced levels of peptidoglycan cross-linking in most isolates studied (Fig. 1) (151, 215, 250, 318, 320). For some VISA isolates a small increase in peptidoglycan cross-linking was found (30, 32, 267). Other common features include a reduced growth rate (62, 250) and reduced whole-cell lysostaphin susceptibility (56, 62, 215). In contrast, purified cell walls of laboratory-induced VISA strains demonstrated increased lysostaphin susceptibility (160).
FIG. 5.
Example of the cell wall and capsule changes that occur in hVISA and VISA strains in paired isolates from patients with persistent infections. The top panel demonstrates significant cell wall thickening in VISA strains compared to VSSA strains, while the bottom panel demonstrates significant increases in the expression of capsule by using an anticapsule type 8 immunoblot and serial dilutions of crude capsule extracts from paired VSSA and VISA strains. (Adapted from references 121 and 122, the latter of which was published under an open-access license agreement.)
The thickened cell wall appears to be the most consistent feature, and although the exact mechanisms leading to thickening have not been determined, the thickened cell wall is thought to prevent the diffusion of vancomycin to its active site in the cytoplasmic membrane in the division septum (250, 318). Recent studies using fluorescent vancomycin and fluorescent ratio imaging microscopy demonstrated that the vancomycin binding capacity was increased in resistant strains, with evidence of a delayed access of vancomycin to the active site in the septum in resistant strains (Fig. 1 and 2) (245).
Autolytic Activity
Reduced autolytic activity is a common feature of hVISA and VISA strains and is a common early phenotypic change in serial isolates obtained during persistent infection (30, 31, 108, 160), although this has not been demonstrated for all isolates (121). Initial studies of Japanese VISA strain Mu50 reported an increase in autolytic activity (362); however, recent data have confirmed that Mu50 demonstrates reduced whole-cell autolytic activity like other VISA strains (362). Some data suggest a possible role for wall teichoic acids of VISA strains suppressing peptidoglycan degradation by autolytic enzymes (318), while other studies suggested that a reduction in the autolytic activity and altered peptidoglycan hydrolase activity of VISA autolysin extracts are responsible for the reduced autolytic activity (160). It was proposed that vancomycin binding in the staphylococcal cell wall directly blocks the activity of a peptidoglycan hydrolase and explains the reduced autolytic activity demonstrated in the presence of vancomycin (321); however, this does not explain the reduced autolytic activity of hVISA and VISA strains in the absence of vancomycin (121). A loss of agr function in S. aureus has also been linked with reduced autolytic activity (295).
Metabolic Changes
An analysis of metabolic changes in highly resistant (vancomycin MIC of 32 μg per ml) in vitro derivatives of VISA strains demonstrated impaired acetate catabolism. Further analysis revealed similar changes in other VISA strains (71% had reduced acetate catabolism, compared to 8% of VSSA strains) (224). The authors of that report made the point that reduced acetate catabolism could lead to altered growth characteristics, antibiotic tolerance, changes in cell death, and increased polysaccharide intercellular adhesin synthesis, which was demonstrated in their study (224).
Molecular Mechanisms of Resistance
A number of studies have demonstrated the absence of the vancomycin resistance genes vanA, vanB, and vanC1 to vanC3 in hVISA and VISA strains (108, 121, 345). Because of the activated cell wall synthesis demonstrated for some hVISA and VISA strains (108), significant interest in regard to the molecular mechanisms of resistance have focused on pathways of cell wall biosynthesis in S. aureus.
Transcriptional changes.
A variety of experimental approaches have been used in an attempt to determine the genetic basis for the intermediate-level vancomycin resistance evident in VISA strains. These approaches include cDNA differential hybridization (169) and, more recently, DNA microarray analyses (57, 122, 168, 199, 201, 212, 303) to determine changes in the transcriptional profile of strains as they acquire the VISA phenotype. A proteomics approach has also been used to identify proteins that are differentially expressed in VISA strains (74, 251, 303). These comparisons have been done between closely related VISA and VSSA strains (57, 169), between laboratory-derived VISA strains and their less sensitive parents (57, 74, 201), between VSSA and VISA strains derived from a mouse model of infection (303), and between sequential VSSA and VISA isolates from the same patient (122, 199). From these studies, it appears that the acquisition of the VISA phenotype is probably a multistep process and that there are likely to be multiple pathways to intermediate vancomycin resistance, even among closely related strains. Conversely, the same genes have often been identified in different laboratories as differentially expressed in VISA strains that have diverse genetic backgrounds.
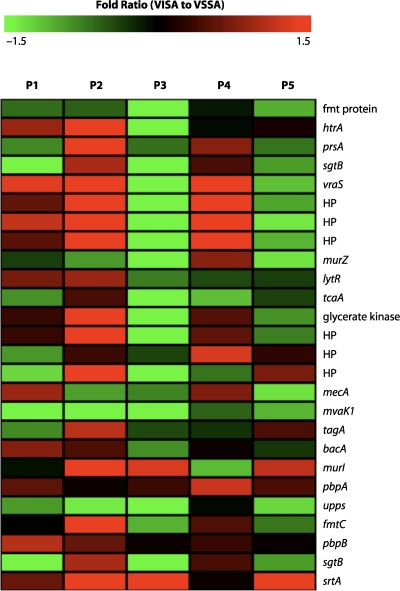
Also of relevance here are the transcriptional profiles of VSSA and VISA strains exposed to vancomycin and other cell wall-active antibiotics (168, 199, 202, 361, 392) or when cell wall synthesis is disrupted by the modulation of the expression of the murF or pbpB gene (92, 330). These experimental conditions lead to a cell envelope stress response (148) and the upregulation of a “cell wall stimulon” that responds to abnormal cell wall synthesis (199, 361). This stimulon contains genes encoding the two-component system VraSR, which in turn positively regulates a number of genes involved in cell wall synthesis (92, 168, 199). It appears that for some VISA strains, this stimulon is permanently upregulated. VraSR was first identified as being upregulated in VISA strains by using cDNA differential hybridization (169) and subsequently by microarray analyses (122, 199). Some of the genes within the stimulon (murZ and sgtB) were also identified by another transcriptional profiling study that involved six different comparisons between VISA and VSSA strains (57). The two genes were upregulated in at least five out of six comparisons, and an overexpression of the gene products resulted in a slight increase in the MIC of vancomycin (57). However, it should be emphasized that the VISA phenotype can be achieved in some strains without the induction of the cell wall stimulon (Fig. 6) (122, 202).
FIG. 6.
Microarray transcriptional heat map analysis of cell wall stimulon activation of hVISA/VISA (defined by population analysis and vancomycin broth MIC) relative to parental VSSA. Five isolate pairs are included (P1 to P5). For each pair, the hVISA/VISA and VSSA strains were isolated from the same patient. The fold ratio of gene transcription for hVISA/VISA compared to VSSA was calculated from pooled microarray data using The Institute for Genomic Research S. aureus arrays. Divergent transcriptional patterns were observed for the five pairs and demonstrate that for some hVISA/VISA strains, the cell wall stimulon is activated compared to parental VSSA strains, while in others, it is not activated compared to parental strains. HP, hypothetical protein. (Adapted from reference 122, which was published under an open-access license agreement.)
The fact that the cell wall stimulon is activated in some VISA strains but not others could also explain some conflicting conclusions. It was suggested that decreased levels of penicillin binding protein 4 (PBP4) were partially responsible for the VISA phenotype (81). Clinical VISA isolates (but not serially passaged laboratory isolates) had no or substantially lowered levels of PBP4, while all VSSA strains examined had detectable levels of this protein. Furthermore, the overproduction of PBP4 in VISA resulted in a lower MIC of vancomycin (81). However, a subsequent, more comprehensive, survey found that many isolates with intermediate-level resistance to vancomycin had normal levels of PBP4 (386). It seems that PBP4 expression levels are strain specific rather than obligatorily linked to the VISA phenotype.
Along similar lines, there has been some debate about the involvement of genes involved in purine biosynthesis in determining the VISA phenotype. A comparison has been made between the transcriptional profiles of two VISA isolates and resistant (MIC of vancomycin of 32 μg per ml) derivatives obtained by three serial passages at increasing vancomycin concentrations (212). It was argued that this comparison might exaggerate the changes that occur in VISA isolates. However, that same study suggested that there are no transcriptional changes between one of the VISA parents and a VSSA derivative obtained by 100 serial passages in the absence of vancomycin. A series of genes involved in purine biosynthesis were upregulated in both resistant mutants.
This was later confirmed by using a proteomics approach (251). The transcription of an operon encoding purine biosynthesis genes is controlled by the PurR regulatory protein. The purR gene of both resistant mutants was sequenced, and in both cases, the same identical mutation had occurred (212). An inactivation of PurR provided an explanation for the overexpression of the operon involved in purine biosynthesis, and the authors of that study suggested that this might be needed to increase ATP production for the increased cell wall thickness observed for VISA strains (212). A subsequent biochemical and genetic study could not confirm a link between purine biosynthesis and vancomycin resistance (89). Neither a mutation of purR nor the overproduction of wild-type or mutated PurR had any effect on the level of vancomycin resistance in VSSA. Furthermore, a panel of VSSA and VISA strains were all passaged to higher levels of vancomycin resistance, and none had a mutation in purR (89).
Among the genes that have been observed to be upregulated in VISA strains are those encoding the GraRS two-component regulatory system, so named for its glycopeptide resistance association (57). The locus has also been called aps because of its more general antimicrobial-peptide-sensing capacity (180). The overexpression of GraR or GraS results in a slight increase in the MIC of vancomycin (57), and a knockout mutation results in hypersensitivity (122, 204). However, in two different instances, point mutations have resulted in increased resistance to vancomycin, presumably by modifying the activity of the proteins (60, 123). The GraRS two-component regulatory system has been shown to control the expression of a large number of genes, including many genes involved in cell wall synthesis (111). Interestingly, among the genes upregulated in a knockout mutant are the genes involved in purine biosynthesis described above. The GraRS two-component regulatory system also positively regulates rot and mgrA, which encode global regulatory proteins that in turn control the expression of many genes encoding virulence determinants and others where the gene product is yet another regulatory protein (111).
In one study, the mgrA gene was found to be upregulated in five out of six VSSA/VISA comparisons (57). The overexpression of MrgA also resulted in a slight increase in the MIC of vancomycin (57). This perhaps unexpected connection between genes involved in endowing the VISA phenotype and the very complex interconnected regulatory pathways controlling virulence in S. aureus has been noticed for many of the experiments comparing the transcriptional profiles of VSSA and VISA strains as well as for associated genetic and biochemical studies. Where it has been investigated, it does not seem that the differential expression of a particular virulence determinant in the VISA strain has a direct impact on the level of vancomycin resistance. For instance, capsule production was shown to be upregulated in VISA strains by several studies, yet mutations that result in the absence of capsule have no effect on the vancomycin resistance of VISA strains (57, 122, 199). Rather, it seems that the genes necessary for capsule production, and one or more genes that directly or indirectly increase vancomycin resistance, are part of the same regulon. Because of the interconnected nature of the regulatory pathways, mutations in multiple genes encoding regulatory proteins could simultaneously result in increased capsule production and increased vancomycin resistance.
Another example involves the alternative sigma factor SigB, which has a positive effect on the expression of a regulon that contains genes controlling pigmentation, among others (23). SigB activity is suppressed by the action of the anti-sigma factor RsbW. The activity of RsbW is in turn suppressed by RsbU, so increased RsbU levels have a positive effect on the expression of SigB-controlled genes. On the other hand, an rsbU mutant will have no expression of SigB-controlled genes, including those involved in pigmentation. Selection for increased resistance to teicoplanin in an rbsU mutant resulted in a strain that not only was pigmented but also had intermediate-level resistance to both teicoplanin and vancomycin (23). The relevant mutations inactivated the rsbW gene, allowing increased SigB activity. In addition, the mutation of genes involved in pigmentation had no effect on glycopeptide resistance (23). This suggests that apart from genes involved in pigmentation, the SigB regulon contains a gene(s) that contributes to glycopeptide resistance in S. aureus (23). Recently, it was demonstrated that the deletion of SpoVG, a downstream regulator controlled by SigB, leads to an absence of capsule production and reduced glycopeptide resistance in a VISA strain (305). In a separate study, the overexpression of RsbU and SigB in a VSSA strain resulted in a slight increase in the MIC of vancomycin (57).
A final example of this complex connection between the VISA phenotype and virulence regulons involves the production of protein A, the product of the spa gene. Perhaps the most consistent transcriptional change observed for VISA strains is the downregulation of the spa gene (74, 122, 169, 199, 201, 251, 303). A possibly incomplete list of genes where the gene product has been shown to affect the transcription of spa includes agr (76, 343), sarA (51, 76, 343), sarS (50), arlSR (85, 86), sarT (304), srrAB (262), tcaR (200), mgrA (138), rot (240), ccpA (307), and sarZ (342). Potentially, a mutation in any of these regulatory genes could simultaneously increase vancomycin resistance and decrease protein A production.
Mutations associated with resistance.
It was proposed early that VISA may have evolved in strains with defects in DNA mismatch repair (i.e., strains with an elevated mutation frequency) (221). Schaaff et al. generated a mutS knockout of laboratory strain RN4220 and demonstrated that the rate of acquisition and level of vancomycin resistance attained for RN4220ΔmutS were higher than those for RN4220 (302). Studies that have determined mutation frequencies for clinical isolates of VISA did not detect an increased mutation frequency (236-238). Additionally, an initial analysis of the Mu50 genome sequence suggested a frameshift mutation in mutS (12), raising the possibility that a loss of mutS function could have contributed to the emergence of vancomycin resistance in Mu50; however, a resequencing of mutS from Mu50 did not confirm this frameshift mutation (236), and the sequencing of parental and VISA laboratory-induced isolates found no mutS mutations (95, 186).
Transcriptomic and proteomic analyses have highlighted that there are multiple pathways to the VISA phenotype. However, the most recent applications of comparative genomics using isogenic VSSA/VISA pairs (or series of strains) isolated from patients before, during, and after antibiotic treatment have pinpointed some of the key S. aureus genes that are involved in vancomycin resistance.
In a landmark study involving the complete genome sequencing of an isolate pair (JH1 and JH9) and mutation detection in sequential S. aureus bloodstream isolates (named JH1, JH2, JH5, JH6, and JH9) collected from a patient during 3 months of antibiotic treatment, Mwangi et al. uncovered a total of 35 point mutations across the five isolates (222). The appearance of each mutation (or groups of mutations) was then correlated with changes in vancomycin susceptibility patterns. While no experimental data were provided to support the contribution of individual changes to vancomycin susceptibility, there were only a limited number of mutations observed between successive isolates. One of the first mutations to arise was a nucleotide change in SA1702 (strain N315 locus tag nomenclature), resulting in a predicted amino acid substitution (H164R). SA1702 is a gene of unknown function within the vraSR operon, but interestingly, its predicted protein product shares 42% amino acid similarity with LiaF from Bacillus subtilis. LiaF is a potent negative regulator of LiaR-dependent gene expression (149), and the LiaRS two-component regulatory (2CR) system in B. subtilis drives the cell envelope stress response, responding to changes in the cycling of undecaprenol and disturbances of the cytoplasmic membrane (149). Furthermore, a liaF deletion in B. subtilis leads to the constitutive activation of LiaR-dependent promoters. It is possible that the H164R mutation of SA1702 has led to the upregulation of vraR, the liaR ortholog in S. aureus. In support of this hypothesis, the overexpression of vraR was shown to increase resistance to vancomycin (169). However, while these systems share amino acid sequence identity, LiaRS and VraSR are not functional orthologs, as VraSR can regulate both early and late steps in peptidoglycan synthesis (92, 168, 361).
Other mutations uncovered by Mwangi et al. in the three subsequent isolates (JH5, JH6, and JH9), where the vancomycin MIC increased stepwise from 4 μg per ml to 8 μg per ml, included eight intragenic and nonsynonymous mutations that caused changes in genes of the agr quorum-sensing system, the WalKR cell wall regulatory operon, and a gene potentially involved in peptidoglycan biosynthesis (Table 4) (222).
TABLE 4.
Genes associated with VISAb
| Gene, predicted mutation | Impact of mutation | Description | Reference(s) |
|---|---|---|---|
| graR, nucleotide substitution | Led to a GraR aa substitution, N197S; expression of the mutant graR allele in hVISA strain Mu3 converted this strain to VISA | Comparative genomic study also with allele-swapping experiments using hVISA strain Mu3, VISA strain Mu50, and VSSA strain N315; looking at the contribution of individual genes to vancomycin susceptibility | 225 |
| graRS, DNA deletion | Deletion of graR led to increased susceptibility to vancomycin | Reverse genetics, using laboratory strains of S. aureus and VISA strain Mu50; this study shows that GraRS and VraFG are important mediators of VISA | 204 |
| vraFG, DNA deletion | Deletion of vraG led to increased susceptibility to vancomycin | ||
| SA1702,a nucleotide substitution | Led to a predicted aa substitution in SA1702, H164R; SA1702 is a protein of unknown function immediately upstream of vraS; this isolate was obtained when the vancomycin MIC increased from 1 to 4 μg per ml | Comparative genomic study of five sequential S. aureus bloodstream isolates obtained from a single patient during 3 mo of antibiotic treatment | 222 |
| SA1249, frameshift | Led to a predicted loss-of-function in SA1249; the function of SA1249 is unknown, but its genomic location suggests that it might be part of the murG operon with a role in peptidoglycan synthesis; this isolate was obtained when the vancomycin MIC increased from 4 to 6 μg per ml | ||
| agrC, frameshift | Led to a predicted loss of function in agrC; this gene is part of the agr quorum-sensing locus; this is one of six mutations affecting protein-coding sequences, where the vancomycin MIC increased from 6 to 8 μg per ml | ||
| yycH, premature stop codon | Led to a predicted loss of function in yycH (90% protein not translated); YycH is a hypothetical protein within an operon containing the two-component regulator that controls cell wall synthesis (WalKR/YycFG) by promoting expression of genes involved in autolysis; this is one of six mutations affecting protein-coding sequences, where the vancomycin MIC increased from 6 to 8 μg per ml | ||
| isdE, nucleotide substitution | Led to a predicted aa substitution in IsdE, A84V; involved in heme-iron transport; this is one of six mutations affecting protein-coding sequences, where the vancomycin MIC increased from 6 to 8 μg per ml | ||
| prsA, frameshift | Led to a possible loss of function in PrsA; PrsA is a putative membrane-linked ribose-phosphate pyrophosphokinase that can chaperone secreted proteins in Gram-positive bacteria; this is one of six mutations affecting protein-coding sequences, where the vancomycin MIC increased from 6 to 8 μg per ml | ||
| SA2094, nucleotide substitution | Led to a predicted aa substitution in SA2094, A94T; the function of SA2094 is unknown, but it is predicted to be membrane associated, and it shares similarity with an Na+/H+ antiporter from B. subtilis; this is one of six mutations affecting protein-coding sequences, where the vancomycin MIC increased from 6 to 8 μg per ml | ||
| graS, nucleotide substitution | Led to an aa substitution in GraS, T136I; this was one of six mutations detected in VISA strain JKD6008 compared to VSSA progenitor strain JKD6009; replacement of the mutant graS allele in VSSA strain JKD6009 resulted in an increase in the MET MIC from 2 to 6 μg per ml | Comparative genomic study with allele-swapping experiments with two S. aureus blood culture isolates (ST239) obtained from a patient before and after 42 days of vancomycin therapy; this study highlights the key role that GraRS plays in the formation of VISA among clinical isolates | 123 |
| graRS, DNA deletion | Led to a loss of GraRS function in VSSA JKD6009 and resulted in increased sensitivity of this strain to vancomycin, with a decrease in the MET MIC from 2 to 1 μg per ml | ||
| vraS, premature stop codon | Led to a predicted loss of VraS function in VSSA strain Mu50Ω; replacement of this disrupted vraS allele with intact vraS from VISA strain Mu50 resulted in an increase in the vancomycin MIC from 0.5 to 3.5 μg per ml | Comparative genomic study also with allele-swapping experiments using VISA isolates Mu50 and Mu50Ω; the latter strain was isolated from the same patient 1.5 yr after Mu50 was isolated | 60 |
| graR, no change compared with other VSSA isolates | Replacement of this graR allele in strain Mu50Ω with both vraS and graR from VISA strain Mu50 resulted in an increase in the vancomycin MIC for Mu50Ω from 0.5 to 6.0 μg per ml; these experiments show that point mutations in vraSR and graRS together are sufficient to induce VISA | ||
| yycFG, IS256 upstream insertion | Insertion of IS256 led to upregulation of yycFG (walKR); overexpression in trans of yycFG led to increase in vancomycin resistance | Comparative transcriptome study with overexpression experiments using hVISA strain SA137/93A and laboratory derivative SA137/93G | 141 |
| mgrA and sarA, DNA deletion | Led to a loss of SarA and MgrA function; SarA and MgrA are negative regulators of murein hydrolases (or autolysins); these enzymes (both dimeric, winged-helix proteins) are required for cell wall turnover; the double mutant showed increased Triton X-100-induced autolysis and increased sensitivity to killing by vancomycin and oxacillin | Used laboratory strains COL, MW2, and derivatives | 353 |
| spoVG, deleted | Deletion of spoVG decreases resistance to oxacillin and teicoplanin, with less impact on vancomycin resistance; loss of spoVG also led to a loss of capsule production; the yabJ/spoVG operon is under the control of the alternative sigma factor (SigB); the SigB regulon has been implicated in glycopeptide resistance | Used laboratory strains COL, Newman, and derivatives | 305 |
| vraS, nucleotide substitution | Led to a predicted aa substitution in VraS, I5N; this mutation was present only in Mu3 and Mu50 and not in other VSSA isolates | Comparative genomic VISA strain, Mu50, hVISA strain Mu3, and VSSA strain N315; no experimental data provided | 234 |
| mprF/fmtC, Tn917 insertion | Insertion of Tn917 led to MprF loss of function and decreased vancomycin resistance; MprF (FmtC) is involved in synthesis of lysyl-phosphatidylglycerol (a major cell wall component), changing the content of the cell wall and increasing the net negative charge | Mutagenesis study using S. aureus RN4220 | 286 |
| mprF/fmtC, Tn551 insertion | Insertion of Tn551 led to MprF loss of function and decreased vancomycin resistance in VISA strains but slightly increased vancomycin resistance in VSSA strains | Mutagenesis study using S. aureus COL and vancomycin-resistant laboratory derivatives | 228 |
| trfA/trfB, insertional inactivation (antibiotic resistance marker) | Loss of this locus led to an increase in susceptibility to teicoplanin, oxacillin, and vancomycin; function of TrfAB not known | Conducted genetic studies with S. aureus RN4220, laboratory-derived teicoplanin-resistant mutants, and clinical strain NRS3 | 270 |
| ccpA, insertional inactivation (antibiotic resistance marker) | Loss of CcpA (carbon catabolite protein A) impacts S. aureus in many ways (reduces growth, carbon metabolism, RNAIII expression, and capsule synthesis); loss of CcpA also reduced resistance to teicoplanin; the effect of this mutation on vancomycin resistance was not reported | Conducted genetic studies with S. aureus strains COL, Newman, RN4220, and laboratory-derived mutants | 307 |
| agr, insertional inactivation (antibiotic resistance marker) | Loss of Agr led to increased probability of hVISA when population was exposed to 1 μg/ml vancomycin | Conducted genetic studies with S. aureus strains RN6390, RN6607, and RN4580 | 294, 296 |
| rsbU, DNA deletion | RsbU is an anti-anti-sigma factor; selection for teicoplanin resistance in an rsbU mutant resulted in GISA, suggesting that a gene(s) (e.g., spoVG) within the SigB regulon can mediates glycopeptide resistance (refer to reference 305) | Used S. aureus strain MRGR3, a teicoplanin-resistant mutant obtained from animal studies of wound infection | 23 |
| pbp4, DNA deletion | Loss of Pbp4 led to increased vancomycin resistance in VSSA, while overexpression of Pbp4 reduced vancomycin resistance in VISA; Pbp4 may be involved in transpeptidation, i.e., formation of peptidoglycan cross-linking | Conducted genetic studies using S. aureus strains COL, RN450M, N315, and their laboratory-derived VISA mutants | 81 |
Gene identifier according to S. aureus N315 locus tags.
aa, amino acids.
Neoh et al. also used comparative genomics to investigate the evolution of VISA from hVISA by comparing the genomes of hVISA strain Mu3 with a related VISA strain isolated from a different patient, strain Mu50 (225). Sixteen point mutations were observed, including a predicted amino acid substitution (N197S) in GraR of Mu50, the response regulator of the above-mentioned GraRS two-component regulatory system. The introduction of GraR N197S from Mu50 into Mu3 converted this hVISA strain into a full VISA strain, accompanied by cell wall thickening and decreased autolysis (225). The introduction of GraR N197S to N315 (VSSA) had no effect upon vancomycin susceptibility, thus indicating that additional genomic changes in N315 were required to develop the VISA phenotype. Microarray studies of Mu3 expressing GraR N197S suggested that at least 14 genes were specifically upregulated by GraR, including the genes encoding the ABC transporters VraFG and VraDE. Interestingly, these experiments also showed that GraR N197S expression in Mu3 led to a downregulation of spa. The reduced level of expression of protein A is a known VISA phenomenon. In contrast, a graRS knockout in Mu3 did not lead to the VISA phenotype or any of the associated cell wall changes. These data suggest that perhaps the N197S amino acid substitution resulted in GraR remaining in an activated “on” state. Most recently, that team used genomics to compare clinical VISA strain Mu50 with a strain with less vancomycin resistance isolated 1.5 years later from the same patient, strain Mu50Ω (60). Mu50Ω had a loss-of-function mutation in vraS. The restoration of intact vraS(I5N) and the replacement of graR(N197S) from Mu50 resulted in Mu50Ω developing the full Mu50 VISA phenotype, clearly demonstrating that point mutations in these two regulatory elements are sufficient for VISA, at least in Mu50 (60).
Another comparative genomics study investigated the evolution of VISA and also implicated the graRS locus in resistance (123). An isogenic pair of bloodstream VSSA and VISA isolates, obtained from a patient before and after 42 days of vancomycin therapy, were fully sequenced and compared. Six mutations were detected, and one of these mutations was in graS, resulting in an amino acid substitution (T136I). This mutation was confirmed to be a key contributor to an increase in vancomycin resistance by the replacement of graS in VSSA strain JKD6009 with graS(T136I) from JKD6008. Significantly, however, the allele swap did not restore the full VISA phenotype, and so presumably, one or more of the remaining five mutations detected in JKD6008 are also required to generate the full VISA phenotype.
Table 4 contains a summary of genes linked to intermediate-level vancomycin resistance in S. aureus. A number of loci are included in Table 4, and a number of methodologies have been used to investigate this problem, but the key message that has emerged from genome-wide studies is that small changes in key regulatory genes involved in cell wall metabolism have a profound impact on vancomycin susceptibility. With the rapidly diminishing cost and concomitant higher throughput of next-generation DNA sequencing, comparative genomics can now be routinely applied to identify mutations associated with resistance in clinical MRSA isolates. However, to go beyond an association and understand the contribution of each mutation to resistance, we need to continue to develop the tools that will facilitate the genetic manipulation of S. aureus. These include tools that permit the rapid creation of unmarked, site-directed mutants in S. aureus, such as the allelic exchange vector pKOR1 (14), and, perhaps most importantly, methods for efficient delivery of DNA to those clinical strains of S. aureus that are refractory to transformation by electroporation.
Other Features of hVISA and VISA
The development of reduced vancomycin susceptibility in S. aureus in many cases appears to be associated with changes in bacteria that are predicted to impact host-pathogen interactions and are not linked directly to the expression of the antibiotic resistance phenotype. Although only limited work has been done in this area, it has demonstrated changes in in vitro biofilm formation, polysaccharide capsule production, surface protein A expression, and agr activation in hVISA and VISA isolates, with preliminary results suggesting an impact on host immune responses (121, 122). Interestingly, it was demonstrated using a rat model of chronic foreign-body infection that reduced teicoplanin and vancomycin susceptibility can occur in vivo without antibiotic exposure (367), suggesting a possible coevolutionary effect where bacterial changes to favor persistent infection may lead to reduced glycopeptide susceptibility, and vice versa.
Surface proteins.
In strain 14-4 (derived from rat tissue cage infection), an increased surface expression of fibronectin binding proteins was associated with increased fibronectin-mediated adherence in an in vitro model (269). As described above, a consistent transcriptional change observed in VISA strains is the downregulation of the spa gene, encoding protein A (74, 122, 169, 199, 201, 251, 303). For a small number of strains, we have demonstrated that reduced spa expression levels in hVISA and VISA strains are associated with decreased levels of cell surface protein A (Fig. 1) (122). Protein A is a very important S. aureus surface protein that is highly conserved and abundantly expressed in infections of the lung (83). It possesses antiphagocytic activity by binding the Fc portion of immunoglobulin (275), acts as a B-cell superantigen (241), activates clotting by binding von Willebrand factor (96, 97), and has been shown to activate tumor necrosis factor (TNF) receptor 1 (TNFR1) and lead to proinflammatory signaling, which is important in the pathogenesis of staphylococcal pneumonia (98). Recently, it was shown that protein A also cleaves TNFR1 from the surface of epithelial cells and macrophages, which limits TNF-α signaling. This process is mediated by a protein A interaction with the epidermal growth factor receptor (EGFR) and by the activation of tumor necrosis factor alpha-converting enzyme (TACE), the TNF1 sheddase (98). Changes in protein A expression are therefore likely to impact host-pathogen interactions in hVISA/VISA strains.
Capsule.
An early investigation of two VISA strains from the United States (MI and NJ) demonstrated an increase in extracellular matrix by electron microscopy (326). Several transcriptional studies demonstrated an upregulation of genes associated with capsule biosynthesis (57, 122, 199), and increased capsule production has been demonstrated for a small number of hVISA and VISA strains that have been tested (Fig. 5). The majority of S. aureus strains produce microcapsules (10, 239, 368). Although 11 serotypes of S. aureus have been found, most clinical isolates produce polysaccharide capsule type 5 or 8 (312). The type 5 and 8 capsules are structurally very similar and differ only in the linkages between sugars and in O-acetylation (239).
The production of polysaccharide capsule by S. aureus has an effect on host immune evasion as well as changing endothelial binding and virulence properties. Although early results were conflicting, capsule expression does protect the bacterium from phagocytic uptake and killing by human polymorphonuclear leukocytes (261, 336). Initial animal studies of virulence in wild-type and capsule knockout strains did not demonstrate a difference (227, 260, 349). Later studies demonstrated that type 5 encapsulated strains of S. aureus were more virulent in a mouse bacteremia model, a renal abscess model, and a septic arthritis model (274). Recently, it was demonstrated that capsule expression reduces clumping factor A-mediated binding to fibrinogen and platelets, suggesting that capsule can mask surface adhesins and prevent binding (274).
d-Alanine esters in teichoic acids.
Mutations, inactivation, and altered expression of graRS have been linked to changes in vancomycin susceptibility in S. aureus (57, 60, 111, 123, 204, 225). GraRS (Aps) is known to control the dltABCD operon, which controls the alanylation of wall teichoic acids in response to antimicrobial challenge, indicating that the structure of teichoic acids can change in response to challenges (111, 180, 181). The Aps and dltABCD pathway is linked to cationic antimicrobial peptide resistance in S. aureus, and the positive charge of d-alanine residues repels positively charged molecules such as defensins (54, 248, 249). There is also evidence of a link between the d-alanylation state of teichoic acids and vancomycin susceptibility in S. aureus, where a dlt mutant strain lacking in d-alanine in teichoic acids was shown to have increased vancomycin susceptibility compared to that of the wild-type strain (249). Wall teichoic acids also have a role in attachment to host cells, with studies demonstrating reduced nasal colonization and reduced binding to endothelial cells in strains deficient in WTA and in strains with a reduced dltABCD-mediated alanylation of teichoic acids (378, 380, 381). It is therefore likely that for isolates of hVISA or VISA where the development of resistance is associated with an increased level of expression of graRS or mutations in the locus, alterations in susceptibility to antimicrobial peptides are likely to occur, favoring resistance to these agents (Fig. 1). Further work is required to clearly define the interplay between hVISA, VISA, and susceptibility to antimicrobial peptides in S. aureus.
Accessory gene regulator (agr).
The agr locus is a quorum-sensing system that consists of a ∼3-kb locus with divergent transcription units, which are driven by two promoters, P2 and P3 (231). The agr locus consists partly of 4 genes (agrBDCA) controlled by the P2 promoter that act like a two-component regulatory system with its autoinducing ligand. This transcript is called RNAII. The autoinducing peptide is encoded by agrD and is processed and secreted by the transmembrane protein AgrB (394). agrC acts as the sensor histidine kinase that is phosphorylated in response to the autoinducing peptide (182), while agrA is the response regulator that is thought to bind and activate the two agr promoters, P2 and P3, thus completing the autoinducing circuit; however, this has been difficult to demonstrate definitively (231).
The agr locus is conserved throughout the staphylococci, with a hypervariable region found between the 3′ end of agrB and the 5′ region of agrC (and including agrD) leading to the designation of four agr specificity groups or types (75). The ability of an autoinducing peptide to activate its receptor is highly sequence specific, and a single amino acid change can alter group specificity (231). Therefore, functional variants of the agr locus would lead to cross-group inhibition rather than cooperative communication (231). The agr types appear to map to different clonal complexes by multilocus sequence typing (144), suggesting that agr may have evolved early in the staphylococcal evolutionary process (390).
An RNA molecule (RNAIII) (∼0.5 kb) is produced by the divergent agr transcript promoted by P3 and overlaps the hld gene, which encodes delta-hemolysin (232). RNAIII is the intracellular effector molecule of the agr locus and is responsible for the direct activation or inhibition of other loci. RNAIII transcription increases during growth and is maximal in the postexponential growth phase (364). It has a long half-life of approximately 15 min (143). RNAIII has a complex secondary structure consisting of 14 hairpins, many of which are conserved in all staphylococcal species (17). RNAIII upregulates the transcription of genes encoding most of the extracellular proteins and downregulates the transcription of genes encoding many surface proteins (232, 301). RNAIII was also shown to act at the level of translation to control the expression of alpha-hemolysin and protein A (135, 232). Although hld is cotranscribed with RNAIII, it does not have any regulatory effect, as the deletion of the genomic region encoding this small 26-amino-acid hemolysin did not lead to an agr-negative phenotype (143).
A number of studies have linked alterations in agr activation or function with vancomycin tolerance, an increased tendency to develop vancomycin resistance, and the presence of the hVISA or VISA phenotype (121, 269, 295, 296, 357). In a number of these studies, determinations of agr activity were performed by the analysis of delta-hemolysin production on sheep blood agar; however, this is only a semiquantitative measure (296). Similar results were found by using microarray transcriptional analysis or real-time quantitative PCR to measure RNAIII transcripts in paired isolates (121, 269). Although mutations have been found in the agr locus in a small number of isolates (296), reduced levels of agr expression occur without mutations in the locus (121, 269). The mechanisms of reduced levels of agr expression in these isolates are not completely understood.
Initially, it appeared that agr group II isolates were overrepresented in VISA strains from around the world (369); however, a subsequent study from Europe demonstrated that many of their VISA and hVISA isolates were of agr group I (295, 296). In addition, it was demonstrated that S. aureus from agr groups I to IV all develop intermediate vancomycin resistance upon in vitro exposure to the drug (358). It was demonstrated that when hVISA or VISA develops from VSSA, there is a reduction in levels of agr expression in the resistant isolates (121, 269). Given the global regulatory role that agr plays in virulence factor expression, in particular the production of exotoxins, it is likely that strains of hVISA and VISA that have reduced agr activity produce fewer exotoxins than parental strains. This has not been tested specifically.
In a number of in vitro studies, reduced agr function has been shown to favor the development of vancomycin resistance (295, 296, 358); however, an association between agr function and teicoplanin resistance development was not demonstrated (281). Assuming that the relationship between agr function and the development of vancomycin resistance exists, it is worth noting that in one study, 48% of hospital MRSA strains were found to have reduced agr function, compared to 3.5% of community-associated MRSA strains (357), suggesting a possible higher tendency for hospital MRSA strains to develop into hVISA or VISA strains. A clinical study also linked agr group II polymorphisms with poor responses to vancomycin therapy for patients with MRSA infections (211). In addition, a loss of agr function was associated with a reduced susceptibility to platelet microbiocidal protein and reduced autolysis, a common feature of hVISA and VISA strains (295, 296).
Interestingly, some studies of persistent S. aureus bacteremia isolates (where hVISA and VISA strains have not been detected or assessed) have demonstrated some changes in isolates similar to those seen with hVISA and VISA. In particular, these changes have included reduced agr function, reduced susceptibility to host antimicrobial peptides including platelet microbiocidal protein and hNP-1, and increased adhesion to fibrinogen, fibronectin, and endothelial cells (88, 297, 391). In a study by Xiong et al., although there was no formal assessment for hVISA, in a rabbit endocarditis model, there was a reduced efficacy of vancomycin in one persistent bacteremia isolate compared to the control (391).
Host immune interactions and virulence.
A few studies have suggested that the acquisition of vancomycin or teicoplanin resistance by S. aureus is associated with changes in bacterial fitness and virulence potential. In a rat infection model, a reduced fitness of a teicoplanin-resistant laboratory-derived strain of S. aureus was demonstrated (201). Using the invertebrate model Galleria mellonella, the impact of reduced vancomycin susceptibility and altered agr function was assessed for a few isolates (244). As the vancomycin MIC increased, virulence in the model was reduced. Additionally, S. aureus with reduced agr function demonstrated reduced virulence.
As a potential clinical correlate of the laboratory studies, a recent clinical study assessed the relative clinical importance of hVISA/VISA compared to VSSA with regard to the likelihood of causing active clinical infection (118). That study included 59 patients with VSSA isolates and 58 patients with hVISA/VISA isolates defined by PAP and microbroth MIC (56 hVISA isolates with a vancomycin MIC of 2 μg per ml and 2 VISA isolates with a vancomycin MIC of 4 μg per ml). The hVISA/VISA isolates were less likely to be associated with clinical infection; in particular, this was due to a decrease in the rate of bacteremia caused by hVISA/VISA compared to the rate of bacteremia caused by VSSA.
Because of the changes in the cell surface of hVISA and VISA isolates that we have studied, we assessed the impact of the hVISA and VISA phenotypes on proinflammatory cytokine stimulation by using an in vitro system (122). In the clinical isolate pairs tested, the hVISA and VISA phenotypes were associated with a significant reduction in levels of NF-κB, TNF-α, and interleukin-1β (IL-1β) expression. In a clinical study of MRSA bacteremia, the risk of shock was lower for patients infected with strains of S. aureus with higher vancomycin MICs, suggesting a possible correlation with in vitro data (332). In that clinical study, proinflammatory cytokine levels were not assessed for the patients with S. aureus bacteremia; however, such a study linking S. aureus vancomycin susceptibility to patient immune responses would provide an interesting insight into the impact of vancomycin resistance on host-pathogen interactions.
LABORATORY DETECTION
Despite significant effort, the genetic determinants of hVISA and VISA have not been completely resolved. Therefore, no molecular-based assays are available for the detection of hVISA and VISA. The accurate detection of hVISA and, to some extent, VISA has been difficult with phenotypic methods; however, increasing amounts of data support a number of methods for the screening for and confirmation of hVISA and VISA infection. Isolates of S. aureus with vancomycin MICs of 4 to 8 μg per ml are rare, while S. aureus isolates with vancomycin MICs of 2 μg per ml are relatively common. For example, Surveillance Network data from the United States for 2005 found that only 0.2% (n = 520) of all S. aureus isolates had a vancomycin MIC of ≥4 μg per ml, while 16.2% (n = 39,223) had a vancomycin MIC of 2 μg per ml (346). A high rate of hVISA isolation can be detected for S. aureus isolates with a vancomycin MIC of ≤2 μg per ml in some settings (118, 175); therefore, the vancomycin MIC result alone is unable to accurately distinguish hVISA from VSSA isolates, and the use of MIC testing alone will fail to detect hVISA strains that are relatively common among isolates of S. aureus with broth MICs of 2 μg per ml (Fig. 4).
The relative levels of resistance to vancomycin and teicoplanin can vary between different VISA isolates. Some authors suggested classifying strains as class A, B, and C glycopeptide resistant (class A, vancomycin intermediate and teicoplanin intermediate; class B, vancomycin intermediate and teicoplanin susceptible; class C, vancomycin susceptible and teicoplanin intermediate) (29); however, this has not been widely adopted.
Colony Morphology of hVISA and VISA
The morphological features of VISA and hVISA strains can be different from those of standard S. aureus cultures on agar plates, although these changes are often subtle variations such as reduced pigmentation or slightly smaller colony size (see “SCV S. aureus and hVISA/VISA”). In some cases, the changes are more obvious. For example, hVISA and VISA isolates can be slow growing and can generate a “mixed” colony morphology where large and small colonies and colonies with different pigmentations are present in a pure culture from a clinical specimen (Fig. 7) (197). It is therefore important to test each of the different morphotypes from a mixed-morphotype culture of S. aureus for vancomycin susceptibility.
FIG. 7.
Examples of changes in colony morphology that can occur with hVISA and VISA strains. (A) Mixed-colony morphologies from a pure culture of S. aureus from a bile specimen demonstrate remarkable heterogeneity in colony size and hemolytic activity. The small colonies were VISA, while the larger colonies were VSSA. (Reprinted from reference 197 with permission.) (B) Mixed-colony pigmentation in a pure culture of S. aureus, where the yellow colonies were VISA and the gray colonies were VSSA. The different morphologies were identical by pulsed-field gel electrophoresis. (C and D) Subtle changes in colony morphology that occur in VISA strains compared to parental VSSA strains. JKD6009 (C) is a VSSA strain, and JKD6008 (D) is a VISA strain isolated from the same patient after failed vancomycin therapy. The pulsed-field gel electrophoresis patterns were also identical for JKD6008 and JKD6009.
VISA and Vancomycin MIC Testing
Vancomycin-intermediate S. aureus is defined by a vancomycin MIC result of 4 to 8 μg per ml (53). Possible methods for defining the vancomycin MIC include CLSI-approved methods (agar dilution and broth MIC), Etest MIC using a 0.5 McFarland standard, 24 h of incubation on Mueller-Hinton agar (MHA), and other commercial tests such as MicroScan and Vitek 2. The 2-McFarland-standard macromethod Etest is a screening tool for hVISA and VISA but is not a true vancomycin or teicoplanin MIC, and the results of the macromethod Etest should not be reported as a true MIC. Subtle but potentially important variability in vancomycin MIC results is obtained with different methods (130, 174, 263, 291, 338). In a study by Leonard et al., 100 hVISA and 50 VSSA isolates were tested by vancomycin broth and Etest MICs. Overall, the MIC result tended to be higher by Etest than by broth microdilution (174). In a study by Prakash et al. (263), 101 MRSA isolates were tested by agar dilution and microbroth MICs according to CLSI criteria as well as by standard vancomycin Etest performed with two different brands of Mueller-Hinton agar (BBL and Remel). While there was reasonable correlation between the CLSI agar dilution and broth microdilution methods, the Etest results were consistently one twofold dilution higher than results for the other methods, changing the modal vancomycin MIC from 1 μg per ml (broth and agar dilution) to 2 μg per ml (Etest) (263). Similar results were recently reported for 1,800 randomly selected MRSA isolates, where Etest MICs were 0.5 to 1.5 log2 dilution steps higher than the reference broth MIC results (291). Even the agar dilution and broth microdilution methods demonstrated some inconsistency, with 20.8% of isolates having a vancomycin MIC of 0.5 μg per ml by broth microdilution and 1% having a vancomycin MIC of 0.5 μg per ml by agar dilution. Swenson et al. investigated a number of commercial assays and reference methods for the detection of VISA (338). A number of the assays (Sensititre, Vitek Legacy, and Vitek 2) tended to categorize VISA strains as susceptible, while others (MicroScan and Phoenix) tended to categorize susceptible strains as VISA. Given these inconsistencies, determinations of vancomycin broth MIC performed according to CLSI criteria should be used to define VISA (53). Additionally, for the purposes of further study and clinical treatment decisions, reporting of vancomycin MIC results should clearly describe the method used to generate the result.
Many of the VISA strains with a vancomycin MIC of 8 μg per ml that have been reported from around the world have developed in patients during failed vancomycin therapy, under vancomycin selective pressure (258, 316). A number of these strains demonstrated lower vancomycin MICs (as low as 2 μg per ml) when subsequently retested, demonstrating the unstable nature of this resistance phenotype and a tendency for some strains to have a reduction in MICs when vancomycin selective pressure is removed (339).
There has been recent interest in the use of vancomycin MIC results (within the susceptible range) to predict outcomes for patients with serious S. aureus infections being treated with vancomycin (184, 298, 332). Generally, these studies demonstrated a higher failure rate for vancomycin treatment of S. aureus strains with higher vancomycin MICs within the susceptible range (see “Clinical Studies”). None of these studies assessed the presence of the hVISA phenotype, and it is possible that this may have been an even better predictor of clinical failure. The three studies used different methods for vancomycin MIC determinations, agar dilution MIC (298), Etest MIC (184), and microdilution MIC (332), making it somewhat difficult to draw direct comparisons between studies. Clearly, given the apparent variability in vancomycin MIC results obtained with the different methods, the use of the vancomycin MIC to predict the outcome of serious S. aureus infections needs to take into account the method used and the results of studies using that particular method.
hVISA
There is currently no standardized method for the accurate detection of hVISA (183), which makes laboratory testing and interpretation of the clinical significance of hVISA difficult. A large number of screening methods have been analyzed, and although a number of them are useful for the detection of VISA, the majority are not adequate for screening for hVISA. Any method to detect the resistant subpopulation of cells present in an hVISA strain is inherently more difficult than standard testing according to CLSI criteria, because the resistant subpopulation can be present in a low ratio compared to the susceptible population (typically 10−5 to 10−6). Given the low inoculum used for CLSI broth and agar dilution methods, the hVISA phenotype will not be detected. Methods to detect hVISA therefore tend to rely on the testing of a higher inoculum (for example, 108 CFU for population analysis and a 2-McFarland-standard inoculum for the MET) and methods to promote the growth of the resistant subpopulation, such as prolonged incubation (usually 48 h) or use of more nutritious media (e.g., brain heart infusion agar [BHIA] or Mueller-Hinton agar [MHA] with blood). From a laboratory point of view, PAP appears to be the best method for the confirmation of the presence of hVISA, while potentially effective screening tests include modified Etest methods (MET or Etest GRD) and screening agar consisting of MHA with 5 μg teicoplanin per ml when population analysis is used as the reference method (see Table 5 for a summary of screening methods).
TABLE 5.
Laboratory detection of hVISA and accuracy of methods compared to those of modified population analysis/area under the curvea
| Method | Sensitivity | Specificity | Reference(s) |
|---|---|---|---|
| Vancomycin broth MICb | 11% | 100% | 372 |
| BHIA + vancomycin at 6 μg per ml, 10 μl of a 0.5-McFarland-standard suspension (BHIA6V)c | 48 h, 4.5-12% | 48 h, 68-100% | 370, 389, 393 |
| MHA + teicoplanin at 5 μg per ml, 10 μl of a 2-McFarland-standard suspension (MHA5T)d | 48 h, 65-79% | 48 h, 35-95% | 82, 252, 370, 389, 393 |
| MHA + teicoplanin at 5 μg per ml, 10 μl of a 2-McFarland-standard suspensione | 48 h, 98% | 48 h, 53% | 82 |
| MHA + vancomycin at 5 μg per ml, 10 μl of a 0.5-McFarland-standard suspension | 48 h, 1-20% | 48 h, 59-99% | 370, 372 |
| Simplified PAPf | 48 h, 71% | 48 h, 88% | 372 |
| Macromethod Etest (MET) | 48 h, 69-98.5% | 48 h, 89-94% | 174, 289, 370, 372, 389 |
| Etest GRD | 24 h, 70-77% | 24 h, 98-100% | 174, 393 |
| 48 h, 93-94% | 48 h, 82-95% |
In all studies, vancomycin population analysis/area under the curve (PAP/AUC) was considered the “gold standard” for calculating sensitivity and specificity.
Evaluation of vancomycin broth MICs included detection of VISA and hVISA. By definition, hVISA will not be detected by determinations of broth MIC.
BHIA6V is the screening plate recommended by the CDC and the Clinical and Laboratory Standards Institute for the detection of VRSA and VISA strains with vancomycin MICs of ≥8 μg per ml (http://www.cdc.gov) (53), which is spot inoculated with 10 μl from a 0.5-McFarland-standard suspension and read at 24 and 48 h. The culture is considered positive if there is growth of 2 or more colonies.
MHA5T is the screening plate recommended by the Comité de l'Antibiogramme de la Société Française de Microbiologie (http://www.sfm.asso.fr),which is spot inoculated with 10 μl from a 2-McFarland-standard suspension and read at 24 and 48 h. The culture is considered positive if there is growth of 1 or more colonies.
This analysis included some isolates with a hetero-teicoplanin-resistant but vancomycin-susceptible phenotype by population analysis.
Simplified PAP consists of inoculating BHIA with 4 μg per ml of vancomycin with 10 μl from a 0.5-McFarland-standard suspension and reading at 24 and 48 h for any growth.
Screening methods.
hVISA strains tend to have higher vancomycin MICs within the susceptible range such that 30 to 49% of S. aureus isolates with MICs of 2 μg per ml determined by Etest are hVISA strains by detailed PAP testing (Fig. 4) (174, 220), while in our experience, approximately 60% of our MRSA isolates with a vancomycin microbroth MIC of 2 μg per ml are hVISA strains (118). Some strains of S. aureus with Etest MICs of 0.75 μg per ml or microbroth MICs of 0.5 to 1 μg per ml are hVISA strains when tested by PAP (174).
Early after the emergence of hVISA and VISA, it became clear that automated susceptibility testing methods and standard disc diffusion were inadequate for the detection of VISA let alone the low-level-resistant subpopulations present in hVISA strains (91, 127, 347). A number of screening tests for hVISA and VISA have since been developed and assessed, although the sensitivity has been generally low for the detection of hVISA (Table 5). A detailed analysis of different screening methods for hVISA detection was reported by Walsh et al. in 2001 (372), and two subsequent multicenter analyses helped to define the optimal screening methods for hVISA by using population analysis as the “gold standard” (370, 389). Significant intra- and interlaboratory variability exists for a number of the proposed screening methods (370).
In the original report by Hiramatsu et al., a simplified population analysis was described (114). This method involved inoculating 10 μl of a suspension of 108 CFU per ml onto BHIA with 4 μg vancomycin per ml. Any growth at 24 h was considered potential VISA and tested by determination of the vancomycin MIC. Any growth after 48 h was considered potential hVISA, and this was confirmed if the vancomycin MIC was 8 μg per ml after selection on vancomycin-containing medium and if the phenotype was maintained for >9 days on drug-free medium. A number of studies used the simplified PAP as a screening method (22, 154, 158, 383); however, the method described for the confirmation of hVISA by Hiramatsu et al. using this approach is not practical, and there have been significant concerns raised about the induction of resistance when testing isolates after growth on vancomycin (128, 129, 344, 372). In addition, other studies found that this method has poor reproducibility and detects Mu3 only 80% of the time (372, 387). A number of variations of this method have been reported, including the use of different media such as MHA with different vancomycin concentrations (2 to 5 μg per ml) (133, 271) and the use of a higher inoculum size on the screening plate (352). Jung et al. performed a one-point population analysis by using BHIA plates with 4 μg vancomycin per ml but found that it was not superior to the original method proposed by Hiramatsu et al. (152). These differences in methodologies make it difficult to interpret the significance of studies of hVISA epidemiology and clinical impact.
Two agar screening plates have been most extensively tested in a number of studies. The CDC recommends BHIA with 6 μg vancomycin per ml (BHIA6V) inoculated with 10 μl of a 0.5-McFarland-standard suspension, aimed primarily at detecting VRSA and possibly VISA. The growth of two or more colonies after 48 h is considered a positive result (344, 372, 393). The agar was originally developed to screen for vancomycin-resistant enterococci that typically have much higher vancomycin MICs (8). An important component of this method is the use of commercially prepared BHIA6V plates, as significant variability in performance exists if in-house medium is used. Not surprisingly, a number of studies demonstrated a very low sensitivity of the BHIA6V method for the detection of hVISA (12%) compared to that of PAP, but detection rates of VISA with an MIC of 8 μg per ml are good (389, 393); however, a recent study detected only 33% of VISA isolates with a vancomycin MIC of 4 μg per ml using this screening agar (338). The Comité de l'Antibiogramme de la Société Française de Microbiologie (http://www.sfm.asso.fr) recommends the use of MHA with 5 μg teicoplanin per ml (MHA5T). This screening plate has been tested by a number of studies using an inoculum of 10 μl of a 2-McFarland-standard suspension for the detection of hVISA and VISA (49, 389, 393). This has been adopted in the European Antimicrobial Resistance Surveillance Scheme; however, the inoculum described in this document is 10 μl of a stationary-phase culture grown overnight in BHI broth (BHIB) (http://www.rivm.nl/earss/). Any growth is considered positive after 48 h of incubation. Although this screening plate has a higher sensitivity than the BHIA6V plate, the sensitivity remains relatively low, at 65 to 79% for the detection of hVISA and VISA (389, 393), and the specificity was reported to be low by some studies (252).
Other screening plates have also been variously tested but have generally proven to have poor sensitivity and specificity for hVISA detection and have not been widely used. These include BHIA with 2 μg vancomycin per ml and BHIA with 5 μg teicoplanin per ml, both inoculated with 100 μl of a bacterial suspension adjusted to a 2 McFarland standard (21), as well as MHA with 5 μg vancomycin per ml inoculated with 10 μl of a 0.5-McFarland-standard suspension, which was aimed at detecting VISA (133), and MHA with 8 μg teicoplanin per ml inoculated with 10 μl of a 0.5-McFarland-standard suspension (242). The use of BHIA with 3 μg vancomycin per ml inoculated with 10 μl of a 0.5-McFarland-standard suspension had a high false-positive rate reported by one study (165). Fitzgibbon et al. assessed a range of screening media for the detection of hVISA (MHA with 5 μg teicoplanin per ml and BHIA with 5 μg teicoplanin per ml), all inoculated with 10 μl of a 0.5-McFarland-standard suspension, a 2-McFarland-standard suspension, and a BHIB stationary-phase culture (82). The hVISA-positive isolates were confirmed by PAP but included isolates that were positive for hetero-teicoplanin resistance but negative for hVISA by PAP. All BHIA plates and MHA inoculated with the stationary-phase culture had a sensitivity of 100%, but specificity was only between 4 and 57%. MHA inoculated with a 0.5 McFarland standard had sensitivity and specificity of 66% and 82%, respectively, while MHA inoculated with a 2-McFarland-standard suspension had sensitivity and specificity of 98% and 53%, respectively. Additional supplementation of BHIA with 5% blood or 20% serum was found to enhance the growth of hVISA in a pilot study, suggesting that such supplementation could improve the routine detection of hVISA (126); however, a more detailed analysis of these media has not confirmed this (118).
The MET utilizes a higher inoculum and prolonged incubation to detect hVISA (Table 5 and Fig. 8). It should be noted that the result of the MET is just a cutoff level and is not a true MIC. Even though this method is clearly defined and the manufacturer recommends the use of an inoculum of 200 μl, various inoculum sizes have been used in different studies, such as 100 μl (370, 389) and 50 μl (194), again potentially limiting the extrapolation and comparison of results.
FIG. 8.
Examples of Etest methodology used to detect hVISA. (A) Etest GRD result for Mu50 after 48 h of incubation. (B and C) Macromethod (2 McFarland standard) Etest result for Mu50 against vancomycin (VA) and teicoplanin (TP) after 48 h of incubation. Note the microcolonies that are best detected using magnification and are important for interpreting the test result.
The recently released Etest GRD provides a new method for the detection of reduced vancomycin susceptibility and, in particular, a potentially useful improvement in result turnaround time, with an initial reading performed after 24 h of incubation. The method involves the use of a double-ended Etest strip that contains vancomycin, teicoplanin, and a nutritional supplement. A standard inoculum (0.5 McFarland standard), rather than the 2 McFarland standard used for the MET, is used and inoculated onto MHA with 5% blood (Fig. 8). Two studies evaluated the Etest GRD for hVISA detection (174, 393). In an initial evaluation, Yusof et al. found the Etest GRD with the nutritional supplement to be almost equivalent to PAP when read at 48 h after growth on MHA with 5% blood (393). While the sensitivity was only 70% at 24 h of incubation, it increased to 94% at 48 h, while the specificity remained high (100% at 24 h and 95% at 48 h). Leonard et al. found similar results for sensitivity at 24 and 48 h (77% and 93%, respectively) but a drop-off in specificity at the 48-h reading (from 98% to 82%); however, the reasons for this difference are unclear (174).
Given the turnaround time for hVISA detection in the routine laboratory, some research groups have been attempting to develop more rapid assays for hVISA and VISA detection. Tajima et al. utilized a chemiluminescence assay induced by active oxygen species to measure bacterial metabolic activity upon exposure to vancomycin or teicoplanin in an assay with a turnaround time of 4 to 6 h. They were able to detect hVISA with high sensitivity and specificity for a well-defined set of hVISA/VISA and VSSA isolates; however, this assay requires further validation in a routine setting (339).
Confirmatory methods.
The vancomycin population analysis profile (PAP) was proposed to be the most accurate method for the detection of hVISA, and this is the method used in our laboratory; however, it is relatively time-consuming and requires the use of a spiral plater. Results are generally not available in a clinically relevant time frame, often taking at least 3 to 5 days. Wootton et al. described a modified PAP method by calculating the area under the curve (AUC) of the standard PAP graph (PAP/AUC) and comparing the result of the test organism to hVISA reference strain Mu3 (Table 6) (387). Those authors defined VSSA as a PAP/AUC ratio of <0.9 and hVISA as a PAP/AUC ratio of ≥0.9 (387). Based upon repeat testing of VISA strain Mu50, a strain with a PAP/AUC ratio of >1.3 has a resistant population similar to that of a VISA strain with a vancomycin MIC of 8 μg per ml (372). A key factor in defining hVISA in this way is the reproducibility of the PAP/AUC for reference strain Mu3. In the original description, Mu3 was tested on 16 occasions and gave very reproducible results (mean AUC of 21.06 ± 2.47) (387). We have tested Mu3 on multiple occasions and have also found the PAP/AUC ratio to be highly reproducible for this strain. At present, given the lack of a simple alternative, the modified PAP method described by Wootton et al. appears to be the best method for the definitive identification of hVISA (387). Whether the hVISA phenotype requires definitive identification in all suspected cases is debatable, and it may be that the use of an accurate screening method is adequate for most laboratories, with the referral of isolates for PAP testing in selected cases.
TABLE 6.
Methodology for screening and confirmation of hVISA and VISA strainsb
| Method | Inoculum | Interpretation | Description | Reference(s) or source |
|---|---|---|---|---|
| Vancomycin MIC | Prepare a 0.5-McFarland-standard direct-colony suspension in water, saline, or MHB, dilute, and inoculate cation-adjusted MHB to final concn of 5 × 105 CFU per ml; incubate a full 24 h at 35°C ± 2°C | Read with unaided eye and determine lowest concn with complete inhibition of growth | Inoculum prepared from overnight growth on nonselective agar; control strain: ATCC 29213 (S. aureus) | 53 |
| MHA + teicoplanin at 5 μg per ml (MHA5T) | Prepare a 2-McFarland-standard direct-colony suspension; 10 μl onto plate; incubate for 48 h at 35°Ca | Any growth positive | Inoculum prepared from growth on blood agar plate overnight | 389, 393 |
| Macromethod Etest (MET) | Pipette 200 μl of a 2-McFarland-standard suspension onto a 90-mm BHI plate and swab evenly; dry the agar surface (15 to 20 min) and then apply Etest strips; incubate for 48 h at 35°C | Use oblique light and magnifying glass or plate microscope to read point of complete inhibition (Fig. 5); positive result, vancomycin MIC of ≥8 and teicoplanin MIC of ≥8 μg per ml or teicoplanin MIC of ≥12 μg per ml | Inoculum prepared from overnight growth on blood agar plate; beware of adherent growth if plate not dry; do not convert a result of 6 μg per ml to the next upper dilution; control strains: ATCC 29213 (S. aureus), ATCC 700699 (Mu50), ATCC 29212 (Enterococcus faecalis) | EAS 003 product information; AB Biodisc, Solna, Sweden |
| Etest GRD | Using sterile cotton swab, inoculate MHA with 5% sheep or horse blood from a 0.5-McFarland-standard suspension in MHB; after dipping cotton swab into broth, press against side of tube to remove excess fluid; dry the agar surface (15 to 20 min), and then apply Etest GRD strip; read after 24 and 48 h at 35°C | Vancomycin or teicoplanin MIC of ≥8 μg per ml | Inoculum prepared from overnight growth on blood agar plate; control strains: ATCC 29213 (S. aureus), ATCC 700699 (Mu50), ATCC 700698 (Mu3) | Product information, AB Biodisc, Solna, Sweden |
| PAP/AUC | Culture grown overnight in TSB diluted to 10−3 and 10−6; inoculate BHIA with vancomycin at 0, 0.5, 1, 2, 2.5, and 4 μg per ml using a spiral plater; incubate for 48 h prior to counting colonies | Plot graph of CFU per ml vs vancomycin concn and calculate AUC; determine ratio of AUC of test organism vs Mu3 (ATCC 700698); if AUC ratio is ≥0.9 and vancomycin MIC is ≤2 μg per ml, the isolate is hVISA | Mu3 should be run in parallel with test organism; software such as GraphPad Prism (San Diego, CA) is available for calculating AUC | 385, 387 |
Note that the European Antimicrobial Resistance Surveillance Scheme suggests an inoculum using 10 μl of a stationary-phase culture grown overnight in BHIB (http://www.rivm.nl/earss/).
TSB, tryptone soy broth.
Practical Approach to hVISA and VISA Detection
The optimal approach for the detection and confirmation of hVISA is yet to be determined; however, PAP is currently considered to be the gold standard for hVISA confirmation. To develop an accurate understanding of hVISA and VISA epidemiology and clinical impact, it is critical that research groups around the world use the same methodology to detect and confirm the presence of hVISA and VISA, including adhering to the correct inoculum size and result interpretation. To this end, the methodologies for the most important and commonly used methods are summarized in Table 6.
A number of screening approaches are available, and the method used will depend on the number of isolates to be tested and work flows and capabilities within the laboratory. For high-throughput screening, the MHA5T screening plate appears to be the best method; however, for laboratories where modified Etest screening is feasible, the MET or the Etest GRD is more accurate. A key factor in deciding which screening approach to use is the ultimate relevance of the result to patient management and whether all isolates or only selected isolates from high-risk patients need to be screened. At Austin Health, we have detected high rates of hVISA (up to 50% of MRSA isolates) when all MRSA isolates are assessed in detail by PAP; however, a significant clinical impact of the hVISA phenotype has not been demonstrated for less severe infections such as superficial-wound infections (118). As highlighted in a recent case report of serious MRSA infection, the early detection of hVISA in a patient that subsequently failed vancomycin and then daptomycin therapy may have alerted the clinician to the potential for treatment failure and a possible alternate approach to therapy (348). At present, we favor the screening of isolates from high-risk patients (those with bacteremia or deep-seated infection) and patients who are failing vancomycin therapy, as indicated by persisting positive cultures upon therapy (Fig. 9).
FIG. 9.
Flow diagram describing the possible approaches to the investigation and management of patients with serious MRSA infections being treated with vancomycin.
Selection of the clinical isolate to test.
As described above, in many cases, the hVISA or VISA phenotype is detected after a prolonged period of infection, which is associated with a failure of glycopeptide therapy (121, 151, 213, 313, 316, 326, 337, 340, 348, 376). The testing of later clinical isolates from patients who have failed glycopeptide treatment appears more likely to yield a positive result for hVISA or VISA. In reality, susceptibility testing is aimed at predicting the response to a particular antibiotic therapy at the onset of clinical infection. Therefore, studies to correlate the presence of reduced vancomycin susceptibility and glycopeptide treatment outcome should clearly define which clinical isolates have been tested: the first available isolate or a later isolate after treatment failure.
Measurement of Vancomycin Bactericidal Activity
Vancomycin bactericidal activity can be assessed in a number of ways. Recently, Sakoulas et al. (298) and Moise et al. (208) determined the impact of reduced bactericidal activity on vancomycin treatment outcome for a select group of patients. Different methods for the determination of vancomycin bactericidal activity were used. Sakoulas et al. used an overnight BHIB culture that was diluted 1:100 in 20 ml of BHIB containing 16 μg vancomycin per ml. Colony counts were performed at time zero and at 72 h. Vancomycin bactericidal activity for each strain was expressed as log10 CFU per ml at 0 h − log10 CFU per ml at 72 h. Moise et al. (208) determined the reduction in log10 CFU per ml with 16 μg vancomycin per ml in MHB after only 24 h of incubation using the same inoculum as that described by Sakoulas et al. (298). Although a correlation between reduced bactericidal activity and treatment outcome was demonstrated by these studies, no further studies using these methods have been published. The laboratory work required to obtain these results is similar to performing a population analysis and is unlikely to be applied in the routine diagnostic laboratory.
IMPACT OF hVISA AND VISA ON TREATMENT OUTCOMES
In Vitro Models and Animal Studies
Hiramatsu demonstrated that the hVISA phenotype associated with a subtle change in the vancomycin MIC could impact the efficacy of vancomycin treatment in vitro. Using Mu3 (hVISA strain with a vancomycin MIC of 2 μg per ml), 10 μg per ml of vancomycin was required to completely suppress growth, whereas in a comparator strain, 87/20 (VSSA strain with a vancomycin MIC of 1 μg per ml), 2 μg per ml of vancomycin suppressed growth to the same degree (113). An in vitro analysis using three VISA strains demonstrated a reduced rate of vancomycin killing but no difference in the final extent of killing due to the VISA phenotype (1); however, vancomycin had no effect against Mu50 in a rabbit endocarditis model (13). Other in vitro models have demonstrated a reduced efficacy of vancomycin against hVISA and VISA (175, 282), in particular in situations where a high inoculum was used (171, 282).
Moore et al. studied isolates from a patient who experienced vancomycin treatment failure for MRSA endocarditis. Although all isolates remained susceptible by MIC testing, the later clinical isolate was determined to be an hVISA isolate by PAP (vancomycin MIC of 2 μg per ml) (213). Using a rabbit endocarditis infection model, the early clinical isolate was eradicated by vancomycin; however, the hVISA strain persisted, suggesting that the hVISA phenotype contributed to treatment failure in this case.
Clinical Studies
The clinical significance of hVISA and VISA has been difficult to clearly determine. This has been due partly to differences in definitions and laboratory detection but predominately because no well-controlled prospective studies have been performed. One of the major outcome measures for patients with serious MRSA infections is the persistence of the infection. Many patients with serious hVISA or VISA infections have persistent bacteremia while on appropriate doses of vancomycin (48, 121, 124, 213); however, many clinicians consider vancomycin to be inferior to beta-lactams for the treatment of S. aureus bacteremia and endocarditis, and patients with apparently vancomycin-susceptible infections can fail vancomycin therapy (46, 324).
A summary of the various definitions used to describe these non-van-operon-containing strains of MRSA with reduced vancomycin susceptibility is provided in Table 2. It should be remembered that hVISA/VISA appears to be able to develop ex vivo from more sensitive strains in a variety of S. aureus lineages. Hence, the provenance of clinical strains linked to treatment failure is critical for determining whether hVISA/VISA is the a priori cause of treatment failure or emerges after treatment failure has already occurred. This important detail is not always described clearly in the clinical literature. The availability of clinical isolates of hVISA and VISA from patients with persistent infection where the earlier clinical isolates are fully vancomycin susceptible has provided an opportunity to understand the mechanisms of hVISA and VISA. However, in determining the clinical impact of the resistance phenotype, the expression of resistance in the first clinical isolate and the impact on treatment outcomes are more relevant to the clinician.
In 1999, Sieradzki et al. described a renal dialysis patient in the United States from whom sequentially more vancomycin-resistant blood isolates of MRSA were obtained. The MIC of vancomycin for the initial isolate was 2 μg per ml but had risen to 8 μg per ml just prior to death. However, this patient also had an infected deep focus (Gore-Tex fistula) that was not removed; hence, the progressive rise in the MIC of the blood culture isolates may represent a secondary adaptation rather than the primary cause of treatment failure (316). At the same time, Smith et al. reported two further patients from the United States, one on renal dialysis and one with diabetes, both of whom had persistent or recurrent invasive MRSA infections. Initial isolates had a vancomycin MIC of 8 to 16 μg per ml (VISA) (326). An outbreak investigation did not identify evidence of lateral transfer of related VISA strains. Both patients were ultimately cured with antibiotic therapy but subsequently died, indicating that they suffered from significant comorbidities, a common theme in many of these reports. In the same year, Ariza et al. (11) reported a case series from Spain of 19 patients with MRSA infections, all with vancomycin MICs of <4 μg per ml, 14 of whom had metallic implants. Strains were also examined by PAP. Although treatment failure occurred for 68% of individuals overall, only 20% (1/5) failed treatment when hVISA was excluded by PAP, compared with 86% (12/14) of individuals whose isolates were confirmed as hVISA. All patients with prosthetic devices and treatment failure in that report were ultimately cured following the removal of prosthetic material (11). These authors also surveyed a larger number of isolates from across Spain and concluded that hVISA appeared to have been present in their hospital since 1990 and that heteroresistance may be a general and long-standing property of their common Iberian MRSA clone. Later, in 1999, a group from Hong Kong reported a case-control study of staphylococci with inducible vancomycin resistance. However, only 3 patients were infected with S. aureus, two of whom died. Initial MICs for these 3 cases were 1 to 2 μg per ml, but selectable subclones with MICs of 8 μg per ml defined these patients as having hVISA infections (383).
In 2001, Fridkin reviewed the U.S. experience with VISA (defined at the time as MRSA clinical isolates with vancomycin MICs of 8 to 16 μg per ml). Six patients were discussed, some previously reported. Five patients died, but only one died directly from MRSA sepsis, and this patient was a dialysis patient with line sepsis. Of the other patients, one patient treated with surgical drainage plus linezolid, trimethoprim-sulfamethoxazole (SXT), and doxycycline survived. The remaining 4 patients either refused surgical treatment or were cured of MRSA before death from another cause (90). For the survivor, weekly serum vancomycin levels were in the range of 2.7 to 4.9 μg per ml during the 10 weeks prior to the detection of VISA in peritoneal fluid (107).
In Brazil in the same year, 140 clinical isolates of MRSA in a single hospital with a burn unit were screened by agar dilution and macrobroth MIC followed by confirmation with a modified PAP. Five isolates with a macrodilution MIC of 8 μg per ml, 4 from patients with burns and 1 from a patient with osteomyelitis, were identified. Initial broth MICs were not provided in that report. The outcome was described as favorable in 4/5 cases despite extensive resistance to other agents and the unavailability of newer alternative agents to vancomycin in Brazil at the time. The patients with burns had been exposed to more than 30 days of vancomycin at the time at which their clinical isolates were obtained (235). In 2003, a case report, also from Brazil, described a patient with MRSA endocarditis where blood culture isolates of MRSA had vancomycin MICs of 4 to 8 μg per ml. Despite very prolonged treatment with vancomycin, with documented trough levels of 43 μg per ml, the patient remained febrile but promptly responded when treated with linezolid. This is a convincing case of vancomycin failure due to a VISA strain that became progressively more resistant during treatment (7).
In 2003, a case-control study described 19 cases and 42 controls in the United States with or without MRSA infections with reduced susceptibility to vancomycin (SA-RVS). SA-RVS was defined as a vancomycin MIC of ≥4 μg per ml (Table 2); isolates from control patients had vancomycin MICs of ≤2 μg per ml. Prior exposure to a glycopeptide and previous history of MRSA infection increased the likelihood of being classified as a case-patient. Case-patients had a higher mortality rate but were also more likely to have bloodstream infections than controls. There were no differences in clinical presentations between cases and controls, and neither renal failure nor dialysis per se predicted definition as a case-patient (91). In a multivariate analysis, bloodstream infections and SA-RVS remained independent predictors of death. An overall attributable mortality of 63% for patients with SA-RVS was reported, but numbers in this study were small, and there remains a possibility that the comparison was confounded by the increased likelihood of bloodstream infections or other unknown factors in case-patients despite statistical adjustment. Isolates from case-patients appeared to be heterogeneous rather than belonging to a predominant clone.
In the same year, Schwaber et al. described a case series from the Beth Israel and Deaconess Medical Centre and Johns Hopkins Hospital in the United States (306). A definition of SA-RVS similar to that provided by Fridkin et al. was employed, but a subanalysis using PAP for hVISA was also included. In that retrospective study, 61 patients with SA-RVS isolates (defined by initial growth on a 4-μg/ml vancomycin screening plate) were compared with 88 controls. No isolates were identified as being hVISA. There was no difference in clinical outcomes between case-patients and controls. Those authors concluded that screening for hVISA was not necessary if MRSA with a low vancomycin MIC was isolated from a clinical specimen (306).
In 2004, an Australian study reported a consecutive series of 53 patients with MRSA blood culture isolates over 12 months. Five patients with infections due to hVISA defined by modified PAP (375) were compared with 48 patients with negative hVISA screens. Patients with hVISA bacteremia were more likely to have high-bacterial-load infections, vancomycin treatment failure (defined as persistent fever and bacteremia for >7 days after the start of therapy), and initially low serum vancomycin levels (48). hVISA was defined by the modified PAP described by Wootton et al. (387). The precise definition of hVISA was a blood culture isolate with a vancomycin MIC of ≤4 μg/ml and a PAP/AUC ratio of ≥0.9 compared with Mu3 (387). All isolates classified as being hVISA also had a standard inoculum vancomycin Etest result of 4 μg per ml, while vancomycin microbroth MICs were 2 to 4 μg per ml. Despite small numbers, that study clearly associated hVISA rather than SA-RVS, VISA, or VRSA with treatment failure, but because the last rather than the first blood culture isolate from each patient was studied, it potentially invokes the post hoc ergo propter hoc problem: did treatment fail because of the a priori presence of hVISA, or did hVISA appear as an epiphenomenon in those who failed treatment for some other reason? This is addressed partly by the rapid clinical response to linezolid in 4 of 5 patients, which is reminiscent of the case from Brazil described by Andrade-Baiocchi et al. (7).
In a subsequent case series from Australia and New Zealand, 25 patients with serious infections due to hVISA were studied (124). Eight patients had endocarditis, 9 had bacteremia associated with deep-seated infection, 6 had osteomyelitis or septic arthritis, and 2 had empyema. All patients had received vancomycin before the isolation of hVISA, and glycopeptide treatment had failed for 19 patients (76%), defined as a blood culture positive for S. aureus after ≥7 days of glycopeptide therapy or a sterile-site isolate positive for S. aureus after ≥21 days of glycopeptide therapy. Twenty-one patients subsequently received alternative antibiotic treatment, including 18 who received linezolid, which was effective for 14 patients (78%), including 4 patients with endocarditis. Twelve patients received a combination of rifampin and fusidic acid. Surgical intervention was required for 15 patients (60%). It appeared from this case series that for patients with glycopeptide treatment failure and hVISA, linezolid with or without rifampin and fusidic acid in conjunction with surgical debulking is effective therapy for the majority of cases, including those with endocarditis (124).
Khosrovaneh et al. described 22 patients with recurrent or persistent MRSA bacteremia in Detroit, MI (155). Patient isolates were specifically examined for hVISA using a PAP/AUC ratio of ≥0.9, and results from that case series are therefore directly comparable with data from studies from our institution. However, Khosrovaneh et al., while observing that isolates with higher initial MICs (in the vicinity of 4 μg per ml) were more likely to produce subcolonies with even higher MICs, detected definite hVISA in only 3 of their 22 patients and concluded that hVISA defined in this way was uncommon and that treatment failure could be explained by other factors without the need to invoke the presence of resistant subpopulations per se (155).
Moise-Broder et al. then reported a different kind of comparative case series, highly enriched for vancomycin treatment failure, as this had been an entry criterion for an earlier related pharmaceutical company trial (211). The key finding in their initial univariate analysis was that while all isolates had an initial MIC of ≤4 μg per ml, as the MIC increased, so did the likelihood of vancomycin treatment failure. However, in the multivariate analysis, only preexisting renal failure and isolates that belonged to agr group II predicted treatment failure. The authors felt that agr group II polymorphism was more likely a marker rather than a cause of vancomycin treatment failure, although agr dysfunction may shift S. aureus in the direction of vancomycin resistance and persistence rather than virulence, as described above (356). The relationship between renal impairment and treatment failure was unexplained, although reduced host defense secondary to platelet dysfunction was postulated. In a related study, Sakoulas et al. reported isolates from 30 patients with MRSA bacteremia and were also able to demonstrate a statistically significant association between increasing vancomycin MICs and treatment failure, although all isolates were within the vancomycin-susceptible range (all isolates had an MIC of ≤2 μg per ml). In that report, the in vitro bactericidal activity of vancomycin was also studied. Considerable strain-to-strain variability (72-h vancomycin killing ranged from 0.17 to 8.6 log10 CFU per ml) existed, and although no association could be found between the bactericidal activity of vancomycin and the vancomycin MIC, multivariate analysis did identify increased vancomycin bactericidal power and decreased MIC as being independent predictors of vancomycin treatment success or failure (298). A further publication developed this theme and concluded that once the vancomycin MIC approached 2 μg per ml, the efficacy of vancomycin for MRSA bacteremia is severely compromised, even though these isolates are defined as being susceptible by national guidelines (208).
In 2006, the CLSI, in response to mounting concern and some evidence, altered its breakpoints for vancomycin susceptibility (Table 2). Tenover and Moellering provided a very clear discussion of the thinking behind this decision (346). The review recognized the possible role of hVISA in some of the reports of vancomycin treatment failure leading up to this decision but pointed out that hVISA and SA-RVS are not identical. In fact, strains of S. aureus with vancomycin MICs as low as 0.5 μg per ml by broth dilution or 1 μg per ml by standard Etest may produce subpopulations with higher MICs in the VISA/VRSA range (i.e., hVISA). However, despite new evidence from some investigators who believed that a vancomycin Etest MIC of ≤1.5 μg per ml may be a better breakpoint for MRSA (184, 185), the review pointed out that a further reduction from the current breakpoint of ≤2 μg per ml would define 16% of S. aureus isolates surveyed in the United States as being vancomycin intermediate, an outcome out of keeping with current clinical experience (346).
Also in 2006, a study from the University of California, Los Angeles, aimed to link the initial MRSA vancomycin MIC to outcome and to compare usual treatment with targeted therapy aimed to achieve an unbound serum vancomycin trough level at least 4 times the MIC of the infecting strain. Hidayat et al. (112) reported results from 95 patients, 54% of whom had isolates with high vancomycin MICs by standard Etest (≥2 μg per ml). The majority of these patients had pneumonia, and only 25% had bloodstream infections. The key findings were that outcome was improved by achieving the target trough regardless of MIC but that poorer APACHE II scores and higher MICs remained predictors of a poor treatment response in a multivariate analysis. Another finding was that renal impairment was more common for patients with serum vancomycin troughs of ≥15 μg per ml. Those authors commented that additional agents (e.g., rifampin, linezolid, and daptomycin) may improve outcome and should be considered for patients with high-vancomycin-MIC MRSA infections (112).
In a retrospective cohort study from Texas, Maclayton et al. studied patients undergoing hemodialysis who developed MRSA bacteremia, and these researchers also attempted to relate outcomes to initial MICs. In the univariate analysis, MICs of <0.5 μg per ml predicted improved survival, but recent surgery and ICU admission were also risk factors for having MRSA isolates with MICs of 2 μg per ml. The MIC method was not defined. In a multivariate analysis that included cost modeling, patients with high-MIC isolates had increased lengths of stay and increased hospital costs compared with patients with low-MIC isolates and uninfected control patients, but mortality was not increased (188). A further recent report suggested that prior glycopeptide exposure is associated with increased MICs and reduced in vitro vancomycin killing in patients who subsequently developed MRSA sepsis (209).
In 2007, Maor et al. reported a case series of 264 patients with MRSA bacteremia from the Sheba Medical Centre, Tel Hashomer, Israel. Isolates were screened by the macromethod Etest to detect hVISA, with no confirmation by PAP (194). Sixteen hVISA cases were identified (6%), and although those authors were not able to directly link outcome to MIC, there was a higher mortality rate in this group, and they reported an hVISA-attributable mortality of 50%. The same group followed up this initial report with a case-control study of 27 case-patients with positive METs with 227 control patients with MRSA bacteremia who had negative hVISA screens. Case-patients were more likely to have prolonged bacteremia, osteomyelitis, and endocarditis and to develop resistance to rifampin. Those authors were not able to relate these complications to low serum vancomycin levels. The infection-attributable mortality for hVISA was similar to that for MRSA bacteremia (193).
In a study from Japan, Neoh et al. reported a 7-year review of hVISA defined by PAP/AUC at a single center. Twenty of 209 cases of MRSA bacteremia were defined as hVISA cases. The vancomycin treatment response characteristics “days until afebrile” and “CRP [C-reactive protein] of ≤30% of maximum” correlated with hVISA, but no relationship to mortality could be established (226).
A recent study from our institution evaluated the clinical importance of hVISA compared to VSSA in relation to the likelihood of causing active infection and also determined treatment outcomes (118). hVISA and VISA strains were less likely to cause invasive disease, and for infected patients who were treated, a statistical difference in cure rates was not identified (however, cure rates were 58% for hVISA and VISA and 80% for VSSA infections; P = 0.08). A larger study, especially including more patients with invasive disease, will be required.
In a large recent study from Spain, Soriano et al. described 414 cases of MRSA bacteremia, covering the period 1991 to 2005, at a single institution (332). Those authors highlighted the use of the first available isolate for MIC testing using standard Etest to subclassify patients. They then examined outcome with a multivariate model based on MIC classification of the infecting isolate (standard-method Etest MICs of 1 μg per ml [n = 109], 1.5 μg per ml [n = 213], and 2 μg per ml [n = 92]). In their univariate analysis, there was no difference in rates of mortality among these 3 groups, but there was a highly significant reduction in the risk of septic shock with higher-MIC isolates. This is initially counterintuitive but aligns with data from other reports suggesting that a downregulation of virulence may be associated with increased vancomycin resistance [see “Accessory gene regulator (agr)” and “Host immune interactions and virulence”]. However, some other conclusions from that study are harder to understand: despite shock being an independent predictor of death, an odds ratio of 7.38 (95% confidence interval [CI], 4.1 to 13.3) and high MIC being protective against shock, the use of vancomycin for patients with an MRSA isolate with an MIC of 2 μg per ml was also associated with an odds ratio for death of 6.39 (95% CI, 1.68 to 24.3). Those authors explained this with reference to a pharmacokinetic study of humans with MRSA pneumonia for which an AUC at 24 h (AUC24)/MIC ratio of ≥350 was predictive of treatment success. The probabilities of achieving this target with an isolate with an MIC of 1 μg per ml with serum vancomycin trough levels of 10 and 15 μg per ml were 40 and 60%, respectively (210). Clearly, doubling the MIC would substantially reduce the chance of achieving this measure. However, lung penetration of vancomycin is relatively poor, meaning that direct extrapolation to MRSA bacteremia may not be valid. In fact, according to data reported by Soriano et al., the odds ratio for death for vancomycin treatment when the MRSA isolate had a vancomycin MIC of 2 μg per ml was higher than that for treatment with a drug not predicted to have any activity against MRSA. Some of these inconsistencies may be oddities of statistics but should lead to some caution in drawing firm conclusions (332).
Twelve years after Hiramatsu's first description of this mode of vancomycin resistance in S. aureus, it seems safe to conclude that patients infected with isolates of MRSA with vancomycin broth microdilution MICs of 8 μg per ml should not be treated with vancomycin. We are fortunate to have new and effective alternatives, but we should also recall that the removal of deep sources and other surgical interventions should be considered in every case, whatever the MIC. There is increasing evidence that isolates with MICs in the 4- to 8-μg/ml range are also likely to fail treatment, and the CLSI has recently adjusted breakpoints to account for this. There is contentious ground in the MIC range of 1 to 2 μg per ml and some evidence that breakpoints should be lowered again. However, such a move would lead immediately to a large increase in the use of more expensive alternative agents, with associated increases in costs and potential toxicity. The relative contribution of the hVISA phenotype in S. aureus strains with low vancomycin MICs to treatment failure has not been definitively defined. Future studies will be required to answer these questions, but the most revealing of potential studies, prospective blinded randomized comparisons of correctly dosed vancomycin with newer agents, stratified by MIC and perhaps PAP/AUC, may never be performed. Future studies should include the use of clearly defined methods for hVISA and VISA detection and should clearly define which clinical isolates are tested.
THERAPEUTIC OPTIONS AND POTENTIAL CROSS-RESISTANCE
Role of Surgery
A feature of many of the reported cases of hVISA and VISA infection has been the association with “high-bacterial-load” infections such as endocarditis, osteomyelitis/septic arthritis, deep abscesses, and infection of prosthetic devices such as joint replacements (48, 91, 124). In a review of 25 cases of serious hVISA and VISA infections, surgical debridement was a key component of therapy for over 60% of patients who were successfully treated (124). An important adjunct to antimicrobial therapy for these patients, therefore, is an adequate surgical debridement of infection (Fig. 9).
Potentially Active Antimicrobials Currently Available
A number of antimicrobials with activity against S. aureus retain activity in vitro against hVISA and VISA. These include older agents such as rifampin and fusidic acid and a number of newer agents, some which are not yet released but for which there is in vitro data or animal model data potentially supporting their use for infections caused by these strains. Ultimately, there are no clinical trials of hVISA and VISA treatment, and only a few patients with serious hVISA or VISA infections that have been successfully treated have been reported in the literature. Table 7 summarizes detailed cases of serious hVISA or VISA infections that have been successfully treated and reported in the literature.
TABLE 7.
Clinical cases of successfully treated hVISA or VISA infectionsc
| Predominant antibiotic therapya | No. of cases | Feature of case | Key feature(s) of treatment | Outcome and description | Reference(s) |
|---|---|---|---|---|---|
| Linezolid | 1 | MRSA endocarditis. Failed VAN (90 days) despite trough levels up to 43 μg per ml | Oral LZD, no surgery | Cured | 7 |
| 1 | MRSA endocarditis and septic thrombophlebitis; failed VAN and then DPT (58 days of bacteremia) | Changed to LZD + FA | Cleared bacteremia; died from candidemia | 131 | |
| 1 | MRSA endocarditis; failed VAN and then DPT (10 wk); developed VISA and reduced DPT susceptibility | Changed to LZD + SXT; also had course of QD | Cured; thrombocytopenia from LZD, severe arthralgias/myalgias from QD | 348 | |
| 9 | 4 endocarditis, 3 OM, 1 bacteremia + OM, 1 empyema; VAN MIC of 2 μg per ml (n = 6) + 4 μg per ml (n = 3) | Surgery (n = 5); linezolid alone | Effective (n = 7), not effective (n = 2) | 124 | |
| 1 | MRSA AV and MV endocarditis with implantable defibrillator; SA-RVS; failed VAN | LZD for 67 days; removal of implantable defibrillator | Infection cleared but developed recurrent MRSA bacteremia 2 mo later | 384 | |
| Linezolid and then rifampin + fusidic acidb | 8 | 3 endocarditis, 2 bacteremia + SA/OM, 2 bacteremia + intra-abdominal infection, 1 prosthetic joint; VAN MIC of 2 μg per ml (n = 5) + 4 μg per ml (n = 3) | Surgery (n = 5) | Effective (n = 6), not effective (n = 2) | 124 |
| Linezolid + other agents | 1 | VISA hepatic abscess (VAN MIC of 8 μg per ml), after 10 wk VAN | Surgery, LZD + SXT + DOX | Infection cleared | 90, 107 |
| 1 | MRSA prosthetic AV valve endocarditis after repeated bacteremia and VAN therapy; failed VAN (MIC of 6 μg per ml by Etest) | LZD + AMI, no surgery | Cured | 176 | |
| 1 | MRSA-infected hip fracture after fixation; failed debridement and VAN therapy (last isolate Etest MIC of 4 μg per ml) | LZD + RIF, further debridement | Infection cleared | 376 | |
| Rifampin + | 3 | 2 bacteremia, 1 OM; VAN MIC of 2 | Surgery (n = 2), oral | Cured (n = 1) | 124 |
| fusidic acid | μg per ml (n = 2) + 4 μg per ml | RIF + FA | Not effective (n = 1) | ||
| (n = 1) | NA (n = 1) | ||||
| Others | 1 | VISA (MIC of 8 μg per ml), bacteremia and vertebral OM; prior 18 wk of VAN | VAN + NAF + GEN | Infection cleared; died from another cause | 90 |
| 1 | Recurrent MRSA bacteremia; VISA (MIC of 8 μg per ml) after 18 wk of VAN | GEN + RIF | Infection cleared; died from candidemia | 326 | |
| 1 | Recurrent peritonitis in peritoneal dialysis pt VISA (MIC of 8 μg per ml) after 18 wk of VAN; failed VAN treatment | SXT + RIF | Infection cleared | 326 |
Defined as the main antistaphylococcal antibiotic therapy used to cure the infection or to obtain control of the infection prior to long-term suppression.
One patient received linezolid followed by fusidic acid plus chloramphenicol. One patient received rifampin plus fusidic acid followed by linezolid.
VAN, vancomycin; DPT, daptomycin; SXT, trimethoprim-sulfamethoxazole; RIF, rifampin; FA, fusidic acid; QD, quinupristin-dalfopristin; DOX, doxycycline; NAF, nafcillin; NA, not applicable; SA, septic arthritis; OM, osteomyelitis; AV, aortic valve; MV, mitral valve; AMI, amikacin; LZD, linezolid; GEN, gentamicin.
There is concern about potential cross-resistance between hVISA/VISA and the lipoglycopeptides that are structurally and functionally similar to vancomycin. At this stage, it is not clear whether the hVISA or VISA phenotype will impact clinical treatment responses to these agents, but this needs to be monitored. In addition, there are increasing numbers of reports of the potential for reduced daptomycin susceptibility in hVISA/VISA strains and a number of reports describing daptomycin treatment failure following vancomycin failure for serious MRSA infections (see below). The clinical implications of this are not yet clear.
Rifampin and fusidic acid.
Rifampin and fusidic acid both possess good in vitro activity against S. aureus and in particular have been useful for oral treatment of multiresistant MRSA infections (134, 360, 395). Resistance develops rapidly with monotherapy with either agent; therefore, these agents should always be used in combination with another effective antistaphylococcal agent (125). Typical combinations include rifampin plus fusidic acid or rifampin plus a quinolone if the S. aureus isolate is susceptible. In Australia and some other parts of the world, the use of oral combination therapy with rifampin and fusidic acid is the mainstay of therapy for complicated MRSA infections requiring prolonged nonparenteral therapy. This combination is not available in the United States because fusidic acid is not approved for use. In a few cases, therapy with oral rifampin and fusidic acid was an important component of therapy for the successful treatment of hVISA and VISA infections in patients who had failed vancomycin therapy (124). Importantly, it appears that vancomycin does not adequately protect against the development of rifampin resistance when this combination alone is used to treat serious MRSA infections (35, 150). In addition, fusidic acid monotherapy should not be used because of the frequent development of fusidic acid resistance in S. aureus (120).
Linezolid.
Linezolid is the first in a new class of completely synthetic antimicrobial agents, the oxazolidinones. Strains of S. aureus and coagulase-negative staphylococci with reduced vancomycin susceptibility retain susceptibility to linezolid, with no change in the MIC90 (146). Although linezolid resistance has been reported for S. aureus (162, 355), rates of resistance remain very low (350). Also, although linezolid is essentially bacteriostatic against S. aureus in vitro, a number of serious cases of MRSA, hVISA, and VISA infections, including endocarditis, have been cured with linezolid (Table 7) (124, 131, 287). We have found linezolid to be very useful for the treatment of hVISA and VISA infections or infections with MRSA where vancomycin has failed. A key issue with linezolid, however, is toxicity. Although a number of large postmarketing studies have found linezolid toxicity rates to be similar to those of comparator drugs (140, 145, 285, 382), high rates of toxicity have been found for complex patients, and prolonged therapy should be used with caution in these cases (24).
Daptomycin.
Daptomycin is a cyclic lipopeptide derived from Streptomyces roseosporus. Reduced daptomycin susceptibility in S. aureus was reported to emerge during therapy and appears to be associated with high-bacterial-load infections (87, 110, 116, 191, 198). In some cases resistance appears to be associated with mutations in mprF and yycG, loci which have been implicated in reduced vancomycin susceptibility in some strains. There has been an association between hVISA and VISA and reduced susceptibility to daptomycin (61, 222, 243, 348, 388). It appears that vancomycin exposure per se can induce low-level daptomycin resistance or daptomycin heteroresistance (131, 283, 293, 348); however, these changes appear to be strain specific and may be unstable (283). Data from in vitro and animal studies suggest that daptomycin may have a lower rate of in vitro killing of hVISA than against VSSA (175); however, daptomycin retains bactericidal activity against hVISA and VISA (2, 40, 196, 283). hVISA and VISA are generally susceptible to daptomycin by MIC testing (299), with an MIC range from 0.125 to 1 μg per ml against 50 hVISA/VISA strains in one study. In that study, minimal bacterial concentrations (MBCs) were equal to MICs. Thirty-two VISA strains from Israel had an MIC of daptomycin of ≤0.5 μg per ml. Some case reports described the failure of daptomycin therapy after failed vancomycin therapy for serious MRSA infections (18, 131, 348), and in one case, persistent MRSA bacteremia was subsequently successfully cleared with linezolid and fusidic acid (131).
The potential relevance of the vancomycin induction of low-level daptomycin resistance is clearly important in the decision-making process for clinicians who are faced with a patient with a serious MRSA infection who has failed vancomycin therapy. It is not clear at present if daptomycin should be avoided in this situation or if additional laboratory studies will help resolve this question.
QD.
Quinupristin (pristinamycin IA) is a group B streptogramin, while dalfopristin (pristinamycin IIA) is a semisynthetic derivative of a group A streptogramin. They have been combined to generate a water-soluble intravenous preparation. Quinupristin-dalfopristin (QD) is active against MRSA and has also demonstrated in vitro activity against a range of hVISA and VISA strains (MIC90 for hVISA and VISA of 1 μg per ml) (146, 177).
Tigecycline.
Tigecycline is the first glycylcycline to be available for clinical use and is a member of the tetracycline family. It has very good in vitro activity against MSSA and MRSA strains (MIC90 of 0.12 to 1 μg per ml), including a small number of VISA strains; however, it has not been extensively tested against hVISA and VISA strains (132).
Potentially Active Antimicrobials in Development
Dalbavancin.
Dalbavancin is a semisynthetic lipoglycopeptide with an extended half-life that enables once-weekly dosing. It has very good in vitro activity against S. aureus (MIC range of ≤0.015 to 0.5 μg per ml; MIC90 of 0.06 μg per ml) with no effect of the methicillin resistance phenotype on MICs (334). It retains good activity against staphylococcal strains that are resistant to vancomycin and other new agents such as linezolid and quinupristin-dalfopristin (335). The MIC range is 0.06 to 2 μg per ml against hVISA and VISA (335). Notably, the dalbavancin MICs are higher for hVISA and VISA strains, and higher concentrations were required to achieve bactericidal activity than for MRSA in an in vitro model (28); however, the clinical implications of this finding are unclear. In an endocarditis model, dalbavancin was equally effective against VISA and teicoplanin-intermediate S. aureus, and the bactericidal results were nearly identical (173).
Oritavancin.
Oritavancin is a lipoglycopeptide antibiotic and is structurally very similar to vancomycin. Despite the structural similarity, oritavancin retains in vitro activity against vancomycin-intermediate and vancomycin-resistant S. aureus strains (33). However, in one study, the MICs were higher for VISA than for MSSA or MRSA, and a time-kill analysis demonstrated reduced bactericidal activity against VISA strains (203).
Telavancin.
Telavancin is a semisynthetic lipoglycopeptide derivative of vancomycin. Against methicillin-susceptible and -resistant S. aureus strains, telavancin has low MICs, ranging between 0.03 and 1 μg per ml (73, 142), with similar results for VISA strains (0.125 to 1 μg per ml) but higher MICs for VRSA strains (1 to 4 μg per ml) (73).
New cephalosporins.
The new cephalosporins with anti-MRSA activity are also active against hVISA and VISA in vitro and in animal models.
Ceftaroline has potent activity against S. aureus strains, including MRSA strains; however, the MIC90s are higher for MRSA strains (MIC90 of 2 μg per ml) than for MSSA strains (MIC90 of 0.25 to 0.5 μg per ml) (290). The MIC90 for 100 strains of hVISA and VISA (19 VISA and 81 hVISA strains) was also 2 μg per ml (range, 0.25 to 4 μg per ml) (290). A few animal studies have shown ceftaroline to be useful for the treatment of MRSA infections, and in mice, ceftaroline was comparable or superior to vancomycin, linezolid, teicoplanin, and arbekacin (136). In a rabbit endocarditis model, ceftaroline was highly bactericidal against MRSA and hVISA compared with linezolid and vancomycin (139).
Ceftobiprole has demonstrated good in vitro activity against MRSA strains, with an MIC90 of 2 μg per ml (27). Ceftobiprole retained good activity against a small number of VISA and VRSA strains (MICs of ≤2 μg per ml) (27). In a rabbit model of endocarditis due to MRSA and VISA, ceftobiprole was as effective as vancomycin against MRSA and superior to vancomycin against VISA (41).
Should Combination Therapy Be Used Routinely with Vancomycin for Serious MRSA Infections?
hVISA and VISA can emerge from VSSA during vancomycin therapy for serious S. aureus infections. Although the molecular determinants of resistance are not completely determined, it is also clear that the sequential acquisition of point mutations can lead to resistance. It could therefore be argued that combination therapy of serious S. aureus infections where vancomycin is being used may be appropriate in an attempt to avoid the selection of resistant mutants. There are no data to support this proposal at present, but further work in this area is warranted.
SUMMARY AND FUTURE DIRECTIONS
Issues surrounding laboratory detection and the clinical impact of reduced vancomycin susceptibility on S. aureus continue to trouble clinical microbiologists and infectious disease specialists. At this point, the optimal approach for the detection of hVISA and VISA strains has not been determined, and appropriate treatment decisions in cases where such strains are detected are not clear. The clinical impact of hVISA is likely to be highest for patients with high-bacterial-load infections, and changes in therapy as well as aggressive surgical debridement appear to be appropriate in these cases.
The use of high-throughput genomics will soon allow a clearer understanding of the genetic determinants of altered vancomycin susceptibility in S. aureus, with the potential to have an impact on clinical definitions and laboratory detection if the range of mutations is not too broad. Future clinical studies using strict definitions and randomized treatment protocols may provide the answers for clinicians keen to understand the best approach to treating patients with serious MRSA infections. Appropriately dosed vancomycin in conjunction with other active agents may still present a relatively safe and cheap option for the treatment of patients with MRSA infections once the clinical isolate has been appropriately tested.
Acknowledgments
A postgraduate medical and dental scholarship and a career development training fellowship from the National Health and Medical Research Council, Australia, supported Benjamin P. Howden and Timothy P. Stinear, respectively. Components of this work were also supported by the National Health and Medical Research Council, Australia, and the Austin Hospital Medical Research Foundation.
We thank Peter Ward for his invaluable assistance. Photographs of agar plates were kindly taken by Danielle Edwards, Clinical Photography, Austin Health.
Benjamin P. Howden was previously a daptomycin advisory board member for Novartis. No other author has any conflict of interest to declare.
Biography
 Benjamin P. Howden graduated from Monash University Medical School in 1993. He is an Infectious Diseases physician and Medical Microbiologist at Austin Health in Melbourne, Australia. He completed his Infectious Diseases and Medical Microbiology training in Melbourne and recently completed his Ph.D. in the Department of Microbiology at Monash University, where he investigated the molecular mechanisms of vancomycin-intermediate Staphylococcus aureus. His research interests include understanding the molecular mechanisms of antimicrobial resistance and pathogenesis in S. aureus as well as the clinical impact and management of multiresistant gram-positive pathogens.
Benjamin P. Howden graduated from Monash University Medical School in 1993. He is an Infectious Diseases physician and Medical Microbiologist at Austin Health in Melbourne, Australia. He completed his Infectious Diseases and Medical Microbiology training in Melbourne and recently completed his Ph.D. in the Department of Microbiology at Monash University, where he investigated the molecular mechanisms of vancomycin-intermediate Staphylococcus aureus. His research interests include understanding the molecular mechanisms of antimicrobial resistance and pathogenesis in S. aureus as well as the clinical impact and management of multiresistant gram-positive pathogens.
 John K. Davies completed his Ph.D. in the Department of Microbiology at the University of Adelaide, Australia. After postdoctoral periods in the United States and Sweden he returned to Australia in 1980 to take up a position as Lecturer in Microbiology at Deakin University. In 1983 he moved to Monash University in Melbourne, Australia, where he is now a Professor in the Department of Microbiology. Since 2004 he has been both Head of the Department of Microbiology and Deputy Head of the School of Biomedical Sciences at Monash University. He has worked on the genetics and gene regulation of Neisseria species for more than 30 years but in the last decade has also developed research projects investigating low-level vancomycin resistance in Staphylococcus aureus and mycobacterial infections. He has extensive experience in bacterial genomics and the regulation of gene expression.
John K. Davies completed his Ph.D. in the Department of Microbiology at the University of Adelaide, Australia. After postdoctoral periods in the United States and Sweden he returned to Australia in 1980 to take up a position as Lecturer in Microbiology at Deakin University. In 1983 he moved to Monash University in Melbourne, Australia, where he is now a Professor in the Department of Microbiology. Since 2004 he has been both Head of the Department of Microbiology and Deputy Head of the School of Biomedical Sciences at Monash University. He has worked on the genetics and gene regulation of Neisseria species for more than 30 years but in the last decade has also developed research projects investigating low-level vancomycin resistance in Staphylococcus aureus and mycobacterial infections. He has extensive experience in bacterial genomics and the regulation of gene expression.
 Paul D. R. Johnson, M.B.B.S., Ph.D., F.R.A.C.P., M.A.S.M., is an Infectious Diseases Physician trained at the University of Melbourne, Fairfield Infectious Diseases Hospital, and the Austin Hospital. He is currently Deputy Director, Infectious Diseases Department, Austin Health, and has honorary appointments at Monash University and the University of Melbourne. After completing physician training, he undertook a Ph.D. in microbiology on Haemophilus influenzae type B at the Royal Children's in Melbourne but at the same time became involved in investigating a community outbreak of Mycobacterium ulcerans infection. This began a long fruitful collaboration with Dr. Tim Stinear, Professor John Davies, and, more recently, Dr. Ben Howden, whereby emerging clinical problems are introduced directly to a molecular pathogenesis research group. His developing interest in staphylococcal epidemiology and resistance arose from necessity—the requirement to control MRSA in hospitals and the growing importance of S. aureus resistance in modern infectious disease medicine.
Paul D. R. Johnson, M.B.B.S., Ph.D., F.R.A.C.P., M.A.S.M., is an Infectious Diseases Physician trained at the University of Melbourne, Fairfield Infectious Diseases Hospital, and the Austin Hospital. He is currently Deputy Director, Infectious Diseases Department, Austin Health, and has honorary appointments at Monash University and the University of Melbourne. After completing physician training, he undertook a Ph.D. in microbiology on Haemophilus influenzae type B at the Royal Children's in Melbourne but at the same time became involved in investigating a community outbreak of Mycobacterium ulcerans infection. This began a long fruitful collaboration with Dr. Tim Stinear, Professor John Davies, and, more recently, Dr. Ben Howden, whereby emerging clinical problems are introduced directly to a molecular pathogenesis research group. His developing interest in staphylococcal epidemiology and resistance arose from necessity—the requirement to control MRSA in hospitals and the growing importance of S. aureus resistance in modern infectious disease medicine.
 Timothy P. Stinear leads a bacterial pathogenesis research group in the Department of Microbiology and Immunology at the University of Melbourne. He received his Ph.D. in Microbiology from Monash University in 2001 followed by a 3-year postdoc with Stewart Cole at the Institut Pasteur, Paris, France. His research interests include using genomics to understand mechanisms of antibiotic resistance in Staphylococcus aureus.
Timothy P. Stinear leads a bacterial pathogenesis research group in the Department of Microbiology and Immunology at the University of Melbourne. He received his Ph.D. in Microbiology from Monash University in 2001 followed by a 3-year postdoc with Stewart Cole at the Institut Pasteur, Paris, France. His research interests include using genomics to understand mechanisms of antibiotic resistance in Staphylococcus aureus.
 M. Lindsay Grayson graduated from Monash University Medical School in 1979 and completed Infectious Diseases training at New England Deaconess Hospital, Harvard Medical School, Boston, MA, in 1991; he completed his M.D. research thesis in 1994 and Master of Science (M.Sc.) in Clinical Epidemiology and Clinical Effectiveness, Harvard School of Public Health, Boston, MA, in 1997. From 1991 to 2000, Dr. Grayson was Deputy Director, and then Director, of Infectious Diseases and Clinical Epidemiology, Monash Medical Centre, Melbourne, Australia. Since 2000, Dr. Grayson has been Director of Infectious Diseases and Microbiology, Austin Health, and Professor of Infectious Diseases, Department of Medicine, University of Melbourne. Professor Grayson is currently Vice Chair of the Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) Program Committee. Dr. Grayson has had a longstanding interest in antimicrobial resistance, particularly related to staphylococci and enterococci, and is supervising editor of the forthcoming ASM text, Kucers' the Use of Antibiotics, 6th Edition.
M. Lindsay Grayson graduated from Monash University Medical School in 1979 and completed Infectious Diseases training at New England Deaconess Hospital, Harvard Medical School, Boston, MA, in 1991; he completed his M.D. research thesis in 1994 and Master of Science (M.Sc.) in Clinical Epidemiology and Clinical Effectiveness, Harvard School of Public Health, Boston, MA, in 1997. From 1991 to 2000, Dr. Grayson was Deputy Director, and then Director, of Infectious Diseases and Clinical Epidemiology, Monash Medical Centre, Melbourne, Australia. Since 2000, Dr. Grayson has been Director of Infectious Diseases and Microbiology, Austin Health, and Professor of Infectious Diseases, Department of Medicine, University of Melbourne. Professor Grayson is currently Vice Chair of the Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) Program Committee. Dr. Grayson has had a longstanding interest in antimicrobial resistance, particularly related to staphylococci and enterococci, and is supervising editor of the forthcoming ASM text, Kucers' the Use of Antibiotics, 6th Edition.
REFERENCES
- 1.Aeschlimann, J. R., E. Hershberger, and M. J. Rybak. 1999. Analysis of vancomycin population susceptibility profiles, killing activity, and postantibiotic effect against vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, R. L., and M. J. Rybak. 2001. Bactericidal activities of two daptomycin regimens against clinical strains of glycopeptide intermediate-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium, and methicillin-resistant Staphylococcus aureus isolates in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 45:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aligholi, M., M. Emaneini, F. Jabalameli, S. Shahsavan, H. Dabiri, and H. Sedaght. 2008. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med. Princ. Pract. 17:432-434. [DOI] [PubMed] [Google Scholar]
- 4.Allen, N. E., D. I. LeTourneau, and J. N. J. Hobbs. 1997. Molecular interactions of a semisynthetic glycopeptide antibiotic with D-alanyl-D-alanine and D-alanyl-D-lactate residues. Antimicrob. Agents Chemother. 41:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, N. E., and T. I. Nicas. 2003. Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiol. Rev. 26:511-532. [DOI] [PubMed] [Google Scholar]
- 6.Alos, J. I., A. Garcia-Canas, P. Garcia-Hierro, and F. Rodriguez-Salvanes. 2008. Vancomycin MICs did not creep in Staphylococcus aureus isolates from 2002 to 2006 in a setting with low vancomycin usage. J. Antimicrob. Chemother. 62:773-775. [DOI] [PubMed] [Google Scholar]
- 7.Andrade-Baiocchi, S., M. C. Tognim, O. C. Baiocchi, and H. S. Sader. 2003. Endocarditis due to glycopeptide-intermediate Staphylococcus aureus: case report and strain characterization. Diagn. Microbiol. Infect. Dis. 45:149-152. [DOI] [PubMed] [Google Scholar]
- 8.Appelbaum, P. C. 2007. Reduced glycopeptide susceptibility in methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 30:398-408. [DOI] [PubMed] [Google Scholar]
- 9.Arakawa, Y., Y. Ike, and M. Nagasawa. 2004. Where has vancomycin-heterogeneously resistant Staphylococcus aureus gone? Lancet 363:1401. [DOI] [PubMed] [Google Scholar]
- 10.Arbeit, R. D., W. W. Karakawa, W. F. Vann, and J. B. Robbins. 1984. Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 2:85-91. [DOI] [PubMed] [Google Scholar]
- 11.Ariza, J., M. Pujol, J. Cabo, C. Pena, N. Fernandez, J. Linares, J. Ayats, and F. Gudiol. 1999. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet 353:1587-1588. [DOI] [PubMed] [Google Scholar]
- 12.Avison, M. B., P. M. Bennett, R. A. Howe, and T. R. Walsh. 2002. Preliminary analysis of the genetic basis for vancomycin resistance in Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 49:255-260. [DOI] [PubMed] [Google Scholar]
- 13.Backo, M., E. Gaenger, A. Burkart, Y. L. Chai, and A. S. Bayer. 1999. Treatment of experimental staphylococcal endocarditis due to a strain with reduced susceptibility in vitro to vancomycin: efficacy of ampicillin-sulbactam. Antimicrob. Agents Chemother. 43:2565-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. [DOI] [PubMed] [Google Scholar]
- 15.Barna, J. C., and D. H. Williams. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38:339-357. [DOI] [PubMed] [Google Scholar]
- 16.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 17.Benito, Y., F. A. Kolb, P. Romby, G. Lina, J. Etienne, and F. Vandenesch. 2000. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6:668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett, J. W., C. K. Murray, R. L. Holmes, J. E. Patterson, and J. H. Jorgensen. 2008. Diminished vancomycin and daptomycin susceptibility during prolonged bacteremia with methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 60:437-440. [DOI] [PubMed] [Google Scholar]
- 19.Bera, A., R. Biswas, S. Herbert, E. Kulauzovic, C. Weidenmaier, A. Peschel, and F. Gotz. 2007. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 189:280-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger-Bachi, B., and M. Tschierske. 1998. Role of fem factors in methicillin resistance. Drug Resist. Updat. 1:325-335. [DOI] [PubMed] [Google Scholar]
- 21.Bernard, L., P. Vaudaux, P. Rohner, E. Huggler, M. Armanet, D. Pittet, D. P. Lew, and J. Schrenzel. 2004. Comparative analysis and validation of different assays for glycopeptide susceptibility among methicillin-resistant Staphylococcus aureus strains. J. Microbiol. Methods 57:231-239. [DOI] [PubMed] [Google Scholar]
- 22.Bierbaum, G., K. Fuchs, W. Lenz, C. Szekat, and H. G. Sahl. 1999. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 18:691-696. [DOI] [PubMed] [Google Scholar]
- 23.Bischoff, M., and B. Berger-Bachi. 2001. Teicoplanin stress-selected mutations increasing sigma(B) activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishop, E., S. Melvani, B. P. Howden, P. G. Charles, and M. L. Grayson. 2006. Good clinical outcomes but high rates of adverse reactions during linezolid therapy for serious infections: a proposed protocol for monitoring therapy in complex patients. Antimicrob. Agents Chemother. 50:1599-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop, E. J., and B. P. Howden. 2007. Treatment of Staphylococcus aureus infections: new issues, emerging therapies and future directions. Expert Opin. Emerg. Drugs 12:1-22. [DOI] [PubMed] [Google Scholar]
- 26.Bobin-Dubreux, S., M. E. Reverdy, C. Nervi, M. Rougier, A. Bolmstrom, F. Vandenesch, and J. Etienne. 2001. Clinical isolate of vancomycin-heterointermediate Staphylococcus aureus susceptible to methicillin and in vitro selection of a vancomycin-resistant derivative. Antimicrob. Agents Chemother. 45:349-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogdanovich, T., L. M. Ednie, S. Shapiro, and P. C. Appelbaum. 2005. Antistaphylococcal activity of ceftobiprole, a new broad-spectrum cephalosporin. Antimicrob. Agents Chemother. 49:4210-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowker, K. E., A. R. Noel, and A. P. MacGowan. 2006. Pharmacodynamics of dalbavancin studied in an in vitro pharmacokinetic system. J. Antimicrob. Chemother. 58:802-805. [DOI] [PubMed] [Google Scholar]
- 29.Boyle-Vavra, S., S. K. Berke, J. C. Lee, and R. S. Daum. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle-Vavra, S., R. B. Carey, and R. S. Daum. 2001. Development of vancomycin and lysostaphin resistance in a methicillin-resistant Staphylococcus aureus isolate. J. Antimicrob. Chemother. 48:617-625. [DOI] [PubMed] [Google Scholar]
- 31.Boyle-Vavra, S., M. Challapalli, and R. S. Daum. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bozdogan, B., D. Esel, C. Whitener, F. A. Browne, and P. C. Appelbaum. 2003. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J. Antimicrob. Chemother. 52:864-868. [DOI] [PubMed] [Google Scholar]
- 34.Brunet, F., G. Vedel, F. Dreyfus, J. F. Vaxelaire, T. Giraud, B. Schremmer, and J. F. Monsallier. 1990. Failure of teicoplanin therapy in two neutropenic patients with staphylococcal septicemia who recovered after administration of vancomycin. Eur. J. Clin. Microbiol. Infect. Dis. 9:145-147. [DOI] [PubMed] [Google Scholar]
- 35.Burnie, J., R. Matthews, A. Jiman-Fatami, P. Gottardello, S. Hodgetts, and S. D'Arcy. 2000. Analysis of 42 cases of septicemia caused by an epidemic strain of methicillin-resistant Staphylococcus aureus: evidence of resistance to vancomycin. Clin. Infect. Dis. 31:684-689. [DOI] [PubMed] [Google Scholar]
- 36.Camargo, I. L., and M. S. Gilmore. 2008. Staphylococcus aureus—probing for host weakness? J. Bacteriol. 190:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cartolano, G. L., M. Cheron, D. Benabid, M. Leneveu, and A. Boisivon. 2004. Methicillin-resistant Staphylococcus aureus (MRSA) with reduced susceptibility to glycopeptides (GISA) in 63 French general hospitals. Clin. Microbiol. Infect. 10:448-451. [DOI] [PubMed] [Google Scholar]
- 38.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. MMWR Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 39.Cercenado, E. 2000. Glycopeptide-intermediate Staphylococcus aureus: rediscovery of an old problem? Clin. Microbiol. Infect. 6:517-518. [DOI] [PubMed] [Google Scholar]
- 40.Cha, R., and M. J. Rybak. 2003. Daptomycin against multiple drug-resistant staphylococcus and enterococcus isolates in an in vitro pharmacodynamic model with simulated endocardial vegetations. Diagn. Microbiol. Infect. Dis. 47:539-546. [DOI] [PubMed] [Google Scholar]
- 41.Chambers, H. F. 2005. Evaluation of ceftobiprole in a rabbit model of aortic valve endocarditis due to methicillin-resistant and vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 49:884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers, S. T. 2005. Diagnosis and management of staphylococcal infections of pacemakers and cardiac defibrillators. Intern. Med. J. 35(Suppl. 2):S63-S71. [DOI] [PubMed] [Google Scholar]
- 45.Chambers, S. T. 2005. Diagnosis and management of staphylococcal infections of vascular grafts and stents. Intern. Med. J. 35(Suppl. 2):S72-S78. [DOI] [PubMed] [Google Scholar]
- 46.Chang, F. Y., J. E. Peacock, Jr., D. M. Musher, P. Triplett, B. B. MacDonald, J. M. Mylotte, A. O'Donnell, M. M. Wagener, and V. L. Yu. 2003. Staphylococcus aureus bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study. Medicine (Baltimore) 82:333-339. [DOI] [PubMed] [Google Scholar]
- 47.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 48.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 49.Chesneau, O., A. Morvan, and N. E. Solh. 2000. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 45:887-890. [DOI] [PubMed] [Google Scholar]
- 50.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 52.Chuard, C., P. E. Vaudaux, R. A. Proctor, and D. P. Lew. 1997. Decreased susceptibility to antibiotic killing of a stable small colony variant of Staphylococcus aureus in fluid phase and on fibronectin-coated surfaces. J. Antimicrob. Chemother. 39:603-608. [DOI] [PubMed] [Google Scholar]
- 53.CLSI. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed. CLSI document M7-A8. CLSI, Wayne, PA.
- 54.Collins, L. V., S. A. Kristian, C. Weidenmaier, M. Faigle, K. P. Van Kessel, J. A. Van Strijp, F. Gotz, B. Neumeister, and A. Peschel. 2002. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186:214-219. [DOI] [PubMed] [Google Scholar]
- 55.Cosgrove, S. E., K. C. Carroll, and T. M. Perl. 2004. Staphylococcus aureus with reduced susceptibility to vancomycin. Clin. Infect. Dis. 39:539-545. [DOI] [PubMed] [Google Scholar]
- 56.Cui, L., A. Iwamoto, J. Q. Lian, H. M. Neoh, T. Maruyama, Y. Horikawa, and K. Hiramatsu. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui, L., J. Q. Lian, H. M. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui, L., H. M. Neoh, M. Shoji, and K. Hiramatsu. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui, L., E. Tominaga, H. M. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daum, R. S., S. Gupta, R. Sabbagh, and W. M. Milewski. 1992. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J. Infect. Dis. 166:1066-1072. [DOI] [PubMed] [Google Scholar]
- 63.Davis, J. S. 2005. Management of bone and joint infections due to Staphylococcus aureus. Intern. Med. J. 35(Suppl. 2):S79-S96. [DOI] [PubMed] [Google Scholar]
- 64.de Lassence, A., N. Hidri, J. F. Timsit, M. L. Joly-Guillou, G. Thiery, A. Boyer, P. Lable, A. Blivet, H. Kalinowski, Y. Martin, J. P. Lajonchere, and D. Dreyfuss. 2006. Control and outcome of a large outbreak of colonization and infection with glycopeptide-intermediate Staphylococcus aureus in an intensive care unit. Clin. Infect. Dis. 42:170-178. [DOI] [PubMed] [Google Scholar]
- 65.de Lencastre, H., and A. Tomasz. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Lencastre, H., S. W. Wu, M. G. Pinho, A. M. Ludovice, S. Filipe, S. Gardete, R. Sobral, S. Gill, M. Chung, and A. Tomasz. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5:163-175. [DOI] [PubMed] [Google Scholar]
- 67.Delgado, A., J. T. Riordan, R. Lamichhane-Khadka, D. C. Winnett, J. Jimenez, K. Robinson, F. G. O'Brien, S. A. Cantore, and J. E. Gustafson. 2007. Hetero-vancomycin-intermediate methicillin-resistant Staphylococcus aureus isolate from a medical center in Las Cruces, New Mexico. J. Clin. Microbiol. 45:1325-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Elia, M. A., M. P. Pereira, Y. S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188:4183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Denis, O., C. Nonhoff, B. Byl, C. Knoop, S. Bobin-Dubreux, and M. J. Struelens. 2002. Emergence of vancomycin-intermediate Staphylococcus aureus in a Belgian hospital: microbiological and clinical features. J. Antimicrob. Chemother. 50:383-391. [DOI] [PubMed] [Google Scholar]
- 70.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 71.Diep, B. A., H. F. Chambers, C. J. Graber, J. D. Szumowski, L. G. Miller, L. L. Han, J. H. Chen, F. Lin, J. Lin, T. H. Phan, H. A. Carleton, L. K. McDougal, F. C. Tenover, D. E. Cohen, K. H. Mayer, G. F. Sensabaugh, and F. Perdreau-Remington. 2008. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann. Intern. Med. 148:249-257. [DOI] [PubMed] [Google Scholar]
- 72.Dmitriev, B. A., F. V. Toukach, O. Holst, E. T. Rietschel, and S. Ehlers. 2004. Tertiary structure of Staphylococcus aureus cell wall murein. J. Bacteriol. 186:7141-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draghi, D. C., M. Jones, C. Thornsberry, R. K. Flamm, and D. F. Sahm. 2005. Telavancin activity against current and diverse Staphylococcus aureus populations, abstr. E-1744. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 74.Drummelsmith, J., E. Winstall, M. G. Bergeron, G. G. Poirier, and M. Ouellette. 2007. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J. Proteome Res. 6:4690-4702. [DOI] [PubMed] [Google Scholar]
- 75.Dufour, P., S. Jarraud, F. Vandenesch, T. Greenland, R. P. Novick, M. Bes, J. Etienne, and G. Lina. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elhag, K. M., A. K. Al Jardani, F. M. Al Yaqubi, and N. Mohsin. 2000. The first glycopeptide-intermediate Staphylococcus aureus in Oman. Clin. Microbiol. Infect. 6:173-174. [DOI] [PubMed] [Google Scholar]
- 78.Fedtke, I., D. Mader, T. Kohler, H. Moll, G. Nicholson, R. Biswas, K. Henseler, F. Gotz, U. Zahringer, and A. Peschel. 2007. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol. Microbiol. 65:1078-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferraz, V., A. G. Duse, M. Kassel, A. D. Black, T. Ito, and K. Hiramatsu. 2000. Vancomycin-resistant Staphylococcus aureus occurs in South Africa. S. Afr. Med. J. 90:1113. [PubMed] [Google Scholar]
- 80.Filice, G., P. Lanzarini, P. Orsolini, L. Soldini, L. Perversi, G. Carnevale, G. Comolli, and F. Castelli. 1990. Effect of teicoplanin and vancomycin on Staphylococcus aureus ultrastructure. Microbiologica 13:231-237. [PubMed] [Google Scholar]
- 81.Finan, J. E., G. L. Archer, M. J. Pucci, and M. W. Climo. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fitzgibbon, M. M., A. S. Rossney, and B. O'Connell. 2007. Investigation of reduced susceptibility to glycopeptides among methicillin-resistant Staphylococcus aureus isolates from patients in Ireland and evaluation of agar screening methods for detection of heterogeneously glycopeptide-intermediate S. aureus. J. Clin. Microbiol. 45:3263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948-958. [DOI] [PubMed] [Google Scholar]
- 84.Foucault, M. L., P. Courvalin, and C. Grillot-Courvalin. 2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fournier, B., and A. Klier. 2004. Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology 150:3807-3819. [DOI] [PubMed] [Google Scholar]
- 86.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 87.Fowler, V. G., Jr., H. W. Boucher, G. R. Corey, E. Abrutyn, A. W. Karchmer, M. E. Rupp, D. P. Levine, H. F. Chambers, F. P. Tally, G. A. Vigliani, C. H. Cabell, A. S. Link, I. DeMeyer, S. G. Filler, M. Zervos, P. Cook, J. Parsonnet, J. M. Bernstein, C. S. Price, G. N. Forrest, G. Fatkenheuer, M. Gareca, S. J. Rehm, H. R. Brodt, A. Tice, and S. E. Cosgrove. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355:653-665. [DOI] [PubMed] [Google Scholar]
- 88.Fowler, V. G., Jr., G. Sakoulas, L. M. McIntyre, V. G. Meka, R. D. Arbeit, C. H. Cabell, M. E. Stryjewski, G. M. Eliopoulos, L. B. Reller, G. R. Corey, T. Jones, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190:1140-1149. [DOI] [PubMed] [Google Scholar]
- 89.Fox, P. M., M. W. Climo, and G. L. Archer. 2007. Lack of relationship between purine biosynthesis and vancomycin resistance in Staphylococcus aureus: a cautionary tale for microarray interpretation. Antimicrob. Agents Chemother. 51:1274-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fridkin, S. K. 2001. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin. Infect. Dis. 32:108-115. [DOI] [PubMed] [Google Scholar]
- 91.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, and F. C. Tenover. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 92.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garnier, F., D. Chainier, T. Walsh, A. Karlsson, A. Bolmstrom, C. Grelaud, M. Mounier, F. Denis, and M. C. Ploy. 2006. A 1 year surveillance study of glycopeptide-intermediate Staphylococcus aureus strains in a French hospital. J. Antimicrob. Chemother. 57:146-149. [DOI] [PubMed] [Google Scholar]
- 94.Gemmell, C. G. 2004. Glycopeptide resistance in Staphylococcus aureus: is it a real threat? J. Infect. Chemother. 10:69-75. [DOI] [PubMed] [Google Scholar]
- 95.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gomez, M. I., A. Lee, B. Reddy, A. Muir, G. Soong, A. Pitt, A. Cheung, and A. Prince. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842-848. [DOI] [PubMed] [Google Scholar]
- 97.Gomez, M. I., M. O'Seaghdha, M. Magargee, T. J. Foster, and A. S. Prince. 2006. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J. Biol. Chem. 281:20190-20196. [DOI] [PubMed] [Google Scholar]
- 98.Gomez, M. I., M. O. Seaghdha, and A. S. Prince. 2007. Staphylococcus aureus protein A activates TACE through EGFR-dependent signaling. EMBO J. 26:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gosbell, I. B. 2005. Diagnosis and management of catheter-related bloodstream infections due to Staphylococcus aureus. Intern. Med. J. 35(Suppl. 2):S45-S62. [DOI] [PubMed] [Google Scholar]
- 100.Gosbell, I. B., D. H. Mitchell, H. Ziochos, and P. B. Ward. 2003. Emergence of hetero-vancomycin-intermediate Staphylococcus aureus (hVISA) in Sydney. Med. J. Aust. 178:354. [DOI] [PubMed] [Google Scholar]
- 101.Gould, I. M. 2008. The epidemiology of antibiotic resistance. Int. J. Antimicrob. Agents 32(Suppl. 1):S2-S9. [DOI] [PubMed] [Google Scholar]
- 102.Graber, C. J., M. K. Wong, H. A. Carleton, F. Perdreau-Remington, B. L. Haller, and H. F. Chambers. 2007. Intermediate vancomycin susceptibility in a community-associated MRSA clone. Emerg. Infect. Dis. 13:491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grabs, A. J., and R. S. Lord. 2002. Treatment failure due to methicillin-resistant Staphylococcus aureus (MRSA) with reduced susceptibility to vancomycin. Med. J. Aust. 176:563. [DOI] [PubMed] [Google Scholar]
- 104.Grundling, A., and O. Schneewind. 2007. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guerin, F., A. Buu-Hoi, J. L. Mainardi, G. Kac, N. Colardelle, S. Vaupre, L. Gutmann, and I. Podglajen. 2000. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a Parisian hospital. J. Clin. Microbiol. 38:2985-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hageman, J. C., J. Patel, P. Franklin, K. Miscavish, L. McDougal, D. Lonsway, and F. N. Khan. 2008. Occurrence of a USA300 vancomycin-intermediate Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 62:440-442. [DOI] [PubMed] [Google Scholar]
- 107.Hageman, J. C., D. A. Pegues, C. Jepson, R. L. Bell, M. Guinan, K. W. Ward, M. D. Cohen, J. A. Hindler, F. C. Tenover, S. K. McAllister, M. E. Kellum, and S. K. Fridkin. 2001. Vancomycin-intermediate Staphylococcus aureus in a home health-care patient. Emerg. Infect. Dis. 7:1023-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 109.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 110.Hayden, M. K., K. Rezai, R. A. Hayes, K. Lolans, J. P. Quinn, and R. A. Weinstein. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5285-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Herbert, S., A. Bera, C. Nerz, D. Kraus, A. Peschel, C. Goerke, M. Meehl, A. Cheung, and F. Gotz. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 3:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hidayat, L. K., D. I. Hsu, R. Quist, K. A. Shriner, and A. Wong-Beringer. 2006. High-dose vancomycin therapy for methicillin-resistant Staphylococcus aureus infections: efficacy and toxicity. Arch. Intern. Med. 166:2138-2144. [DOI] [PubMed] [Google Scholar]
- 113.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 114.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 115.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 116.Hirschwerk, D., C. C. Ginocchio, M. Bythrow, and S. Condon. 2006. Diminished susceptibility to daptomycin accompanied by clinical failure in a patient with methicillin-resistant Staphylococcus aureus bacteremia. Infect. Control Hosp. Epidemiol. 27:315-317. [DOI] [PubMed] [Google Scholar]
- 117.Holmes, R. L., and J. H. Jorgensen. 2008. Inhibitory activities of 11 antimicrobial agents and bactericidal activities of vancomycin and daptomycin against invasive methicillin-resistant Staphylococcus aureus isolates obtained from 1999 through 2006. Antimicrob. Agents Chemother. 52:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Horne, K. C., B. P. Howden, E. A. Grabsch, M. Graham, P. D. Ward, S. Xie, B. C. Mayall, P. D. R. Johnson, and M. L. Grayson. 2009. Prospective comparison of the clinical impact of heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-susceptible MRSA. Antimicrob. Agents Chemother. 53:3447-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Howden, B. P. 2005. Recognition and management of infections caused by vancomycin-intermediate Staphylococcus aureus (VISA) and heterogenous VISA (hVISA). Intern. Med. J. 35(Suppl. 2):S136-S140. [DOI] [PubMed] [Google Scholar]
- 120.Howden, B. P., and M. L. Grayson. 2006. Dumb and dumber—the potential waste of a useful antistaphylococcal agent: emerging fusidic acid resistance in Staphylococcus aureus. Clin. Infect. Dis. 42:394-400. [DOI] [PubMed] [Google Scholar]
- 121.Howden, B. P., P. D. Johnson, P. B. Ward, T. P. Stinear, and J. K. Davies. 2006. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 50:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Howden, B. P., D. J. Smith, A. Mansell, P. D. Johnson, P. B. Ward, T. P. Stinear, and J. K. Davies. 2008. Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol. 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Howden, B. P., T. P. Stinear, D. L. Allen, P. D. Johnson, P. B. Ward, and J. K. Davies. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3755-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Howden, B. P., P. B. Ward, P. G. Charles, T. M. Korman, A. Fuller, P. du Cros, E. A. Grabsch, S. A. Roberts, J. Robson, K. Read, N. Bak, J. Hurley, P. D. Johnson, A. J. Morris, B. C. Mayall, and M. L. Grayson. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521-528. [DOI] [PubMed] [Google Scholar]
- 125.Howden, B. P., P. B. Ward, P. D. Johnson, P. G. Charles, and M. L. Grayson. 2005. Low-level vancomycin resistance in Staphylococcus aureus—an Australian perspective. Eur. J. Clin. Microbiol. Infect. Dis. 24:100-108. [DOI] [PubMed] [Google Scholar]
- 126.Howden, B. P., P. B. Ward, S. Xie, P. D. Johnson, P. G. Charles, and M. L. Grayson. 2004. Identification of a new agar dilution screening method for the accurate detection of heterogenous vancomycin-intermediate Staphylococcus, abstr. D59. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 126a.Howe, R. A., A. Monk, M. Wootton, T. R. Walsh, and M. C. Enright. 2004. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 10:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Howe, R. A., K. E. Bowker, T. R. Walsh, T. G. Feest, and A. P. MacGowan. 1998. Vancomycin-resistant Staphylococcus aureus. Lancet 351:602. [DOI] [PubMed] [Google Scholar]
- 128.Howe, R. A., M. Wootton, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 1999. Expression and detection of hetero-vancomycin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 44:675-678. [DOI] [PubMed] [Google Scholar]
- 129.Howe, R. A., M. Wootton, T. R. Walsh, P. M. Bennett, and A. P. Macgowan. 2000. Heterogeneous resistance to vancomycin in Staphylococcus aureus. J. Antimicrob. Chemother. 45:130-132. [DOI] [PubMed] [Google Scholar]
- 130.Hsu, D. I., L. K. Hidayat, R. Quist, J. Hindler, A. Karlsson, A. Yusof, and A. Wong-Beringer. 2008. Comparison of method-specific vancomycin minimum inhibitory concentration values and their predictability for treatment outcome of meticillin-resistant Staphylococcus aureus (MRSA) infections. Int. J. Antimicrob. Agents 32:378-385. [DOI] [PubMed] [Google Scholar]
- 131.Huang, Y. T., C. H. Hsiao, C. H. Liao, C. W. Lee, and P. R. Hsueh. 2008. Bacteremia and infective endocarditis caused by a non-daptomycin-susceptible, vancomycin-intermediate, and methicillin-resistant Staphylococcus aureus strain in Taiwan. J. Clin. Microbiol. 46:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang, Y. T., C. H. Liao, L. J. Teng, and P. R. Hsueh. 2008. Comparative bactericidal activities of daptomycin, glycopeptides, linezolid and tigecycline against blood isolates of Gram-positive bacteria in Taiwan. Clin. Microbiol. Infect. 14:124-129. [DOI] [PubMed] [Google Scholar]
- 133.Hubert, S. K., J. M. Mohammed, S. K. Fridkin, R. P. Gaynes, J. E. McGowan, Jr., and F. C. Tenover. 1999. Glycopeptide-intermediate Staphylococcus aureus: evaluation of a novel screening method and results of a survey of selected U.S. hospitals. J. Clin. Microbiol. 37:3590-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huebner, J., A. Kropec, I. Engels, and F. Daschner. 1992. In vitro susceptibility of methicillin-resistant Staphylococcus aureus and slime-producing and non-slime-producing coagulase-negative staphylococci to fusidic acid. Chemotherapy 38:206-210. [DOI] [PubMed] [Google Scholar]
- 135.Huntzinger, E., S. Boisset, C. Saveanu, Y. Benito, T. Geissmann, A. Namane, G. Lina, J. Etienne, B. Ehresmann, C. Ehresmann, A. Jacquier, F. Vandenesch, and P. Romby. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iizawa, Y., J. Nagai, T. Ishikawa, S. Hashiguchi, M. Nakao, A. Miyake, and K. Okonogi. 2004. In vitro antimicrobial activity of T-91825, a novel anti-MRSA cephalosporin, and in vivo anti-MRSA activity of its prodrug, TAK-599. J. Infect. Chemother. 10:146-156. [DOI] [PubMed] [Google Scholar]
- 137.Ike, Y., Y. Arakawa, X. Ma, K. Tatewaki, M. Nagasawa, H. Tomita, K. Tanimoto, and S. Fujimoto. 2001. Nationwide survey shows that methicillin-resistant Staphylococcus aureus strains heterogeneously and intermediately resistant to vancomycin are not disseminated throughout Japanese hospitals. J. Clin. Microbiol. 39:4445-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jacqueline, C., J. Caillon, V. Le Mabecque, A. F. Miegeville, A. Hamel, D. Bugnon, J. Y. Ge, and G. Potel. 2007. In vivo efficacy of ceftaroline (PPI-0903), a new broad-spectrum cephalosporin, compared with linezolid and vancomycin against methicillin-resistant and vancomycin-intermediate Staphylococcus aureus in a rabbit endocarditis model. Antimicrob. Agents Chemother. 51:3397-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jaksic, B., G. Martinelli, J. Perez-Oteyza, C. S. Hartman, L. B. Leonard, and K. J. Tack. 2006. Efficacy and safety of linezolid compared with vancomycin in a randomized, double-blind study of febrile neutropenic patients with cancer. Clin. Infect. Dis. 42:597-607. [DOI] [PubMed] [Google Scholar]
- 141.Jansen, A., M. Turck, C. Szekat, M. Nagel, I. Clever, and G. Bierbaum. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 297:205-215. [DOI] [PubMed] [Google Scholar]
- 142.Jansen, W. T. M., A. Verel, J. Verhoef, and D. Milatovic. 2007. In vitro activity of telavancin against Gram-positive clinical isolate recently obtained in Europe. Antimicrob. Agents Chemother. 51:3420-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jauregui, L. E., S. Babazadeh, E. Seltzer, L. Goldberg, D. Krievins, M. Frederick, D. Krause, I. Satilovs, Z. Endzinas, J. Breaux, and W. O'Riordan. 2005. Randomized, double-blind comparison of once-weekly dalbavancin versus twice-daily linezolid therapy for the treatment of complicated skin and skin structure infections. Clin. Infect. Dis. 41:1407-1415. [DOI] [PubMed] [Google Scholar]
- 146.Jevitt, L. A., A. J. Smith, P. P. Williams, P. M. Raney, J. E. McGowan, Jr., and F. C. Tenover. 2003. In vitro activities of daptomycin, linezolid, and quinupristin-dalfopristin against a challenge panel of staphylococci and enterococci, including vancomycin-intermediate Staphylococcus aureus and vancomycin-resistant Enterococcus faecium. Microb. Drug Resist. 9:389-393. [DOI] [PubMed] [Google Scholar]
- 147.Jones, R. N. 2006. Microbiological features of vancomycin in the 21st century: minimum inhibitory concentration creep, bactericidal/static activity, and applied breakpoints to predict clinical outcomes or detect resistant strains. Clin. Infect. Dis. 42(Suppl. 1):S13-S24. [DOI] [PubMed] [Google Scholar]
- 148.Jordan, S., M. I. Hutchings, and T. Mascher. 2007. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 149.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ju, O., M. Woolley, and D. Gordon. 2006. Emergence and spread of rifampicin-resistant, methicillin-resistant Staphylococcus aureus during vancomycin-rifampicin combination therapy in an intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 25:61-62. [DOI] [PubMed] [Google Scholar]
- 151.Julian, K., K. Kosowska-Shick, C. Whitener, M. Roos, H. Labischinski, A. Rubio, L. Parent, L. Ednie, L. Koeth, T. Bogdanovich, and P. C. Appelbaum. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jung, S. I., S. Kiem, N. Y. Lee, Y. S. Kim, W. S. Oh, H. L. Cho, K. R. Peck, and J. H. Song. 2002. One-point population analysis and effect of osmolarity on detection of hetero-vancomycin-resistant Staphylococcus aureus. J. Clin. Microbiol. 40:1493-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaatz, G. W., S. M. Seo, N. J. Dorman, and S. A. Lerner. 1990. Emergence of teicoplanin resistance during therapy of Staphylococcus aureus endocarditis. J. Infect. Dis. 162:103-108. [DOI] [PubMed] [Google Scholar]
- 154.Kantzanou, M., P. T. Tassios, A. Tseleni-Kotsovili, N. J. Legakis, and A. C. Vatopoulos. 1999. Reduced susceptibility to vancomycin of nosocomial isolates of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 43:729-731. [DOI] [PubMed] [Google Scholar]
- 155.Khosrovaneh, A., K. Riederer, S. Saeed, M. S. Tabriz, A. R. Shah, M. M. Hanna, M. Sharma, L. B. Johnson, M. G. Fakih, and R. Khatib. 2004. Frequency of reduced vancomycin susceptibility and heterogeneous subpopulation in persistent or recurrent methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 38:1328-1330. [DOI] [PubMed] [Google Scholar]
- 156.Kim, H. B., W. B. Park, K. D. Lee, Y. J. Choi, S. W. Park, M. D. Oh, E. C. Kim, and K. W. Choe. 2003. Nationwide surveillance for Staphylococcus aureus with reduced susceptibility to vancomycin in Korea. J. Clin. Microbiol. 41:2279-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim, M. N., S. H. Hwang, Y. J. Pyo, H. M. Mun, and C. H. Pai. 2002. Clonal spread of Staphylococcus aureus heterogeneously resistant to vancomycin in a university hospital in Korea. J. Clin. Microbiol. 40:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kim, M. N., C. H. Pai, J. H. Woo, J. S. Ryu, and K. Hiramatsu. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 160.Koehl, J. L., A. Muthaiyan, R. K. Jayaswal, K. Ehlert, H. Labischinski, and B. J. Wilkinson. 2004. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3749-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kojima, N., Y. Araki, and E. Ito. 1985. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J. Bacteriol. 161:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kola, A., P. Kirschner, B. Gohrbandt, I. F. Chaberny, F. Mattner, M. Struber, P. Gastmeier, and S. Suerbaum. 2007. An infection with linezolid-resistant S. aureus in a patient with left ventricular assist system. Scand. J. Infect. Dis. 39:463-465. [DOI] [PubMed] [Google Scholar]
- 163.Koprivnjak, T., C. Weidenmaier, A. Peschel, and J. P. Weiss. 2008. Wall teichoic acid deficiency in Staphylococcus aureus confers selective resistance to mammalian group IIA phospholipase A(2) and human beta-defensin 3. Infect. Immun. 76:2169-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kornblum, J., B. J. Hartman, R. P. Novick, and A. Tomasz. 1986. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur. J. Clin. Microbiol. 5:714-718. [DOI] [PubMed] [Google Scholar]
- 165.Kosowska-Shick, K., L. M. Ednie, P. McGhee, K. Smith, C. D. Todd, A. Wehler, and P. C. Appelbaum. 2008. Incidence and characteristics of vancomycin nonsusceptible strains of methicillin-resistant Staphylococcus aureus at Hershey Medical Center. Antimicrob. Agents Chemother. 52:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Reference deleted.
- 167.Krzyszton-Russjan, J., M. Gniadkowski, H. Polowniak-Pracka, E. Hagmajer, and W. Hryniewicz. 2002. The first Staphylococcus aureus isolates with reduced susceptibility to vancomycin in Poland. J. Antimicrob. Chemother. 50:1065-1069. [DOI] [PubMed] [Google Scholar]
- 168.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 169.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 170.Labischinski, H., G. Barnickel, H. Bradaczek, and P. Giesbrecht. 1979. On the secondary and tertiary structure of murein. Low and medium-angle X-ray evidence against chitin-based conformations of bacterial peptidoglycan. Eur. J. Biochem. 95:147-155. [DOI] [PubMed] [Google Scholar]
- 171.LaPlante, K. L., and M. J. Rybak. 2004. Impact of high-inoculum Staphylococcus aureus on the activities of nafcillin, vancomycin, linezolid, and daptomycin, alone and in combination with gentamicin, in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 48:4665-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 173.Lefort, A., J. Pavie, L. Garry, F. Chau, and B. Fantin. 2004. Activities of dalbavancin in vitro and in a rabbit model of experimental endocarditis due to Staphylococcus aureus with or without reduced susceptibility to vancomycin and teicoplanin. Antimicrob. Agents Chemother. 48:1061-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Leonard, S. N., K. L. Rossi, K. L. Newton, and M. J. Rybak. 2009. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J. Antimicrob. Chemother. 63:489-492. [DOI] [PubMed] [Google Scholar]
- 175.Leonard, S. N., and M. J. Rybak. 2009. Evaluation of vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus and heterogeneously vancomycin-intermediate S. aureus in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations. J. Antimicrob. Chemother. 63:155-160. [DOI] [PubMed] [Google Scholar]
- 176.Leung, K. T., M. K. Tong, Y. P. Siu, C. S. Lam, H. L. Ng, and H. K. Lee. 2004. Treatment of vancomycin-intermediate Staphylococcus aureus endocarditis with linezolid. Scand. J. Infect. Dis. 36:483-485. [DOI] [PubMed] [Google Scholar]
- 177.Leuthner, K. D., C. M. Cheung, and M. J. Rybak. 2006. Comparative activity of the new lipoglycopeptide telavancin in the presence and absence of serum against 50 glycopeptide non-susceptible staphylococci and three vancomycin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 58:338-343. [DOI] [PubMed] [Google Scholar]
- 178.Levine, D. P. 2006. Vancomycin: a history. Clin. Infect. Dis. 42(Suppl. 1):S5-S12. [DOI] [PubMed] [Google Scholar]
- 179.Lew, D. P., and F. Waldvogel. 2004. Osteomyelitis. Lancet 364:369-379. [DOI] [PubMed] [Google Scholar]
- 180.Li, M., D. J. Cha, Y. Lai, A. E. Villaruz, D. E. Sturdevant, and M. Otto. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66:1136-1147. [DOI] [PubMed] [Google Scholar]
- 181.Li, M., Y. Lai, A. E. Villaruz, D. J. Cha, D. E. Sturdevant, and M. Otto. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104:9469-9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 183.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lodise, T. P., J. Graves, A. Evans, E. Graffunder, M. Helmecke, B. M. Lomaestro, and K. Stellrecht. 2008. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 52:3315-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lodise, T. P., C. D. Miller, J. Graves, A. Evans, E. Graffunder, M. Helmecke, and K. Stellrecht. 2008. Predictors of high vancomycin MIC values among patients with methicillin-resistant Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 62:1138-1141. [DOI] [PubMed] [Google Scholar]
- 186.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 187.MacKenzie, F. M., P. Greig, D. Morrison, G. Edwards, and I. M. Gould. 2002. Identification and characterization of teicoplanin-intermediate Staphylococcus aureus blood culture isolates in NE Scotland. J. Antimicrob. Chemother. 50:689-697. [DOI] [PubMed] [Google Scholar]
- 188.Maclayton, D. O., K. J. Suda, K. A. Coval, C. B. York, and K. W. Garey. 2006. Case-control study of the relationship between MRSA bacteremia with a vancomycin MIC of 2 microg/mL and risk factors, costs, and outcomes in inpatients undergoing hemodialysis. Clin. Ther. 28:1208-1216. [DOI] [PubMed] [Google Scholar]
- 189.Mainardi, J. L., D. M. Shlaes, R. V. Goering, J. H. Shlaes, J. F. Acar, and F. W. Goldstein. 1995. Decreased teicoplanin susceptibility of methicillin-resistant strains of Staphylococcus aureus. J. Infect. Dis. 171:1646-1650. [DOI] [PubMed] [Google Scholar]
- 190.Mallaval, F. O., A. Carricajo, F. Delavenna, C. Recule, N. Fonsale, G. Manquat, D. Raffenot, O. Rogeaux, G. Aubert, and J. Tous. 2004. Detection of an outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a French hospital. Clin. Microbiol. Infect. 10:459-461. [DOI] [PubMed] [Google Scholar]
- 191.Mangili, A., I. Bica, D. R. Snydman, and D. H. Hamer. 2005. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 40:1058-1060. [DOI] [PubMed] [Google Scholar]
- 192.Manquat, G., J. Croize, J. P. Stahl, M. Meyran, P. Hirtz, and M. Micoud. 1992. Failure of teicoplanin treatment associated with an increase in MIC during therapy of Staphylococcus aureus septicaemia. J. Antimicrob. Chemother. 29:731-732. [DOI] [PubMed] [Google Scholar]
- 193.Maor, Y., M. Hagin, N. Belausov, N. Keller, D. Ben-David, and G. Rahav. 2009. Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J. Infect. Dis. 199:619-624. [DOI] [PubMed] [Google Scholar]
- 194.Maor, Y., G. Rahav, N. Belausov, D. Ben-David, G. Smollan, and N. Keller. 2007. Prevalence and characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia in a tertiary care center. J. Clin. Microbiol. 45:1511-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marchese, A., G. Balistreri, E. Tonoli, E. A. Debbia, and G. C. Schito. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Marco, F., C. G. de la Maria, Y. Armero, E. Amat, D. Soy, A. Moreno, A. del Rio, M. Almela, C. A. Mestres, J. M. Gatell, M. T. Jimenez de Anta, and J. M. Miro. 2008. Daptomycin is effective in treatment of experimental endocarditis due to methicillin-resistant and glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Marlowe, E. M., M. D. Cohen, J. F. Hindler, K. W. Ward, and D. A. Bruckner. 2001. Practical strategies for detecting and confirming vancomycin-intermediate Staphylococcus aureus: a tertiary-care hospital laboratory's experience. J. Clin. Microbiol. 39:2637-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Marty, F. M., W. W. Yeh, C. B. Wennersten, L. Venkataraman, E. Albano, E. P. Alyea, H. S. Gold, L. R. Baden, and S. K. Pillai. 2006. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J. Clin. Microbiol. 44:595-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.McAleese, F., S. W. Wu, K. Sieradzki, P. Dunman, E. Murphy, S. Projan, and A. Tomasz. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McCallum, N., M. Bischoff, H. Maki, A. Wada, and B. Berger-Bachi. 2004. TcaR, a putative MarR-like regulator of sarS expression. J. Bacteriol. 186:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.McCallum, N., H. Karauzum, R. Getzmann, M. Bischoff, P. Majcherczyk, B. Berger-Bachi, and R. Landmann. 2006. In vivo survival of teicoplanin-resistant Staphylococcus aureus and fitness cost of teicoplanin resistance. Antimicrob. Agents Chemother. 50:2352-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.McCallum, N., G. Spehar, M. Bischoff, and B. Berger-Bachi. 2006. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim. Biophys. Acta 1760:1475-1481. [DOI] [PubMed] [Google Scholar]
- 203.McKay, G. A., S. Beaulieu, F. F. Arhin, A. Belley, I. Sarmiento, T. Parr, Jr., and G. Moeck. 2009. Time-kill kinetics of oritavancin and comparator agents against Staphylococcus aureus, Enterococcus faecalis and Enterococcus faecium. J. Antimicrob. Chemother. 63:1191-1199. [DOI] [PubMed] [Google Scholar]
- 204.Meehl, M., S. Herbert, F. Gotz, and A. Cheung. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2679-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Memmi, G., S. R. Filipe, M. G. Pinho, Z. Fu, and A. Cheung. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Menezes, G. A., B. N. Harish, S. Sujatha, K. Vinothini, and S. C. Parija. 2008. Emergence of vancomycin-intermediate Staphylococcus species in southern India. J. Med. Microbiol. 57:911-912. [DOI] [PubMed] [Google Scholar]
- 207.Mitchell, D. H., and B. P. Howden. 2005. Diagnosis and management of Staphylococcus aureus bacteraemia. Intern. Med. J. 35(Suppl. 2):S17-S24. [DOI] [PubMed] [Google Scholar]
- 208.Moise, P. A., G. Sakoulas, A. Forrest, and J. J. Schentag. 2007. Vancomycin in vitro bactericidal activity and its relationship to efficacy in clearance of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 51:2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Moise, P. A., D. S. Smyth, N. El-Fawal, D. A. Robinson, P. N. Holden, A. Forrest, and G. Sakoulas. 2008. Microbiological effects of prior vancomycin use in patients with methicillin-resistant Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 61:85-90. [DOI] [PubMed] [Google Scholar]
- 210.Moise-Broder, P. A., A. Forrest, M. C. Birmingham, and J. J. Schentag. 2004. Pharmacodynamics of vancomycin and other antimicrobials in patients with Staphylococcus aureus lower respiratory tract infections. Clin. Pharmacokinet. 43:925-942. [DOI] [PubMed] [Google Scholar]
- 211.Moise-Broder, P. A., G. Sakoulas, G. M. Eliopoulos, J. J. Schentag, A. Forrest, and R. C. Moellering, Jr. 2004. Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 38:1700-1705. [DOI] [PubMed] [Google Scholar]
- 212.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Moore, M. R., F. Perdreau-Remington, and H. F. Chambers. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Moreillon, P., Y. A. Que, and M. Glauser. 2005. Staphylococcus aureus (including staphylococcal toxic shock), p. 2321-2351. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas and Bennett's principles and practice of infectious diseases, vol. 2. Elsevier, Philadelphia, PA. [Google Scholar]
- 215.Moreira, B., S. Boyle-Vavra, B. L. deJonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Munckhof, W. J., S. L. Kleinschmidt, and J. D. Turnidge. 2004. Resistance development in community-acquired strains of methicillin-resistant Staphylococcus aureus: an in vitro study. Int. J. Antimicrob. Agents 24:605-608. [DOI] [PubMed] [Google Scholar]
- 217.Murakami, K., and A. Tomasz. 1989. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 171:874-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Murray, R. J. 2005. Staphylococcus aureus infective endocarditis: diagnosis and management guidelines. Intern. Med. J. 35(Suppl. 2):S25-S44. [DOI] [PubMed] [Google Scholar]
- 219.Murray, R. J., K. Sieunarine, P. B. Ward, and J. W. Pearman. 2004. Emergence of heteroresistant vancomycin-intermediate Staphylococcus aureus (hVISA) infection in Western Australia. Med. J. Aust. 181:227-228. [DOI] [PubMed] [Google Scholar]
- 220.Musta, A. C., K. Riederer, S. Shemes, P. Chase, J. Jose, L. B. Johnson, and R. Khatib. 2009. Vancomycin MIC plus heteroresistance and the outcome of methicillin-resistant Staphylococcus aureus bacteremia: trends over 11 years. J. Clin. Microbiol. 47:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Muthaiyan, A., R. K. Jayaswal, and B. J. Wilkinson. 2004. Intact mutS in laboratory-derived and clinical glycopeptide-intermediate Staphylococcus aureus strains. Antimicrob. Agents Chemother. 48:623-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 104:9451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Naimi, T. S., D. Anderson, C. O'Boyle, D. J. Boxrud, S. K. Johnson, F. C. Tenover, and R. Lynfield. 2003. Vancomycin-intermediate Staphylococcus aureus with phenotypic susceptibility to methicillin in a patient with recurrent bacteremia. Clin. Infect. Dis. 36:1609-1612. [DOI] [PubMed] [Google Scholar]
- 224.Nelson, J. L., K. C. Rice, S. R. Slater, P. M. Fox, G. L. Archer, K. W. Bayles, P. D. Fey, B. N. Kreiswirth, and G. A. Somerville. 2007. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob. Agents Chemother. 51:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Neoh, H. M., L. Cui, H. Yuzawa, F. Takeuchi, M. Matsuo, and K. Hiramatsu. 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Neoh, H. M., S. Hori, M. Komatsu, T. Oguri, F. Takeuchi, L. Cui, and K. Hiramatsu. 2007. Impact of reduced vancomycin susceptibility on the therapeutic outcome of MRSA bloodstream infections. Ann. Clin. Microbiol. Antimicrob. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Nilsson, I. M., J. C. Lee, T. Bremell, C. Ryden, and A. Tarkowski. 1997. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect. Immun. 65:4216-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nishi, H., H. Komatsuzawa, T. Fujiwara, N. McCallum, and M. Sugai. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4800-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Nishi, H., H. Komatsuzawa, S. Yamada, T. Fujiwara, M. Ohara, K. Ohta, M. Sugiyama, T. Ishikawa, and M. Sugai. 2003. Moenomycin-resistance is associated with vancomycin-intermediate susceptibility in Staphylococcus aureus. Microbiol. Immunol. 47:927-935. [DOI] [PubMed] [Google Scholar]
- 230.Nonhoff, C., O. Denis, and M. J. Struelens. 2005. Low prevalence of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in Belgian hospitals. Clin. Microbiol. Infect. 11:214-220. [DOI] [PubMed] [Google Scholar]
- 231.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 232.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ogston, A. 1882. Micrococcus poisoning. J. Anat. 17:24-58. [PMC free article] [PubMed] [Google Scholar]
- 234.Ohta, T., H. Hirakawa, K. Morikawa, A. Maruyama, Y. Inose, A. Yamashita, K. Oshima, M. Kuroda, M. Hattori, K. Hiramatsu, S. Kuhara, and H. Hayashi. 2004. Nucleotide substitutions in Staphylococcus aureus strains, Mu50, Mu3, and N315. DNA Res. 11:51-56. [DOI] [PubMed] [Google Scholar]
- 235.Oliveira, G. A., A. M. Dell'Aquila, R. L. Masiero, C. E. Levy, M. S. Gomes, L. Cui, K. Hiramatsu, and E. M. Mamizuka. 2001. Isolation in Brazil of nosocomial Staphylococcus aureus with reduced susceptibility to vancomycin. Infect. Control Hosp. Epidemiol. 22:443-448. [DOI] [PubMed] [Google Scholar]
- 236.O'Neill, A. J., and I. Chopra. 2002. Insertional inactivation of mutS in Staphylococcus aureus reveals potential for elevated mutation frequencies, although the prevalence of mutators in clinical isolates is low. J. Antimicrob. Chemother. 50:161-169. [DOI] [PubMed] [Google Scholar]
- 237.O'Neill, A. J., and I. Chopra. 2003. Lack of evidence for involvement of hypermutability in emergence of vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1484-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.O'Neill, A. J., J. H. Cove, and I. Chopra. 2001. Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus. J. Antimicrob. Chemother. 47:647-650. [DOI] [PubMed] [Google Scholar]
- 239.O'Riordan, K., and J. C. Lee. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Oscarsson, J., C. Harlos, and S. Arvidson. 2005. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int. J. Med. Microbiol. 295:253-266. [DOI] [PubMed] [Google Scholar]
- 241.O'Seaghdha, M., C. J. van Schooten, S. W. Kerrigan, J. Emsley, G. J. Silverman, D. Cox, P. J. Lenting, and T. J. Foster. 2006. Staphylococcus aureus protein A binding to von Willebrand factor A1 domain is mediated by conserved IgG binding regions. FEBS J. 273:4831-4841. [DOI] [PubMed] [Google Scholar]
- 242.Park, Y. J., J. J. Park, S. O. Lee, E. J. Oh, and B. K. Kim. 2001. Low-level resistance to glycopeptides amongst Staphylococcus species: surveillance in a university hospital and evaluation of a vancomycin screening agar. Diagn. Microbiol. Infect. Dis. 41:155-159. [DOI] [PubMed] [Google Scholar]
- 243.Patel, J. B., L. A. Jevitt, J. Hageman, L. C. McDonald, and F. C. Tenover. 2006. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42:1652-1653. [DOI] [PubMed] [Google Scholar]
- 244.Peleg, A. Y., D. Monga, S. Pillai, E. Mylonakis, R. C. Moellering, Jr., and G. M. Eliopoulos. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pereira, P. M., S. R. Filipe, A. Tomasz, and M. G. Pinho. 2007. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3627-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pereira, S. F., A. O. Henriques, M. G. Pinho, H. de Lencastre, and A. Tomasz. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189:3525-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Perichon, B., and P. Courvalin. 2009. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:4580-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 249.Peschel, A., C. Vuong, M. Otto, and F. Gotz. 2000. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pieper, R., C. L. Gatlin-Bunai, E. F. Mongodin, P. P. Parmar, S. T. Huang, D. J. Clark, R. D. Fleischmann, S. R. Gill, and S. N. Peterson. 2006. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6:4246-4258. [DOI] [PubMed] [Google Scholar]
- 252.Pierard, D., H. Vandenbussche, I. Verschraegen, and S. Lauwers. 2004. Screening for Staphylococcus aureus with a reduced susceptibility to vancomycin in a Belgian hospital. Pathol. Biol. (Paris) 52:486-488. (In French.) [DOI] [PubMed] [Google Scholar]
- 253.Pina, P., C. Marliere, F. Vandenesch, J. P. Bedos, J. Etienne, and P. Y. Allouch. 2000. An outbreak of Staphylococcus aureus strains with reduced susceptibility to glycopeptides in a French general hospital. Clin. Infect. Dis. 31:1306-1308. [DOI] [PubMed] [Google Scholar]
- 254.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 182:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Pinho, M. G., and J. Errington. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871-881. [DOI] [PubMed] [Google Scholar]
- 256.Pinho, M. G., and J. Errington. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799-807. [DOI] [PubMed] [Google Scholar]
- 257.Plipat, N., G. Livni, H. Bertram, and R. B. Thomson, Jr. 2005. Unstable vancomycin heteroresistance is common among clinical isolates of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:2494-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ploy, M. C., C. Grelaud, C. Martin, L. de Lumley, and F. Denis. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 259.Pootoolal, J., J. Neu, and G. D. Wright. 2002. Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 42:381-408. [DOI] [PubMed] [Google Scholar]
- 260.Portoles, M., K. B. Kiser, N. Bhasin, K. H. Chan, and J. C. Lee. 2001. Staphylococcus aureus Cap5O has UDP-ManNAc dehydrogenase activity and is essential for capsule expression. Infect. Immun. 69:917-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Poutrel, B., P. Rainard, and P. Sarradin. 1997. Heterogeneity of cell-associated CP5 expression on Staphylococcus aureus strains demonstrated by flow cytometry. Clin. Diagn. Lab. Immunol. 4:275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Prakash, V., J. S. Lewis II, and J. H. Jorgensen. 2008. Vancomycin MICs for methicillin-resistant Staphylococcus aureus isolates differ based upon the susceptibility test method used. Antimicrob. Agents Chemother. 52:4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Proctor, R. A., B. Kahl, C. von Eiff, P. E. Vaudaux, D. P. Lew, and G. Peters. 1998. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 27(Suppl. 1):S68-S74. [DOI] [PubMed] [Google Scholar]
- 265.Proctor, R. A., and G. Peters. 1998. Small colony variants in staphylococcal infections: diagnostic and therapeutic implications. Clin. Infect. Dis. 27:419-422. [DOI] [PubMed] [Google Scholar]
- 266.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 267.Reipert, A., K. Ehlert, T. Kast, and G. Bierbaum. 2003. Morphological and genetic differences in two isogenic Staphylococcus aureus strains with decreased susceptibilities to vancomycin. Antimicrob. Agents Chemother. 47:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Renzoni, A., C. Barras, P. Francois, Y. Charbonnier, E. Huggler, C. Garzoni, W. L. Kelley, P. Majcherczyk, J. Schrenzel, D. P. Lew, and P. Vaudaux. 2006. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Renzoni, A., P. Francois, D. Li, W. L. Kelley, D. P. Lew, P. Vaudaux, and J. Schrenzel. 2004. Modulation of fibronectin adhesins and other virulence factors in a teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Renzoni, A., W. L. Kelley, C. Barras, A. Monod, E. Huggler, P. Francois, J. Schrenzel, R. Studer, P. Vaudaux, and D. P. Lew. 2009. Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 53:903-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Reverdy, M. E., S. Jarraud, S. Bobin-Dubreux, E. Burel, P. Girardo, G. Lina, F. Vandenesch, and J. Etienne. 2001. Incidence of Staphylococcus aureus with reduced susceptibility to glycopeptides in two French hospitals. Clin. Microbiol. Infect. 7:267-272. [DOI] [PubMed] [Google Scholar]
- 272.Reynolds, P. 1961. Studies on the mode of action of vancomycin. Biochim. Biophys. Acta 52:403-405. [DOI] [PubMed] [Google Scholar]
- 273.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 274.Risley, A. L., A. Loughman, C. Cywes-Bentley, T. J. Foster, and J. C. Lee. 2007. Capsular polysaccharide masks clumping factor A-mediated adherence of Staphylococcus aureus to fibrinogen and platelets. J. Infect. Dis. 196:919-927. [DOI] [PubMed] [Google Scholar]
- 275.Roben, P. W., A. N. Salem, and G. J. Silverman. 1995. VH3 family antibodies bind domain D of staphylococcal protein A. J. Immunol. 154:6437-6445. [PubMed] [Google Scholar]
- 276.Robert, J., R. Bismuth, and V. Jarlier. 2006. Decreased susceptibility to glycopeptides in methicillin-resistant Staphylococcus aureus: a 20 year study in a large French teaching hospital, 1983-2002. J. Antimicrob. Chemother. 57:506-510. [DOI] [PubMed] [Google Scholar]
- 277.Roberts, S., and S. Chambers. 2005. Diagnosis and management of Staphylococcus aureus infections of the skin and soft tissue. Intern. Med. J. 35(Suppl. 2):S97-S105. [DOI] [PubMed] [Google Scholar]
- 278.Rohrer, S., and B. Berger-Bachi. 2003. Application of a bacterial two-hybrid system for the analysis of protein-protein interactions between FemABX family proteins. Microbiology 149:2733-2738. [DOI] [PubMed] [Google Scholar]
- 279.Rohrer, S., K. Ehlert, M. Tschierske, H. Labischinski, and B. Berger-Bachi. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. U. S. A. 96:9351-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rohrer, S., H. Maki, and B. Berger-Bachi. 2003. What makes resistance to methicillin heterogeneous? J. Med. Microbiol. 52:605-607. [DOI] [PubMed] [Google Scholar]
- 281.Rose, W. E., G. W. Kaatz, G. Sakoulas, and M. J. Rybak. 2008. Teicoplanin pharmacodynamics in reference to the accessory gene regulator (agr) in Staphylococcus aureus using an in vitro pharmacodynamic model. J. Antimicrob. Chemother. 61:1099-1102. [DOI] [PubMed] [Google Scholar]
- 282.Rose, W. E., S. N. Leonard, K. L. Rossi, G. W. Kaatz, and M. J. Rybak. 2009. Impact of inoculum size and heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) on vancomycin activity and emergence of VISA in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 53:805-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Rose, W. E., S. N. Leonard, G. Sakoulas, G. W. Kaatz, M. J. Zervos, A. Sheth, C. F. Carpenter, and M. J. Rybak. 2008. Daptomycin activity against Staphylococcus aureus following vancomycin exposure in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 52:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Rubinstein, E., R. Isturiz, H. C. Standiford, L. G. Smith, T. H. Oliphant, S. Cammarata, B. Hafkin, V. Le, and J. Remington. 2003. Worldwide assessment of linezolid's clinical safety and tolerability: comparator-controlled phase III studies. Antimicrob. Agents Chemother. 47:1824-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ruzin, A., A. Severin, S. L. Moghazeh, J. Etienne, P. A. Bradford, S. J. Projan, and D. M. Shlaes. 2003. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim. Biophys. Acta 1621:117-121. [DOI] [PubMed] [Google Scholar]
- 287.Rybak, M. J., D. M. Cappelletty, T. Moldovan, J. R. Aeschlimann, and G. W. Kaatz. 1998. Comparative in vitro activities and postantibiotic effects of the oxazolidinone compounds eperezolid (PNU-100592) and linezolid (PNU-100766) versus vancomycin against Staphylococcus aureus, coagulase-negative staphylococci, Enterococcus faecalis, and Enterococcus faecium. Antimicrob. Agents Chemother. 42:721-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Rybak, M. J., R. Cha, C. M. Cheung, V. G. Meka, and G. W. Kaatz. 2005. Clinical isolates of Staphylococcus aureus from 1987 and 1989 demonstrating heterogeneous resistance to vancomycin and teicoplanin. Diagn. Microbiol. Infect. Dis. 51:119-125. [DOI] [PubMed] [Google Scholar]
- 289.Rybak, M. J., S. N. Leonard, K. L. Rossi, C. M. Cheung, H. S. Sader, and R. N. Jones. 2008. Characterization of vancomycin-heteroresistant Staphylococcus aureus from the metropolitan area of Detroit, Michigan, over a 22-year period (1986 to 2007). J. Clin. Microbiol. 46:2950-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sader, H. S., T. R. Fritsche, K. Kaniga, Y. Ge, and R. N. Jones. 2005. Antimicrobial activity and spectrum of PPI-0903M (T-91825), a novel cephalosporin, tested against a worldwide collection of clinical strains. Antimicrob. Agents Chemother. 49:3501-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sader, H. S., P. R. Rhomberg, and R. N. Jones. 2009. Nine-hospital study comparing broth microdilution and Etest method results for vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:3162-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Saha, B., A. K. Singh, A. Ghosh, and M. Bal. 2008. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 57:72-79. [DOI] [PubMed] [Google Scholar]
- 293.Sakoulas, G., J. Alder, C. Thauvin-Eliopoulos, R. C. Moellering, Jr., and G. M. Eliopoulos. 2006. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Sakoulas, G., G. M. Eliopoulos, V. G. Fowler, Jr., R. C. Moellering, Jr., R. P. Novick, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2005. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49:2687-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., R. P. Novick, L. Venkataraman, C. Wennersten, P. C. DeGirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929-938. [DOI] [PubMed] [Google Scholar]
- 296.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sakoulas, G., H. S. Gold, R. A. Cohen, L. Venkataraman, R. C. Moellering, and G. M. Eliopoulos. 2006. Effects of prolonged vancomycin administration on methicillin-resistant Staphylococcus aureus (MRSA) in a patient with recurrent bacteraemia. J. Antimicrob. Chemother. 57:699-704. [DOI] [PubMed] [Google Scholar]
- 298.Sakoulas, G., P. A. Moise-Broder, J. Schentag, A. Forrest, R. C. Moellering, Jr., and G. M. Eliopoulos. 2004. Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 42:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Samra, Z., O. Ofir, and H. Shmuely. 2007. In vitro susceptibility of methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecium to daptomycin and other antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 26:363-365. [DOI] [PubMed] [Google Scholar]
- 300.Sancak, B., S. Ercis, D. Menemenlioglu, S. Colakoglu, and G. Hascelik. 2005. Methicillin-resistant Staphylococcus aureus heterogeneously resistant to vancomycin in a Turkish university hospital. J. Antimicrob. Chemother. 56:519-523. [DOI] [PubMed] [Google Scholar]
- 301.Saravia-Otten, P., H. P. Muller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Schaaff, F., A. Reipert, and G. Bierbaum. 2002. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Scherl, A., P. Francois, Y. Charbonnier, J. M. Deshusses, T. Koessler, A. Huyghe, M. Bento, J. Stahl-Zeng, A. Fischer, A. Masselot, A. Vaezzadeh, F. Galle, A. Renzoni, P. Vaudaux, D. Lew, C. G. Zimmermann-Ivol, P. A. Binz, J. C. Sanchez, D. F. Hochstrasser, and J. Schrenzel. 2006. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics 7:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Schulthess, B., S. Meier, D. Homerova, C. Goerke, C. Wolz, J. Kormanec, B. Berger-Bachi, and M. Bischoff. 2009. Functional characterization of the sigmaB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 53:1832-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Schwaber, M. J., S. B. Wright, Y. Carmeli, L. Venkataraman, P. C. DeGirolami, A. Gramatikova, T. M. Perl, G. Sakoulas, and H. S. Gold. 2003. Clinical implications of varying degrees of vancomycin susceptibility in methicillin-resistant Staphylococcus aureus bacteremia. Emerg. Infect. Dis. 9:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Seidl, K., M. Stucki, M. Ruegg, C. Goerke, C. Wolz, L. Harris, B. Berger-Bachi, and M. Bischoff. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sheagren, J. N. 1985. Staphylococcal infections of the skin and skin structures. Cutis 36:2-6. [PubMed] [Google Scholar]
- 309.Sheagren, J. N. 1999. Staphylococcus aureus infections in trauma patients. Crit. Care Med. 27:692-693. [DOI] [PubMed] [Google Scholar]
- 310.Sheagren, J. N. 1984. Staphylococcus aureus. The persistent pathogen (first of two parts). N. Engl. J. Med. 310:1368-1373. [DOI] [PubMed] [Google Scholar]
- 311.Sheagren, J. N. 1984. Staphylococcus aureus. The persistent pathogen (second of two parts). N. Engl. J. Med. 310:1437-1442. [DOI] [PubMed] [Google Scholar]
- 312.Shinefield, H., S. Black, A. Fattom, G. Horwith, S. Rasgon, J. Ordonez, H. Yeoh, D. Law, J. B. Robbins, R. Schneerson, L. Muenz, S. Fuller, J. Johnson, B. Fireman, H. Alcorn, and R. Naso. 2002. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med. 346:491-496. [DOI] [PubMed] [Google Scholar]
- 313.Shlaes, D. M., J. H. Shlaes, S. Vincent, L. Etter, P. D. Fey, and R. V. Goering. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 316.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 317.Sieradzki, K., and A. Tomasz. 1996. A highly vancomycin-resistant laboratory mutant of Staphylococcus aureus. FEMS Microbiol. Lett. 142:161-166. [DOI] [PubMed] [Google Scholar]
- 318.Sieradzki, K., and A. Tomasz. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sieradzki, K., and A. Tomasz. 2006. Inhibition of the autolytic system by vancomycin causes mimicry of vancomycin-intermediate Staphylococcus aureus-type resistance, cell concentration dependence of the MIC, and antibiotic tolerance in vancomycin-susceptible S. aureus. Antimicrob. Agents Chemother. 50:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Sieradzki, K., S. W. Wu, and A. Tomasz. 1999. Inactivation of the methicillin resistance gene mecA in vancomycin-resistant Staphylococcus aureus. Microb. Drug Resist. 5:253-257. [DOI] [PubMed] [Google Scholar]
- 323.Sievert, D. M., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin. Infect. Dis. 46:668-674. [DOI] [PubMed] [Google Scholar]
- 324.Small, P. M., and H. F. Chambers. 1990. Vancomycin for Staphylococcus aureus endocarditis in intravenous drug users. Antimicrob. Agents Chemother. 34:1227-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Smith, G. 1982. Ogston's coccus: 102 years and still going strong. South. Med. J. 75:1559-1562. [PubMed] [Google Scholar]
- 326.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureus Working Group. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 327.Sng, L. H., T. H. Koh, G. C. Wang, L. Y. Hsu, M. Kapi, and K. Hiramatsu. 2005. Heterogeneous vancomycin-resistant Staphylococcus aureus (hetero-VISA) in Singapore. Int. J. Antimicrob. Agents 25:177-179. [DOI] [PubMed] [Google Scholar]
- 328.Snowden, M. A., H. R. Perkins, A. W. Wyke, M. V. Hayes, and J. B. Ward. 1989. Cross-linking and O-acetylation of newly synthesized peptidoglycan in Staphylococcus aureus H. J. Gen. Microbiol. 135:3015-3022. [DOI] [PubMed] [Google Scholar]
- 329.Sobral, R. G., A. E. Jones, S. G. Des Etages, T. J. Dougherty, R. M. Peitzsch, T. Gaasterland, A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2007. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189:2376-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Sobral, R. G., A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2006. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J. Bacteriol. 188:2543-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Song, J. H., K. Hiramatsu, J. Y. Suh, K. S. Ko, T. Ito, M. Kapi, S. Kiem, Y. S. Kim, W. S. Oh, K. R. Peck, and N. Y. Lee. 2004. Emergence in Asian countries of Staphylococcus aureus with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 48:4926-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Soriano, A., F. Marco, J. A. Martinez, E. Pisos, M. Almela, V. P. Dimova, D. Alamo, M. Ortega, J. Lopez, and J. Mensa. 2008. Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 46:193-200. [DOI] [PubMed] [Google Scholar]
- 333.Steinkraus, G., R. White, and L. Friedrich. 2007. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001-05. J. Antimicrob. Chemother. 60:788-794. [DOI] [PubMed] [Google Scholar]
- 334.Streit, J. M., T. R. Fritsche, H. S. Sader, and R. N. Jones. 2004. Worldwide assessment of dalbavancin activity and spectrum against over 6,000 clinical isolates. Diagn. Microbiol. Infect. Dis. 48:137-143. [DOI] [PubMed] [Google Scholar]
- 335.Streit, J. M., H. S. Sader, T. R. Fritsche, and R. N. Jones. 2005. Dalbavancin activity against selected populations of antimicrobial-resistant Gram-positive pathogens. Diagn. Microbiol. Infect. Dis. 53:307-310. [DOI] [PubMed] [Google Scholar]
- 336.Stringfellow, W. T., B. Dassy, M. Lieb, and J. M. Fournier. 1991. Staphylococcus aureus growth and type 5 capsular polysaccharide production in synthetic media. Appl. Environ. Microbiol. 57:618-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Sugino, Y., Y. Iinumab, S. Ichiyama, Y. Ito, S. Ohkawa, N. Nakashima, K. Shimokata, and Y. Hasegawa. 2000. In vivo development of decreased susceptibility to vancomycin in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 38:159-167. [DOI] [PubMed] [Google Scholar]
- 338.Swenson, J. M., K. F. Anderson, D. R. Lonsway, A. Thompson, S. K. McAllister, B. M. Limbago, R. B. Carey, F. C. Tenover, and J. B. Patel. 2009. Accuracy of commercial and reference susceptibility testing methods for detecting vancomycin-intermediate Staphylococcus aureus. J. Clin. Microbiol. 47:2013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Tajima, Y., M. Komatsu, T. Ito, and K. Hiramatsu. 2007. Rapid detection of Staphylococcus aureus strains having reduced susceptibility to vancomycin using a chemiluminescence-based drug-susceptibility test. J. Microbiol. Methods 70:434-441. [DOI] [PubMed] [Google Scholar]
- 340.Takayama, Y., H. Hanaki, K. Irinoda, H. Kokubun, K. Yoshida, and K. Sunakawa. 2003. Investigation of methicillin-resistant Staphylococcus aureus showing reduced vancomycin susceptibility isolated from a patient with infective endocarditis. Int. J. Antimicrob. Agents 22:567-573. [DOI] [PubMed] [Google Scholar]
- 341.Tallent, S. M., T. Bischoff, M. Climo, B. Ostrowsky, R. P. Wenzel, and M. B. Edmond. 2002. Vancomycin susceptibility of oxacillin-resistant Staphylococcus aureus isolates causing nosocomial bloodstream infections. J. Clin. Microbiol. 40:2249-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Tamber, S., and A. L. Cheung. 2009. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 77:419-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 344.Tenover, F. C., J. W. Biddle, and M. V. Lancaster. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, S. K. McAllister, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Tenover, F. C., and R. C. Moellering, Jr. 2007. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin. Infect. Dis. 44:1208-1215. [DOI] [PubMed] [Google Scholar]
- 347.Tenover, F. C., M. J. Mohammed, J. Stelling, T. O'Brien, and R. Williams. 2001. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organization's external quality assurance system for antimicrobial susceptibility testing. J. Clin. Microbiol. 39:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Tenover, F. C., S. W. Sinner, R. E. Segal, V. Huang, S. S. Alexandre, J. E. McGowan, Jr., and M. P. Weinstein. 2009. Characterisation of a Staphylococcus aureus strain with progressive loss of susceptibility to vancomycin and daptomycin during therapy. Int. J. Antimicrob. Agents 33:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Thakker, M., J. S. Park, V. Carey, and J. C. Lee. 1998. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Tillotson, G. S., D. C. Draghi, D. F. Sahm, K. M. Tomfohrde, T. Del Fabro, and I. A. Critchley. 2008. Susceptibility of Staphylococcus aureus isolated from skin and wound infections in the United States 2005-07: laboratory-based surveillance study. J. Antimicrob. Chemother. 62:109-115. [DOI] [PubMed] [Google Scholar]
- 351.Tomasz, A. 2006. The staphylococcal cell wall, p. 443-455. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 352.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Dhiraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39:591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Trotonda, M. P., Y. Q. Xiong, G. Memmi, A. S. Bayer, and A. L. Cheung. 2009. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J. Infect. Dis. 199:209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Tsakris, A., E. Papadimitriou, J. Douboyas, F. Stylianopoulou, and E. Manolis. 2002. Emergence of vancomycin-intermediate Staphylococcus aureus and S. sciuri, Greece. Emerg. Infect. Dis. 8:536-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 356.Tsuji, B. T., Y. Harigaya, A. J. Lesse, G. Sakoulas, and J. M. Mylotte. 2009. Loss of vancomycin bactericidal activity against accessory gene regulator (agr) dysfunctional Staphylococcus aureus under conditions of high bacterial density. Diagn. Microbiol. Infect. Dis. 64:220-224. [DOI] [PubMed] [Google Scholar]
- 357.Tsuji, B. T., M. J. Rybak, C. M. Cheung, M. Amjad, and G. W. Kaatz. 2007. Community- and health care-associated methicillin-resistant Staphylococcus aureus: a comparison of molecular epidemiology and antimicrobial activities of various agents. Diagn. Microbiol. Infect. Dis. 58:41-47. [DOI] [PubMed] [Google Scholar]
- 358.Tsuji, B. T., M. J. Rybak, K. L. Lau, and G. Sakoulas. 2007. Evaluation of accessory gene regulator (agr) group and function in the proclivity towards vancomycin intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:1089-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Tsuji, B. T., C. von Eiff, P. A. Kelchlin, A. Forrest, and P. F. Smith. 2008. Attenuated vancomycin bactericidal activity against Staphylococcus aureus hemB mutants expressing the small-colony-variant phenotype. Antimicrob. Agents Chemother. 52:1533-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Turnidge, J. D., G. R. Nimmo, and G. Francis. 1996. Evolution of resistance in Staphylococcus aureus in Australian teaching hospitals. Australian Group on Antimicrobial Resistance (AGAR). Med. J. Aust. 164:68-71. [PubMed] [Google Scholar]
- 361.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 362.Utaida, S., R. F. Pfeltz, R. K. Jayaswal, and B. J. Wilkinson. 2006. Autolytic properties of glycopeptide-intermediate Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 50:1541-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i(8575):57-58. [DOI] [PubMed] [Google Scholar]
- 364.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Van Griethuysen, A., A. Van't Veen, A. Buiting, T. Walsh, and J. Kluytmans. 2003. High percentage of methicillin-resistant Staphylococcus aureus isolates with reduced susceptibility to glycopeptides in The Netherlands. J. Clin. Microbiol. 41:2487-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Vaudaux, P., P. Francois, B. Berger-Bachi, and D. P. Lew. 2001. In vivo emergence of subpopulations expressing teicoplanin or vancomycin resistance phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 47:163-170. [DOI] [PubMed] [Google Scholar]
- 368.Verdier, I., G. Durand, M. Bes, K. L. Taylor, G. Lina, F. Vandenesch, A. I. Fattom, and J. Etienne. 2007. Identification of the capsular polysaccharides in Staphylococcus aureus clinical isolates by PCR and agglutination tests. J. Clin. Microbiol. 45:725-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Verdier, I., M. E. Reverdy, J. Etienne, G. Lina, M. Bes, and F. Vandenesch. 2004. Staphylococcus aureus isolates with reduced susceptibility to glycopeptides belong to accessory gene regulator group I or II. Antimicrob. Agents Chemother. 48:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Voss, A., J. W. Mouton, E. P. van Elzakker, R. G. Hendrix, W. Goessens, J. A. Kluytmans, P. F. Krabbe, H. J. de Neeling, J. H. Sloos, N. Oztoprak, R. A. Howe, and T. R. Walsh. 2007. A multi-center blinded study on the efficiency of phenotypic screening methods to detect glycopeptide intermediately susceptible Staphylococcus aureus (GISA) and heterogeneous GISA (h-GISA). Ann. Clin. Microbiol. Antimicrob. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wada, A., and H. Watanabe. 1998. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J. Bacteriol. 180:2759-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 374.Wang, G., J. F. Hindler, K. W. Ward, and D. A. Bruckner. 2006. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-year period. J. Clin. Microbiol. 44:3883-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Ward, P. B., P. D. Johnson, E. A. Grabsch, B. C. Mayall, and M. L. Grayson. 2001. Treatment failure due to methicillin-resistant Staphylococcus aureus (MRSA) with reduced susceptibility to vancomycin. Med. J. Aust. 175:480-483. [DOI] [PubMed] [Google Scholar]
- 376.Webster, D., R. P. Rennie, C. L. Brosnikoff, L. Chui, and C. Brown. 2007. Methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Canada. Diagn. Microbiol. Infect. Dis. 57:177-181. [DOI] [PubMed] [Google Scholar]
- 377.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
- 378.Weidenmaier, C., J. F. Kokai-Kun, E. Kulauzovic, T. Kohler, G. Thumm, H. Stoll, F. Gotz, and A. Peschel. 2008. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int. J. Med. Microbiol. 298:505-513. [DOI] [PubMed] [Google Scholar]
- 379.Weidenmaier, C., and A. Peschel. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6:276-287. [DOI] [PubMed] [Google Scholar]
- 380.Weidenmaier, C., A. Peschel, V. A. Kempf, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2005. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect. Immun. 73:8033-8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Weidenmaier, C., A. Peschel, Y. Q. Xiong, S. A. Kristian, K. Dietz, M. R. Yeaman, and A. S. Bayer. 2005. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 191:1771-1777. [DOI] [PubMed] [Google Scholar]
- 382.Weigelt, J., K. Itani, D. Stevens, W. Lau, M. Dryden, and C. Knirsch. 2005. Linezolid versus vancomycin in treatment of complicated skin and soft tissue infections. Antimicrob. Agents Chemother. 49:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Wong, S. S., P. L. Ho, P. C. Woo, and K. Y. Yuen. 1999. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin. Infect. Dis. 29:760-767. [DOI] [PubMed] [Google Scholar]
- 384.Woods, C. W., A. C. Cheng, V. G. Fowler, Jr., M. Moorefield, J. Frederick, G. Sakoulas, V. G. Meka, F. C. Tenover, P. Zwadyk, and K. H. Wilson. 2004. Endocarditis caused by Staphylococcus aureus with reduced susceptibility to vancomycin. Clin. Infect. Dis. 38:1188-1191. [DOI] [PubMed] [Google Scholar]
- 385.Wootton, M. 2006. The need for accuracy in performing vancomycin intermediate resistant Staphylococcus aureus (VISA) and hetero-VISA detection methods. Int. J. Antimicrob. Agents 28:586. [DOI] [PubMed] [Google Scholar]
- 386.Wootton, M., P. M. Bennett, A. P. MacGowan, and T. R. Walsh. 2005. Strain-specific expression levels of pbp4 exist in isolates of glycopeptide-intermediate Staphylococcus aureus (GISA) and heterogeneous GISA. Antimicrob. Agents Chemother. 49:3598-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]
- 388.Wootton, M., A. P. MacGowan, and T. R. Walsh. 2006. Comparative bactericidal activities of daptomycin and vancomycin against glycopeptide-intermediate Staphylococcus aureus (GISA) and heterogeneous GISA isolates. Antimicrob. Agents Chemother. 50:4195-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Wright, J. S., III, K. E. Traber, R. Corrigan, S. A. Benson, J. M. Musser, and R. P. Novick. 2005. The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187:5585-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Xiong, Y. Q., V. G. Fowler, M. R. Yeaman, F. Perdreau-Remington, B. N. Kreiswirth, and A. S. Bayer. 2009. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J. Infect. Dis. 199:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Yin, S., R. S. Daum, and S. Boyle-Vavra. 2006. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Yusof, A., A. Engelhardt, A. Karlsson, L. Bylund, P. Vidh, K. Mills, M. Wootton, and T. R. Walsh. 2008. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J. Clin. Microbiol. 46:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Zhang, L., L. Gray, R. P. Novick, and G. Ji. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277:34736-34742. [DOI] [PubMed] [Google Scholar]
- 395.Zinn, C. S., H. Westh, and V. T. Rosdahl. 2004. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb. Drug Resist. 10:160-168. [DOI] [PubMed] [Google Scholar]